Abstract
There are fifteen different DNA polymerases encoded in mammalian genomes, which are specialized for replication, repair or the tolerance of DNA damage. New evidence is emerging for lesion-specific and tissue-specific functions of DNA polymerases. Many point mutations that occur in cancer cells arise from the error-generating activities of DNA polymerases. However, the ability of some of these enzymes to bypass DNA damage may actually defend against chromosome instability in cells and at least one DNA polymerase, POLζ, is a suppressor of spontaneous tumorigenesis. Because DNA polymerases can help cancer cells tolerate DNA damage, some of these enzymes may be viable targets for therapeutic strategies.
DNA polymerases are enzymes that synthesize DNA. These proteins have an essential role in genome duplication, but they are also critical for protecting the cell against the effects of DNA damage. In both normal and cancer cells, DNA is subjected to damage from many sources. Water-catalyzed reactions and attack by reactive oxygen species (ROS) are inescapable and inflict continual damaging alterations in DNA. Other ubiquitous sources of lesions include naturally occurring ionizing radiation, ultraviolet (UV) radiation from the sun, and reactive chemicals that are present in the environment or arise as natural metabolites. The toxic and mutagenic consequences of such damage are minimized by distinct DNA repair pathways including base excision repair (BER) and nucleotide excision repair (NER) (Box 1). These repair mechanisms rely on a DNA polymerase to fill gaps in the DNA left by the removal of damaged bases. If the DNA damage is unrepaired, cells can often tolerate it by using a specialized DNA polymerase during DNA replication to insert a base opposite a lesion and bypass the damage, in a process called translesion DNA synthesis (TLS) (Figures 1 and 2). This process is responsible for many of the point mutations in cells1, and is particularly relevant to the large increase in point mutations found in cancer genomes compared with normal tissues2. Another method by which cells tolerate DNA damage encountered at replication forks is template switching, which can be mediated by recombination and is primarily error-free3. Finally, breaks and gaps can arise in DNA, and repair mechanisms must be called upon, including homologous recombination (HR) and various non-homologous end-joining processes. DNA polymerases are also essential components of these pathways (Box 1).
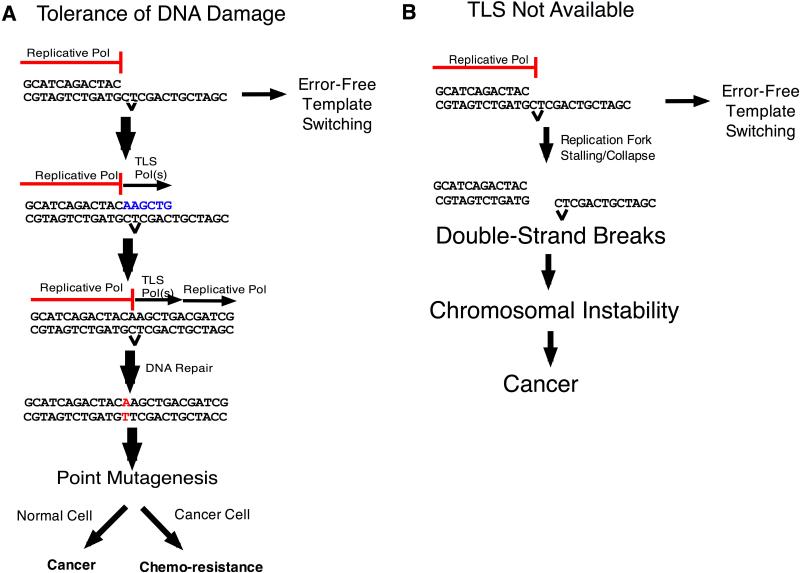
Figure 1. DNA damage tolerance and carcinogenesis.
A) Replication forks can be blocked by lesions in DNA. When a translesion DNA synthesis (TLS) polymerase is available, a lesion can be bypassed by TLS, potentially causing point mutagenesis, or template switching can mediate bypass in an error-free manner (see Figure 2 for details). Point mutagenesis can lead to cancer formation in normal cells, or to resistance to therapeutic agents in cancer cells. B) In the complete absence of a TLS polymerase, no translesion bypass occurs (although there will still be some template switching) and replication forks collapse, leading to double-strand breaks and chromosome instability, which increases the incidence of cancer.
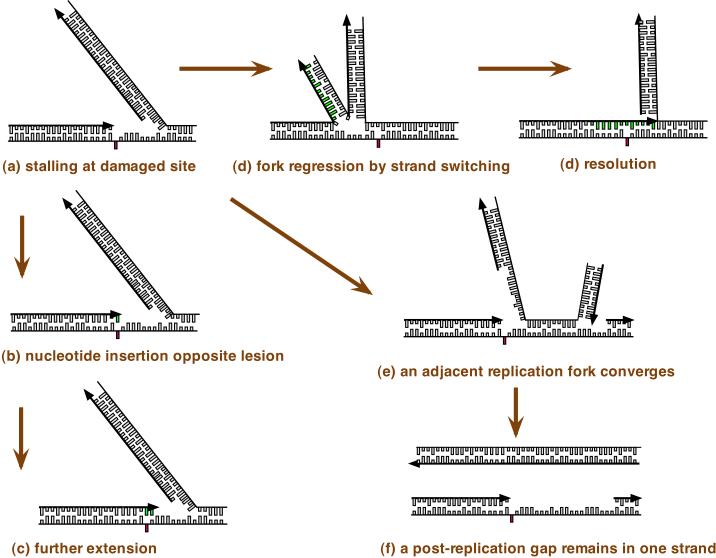
Figure 2. Strategies for translesion DNA synthesis.
Some types of DNA damage, if not repaired, will block the progression of a DNA replication fork. When a site of DNA damage on the leading strand is encountered by the DNA replication machinery and this prevents normal base pairing (red rectangle), replication is blocked. The lagging strand may continue replication, but the leading strand on which the replication machinery is blocked is fragile. Replication on the two strands can uncouple and dissociation of the DNA replication machinery causes ‘collapse’ of the DNA replication fork, eventually leading to a DNA break (Figure 1). Several possible strategies to overcome this block to replication may be activated. One strategy (part a) is to carry out translesion DNA synthesis (TLS) by successive steps. The replication machinery switches to a specialized DNA polymerase for insertion of a base. This step is potentially mutagenic because the wrong base will sometimes be incorporated. A switch to a second specialized DNA polymerase may take place to extend the nonstandard terminus opposite the damage, and finally there is a switch to a replicative DNA polymerase (POLε or POLδ). DNA polymerase switching is facilitated by post-translational modifications of DNA polymerases and their accessory factors, as summarized in the text and reviewed in depth elsewhere 1, 3, 98, 99. A second strategy (b) is DNA replication fork regression. Here, the blocked leading strand switches templates and begins to copy the already-replicated lagging strand. The newly-replicated bases are shown in green. The regressed fork resembles a four-way junction that can be processed by homologous recombination enzymes and resolved. This pathway avoids errors, as it makes use of genetic information from the undamaged strand. A third strategy is illustrated in part c. If the replication fork remains stalled for long enough, an adjacent replication fork will converge with it. This allows one strand to replicate fully, while one strand will contain a gap. This gap will then remain through to late S phase or G2 phase of the cell cycle. The gap is then filled by DNA synthesis. During gap filling, two different specialized DNA polymerases may also be needed to accomplish synthesis across from a lesion, for insertion and extension, and this is potentially mutagenic. Gaps could also conceivably arise by re-initiation of DNA synthesis on the other side of a DNA adduct. Arrows indicate the direction of DNA replication, which is 5’ to 3’ with respect to the deoxyribose sugar-phosphate.
DNA strand breaks, particularly double-strand breaks (DSBs), seem to be formed more frequently in cancer cells than in normal cells. This is probably a consequence of the frequent disruption of normal controls on DNA replication in cancer cells4. For example, loss of p53 function, which is frequent in cancer cells, weakens a checkpoint control that would normally prohibit cells from initiating DNA replication when breaks are present. Many cancer cells exhibit a heightened basal level of activation of some responses to DNA breaks, accumulating HR and other repair proteins on chromatin. This may be caused by the expression of oncoproteins, which is hypothesized to lead to recurrent initiation and collision of DNA replication forks, thereby resulting in increased numbers of DNA4.
Fifteen mammalian DNA polymerases have been identified (Table 1). Some of them function in replication of the genome, but most of them participate in DNA repair and TLS. Here we describe how DNA polymerases are specialized for distinct cellular mechanisms, focusing on the roles of DNA polymerases in carcinogenesis and tumorigenesis. Insights into these roles are anticipated to provide new approaches for cancer prevention, diagnosis and treatment. Much background understanding of DNA polymerases has come from research done with the yeast Saccharomyces cerevisiae, providing knowledge that has proven to be predictive of the situation in other eukaryotes. Here, we primarily focus on DNA polymerases in vertebrates.
Table 1.
Mammalian DNA polymerases
| DNA polymerase | Gene (human protein size) and protein domain structure | Function | Family |
|---|---|---|---|
| POLα |
|
DNA replication priming | B |
| POLδ |
|
DNA replication, NER, MMR | B |
| POLε |
|
DNA replication, NER, MMR | B |
| POLγ |
|
Mitochondrial DNA replication and repair | A |
| POLβ |
|
BER and meiotic recombination | X |
| POLλ |
|
V(D)J recombination; possibly end-joining and BER | X |
| POLμ |
|
V(D)J recombination; possibly end-joining | X |
| TDT |
|
Immunoglobulin diversity at junctions of coding regions | X |
| POLζ |
|
TLS and mutagenesis | B |
| REV1 |
|
TLS and mutagenesis, anchor for several DNA polymerases | Y |
| POLη |
|
Bypass of UV radiation-induced DNA adducts, especially CPDs | Y |
| POLι |
|
Backup enzyme for bypass of UV radiation-induced DNA adducts and BER | Y |
| POLκ |
|
Bypass of bulky adducts, backup enzyme for NER | Y |
| POLθ |
|
Defence against ionizing radiation-induced DNA damage | A |
| POLν |
|
ICL repair or testis-specific function? | A |
BER, base excision repair; CPD, cyclobutane pyrimidine dimer; ICL, interstrand crosslink; MMR, mismatch repair; NER, nucleotide excision repair; NHEJ, non-homologous end-joining; TLS, translesion DNA synthesis; TDT, terminal deoxynucleotidyltransferase.  : DNA polymerase domain,
: DNA polymerase domain,  : exonuclease domain,
: exonuclease domain,  : dRP lyase domain,
: dRP lyase domain,  : dRP lyase activity,
: dRP lyase activity,  : BRCT domain,
: BRCT domain,  : helicase-like domain. Most eukaryotic DNA polymerase proteins are named with Greek letters (α, β, γ, etc.) and the genes are named with the corresponding roman letter. In mammalian cells, these enzymes fall into four distinct families, designated A, B, X and Y, based on amino acid sequence relationships.
: helicase-like domain. Most eukaryotic DNA polymerase proteins are named with Greek letters (α, β, γ, etc.) and the genes are named with the corresponding roman letter. In mammalian cells, these enzymes fall into four distinct families, designated A, B, X and Y, based on amino acid sequence relationships.
Replicative DNA polymerases
Genomic DNA in the nucleus is normally replicated accurately by DNA polymerases α, ™ δ and ε. POLα initiates DNA synthesis on both the leading strands and lagging strands by providing an RNA primer and synthesizing approximately 20-30 bases of DNA5. POLε and POLδ elongate these primers. In S. cerevisiae, POLε may be especially important for leading strand synthesis and POLδ for lagging strand synthesis6, 7. The base substitution error rates of POLδ and POLε are approximately 10−5, the lowest among all of the characterized DNA polymerases7, and when they do occasionally misincorporate a nucleotide it is usually removed by a 3’-5’ exonuclease associated with these DNA polymerases8. Errors that escape such proofreading can be corrected by the DNA mismatch repair (MMR) pathway (Box 1), so that the spontaneous mutation rate during nuclear DNA replication is very low at less than 10−9 per base pair per cell division9.
The proofreading 3’-5’ exonuclease activities of POLδ and POLε are critical for preventing mutations; cells from Pold1 or Pole1 exonuclease-deficient (exo) mice have a 10-fold increased frequency of mutagenesis, and these mutations have been shown to drive carcinogenesis. Pold1 mutant mice either die by 8 months of age from thymic lymphomas, or they develop skin tumors, lung adenocarcinomas or teratomas10, 11. Pole1exo/exo-deficient mice die prematurely of intestinal adenomas and adenocarcinomas12. Pole1exo/exo; Pold1exo/exo double exonuclease-mutant mice die even more rapidly from thymic lymphomas than single mutant mice 12. Mice carrying a mutator allele of Pold1 (which confers an increase in nucleotide misincorporation and genomic instability) are not viable in the homozygous state, emphasizing the importance of high fidelity DNA replication for survival of an organism13. In view of these results in mouse models, it is intriguing that sporadic sequence changes have been found in POLD1 in human colon cancer cell lines and patient tumour tissue samples14. Most of these changes appear to have no functional effect; however, an R689W mutation caused lethality when modelled as a homologous change in pol3, which encodes the catalytic subunit of POLδ in S. cerevisiae15. The expression of low levels of normal POLδ rescued this lethality, but was associated with an increased mutation rate. It is consequently possible that some mutations in POLD1 or POLE1 might contribute to tumorigenesis or tumour progression in humans by increasing mutation rates.
TLS
Despite the existence of DNA repair mechanisms, some DNA damage escapes repair and can stall the replication machinery. Stalled DNA replication forks appear to be fragile, and if not resolved they can collapse into structures that cause a DSB to be formed, thereby increasing genomic instability (Figure 1).
The main strategy by which cells are able to tolerate DNA damage during replication is by synthesizing DNA past damaged bases (Figure 2). The replicative DNA polymerases have exquisite specificity for normal DNA base pairs, but very little capacity for replication opposite damaged bases8. Mammalian cells have at least seven enzymes with substantial TLS activity. These include four Y-family polymerases (POLη (also known as POLH), POLι (also known as POLI), POLκ (also known as POLK) and REV1), one B-family polymerase (POLη, catalytic subunit REV3L), and two A-family polymerases (POLθ (also known as POLQ) and POLν (also known as POLN). None of the TLS DNA polymerases have proofreading exonuclease activity, and they possess unique DNA damage bypass and fidelity profiles. In the context of TLS, these DNA polymerases are not DNA repair enzymes, but are DNA damage tolerance factors. As described below, physiological roles in lesion bypass are established for only some of these enzymes16, 17.
Bypass of DNA damage caused by UV radiation
The most notable TLS polymerase for bypass of UV radiation-induced DNA damage is POLη. Currently, POLη is the only polymerase for which a deficiency is known to predispose humans to cancer18. The inherited disorder xeroderma pigmentosum is associated with a greatly increased risk of sunlight-induced carcinomas of the skin, and individuals with the variant type of the condition, XP-V, have disabling mutations in POLη18, 19. POLη can bypass a TT cyclobutane pyrimidine dimer (CPD) – the major form of DNA damage induced by UV radiation – with high efficiency and fidelity. Purified human POLη usually correctly inserts A deoxynucleotides opposite the linked bases of a TT-CPD20. Co-crystal structures show that POLη is exquisitely suited for bypassing TT-CPDs as it makes many specific contacts with the lesion within the spacious active site21, 22. Mutations in POLH that occur in individuals with XP-V disable specific interactions of the enzyme with the TT-CPD21, 22.
Polh−/− mice mirror the XP-V phenotype, developing UV radiation-induced tumors rapidly. UV-irradiated heterozygous Polh−/+ mice are also susceptible to UV radiation-induced skin carcinogenesis, at a lesser rate23. UV irradiation of XP-V cells causes DNA DSBs to form because the absence of the TLS function of POLη causes DNA replication forks to stall and collapse at sites of DNA damage on the template strand24. Prolonged replication delay in the absence of POLη may also inhibit DNA repair of UV radiation-induced lesions25. Unirradiated POLH−/− cells have been reported to have more chromatid breaks than normal, including breaks at a common fragile site26.
Patients with XP-V have an increased incidence of squamous cell carcinoma of the skin, and so experiments have been conducted to determine if there are mutations in POLH associated with sporadic skin carcinomas27 or other human cancers. No mutations affecting the function of POLη have yet been identified.
Another frequently formed type of UV radiation-induced DNA damage is the (6-4) photoproduct28, 29. This adduct is more distorting to the DNA double helix than CPDs and is not handled efficiently by POLη30; the enzyme mostly misincorporates one G residue (instead of two As) and cannot continue replicating. POLι is more suited to insert the correct bases opposite UV radiation-induced (6-4) photoproducts29, 31. POLι can also insert bases opposite a TT-CPD, particularly when POLη is absent, although this bypass is mutagenic32. When allowed to replicate undamaged DNA, POLι may be the most error-generating DNA polymerase, commonly misincorporating G opposite a template T before stalling31.
Poli−/− cells are no more sensitive than wild-type cells to DNA damaging agents. However, Poli−/−; Polh−/− double knockout mice have increased rates of UV radiation-induced skin carcinogenesis, compared with Polh−/− mice33, 34. The increase in UV radiation-induced mutagenesis that is observed in Polh−/− mice and cells is suppressed when POLι function is ablated33, 35, suggesting that POLι is an error-generating backup polymerase for the bypass of UV radiation-induced DNA lesions, and may be responsible for skin tumorigenesis in XP-V patients.
Bypass of bulky adducts in DNA
Bulky or helix-distorting DNA adducts are clinically relevant because they are caused by both carcinogenic compounds (such as benzo[a]pyrene diol epoxide (BPDE)) and chemotherapeutic agents (such as cisplatin and mitomycin C (MMC)). Several lines of evidence suggest specialized roles for POLκ in bypassing bulky adducts in DNA. In vitro, POLκ is adept at bypass of adducts formed with the N2 of G in the minor groove of DNA, including BPDE adducts and cross-linked peptides36-38. The promoter region of POLK contains aryl-hydrocarbon receptor binding sites, and its expression can be increased by treatment with the carcinogenic polycyclic aromatic hydrocarbon 3-methylcholanthrene39. Polk−/− mouse embryonic fibroblasts are sensitive to BPDE40 and depletion of POLκ sensitizes cells to apoptosis and chromosome aberrations caused by MMC, which can crosslink N2-Gs38.
Polk−/− mice are viable, although survival is shortened compared with Polk+/− and Polk+/+ mice. Polk−/− mice have a spontaneous mutator phenotype in tissues including kidney, liver and lung, and it has been suggested that POLκ has a role in tolerating bulky DNA adducts generated endogenously, for example by cholesterol metabolism41. Despite its low fidelity when copying undamaged DNA, POLκ appears to protect against spontaneous mutagenesis.
POLη is another polymerase that, to some extent, can incorporate a base opposite bulky types of DNA damage that are produced by carcinogens and chemotherapeutic agents. In addition to its ability to bypass UV radiation-induced DNA damage, POLη can bypass cisplatin-GG adducts20, acetylaminofluorene-G42, lesions induced by ionizing radiation including 8-oxoG43, 44, and a major G adduct, γ-hydroxypropanodeoxyguanosine, formed with the mutagen acrolein45. In experiments where DNA containing a single site of damage is transfected into mammalian cells, POLη participates in the bypass of BPDE-G and cisplatin-GG adducts46 as well as CPDs. Furthermore, XP-V cells are more sensitive than normal cells to cisplatin47. POLη also contributes to replication through naturally occurring alternative DNA structures, such as G-quartets48.
Bypass of AP sites and thymine glycol
Abasic sites (apurinic or apyrimidinic (AP) sites) are the most frequent spontaneous lesions in DNA. They are formed by the release of bases from the sugar-phosphate backbone, and as intermediates in DNA repair. Many DNA polymerases can insert a base opposite an AP site49. POLη and POLι can do so, for example31, 42, but their activities are not needed for AP site bypass in human cells50.
Mammalian POLθ is unique in its ability to both efficiently insert an A opposite an AP site, and extend past it51. It is more proficient at this TLS reaction in vitro than any of the Y-family DNA polymerases. On undamaged template DNA, POLθ has a much lower fidelity than prokaryotic A-family DNA polymerases and a tendency to delete or add single bases during DNA synthesis51, 52.
Thymine glycol residues are a major product of ROS-induced damage to DNA and are frequently caused by ionizing radiation. Several DNA polymerases can bypass thymine glycols, including POLs θ51, η53, κ54, and ν. POLν 55 is unusually proficient at accurate bypass of 5S-thymine glycol56, 57 and of bulky major groove DNA lesions such as N6-A crosslinks58. Further distinctive properties of POLν include its strong strand displacement activity56 and low fidelity56, 59. Physiological roles have been suggested for POLν in TLS past a short olignucleotide crosslinked to DNA, representing an intermediate in interstrand crosslink (ICL) repair, or in HR60, 61. However, POLN−/− chicken DT40 cells are not sensitive to the chemotherapeutic agents MMC, cisplatin62 or camptothecin63. POLN is preferentially expressed in testis, and is expressed weakly in other tissues or in cultured mammalian cells and POLN is located near the telomere of human chromosome 4p, an area of frequent loss in some human cancers64.
POLζ and REV1, master control proteins for TLS
In S. cerevisiae, most mutagenesis induced by DNA damaging agents is dependent on the action of POLζ65-67. Biochemical assays show that yeast POLζ (comprising the catalytic subunit Rev3 and an accessory subunit, Rev7) can facilitate bypass of several types of DNA damage. The closest mammalian homolog of S. cerevisiae Rev3 is Rev3-like (REV3L), but with over 3000 amino acid residues, it is twice the size of the yeast protein68. Mammalian REV7 (also known as MAD2L2) is thought to be a binding partner of REV3L.
Knowledge of the in vitro biochemical activity of POLζ comes from studies done in S. cerevisiae because an active form of mammalian POLζ has not yet been purified. However, from work in knockout and knockdown cell lines, information has been gathered about the role of POLζ in mammalian and other higher eukaryotic cells. Experiments with cell lines in which REV3L function is disrupted indicate that POLζ is involved in the bypass of many types of clinically relevant DNA lesions, including cisplatin-GG46, BPDE-GG46, 6-4 photoproducts30, 46, 69, AP sites46 and thymine glycols54. The role of POLζ in the bypass of these lesions is proposed to be primarily as an extender, after another polymerase has inserted a nucleotide opposite a damaged site70. Mammalian POLζ may operate principally to fill single-stranded DNA gaps following DNA replication (Figure 2), rather than directly at the replication fork69.
Yeast rev3 knockouts are viable, but Rev3l−/− mice die at embryonic days 9.5-15.571-75, and Rev3l−/− blastocysts are not viable in cell culture. This role in maintaining viability is unique among DNA polymerases involved in TLS. Rev3l−/− cell lines can only proliferate if p53 function is compromised. This is a necessary but not a sufficient condition, as loss of p53 does not rescue the embryonic lethality of Rev3l deletion in mice73-75. The frequency of point mutations was decreased by up to 90% in mouse and human Rev3l knockout and knockdown cells, and the knockout cells are sensitive to DNA damaging agents68, 69, 76-83. However, Rev3l−/− cells have a high frequency of spontaneous chromosome aberrations, particularly translocations74, 78, 80-82.
The consequences of POLζ disruption for tumorigenesis have been explored in a viable mouse model in which one allele of Rev3l is deleted and the second allele contains loxP sites. The second allele can be deleted upon expression of Cre recombinase from the MMTV promoter, which is active in a substantial fraction of epithelial and hematopoietic cells84. On a Trp53−/− background, such mosaic mice conditionally deleting Rev3l developed thymic lymphomas more rapidly than Trp53−/− mice with a wild-type copy of Rev3l. Importantly, most of the tumors arose from Rev3l−/− cells, despite an initial growth disadvantage caused by Rev3l deletion84. Moreover, these tumors were frequently multiclonal, indicating that a loss of REV3L confers a selective advantage for tumor formation. In Trp53−/− and Trp53+/− backgrounds, generation of Rev3l−/− cells predisposed mosaic mice to the development of spontaneous mammary tumors. Most of these tumors were dominated by Rev3l−/− cells, were multifocal, and were associated with mammary intraepithelial neoplasia84. While there is no direct evidence that these tumors had increased chromosome aberrations, extensive data from REV3L defective cell lines, as well as from another conditional Rev3l deletion mouse model74, 78, 80-82, 85, strongly suggests the production of genomic instability in Rev3l-deleted tumors. These observations indicate that if a preneoplastic cell can overcome checkpoint responses that are induced on deletion of Rev3l, an accelerated rate of gross genetic change can drive tumorigenesis (Figure 1). It is interesting that the human REV3L gene is located at a fragile site (FRA6F) on chromosome 6q2181,86, a common region of deletion in human hematopoietic neoplasms 83.
In S. cerevisiae, Rev1 functions together with POLζ87, and rev1 mutants have dramatically reduced frequencies of mutagenesis induced by DNA damaging agents88. Mouse and chicken cells with mutations causing Rev1 deficiency have decreased damage-induced mutagenesis and are more sensitive to DNA damaging agents, but they also have an increased incidence of chromosome aberrations89-91. Deletion of Rev1 from mouse cells does not affect cellular viability90, 92-94. A knockout of Rev1 induces cell cycle defects in chicken DT40 cells89, but not in mouse embryonic fibroblasts91.
REV1 is a DNA polymerase in only a very limited sense, as it cannot synthesize a DNA polymer, but can add a single C residue to a primer end in special cases. Human REV1 preferentially inserts C opposite a template G, U, or an AP site95. Mammalian REV1 is involved in the bypass of UV radiation-induced damage91 and crosslinks96, but this role does not involve its deoxycytidine monophosphate (dCMP) transferase activity. The cellular sensitivity of Rev1 knockout cells to some DNA damaging agents can be rescued with a catalytically inactive REV1 mutant protein97. The primary role of REV1 during TLS may be in DNA polymerase switching, as described in the next section.
DNA polymerase choice and switching during TLS
The structure of Y-family DNA polymerases such as POLη is specially adapted for DNA damage bypass (Box 2). However, the capacious active site of Y-family DNA polymerases is not suitable for accurate DNA replication of undamaged templates (the base substitution error rates of Y-family enzymes are more than 10−3, which is about 10-100 times higher than those of replicative DNA polymerases7). Therefore, the access of Y-family polymerases to DNA is tightly controlled by post-translational modifications of interacting proteins. Each Y-family polymerase has a ubiquitin-binding domain that interacts with ubiquitylated forms of the sliding clamp proliferating cell nuclear antigen (PCNA)98. Blocks to DNA replication trigger monoubiquitylation of PCNA by E3 ubiquitin ligases including RAD1899, 100. This creates a binding site for Y-family polymerases and, because PCNA is in contact with DNA this thereby facilitates the interaction of these polymerases with DNA. There is also evidence that POLη is ubiquitylated in response to DNA damage101. Although not so intensively studied, access of A-, B-, and X-family polymerases to DNA is also likely to be tightly regulated.
Monoubiquitylated PCNA is not the only factor responsible for regulating TLS102. The extreme carboxy-terminus of REV1 interacts with Y-family polymerases: POLη, POLι and POLκ103-106. Through distinct ubiquitin-binding motifs, REV1 also interacts with ubiquitylated PCNA97, 107. These interactions may allow REV1 to help regulate DNA polymerase switching and this may be the primary role of REV1 in the cell98, 108. It will be interesting to learn whether the DNA damage-sensing clamp RAD9-HUS1-RAD1 (known as the 9-1-1 complex) is also involved in TLS polymerase loading and switching.
The mechanism of selection of a particular TLS polymerase is under investigation. Perhaps the polymerase that catalyzes the most thermodynamically favourable reaction at a particular lesion is most likely to mediate bypass of that lesion109. The abundance of a polymerase may also play a role110. In addition, in yeast, TLS polymerases may be employed in lesion bypass when cells are in G2 phase of the cell cycle, when replication is complete111, 112. A gap can arise in DNA when an adjacent replication origin converges on a stalled replication fork (Figure 2), or if the replication apparatus can reprime on the DNA template downstream of a DNA lesion111. There is evidence for post-replication gaps in mouse cells in which Rev1 and Rev3l are deleted, suggesting a role for these polymerases in bypassing DNA lesions during G269, 91. The relative contribution of different TLS polymerases during replication and gap filling is actively discussed69, 113, 114, and many of the same proteins that enable the polymerase switch are also required for gap filling115, 116.
BER
BER (Box 1) is responsible for repairing many DNA adducts generated by ROS, hydrolytic reactions and alkylating agents. DNA polymerase β (also known as POLB) is the primary enzyme used for gap-filling DNA synthesis during BER in the nucleus117. POLβ also has a 5’-deoxyribose phosphate (dRP) lyase activity that is normally required to remove the sugar-phosphate residue produced by the action of an AP endonuclease during BER. In mitochondria, the replicative polymerase POLγ is responsible for gap-filling in BER118.
In some human tumors, sporadic sequence changes have been reported in POLB cDNA. In only a few cases have these changes been confirmed in genomic DNA119. These modifications usually encode single amino acid substitutions, and the expression of some of them can transform cultured cells, allowing growth on soft agar120. It remains to be seen whether POLB mutations will be found by cancer genome sequencing projects and whether such mutations contribute to a mutator phenotype. Mice that are Polb+/− develop normally, but have an increased incidence of lymphomas121.
Other DNA polymerases have been proposed to be backup enzymes for BER. A dRP lyase activity is present in POLι122, POLλ (also known as POLL)123, and POLθ124. It has been reported that BER is defective in cells in which POLI is knocked down125, and POLI can compensate for the loss of POLB in the removal of uracil by BER in cell extracts126. However, the mouse 129 strain carries a homozygous nonsense mutation in Poli, and has no obvious indications of a BER defect. POLλ-deficient cells are sensitive to H2O2 (but not to alkylating agents)123, so POLλ might act as a backup for repair of a subset of lesions in BER127. A knockout of POLQ in the chicken DT40 B-cell line causes increased sensitivity to H2O2, and it has been suggested that POLθ might function in BER62. Cells from Polq−/− mice, however, do not have higher than normal sensitivity to H2O2 128. Mammalian POLθ may instead help cells tolerate DNA damage, perhaps by TLS of lesions such as thymine glycol or by participating in DNA end-joining, as described below.
NER
NER is a pathway responsible for the repair of many types of DNA damage caused by carcinogenic and chemotherapeutic agents, such as BPDE, UV radiation and cisplatin. In mammalian cell extracts in vitro, either POLΣ or POL™ can synthesize DNA to fill the gap of about 27 nucleotides produced during NER. This reaction is dependent on PCNA and the single-stranded DNA binding protein replication protein A (RPA). POLΣ appears to be well-suited to fill gaps that can be sealed by DNA ligase I (LIG1)129. A second mode of NER gap filling utilizes a combination of POL™ and POLκ with LIG3 to seal the nick. This mode may be utilized preferentially when deoxynucleotide concentrations are low, as is the case in non-cycling cells130. Inherited defects in components of the NER pathway that recognize and incise damaged DNA are the main cause of xeroderma pigmentosum131. However, defects in POLΣ, POL™, or POLκ have not been associated with this skin cancer-prone syndrome, or with other cancer predispositions. As mentioned above, mutations in POLη give rise to the XP-V subtype of xeroderma pigmentosum and cells from afflicted individuals are proficient in NER but defective in TLS.
Repair of DNA breaks
Breaks in the DNA can be caused by many agents, including ionizing radiation, antibiotics such as bleomycin, or chemotherapeutic agents like etoposide. DNA polymerases β, ζ, η, θ, λ, μ, and terminal deoxynucleotidyltransferase (TDT) have been implicated in different aspects of DNA break repair by HR or NHEJ. POLβ is necessary for normal meiotic synapsis and functions in an early stage in the processing of DSBs that initiate HR during meiosis132. POLη has been suggested to be involved in some aspects of DSB repair as POLH-deficient chicken DT40 cells have been reported to have a reduced frequency of the DSB-initiated HR-mediated gene conversion events that create immunoglobulin diversity133. Furthermore, POLη can mediate extension of a D-loop (an HR intermediate) and second strand capture in vitro, and the polymerase interacts with RAD51 and RAD52, both of which are major components of the HR pathway134. The multiple roles of POLη in DNA repair and TLS might mean that several factors could contribute to the phenotype of cells from patients with XP-V, including a defect in TLS that causes replication fork collapse and consequently the formation of DSBs, a deficiency in DSB repair, and a replacement of the TLS function of POLη with an error-generating polymerase that increases point mutagenesis. There is also evidence that POLζ may be involved in some aspects of DSB repair65, 67, 78-80, 135 and ICL repair136, 137.
DNA end-joining
NHEJ is a major mode of DSB repair in mammalian somatic cells and it is also a central process for generating diversity in the immune system through V(D)J recombination. The X-family enzymes POLλ, POLμ (POLM), and TDT have all been implicated in immunogloblin V(D)J gene recombination. These DNA polymerases contain a BRCT domain (Table 1) that binds to NHEJ factors such as KU80-KU70 and the XRCC4-LIG4 complex138, 139. POLμ and TDT are closely related and contain a protein domain called “loop1” that is proposed to preclude binding to a template DNA strand. Both enzymes can add bases to the end of a DNA strand in the absence of a template140,141a. TDT is the key enzyme for generating diversity during V(D)J recombination by adding random nucleotides at the junctions between the V, D, and J elements141. POLλ and POLμ are also implicated in gap filling during V(D)J recombination142. There is evidence for increased IR sensitivity and reduced DSB repair in cells of Polm−/− mice142a, 142b. This suggests a function for POLM in general non-homologous end-joining, in addition to a specialized function in V(D)J recombination. Hypersensitivity to IR or bleomycin has not been detected in other studies of Polm−/− or Poll−/− mice142, 143.
POLθ and defence against breaks caused by ionizing radiation
Evidence is emerging for the importance of POLθ in the defence against DNA damage caused by ionizing radiation. Mouse bone marrow cell lines in which Polq (which encodes POLθ) is deleted are more sensitive than normal cells to ionizing radiation and bleomycin, but not to the ROS-generating agents H2O2 and paraquat128. This suggests that cells may rely on POLθ for repair of some DSBs in DNA128. Knockdown of POLQ by short interfering RNA (siRNA) in human tumour cell lines also increases the sensitivity to ionizing radiation and causes persistent phosphorylation of histone H2AX144. Polq−/− mice develop normally, but have increased frequencies of spontaneous and radiation-induced micronuclei, in erythrocytes, which can arise from chromosome breakage145. The viability of Polq−/− mice is severely compromised by an additional mutation in ataxiatelangiectasia mutated (Atm), suggesting that POLθ is particularly critical in the absence of the checkpoint and repair functions of ATM145. These results indicate that POLθ is involved in a DSB repair process, or that it prevents generation of DSBs at stalled DNA replication forks by mediating TLS opposite some lesions generated by ionizing radiation. A homolog of POLθ in Drosophila melanogaster, called mutagen-sensitive 308 (MUS308), is involved in a microhomology-mediated pathway of DNA end-joining146, 147. It is possible that POLθ is involved in a similar pathway in mammalian cells, but it is not yet certain that POLθ and D. melanogaster MUS308 are functionally orthologous. Although MUS308 and mammalian POLθ share an unusual helicase-polymerase domain organization, they differ significantly in their polymerase motifs51.
DNA polymerase gene expression changes in cancer
Many studies have been conducted to determine changes in gene expression of DNA polymerases in human cancers. A pattern of expression in tumors has been difficult to discern. For example, the expression levels of the human REV3L, POLK and POLI genes have been explored in various types of cancer, but no consistent picture has emerged151-154. Poli was proposed as a candidate for the pulmonary adenoma resistance locus designated Par2 in mice, but further analysis excluded Poli and identified (also known as Las2), which is adjacent to Poli, as the tumor susceptibility gene that is mutated in Par2 mice155.
Some polymerases do show a trend in gene expression in tumors. The expression of POLH is reduced in lung and stomach cancers in comparison to paired normal tissues150. In non-small-cell lung cancer156, higher POLH expression correlated with poorer outcome156. Increased expression of POLB has been reported in gastric152,157, uterine152, prostate152, ovarian152 and thyroid158 carcinomas, and forced overexpression of POLB in cells can interfere with normal replication, causing mutagenesis159, 160. Transgenic mice overexpressing Polb have a number of pathologies, including increased development of osteosarcoma158. This suggests that proper regulation of POLB expression is important for preventing tumorigenesis.
In addition, POLQ expression may be higher in tumour cells than in normal cells. POLQ expression is higher in colorectal cancer tumour samples compared with matched surrounding normal tissues, independent of the proliferation state of the tumours161, 162. When the expression profiles for all of the human nuclear DNA polymerase genes were examined in patients with previously untreated primary breast cancers, POLQ was the only one significantly higher in breast cancer tumour samples compared with normal breast tissues and higher expression was correlated with poor clinical outcome151, 163. Ectopic overexpression of POLQ in human cell lines impairs DNA replication fork progression and causes chromosome damage151. Error-generating, low processivity DNA polymerases are normally expressed at low levels in cells and overexpression might lead to a more frequent imposition of these enzymes at replication forks (temporarily replacing higher processivity, error-free replicative polymerases), and leading to mutagenesis and replication fork stalling.
DNA polymerases as therapeutic targets
Drugs that inhibit DNA replication, such as folate inhibitors, pyrimidine analogs and hydroxyurea, are in standard use for chemotherapy, but some of these therapies are highly toxic as they do not selectively target cancer cells. This is promoting searches for other drugs to target DNA polymerases for chemotherapeutic benefit. For example, derivatives of the antibiotic dehydroaltenusin inhibit POLα but not POLβ, δ, ε, or γ164, 165. The drug suppresses HeLa cell xenograft tumor growth in mice166. Eicosapentaenoic acid radiosensitizes cells, and is also an inhibitor of POLβ, δ, and ε167.
POLβ, the principal DNA polymerase for gap-filling DNA synthesis in BER, is being explored as a target for therapeutics. Several small molecule inhibitors of POLβ have been found168, including some that can potentiate the toxic effects of the chemotherapeutic agents bleomycin169 and temozolomide170, 171. Reduction of POLB expression also increases sensitivity to the chemotherapeutic agent oxaliplatin172. The mechanism for some of these effects is unclear, as the specific function of BER in response to the DNA damage caused by oxaliplatin or bleomycin is not yet known.
TLS DNA polymerases are also possible targets for enhancing DNA damaging therapies, and inhibitors of some of these are beginning to emerge173, 174. Studies on DNA polymerase inhibitors are intriguing but are still in their early stages. As strategies for small molecule screens are being developed174, they will have to be coupled with screens for the most pharmacologically effective compounds. For example, the most potent DNA polymerase inhibitors may not be transported efficiently into cells164. As drugs are found that inhibit specific polymerases and are preferentially toxic to tumor cells, it will be critical to determine whether the inhibition of DNA polymerase activity is mechanistically responsible for mediating any anti-tumor effects.
Additionally, suppression of Rev1 and Rev3l expression is being explored in mouse cancer models. A Rev1-specific ribozyme was delivered to the lungs of BPDE-treated mice, and it decreased BPDE-induced lung tumors by 50%175, presumably by decreasing point mutagenesis. Recent work also demonstrates that lymphomas with decreased expression of Rev1 or Rev3l are more sensitive to cisplatin and cyclophosphamide in a xenograft model, and tumors in which Rev1 expression is knocked down do not develop resistance to cyclophosphamide176. Rev3l downregulation increased the sensitivity to cisplatin in a xenograft model of non-small-cell lung cancer 177. Vigilance will need to be employed in the development of drugs targeting REV1 or REV3L, because the loss of Rev1 or Rev3l in cells and deletion of Rev3l in mice results in chromosome instability, as described above. Even though Rev3l is essential for base change mutagenesis, loss of Rev3l in mice accelerated spontaneous cancer formation84. Furthermore, mice carrying disruptions of the BRCT domain of REV1 in Xpc−/− mice show accelerated UV radiation-induced skin tumorigenesis94. This disruption of the BRCT domain of REV1 does not confer a marked defect in UV radiation-induced mutagenesis in mouse skin, but it substantially enhances the toxicity of UV radiation, resulting in greatly increasing inflammation that contributes to accelerated formation of skin cancer. Consequently, REV3L and REV1 may be problematic targets for cancer therapeutics, because their suppression may lead to additional genomic instability and inflammatory responses in surviving cells.
Conclusions and perspectives
The fifteen DNA polymerases encoded by mammalian genomes are specialized for different functions, including DNA replication, DNA repair, recombination and TLS. Genetic and biochemical experiments are showing that different DNA polymerases are suited for TLS of particular types of DNA damage. For some DNA polymerases that can perform TLS, firm identification of the most biologically relevant substrates remains a challenge. Newly obtained structures of DNA polymerases in complexes with lesions show how the architecture of an active site can accommodate relatively bulky adducts, and different enzymes seem to solve this problem in different ways21, 22, 178, 179. Nevertheless, evidence is accumulating that even those DNA polymerases that are especially adept at TLS have additional and unexpected roles in human cells.
It appears that some DNA polymerases are more significant in particular tissues than others, and more analysis is needed on tissue-specific and developmental expression patterns. The spectrum of available DNA polymerases in stem cells is worthy of attention as it is imperative that these cells maintain stable genomes. One specialized DNA polymerase, POLζ, has now been shown to act effectively as a suppressor of spontaneous tumorigenesis in mice84. However, other than POLH in XP-V cells, no definitive evidence has yet emerged in human cancer cells for frequent homozygous loss or inactivating mutation of any gene encoding a DNA polymerase. As whole-genome cancer analyses continue, it will be interesting to monitor the loss, rearrangement or mutation of genes encoding specialized DNA polymerases. Because tumors are genetically heterogeneous, sequencing of individual cell genomes may be required to achieve this. New evidence indicates that POLQ is the DNA polymerase gene with the greatest difference in expression between breast tumors and non-tumor cells180. Further analysis will be needed to determine whether this is also the case for other types of cancer, and if DNA polymerase expression is regulated or sometimes silenced by DNA methylation, microRNAs or other mechanisms. Investigation of DNA polymerase protein levels in tumors is also needed, as nearly all work in this area has only explored variations in gene expression. Numerous single-nucleotide polymorphisms exist in DNA polymerases, reflecting differences between individuals, and it is of interest to determine whether any of them have a significant functional effect on activity and predisposition to developing cancer.
Because many non-surgical cancer therapies work by damaging DNA, consideration of DNA repair capacity is an ongoing concern in improving responses to treatments. In a few cases, knowledge of the DNA repair status of a tumor allows significant tailoring of therapy. For example, patients with glioblastoma benefit from temozolomide treatment when their tumors contain a methylated promoter in the gene MGMT, encoding the DNA repair enzyme methylated-DNA-protein-cysteine methyltransferase180a.
The more that is known about associations between the expression of genes encoding DNA repair proteins and others involved in maintaining genomic stability and prognosis, the more progress can be made in this area. This requires highly specific reagents and careful quality control181. Like other gene products involved in processing DNA damage, DNA polymerases are potential targets. Specific inhibitors of a DNA polymerase important for tolerating particular types of damage could be useful as an adjuvant for DNA damaging radiation or chemotherapy. As more is known about the active sites of specialized DNA polymerases, more specific small molecule inhibitors may be selected or designed. Many surveys of small-molecule libraries for DNA polymerase inhibitors are underway, but no studies have yet screened all fifteen mammalian polymerases simultaneously in order to determine specificity. Alternatively, the forced overexpression of a DNA polymerase in a tumor might also cause desired toxic effects by interfering with replication. Increasing the expression of a DNA polymerase to intentionally cause an increase in mutagenesis and the potential loss of essential genes is another suggested strategy16. Toxicity is apparent when POLβ, POLκ or POLθ are overexpressed at levels much higher than the variations in expression seen in human tumor samples159; cells that are engineered to overexpress POLη do not show obvious toxicity182.
It is possible that further enzymes with DNA polymerase activity may be discovered in mammalian genomes, and it seems certain that additional accessory proteins will be found that modulate the activity of the known DNA polymerases. Improved knowledge of these factors may also have implications for tumorigenesis and for designing anti-cancer strategies.
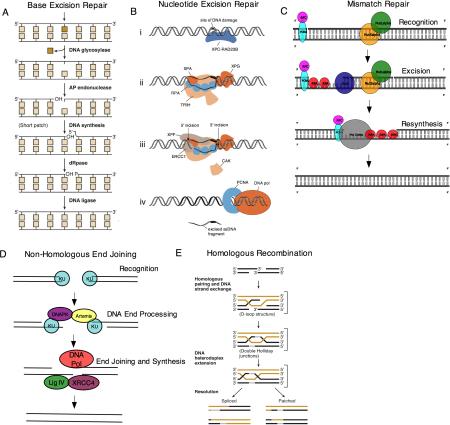
Box 1 – DNA repair mechanisms.
DNA repair mechanisms generally involve the removal of damaged or incorrect bases, and require a DNA polymerase to resynthesize DNA, using the undamaged strand as a template.
Base excision repair (BER) typically mediates the removal and replacement of a single base residue (part a of the figure). Substrates include uracil residues in DNA and damaged bases caused by reactive oxygen species, hydrolytic reactions and methylation. BER is initiated by a DNA glycosylase specific for a damaged base, producing an abasic site (apurinic or apyrimidinic (AP) site) that is incised by an AP endonuclease. The 5’-deoxyribose-phosphate (dRP) residue is removed by a dRP lyase, leaving a 1 nt gap, which is filled in by DNA polymerase β (POLβ).
Nucleotide excision repair (NER) can remove various helix-distorting adducts including those caused by ultraviolet (UV) radiation, cisplatin and polycyclic aromatic hydrocarbons (part b of the figure). A distorted region is recognized, and two incisions are made on either side of the adduct to excise the damaged DNA. The resulting 27-29 nucleotide gap is filled by POLδ or POLε, and under some circumstances, POLκ.
Mismatch repair (MMR) is an excision repair process that removes mismatched bases (part c of the figure). It is initiated by mismatch recognition proteins, and a segment of DNA is excised between the mismatch and a nearby nick. The gap that is left in the DNA is filled by POLδ.
DNA double strand breaks (DSBs) can be repaired by non-homologous end-joining (NHEJ) (part d of the figure). Strand breaks caused by ionizing radiation or by enzymes that cleave DNA usually do not yield DNA ends that can be ligated directly. End-trimming and resynthesis of bases is therefore required to join breaks.
An alternative strategy for DSB repair is homologous recombination (HR) (part e of the figure). HR only operates when a double-stranded copy of the sequence is available, for example as a sister chromatid in late S or G2 phase of the cell cycle.
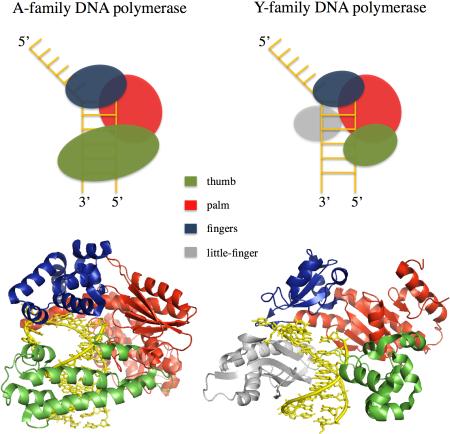
Box 2 – DNA polymerase structure.
DNA polymerases have structural domains regularly likened to a right hand, in which DNA lies in the “palm” and is surrounded by “fingers” and “thumb” domains. Y-family DNA polymerases have significantly smaller finger and thumb domains than those of replicative DNA polymerases, and spacious active sites that enable them to bypass bulky DNA adducts183. The “little-finger” domain that is uniquely encoded in Y-family enzymes not only helps the short finger and thumb domains to stably bind template DNA, but also influences fidelity and bypass activity184. The figure shows a structural comparison of the large fragment of Thermus aquaticus DNA polymerasel a member of the A-family; protein data bank (PDB) ID code 3M8R184a) and POLη (a member of the Y-family; PDB ID code 3MR322). The thumb domain is shown in blue, the palm in red, the fingers in green and the little-finger in gray. Figures were generated with PyMol.
Box 3 – Somatic hypermutation of antibody genes.
Following V(D)J recombination, antibody variable genes undergo a process of mutagenesis in B cells, initiated when activation-induced cytidine deaminase (AID; also known as AICDA) mediates cytosine deamination. The uracil-DNA glycosylase removes the resulting uracil to form an apurinic or apyrimidinic (AP) site and this is followed by repair of the AP site by BER and an error-generating DNA polymerase for DNA repair synthesis. Alternatively, translesion DNA synthesis (TLS) might be used to bypass the AP site. This process of somatic hypermutation involves DNA polymerase η (POLη)185, 186 as well as REV1 and Rev3-like (REV3L)85, 89, 90, 92, 93. The AP site bypass activity of POLθ has encouraged the suggestion that it might be involved in somatic hypermutation63, but it appears to play only a minor role in this process in mammalian cells187. In the chicken DT40 cell line, both immunoglobulin gene diversification by recombination and somatic hypermutation are reported to depend on POLη, POLθ, POLν and REV163, 188.
At-a-glance.
Fifteen DNA polymerases are encoded in mammalian genomes. Some of them function in replication of the genome, but most participate in specialized DNA repair and DNA damage tolerance processes. The activity of these DNA polymerases will affect the response of a cell to DNA-damaging carcinogens and chemotherapeutic agents.
Some DNA polymerases catalyze DNA synthesis on damaged sites in DNA, helping cells tolerate DNA damage by translesion DNA synthesis (TLS). TLS polymerases are specialized for bypass of different DNA adducts. Defects in POLη (also known as POLH) are responsible for the variant type of xeroderma pigmentosum (XP-V).
POLζ (the catalytic subunit of which is REV3L) and REV1 are required for nearly all damage-induced base change mutagenesis in mammalian cells. Reduction of their activities sensitizes cells, including tumor cells, to DNA damaging agents. However, chromosome rearrangements and inflammation can increase in the absence of these proteins, promoting carcinogenesis.
The expression of some genes encoding DNA polymerases may be altered in some cancers. In breast cancers, levels of POLQ (which encodes POLθ) appear to be the most elevated compared with normal levels of expression. Comprehensive studies of DNA polymerase protein levels in cancer remain to be carried out.
The inhibition of DNA polymerase activities could be useful as an adjuvant to DNA damaging therapies, and inhibitors for some polymerases have been found. Pharmacologically effective inhibitors highly specific for a single DNA polymerase remain to be identified.
Whole genome analyses of cancers have not yet revealed cancer-associated alterations in DNA polymerase genes. It seems likely, however, that at least some cells in a tumor will have relevant alterations. Some DNA polymerases can be considered as tumor suppressors (POLζ, REV1, POLη, POLι, POLκ, POLδ and POLε).
Table 2.
DNA polymerase knockout mouse phenotypes
| DNA polymerase | Gene for catalytic subunit | Phenotype of knockout mouse |
|---|---|---|
| α | Pola1 | Embryonic lethality? |
| δ | Pold1 | Embryonic lethality after E4.5 Tumorigenesis in Pold1exo/exo and Pold1exo/− mice11, 12 |
| ε | Pole1 | Embryonic lethality? Tumorigenesis in Pole1exo/exo mice12 |
| γ | Polg1 | Embryonic lethality after E7.5189 |
| β | Polb | Exhibit apoptosis in post mitotic neuronal cells and die at birth. Loss of p53 rescues lethality190, 191,192 |
| λ | Poll | Viable, fertile, and display reduced immunoglobulin heavy chain junction variability142 |
| μ | Polm | Viable, fertile, and display reduced immunoglobulin light chain junction variability143 |
| TDT | Dntt | Viable, fertile, and display reduced immunoglobulin heavy chain junction variability141 |
| ξ | Rev3l | Embryonic lethality after E9.5. p53 deficiency cannot rescue the lethality71-73. Spontaneous tumor development in conditional knockout mice84 |
| REV1 | Rev1 | Inviable on the C57BL/6 background, but viable on the 129/OLA background92. Loss of C:G transversions during somatic hypermutation. |
| η | Polh | Viable and fertile, susceptible to sunlight-induced skin cancer. Generation of A:T mutations during the somatic hypermutation of Ig genes193. |
| ι | Poli | Viable and fertile. The 129/OLA strain of mice has a naturally-occurring nonsense mutation in Poli. Polh−/−; Poli−/− mice show slightly earlier onset of skin tumor formation194,193 |
| κ | Polk | Viable and fertile40, 195 Increased mutation frequencies in tissues41 |
| θ | Polq | Viable and fertile, high micronuclei frequency in reticulocytes Very low viability of Polq−/−; Atm−/− mice145, 196 |
| ν | Poln | Unknown |
Atm, ataxia-telangiectasia mutated; E, embryonic day; TDT, terminal deoxynucleotidyltransferase.
A database of knockout mouse strains related to DNA damage processing is available197
Acknowledgements
Research on DNA polymerases by the authors is supported by NIH grants CA09717 and CA132840 from the National Cancer Institute, by grant P30ES007784 from the National Institute of Environmental Health Sciences and by NIH Cancer Center Support Grant P30-CA016672 (University of Texas MD Anderson Cancer Center).
Glossary
- DNA damage
Encompasses the many types of chemical alterations that can change the structure of DNA. Damage can be caused by reactions that disrupt bonds in the nucleobases, the deoxyribose sugar ring, or the phosphate groups of DNA or by addition of chemical moieties such as hydroxyl groups, methyl groups, or even bulkier groups derived from polycyclic molecules.
- Leading strand
DNA synthesis can only add nucleotides to the terminal 3’-OH group of a growing polymer. The strand synthesized continuously during DNA replication is the leading strand.
- Lagging strand
The strand of DNA that is synthesized in discontinuous segments.
Footnotes
Links on the Web
Information on DNA polymerase genes and 150 other genes related to DNA repair are accessible in public databases, conveniently accessed through the portal page “Human DNA Repair Genes”.
Classification of DNA polymerases:
DNA-dependent DNA polymerases are classified into six families based on primary amino acid sequence similarity in the enzyme active site: A, B, C, D, X, and Y. Representatives of four of these families, A, B, X, and Y, are encoded by vertebrate genomes.
The A-family DNA polymerase domain is similar to Escherichia coli DNA polymerase I, encoded by the bacterial PolA gene.
B-family DNA polymerases show similarity to E. coli DNA polymerase II (pol II).
The prototypes for Y-family polymerases are the E. coli TLS DNA polymerases DinB (pol IV) and UmuC (pol V).
X-family polymerases in mammalian cells are POLβ, POLλ, POLμ, and TDT
Abasic (AP) site – A site in a DNA chain that is missing a pyrimidine or purine base residue, but where the phosphodiester backbone remains intact. Such sites can arise when a base-sugar bond is cleaved by a DNA glycosylase during BER, or by a spontaneous hydrolytic reaction.
Alkylating agent – An electrophilic compound that can covalently add an alkyl group to a DNA base, or to other biological macromolecules. These compounds act both as carcinogens (e.g. methyl chloride) and as chemotherapeutic agents (e.g. mechloroethamine).
Checkpoint – A control mechanism to verify whether each phase of the cell cycle has been completed accurately. If DNA damage is present, some checkpoint controls prevent or delay progression through the cell cycle, for example from G1 to S phase or from G2 phase to mitosis.
Cyclobutane pyrimidine dimer (CPD)– The most frequent ultraviolet radiation-induced DNA lesion, formed by the covalent linkage of the 5, 6 bonds of adjacent pyrimidines to form a cyclobutane ring, without directly altering the base pairing faces of the dimerized bases. Such dimers are formed most commonly between adjacent thymines, but also between thymine and cytosine or two adjacent cytosines.
(6-4) photoproduct – The second most common type of ultraviolet radiation-induced DNA damage, involving linkage of the C6 position of a 5’ pyrimidine base to the C4 position of a 3’ adjacent pyrimidine base. (6-4) photoproducts distort the DNA helix more than a CPD, and form most often at 5’ thymine-cytosine-3’ sequences.
DNA damage – A term that encompasses the many types of chemical alterations that can change the structure of DNA. Damage can be caused by reactions that disrupt bonds in the nucleobases, the deoxyribose sugar ring, or the phosphate groups of DNA or by addition of chemical moieties such as hydroxyl groups, methyl groups, or even bulkier groups derived from polycyclic molecules.
Exonuclease – An exonuclease cleaves DNA phosphodiester bonds to release nucleotides from one end of a polynucleotide chain. DNA polymerases synthesize DNA in a 5'-3' direction and some DNA polymerases have an intrinsic 3’- 5’ exonuclease activity that enables proofreading of their own mistakes.
Fragile site – Heritable regions on chromosomes that are associated with an increased frequency of chromosome breaks, gaps and other aberrations. Fragile sites, and the genes that they contain, are frequently rearranged or deleted in cancer cells.
Hydrolytic reaction – Decomposition of a chemical compound or a molecular bond by reaction with water.
Interstrand crosslink – An interstrand crosslink (ICL) covalently links the two complementary strands of duplex DNA. Such crosslinks are formed by some carcinogenic and chemotherapeutic agents, and they are especially toxic because they block the complementary DNA strand separation that is necessary for DNA replication and transcription.
Lagging strand – The strand of DNA that is synthesized in discontinuous segments.
Leading strand – DNA synthesis can only add nucleotides to the terminal 3’-OH group of a growing polymer. The strand synthesized continuously during DNA replication is the leading strand.
Micronuclei – A piece of DNA that resides outside of the nucleus, caused by chromosomal breakage leading to accentric chromosome fragments that lack spindle attachments, or by chromosomal mis-segregation during mitosis. Micronuclei are most easily detected in mature erythrocytes that lack nuclear DNA.
Nonsense mutation – A change in the codon for an amino acid that results in a stop codon. Nonsense mutations cause protein truncation and often nonsense-mediated decay of the encoding messenger RNA.
Ribozyme - A ribozyme is a catalytic enzyme made entirely of RNA. Some ribozymes are nucleases and can include base-pairing regions that enable specific binding and cleavage of a target RNA molecule.
Sliding clamp – A mobile platform for DNA replication and repair machinery. The eukaryotic sliding clamp, Proliferating Cell Nuclear Antigen (PCNA) binds to DNA polymerases and is crucial for the switching of polymerases during TLS and DNA repair.
Template switching – an error-avoiding strategy for DNA damage tolerance that uses the newly synthesized, undamaged strand of a sister chromatid for bypass replication (see Figure 2).
Terminal deoxynucleotidyltransferase (also known as DNA nucleotidylexotransferase) – A template-independent DNA synthesis activity that catalyzes addition of nucleotides to the 3' terminus end of DNA. The TDT enzyme in human cells contributes to immune diversity by adding nucleotides of varying lengths between gene segments during V(D)J recombination.
V(D)J recombination – V(D)J recombination assembles immunoglobulin and T-cell receptor genes from different segments. The RAG1-RAG2 nuclease introduces DNA double-strand breaks to produce segments that are joined by non-homologous end-joining (NHEJ) repair of DNA.
Xeroderma pigmentosum – An inherited human syndrome characterized by severe photosensitivity, a high incidence of skin cancer and neurological abnormalities. The disorder is caused by a deficiency in nucleotide excision repair genes (XPA though XPG), or in translesion synthesis past ultraviolet irradiation-induced DNA damage (the XP group V, caused by mutations in the POLH gene).
References
- 1.Waters LS, et al. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol Mol Biol Rev. 2009;73:134–54. doi: 10.1128/MMBR.00034-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bielas JH, Loeb KR, Rubin BP, True LD, Loeb LA. Human cancers express a mutator phenotype. Proc Natl Acad Sci U S A. 2006;103:18238–42. doi: 10.1073/pnas.0607057103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang DJ, Cimprich KA. DNA damage tolerance: when it's OK to make mistakes. Nat Chem Biol. 2009;5:82–90. doi: 10.1038/nchembio.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–5. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- 5.Hubscher U, Maga G, Spadari S. Eukaryotic DNA polymerases. Annu Rev Biochem. 2002;71:133–63. doi: 10.1146/annurev.biochem.71.090501.150041. [DOI] [PubMed] [Google Scholar]
- 6.Pursell ZF, Isoz I, Lundstrom EB, Johansson E, Kunkel TA. Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science. 2007;317:127–30. doi: 10.1126/science.1144067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCulloch SD, Kunkel TA. The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Res. 2008;18:148–161. doi: 10.1038/cr.2008.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmitt MW, Matsumoto Y, Loeb LA. High fidelity and lesion bypass capability of human DNA polymerase delta. Biochimie. 2009;91:1163–72. doi: 10.1016/j.biochi.2009.06.007. [A discussion of how a replicative DNA polymerase balances high fidelity at the expense of bypass activity.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCulloch SD, Kunkel TA. The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Res. 2008 doi: 10.1038/cr.2008.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldsby RE, et al. High incidence of epithelial cancers in mice deficient for DNA polymerase delta proofreading. Proc Natl Acad Sci U S A. 2002;99:15560–5. doi: 10.1073/pnas.232340999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uchimura A, Hidaka Y, Hirabayashi T, Hirabayashi M, Yagi T. DNA polymerase delta is required for early mammalian embryogenesis. PLoS One. 2009;4:e4184. doi: 10.1371/journal.pone.0004184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Albertson TM, et al. DNA polymerase epsilon and delta proofreading suppress discrete mutator and cancer phenotypes in mice. Proc Natl Acad Sci U S A. 2009;106:17101–4. doi: 10.1073/pnas.0907147106. [References 11 and 12 show that loss of proofreading exonuclease activity in either Polδ or Polε leads to an increase in spontaneous cancers. The types of cancers are different, suggesting tissue-specific requirements for DNA replication fidelity.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Venkatesan RN, et al. Mutation at the polymerase active site of mouse DNA polymerase delta increases genomic instability and accelerates tumorigenesis. Mol Cell Biol. 2007;27:7669–82. doi: 10.1128/MCB.00002-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flohr T, et al. Detection of mutations in the DNA polymerase delta gene of human sporadic colorectal cancers and colon cancer cell lines. Int J Cancer. 1999;80:919–29. doi: 10.1002/(sici)1097-0215(19990315)80:6<919::aid-ijc19>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 15.Daee DL, Mertz TM, Shcherbakova PV. A cancer-associated DNA polymerase delta variant modeled in yeast causes a catastrophic increase in genomic instability. Proc Natl Acad Sci U S A. 2010;107:157–62. doi: 10.1073/pnas.0907526106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loeb LA, Monnat RJ., Jr. DNA polymerases and human disease. Nat Rev Genet. 2008;9:594–604. doi: 10.1038/nrg2345. [DOI] [PubMed] [Google Scholar]
- 17.Hübscher U, Spadari S, Villani G, Maga G. DNA Polymerases: Discovery, characterization and functions in cellular DNA transactions. World Scientific; Singapore: 2010. [Google Scholar]
- 18.Masutani C, et al. Xeroderma pigmentosum variant (XP-V) correcting protein from HeLa cells has a thymine dimer bypass DNA polymerase activity. EMBO J. 1999;18:3491–501. doi: 10.1093/emboj/18.12.3491. [Biochemical identification of polη as a DNA polymerase for bypass of UV radiation induced cyclobutane pyrimidine dimers.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inui H, et al. Xeroderma pigmentosum-variant patients from America, Europe, and Asia. J Invest Dermatol. 2008;128:2055–68. doi: 10.1038/jid.2008.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson RE, Washington MT, Prakash S, Prakash L. Fidelity of human DNA polymerase eta. J Biol Chem. 2000;275:7447–50. doi: 10.1074/jbc.275.11.7447. [DOI] [PubMed] [Google Scholar]
- 21.Silverstein TD, et al. Structural basis for the suppression of skin cancers by DNA polymerase eta. Nature. 2010;465:1039–43. doi: 10.1038/nature09104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Biertumpfel C, et al. Structure and mechanism of human DNA polymerase eta. Nature. 2010;465:1044–8. doi: 10.1038/nature09196. [References 21 and 22 provide a structural explanation for the efficient and error-avoiding bypass activity of polη for a cyclobutane pyrimidine dimer.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin Q, et al. Increased susceptibility to UV-induced skin carcinogenesis in polymerase eta-deficient mice. Cancer Res. 2006;66:87–94. doi: 10.1158/0008-5472.CAN-05-1862. [DOI] [PubMed] [Google Scholar]
- 24.Limoli CL, Giedzinski E, Bonner WM, Cleaver JE. UV-induced replication arrest in the xeroderma pigmentosum variant leads to DNA double-strand breaks, gamma - H2AX formation, and Mre11 relocalization. Proc Natl Acad Sci U S A. 2002;99:233–8. doi: 10.1073/pnas.231611798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Auclair Y, Rouget R, Belisle JM, Costantino S, Drobetsky EA. Requirement for functional DNA polymerase eta in genome-wide repair of UV-induced DNA damage during S phase. DNA Repair (Amst) 2010 doi: 10.1016/j.dnarep.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 26.Rey L, et al. Human DNA polymerase eta is required for common fragile site stability during unperturbed DNA replication. Mol Cell Biol. 2009;29:3344–54. doi: 10.1128/MCB.00115-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Glick E, et al. Mutations in DNA polymerase eta are not detected in squamous cell carcinoma of the skin. Int J Cancer. 2006;119:2225–7. doi: 10.1002/ijc.22099. [DOI] [PubMed] [Google Scholar]
- 28.Johnson RE, Haracska L, Prakash S, Prakash L. Role of DNA polymerase zeta in the bypass of a (6-4) TT photoproduct. Mol Cell Biol. 2001;21:3558–63. doi: 10.1128/MCB.21.10.3558-3563.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamamoto J, et al. Translesion synthesis across the (6-4) photoproduct and its Dewar valence isomer by the Y-family and engineered DNA polymerases. Nucleic Acids Symp Ser (Oxf) 2008:339–40. doi: 10.1093/nass/nrn171. [DOI] [PubMed] [Google Scholar]
- 30.Szuts D, Marcus AP, Himoto M, Iwai S, Sale JE. REV1 restrains DNA polymerase zeta to ensure frame fidelity during translesion synthesis of UV photoproducts in vivo. Nucleic Acids Res. 2008;36:6767–80. doi: 10.1093/nar/gkn651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson RE, Washington MT, Haracska L, Prakash S, Prakash L. Eukaryotic polymerases iota and zeta act sequentially to bypass DNA lesions. Nature. 2000;406:1015–9. doi: 10.1038/35023030. [DOI] [PubMed] [Google Scholar]
- 32.Vaisman A, et al. Sequence context-dependent replication of DNA templates containing UV-induced lesions by human DNA polymerase iota. DNA Repair (Amst) 2003;2:991–1006. doi: 10.1016/s1568-7864(03)00094-6. [DOI] [PubMed] [Google Scholar]
- 33.Dumstorf CA, et al. Participation of mouse DNA polymerase iota in strand-biased mutagenic bypass of UV photoproducts and suppression of skin cancer. Proc Natl Acad Sci U S A. 2006;103:18083–8. doi: 10.1073/pnas.0605247103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohkumo T, et al. UV-B radiation induces epithelial tumors in mice lacking DNA polymerase eta and mesenchymal tumors in mice deficient for DNA polymerase iota. Mol Cell Biol. 2006;26:7696–706. doi: 10.1128/MCB.01076-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Y, et al. Evidence that in xeroderma pigmentosum variant cells, which lack DNA polymerase eta, DNA polymerase iota causes the very high frequency and unique spectrum of UV-induced mutations. Cancer Res. 2007;67:3018–26. doi: 10.1158/0008-5472.CAN-06-3073. [References 33-35 show that both POLH and POLI are important in defending organisms against the carcinogenic effects of UV radiation.] [DOI] [PubMed] [Google Scholar]
- 36.Jarosz DF, Godoy VG, Delaney JC, Essigmann JM, Walker GC. A single amino acid governs enhanced activity of DinB DNA polymerases on damaged templates. Nature. 2006;439:225–8. doi: 10.1038/nature04318. [DOI] [PubMed] [Google Scholar]
- 37.Minko IG, et al. Replication bypass of the acrolein-mediated deoxyguanine DNA-peptide cross-links by DNA polymerases of the DinB family. Chem Res Toxicol. 2008;21:1983–90. doi: 10.1021/tx800174a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Minko IG, et al. Role for DNA polymerase kappa in the processing of N2-N2-guanine interstrand cross-links. J Biol Chem. 2008;283:17075–82. doi: 10.1074/jbc.M801238200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ogi T, et al. Expression of human and mouse genes encoding polkappa: testis-specific developmental regulation and AhR-dependent inducible transcription. Genes Cells. 2001;6:943–53. doi: 10.1046/j.1365-2443.2001.00478.x. [DOI] [PubMed] [Google Scholar]
- 40.Ogi T, Shinkai Y, Tanaka K, Ohmori H. Pol kappa protects mammalian cells against the lethal and mutagenic effects of benzo[a]pyrene. Proc Natl Acad Sci U S A. 2002;99:15548–53. doi: 10.1073/pnas.222377899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stancel JN, et al. Polk mutant mice have a spontaneous mutator phenotype. DNA Repair (Amst) 2009;8:1355–62. doi: 10.1016/j.dnarep.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Masutani C, Kusumoto R, Iwai S, Hanaoka F. Mechanisms of accurate translesion synthesis by human DNA polymerase eta. EMBO J. 2000;19:3100–9. doi: 10.1093/emboj/19.12.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Colis LC, Raychaudhury P, Basu AK. Mutational specificity of gamma-radiation-induced guanine-thymine and thymine-guanine intrastrand cross-links in mammalian cells and translesion synthesis past the guanine-thymine lesion by human DNA polymerase eta. Biochemistry. 2008;47:8070–9. doi: 10.1021/bi800529f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee DH, Pfeifer GP. Translesion synthesis of 7,8-dihydro-8-oxo-2'-deoxyguanosine by DNA polymerase eta in vivo. Mutat Res. 2008;641:19–26. doi: 10.1016/j.mrfmmm.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Minko IG, et al. Translesion synthesis past acrolein-derived DNA adduct, gamma - hydroxypropanodeoxyguanosine, by yeast and human DNA polymerase eta. J Biol Chem. 2003;278:784–90. doi: 10.1074/jbc.M207774200. [DOI] [PubMed] [Google Scholar]
- 46.Shachar S, et al. Two-polymerase mechanisms dictate error-free and error-prone translesion DNA synthesis in mammals. EMBO J. 2009;28:383–393. doi: 10.1038/emboj.2008.281. [The experiments show that different combinations of translesion DNA polymerases act to bypass lesions in mammalian cells, depending on the type of DNA damage. Bypass of most lesions tested was dependent on REV3L (pol ζ) and at least one additional DNA polymerase. The data fit a model whereby DNA polymerases work sequentially to bypass adducts in DNA.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Albertella MR, Green CM, Lehmann AR, O'Connor MJ. A role for polymerase eta in the cellular tolerance to cisplatin-induced damage. Cancer Res. 2005;65:9799–806. doi: 10.1158/0008-5472.CAN-05-1095. [DOI] [PubMed] [Google Scholar]
- 48.Betous R, et al. Role of TLS DNA polymerases eta and kappa in processing naturally occurring structured DNA in human cells. Mol Carcinog. 2009;48:369–78. doi: 10.1002/mc.20509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Obeid S, et al. Replication through an abasic DNA lesion: structural basis for adenine selectivity. EMBO J. 2010 doi: 10.1038/emboj.2010.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Avkin S, Adar S, Blander G, Livneh Z. Quantitative measurement of translesion replication in human cells: evidence for bypass of abasic sites by a replicative DNA polymerase. Proc Natl Acad Sci U S A. 2002;99:3764–9. doi: 10.1073/pnas.062038699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seki M, et al. High-efficiency bypass of DNA damage by human DNA polymerase Q. EMBO J. 2004;23:4484–94. doi: 10.1038/sj.emboj.7600424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arana ME, Seki M, Wood RD, Rogozin IB, Kunkel TA. Low-fidelity DNA synthesis by human DNA polymerase theta. Nucleic Acids Res. 2008;36:3847–56. doi: 10.1093/nar/gkn310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kusumoto R, Masutani C, Iwai S, Hanaoka F. Translesion synthesis by human DNA polymerase eta across thymine glycol lesions. Biochemistry. 2002;41:6090–9. doi: 10.1021/bi025549k. [DOI] [PubMed] [Google Scholar]
- 54.Yoon JH, Bhatia G, Prakash S, Prakash L. Error-free replicative bypass of thymine glycol by the combined action of DNA polymerases kappa and zeta in human cells. Proc Natl Acad Sci U S A. 2010;107:14116–21. doi: 10.1073/pnas.1007795107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marini F, Kim N, Schuffert A, Wood RD. POLN, a nuclear PolA family DNA polymerase homologous to the DNA cross-link sensitivity protein Mus308. J Biol Chem. 2003;278:32014–9. doi: 10.1074/jbc.M305646200. [DOI] [PubMed] [Google Scholar]
- 56.Takata KI, Shimizu T, Iwai S, Wood RD. Human DNA polymerase N (POLN) is a low-fidelity enzyme capable of error-free bypass of 5S-thymine glycol. J Biol Chem. 2006;281:23445–23455. doi: 10.1074/jbc.M604317200. [DOI] [PubMed] [Google Scholar]
- 57.Takata KI, Arana ME, Seki M, Kunkel TA, Wood RD. Evolutionary conservation of residues in vertebrate DNA polymerase N conferring low fidelity and bypass activity. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yamanaka K, et al. Novel Enzymatic Function of DNA Polymerase nu in Translesion DNA Synthesis Past Major Groove DNA-Peptide and DNA-DNA Cross-Links. Chem Res Toxicol. 2010;23:689–695. doi: 10.1021/tx900449u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Arana ME, Takata K, Garcia-Diaz M, Wood RD, Kunkel TA. A unique error signature for human DNA polymerase v. DNA Repair (Amst) 2007;6:213–23. doi: 10.1016/j.dnarep.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zietlow L, Smith LA, Bessho M, Bessho T. Evidence for the involvement of human DNA polymerase N in the repair of DNA interstrand cross-links. Biochemistry. 2009;48:11817–24. doi: 10.1021/bi9015346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moldovan GL, et al. DNA polymerase POLN participates in cross-link repair and homologous recombination. Mol Cell Biol. 30:1088–96. doi: 10.1128/MCB.01124-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yoshimura M, et al. Vertebrate POLQ and POLbeta cooperate in base excision repair of oxidative DNA damage. Mol Cell. 2006;24:115–25. doi: 10.1016/j.molcel.2006.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kohzaki M, et al. DNA polymerases nu and theta are required for efficient immunoglobulin V gene diversification in chicken. J Cell Biol. 2010;189:1117–27. doi: 10.1083/jcb.200912012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shivapurkar N, et al. Multiple regions of chromosome 4 demonstrating allelic losses in breast carcinomas. Cancer Res. 1999;59:3576–80. [PubMed] [Google Scholar]
- 65.Holbeck SL, Strathern JN. A role for REV3 in mutagenesis during double-strand break repair in Saccharomyces cerevisiae. Genetics. 1997;147:1017–24. doi: 10.1093/genetics/147.3.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Abdulovic AL, Jinks-Robertson S. The in vivo characterization of translesion synthesis across UV-induced lesions in Saccharomyces cerevisiae: insights into Pol zeta- and Pol eta-dependent frameshift mutagenesis. Genetics. 2006;172:1487–98. doi: 10.1534/genetics.105.052480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hirano Y, Sugimoto K. ATR homolog Mec1 controls association of DNA polymerase zeta-Rev1 complex with regions near a double-strand break. Curr Biol. 2006;16:586–90. doi: 10.1016/j.cub.2006.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gibbs PE, McGregor WG, Maher VM, Nisson P, Lawrence CW. A human homolog of the Saccharomyces cerevisiae REV3 gene, which encodes the catalytic subunit of DNA polymerase zeta. Proc Natl Acad Sci U S A. 1998;95:6876–80. doi: 10.1073/pnas.95.12.6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jansen JG, et al. Mammalian polymerase zeta is essential for post-replication repair of UV-induced DNA lesions. DNA Repair (Amst) 2009 doi: 10.1016/j.dnarep.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 70.Takata K, Wood RD. Bypass specialists work together. EMBO J. 2009;28:313–4. doi: 10.1038/emboj.2008.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bemark M, Khamlichi AA, Davies SL, Neuberger MS. Disruption of mouse polymerase zeta (Rev3) leads to embryonic lethality and impairs blastocyst development in vitro. Curr Biol. 2000;10:1213–6. doi: 10.1016/s0960-9822(00)00724-7. [DOI] [PubMed] [Google Scholar]
- 72.Esposito G, et al. Disruption of the Rev3l-encoded catalytic subunit of polymerase zeta in mice results in early embryonic lethality. Curr Biol. 2000;10:1221–4. doi: 10.1016/s0960-9822(00)00726-0. [DOI] [PubMed] [Google Scholar]
- 73.Wittschieben J, et al. Disruption of the developmentally regulated Rev3l gene causes embryonic lethality. Curr Biol. 2000;10:1217–20. doi: 10.1016/s0960-9822(00)00725-9. [DOI] [PubMed] [Google Scholar]
- 74.Van Sloun PP, et al. Involvement of mouse Rev3 in tolerance of endogenous and exogenous DNA damage. Mol Cell Biol. 2002;22:2159–69. doi: 10.1128/MCB.22.7.2159-2169.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.O-Wang J, et al. An essential role for REV3 in mammalian cell survival: absence of REV3 induces p53-independent embryonic death. Biochem Biophys Res Commun. 2002;293:1132–7. doi: 10.1016/S0006-291X(02)00341-8. [DOI] [PubMed] [Google Scholar]
- 76.Li Z, et al. hREV3 is essential for error-prone translesion synthesis past UV or benzo[a]pyrene diol epoxide-induced DNA lesions in human fibroblasts. Mutat Res. 2002;510:71–80. doi: 10.1016/s0027-5107(02)00253-1. [DOI] [PubMed] [Google Scholar]
- 77.Diaz M, et al. Decreased frequency and highly aberrant spectrum of ultraviolet-induced mutations in the hprt gene of mouse fibroblasts expressing antisense RNA to DNA polymerase zeta. Mol Cancer Res. 2003;1:836–47. [PubMed] [Google Scholar]
- 78.Sonoda E, et al. Multiple roles of Rev3, the catalytic subunit of polzeta in maintaining genome stability in vertebrates. Embo J. 2003;22:3188–97. doi: 10.1093/emboj/cdg308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wu F, Lin X, Okuda T, Howell SB. DNA polymerase zeta regulates cisplatin cytotoxicity, mutagenicity, and the rate of development of cisplatin resistance. Cancer Res. 2004;64:8029–35. doi: 10.1158/0008-5472.CAN-03-3942. [DOI] [PubMed] [Google Scholar]
- 80.Okada T, et al. Multiple roles of vertebrate REV genes in DNA repair and recombination. Mol Cell Biol. 2005;25:6103–11. doi: 10.1128/MCB.25.14.6103-6111.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wittschieben JP, Reshmi SC, Gollin SM, Wood RD. Loss of DNA polymerase zeta causes chromosomal instability in mammalian cells. Cancer Res. 2006;66:134–42. doi: 10.1158/0008-5472.CAN-05-2982. [DOI] [PubMed] [Google Scholar]
- 82.Gueranger Q, et al. Role of DNA polymerases eta, iota and zeta in UV resistance and UV-induced mutagenesis in a human cell line. DNA Repair (Amst) 2008;7:1551–62. doi: 10.1016/j.dnarep.2008.05.012. [DOI] [PubMed] [Google Scholar]
- 83.Roos WP, et al. The translesion polymerase Rev3L in the tolerance of alkylating anticancer drugs. Mol Pharmacol. 2009;76:927–34. doi: 10.1124/mol.109.058131. [DOI] [PubMed] [Google Scholar]
- 84.Wittschieben JP, et al. Loss of DNA polymerase zeta enhances spontaneous tumorigenesis. Cancer Res. 2010;70:2770–8. doi: 10.1158/0008-5472.CAN-09-4267. [Loss of pol zeta provides a selective advantage for the formation of tumor cells, with enhanced spontaneous tumorigenesis in the presence and absence of p53 function.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schenten D, et al. Pol zeta ablation in B cells impairs the germinal center reaction, class switch recombination, DNA break repair, and genome stability. J Exp Med. 2009;206:477–90. doi: 10.1084/jem.20080669. [B-cells deleting Rev3L display pronounced chromosome instability in mice.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Morelli C, et al. Cloning and characterization of the common fragile site FRA6F harboring a replicative senescence gene and frequently deleted in human tumors. Oncogene. 2002;21:7266–76. doi: 10.1038/sj.onc.1205573. [DOI] [PubMed] [Google Scholar]
- 87.Nelson JR, Lawrence CW, Hinkle DC. Thymine-thymine dimer bypass by yeast DNA polymerase zeta. Science. 1996;272:1646–9. doi: 10.1126/science.272.5268.1646. [DOI] [PubMed] [Google Scholar]
- 88.Lemontt JF. Mutants of Yeast Defective in Mutation Induced by Ultraviolet Light. Genetics. 1971;68:21–33. doi: 10.1093/genetics/68.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Simpson LJ, Sale JE. Rev1 is essential for DNA damage tolerance and nontemplated immunoglobulin gene mutation in a vertebrate cell line. Embo J. 2003;22:1654–64. doi: 10.1093/emboj/cdg161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jansen JG, et al. The BRCT domain of mammalian Rev1 is involved in regulating DNA translesion synthesis. Nucleic Acids Res. 2005;33:356–65. doi: 10.1093/nar/gki189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jansen JG, et al. Separate domains of Rev1 mediate two modes of DNA damage bypass in mammalian cells. Mol Cell Biol. 2009;29:3113–23. doi: 10.1128/MCB.00071-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jansen JG, et al. Strand-biased defect in C/G transversions in hypermutating immunoglobulin genes in Rev1-deficient mice. J Exp Med. 2006;203:319–23. doi: 10.1084/jem.20052227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Masuda K, et al. A critical role for REV1 in regulating the induction of C:G transitions and A:T mutations during Ig gene hypermutation. J Immunol. 2009;183:1846–50. doi: 10.4049/jimmunol.0901240. [DOI] [PubMed] [Google Scholar]
- 94.Tsaalbi-Shtylik A, et al. Error-prone translesion replication of damaged DNA suppresses skin carcinogenesis by controlling inflammatory hyperplasia. Proc Natl Acad Sci U S A. 2009;106:21836–41. doi: 10.1073/pnas.0909507106. [Partial ablation of REV1 function sensitizes cells to UV radiation and greatly increases pro-carcinogenic inflammation.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lin W, et al. The human REV1 gene codes for a DNA template-dependent dCMP transferase. Nucleic Acids Res. 1999;27:4468–75. doi: 10.1093/nar/27.22.4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hlavin EM, Smeaton MB, Noronha AM, Wilds CJ, Miller PS. Cross-link structure affects replication-independent DNA interstrand cross-link repair in mammalian cells. Biochemistry. 2010;49:3977–88. doi: 10.1021/bi902169q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ross AL, Simpson LJ, Sale JE. Vertebrate DNA damage tolerance requires the C-terminus but not BRCT or transferase domains of REV1. Nucleic Acids Res. 2005;33:1280–9. doi: 10.1093/nar/gki279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lehmann AR, et al. Translesion synthesis: Y-family polymerases and the polymerase switch. DNA Repair (Amst) 2007;6:891–9. doi: 10.1016/j.dnarep.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 99.Andersen PL, Xu F, Xiao W. Eukaryotic DNA damage tolerance and translesion synthesis through covalent modifications of PCNA. Cell Res. 2007 doi: 10.1038/cr.2007.114. [DOI] [PubMed] [Google Scholar]
- 100.Terai K, Abbas T, Jazaeri AA, Dutta A. CRL4(Cdt2) E3 ubiquitin ligase monoubiquitinates PCNA to promote translesion DNA synthesis. Mol Cell. 2010;37:143–9. doi: 10.1016/j.molcel.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bienko M, et al. Regulation of translesion synthesis DNA polymerase eta by monoubiquitination. Mol Cell. 2010;37:396–407. doi: 10.1016/j.molcel.2009.12.039. [DOI] [PubMed] [Google Scholar]
- 102.Pastushok L, Hanna M, Xiao W. Constitutive fusion of ubiquitin to PCNA provides DNA damage tolerance independent of translesion polymerase activities. Nucleic Acids Res. doi: 10.1093/nar/gkq239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Guo C, et al. Mouse Rev1 protein interacts with multiple DNA polymerases involved in translesion DNA synthesis. EMBO J. 2003;22:6621–30. doi: 10.1093/emboj/cdg626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ohashi E, et al. Interaction of hREV1 with three human Y-family DNA polymerases. Genes Cells. 2004;9:523–31. doi: 10.1111/j.1356-9597.2004.00747.x. [DOI] [PubMed] [Google Scholar]
- 105.Yuasa MS, et al. A human DNA polymerase eta complex containing Rad18, Rad6 and Rev1; proteomic analysis and targeting of the complex to the chromatin-bound fraction of cells undergoing replication fork arrest. Genes Cells. 2006;11:731–44. doi: 10.1111/j.1365-2443.2006.00974.x. [DOI] [PubMed] [Google Scholar]
- 106.Ohashi E, et al. Identification of a novel REV1-interacting motif necessary for DNA polymerase kappa function. Genes Cells. 2009;14:101–11. doi: 10.1111/j.1365-2443.2008.01255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Guo C, et al. REV1 protein interacts with PCNA: significance of the REV1 BRCT domain in vitro and in vivo. Mol Cell. 2006;23:265–71. doi: 10.1016/j.molcel.2006.05.038. [DOI] [PubMed] [Google Scholar]
- 108.Edmunds CE, Simpson LJ, Sale JE. PCNA ubiquitination and REV1 define temporally distinct mechanisms for controlling translesion synthesis in the avian cell line DT40. Mol Cell. 2008;30:519–29. doi: 10.1016/j.molcel.2008.03.024. [DOI] [PubMed] [Google Scholar]
- 109.McCulloch SD, Kokoska RJ, Kunkel TA. Efficiency, fidelity and enzymatic switching during translesion DNA synthesis. Cell Cycle. 2004;3:580–3. [PubMed] [Google Scholar]
- 110.Ogawara D, et al. Near-full-length REV3L appears to be a scarce maternal factor in Xenopus laevis eggs that changes qualitatively in early embryonic development. DNA Repair (Amst) 2010;9:90–5. doi: 10.1016/j.dnarep.2009.10.004. [A quantitative assessment of the abundance of DNA polymerases in Xenopus eggs, showing that TLS DNA polymerases have low abundance, particularly pol zeta.] [DOI] [PubMed] [Google Scholar]
- 111.Lopes M, Foiani M, Sogo JM. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol Cell. 2006;21:15–27. doi: 10.1016/j.molcel.2005.11.015. [Molecular and genetic analysis of the interplay between TLS, strand switching and recombination at a site of DNA damage.] [DOI] [PubMed] [Google Scholar]
- 112.Daigaku Y, Davies AA, Ulrich HD. Ubiquitin-dependent DNA damage bypass is separable from genome replication. Nature. 2010 doi: 10.1038/nature09097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Daigaku Y, Davies AA, Ulrich HD. Ubiquitin-dependent DNA damage bypass is separable from genome replication. Nature. 2010;465:951–955. doi: 10.1038/nature09097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Waters LS, Walker GC. The critical mutagenic translesion DNA polymerase Rev1 is highly expressed during G(2)/M phase rather than S phase. Proc Natl Acad Sci U S A. 2006;103:8971–6. doi: 10.1073/pnas.0510167103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Putnam CD, Hayes TK, Kolodner RD. Post-replication repair suppresses duplication-mediated genome instability. PLoS Genet. 2010;6:e1000933. doi: 10.1371/journal.pgen.1000933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Karras GI, Jentsch S. The RAD6 DNA damage tolerance pathway operates uncoupled from the replication fork and is functional beyond S phase. Cell. 2010;141:255–67. doi: 10.1016/j.cell.2010.02.028. [DOI] [PubMed] [Google Scholar]
- 117.Almeida KH, Sobol RW. A unified view of base excision repair: Lesion-dependent protein complexes regulated by post-translational modification. DNA Repair (Amst) 2007;6:695–711. doi: 10.1016/j.dnarep.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Copeland WC. The mitochondrial DNA polymerase in health and disease. Subcell Biochem. 2010;50:211–22. doi: 10.1007/978-90-481-3471-7_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Starcevic D, Dalal S, Sweasy JB. Is there a link between DNA polymerase beta and cancer? Cell Cycle. 2004;3:998–1001. [PubMed] [Google Scholar]
- 120.Sweasy JB, et al. Expression of DNA polymerase {beta} cancer-associated variants in mouse cells results in cellular transformation. Proc Natl Acad Sci U S A. 2005;102:14350–5. doi: 10.1073/pnas.0505166102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Cabelof DC, et al. Haploinsufficiency in DNA polymerase beta increases cancer risk with age and alters mortality rate. Cancer Res. 2006;66:7460–5. doi: 10.1158/0008-5472.CAN-06-1177. [DOI] [PubMed] [Google Scholar]
- 122.Bebenek K, et al. 5'-Deoxyribose phosphate lyase activity of human DNA polymerase iota in vitro. Science. 2001;291:2156–9. doi: 10.1126/science.1058386. [DOI] [PubMed] [Google Scholar]
- 123.Braithwaite EK, et al. DNA polymerase lambda mediates a back-up base excision repair activity in extracts of mouse embryonic fibroblasts. J Biol Chem. 2005;280:18469–75. doi: 10.1074/jbc.M411864200. [DOI] [PubMed] [Google Scholar]
- 124.Prasad R, et al. Human DNA polymerase {theta} possesses 5'-dRP lyase activity and functions in single-nucleotide base excision repair in vitro. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Petta TB, et al. Human DNA polymerase iota protects cells against oxidative stress. EMBO J. 2008;27:2883–95. doi: 10.1038/emboj.2008.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Prasad R, et al. Localization of the deoxyribose phosphate lyase active site in human DNA polymerase iota by controlled proteolysis. J Biol Chem. 2003;278:29649–54. doi: 10.1074/jbc.M305399200. [DOI] [PubMed] [Google Scholar]
- 127.van Loon B, Markkanen E, Hubscher U. Oxygen as a friend and enemy: How to combat the mutational potential of 8-oxo-guanine. DNA Repair (Amst) 2010;9:604–16. doi: 10.1016/j.dnarep.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 128.Goff JP, et al. Lack of DNA polymerase theta (POLQ) radiosensitizes bone marrow stromal cells in vitro and increases reticulocyte micronuclei after total-body irradiation. Radiation Research. 2009;172:165–74. doi: 10.1667/RR1598.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Shivji MK, Podust VN, Hubscher U, Wood RD. Nucleotide excision repair DNA synthesis by DNA polymerase epsilon in the presence of PCNA, RFC, and RPA. Biochemistry. 1995;34:5011–7. doi: 10.1021/bi00015a012. [DOI] [PubMed] [Google Scholar]
- 130.Ogi T, et al. Three DNA polymerases, recruited by different mechanisms, carry out NER repair synthesis in human cells. Mol Cell. 2010;37:714–27. doi: 10.1016/j.molcel.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 131.Friedberg EC, et al. DNA repair and mutagenesis. 2nd edition ASM Press; Washington, DC: 2006. [Google Scholar]
- 132.Kidane D, et al. DNA polymerase beta is critical for mouse meiotic synapsis. EMBO J. 2010;29:410–23. doi: 10.1038/emboj.2009.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kawamoto T, et al. Dual roles for DNA polymerase eta in homologous DNA recombination and translesion DNA synthesis. Mol Cell. 2005;20:793–9. doi: 10.1016/j.molcel.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 134.McIlwraith MJ, West SC. DNA repair synthesis facilitates RAD52-mediated second-end capture during DSB repair. Mol Cell. 2008;29:510–6. doi: 10.1016/j.molcel.2007.11.037. [DOI] [PubMed] [Google Scholar]
- 135.Rattray AJ, Shafer BK, McGill CB, Strathern JN. The roles of REV3 and RAD57 in double-strand-break-repair-induced mutagenesis of Saccharomyces cerevisiae. Genetics. 2002;162:1063–77. doi: 10.1093/genetics/162.3.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Shen X, et al. REV3 and REV1 play major roles in recombination-independent repair of DNA interstrand cross-links mediated by monoubiquitinated proliferating cell nuclear antigen (PCNA). J Biol Chem. 2006;281:13869–72. doi: 10.1074/jbc.C600071200. [DOI] [PubMed] [Google Scholar]
- 137.Zhang N, Liu X, Li L, Legerski R. Double-strand breaks induce homologous recombinational repair of interstrand cross-links via cooperation of MSH2, ERCC1-XPF, REV3, and the Fanconi anemia pathway. DNA Repair (Amst) 2007;6:1670–8. doi: 10.1016/j.dnarep.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Mahajan KN, et al. Association of terminal deoxynucleotidyl transferase with Ku. Proc Natl Acad Sci U S A. 1999;96:13926–31. doi: 10.1073/pnas.96.24.13926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ma Y, et al. A biochemically defined system for mammalian nonhomologous DNA end joining. Mol Cell. 2004;16:701–13. doi: 10.1016/j.molcel.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 140.Bebenek K, Garcia-Diaz M, Zhou RZ, Povirk LF, Kunkel TA. Loop 1 modulates the fidelity of DNA polymerase {lambda}. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140a.Andrade P, Martin MJ, Juarez R, Lopez de Saro F, Blanco L. Limited terminal transferase in human DNA polymerase mu defines the required balance between accuracy and efficiency in NHEJ. Proc Natl Acad Sci U S A. 2009;106:16203–8. doi: 10.1073/pnas.0908492106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Desiderio SV, et al. Insertion of N regions into heavy-chain genes is correlated with expression of terminal deoxytransferase in B cells. Nature. 1984;311:752–5. doi: 10.1038/311752a0. [DOI] [PubMed] [Google Scholar]
- 142.Bertocci B, De Smet A, Weill JC, Reynaud CA. Nonoverlapping functions of DNA polymerases mu, lambda, and terminal deoxynucleotidyltransferase during immunoglobulin V(D)J recombination in vivo. Immunity. 2006;25:31–41. doi: 10.1016/j.immuni.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 142a.Lucas D, et al. Altered hematopoiesis in mice lacking DNA polymerase mu is due to inefficient double-strand break repair. PLoS Genet. 2009;5:e1000389. doi: 10.1371/journal.pgen.1000389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142b.Chayot R, Danckaert A, Montagne B, Ricchetti M. Lack of DNA polymerase mu affects the kinetics of DNA double-strand break repair and impacts on cellular senescence. DNA Repair (Amst) 2010;9:1187–99. doi: 10.1016/j.dnarep.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 143.Bertocci B, De Smet A, Berek C, Weill JC, Reynaud CA. Immunoglobulin kappa light chain gene rearrangement is impaired in mice deficient for DNA polymerase mu. Immunity. 2003;19:203–11. doi: 10.1016/s1074-7613(03)00203-6. [DOI] [PubMed] [Google Scholar]
- 144.Higgins GS, et al. A small interfering RNA screen of genes involved in DNA repair identifies tumor-specific radiosensitization by POLQ knockdown. Cancer Res. 2010;70:2984–93. doi: 10.1158/0008-5472.CAN-09-4040. [POLQ is identified as a gene conferring radioprotection to human tumor cells.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Shima N, Munroe RJ, Schimenti JC. The mouse genomic instability mutation chaos1 is an allele of Polq that exhibits genetic interaction with Atm. Mol Cell Biol. 2004;24:10381–9. doi: 10.1128/MCB.24.23.10381-10389.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Yu AM, McVey M. Synthesis-dependent microhomology-mediated end joining accounts for multiple types of repair junctions. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Chan SH, Yu AM, McVey M. Dual Roles for DNA Polymerase Theta in Alternative End-Joining Repair of Double-Strand Breaks in Drosophila. PLoS Genet. 2010;6:e1001005. doi: 10.1371/journal.pgen.1001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Murakumo Y, et al. Interactions in the error-prone postreplication repair proteins hREV1, hREV3, and hREV7. J Biol Chem. 2001;276:35644–51. doi: 10.1074/jbc.M102051200. [DOI] [PubMed] [Google Scholar]
- 149.Brondello JM, et al. Novel evidences for a tumor suppressor role of Rev3, the catalytic subunit of Pol zeta. Oncogene. 2008;27:6093–101. doi: 10.1038/onc.2008.212. [DOI] [PubMed] [Google Scholar]
- 150.Pan Q, Fang Y, Xu Y, Zhang K, Hu X. Down-regulation of DNA polymerases kappa, eta, iota, and zeta in human lung, stomach, and colorectal cancers. Cancer Lett. 2005;217:139–47. doi: 10.1016/j.canlet.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 151.Lemée F, et al. POLQ up-regulation is associated with poor survival in breast cancer, perturbs DNA replication and promotes genetic instability. Proc Natl Acad Sci (USA) 2010;107:13390–13395. doi: 10.1073/pnas.0910759107. [A screen showing that POLQ gene expression is enhanced in breast cancer samples compared to normal tissue.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Albertella MR, Lau A, O'Connor MJ. The overexpression of specialized DNA polymerases in cancer. DNA Repair (Amst) 2005;4:583–93. doi: 10.1016/j.dnarep.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 153.O-Wang J, et al. DNA polymerase kappa, implicated in spontaneous and DNA damage-induced mutagenesis, is overexpressed in lung cancer. Cancer Res. 2001;61:5366–9. [PubMed] [Google Scholar]
- 154.Wang H, et al. Analysis of specialized DNA polymerases expression in human gliomas: association with prognostic significance. Neuro Oncol. 2010 doi: 10.1093/neuonc/nop074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Liu PY, et al. Identification of Las2, a major modifier gene affecting the Pas1 mouse lung tumor susceptibility locus. Cancer Res. 2009;69:6290–8. doi: 10.1158/0008-5472.CAN-09-0782. [DOI] [PubMed] [Google Scholar]
- 156.Ceppi P, et al. Polymerase eta mRNA expression predicts survival of non-small cell lung cancer patients treated with platinum-based chemotherapy. Clin Cancer Res. 2009;15:1039–45. doi: 10.1158/1078-0432.CCR-08-1227. [DOI] [PubMed] [Google Scholar]
- 157.Tan XH, et al. Frequent mutation related with overexpression of DNA polymerase beta in primary tumors and precancerous lesions of human stomach. Cancer Lett. 2005;220:101–14. doi: 10.1016/j.canlet.2004.07.049. [DOI] [PubMed] [Google Scholar]
- 158.Yoshizawa K, et al. Gastrointestinal hyperplasia with altered expression of DNA polymerase beta. PLoS One. 2009;4:e6493. doi: 10.1371/journal.pone.0006493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Pillaire MJ, et al. Upregulation of error-prone DNA polymerases beta and kappa slows down fork progression without activating the replication checkpoint. Cell Cycle. 2007;6:471–7. doi: 10.4161/cc.6.4.3857. [DOI] [PubMed] [Google Scholar]
- 160.Chan K, et al. Overexpression of DNA polymerase beta results in an increased rate of frameshift mutations during base excision repair. Mutagenesis. 2007;22:183–8. doi: 10.1093/mutage/gel070. [DOI] [PubMed] [Google Scholar]
- 161.Kawamura K, et al. DNA polymerase θ is preferentially expressed in lymphoid tissues and upregulated in human cancers. Int J Cancer. 2004;109:9–16. doi: 10.1002/ijc.11666. [DOI] [PubMed] [Google Scholar]
- 162.Pillaire MJ, et al. A ‘DNA replication’ signature of progression and negative outcome in colorectal cancer. Oncogene. 2010;29:876–87. doi: 10.1038/onc.2009.378. [DOI] [PubMed] [Google Scholar]
- 163.Higgins GS, et al. Overexpression Of POLQ Confers a Poor Prognosis In Early Breast Cancer Patients. Oncotarget. 2010;1:175–184. doi: 10.18632/oncotarget.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kuriyama I, et al. Effect of dehydroaltenusin-C12 derivative, a selective DNA polymerase alpha inhibitor, on DNA replication in cultured cells. Molecules. 2008;13:2948–61. doi: 10.3390/molecules13122948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kuramochi K, et al. Synthesis and structure-activity relationships of dehydroaltenusin derivatives as selective DNA polymerase alpha inhibitors. Bioorg Med Chem. 2009;17:7227–38. doi: 10.1016/j.bmc.2009.08.051. [DOI] [PubMed] [Google Scholar]
- 166.Maeda N, et al. Anti-tumor effects of dehydroaltenusin, a specific inhibitor of mammalian DNA polymerase alpha. Biochem Biophys Res Commun. 2007;352:390–6. doi: 10.1016/j.bbrc.2006.11.021. [DOI] [PubMed] [Google Scholar]
- 167.Kumamoto-Yonezawa Y, et al. Enhancement of human cancer cell radiosensitivity by conjugated eicosapentaenoic acid - a mammalian DNA polymerase inhibitor. Int J Oncol. 2010;36:577–84. doi: 10.3892/ijo_00000532. [DOI] [PubMed] [Google Scholar]
- 168.Mizushina Y, et al. The inhibitory action of kohamaic acid A derivatives on mammalian DNA polymerase beta. Molecules. 2009;14:102–21. doi: 10.3390/molecules14010102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Gao Z, Maloney DJ, Dedkova LM, Hecht SM. Inhibitors of DNA polymerase beta: activity and mechanism. Bioorg Med Chem. 2008;16:4331–40. doi: 10.1016/j.bmc.2008.02.071. [DOI] [PubMed] [Google Scholar]
- 170.Stachelek GC, et al. Potentiation of temozolomide cytotoxicity by inhibition of DNA polymerase beta is accentuated by BRCA2 mutation. Cancer Res. 2010;70:409–17. doi: 10.1158/0008-5472.CAN-09-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Jaiswal AS, et al. A novel inhibitor of DNA polymerase beta enhances the ability of temozolomide to impair the growth of colon cancer cells. Mol Cancer Res. 2009;7:1973–83. doi: 10.1158/1541-7786.MCR-09-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Yang J, et al. Cells deficient in the base excision repair protein, DNA polymerase beta, are hypersensitive to oxaliplatin chemotherapy. Oncogene. 2010;29:463–8. doi: 10.1038/onc.2009.327. [DOI] [PubMed] [Google Scholar]
- 173.Mizushina Y, et al. 3-O-methylfunicone, a selective inhibitor of mammalian Y-family DNA polymerases from an Australian sea salt fungal strain. Mar Drugs. 2009;7:624–39. doi: 10.3390/md7040624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Dorjsuren D, et al. A real-time fluorescence method for enzymatic characterization of specialized human DNA polymerases. Nucleic Acids Res. 2009;37:e128. doi: 10.1093/nar/gkp641. [Describes an example of an assay for small molecule inhibitor, and reports nanomolar inhibitors of DNA polymerases iota and eta.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Dumstorf CA, Mukhopadhyay S, Krishnan E, Haribabu B, McGregor WG. REV1 is implicated in the development of carcinogen-induced lung cancer. Mol Cancer Res. 2009;7:247–54. doi: 10.1158/1541-7786.MCR-08-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Xie K, Doles J, Hemann MT, Walker GC. Error-prone translesion synthesis mediates acquired chemoresistance. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.1011412107. [In a mouse cancer model, the activity of Rev1 in mutagenesis plays a critical role in the development of acquired cyclophosphamide resistance.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Doles J, et al. Suppression of Rev3, the catalytic subunit of Pol{zeta}, sensitizes drug-resistant lung tumors to chemotherapy. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.1011409107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Swan MK, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Structure of the human Rev1-DNA-dNTP ternary complex. J Mol Biol. 2009;390:699–709. doi: 10.1016/j.jmb.2009.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Nair DT, Johnson RE, Prakash S, Prakash L, Aggarwal AK. Replication by human DNA polymerase-iota occurs by Hoogsteen base-pairing. Nature. 2004;430:377–80. doi: 10.1038/nature02692. [DOI] [PubMed] [Google Scholar]
- 180.Lemée F, et al. POLQ up-regulation is associated with poor survival in breast cancer, perturbs DNA replication and promotes genetic instability. Proc Natl Acad Sci (USA) 2010;107:13390–13395. doi: 10.1073/pnas.0910759107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180a.Hegi ME, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 181.Bhagwat NR, et al. Immunodetection of DNA repair endonuclease ERCC1-XPF in human tissue. Cancer Res. 2009;69:6831–8. doi: 10.1158/0008-5472.CAN-09-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.King NM, et al. Overproduction of DNA polymerase eta does not raise the spontaneous mutation rate in diploid human fibroblasts. DNA Repair (Amst) 2005;4:714–24. doi: 10.1016/j.dnarep.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 183.Ohmori H, Hanafusa T, Ohashi E, Vaziri C. Separate Roles of Structured and Unstructured Regions of Y-Family DNA Polymerases. Advances in Protein Chemistry and Structural Biology. 2009;78:99–146. doi: 10.1016/S1876-1623(08)78004-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Boudsocq F, et al. Investigating the role of the little finger domain of Y-family DNA polymerases in low fidelity synthesis and translesion replication. J Biol Chem. 2004;279:32932–40. doi: 10.1074/jbc.M405249200. [DOI] [PubMed] [Google Scholar]
- 184a.Betz K, et al. Structures of DNA polymerases caught processing size-augmented nucleotide probes. Angew Chem Int Ed Engl. 2010;49:5181–4. doi: 10.1002/anie.200905724. [DOI] [PubMed] [Google Scholar]
- 185.Zeng X, et al. DNA polymerase eta is an A-T mutator in somatic hypermutation of immunoglobulin variable genes. Nat Immunol. 2001;2:537–41. doi: 10.1038/88740. [DOI] [PubMed] [Google Scholar]
- 186.Delbos F, Aoufouchi S, Faili A, Weill JC, Reynaud CA. DNA polymerase eta is the sole contributor of A/T modifications during immunoglobulin gene hypermutation in the mouse. J Exp Med. 2007;204:17–23. doi: 10.1084/jem.20062131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Martomo SA, Saribasak H, Yokoi M, Hanaoka F, Gearhart PJ. Reevaluation of the role of DNA polymerase theta in somatic hypermutation of immunoglobulin genes. DNA Repair (Amst) 2008;7:1603–8. doi: 10.1016/j.dnarep.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Ross AL, Sale JE. The catalytic activity of REV1 is employed during immunoglobulin gene diversification in DT40. Mol Immunol. 2006;43:1587–94. doi: 10.1016/j.molimm.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 189.Hance N, Ekstrand MI, Trifunovic A. Mitochondrial DNA polymerase gamma is essential for mammalian embryogenesis. Hum Mol Genet. 2005;14:1775–83. doi: 10.1093/hmg/ddi184. [DOI] [PubMed] [Google Scholar]
- 190.Gu H, Marth JD, Orban PC, Mossmann H, Rajewsky K. Deletion of a DNA polymerase β gene segment in T cells using cell type-specific gene targeting. Science. 1994;265:103–106. doi: 10.1126/science.8016642. [DOI] [PubMed] [Google Scholar]
- 191.Sugo N, Aratani Y, Nagashima Y, Kubota Y, Koyama H. Neonatal lethality with abnormal neurogenesis in mice deficient in DNA polymerase beta. EMBO J. 2000;19:1397–404. doi: 10.1093/emboj/19.6.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Sugo N, Niimi N, Aratani Y, Takiguchi-Hayashi K, Koyama H. p53 Deficiency rescues neuronal apoptosis but not differentiation in DNA polymerase beta-deficient mice. Mol Cell Biol. 2004;24:9470–7. doi: 10.1128/MCB.24.21.9470-9477.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Delbos F, et al. Contribution of DNA polymerase eta to immunoglobulin gene hypermutation in the mouse. J Exp Med. 2005;201:1191–6. doi: 10.1084/jem.20050292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.McDonald JP, et al. 129-derived strains of mice are deficient in DNA polymerase iota and have normal immunoglobulin hypermutation. J Exp Med. 2003;198:635–43. doi: 10.1084/jem.20030767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Schenten D, et al. DNA polymerase kappa deficiency does not affect somatic hypermutation in mice. Eur J Immunol. 2002;32:3152–60. doi: 10.1002/1521-4141(200211)32:11<3152::AID-IMMU3152>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 196.Shima N, et al. Phenotype-based identification of mouse chromosome instability mutants. Genetics. 2003;163:1031–40. doi: 10.1093/genetics/163.3.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Friedberg EC, Meira LB. Database of mouse strains carrying targeted mutations in genes affecting biological responses to DNA damage Version 7. DNA Repair (Amst) 2006;5:189–209. doi: 10.1016/j.dnarep.2005.09.009. [DOI] [PubMed] [Google Scholar]