To the Editor: Bats play a critical role in the transmission and origin of zoonotic diseases, primarily viral zoonoses associated with high case-fatality rates, including those caused by Nipah virus (NiV) and severe acute respiratory syndrome (SARS)–like coronavirus (CoV) infections (1). Recently, the World Health Organization (WHO) reported 44 confirmed cases of human infection with Middle East respiratory syndrome CoV, resulting in 22 deaths. Full-genome and phylogenetic analyses of these Middle East respiratory syndrome CoVs have been published elsewhere (2). The identified viruses from 2 patients (previously referred to as England/Qatar/2012 and EMC/2012) are genetically related and belong to group C betacoronavirus, which is most related to CoVs from Nycteris bats in Ghana and Pipistrellus bats in Europe (2,3). In addition, bat CoVs HKU4 and HKU5 originated from Tylonycteris pachypus and Pipistrellus abramus bats, respectively, in the People’s Republic of China (4). Bats are also known to harbor and transmit nonviral zoonotic pathogens, including the fungal pathogen Histoplasma capsulatum, which causes histoplasmosis in humans (5).
Bat guano is sold for use as a fertilizer in several countries, including Thailand, Indonesia, Mexico, Cuba, and Jamaica. The practice of collecting and harvesting bat guano may pose a considerable health risk because guano miners have a high level of contact and potential exposure to bat-borne pathogens. To assess pathogens in bat guano, we examined bat guano from a cave in the Khao Chong Phran Non-hunting Area (KCP-NHA) in Ratchaburi Province, Thailand, where bat guano was sold as agricultural fertilizer, for the presence of NiV, CoV, and H. capsulatum fungi. Bats from 14 species in 7 families have been found roosting within this area. Tadarida plicata bats are the most abundant species (2,500,000 bats), and 3 other species of bats found at the site each had thousands of members: Taphozous melanopogon, Taphozous theobaldi, and Hipposideros larvatus.
A random sample of dry bat guano, ≈100 g, was collected in a sterile plastic bag weekly from the main cave at KCP-NHA from September 2006 through August 2007. The specimens were sent for analysis by express mail (at room temperature within 2–3 days) to the WHO Collaborating Centre for Research and Training in the Viral Zoonoses Laboratory at Chulalongkorn University. Samples were frozen immediately at –80°C until nucleic acids were extracted and PCR assays were run. A total of 52 collected bat guano specimens were examined in this study.
Two aliquots of feces from each weekly specimen (104 samples total) were screened for CoV, NiV, and H. capsulatum by PCR. RNA was extracted from 10 mg of fecal pellet by using the QIAamp Viral RNA Mini Kit (QIAGEN, Hilden, Germany). CoV RNA was detected by using nested reverse transcription PCR with the degenerated primers to amplify the RNA-dependent RNA polymerase (RdRp) gene (6). NiV RNA was detected by duplex nested reverse transcription PCR (7). To detect H. capsulatum and other fungi, we extracted genomic DNA directly from bat guano by using the silica-guanidine thiocyanate protocol, NucliSense Isolation Reagent (bioMérieux, Boxtel, the Netherlands), according to the manufacturer’s protocol. We tested for fungal ribosomal DNA (rDNA) in extracted total nucleic acid specimens by using the PCR protocol designed to amplify all rDNA from 4 major fungus phyla at the internal transcribed spacer 1 and 2 regions (8).
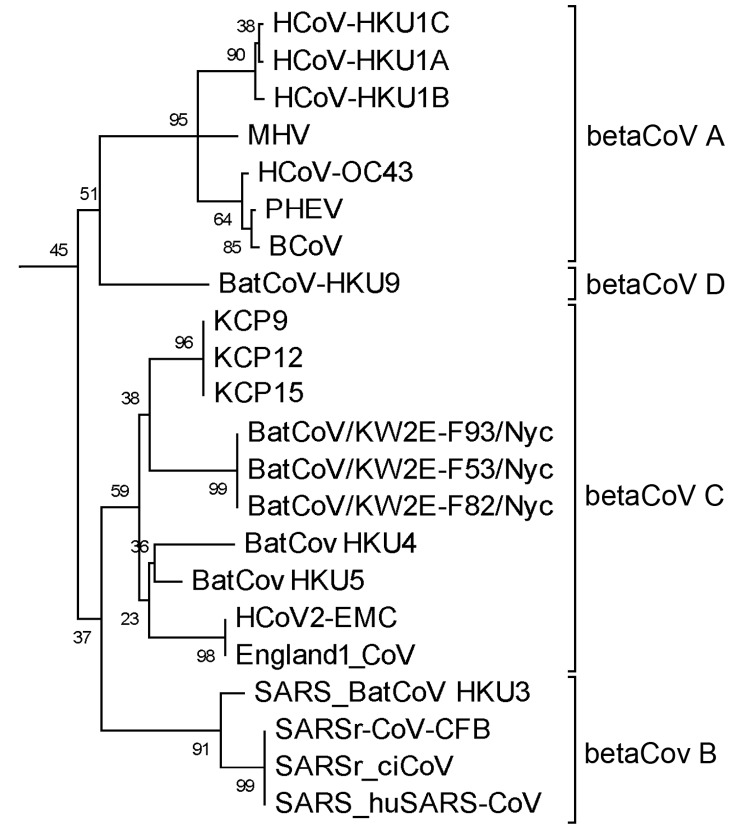
Four (3.8%) of 104 samples were positive for CoV. They were collected on September 2, 2006 (KCP9), October 26, 2006 (KCP12), November 14, 2006 (KCP15), and March 4, 2007 (KCP31). Three of the 4 positive CoV sequences (KCP9, KCP12, and KCP15) were identical at 152 nt of the RdRp region (ATCGTGCTATGCCTAATATGTGTAGGATTTTTGCATCTCTCATATTAGCTCGTAAACACAATACTTGTTGTAGTGTTTCAGACCGCTtTtATAGACTTGCaAACGAGTGTGCGCAAGTCTTGAGTGAGTATGTGCTATGTGGTGGTGGCTAT) and phylogenetically clustered with the group C betacoronavirus (Figure), with 76%, 80%, and 77% nt identity to bat CoV HKU4, bat CoV HKU5, and human CoV EMC and England1_CoV, respectively. The other CoV sequence (KCP31: ATCGTGCACTTCCCAATATGATACGCATGATTTCCGCCATGATTTTGGGATCAAAGCATGTTACTTGCTGTGACACATCTGATAAGTATTACCGTCTTTGTAATGAGCTtGCACAAGTTTTGACAGAGGTTGTTTATTCTAATGGTGGTTTC) showed 82% nt identity with bat CoV HKU8, an alphacoronavirus. Although we recognize that longer sequences or full genomes may alter the topology of the phylogeny slightly and give stronger branch support, we expect that the overall topology and placement of these CoVs would remain consistent. Samples from particular bat species could not be identified because bats of different species roost in this cave, and samples were pooled during collection for bat guano fertilizer. The detection of CoVs in bat guano from the KCP-NHA cave in Ratchaburi was consistent with the previous finding of alphacoronavirus from Hipposideros armiger bats from the same province in 2007, but those researchers tested fresh bat feces (9).
Figure.
Phylogenetic tree of 4 coronaviruses (CoVs) isolated from bat guano collected in this study (KCP9, KCP12, KCP15, and KCP31); 35 additional human and animal CoVs from the National Center for Biotechnology Information database are included. Construction of the tree was based on 152 nt of the RNA-dependent RNA polymerase gene region by maximum-likelihood method and GTR+I model with the 1,000 bootstrap resampling method implemented in MEGA5 (http://megasoftware.net/). Numbers on branches indicate percentages of bootstrap support from 1,000 replicates. The scale bar indicates the estimated 0.1 nt substitutions per site. CoV genera (Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus) based on International Committee on Taxonomy of Viruses recommendation are indicated (alphaCoV, betaCoV, gammaCoV, and deltaCoV, respectively). BatCoV_HKU8, Miniopterus bat CoV HKU8 (EU420139); HCoV-NL63, human CoV NL63 (JX524171); PEDV, porcine epidemic diarrhea virus (NC_003436); BatCoV 512, Scotophilus bat CoV 512 (DQ648858); HCoV-229E, human CoV 229E (NC_002645); BatCoV-HKU2, Rhinolophus bat CoV HKU2 (EF203064); TGEV, transmissible gastroenteritis virus (AJ271965); FIPV, feline CoV (DQ010921); HCoV-HKU1C, human CoV HKU1C (DQ415913); HCoV-HKU1A, human CoV HKU1A (DQ415903); HCoV-HKU1B, human CoV HKU1B (AY884001); MHV, murine hepatitis virus (NC001846); HCoV-OC43, human CoV OC43 (AY585229); PHEV, porcine hemagglutinating encephalomyelitis virus (DQ011855); BCoV, bovine CoV (AF391541); BatCoV-HKU9, Rousettus bat CoV HKU9 (NC009021); BatCoV/KW2E-F93/Nyc, Nycteris bat CoV (JX899383); BatCoV/KW2E-F53/Nyc, Nycteris bat CoV (JX899384); BatCoV/KW2E-F82/Nyc, Nycteris bat CoV (JX899382); BatCoV HKU4, Tylonycteris bat CoV HKU4 (NC009019); BatCoV HKU5, Pipistrellus bat CoV HKU5 (NC_009020); HCoV-EMC, human betacoronavirus 2c EMC/2012 (JX869059); England1_CoV, human betacoronavirus England 1 (NC_019843); SARS_BatCoV HKU3, severe acute respiratory syndrome (SARS)–related Rhinolophus bat CoV HKU3 (DQ022305); SARSr-CoV-CFB, SARS-related Chinese ferret badger CoV (AY545919); SARSr-ciCoV, SARS-related palm civet CoV (AY304488); SARS_huSARS-CoV, SARS human CoV (NC_004718); IBV_Partridge, avian infectious bronchitis virus partridge (AY646283.1); IBV_Peafowl, avian infectious bronchitis virus isolate Peafowl (AY641576); IBV_Beaudette, infectious bronchitis virus strain Beaudette CK (AJ311317); WiCoV HKU20, wigeon CoV HKU20 (NC_016995); NHCoV HKU19, night heron CoV HKU19 (NC_016994); ThCoV HKU12, thrush CoV HKU12 (FJ376621); PorCoV HKU15, porcine CoV HKU15 (NC_016990); MRCoV HKU18, magpie robin CoV HKU18 (NC_016993).
All bat guano samples screened by PCR were negative for NiV and Histoplasma spp. but were positive for group C betacoronavirus. The natural reservoir and complete geographic distribution of this CoV are currently unknown. Although we did not isolate live virus from these samples, the detection of nucleic acid and previous isolation of viruses from bat feces and urine (10) warrants some concern that guano miners might be exposed to bat pathogens in fresh excreta as well as in soil substances. We suggest that guano miners use preventive measures of personal hygiene and improved barrier protection to reduce the possibility of exposure to zoonotic pathogens.
Acknowledgments
This research was supported by the Thailand Research Fund (DBG5180026, RDG5420089); the Higher Education Research Promotion and National Research University Project of Thailand, Office of the Higher Education Commission (HR1160A-55); the Thai Red Cross Society, Thailand; the Naval Health Research Center BAA-10-93 under Cooperative Agreement no. W911NF-11-2-004; the Centre National de la Recherche Scientifique; a nonbiodefense Emerging Infectious Diseases Research Opportunities Award from the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (1 R01 AI079231-01); and the United States Agency for International Development Emerging Pandemic Threats Program, PREDICT project, under the terms of Cooperative Agreement no. GHN-A-OO-09-00010-00.
Footnotes
Suggested citation for this article: Wacharapluesadee S, Sintunawa C, Kaewpom T, Khongnomnan K, Olival KJ, Epstein JH, et al. Group C betacoronavirus in bat guano fertilizer, Thailand [letter]. Emerg Infect Dis [Internet]. 2013 Aug [date cited]. http://dx.doi.org/10.3201/eid1908.130119
References
- 1.Olival KJ, Epstein JH, Wang LF, Field HE, Daszak P. Are bats unique viral reservoirs? In: Aguirre AA, Ostfeld RS, Daszak P, editors. New directions in conservation medicine: applied cases of ecological health. 2nd ed. Oxford: Oxford University Press; 2012. p. 195–212. [Google Scholar]
- 2.Cotten M, Lam TT, Watson SJ, Palser AL, Petrova V, Grant P, et al. Full-genome deep sequencing and phylogenetic analysis of novel human betacoronavirus. Emerg Infect Dis. 2013;19:736–42. 10.3201/eid1905.130057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Annan A, Baldwin HJ, Corman VM, Klose SM, Owusu M, Nkrumah EE, et al. Human betacoronavirus 2c EMC/2012–related viruses in bats, Ghana and Europe. Emerg Infect Dis. 2013;19:456–9 . 10.3201/eid1903.121503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woo PC, Lau SK, Li KS, Poon RW, Wong BH, Tsoi HW, et al. Molecular diversity of coronaviruses in bats. Virology. 2006;351:180–7. 10.1016/j.virol.2006.02.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lyon GM, Bravo AV, Espino A, Lindsley MD, Gutierrez RE, Rodriguez I, et al. Histoplasmosis associated with exploring a bat-inhabited cave in Costa Rica, 1998–1999. Am J Trop Med Hyg. 2004;70:438–42 . [PubMed] [Google Scholar]
- 6.Tong S, Conrardy C, Ruone S, Kuzmin IV, Guo X, Tao Y, et al. Detection of novel SARS-like and other coronaviruses in bats from Kenya. Emerg Infect Dis. 2009;15:482–5. 10.3201/eid1503.081013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wacharapluesadee S, Hemachudha T. Duplex nested RT-PCR for detection of Nipah virus RNA from urine specimens of bats. J Virol Methods. 2007;141:97–101. 10.1016/j.jviromet.2006.11.023 [DOI] [PubMed] [Google Scholar]
- 8.White TJ, Bruns T, Lee S, Taylor JW. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. New York: Academic Press, Inc.; 1990. p. 315–22. [Google Scholar]
- 9.Gouilh MA, Puechmaille SJ, Gonzalez JP, Teeling E, Kittayapong P, Manuguerra JC. SARS-coronavirus ancestor’s foot-prints in South-East Asian bat colonies and the refuge theory. Infect Genet Evol. 2011;11:1690–702. 10.1016/j.meegid.2011.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chua KB, Koh CL, Hooi PS, Wee KF, Khong JH, Chua BH, et al. Isolation of Nipah virus from Malaysian Island flying-foxes. Microbes Infect. 2002;4:145–51. 10.1016/S1286-4579(01)01522-2 [DOI] [PubMed] [Google Scholar]