Abstract
During 2010, we evaluated the presence of extended-spectrum β-lactamase– and AmpC-producing enterobacteria in broiler chickens at slaughter. Samples (70 carcasses and 51 ceca) from 4 flocks were analyzed by direct plating and after enrichment. Extended-spectrum β-lactamase producers were found in 88.6% and 72.5% of carcasses and ceca, respectively; AmpC producers were found in 52.9% and 56.9% of carcasses and ceca, respectively. Most isolates were identified as Escherichia coli; Enterobacter cloacae (cecum) and Proteus mirabilis (carcass) were found in 2 samples each. Molecular characterization revealed the domination of CTX-M genes; plasmidic AmpC was CIT-like. Phylogenetic grouping of E. coli showed types A (31.5%), B1 (20.2%), B2 (13.5%), and D (34.8%). These findings provide evidence that healthy broilers in Germany are a source for the dissemination of transmissible resistance mechanisms in enterobacteria brought from the rearing environment into the food chain during slaughtering.
Keywords: ESBL, Enterobacteriaceae, AmpC, slaughterhouse, Proteus mirabilis, Enterobacter cloacae, Germany, bacteria, foodborne diseases, zoonoses, chickens
Antimicrobial drug resistance is a threat for therapy failure in human medicine. The presence of enterobacteria, especially Escherichia coli that produces extended-spectrum β-lactamases (ESBLs), has increased during past decades in terms of the worldwide distribution of such resistance traits and of the evolution of different genes (1). Resistance genes of the ESBL type are mostly plasmid associated and therefore can spread among bacteria. Although chromosomal AmpC genes exist in several enterobacteria and E. coli, plasmid-bound types also exist that can be transferred among bacteria. These types can lead to the overall distribution of antimicrobial resistance, although the carrier bacteria are not pathogenic per se but might lead to opportunistic infections in predisposed patients because ESBL-producing E. coli are associated with, for example, urinary tract infection and severe systemic disease. E. coli infections can be nosocomial, community acquired, or foodborne. The main ESBL types are TEM, SHV, and CTX-M. Rates of CTX-M infections have increased during the last decade compared with rates of TEM and SHV infections. These enzymes confer resistance to β-lactam antibacterial drugs, particularly cephalosporins, and may be accompanied by co-resistance to drugs of other classes (1,2). Because of the ESBL resistance and associated co-resistance, empiric oral antibacterial therapy appears to be limited, especially in the community setting (3,4).
Sources of infection can be diverse. In addition to human sources of transmission in hospitals and communities, animals pose a reservoir for different pathogenic bacteria with zoonotic potential. Especially with food-producing animals, animals and humans are directly linked. Foodborne pathogens usually do not result in clinical infection of the animal host. Thus identification of sources is possible only by extensive field research in primary production and regular testing of end products. ESBL-producing enterobacteria were shown in different sources of food-producing animals at the farm and from products (5–7).
Several studies have focused on the characterization of ESBL or AmpC producers from food-production animals by testing flocks at the farm; others have focused only on fecal samples at later production steps (8–11). Less is known about the actual prevalence or diversity within single healthy broiler flocks at the slaughterhouse and the effect on meat contamination. The processing of meat contributes to overall transmission of bacteria from contamination during slaughtering and dressing, including transmission of resistant bacteria introduced at slaughter by colonized animals onto the meat product.
Our objective was to assess the prevalence of ESBL and AmpC producers in the broiler chicken–production chain in Germany in different species of the Enterobacteriaceae family. We focused on individual broiler flocks at the slaughterhouse level to show the introduction of enterobacteria to the slaughtering operation and transmission to the product. In addition to determining different resistance phenotypes and molecular characterization of the isolates, we evaluated the number of presumptive ESBL producers found in meat.
Materials and Methods
Sampling
From August through November 2010, we collected samples from housing of broiler chicken flocks at 2 different rearing sites (farms A and B) in Germany during fattening in 2 rearing periods (cycles). Each of the 4 flocks comprised up to 40.000 birds per house. To estimate flock status, we collected 2 pairs of sock swab specimens per housing 1 week before slaughter. In brief, moistened boot covers were put on the boots of specimen collectors, and specimens were collected by walking through the housing.
At the slaughterhouse, broiler carcasses and cecal samples were collected from the same flocks; the carcasses were collected after being chilled. Each flock was slaughtered at day 42 ± 1 d. For farm A, a total of 39 cecal samples and 40 carcasses were collected; for farm B, 12 cecal samples were available for cycle 1, and a total of 30 carcasses were collected for both cycles.
Isolation
Sample preparation and isolation of ESBL- or AmpC-producing enterobacteria were conducted as follows. Sock swabs were rinsed in peptone water (Merck, Darmstadt, Germany). Cecal contents were diluted at a ratio of 1 to 10 in peptone water. Broiler carcasses were rinsed in 500 mL maximum recovery diluent (Oxoid, Wesel, Germany). Rinsates (0.1 mL) were plated onto MacConkey agar (Merck) containing 1 mg/L cefotaxime or ceftazidime (Sigma-Aldrich, Munich, Germany) and onto Brilliance ESBL Agar (Oxoid) directly; plates were incubated aerobically for 24 h at 37°C ± 0.5°C. We mixed 100 mL of carcass rinse with 100 mL of double-strength peptone water (Merck). Samples in peptone water were incubated overnight at 37°C ± 0.5°C. Enrichment cultures were streaked onto MacConkey agar and Brilliance ESBL Agar and incubated as before.
Lactose-fermenting or -nonfermenting colonies of different morphology were collected from MacConkey agar and Brilliance ESBL Agar and screened for ESBL production by disk diffusion by using cefpodoxime (10 μg), aztreonam (30 μg), ceftazidime (30 μg), cefotaxime (30 μg), and ceftriaxone (30 μg) disks (Oxoid) and confirmed by microdilution test according to Clinical Laboratory Standards Institute (CLSI) methods (12) by using Micronaut-S β-lactamase V test plates (Merlin Diagnostika, Bornheim-Hersel, Germany). These plates contained cefepime, cefpodoxime, ceftazidime, and cefotaxime, each with and without the addition of clavulanic acid, aztreonam, piperacillin/tazobactam, and meropenem, and meropenem/EDTA. Isolates were identified to species level by using the API 20E test kit (BioMérieux, Nürtingen, Germany).
Isolates showing cefoxitin resistance were considered presumptive AmpC producers. Production of AmpC was confirmed according to methods of Black et al. (13). Co-production of ESBL was confirmed by adding 200 mg/L cloxacillin (Sigma-Aldrich, Munich, Germany) to Mueller-Hinton agar (Oxoid) using disks (Oxoid) according to CLSI ESBL confirmatory tests with disks of cefotaxime and ceftazidime, with or without clavulanic acid. An increase of the inhibition zone around disks containing cephalosporine and clavulanic acid by at least 5 mm confirmed ESBL production (14). Other antimicrobial agents tested by microdilution (Merlin Diagnostika, Bornheim-Hersel, Germany) were ciprofloxacin, nalidixic acid, chloramphenicol, tetracycline, trimethoprim–sulfamethoxazole, gentamicin, and streptomycin.
MICs were interpreted according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) (15) clinical breakpoints and, for cefoxitin, nalidixic acid, and tetracycline, according to CLSI breakpoints (12). Epidemiologic cutoff values dividing wild-type from non–wild-type strains were evaluated according to EUCAST (16). The EUCAST breakpoints were chosen because of more conservative values (17).
Molecular Characterization of Isolates
A subset of 76 E. coli isolates, confirmed ESBL or AmpC by phenotypic methods, was grouped phylogenetically by using modified triplex PCR (18). In brief, gene fragments of chuA and yjaA and a DNA fragment (TSPE4.C2) were amplified with primers ChuA.1 (5′-GACGAACCAACGGTCAGGAT-3′) and ChuA.2 (5′-TGCCGCCAGTACCAAAGACA-3′), fragment 279 bp; YjaA.1 (5′-TGAAGTGTCAGGAGACGCTG-3′), and YjaA.2 (5′-ATGGAGAATGCGTTCCTCAAC-3′), fragment 211 bp; and TspE4C2.1 (5′-GAGTAATGTCGGGGCATTCA-3′) and TspE4C2.2 (5′-CGCGCCAACAAAGTATTACG-3′), fragment 152 bp. PCR conditions were 3 min of initial denaturation, followed by 35 cycles at 94°C for 15 s, 60°C for 30 s, and 72°C for 45 s and final extension at 72°C for 5 min.
A representative selection of 78 isolates from cecal contents and carcasses, depending on sample type and phenotype was analyzed. The isolates were confirmed to show either an ESBL or an AmpC phenotype in the antimicrobial susceptibility tests. The presence of β-lactamase genes was confirmed by PCR with primers and conditions as reported: blaTEM TEM-F: ATAAAATTCTTGAAGACGAAA, TEM-R: GACAGTTACCAATGCTTAATC, fragment 1,080 bp, annealing 50°C (19); blaSHV SHV-F: GGGTTATTCTTATTTGTCGC, SHV-R: TTAGCGTTGCCAGTGCTC, fragment 930 bp, annealing 56°C (20); and blaCTX-M CTX-M-F: SCSATGTGCAGYACCAGTAA, CTX-M-R: ACCAGAAYVAGCGGBGC fragment 585 bp, annealing 58°C (21). AmpC presumptive isolates were tested for the presence of AmpC gene groups by multiplex PCR according to Pérez-Pérez and Hanson (22): MOX-MF: GCTGCTCAAGGAGCACAGGAT, MOX-MR: CACATTGACATAGGTGTGGTGC, fragment 520 bp; CIT-MF: TGGCCAGAACTGACAGGCAAA, CIT-MR: TTTCTCCTGAACGTGGCTGGC, fragment 462 bp; DHA-MF: AACTTTCACAGGTGTGCTGGGT, DHA-MR: CCGTACGCATACTGGCTTTGC, fragment 405 bp; ACC-MF: AACAGCCTCAGCAGCCGGTTA, ACC-MR: TTCGCCGCAATCATCCCTAGC, fragment 346 bp; EBC-MF: TCGGTAAAGCCGATGTTGCGG, EBC-MR: CTTCCACTGCGGCTGCCAGTT, fragment 302 bp; FOX-MF: AACATGGGGTATCAGGGAGATG, FOX-MR: CAAAGCGCGTAACCGGATTGG, fragment 190 bp; annealing 64°C.
Statistical Analysis
The 78 isolates were analyzed statistically to define phenotypic clusters, according to the resistance pattern. Isolates were characterized as susceptible, non–wild type, or clinically resistant according to aforementioned breakpoints for the individual antimicrobial agents and presence or absence of bla genes (TEM, SHV, CTX-M) and plasmidic AmpC genes. Data were analyzed by using BioNumerics software, version 5.1 (Applied Maths, Sint-Martens-Latem, Belgium) applying Pearson correlation and clustering with UPGMA (unweighted pair-group method with arithmetic mean). The cutoff for similarity was set to 80%.
Results
All 4 broiler flocks tested positive for ESBL- or AmpC-producing enterobacteria in 2 consecutive rearing cycles. All farm-level ESBL-positive isolates were identified as E. coli; in AmpC-producing enterobacteria, we found E. coli, Enterobacter cloacae, or Proteus mirabilis. The rate of ESBL-positive isolates was higher than AmpC-positive isolates in 3 flocks. Only in flock 1 on farm B was the rate of ESBL-confirmed isolates comparatively low (1 isolate was identified as an ESBL producer); AmpC-producers dominated that cycle (data not shown). These results identify healthy broiler flocks as reservoirs of antimicrobial-resistant enterobacteria for further distribution along the food production chain in Germany.
ESBL and AmpC Producers at Slaughter
At slaughter, ESBL- and AmpC-producing enterobacteria were found in 88.6% and 52.9% of the 70 carcasses and 72.5% and 56.9% of the 51 ceca, respectively. Most isolates were identified as E. coli; an AmpC-producing P. mirabilis isolate was detected in 1 carcass from each farm. The isolate from farm A could not be characterized further because the isolate died during storage. Furthermore, during the first cycle of farm B, E. cloacae strains were isolated from 2 cecal samples. Those isolates shared the same phenotype; thus only 1 isolate was characterized. At the flock level, up to 100% of the analyzed carcasses and cecal samples tested positive for ESBL-producing E. coli (Table 1). Overall, the presence of ESBL- or AmpC-positive enterobacteria on carcasses mirrored the presence in cecal contents.
Table 1. Prevalence of ESBL- or AmpC-producing enterobacteria in broiler chickens at slaughter, Germany, 2010*.
| Farm | Carcass |
Cecum |
|||||
|---|---|---|---|---|---|---|---|
| No. samples | ESBL, no. (%) | AmpC, no. (%) | No. samples | ESBL, no. (%) | AmpC, no. (%) | ||
| A, 1 | 20 | 20 (100) | 11 (55) | 20 | 16 (80) | 10 (50) | |
| A, 2 | 20 | 17 (85) | 9 (45) | 19 | 19 (100) | 7 (36.8) | |
| B, 1 | 10 | 6 (60) | 3 (30) | 12 | 2 (16.7) | 12 (100) | |
| B, 2 | 20 | 19 (95) | 14 (70) | 0 | 0 | 0 | |
*ESBL, extended-spectrum β-lactamase.
We tested samples of carcasses after direct plating and after enrichment. Direct plating allowed us to estimate numbers of suspected ESBL-producing E. coli isolates. After direct plating, 24 (38.7%) carcasses were positive. The calculated limit of detection for the direct plating was 3.7 log CFU/carcass, compared with 0.7 log CFU/carcass for the enrichment culture. The number of presumptive ESBL-producing E. coli isolates in samples from direct plating was 3.7–4.2 log CFU/carcass.
Antimicrobial Resistance, β-Lactamase Genes, and Phylogenetic Typing
Testing of antimicrobial agents found 3 different resistance phenotypes. The first phenotype was ESBL only, with reduced susceptibility to at least 1 of the tested cephalosporins with sensitivity to clavulanic acid. The second was AmpC only, with cefoxitin resistance and absence of a clavulanic acid MIC reduction. The third was ESBL in the presence of AmpC production, which could not be identified directly but was shown by confirmation of the clavulanic acid effect on Mueller-Hinton agar containing 200 mg/L cloxacillin.
The resistance phenotype differed by mechanism of resistance. For all isolates, we demonstrated resistance to cefpodoxime and breakpoints above the epidemiologic cutoff value for cefotaxime and ceftazidime. Although clinical resistance to cefotaxime was more prevalent in ESBL producers, the rate of ceftazidime resistance was higher in AmpC producers and in SHV-containing isolates. Some CTX-M gene–carrying isolates only were resistant to cefepime (Table 2, Appendix; Figure, Appendix). Clinical resistance and indication of other mechanisms according to cutoff values for non–β-lactam agents differed in the isolates. Clinical resistance for tetracycline was found in >50% of isolates and to nalidixic acid and chloramphenicol in 47% and 38% of E. coli isolates, respectively. Because no clinical MIC for streptomycin is defined by CLSI or EUCAST (12,15), isolates were considered resistant according to epidemiologic cutoff values. Resistance was found in 76 (60%) E. coli isolates. Multiresistant isolates showed resistance to nalidixic acid, tetracycline, and chloramphenicol (streptomycin) in most cases.
Table 2. MIC50 and MIC90distribution in Escherichia coli of ESBL or AmpC producers in broiler chickens, Germany, 2010*.
| FEP | FEP/CLA | CTX | CTX/CLA | FOX | CPD | CPD/CLA | CAZ | CAZ/CLA | ATM | TZP | CIP | NAL | CHL | TET | TMP/SXT | MEM | GEN | STR | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Breakpoint, µg/mL | >8 | – | >4 | >4 | >32† | >2 | – | >8 | – | >8 | >32 | >2 | >32 | >16 | >16† | >8 | >16 | >8 | – |
| ECOFF |
<0.125 |
– |
<0.25 |
<0.25 |
<8 |
<2 |
– |
<0.5 |
– |
<0.25 |
<8 |
<0.064 |
<16 |
<16 |
<8 |
<1 |
<0.125 |
<2 |
<16 |
| Total, n = 76 | |||||||||||||||||||
| MIC50 | 2 | <0.25 | 16 | 1 | 8 | >32 | 1 | 16 | 0.5 | 2 | 2 | <0.06 | 8 | 8 | 16 | 0.25 | <0.063 | 0.5 | 32 |
| MIC90 |
32 |
1 |
>32 |
16 |
>64 |
>32 |
>32 |
>32 |
32 |
8 |
4 |
1 |
>128 |
64 |
>16 |
>8 |
<0.063 |
1 |
>64 |
| ESBL, n = 42 | |||||||||||||||||||
| MIC50 | 8 | <0.25 | 32 | <0.25 | 4 | >32 | <0.25 | 8 | <0.25 | 2 | <1 | 0.125 | 16 | 8 | >16 | 0.25 | <0.063 | 0.5 | 32 |
| MIC90 |
>32 |
<0.25 |
>32 |
1 |
8 |
>32 |
1 |
32 |
0.5 |
4 |
2 |
2 |
>128 |
64 |
>16 |
>8 |
<0.063 |
1 |
>64 |
| ESBL, AmpC, n = 13 | |||||||||||||||||||
| MIC50 | 2 | <0.25 | 16 | 16 | 64 | >32 | >32 | 32 | 16 | 4 | 4 | 0.25 | 128 | 32 | >16 | 0.125 | <0.063 | 0.5 | >64 |
| MIC90 |
8 |
0.5 |
32 |
32 |
>64 |
>32 |
>32 |
>32 |
32 |
8 |
4 |
0.5 |
128 |
64 |
>16 |
>8 |
<0.063 |
1 |
>64 |
| AmpC, n = 21 | |||||||||||||||||||
| MIC50 | 0.5 | 0.5 | 16 | 16 | 64 | >32 | >32 | 32 | 16 | 2 | 4 | <0.063 | 2 | 4 | 1 | <0.063 | <0.063 | 0.5 | 4 |
| MIC90 | 1 | 1 | 32 | 16 | >64 | >32 | >32 | 32 | 32 | >8 | 8 | 0.25 | 128 | 16 | 16 | 0.25 | <0.063 | 1 | >64 |
*ESBL, extended spectrum β-lactamase; FEP, cefepime; FEP/CLA, cefepime/clavulanic acid; CTX, cefotaxime; CTX/CLA, cefotaxime/clavulanic acid; FOX, cefoxitin; CPD, cefpodoxime; CPD/CLA, cefpodoxime/clavulanic acid; CAZ, ceftazidime; CAZ/CLA, ceftazidime/clavulanic acid; ATM, aztreonam; TZP, piperacilin/tazobactam; CIP, ciprofloxacin; NAL, nalidixic acid; CHL, chloramphenicol; TET, tetracycline; TMP/SXT, trimethoprim/sulfamethoxazole; MEM, meropenem; GEN, gentamicin; STR, streptomycin; ECOFF, epidemiologic cutoff value.
Figure.
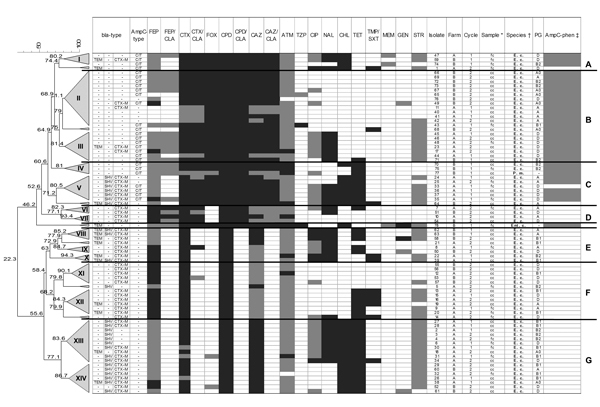
Phenotype distribution and dendrogram of 78 enterobacteria isolates from broiler chickens at the slaughterhouse, Germany, 2010. The dendrogram was generated by the unweighted pair-group method with arithmetic mean and Pearson correlation; trees were collapsed at a cutoff value of 80%. CEP, cefepime; FEP/CLA, cefepime/clavulanic acid; CTX, cefotaxime; COX, cefoxitin; CPP, cefpodoxime; CTX/CLA, cefpodoxime/clavulanic acid; CAZ, ceftazidime; CAZ/CLA, ceftazidime/clavulanic acid; ATM, aztreonam; TZP, piperacilin/tazobactam; CIP, ciprofloxacin; NAL, nalidixic acid; CHL, chloramphenico; TET, tetracycline; TMP/SXT, trimethoprim/sulfamethoxazole; MER, meropenem; GEN, gentamicin; STR, streptomycin; TEM, CTX-M, and SHV are extended spectrum β-lactamase (ESBL) types. Light gray, non–wild-type according to ESBL; ECOFF, epidemiologic cutoff value (16); dark gray, clinically resistant (12,15); *fc, cecal content; cc, carcass after chilling; †E.c., Escherichia coli; P.m., Proteus mirabilis; En.c., Enterobacter cloacae; ‡AmpC-test (13); gray, positive test; ESBL confirmation test with cloxacillin containing agar; A–G: clades; I–XIV, clusters.
We analyzed bla genes by PCR. We found all 3 bla genes (blaTEM, blaSHV, and blaCTX-M), with blaCTX-M being the most prevalent.
A total of 78 (95%) isolates tested by PCR were positive for at least 1 resistance gene. A single resistance gene was detected in 42 (54%) isolates; 32 (41%) isolates showed 2 or 3 resistance genes.
We identified ESBL producers that carried an additional plasmidic AmpC in 9 (12%) isolates from different samples. The blaCTX-M was identified in 55 (89%) typed ESBL producers; blaTEM and blaSHV were found in 27% and 47%, respectively. In AmpC producers, only CIT-like genes were found in 36 (75%) cefoxitin-resistant or AmpC-positive isolates. The CIT group of AmpC β-lactamases includes CMY with CMY-2 as the most prevalent member (23,24). Eight isolates of E. coli indicating an AmpC phenotype tested negative for plasmidic AmpC.
Phylogenetic typing revealed the 4 groups of E. coli. The types found most often were D and A, followed by B1 and, to a lesser extent, B2 (Table 3). The distribution was similar in carcasses of both flocks. In ceca, this distribution also was reflected for farm A; the limited number of isolates from farm B provided evidence only of B2 and D.
Table 3. Phylogenetic group distribution of extended spectrum β-lactamases– and AmpC-producing Escherichia coli in broiler chickens, Germany, 2010.
| Phylogenetic group | At farm, no. (%) |
At slaughter, no. (%) |
Total, no. (%) | |||||
|---|---|---|---|---|---|---|---|---|
| Ceca |
Carcass |
|||||||
| Farm A | Farm B | Farm A | Farm B | Farm A | Farm B | |||
| A | 4 (44) | 1 (25) | 4 (22) | 0 | 9 (30) | 10 (40) | 28 (32) | |
| B1 | 2 (22) | 2 (50) | 5 (28) | 0 | 8 (27) | 1 (4) | 18 (20) | |
| B2 | 1 (11) | 0 | 2 (11) | 2 (67) | 3 (10) | 4 (16) | 12 (14) | |
| D | 2 (22) | 1 (25) | 7 (39) | 1 (33) | 10 (33) | 10 (40) | 31 (35) | |
Comparison of isolates showed 2 major groups with 7 clades (A–G) and 14 clusters at a cutoff at 80% similarity (I–XIV) and 7 isolates that did not cluster with other strains (Figure, Appendix). The major groups were identified by the phenotype of resistance or susceptibility to clavulanic acid. The clades included strains of different origin because isolates from both farms and/or different sample types clustered.
The 2 other enterobacteria found clustered in separate branches. P. mirabilis isolates clustered with E. coli isolates in cluster IV, which might be due to the resistance inferred by the same resistance gene of CIT type possibly resulting from interspecies plasmid transfer. The single E. cloacae strain did not cluster with other isolates but clustered in the group with clavulanic acid–resistant isolates. This strain was positive in the AmpC test. Absence of plasmidic AmpC indicates inducible enzymes. In addition, the strain contained TEM β-lactamase.
Discussion
Bacteria in food-producing animals are spread through the food chain, which is important in terms of food shelf life and for transmission of pathogenic bacteria to the consumer. High numbers of bacteria can reduce shelf life and increase early food spoilage. Foodborne pathogens and bacteria with zoonotic potential are in focus worldwide because of immense health loss and costs that arise from foodborne infection associated with bacteria, such as Salmonella sp. and Campylobacter spp. (25).
Antimicrobial resistance is also of concern because of the limitation and even the risk for loss of effective antimicrobial treatment of infections; evidence exists that resistance from enterobacteria, such as Salmonella sp. and E. coli of animal origin can be transferred to humans (26,27). Food-producing animals may play a role in this process, and the food production chain needs to be evaluated to identify the potential for transmission pathways. Animals infected with antimicrobial-resistant strains of bacteria seem to be linked directly with human bacterial strains of E. coli (28–30).
Studies that focus on ESBL-producing E. coli at slaughter have found prevalences of 42.1% in feces of broilers and of 60% in chicken carcasses at the retail level in Portugal, but these studies have not included a flock attribution analysis (9,31). Results from different studies are sometimes difficult to compare because of different isolation and testing methods.
Total E. coli or coliforms usually are found in 100% of broiler carcasses at slaughter, at concentrations of 2.5 log CFU/mL for E. coli and 2.8 log CFU/mL for coliforms in rinse (100 mL/carcass), resulting in ≈4–5 log CFU/carcass (32). The percentage of samples positive for ESBL enterobacteria after direct plating and their numbers in this study were lower than those in reports of total E. coli, but in the view of possible genetic transfer between bacteria, even low numbers of bacteria harboring mechanisms of antibacterial drug resistance are relevant.
Moreno et al. (11) compared the proportion of E. coli with reduced susceptibility to expanded-spectrum cephalosporins with the total E. coli in the feces of healthy food animals. The resistant population in broilers was 4.3%. Horton et al. (33) estimated shedding densities of presumptive CTX-M E. coli for cattle, pigs, and chicken in the United Kingdom, where the latter showed higher shedding rates than did the red meat species. Although those studies described the numbers or proportions of resistant E. coli in feces of healthy animals, our study focused on total enterobacteria expressing the ESBL phenotype on carcasses at the end of slaughtering.
Fecal contamination, a known problem during broiler production, can lead to contamination of the meat with foodborne pathogens, such as Salmonella sp. and Campylobacter spp. (34,35). The role of shedding of bacteria through feces, which leads to contamination of carcasses during slaughtering, was evident in our study. Consequently, a high prevalence of ESBL-producing bacteria in colonized flocks could be shown. As a result, a considerable proportion of broilers were surface contaminated with ESBL-producing bacteria during slaughter.
In connection with the farm isolates, it is evident that strains from the 4 phylogenetic groups were already present during rearing of the poultry. A similar distribution also was found in other countries, where type A or type D dominated in poultry, and type B2 was present at a lower rate (29,36). Especially groups B2 and, to a lesser extent, type D are of public health concern. Those are supposed to contain strains of higher pathogenic character resulting from more virulence traits. B2 strains can be found in diseased and in healthy poultry and could have zoonotic potential through direct bird-to-human transmission or as genetic reservoir (37,38). This fact is even more important when increased virulence is paired with antibacterial drug resistance.
The sampled farms were geographically separate, and their poultry were supplied by different companies; however the farms were situated in a region with a high density of food animal production, which could indicate a common source, or selective pressure. At the cluster level, the picture was more diverse, and several clusters contained isolates unique to the individual farms. Isolates from the different sample types (cecal contents, carcasses) clustered, which indicates fecal contamination during the slaughtering process.
Co-resistance to non–β-lactam antibacterial drugs was most often associated with ESBL genes and was less prevalent in AmpC isolates (Table 2, Appendix; Figure, Appendix). Resistance to nalidixic acid found in several clusters (I, III, V, VIII–IX, and XIII), was usually combined with reduced susceptibility to ciprofloxacin. This resistance was present in isolates of various ESBL or AmpC gene combinations. High levels of nalidixic acid–resistant isolates are considered a first step in mutation to fluorquinolone-resistant strains (8). Reduced susceptibility to ciprofloxacin in the tested isolates, together with nalidixic acid resistance, indicates this process. Resistance to chloramphenicol and tetracycline, on the other hand, was most often linked with isolates carrying blaSHV alone or in combination with resistance to nalidixic acid. The co-resistance and the rate of reduced susceptibility are comparable to findings in a recent study by Dierikx et al. (5) and may be caused by similar treatment regimens in conventionally reared broilers in Germany and the Netherlands. Phylogenetic groups were not linked to definite resistance patterns, but isolates of the clinically important B2 group were found with either SHV- or CIT-like genes. Overall, the clustering showed a diversity of resistance phenotypes and bla gene combinations. These were present in different E. coli isolates according to the phylogenetic typing.
The ESBL gene families identified showed distributions comparable with distributions recently reported in Europe and other continents. TEM-52, SHV-12, and CTX-M-1 are the most often reported types from the food animal reservoir (2,39). CIT-like was the only AmpC type found. The absence of AmpC genes in some phenotype-confirmed isolates might indicate a different mechanism of resistance, probably attributable to overexpression of chromosomal AmpC, which usually results from mutations in the promoter/attenuator region (40).
Cephalosporin-resistant enterobacteria isolates were prevalent in the broiler flocks studied. Furthermore, colonization of broilers during rearing correlated with considerable contamination of broiler meat at the slaughterhouse. Isolates in the animals’ feces are distributed to the carcasses during the slaughtering operation by fecal contamination. This is a vital point when assessing the transmission potential through the food chain. Therefore, broilers seem to be an important reservoir for enterobacteria with transmissible mechanisms of resistance. In addition to ESBL-producing strains, a considerable number of isolates contained plasmidic AmpC of CIT type.
Phylogenetic characterization of E. coli isolates identified possible extraintestinal pathogenic group B2 strains with low prevalence. Their presence, together with the various resistance phenotype, should be observed further to evaluate the distribution and effect on public health.
Biography
Dr Reich is a scientist at the University of Veterinary Medicine Hanover, Institute of Food Quality and Food Safety. His research interest includes food microbiology with a focus on foodborne zoonoses.
Footnotes
Suggested citation for this article: Reich F, Atanassova V, Klein G. Extended-spectrum β-lactamase– and AmpC-producing enterobacteria in healthy broiler chickens, Germany. Emerg Infect Dis [Internet]. 2013 Aug [date cited]. http://dx.doi.org/10.3201/eid1908.120879
References
- 1.Paterson DL, Bonomo RA. Extended-spectrum beta-lactamases: a clinical update. Clin Microbiol Rev. 2005;18:657–86 . 10.1128/CMR.18.4.657-686.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cantón R, Coque TM. The CTX-M beta-lactamase pandemic. Curr Opin Microbiol. 2006;9:466–75 and. 10.1016/j.mib.2006.08.011 [DOI] [PubMed] [Google Scholar]
- 3.Falagas ME, Karageorgopoulos DE. Extended-spectrum beta-lactamase–producing organisms. J Hosp Infect. 2009;73:345–54 and. 10.1016/j.jhin.2009.02.021 [DOI] [PubMed] [Google Scholar]
- 4.Lavilla S, Gonzalez-Lopez JJ, Miro E, Dominguez A, Llagostera M, Bartolome RM, et al. Dissemination of extended-spectrum beta-lactamase–producing bacteria: the food-borne outbreak lesson. J Antimicrob Chemother. 2008;61:1244–51 and. 10.1093/jac/dkn093 [DOI] [PubMed] [Google Scholar]
- 5.Dierikx C, van Essen-Zandbergen A, Veldman K, Smith H, Mevius D. Increased detection of extended spectrum beta-lactamase producing Salmonella enterica and Escherichia coli isolates from poultry. Vet Microbiol. 2010;145:273–8 . 10.1016/j.vetmic.2010.03.019 [DOI] [PubMed] [Google Scholar]
- 6.Geser N, Stephan R, Kuhnert P, Zbinden R, Kaeppeli U, Cernela N, et al. Fecal carriage of extended-spectrum beta-lactamase–producing Enterobacteriaceae in swine and cattle at slaughter in Switzerland. J Food Prot. 2011;74:446–9 and. 10.4315/0362-028X.JFP-10-372 [DOI] [PubMed] [Google Scholar]
- 7.Leverstein-van Hall MA, Dierikx CM, Cohen Stuart J, Voets GM, van den Munckhof MP, van Essen-Zandbergen A, et al. Dutch patients, retail chicken meat and poultry share the same ESBL genes, plasmids and strains. Clin Microbiol Infect. 2011;17:873–80 and. 10.1111/j.1469-0691.2011.03497.x [DOI] [PubMed] [Google Scholar]
- 8.Smet A, Martel A, Persoons D, Dewulf J, Heyndrickx M, Catry B, et al. Diversity of extended-spectrum beta-lactamases and class C beta-lactamases among cloacal Escherichia coli isolates in Belgian broiler farms. Antimicrob Agents Chemother. 2008;52:1238–43 and. 10.1128/AAC.01285-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costa D, Vinue L, Poeta P, Coelho AC, Matos M, Saenz Y, et al. Prevalence of extended-spectrum beta-lactamase–producing Escherichia coli isolates in faecal samples of broilers. Vet Microbiol. 2009;138:339–44 . 10.1016/j.vetmic.2009.03.029 [DOI] [PubMed] [Google Scholar]
- 10.Randall LP, Clouting C, Horton RA, Coldham NG, Wu G, Clifton-Hadley FA, et al. Prevalence of Escherichia coli carrying extended-spectrum beta-lactamases (CTX-M and TEM-52) from broiler chickens and turkeys in Great Britain between 2006 and 2009. J Antimicrob Chemother. 2011;66:86–95 and. 10.1093/jac/dkq396 [DOI] [PubMed] [Google Scholar]
- 11.Moreno MA, Teshager T, Porrero MA, Garcia M, Escudero E, Torres C, et al. Abundance and phenotypic diversity of Escherichia coli isolates with diminished susceptibility to expanded-spectrum cephalosporins in faeces from healthy food animals after slaughter. Vet Microbiol. 2007;120:363–9 and. 10.1016/j.vetmic.2006.10.032 [DOI] [PubMed] [Google Scholar]
- 12.Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; 19th informational supplement. CLSI document M100–S19. Wayne (PA): The Institute; 2009. [Google Scholar]
- 13.Black JA, Moland ES, Thomson KS. AmpC disk test for detection of plasmid-mediated AmpC beta-lactamases in Enterobacteriaceae lacking chromosomal AmpC beta-lactamases. J Clin Microbiol. 2005;43:3110–3 and. 10.1128/JCM.43.7.3110-3113.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thomson KS. Extended-spectrum-beta-lactamase, AmpC, and carbapenemase issues. J Clin Microbiol. 2010;48:1019–25 and. 10.1128/JCM.00219-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.European Committee on Antimicrobial Susceptibility Testing. Clinical breakpoints—bacteria (v 3.1). 2013. 2013.02.11 [cited 2013 Feb 15]. http://www.eucast.org/clinical_breakpoints
- 16.European Committee on Antimicrobial Susceptibility Testing. Antimicrobial wild type distributions of microorganisms EUCAST version 5.13 [cited 2013 Feb 15]. http://mic.eucast.org/Eucast2/
- 17.Rodríguez-Baño J, Picón E, Navarro MD, López-Cerero L, Pascual A. ESBL-REIPI Group. Impact of changes in CLSI and EUCAST breakpoints for susceptibility in bloodstream infections due to extended-spectrum beta-lactamase–producing Escherichia coli. Clin Microbiol Infect. 2012;18:894–900 and. 10.1111/j.1469-0691.2011.03673.x [DOI] [PubMed] [Google Scholar]
- 18.Higgins J, Hohn C, Hornor S, Frana M, Denver M, Joerger R. Genotyping of Escherichia coli from environmental and animal samples. J Microbiol Methods. 2007;70:227–35 and. 10.1016/j.mimet.2007.04.009 [DOI] [PubMed] [Google Scholar]
- 19.Weill FX, Lailler R, Praud K, Kerouanton A, Fabre L, Brisabois A, et al. Emergence of extended-spectrum-beta-lactamase (CTX-M-9) –producing multiresistant strains of Salmonella enterica serotype Virchow in poultry and humans in France. J Clin Microbiol. 2004;42:5767–73 and. 10.1128/JCM.42.12.5767-5773.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Machado E, Canton R, Baquero F, Galan JC, Rollan A, Peixe L, et al. Integron content of extended-spectrum-beta-lactamase–producing Escherichia coli strains over 12 years in a single hospital in Madrid, Spain. Antimicrob Agents Chemother. 2005;49:1823–9 and. 10.1128/AAC.49.5.1823-1829.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sundsfjord A, Simonsen GS, Haldorsen BC, Haaheim H, Hjelmevoll SO, Littauer P, et al. Genetic methods for detection of antimicrobial resistance. APMIS. 2004;112:815–37 and. 10.1111/j.1600-0463.2004.apm11211-1208.x [DOI] [PubMed] [Google Scholar]
- 22.Pérez-Pérez FJ, Hanson ND. Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol. 2002;40:2153–62 and. 10.1128/JCM.40.6.2153-2162.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin LC, Weir EK, Poppe C, Reid-Smith RJ, Boerlin P. Characterization of blaCMY-2 plasmids in Salmonella and Escherichia coli isolates from food animals in Canada. Appl Environ Microbiol. 2012;78:1285–7 and. 10.1128/AEM.06498-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park YS, Adams-Haduch JM, Rivera JI, Curry SR, Harrison LH, Doi Y. Escherichia coli producing CMY-2 beta-lactamase in retail chicken, Pittsburgh, Pennsylvania, USA. Emerg Infect Dis. 2012;18:515–6 and. 10.3201/eid1803.111434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scharff RL. Economic burden from health losses due to foodborne illness in the United States. J Food Prot. 2012;75:123–31 and. 10.4315/0362-028X.JFP-11-058 [DOI] [PubMed] [Google Scholar]
- 26.Marshall BM, Levy SB. Food animals and antimicrobials: impacts on human health. Clin Microbiol Rev. 2011;24:718–33 and. 10.1128/CMR.00002-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smet A, Martel A, Persoons D, Dewulf J, Heyndrickx M, Herman L, et al. Broad-spectrum beta-lactamases among Enterobacteriaceae of animal origin: molecular aspects, mobility and impact on public health. FEMS Microbiol Rev. 2010;34:295–316 . 10.1111/j.1574-6976.2009.00198.x [DOI] [PubMed] [Google Scholar]
- 28.Manges AR, Smith SP, Lau BJ, Nuval CJ, Eisenberg JN, Dietrich PS, et al. Retail meat consumption and the acquisition of antimicrobial resistant Escherichia coli causing urinary tract infections: a case–control study. Foodborne Pathog Dis. 2007;4:419–31 . 10.1089/fpd.2007.0026 [DOI] [PubMed] [Google Scholar]
- 29.Johnson JR, Sannes MR, Croy C, Johnston B, Clabots C, Kuskowski MA, et al. Antimicrobial drug–resistant Escherichia coli from humans and poultry products, Minnesota and Wisconsin, 2002–2004. Emerg Infect Dis. 2007;13:838–46 and. 10.3201/eid1306.061576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jakobsen L, Kurbasic A, Skjot-Rasmussen L, Ejrnaes K, Porsbo LJ, Pedersen K, et al. Escherichia coli isolates from broiler chicken meat, broiler chickens, pork, and pigs share phylogroups and antimicrobial resistance with community-dwelling humans and patients with urinary tract infection. Foodborne Pathog Dis. 2010;7:537–47 and. 10.1089/fpd.2009.0409 [DOI] [PubMed] [Google Scholar]
- 31.Machado E, Coque TM, Canton R, Sousa JC, Peixe L. Antibiotic resistance integrons and extended-spectrum β-lactamases among Enterobacteriaceae isolates recovered from chickens and swine in Portugal. J Antimicrob Chemother. 2008;62:296–302 and. 10.1093/jac/dkn179 [DOI] [PubMed] [Google Scholar]
- 32.Huezo R, Northcutt JK, Smith DP, Fletcher DL, Ingram KD. Effect of dry air or immersion chilling on recovery of bacteria from broiler carcasses. J Food Prot. 2007;70:1829–34 . [DOI] [PubMed] [Google Scholar]
- 33.Horton RA, Randall LP, Snary EL, Cockrem H, Lotz S, Wearing H, et al. Fecal carriage and shedding density of CTX-M extended-spectrum β-lactamase–producing Escherichia coli in cattle, chickens, and pigs: implications for environmental contamination and food production. Appl Environ Microbiol. 2011;77:3715–9 and. 10.1128/AEM.02831-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bryan FL, Doyle MP. Health risks and consequences of Salmonella and Campylobacter jejuni in raw poultry. J Food Prot. 1995;03:326–44. [DOI] [PubMed] [Google Scholar]
- 35.Reich F, Atanassova V, Haunhorst E, Klein G. The effects of Campylobacter numbers in caeca on the contamination of broiler carcasses with Campylobacter. Int J Food Microbiol. 2008;127:116–20 and. 10.1016/j.ijfoodmicro.2008.06.018 [DOI] [PubMed] [Google Scholar]
- 36.Asai T, Masani K, Sato C, Hiki M, Usui M, Baba K, et al. Phylogenetic groups and cephalosporin resistance genes of Escherichia coli from diseased food-producing animals in Japan. Acta Vet Scand. 2011;53:52 and. 10.1186/1751-0147-53-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ewers C, Antao EM, Diehl I, Philipp HC, Wieler LH. Intestine and environment of the chicken as reservoirs for extraintestinal pathogenic Escherichia coli strains with zoonotic potential. Appl Environ Microbiol. 2009;75:184–92 and. 10.1128/AEM.01324-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cortés P, Blanc V, Mora A, Dahbi G, Blanco JE, Blanco M, et al. Isolation and characterization of potentially pathogenic antimicrobial-resistant Escherichia coli strains from chicken and pig farms in Spain. Appl Environ Microbiol. 2010;76:2799–805 and. 10.1128/AEM.02421-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coque TM, Baquero F, Canton R. Increasing prevalence of ESBL-producing Enterobacteriaceae in Europe. Euro Surveill. 2008;13:19–29 . [PubMed] [Google Scholar]
- 40.Peter-Getzlaff S, Polsfuss S, Poledica M, Hombach M, Giger J, Bottger EC, et al. Detection of AmpC beta-lactamase in Escherichia coli: comparison of three phenotypic confirmation assays and genetic analysis. J Clin Microbiol. 2011;49:2924–32 and. 10.1128/JCM.00091-11 [DOI] [PMC free article] [PubMed] [Google Scholar]


