Abstract
Proteomic technologies have undergone significant development in recent years, which has led to extensive advances in protein research. Currently, proteomic approaches have been applied to many scientific areas, including basic research, various disease and malignant tumour diagnostics, biomarker discovery and other therapeutic applications. In addition, proteomics-driven research articles examining reproductive biology and medicine are becoming increasingly common. The key challenge for this field is to move from lists of identified proteins to obtaining biological information regarding protein function. The present article reviews the available scientific literature related to spermatogenesis. In addition, this study uses two-dimensional electrophoresis mass spectrometry (2DE-MS) and liquid chromatography (LC)-MS to construct a series of proteome profiles describing spermatogenesis. This large-scale identification of proteins provides a rich resource for elucidating the mechanisms underlying male fertility and infertility.
Keywords: infertility, male fertility, proteomics, spermatogenesis
Introduction
Spermatogenesis is the process of the development of sperm from undifferentiated germ cells. It involves successive mitotic, meiotic and post-meiotic phases. During this complex process, round undifferentiated spermatogonia become elongated, terminally differentiated spermatozoa, which are highly specialized cells responsible for delivering the paternal genome to the oocyte. Spermatogenesis requires a precise and well-coordinated system that regulates constantly changing patterns of gene and protein expression.1, 2
In the last 25 years, advances in molecular biology and genomics have significantly improved our knowledge of spermatogenesis by identifying numerous genes essential for the process.3, 4, 5, 6 However, these transcriptomic databases do not provide crucial information on the post-transcriptional control of gene expression, changes in protein expression levels or protein modifications. Moreover, transcriptomics poses certain limitations as the expression levels of the majority of mRNAs and proteins are not always correlated.7, 8 This limitation is particularly distinct during spermatogenesis for the following reasons: (i) male germ cell-specific proteins may be synthesized from specific alternate transcripts; (ii) transcription is arrested during spermiogenesis and pre-existing mRNAs are stored for several days; (ii) for the completion of spermatogenesis, the appropriate temporal activation of stored messages is required, and translational activation must occur independently of transcriptional control.9, 10 Therefore, it is important to explore changes in protein expression directly and systematically.11, 12
The term ‘proteomics' was first used in 1995 and was defined as the large-scale characterisation of the entire protein complement of a cell line, tissue or organism.13 Generally, there are two strategies for proteomics: protein expressional proteomics and functional proteomics.14 The former identifies the entire protein expression level in samples via high throughput technology designed to generate a global view of the organism's biology. The latter is a dynamic proteome and provides a comparative view of different biological processes, which allows a better understanding of the physiological and pathological states of an organism. This approach allows a selected group of proteins to be characterized and can provide important information about protein signalling, post-translational modification (PTM), protein–protein interactions and disease mechanisms.13, 15
In male reproductive biology, proteomics is emerging as a tool for defining specific protein profiles. Searching for terms such as ‘testis proteome' or ‘sperm proteome', one finds more than 150 publications in the National Centre for Biotechnology Information PubMed database (http://www.ncbi.nlm.nih.gov/pubmed/). Among these publications, our laboratory contributed nine papers published in Journal of Proteome Research, Proteomics and Fertility and Sterility, and others. The present review focuses on spermatogenesis-related proteomics in our laboratory and complements other reviews.16, 17, 18, 19, 20 This review aims to improve our understanding of normal spermatogenesis and the cause of abnormalities in spermatogenesis and offers an interpretation of the regulatory mechanisms underlying male fertility and infertility.
Research strategies
We focused on the whole testis from mice and humans to identify the global view of protein expression using different proteomic technologies (expressional proteome of the testis). Subsequently, different germ cells, including tetraploid and haploid germ cells, were separated to identify whole-protein expression profiles (expressional proteome of germ cells). To further understand the regulation of spermatogenesis, we then performed functional proteomics using two models: a mouse model of normal spermatogenesis and a human model of abnormal spermatogenesis. These investigations established a series of proteome profiles of spermatogenesis and provided several important protein lists to understand the regulation of male fertility and infertility (Figure 1).
Figure 1.
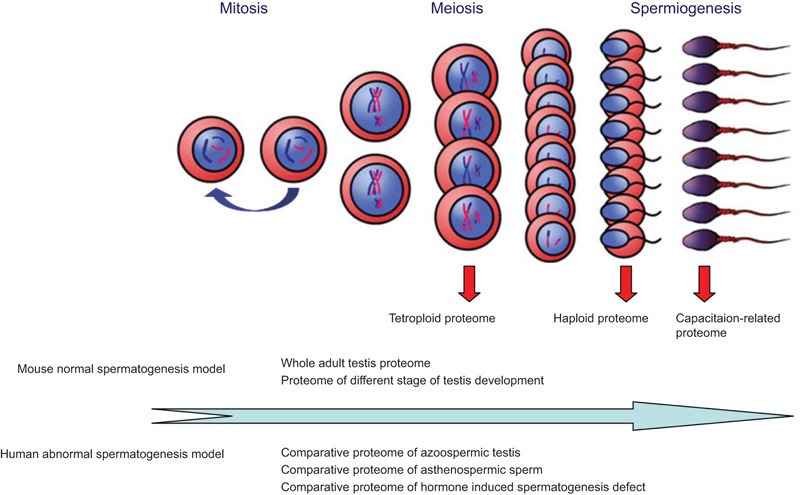
Research strategies in our laboratory for designing spermatogenesis-related proteomics.
Expressional proteome of the testis
The testis is the primary male sex gland responsible for the production of sperm and the secretion of testosterone (T). To identify the whole-protein expression profile of the testis, Zhu et al.,21 in our laboratory, established a proteome reference map containing 504 identified proteins from adult mouse testis using two-dimensional electrophoresis (2DE) and mass spectrometry (MS), which was linked to a nationwide proteome database (full data sets are available on http://reprod.njmu.edu.cn/cgi-bin/2d/2d.cgi). The online reference map hyperlinked the identified protein spots to the individual protein entries. Each protein entry contained identification information including accession number, name, description, spot ID number, organism, isoelectric point (pI), molecular weight (Mr), MS data and links to relevant entries in other online databases (Figure 2). This database can be accessed by WORLD-2DPAGE (http://cn.expasy.org/ch2d/2d-index.html).
Figure 2.

Partial graphs of two-dimensional (2DE) results from mouse adult testis. The red arrow indicates the protein spots with detailed information in the box.
Then, on the basis of rodent data, Guo et al.22 constructed a reference map for normal human testis. This map included 847 protein spots corresponding to 462 unique proteins identified using 2DE and MS (http://reprod.njmu.edu.cn/cgi-bin/2d/2d.cgi). The heterogeneity of the proteins expressed in human testis has been demonstrated by an average of two spots for each protein. The 2DE technique allows the identification of a protein in more than one spot or in a spot with an unexpected molecular mass or pI, which can lead to the characterisation of PTMs.23 Using bioinformatics analysis, SP_PIR_Keywords (http://www.uniprot.org/keywords/) suggested that alternative initiation sites and various forms of PTM may also contribute to this heterogeneity. To verify these results, we next used Pro-Q Diamond phosphostain (Invitrogen, Carlsbad, CA, USA) as a unique fluorescence-based detection system for the specific and sensitive analysis of protein and peptide phosphorylation status.24 A total of 68 spots representing 52 proteins were stained, thereby confirming the presence of phosphorylated forms of these proteins in human testis.22
Novel proteomics techniques are making it possible to identify large numbers of proteins in complex tissues. Combinations of 2DE with MS enable the separation of proteins in a polyacrylamide gel followed by identification using MS and are the most common and widely used techniques for this type of research. Another combination is liquid chromatography (LC) with MS, suitable for high throughput studies of peptides. Using LC-MS, we obtained many more proteins (1430) expressed in human testis compared to 2DE-MS techniques. Detailed bioinformatics analysis revealed the presence of many testis-specific proteins and a number of novel proteins in the testis.25
Integrating these two proteome profiles of human testis, 1495 proteins were identified. All of the above proteins identified in human and mouse testis could serve as references for future studies on the function of testis and mechanisms underlying male fertility and infertility.
Expressional proteome of germ cells
Spermatogenesis is a cyclic process in which diploid spermatogonia differentiate into tetraploid primary spermatocytes and then mature haploid spermatozoa.26 To identify the specific protein expression profiles of different germ cells using flow cytometry, we purified tetraploid and haploid germ cells from adult mouse testis that covered the entire process of meiosis and spermiogenesis, respectively. Subsequently, we constructed a large-scale tetraploid and haploid proteome via LC-MS. A total of 3507 proteins were identified in the tetraploid germ cell, and further analysis characterized 216 homologues of yeast proteins involved in the meiosis process (data unpublished). In all, 2116 proteins from mouse haploid germ cells participated in spermiogenesis. Spermiogenesis is a unique process that involves the formation of the acrosome with a large vesicle structure, condensation of the nucleus, generation of the sperm tail and removal of most of the cytoplasm as a residual body.27, 28, 29 Furthermore, bioinformatics analysis identified 84 proteins annotated to be involved in vesicle-related events, 22 proteins annotated to function in chromatin assembly, 39 proteins annotated to be involved in nucleocytoplasmic transport and 231 proteins having orthologues in human cilia.30
The large-scale proteome that appears to be involved in unique processes during meiosis and spermiogenesis provides more detailed and accurate information to improve our understanding of the mechanisms that cause male infertility, such as in teratozoaspermia and asthenospermia, which are two major types of male infertility. In addition, these findings provide candidate molecules for drug targets for male contraception and male infertility.
Functional proteomics of a mouse model of normal spermatogenesis
During the first wave of spermatogenesis, germ cells multiply and differentiate in a synchronous manner in the testis. During different time points of testis development, the germ cell populations are pure and enriched. Consistent with previous literature,31 haematoxylin and eosin staining demonstrated that the mitotic phase of mouse testis lasts for ∼11 days, the meiotic phase for ∼10 days and the post-meiotic phase for ∼14 days. Accordingly, 2DE and MS technologies were utilized to construct a comparative proteome profile for mouse testis at specific time points (days 0, 7, 14, 21, 28 and 60 postpartum) and identified 362 differential protein spots corresponding to 257 different proteins involved in the initiation of mouse spermatogenesis. Furthermore, cluster analysis revealed six expression patterns, and bioinformatics analysis revealed that each pattern was related to many specific cell processes.32
In fact, Paz et al.33 in 2006 used the same technology to obtain proteome profile changes during mouse testis development. In their work, testes from 8-, 18- and 45-day-old postnatal mice were comparatively studied and 44 proteins or variant forms were identified. However, these time points representing the first wave of spermatogenesis were not exact enough and this time span may have been too large. So, additional time points were chosen using haematoxylin and eosin staining. Comparing these two profiles, numerous proteins were examined and 25 proteins from 44 spots were identified in our laboratory.32 Among them, AOPA1 and GSTM2 were both downregulated during testis development. In contrast, PGK2 and PRDX4 were both upregulated during development (Figure 3).
Figure 3.
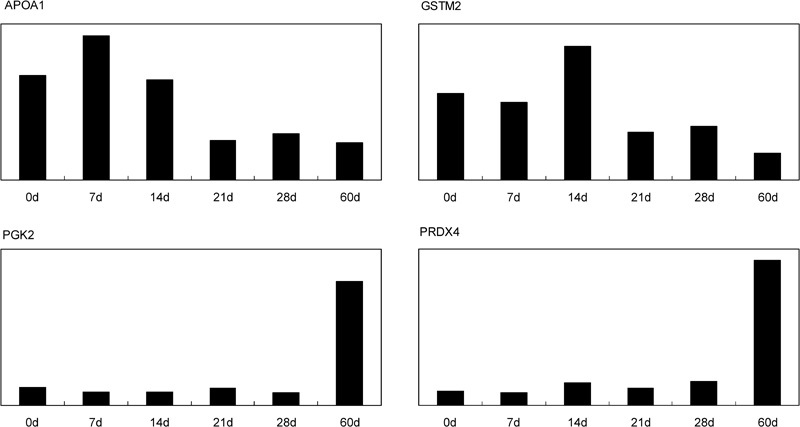
Expression tendency of four proteins in our proteome profile respectively. APOA1 was highly expressed in spermatogonia, whereas it was downregulated during spermatogenesis. GSTM2 was highly expressed in primary spermatocytes, whereas the expression level was reduced with spermiogenesis. PGK2 and PRDX4 were upregulated during mouse spermatogenesis, and the highest expression level was found in spermatozoa. The vertical axis indicates expression levels of these proteins in our studies. The horizontal axis indicates different time points of mouse development stages. d, day.
For many years, it was thought that protein translation did not occur in mature sperm cells because they are terminally differentiated cells in vitro and do not contain sufficient 80S ribosomal complexes, which are degraded or removed during spermatozoal maturation.34 However, it has recently been demonstrated that there is protein translation in mature sperm.35 The incorporation of labelled amino acids into polypeptides during sperm capacitation was completely inhibited by the mitochondrial translation inhibitor 𝒹-chloramphenicol but not by the cytoplasmic translation inhibitor cycloheximide, and inhibition of protein translation significantly reduced sperm motility, capacitation and in vitro fertilization rates.36 Thus, to determine which proteins were translated and to examine the importance of translation during capacitation, we studied the changes in protein expression during capacitation and determined which proteins were newly translated by mitochondrial-type ribosomes using proteomic approaches. A total of 44 proteins were identified with reduced expression in chloramphenicol-treated sperm in comparison with capacitated sperm, and a total of 26 of 44 proteins were involved in some critical processes correlated to sperm–egg interaction events via bioinformatics analysis.37
Functional proteomics of a human model of abnormal spermatogenesis
Azoospermic testes model
Testicular tissues with spermatogenic failure can be used as an alternative model to study spermatogenesis and to identify the genes and proteins involved in this process. Clinically, testicular azoospermia can be classified into several different types according to their pathomorphologies, such as Sertoli cell-only syndrome, germinal arrest, sloughing and disorganisation of germ cells, and hypospermatogenesis.38 Here, we chose pathological testes with the sloughing and disorganisation of germ cells for further studies. 2DE and MS were used to compare the proteome differences between normozoospermic testes and azoospermic testes. In all, 12 spots corresponding to 10 proteins were identified as differentially expressed. Among them, seven proteins (IDH3A, THIC, GPX4, HSP27, SAMP, CTSD and PRX4) were downregulated and three proteins (BLVRB, CAH1 and PRX2) were upregulated in azoospermic testes. Then, three proteins (PRX4, HSP27 and CTSD) were further verified for expression level changes via immunohistochemical analysis. The above proteins may be useful molecular markers to predict clinical parameters of azoospermic patients.39
Asthenospermic sperm model
With no physiologically active transcription or translation, spermatozoa are ideal cells to study from a proteomic perspective.40, 41 There have been several reports using 2DE-MS or LC-MS to identify a limited number of proteins from human spermatozoa. In 2005, Johnston et al.42 used an LC-MS approach to identify 1760 proteins in human sperm. Then in 2006, Martínez-Heredia et al.43 provided a reference two-dimensional map of the mature human sperm proteome with 131 proteins identified. Other studies have also revealed a similar number of proteins.44, 45, 46 These recent global analyses of human sperm are landmark studies and provide rich resources for exploring normal sperm physiology, including sperm motility.
Asthenozoospermia is one of the most common findings with reduced sperm motility in infertile males; however, the molecular basis of asthenozoospermia is not fully understood.47, 48 In 2006, our laboratory used 2DE and MS to compare sperm protein expression profiles in asthenozoospermic patients with those of normozoospermic donors. We selected eight controls with normal sperm motility and eight asthenozoospermic patients with rapid motility (grade A) of 0–3% and progressive motility (grades A+B) of 5–20% within 60 min after ejaculation. In all, 14 differentially expressed protein spots corresponding to 10 proteins were identified. Half of these proteins were enzymes associated with sperm energy metabolism. We identified isocitrate dehydrogenase subunit α as two protein spots that exhibited lower expression in asthenozoospermic patients, whereas three other enzymes (phosphoglycerate mutase 2, triosephosphate isomerase and glutamate oxaloacetate transaminase-1) were highly expressed in most of the patients.49 Among these findings, those relating to triosephosphate isomerase were recently validated by Siva et al..50 Their study used the same strategy to analyse differentially expressed proteins in asthenozoospermic patients, and eight proteins with higher triosephosphate isomerase were identified in patients. We propose that in the case of reduced motile sperm, the overexpression of this enzyme may have been a compensatory reaction.
Semenogelin I precursor was identified as four spots in our study.49 Two of these spots represented 14 kDa fragments and the other two were 17 kDa fragments. All of these spots were highly expressed in asthenozoospermic patients. In 2008, Martínez-Heredia et al.51 also used 2DE-MS to identify 17 protein spots with differential expression levels between asthenozoospermic samples and controls. Notably, semenogelin has been shown to increase in asthenozoospermic samples, which was in good agreement with our present proteomic results. Yoshida et al.52 demonstrated that a 14 kDa fragment of semenogelin has an inhibitory effect on ejaculated spermatozoa. Overall, proteomic analysis techniques were a valuable tool for identifying semenogelin as a sperm motility inhibitor.
As to the above three comparative proteomic studies between asthenozoospermic patients and normozoospermic controls, our inclusion criteria may have been stricter in selecting patients. In the other studies,50, 51 asthenozoospermic samples were defined as those with 25% progressive motility (grade A) or 50% motile sperm (grades A+B). The samples in our study49 were from patients with severe asthenozoospermia, and we think that the proteins identified represent clinically relevant molecular targets for sperm motility that will provide a better understanding of male infertility.
Hormone-induced suppression model of spermatogenesis
The exogenous administration of testosterone alone or in combination with a progestin induces the suppression of spermatogenesis in rats, monkeys and humans.53, 54, 55 In Chinese men, injectable testosterone undecanoate (TU) alone or in combination with oral levonorgestrel (LNG) resulted in the relatively slow suppression of spermatogenesis.56 Thus, this study provides an ideal model with hormone-induced suppression of spermatogenesis. We investigated differential protein expression via 2DE-MS in human testicular biopsies at 2 weeks after treatment with TU alone or TU+LNG. A total of 17 and 46 protein spots were found to exhibit significant differential expression after treatment with TU alone or TU+LNG.57 These key proteins may be the early molecular targets responsible for the suppression of spermatogenesis induced by hormone treatment and may provide new avenues for the management of human infertility and novel targets for contraceptive development.
Perspective
2DE-MS is a well-established technique with well-developed reagents and instrumentation. The trademark of 2DE is the ability to separate proteins according to isoelectric point and molecular mass. This technique is often combined with MS and is often used in expression proteomics and for the analysis of PTMs and protein complexes. Nevertheless, the technique has its limitations, such as poor reproducibility and low sensitivity in identifying proteins with pH values that are too low (pH<3) or too high (pH>10) and molecular masses that are too small (Mr<10 kDa) or too large (Mr>150 kDa).58, 59 However, 2DE-MS is especially useful in studying PTMs. There are two ways to study PTMs by means of 2DE. First, PTMs that alter the molecular weight and/or pI of a protein are characterized by a shift in the location of the corresponding protein spot in a gel. Second, with western blotting, antibodies specific for PTMs can identify spots in a gel containing proteins with these modifications.60 In our studies,21, 22, 32, 37, 39, 49, 57 most of the proteome profiles related to spermatogenesis were established by 2DE-MS, and many proteins were represented by two or more spots in the gel, which indicated PTMs such as phosphorylation.
Recently, fluorescence two-dimensional difference gel electrophoresis has been developed, which allows proteins from up to three samples to be labelled with one of three fluorescent dyes (cyanines 2, 3 and 5). These differentially labelled samples can be mixed and loaded together on one single gel, thereby allowing the quantitative comparative analysis of three samples using a single gel.61 In 2006, Rolland et al.62 used two-dimensional difference gel electrophoresis to compare specific protein profiles of different germ cell types from rat and to identify a first set of 123 unique proteins differentially expressed within the rat male germ lineage. The difference gel electrophoresis technique has dramatically improved the reproducibility and accuracy of quantification; however, its labelling chemistry has some limitations. For example, proteins without lysine cannot be labelled, and they require special equipment for visualisation. Additionally, these fluorophores are very expensive;63, 64 hence, this technique is not widely used.
LC with MS is a gel-free technique where non-separated proteins are digested and a mixture of peptides from all proteins is subjected to LC and then MS. The combination of LC-MS is particularly suitable for high throughput studies because it allows highly automated identification of proteins and specific sites of PTMs. In our studies,25, 30 we used LC-MS to identify proteins from human testis and tetraploid and haploid germ cells in mouse testis. However, LC-MS cannot match proteins identified by software with their Mr and pI, and another major limitation is that this technique is not suitable for quantitative analysis.59, 65
To overcome the shortcomings of 2DE and LC as individual techniques, this study21, 30, 32 combined 2DE and LC to explore protein changes in cells and tissues. In mouse testis, we used 2DE to construct an overall proteome profile and comparative proteome profiles of different stages of spermatogenesis, which revealed that many proteins appeared as two or more spots. Using LC, we identified a number of proteins in the tetraploid and haploid germ cells. Thus, using a combination of 2DE and LC, we obtained a complete protein profile and protein–protein interaction map for testicular development and spermatogenesis. Proteome profiles related to spermatogenesis will help clarify the molecular mechanisms underlying male fertility and infertility.
The authors declare no competing financial interests.
References
- Baccetti B, Afzelius BA. The biology of the sperm cell. Monogr Dev Biol. 1976;10:1–254. [PubMed] [Google Scholar]
- Eddy EM. Male germ cell gene expression. Recent Prog Horm Res. 2002;57:103–28. doi: 10.1210/rp.57.1.103. [DOI] [PubMed] [Google Scholar]
- Anway MD, Li Y, Ravindranath N, Dym M, Griswold MD. Expression of testicular germ cell genes identified by differential display analysis. J Androl. 2003;24:173–84. doi: 10.1002/j.1939-4640.2003.tb02660.x. [DOI] [PubMed] [Google Scholar]
- Sha J, Zhou Z, Li J, Yin L, Yang H, et al. Identification of testis development and spermatogenesis-related genes in human and mouse testes using cDNA arrays. Mol Hum Reprod. 2002;8:511–7. doi: 10.1093/molehr/8.6.511. [DOI] [PubMed] [Google Scholar]
- Guo R, Yu Z, Guan J, Ge Y, Ma J, et al. Stage-specific and tissue-specific expression characteristics of differentially expressed genes during mouse spermatogenesis. Mol Reprod Dev. 2004;67:264–72. doi: 10.1002/mrd.20026. [DOI] [PubMed] [Google Scholar]
- Schlecht U, Demougin P, Koch R, Hermida L, Wiederkehr C, et al. Expression profiling of mammalian male meiosis and gametogenesis identifies novel candidate genes for roles in the regulation of fertility. Mol Biol Cell. 2004;15:1031–43. doi: 10.1091/mbc.E03-10-0762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gygi SP, Rochon Y, Franza BR, Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol Cell Biol. 1999;19:1720–30. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrads KA, Yi M, Simpson KA, Lucas DA, Camalier CE, et al. A combined proteome and microarray investigation of inorganic phosphate-induced pre-osteoblast cells. Mol Cell Proteomics. 2005;4:1284–96. doi: 10.1074/mcp.M500082-MCP200. [DOI] [PubMed] [Google Scholar]
- Venables JP. Alternative splicing in the testes. Curr Opin Genet Dev. 2002;12:615–9. doi: 10.1016/s0959-437x(02)00347-7. [DOI] [PubMed] [Google Scholar]
- DeJong J. Basic mechanisms for the control of germ cell gene expression. Gene. 2006;366:39–50. doi: 10.1016/j.gene.2005.10.012. [DOI] [PubMed] [Google Scholar]
- Tyers M, Mann M. From genomics to proteomics. Nature. 2003;422:193–7. doi: 10.1038/nature01510. [DOI] [PubMed] [Google Scholar]
- Cox J, Mann M. Is proteomics the new genomics. Cell. 2007;130:395–8. doi: 10.1016/j.cell.2007.07.032. [DOI] [PubMed] [Google Scholar]
- Wilkins MR, Sanchez JC, Gooley AA, Appel RD, Humphery-Smith I, et al. Progress with proteome projects: why all proteins expressed by a genome should be identified and how to do it. Biotechnol Genet Eng Rev. 1995;13:19–50. doi: 10.1080/02648725.1996.10647923. [DOI] [PubMed] [Google Scholar]
- Graves PR, Haystead TA. Molecular biologist's guide to proteomics. Microbiol Mol Biol Rev. 2002;66:39–63. doi: 10.1128/MMBR.66.1.39-63.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futcher B, Latter GI, Monardo P, McLaughlin CS, Garrels JI. A sampling of the yeast proteome. Mol Cell Biol. 1999;19:7357–68. doi: 10.1128/mcb.19.11.7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitken RJ, Baker MA. The role of proteomics in understanding sperm cell biology. Int J Androl. 2008;31:295–302. doi: 10.1111/j.1365-2605.2007.00851.x. [DOI] [PubMed] [Google Scholar]
- Aitken RJ, Nixon B, Lin M, Koppers AJ, Lee YH, et al. Proteomic changes in mammalian spermatozoa during epididymal maturation. Asian J Androl. 2007;9:554–64. doi: 10.1111/j.1745-7262.2007.00280.x. [DOI] [PubMed] [Google Scholar]
- Oliva R, de Mateo S, Estanyol JM. Sperm cell proteomics. Proteomics. 2009;9:1004–17. doi: 10.1002/pmic.200800588. [DOI] [PubMed] [Google Scholar]
- Oliva R, Martínez-Heredia J, Estanyol JM. Proteomics in the study of the sperm cell composition, differentiation and function. Syst Biol Reprod Med. 2008;54:23–36. doi: 10.1080/19396360701879595. [DOI] [PubMed] [Google Scholar]
- Calvel P, Rolland AD, Jégou B, Pineau C. Testicular postgenomics: targeting the regulation of spermatogenesis. Philos Trans R Soc Lond B Biol Sci. 2010;365:1481–500. doi: 10.1098/rstb.2009.0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu YF, Cui YG, Guo XJ, Wang L, Bi Y, et al. Proteomic analysis of effect of hyperthermia on spermatogenesis in adult male mice. J Proteome Res. 2006;5:2217–25. doi: 10.1021/pr0600733. [DOI] [PubMed] [Google Scholar]
- Guo X, Zhao C, Wang F, Zhu Y, Cui Y, et al. Investigation of human testis protein heterogeneity using 2-dimensional electrophoresis. J Androl. 2010;31:419–29. doi: 10.2164/jandrol.109.007534. [DOI] [PubMed] [Google Scholar]
- Conrotto P, Souchelnytskyi S. Proteomic approaches in biological and medical sciences: principles and applications. Exp Oncol. 2008;30:171–80. [PubMed] [Google Scholar]
- Steinberg TH, Agnew BJ, Gee KR, Leung WY, Goodman T, et al. Global quantitative phosphoprotein analysis using Multiplexed Proteomics technology. Proteomics. 2003;3:1128–44. doi: 10.1002/pmic.200300434. [DOI] [PubMed] [Google Scholar]
- Guo XJ, Zhang P, Huo R, Zhou ZM, Sha JH. Analysis of the human testis proteome by mass spectrometry and bioinformatics. Proteomics Clin Appl. 2008;2:1651–7. doi: 10.1002/prca.200780120. [DOI] [PubMed] [Google Scholar]
- Berruti G. Signaling events during male germ cell differentiation: bases and perspectives. Front Biosci. 1998;3:D1097–108. doi: 10.2741/a347. [DOI] [PubMed] [Google Scholar]
- Moreno RD, Alvarado CP. The mammalian acrosome as a secretory lysosome: new and old evidence. Mol Reprod Dev. 2006;73:1430–4. doi: 10.1002/mrd.20581. [DOI] [PubMed] [Google Scholar]
- Toshimori K, Ito C. Formation and organization of the mammalian sperm head. Arch Histol Cytol. 2003;66:383–96. doi: 10.1679/aohc.66.383. [DOI] [PubMed] [Google Scholar]
- Escalier D. Knockout mouse models of sperm flagellum anomalies. Hum Reprod Update. 2006;12:449–61. doi: 10.1093/humupd/dml013. [DOI] [PubMed] [Google Scholar]
- Guo X, Shen J, Xia Z, Zhang R, Zhang P, et al. Proteomic analysis of proteins involved in spermiogenesis in mouse. J Proteome Res. 2010;9:1246–56. doi: 10.1021/pr900735k. [DOI] [PubMed] [Google Scholar]
- Clermont Y, Trott M. Duration of the cycle of the seminiferous epithelium in the mouse and hamster determined by means of 3H-thymidine and radioautography. Fertil Steril. 1969;20:805–17. doi: 10.1016/s0015-0282(16)37153-9. [DOI] [PubMed] [Google Scholar]
- Huang XY, Guo XJ, Shen J, Wang YF, Chen L, et al. Construction of a proteome profile and functional analysis of the proteins involved in the initiation of mouse spermatogenesis. J Proteome Res. 2008;7:3435–46. doi: 10.1021/pr800179h. [DOI] [PubMed] [Google Scholar]
- Paz M, Morín M, del Mazo J. Proteome profile changes during mouse testis development. Comp Biochem Physiol Part D Genomics Proteomics. 2006;1:404–15. doi: 10.1016/j.cbd.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Miller D, Ostermeier GC. Spermatozoal RNA: why is it there and what does it do. Gynecol Obstet Fertil. 2006;34:840–6. doi: 10.1016/j.gyobfe.2006.07.013. [DOI] [PubMed] [Google Scholar]
- Gur Y, Breitbart H. Protein synthesis in sperm: dialog between mitochondria and cytoplasm. Mol Cell Endocrinol. 2008;282:45–55. doi: 10.1016/j.mce.2007.11.015. [DOI] [PubMed] [Google Scholar]
- Gur Y, Breitbart H. Mammalian sperm translate nuclear encoded proteins by mitochondrial-type ribosomes. Genes Dev. 2006;20:411–6. doi: 10.1101/gad.367606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Guo XJ, Shi ZH, Wang FQ, Huang XY, et al. Role of translation by mitochondrial-type ribosomes during sperm capacitation: an analysis based on a proteomic approach. Proteomics. 2009;9:1385–99. doi: 10.1002/pmic.200800353. [DOI] [PubMed] [Google Scholar]
- Sharif K. Reclassification of azoospermia: the time has come. Hum Reprod. 2000;15:237–8. doi: 10.1093/humrep/15.2.237. [DOI] [PubMed] [Google Scholar]
- Huo R, He Y, Zhao C, Guo XJ, Lin M, et al. Identification of human spermatogenesis-related proteins by comparative proteomic analysis: a preliminary study. Fertil Steril. 2008;90:1109–18. doi: 10.1016/j.fertnstert.2007.07.1342. [DOI] [PubMed] [Google Scholar]
- Boerke A, Dieleman SJ, Gadella BM. A possible role for sperm RNA in early embryo development. Theriogenology. 2007;68:S147–55. doi: 10.1016/j.theriogenology.2007.05.058. [DOI] [PubMed] [Google Scholar]
- Barratt CL. The human sperm proteome: the potential for new biomarkers of male fertility and a transformation in our understanding of the spermatozoon as a machine: commentary on the article ‘Identification of proteomic differences in asthenozoospermic sperm samples' by Martinez et al. Hum Reprod. 2008;23:1240–1. doi: 10.1093/humrep/den019. [DOI] [PubMed] [Google Scholar]
- Johnston DS, Wooters J, Kopf GS, Qiu Y, Roberts KP. Analysis of the human sperm proteome. Ann NY Acad Sci. 2005;1061:190–202. doi: 10.1196/annals.1336.021. [DOI] [PubMed] [Google Scholar]
- Martínez-Heredia J, Estanyol JM, Ballescà JL, Oliva R. Proteomic identification of human sperm proteins. Proteomics. 2006;6:4356–69. doi: 10.1002/pmic.200600094. [DOI] [PubMed] [Google Scholar]
- Pixton KL, Deeks ED, Flesch FM, Moseley FL, Bjorndahl L, et al. Sperm proteome mapping of a patient who experienced failed fertilization at IVF reveals altered expression of at least 20 proteins compared with fertile donors: case report. Hum Reprod. 2004;19:1438–47. doi: 10.1093/humrep/deh224. [DOI] [PubMed] [Google Scholar]
- Naaby-Hansen S, Flickinger CJ, Herr JC. Two-dimensional gel electrophoretic analysis of vectorially labeled surface proteins of human spermatozoa. Biol Reprod. 1997;56:771–87. doi: 10.1095/biolreprod56.3.771. [DOI] [PubMed] [Google Scholar]
- Secciani F, Bianchi L, Ermini L, Cianti R, Armini A, et al. Protein profile of capacitated versus ejaculated human sperm. J Proteome Res. 2009;8:3377–89. doi: 10.1021/pr900031r. [DOI] [PubMed] [Google Scholar]
- Luconi M, Forti G, Baldi E. Pathophysiology of sperm motility. Front Bios. 2006;11:1433–47. doi: 10.2741/1894. [DOI] [PubMed] [Google Scholar]
- Kierszenbaum AL, Tres LL. The acrosome–acroplaxome–manchette complex and the shaping of the spermatid head. Arch Histol Cytol. 2004;67:271–84. doi: 10.1679/aohc.67.271. [DOI] [PubMed] [Google Scholar]
- Zhao C, Huo R, Wang FQ, Lin M, Zhou ZM, et al. Identification of several proteins involved in regulation of sperm motility by proteomic analysis. Fertil Steril. 2007;87:436–8. doi: 10.1016/j.fertnstert.2006.06.057. [DOI] [PubMed] [Google Scholar]
- Siva AB, Kameshwari DB, Singh V, Pavani K, Sundaram CS, et al. Proteomics-based study on asthenozoospermia: differential expression of proteasome alpha complex. Mol Hum Reprod. 2010;16:452–62. doi: 10.1093/molehr/gaq009. [DOI] [PubMed] [Google Scholar]
- Martínez-Heredia J, de Mateo S, Vidal-Taboada JM, Ballescà JL, Oliva R. Identification of proteomic differences in asthenozoospermic sperm samples. Hum Reprod. 2008;23:783–91. doi: 10.1093/humrep/den024. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Yamasaki T, Yoshiike M, Takano S, Sato I, et al. Quantification of seminal plasma motility inhibitor/semenogelin in human seminal plasma. J Androl. 2003;24:878–84. doi: 10.1002/j.1939-4640.2003.tb03139.x. [DOI] [PubMed] [Google Scholar]
- Lue Y, Hikim AP, Wang C, Im M, Leung A, et al. Testicular heat exposure enhances the suppression of spermatogenesis by testosterone in rats: the “two-hit” approach to male contraceptive development. Endocrinology. 2000;141:1414–24. doi: 10.1210/endo.141.4.7416. [DOI] [PubMed] [Google Scholar]
- Lue Y, Wang C, Liu YX, Hikim AP, Zhang XS, et al. Transient testicular warming enhances the suppressive effect of testosterone on spermatogenesis in adult cynomolgus monkeys (Macaca fascicularis) J Clin Endocrinol Metab. 2006;91:539–45. doi: 10.1210/jc.2005-1808. [DOI] [PubMed] [Google Scholar]
- Wang C, Wang XH, Nelson AL, Lee KK, Cui YG, et al. Levonorgestrel implants enhanced the suppression of spermatogenesis by testosterone implants: comparison between Chinese and non-Chinese men. J Clin Endocrinol Metab. 2006;91:460–70. doi: 10.1210/jc.2005-1743. [DOI] [PubMed] [Google Scholar]
- Wang C, Cui YG, Wang XH, Jia Y, Sinha Hikim AP, et al. Transient testicular hyperthermia enhanced spermatogenesis suppression by testosterone in men. J Clin Endocrinol Metab. 2007;92:3202–4. doi: 10.1210/jc.2007-0367. [DOI] [PubMed] [Google Scholar]
- Cui Y, Zhu H, Zhu Y, Guo X, Huo R, et al. Proteomic analysis of testis biopsies in men treated with injectable testosterone undecanoate alone or in combination with oral levonorgestrel as potential male contraceptive. J Proteome Res. 2008;7:3984–93. doi: 10.1021/pr800259t. [DOI] [PubMed] [Google Scholar]
- Baltimore D. Our genome unveiled. Nature. 2001;409:814–6. doi: 10.1038/35057267. [DOI] [PubMed] [Google Scholar]
- Conrotto P, Souchelnytskyi S. Proteomic approaches in biological and medical sciences: principles and applications. Exp Oncol. 2008;30:171–80. [PubMed] [Google Scholar]
- Boyd-Kimball D, Poon HF, Lynn BC, Cai J, Pierce WM, Jr, et al. Proteomic identification of proteins specifically oxidized in Caenorhabditis elegans expressing human Abeta(1–42): implications for Alzheimer's disease. Neurobiol Aging. 2006;27:1239–49. doi: 10.1016/j.neurobiolaging.2005.07.001. [DOI] [PubMed] [Google Scholar]
- van den Bergh G, Arckens L. Recent advances in 2D electrophoresis: an array of possibilities. Expert Rev Proteomics. 2005;2:243–52. doi: 10.1586/14789450.2.2.243. [DOI] [PubMed] [Google Scholar]
- Rolland AD, Evrard B, Guitton N, Lavigne R, Calvel P, et al. Two-dimensional fluorescence difference gel electrophoresis analysis of spermatogenesis in the rat. J Proteome Res. 2007;6:683–97. doi: 10.1021/pr060436z. [DOI] [PubMed] [Google Scholar]
- Marouga R, David S, Hawkins E. The development of the DIGE system: 2D fluorescence difference gel analysis technology. Anal Bioanal Chem. 2005;382:669–78. doi: 10.1007/s00216-005-3126-3. [DOI] [PubMed] [Google Scholar]
- van den Bergh G, Arckens L. Fluorescent two dimensional difference gel electrophoresis unveils the potential of gel-based proteomics. Curr Opin Biotechnol. 2004;15:38–43. doi: 10.1016/j.copbio.2003.12.001. [DOI] [PubMed] [Google Scholar]
- Wingren C, James P, Borrebaeck CA. Strategy for surveying the proteome using affinity proteomics and mass spectrometry. Proteomics. 2009;9:1511–7. doi: 10.1002/pmic.200800802. [DOI] [PubMed] [Google Scholar]


