Abstract
Multiplex polymerase chain reaction (PCR) has been widely used to detect Y-chromosome microdeletions, which is one of the major causes of male infertility. Both the European Academy of Andrology (EAA) and the European Molecular Genetics Quality Network (EMQN) have recommended the use of sY84 and sY86 markers for the detection of azoospermia factor a (AZFa) microdeletion during DNA testing for male infertility. In this study, a large-scale analysis of AZF microdeletion in a total of 630 Chinese males, including healthy semen donors (n=200), infertile males with normal sperm count (n=226) and patients with either nonobstructive azoospermia or severe oligozoospermia (n=204), was performed. A series of nine sequence-tagged site (STS) markers from the AZF region of the Y chromosome was used to detect microdeletions. All primers were designed based on the recommendations of the National Center for Biotechnology Information. An unusually high incidence (73/630, 11.6%) of sY84-absent but sY86-present genotypes was observed in the AZFa microdeletion screening. Sequencing the sY84-flanking region revealed a total of 73 patients with sY84-absent but sY86-present genotypes have a T-to-G transversion at the fifth base from the 5′ end of the reverse sY84 primer. These prevalent false positives, which were not only observed in infertile men, but also observed in donors, resulted from a single-nucleotide polymorphism (SNP) named rs72609647 in the targeting sequence of the reverse sY84 primer. Our study suggests that a pre-screening of existence of rs72609647 polymorphism can prevent the frequent false positive results of AZFa microdeletions detection in the infertile Chinese males. Given the SNP rs72609647 was recently found in a deep sequencing of a Chinese individual, the current EAA and EMQN standards may need to be scrutinized among different populations to avoid the potential genetic variations in the primer binding sequences.
Keywords: male infertility, multiplex polymerase chain reaction, rs72609647, single-nucleotide polymorphism, sY84, Y-chromosome microdeletion
Introduction
Microdeletions in the Y chromosome at the azoospermia factor (AZF) locus comprise a loss of three major subregions: AZFa, AZFb and AZFc.1 The AZFa subregion harbours two genes, ubiquitin-specific proteinase 9Y (USP9Y) and DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked (DDX3Y). Both are candidate genes for the susceptibility of male infertility.2 Studies have shown carriers of AZFa microdeletion are affected by azoospermia disorders, such as Sertoli cell-only syndrome.3, 4, 5 AZFb microdeletion is frequently associated with spermatogenic arrest, while men with AZFc microdeletions may suffer from severe oligozoospermia to nonobstructive azoospermia.6
Both the European Academy of Andrology and the European Molecular Genetics Quality Network have recommended the inclusion of sY84 and sY86 markers for the detection of AZFa microdeletion, sY127 and sY134 markers for AZFb microdeletion, and sY254 and sY255 markers for AZFc microdeletion in the multiplex polymerase chain reaction (PCR) DNA screening for male infertility.7 sY84 and sY86 are located upstream of the USP9Y and DDX3Y genes, respectively. Given the simplicity and high sensitivity of the sY84 and sY86 markers in the detection of AZFa microdeletion and the azoospermia phenotype, sY84 and sY86 have become the most widely used markers for the diagnosis of the AZFa microdeletion.8
The incidences of Y-chromosome microdeletions in azoospermia and severe oligozoospermia are 10%–15% and 5%–10%, respectively.9 Patients with azoospermia and severe oligozoospermia are more often carriers of AZFb and AZFc microdeletions than the AZFa microdeletion.10 However, Our laboratory found that sY84 microdeletion in the AZFa subregion was frequently detected among patients with severe oligozoospermia and nonobstructive azoospermia and even among males with normal sperm count and healthy donors. Moreover, during detection of the AZFa microdeletion, we observed the absence of the sY84 locus with the presence of sY86 in the same individual. Thus, the identification of AZFa microdeletion becomes ambiguous by the conflicts between sY84 and sY86 markers.
According to the current literature, deletion of the entire AZFa subregion is confirmed only when both sY84 and sY86 multiplex PCR products are absent in gel electrophoretic analysis. Additionally, almost all patients with this confirmed AZFa microdeletion are azoospermic.8 In order to address the question of whether the conflicts between sY84 and sY86 markers is due to a true AZFa microdeletion or just a PCR amplification failure, we performed a comprehensive analysis of the sY84 microdeletion in samples comprising nonobstructive azoospermic males, severe oligozoospermic males, infertile males with normal sperm count, and healthy semen donors.
Materials and methods
Collection of patient samples
A total of 430 patients were recruited from Ruijin Hospital Reproductive Medical Centre, Shanghai Jiai Genetics and IVF Institute (Shanghai, China) and Changhai Hospital Reproductive Medical Centre, (Shanghai, China). In addition, 200 healthy semen donors—the Donor group—with sperm counts ≥60×106 ml−1 were recruited from Shanghai Human Sperm Bank (Shanghai, China). All males are from east coast of China, including Shanghai City, Jiangsu Province and Zhejiang Province. All individuals involved in the study gave informed consent. Based on the results of routine semen tests, 430 patients were divided into two groups: Group 1 contained 226 infertile males with normal sperm count (sperm count ≥15×106 ml−1) and Group 2 contained 204 patients with either nonobstructive azoospermia or severe oligozoospermia (sperm count less than 5×106 ml−1). Sperm concentrations were evaluated at least twice according to World Health Organization guidelines.11 Genomic DNA was extracted from peripheral blood lymphocytes using DNA-isolation kits (TaKaRa Co., Ostu, Japan).
Multiplex PCR
A series of nine sequence-tagged site (STS) markers from the AZF region on the long arm of the Y chromosome were used to detect microdeletions. All primers were designed based on the recommendations of the National Center for Biotechnology Information. These STS markers included sY84 and sY86 for AZFa; sY127 and sY134 for AZFb; sY254, sY157, sY255, sY145 and sY152 for AZFc. Additionally, sY14 (an STS located within the SRY gene) was used as an internal positive control. Fertile male and female samples were used as positive and negative controls, respectively. A blank control was also included in the assay. Nine pairs of primers were pooled into four mixtures. Mix 1 included primers for sY84 (AZFa), sY127 (AZFb), sY152 (AZFc) and a pair of primers as an internal control. Mix 2 contained the same internal control and sY134 (AZFb). sY86 (AZFa), sY145 (AZFc) and an internal control were in Mix 3. Mix 4 included sY254 (AZFc), sY157 (AZFc), sY255 (AZFc) and an internal control. Cycling Multiplex PCR amplifications were carried out in a total volume of 20 µl buffered solution containing the primer mixture, 150 ng genomic DNA, 800 µmol l−1 deoxyribonucleotide triphosphates, 1.5 mmol l−1 Mg2+ and 2.0 U Taq polymerase. The cycling conditions were as follows: 94 °C for 10 min followed by 94 °C for 30 s, 58 °C for 30 s and 72 °C for 45 s for 30 cycles with a final extension at 72 °C for 10 min.
Sequencing of the sY84 locus
According to the genomic DNA sequence of sY84, a new pair of primers (forward: 5′-GCTGAGGAGTTGTGGAGACC-3′ reverse: 5′-GCAAGGACATTCCAGGGTTA-3′) upstream and downstream of the sY84 locus were used to amplify and sequence a 621-bp DNA fragment generated from the Y chromosome that contains the entire sY84 STS region. PCR amplifications were carried out as described above, and products of the amplification were sequenced by Invitrogen Limited (Shanghai, China).
Analysis of PCR products
All reaction products were separated on 2% agarose gels and visualized with ethidium bromide. A molecular weight marker in the range of 100bp–600 bp was included in at least one lane of each gel.
Statistical analysis
The difference in false-positive rates for the sY84 microdeletion among the three groups (Group 1, Group 2 and Donor group) was evaluated using the χ2 test. All analyses were performed using SAS version 9.1 (SAS Inc., Cary, NC, USA).
Results
The incidence rate of false positives in the sY84 screening contributed by single-nucleotide polymorphism rs72609647
DNA samples isolated from 630 males were subjected to four sets of multiplex PCR reactions. Figure 1 shows representative PCR products generated from four sets of primer mixtures in two different patients. PCR products for AZFb, AZFc were observed in both patients. As shown in the results for Mix 1 in Figure 1, a PCR product of sY84 for the AZFa microdeletion was observed in patient 1 but not in patient 2. However, PCR products of sY86 for the AZFa microdeletion were detected in both patients (see the results of Mix 3 in Figure 1). This suggests that patient 1 is not a carrier of AZF microdeletions and patient 2 has an AZFa microdeletion if he were only screened by the sY84 primer set for AZFa microdeletion. In other words, the AZFa screening results of patient 2 are confounded by the inconsistencies between sY84 and sY86.
Figure 1.
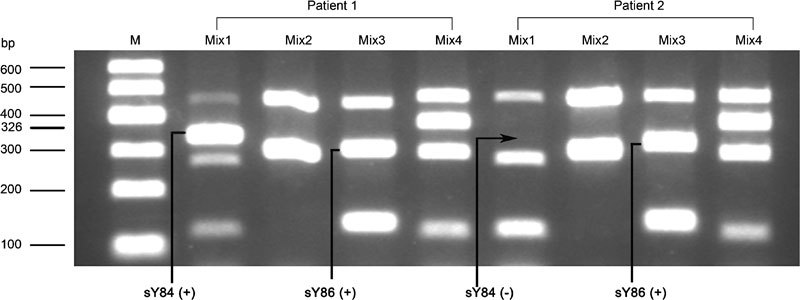
Electrophoresis analysis of the false-positive results from sY84 (326bp) microdeletion detection by multiplex PCR. PCR products generated from two patients using four groups of primer mixtures were analysed using gel electrophoresis. The arrow indicates the PCR products for sY84, which was not detected in patient 2 using Mix 1 primers. ‘M' represents DNA Marker I. Mix 1: sY14, sY84, sY127 and sY152; Mix 2: sY14 and sY134; Mix 3: sY14, sY86 and sY145; Mix 4: sY14, sY254, sY157 and sY255.
This sY84-absent but sY86-present phenomenon was further investigated in three groups of males separated by their semen sperm concentration. As shown in Table 1, the sY84 PCR product was absent in 11.5% of infertile males with sperm count ≥15×106 ml−1 and 9.8% of patients with nonobstructive azoospermia or severe oligozoospermia. Interestingly, a similar high incidence (13.5%) of sY84 absence was also observed in healthy semen donors. A total of 73 males (11.6%) had sY84-absent but sY86-present results in the multiplex PCR (Table 1). The rates of sY84-absent but sY86-present were not different among the three groups (P=0.509, χ2 test). Sequencing of the entire sY84 STS was performed in all 73 males. The results suggest the observation of sY84-absent but sY86-present in the Mix 1 multiplex PCR was a false-positive signal for sY84 microdeletion, which was caused by a T-to-G transversion at the fifth base from the 5′ end of the reverse sY84 primer (Figure 2). This T-to-G transversion was catalogued as rs72609647 in the single-nucleotide polymorphism (SNP) databank from the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/snp).
Table 1. Distribution of false-positive results of sY84 microdeletion in different groups of males classified by sperm count.
| Donor group | Group 1 | Group 2 | Total | P | |
|---|---|---|---|---|---|
| Number | 200 | 226 | 204 | 630 | |
| sY84 absent | 27 | 26 | 20 | 73 | 0.509 |
| Incidence (%) | 13.5 | 11.5 | 9.8 | 11.6 |
Donor group: healthy semen donors with sperm count ≥60×106 ml−1.
Group 1: infertile males with sperm count ≥15×106 ml−1.
Group 2: infertile males with azoospermia or severe oligozoospermia (sperm count less than 5×106 ml−1).
P<0.05 was regarded as the level of significance, X2 test.
Figure 2.
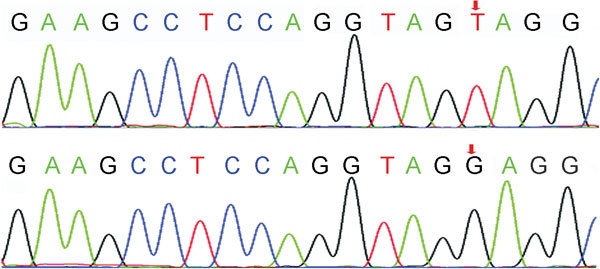
Sequencing analysis of the T-to-G transversion at the fifth base from the 5′ end of the reverse sY84 primer in normal control (upper panel) and a patient with false-positive sY84 microdeletion (bottom panel).
The result of AZF microdeletion analysis
Additionally, 15 patients with AZFc microdeletion and 2 patients with AZFb+c microdeletion were identified in the group of 204 patients with either nonobstructive azoospermia or severe oligozoospermia. Microdeletions of AZFb, AZFc were not found in infertile patients with normal sperm count or healthy donors. We did not find patient with AZFa microdeletion in this study.
Discussion
Y-chromosome microdeletions have been observed for decades, and many studies have reported the prevalence of AZF microdeletions in the Y chromosome.10, 12 As detected by multiplex PCR, AZF microdeletions have been widely identified among patients with nonobstructive azoospermia and severe oligozoospermia.10, 12 Diagnosis of complete deletion of AZFa subregion implies that most patients suffer from Sertoli cell-only syndrome, and retrieval of testicular sperm from these patients for intracytoplasmic sperm injection is virtually impossible.8, 13
Using sY84 and sY86 as markers for the diagnosis of AZFa microdeletions,8 Stahl et al. reported the rate of AZFa microdeletions in 1591 patients with azoospermia and severe oligozoospermia is 0.3%.10 In our study, none of the 204 Chinese patients with nonobstructive azoospermia or severe oligozoospermia had AZFa microdeletion, although we did observe the sY84-absent but sY86-present results in 20 out of 204 azoospermic or severe oligozoospermic males.
Furthermore, a high incidence of sY84 absent in AZFa was observed in infertile males with normal sperm count as well as in normal healthy donors. Similar findings of single sY84 microdeletions have been published by Tseng and Wang.14 who they performed a sY84 screening in 141 patients and showed that 6 in 51 males of normal control, 10 in 58 azoospermic patients and 4 in 32 oligozoospermic patients were had a deletion of the sY84 locus. As the incidence of the sY84-absent phenotype was not statistically different among groups, the authors claimed that the sY84 locus is not associated with spermatogenesis.14 In 2002, Thornhill et al.15 observed the sY84-absent but sY86-present result in a fertile Asian male. After further investigation of the mechanism of sY84 microdeletion, it turned out to be caused by an alteration of a nucleotide (T to G) within one of the sY84 primers.15
In the present study, we showed the sY84-absent but sY86-present results were found not only in 46 infertile males but also in 27 healthy semen donors. We further confirmed that this was caused by the T-to-G transversion in the target sequence of the reverse sY84 primer. Our observations agree with those reported by Thornhill et al.15 and suggest that false positives for the sY84 microdeletion are prevalent in the Chinese population, a finding that is also implied from the plausible findings of Tseng and Wang.14
The single-nucleotide alteration of T to G in the sY84 locus was further identified as an SNP named rs72609647 in National Center for Biotechnology Information website SNP databank. Given that a minor allele frequency ranging from 9.8% to 13.5% was found in both infertile and normal healthy Chinese men, this SNP is likely more prevalent in Chinese males than other ethnic groups. As the European Academy of Andrology guidelines state that the choice of primers for detecting AZF microdeletion is based on the principle of avoiding polymorphisms in the Caucasian population,7 the primers recommended by the guidelines may not apply to other ethnic populations, such as people from Shanghai, Jiangsu, Zhejiang and Taiwan Provinces, China.14 Thus, results generated from these primers of the sY84 locus in non-Caucasian populations should be interpreted with caution.
With the advent of next generation sequencing technology, the catalogue of human genetic variations has been greatly enriched by a number of sequencing projects including the James Watson's genome,16 the first Chinese project,17, 18 the first Korean genome19 and the 1000 genome project (available at: http://www.1000genomes.org). Because the primers for AZF in the multiplex PCR assay were designed almost two decades ago, it is worth checking whether there are other genetic variants affecting the efficiency of PCR amplification.
In summary, our study suggests the genotyping of this SNP may be necessary to comfirm if sY84 is absent in the AZF screening before clinical treatment, especially for the patients in China. We also confirm that two sets of markers (sY84 and sY86) are needed in the detection of AZFa microdeletion. When we use European guidelines as the reference for AZF screening, we might need to look up in the latest human SNP database and check whether there are SNPs in the binding regions of the primers.
Author contributions
QW designed the study, carried out the multiplex PCR and drafted the manuscript. GWC, TFY and HW collected the samples and extracted genomic DNA from peripheral blood lymphocytes. YLL and FS prepared the samples for sequencing and analysed the PCR results. SWD performed the statistical analysis and helped to draft the manuscript. ZL collected the samples and helped to draft the manuscript. YF and HJS designed the study and participated in its coordination. All authors read and approved the final manuscript.
Acknowledgments
This research was supported by the Major State Basic Research Development Program of China (973 Program, Nos. 2006GB504005 and 2009CB941700), the National Natural Science Foundation of China (No. 30872765) and the Basic Research Key Program of Shanghai (10JC1410800). Shi-Wei Duan is sponsored partly by the K. C. Wong Magna Fund of Ningbo University. We thank Dr Ching-Ling Chen for kind suggestions regarding English in drafting this paper.
The authors have nothing to disclose.
References
- Vogt PH, Edelman A, Kirsch S, Henegariu1 O, Hirschmann P, et al. Human Y chromosome azoospermia factors (AZF) mapped to different subregions in Yq11. Hum Mol Genet. 1999;5:933–45. doi: 10.1093/hmg/5.7.933. [DOI] [PubMed] [Google Scholar]
- Vogt PH. Azoospermia factor (AZF) in Yq11: towards a molecular understanding of its function for human male fertility and spermatogenesis. Reprod Biomed Online. 2005;10:81–93. doi: 10.1016/s1472-6483(10)60807-3. [DOI] [PubMed] [Google Scholar]
- Hopps CV, Mielnik A, Goldstein M, Palermo GD, Rosenwaks Z, et al. Detection of sperm in men with Y chromosome microdeletions of the AZFa, AZFb and AZFc regions. Hum Reprod. 2003;18:1660–65. doi: 10.1093/humrep/deg348. [DOI] [PubMed] [Google Scholar]
- Kamp C, Huellen K, Fernandes S, Sousa M, Schlegel PN, et al. High deletion frequency of the complete AZFa sequence in men with Sertoli-cell-only syndrome. Mol Hum Reprod. 2001;7:987–94. doi: 10.1093/molehr/7.10.987. [DOI] [PubMed] [Google Scholar]
- Nuti F, Krausz C. Gene polymorphisms/mutations relevant to abnormal spermatogenesis. Reprod Biomed Online. 2008;6:504–13. doi: 10.1016/s1472-6483(10)60457-9. [DOI] [PubMed] [Google Scholar]
- O' Flynn O' Brien KL, Varghese AC, Agarwal A. The genetic causes of male factor infertility: a review. Fertil Steril. 2010;93:1–12. doi: 10.1016/j.fertnstert.2009.10.045. [DOI] [PubMed] [Google Scholar]
- Simoni M, Bakker E, Eurlings MC, Matthijs G, Moro E, et al. Laboratory guidelines for molecular diagnosis of Y-chromosomal microdeletions. In J Androl. 1999;22:292–9. doi: 10.1046/j.1365-2605.1999.00193.x. [DOI] [PubMed] [Google Scholar]
- Simoni M, Bakker E, Krausz C. EAA/EMQN best practice guidelines for molecular diagnosis of Y-chromosomal microdeletions. State of the art 2004. In J Androl. 2004;27:240–9. doi: 10.1111/j.1365-2605.2004.00495.x. [DOI] [PubMed] [Google Scholar]
- Foresta C, Moro E, Ferlin A. Y chromosome microdeletions and alterations of spermatogenesis. Endocr Rev. 2001;22:226–39. doi: 10.1210/edrv.22.2.0425. [DOI] [PubMed] [Google Scholar]
- Stahl PJ, Masson P, Mielnik A, Marean MB, Schlegel PN, et al. A decade of experience emphasizes that testing for Y microdeletions is essential in American men with azoospermia and severe oligozoospermia. Fertil Steril. 2010;94:1753–6. doi: 10.1016/j.fertnstert.2009.09.006. [DOI] [PubMed] [Google Scholar]
- World Health Organization . Geneva; World Health Organization Press; 2010. WHO Laboratory Manual for the Examination of Human Semen, 5 ed. [Google Scholar]
- Ferlin A, Arredi B, Speltra E, Cazzadore C, Selice R, et al. Molecular and clinical characterization of Y chromosome microdeletions in infertile men: a 10-year experience in Italy. J Clin Endocrinol Metab. 2007;92:762–70. doi: 10.1210/jc.2006-1981. [DOI] [PubMed] [Google Scholar]
- Krausz C, Quintana-Murci L, Mc Elreavey K. Prognostic value of Y deletion analysis: what is the clinical prognostic value of Y chromosome microdeletion analysis. Hum Reprod. 2000;15:1431–4. doi: 10.1093/humrep/15.7.1431. [DOI] [PubMed] [Google Scholar]
- Tseng SL, Wang YC. The locus of sY84 is not associated with spermatogenesis—“Is it better to find Adam or study spermatogenesis?—a different opinion”. Fertil Steril. 1999;72:374–5. [PubMed] [Google Scholar]
- Thornhill AR, Guenther AJ, Barbarotto GM, Session DR, Damario MA, et al. False-positive Y-microdeletion result for a fertile male caused by an alteration under a PCR primer. In J Androl. 2002;25:352–7. doi: 10.1046/j.1365-2605.2002.00377.x. [DOI] [PubMed] [Google Scholar]
- Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–6. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- Wang J, Wang W, Li R, Li Y, Tian G, et al. The diploid genome sequence of an Asian individual. Nature. 2008;456:60–5. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Li Y, Fang X, Yang H, Wang J, et al. SNP detection for massively parallel whole-genome resequencing. Genome Res. 2009;19:1124–32. doi: 10.1101/gr.088013.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn SM, Kim TH, Lee S, Kim D, Ghang H, et al. The first Korean genome sequence and analysis: full genome sequencing for a socio-ethnic group. Genome Res. 2009;19:1622–9. doi: 10.1101/gr.092197.109. [DOI] [PMC free article] [PubMed] [Google Scholar]


