Abstract
A wide variety of techniques for the preparation of sperm are currently available, of which the most commonly employed are density–gradient centrifugation (DGC) and swim-up (SUP). To date, these methods appear to be effective in selecting functional sperm for assisted reproduction techniques (ART), but they may have negative effects on sperm DNA. In this study, the ability of these semen processing techniques to eliminate spermatozoa containing single- and double-strand DNA damage was assessed by the two-tailed comet assay and the sperm chromatin dispersion test in 157 semen samples from patients seeking assisted reproduction treatment. Our results indicated that SUP and DGC are equally efficient in eliminating spermatozoa containing double-strand DNA damage and sperm with highly damaged (degraded) DNA, as characterized by the presence of both single- and double-strand DNA breaks. However, DGC is more efficient than SUP in selecting spermatozoa that are free from single-strand DNA damage. Future studies should characterise the importance of the various types of DNA damage and examine the sperm processing protocols used in each laboratory to determine their ability to eliminate DNA damage and hence, prevent the potential transmission of genetic mutations via ART.
Keywords: density–gradient centrifugation, male factor, sperm chromatin dispersion, sperm DNA fragmentation, swim-up
Introduction
Sperm washing is routinely performed during in vitro fertilisation (IVF) therapy before assisted reproduction techniques (ART). Although a wide variety of sperm preparation methods are available, gentle sperm-processing protocols, such as density–gradient centrifugation (DGC) and swim-up (SUP), are the most commonly used.1 The SUP technique separates a sample into motile and non motile fractions, allowing the spermatozoa with the greatest motility to “swim up” into the culture medium while slow and immotile sperm remain behind, along with most debris in the semen pellet. The DGC method separates spermatozoa according to their density and favours the isolation of motile and morphologically normal spermatozoa. Although several studies have investigated the effectiveness of these two methods, the possibility that these techniques might cause damage to sperm DNA has not been fully elucidated.1, 2, 3Some reports have compared the effects of SUP and DGC on sperm chromatin; however, the results obtained are controversial.4, 5, 6, 7, 8, 9 To date, no consensus has been achieved regarding the effect of these techniques on the DNA integrity of sperm. Several investigations have shown that sperm preparation techniques considerably reduce the level of DNA damage present in a semen sample,4, 5, 6, 7 while other studies have reported that these methods do not recover spermatozoa with lower incidence of DNA damage.8 Some groups have reported that the DGC technique appreciably reduces the percentage of sperm with DNA damage, whereas the SUP method shows no significant improvement.9 Other researchers, in contrast, found that the percentage of sperm with fragmented DNA was reduced drastically in SUP-recovered fractions but not in the DGC-recovered sperm.5 These differences can be partially described by the variety of methods for the assessment of DNA damage and the fact that the methods applied in those studies are unable to differentiate between single-stranded DNA damage (ssDD) and double-stranded DNA damage (dsDD) in the same sperm cell. A method to analyse simultaneously, in the same spermatozoid, the presence of both dsDD and ssDD has been recently developed. This method is called the two-tailed comet (TTC) assay and combines the alkaline and neutral versions of the original comet assay to provide information about both the amount and the type of DNA damage present in a sperm cell.10 The type of DD present in the recovered fractions of sperm after SUP and DGC has not been examined. In the present study, we aim to examine and compare the effects of the DGC and SUP processing techniques on DNA integrity using the TTC assay and the sperm chromatin dispersion (SCD) test.11
Material and methods
Patient selection
One hundred and fifty-seven human semen samples from patients seeking ART treatments at the Hospital Quirón, Madrid, Spain, between January 2008 and May 2009, were used in this study. Informed consent for the use of spermatozoa was obtained.
Sample collection and preparation
Samples were obtained by masturbation after 48 h of sexual abstinence and allowed to liquefy at 37 °C and 5% CO2 for 10–15 min. Conventional semen quality parameters such as volume, concentration and motility were immediately assessed in fresh ejaculates (neat semen samples) following the procedures described elsewhere.12 An aliquot of 50 µl of each neat semen sample was used within 1 h of ejaculation for sperm DNA fragmentation (SDF) index assessment.
DGC
SpermGrad (Vitrolife, Göteborg, Sweden) gradients of 90% (500 µl, lower layer) and 50% (500 µl, upper layer), which were diluted in IVF plus medium (Vitrolife), were used for this study. One millilitre of the liquefied neat semen sample was placed on top of the upper layer and the tube was centrifuged for 20 min at 300g. The upper and lower layers were carefully aspirated without disturbing the pellet. Finally, the pellet was resuspended in 1 ml of IVF plus. Sperm concentration and motility were estimated in the recovered fractions according to World Health Organization guidelines.12 An aliquot of 50 µl of each DGC-recovered semen sample was used for the SDF assessment. To avoid the undesirable effect of iatrogenic DNA damage to the sperm after processing, which produces a variable and dynamic increase in SDF values,13, 14 all sperm samples were assessed for SDF immediately after sperm selection.
SUP
Samples were diluted with 1× volume of IVF plus medium (Vitrolife) and centrifuged at 410g for 10 min. The supernatants were carefully aspirated and discarded and 400 µl of fresh IVF plus medium was added to the sperm pellet. The samples were maintained in an incubator at 37 °C under an atmosphere of 5% CO2 in air for 1 h to allow motile sperm to ‘swim up' from the pellet. After the incubation period, 0.5 ml of the supernatant was aspirated and the concentration and motility of the sperm were estimated in the recovered fractions according to the guidelines of the World Health Organization.12 An aliquot of 50 µl of each SUP recovered semen sample was used for SDF assessment. To avoid the undesirable effects of iatrogenic DNA damage to the sperm after processing, which produces a variable and dynamic increase in SDF values,13, 14 all sperm samples were assessed for SDF immediately after sperm selection.
Sperm DNA fragmentation analysis
SCD
To determine the SDF of the samples studied, the Halosperm kit (Halotech DNA, Madrid, Spain) was used. Briefly, 25 µl of sperm cells diluted to a concentration of 10×106 spermatozoa ml−1 was added to a vial with low melting point agarose (Sigma, St Louis, MO, USA) and mixed. Pre-treated slides (Halotech DNA) were placed onto a metallic plate previously cooled to 4 °C and a drop of the cell suspension was deposited onto the treated face of the slide, covered with a glass coverslip and allowed to solidify for 5 min at 4 °C. The coverslip was smoothly removed and the slide was horizontally placed in 10 ml of the lysis solution provided in the kit (Halosperm kit). Finally, the slides were washed in distilled water, dehydrated in sequential baths of 70%, 90% and 100% ethanol (Merck, Darmstadt, Germany) and stained with propidium iodide (2 µg ml−1; Sigma) in Vectashield (Vector Laboratories, Burlingame, CA, USA). At that point, the samples could either be immediately analysed or stored at room temperature in the dark until needed. The SDF was established as the percentage of fragmented sperm cells in a semen sample. The SDF-d was defined as the percentage of degraded sperm cells in a semen sample15 (Figure 1a). Both the SDF and the SDF-d were calculated by assessing at least 500 sperm cells per slide.
Figure 1.
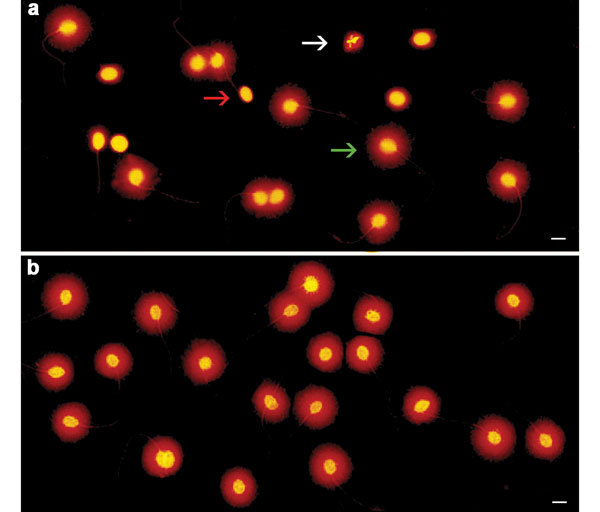
SDF as determined by the SCD test. (a) Neat semen sample showing six sperm cells with fragmented DNA in the microscope field, as evidenced by the absence of a halo (red arrow), one sperm cell with degraded DNA, without a halo and weak or irregular staining (white arrow) and 11 sperm cells with intact DNA and a halo of dispersed DNA loops (green arrow). (b) SUP processed sample showing 20 sperm cells with intact DNA in the microscopic field, as evidenced by the presence of haloes. Scale bars=5 µm. SDF, sperm DNA fragmentation; SCD, sperm chromatin dispersion; SUP, swim-up.
TTC
In this assay, 25 µl of sperm cells diluted to a concentration of 10×106 spermatozoa ml−1 was mixed at 37 °C with 50 µl of freshly prepared 1% low melting point agarose (Sigma). Next, 15 µl of the mixture was deposited on a slide that was pre-treated for gel adhesion (Halotech DNA), covered with a coverslip and transferred to an ice-cold plate to promote fast gelling. As soon as the gel solidified, coverslips were smoothly removed and the slides were submerged sequentially in two lysis solutions: lysis solution 1 (0.4 mol l−1 Tris-HCl (Sigma), 0.8 mol l−1 dithiothreitol (Sigma), 1% SDS (Invitrogen), pH 7.5) for 30 min, followed by lysis solution 2 (0.4 mol l−1 Tris-HCl (Sigma), 2 mol l−1 NaCl (Sigma), 1% SDS (Invitrogen), 0.05 mol l−1 EDTA (Merck), pH 7.5) for 30 min. Then, slides were rinsed in TBE buffer (0.09 mol l−1 Tris-Borate (Merck), 0.002 mol l−1 EDTA (Merck), pH 7.5) for 10 min and transferred to an electrophoresis tank. Electrophoresis was then performed at 20 V (1 V cm−1) and 12 mA for 12 min in fresh TBE buffer. Subsequently, the slides were unwound in an alkaline solution (0.03 mol l−1 NaOH (Sigma) and 1 mol l−1 NaCl (Sigma)) for 2.5 min and transferred to an electrophoresis tank in a position perpendicular to the first neutral run. Electrophoresis was performed at 20 V (1 V cm−1) and 12 mA for 4 min in fresh alkaline buffer (0.03 mol l−1 NaOH (Sigma)). Finally, the slides were rinsed once in a neutralisation buffer (0.4 mol l−1 Tris-HCl (Sigma), pH 7.5) for 5 min, briefly washed in TBE buffer, dehydrated in increasing concentrations of ethanol (Merck) and air dried.
DNA was stained with SYBR Green I (Molecular Probes, Leiden, The Netherlands) at a 1∶3000 dilution in Vectashield medium (Vector Laboratories). At that point, the samples could be immediately analysed or stored at room temperature in the dark until needed. Sperm cell comets were assessed by visual scoring. The frequency of sperm cells with SDF was established by measuring at least 500 sperm cells per slide. Cells were classified as undamaged, ssDD or dsDD cells according to the comet assay's principle that the size of the tail and/or the density of DNA in the tail positively correlates with the extent of DNA damage. The comet tail oriented along the X axis is representative of migrated double-stranded DNA fragments, and the comet tail oriented along the Y axis is composed of single-stranded DNA fragments.10
Microscopic analysis and image capture
For SCD and TTC, a Leica DMLA motorized epifluorescence microscope that was controlled with software for automatic scanning and image digitisation (Leica Microsystems, Barcelona, Spain) was used. The microscope was equipped with a Leica EL6000 fluorescence metal halide lamp light and Plan-Fluotar ×40 objectives for routine analysis and Plan-Fluotar ×60 objectives for image capturing. Images were captured via a cooled Photometrics CCD (Roper Industries, Q3 170 Inc., Duluth, GA, USA) for direct colour 16-bit captures and a CCD DFC-350-FX Leica for 8-bit grey-level imaging (Leica Microsystems). A CY34040B fluorescence filter block was used for propidium iodide visualisation, while for SYBR Green I imaging, a RCT3540B single band set was used, both of which were manufactured by Semrock (Rochester, NY, USA).
Statistical analysis
Data analysis was performed using the SPSS software (SPSS v.17; SPSS Inc., Chicago, IL, USA). The Mann–Whitney U test (P<0.05) was used to detect significant differences in the conventional semen quality parameters and SDF index between neat and DGC/SUP-processed semen samples.
Results
Semen samples processed through the SCD test protocol gave rise to five types of nucleoids: (i) sperm nucleoids with large haloes of dispersed DNA loops; (ii) sperm nucleoids with medium haloes of DNA loops; (iii) sperm nucleoids with small haloes of dispersed DNA loops; (iv) sperm nucleoids without haloes; and (v) sperm nucleoids without haloes and faintly stained (degraded) (Figure 1). According to Fernández et al.,11, 16 sperm containing intact DNA or low levels of DNA damage release their DNA loops, forming large haloes, whereas those sperm nuclei that contain fragmented DNA produce small or nonexistent haloes of dispersed DNA; spermatozoa without haloes and exhibiting faint staining contain a high level of DNA damage.
The TTC assay induces DNA migration in two perpendicular axes. The comet tail positioned on the X axis, which forms as result of a neutral lysis and electrophoresis, contains double-stranded DNA. The comet tail positioned on the Y axis is formed as a result of alkaline lysis and electrophoresis and consists of single-stranded DNA. According to these parameters and to the assumed principle that the size of the tail and/or the density of the DNA in the tail positively correlates with the extent of the DNA damage,17 the TTC types observed in a semen sample can be classified by the criteria described by Enciso et al.10 (Figure 2).
Figure 2.
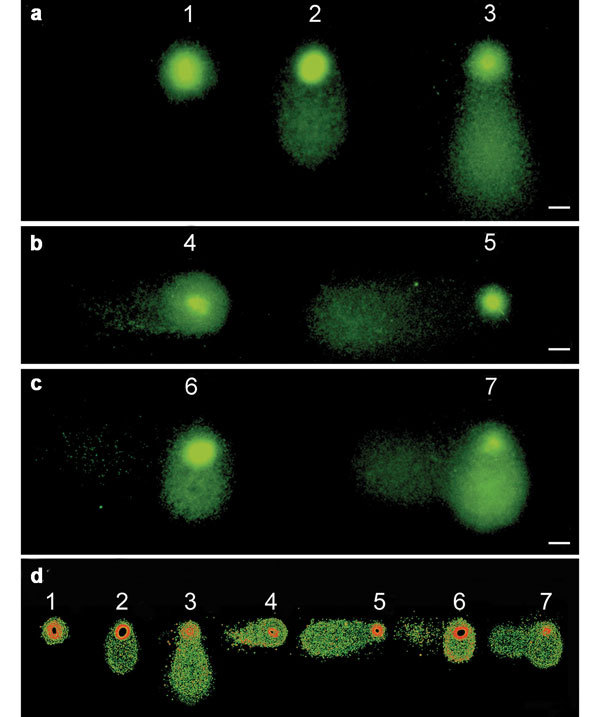
The TTC assay produces DNA migration in two perpendicular axes. The comet tail positioned on the X axis is formed as result of neutral lysis and electrophoresis and contains dsDNA, while the comet tail positioned on the Y axis is formed as a result of alkaline lysis and electrophoresis and consists of ssDNA. Seven TTC types are found in a semen sample. (a) (1) undamaged; (2) low level of ssDD; (3) high level of ssDD; (b) (4) low level of dsDD; (5) high level of dsDD; (c) (6) low level of ssDD and dsDD; (7) high level of ss- and dsDD. (d) The same images after the application of a common electronic filter. Scale bars=5 µm. dsDD, double-stranded DNA damage; dsDNA, double-stranded DNA; ssDD, single-stranded DNA damage; ssDNA, single-stranded DNA; TTC, two-tailed comet.
The results of the DNA damage analyses performed on the semen samples with the SCD test and the TTC assay are shown in Table 1. The mean sperm progressive motility of the samples improved significantly (Mann–Whitney U test, P<0.05) after processing with the DGC and SUP protocols. No significant differences in progressive motility were observed between DGC and SUP selected sperm. The percentage of spermatozoa containing damaged DNA, as assessed by the SCD test (SCD-SDF), was significantly (Mann–Whitney U test, P<0.05) reduced in both the DGC and the SUP fractions compared with that of neat semen (Table 1). Also, the percentage of highly damaged and degraded spermatozoa (SCD-SDF-d) was reduced in both the DGC and the SUP fractions compared with that of neat semen (Mann–Whitney U test, P<0.05) (Table 1). The percentage of spermatozoa with dsDD assessed by the TTC assay was also significantly (Mann–Whitney U test, P<0.05) reduced in fractions recovered by both DGC and SUP (Table 1). In contrast, the percentage of spermatozoa with ssDD was significantly (Mann–Whitney U test, P<0.05) reduced in DGC selected spermatozoa, but not in SUP recovered spermatozoa compared with that of neat semen. Sperm cells recovered by the SUP method did not present a frequency of ssDD that was significantly different from that of neat semen (Table 1).
Table 1. Semen quality parameters of the samples analysed before and after the DGC and SUP techniques were employed.
| Semen parameters | Neat semen | DGC | SUP |
|---|---|---|---|
| Total no. of spermatozoa (×106) | 168.22±9.17 | 8.59±0.84* | 14.18±0.71* |
| Progressive motility (%) | 52.84±1.23 | 71.38±9.70* | 82.74±0.97* |
| SCD-SDF (%) | 30.73±1.31 | 19.80±3.43* | 16.08±1.60* |
| SCD-SDF-d (%) | 8.47±0.47 | 4.75±0.83* | 2.94±0.41* |
| ssDD-TTC (%) | 46.75±3.00 | 25.54±5.84* | 39.44±5.55 |
| dsDD-TTC (%) | 16.66±1.48 | 4.20±0.80* | 5.98±0.97* |
Abbreviations: d, degraded spermatozoa; DGC, density gradient centrifugation; dsDD, double-strand DNA damage; SCD, sperm chromatin dispersion; SDF, sperm DNA fragmentation; ssDD, single-strand DNA damage; SUP, swim-up.
Values are expressed as mean±s.e.m.
*P<0.05, compared with neat semen, Mann–Whitney U test.
Discussion
Despite various advances in the methodology to assess sperm DNA damage and the publication of several studies examining the effectiveness of various semen preparation methods, the recovery rate of fully functional, DNA undamaged spermatozoa after DGC and SUP processing remains unclear.18, 19, 20, 21 In our study, sperm selected after DGC and SUP exhibited sperm subpopulations from which highly motile spermatozoa were collected from the overlying buffer. Similar results have been previously reported by Ricci et al.22 and Boomsma et al.1 However, this improvement in sperm motility was not observed to be consistently associated with a similar improvement in sperm DNA integrity.23, 24 The percentage of spermatozoa with dsDD recovered after DGC and SUP processing was significantly (P<0.05) lower than that of the neat semen sample. Both techniques are equally efficient in recovering spermatozoa with reduced levels of dsDD, but DCG is more efficient than SUP in selecting spermatozoa that are free from ssDD. Previous studies obtained similar results to those reported in this study, indicating that the DGC and SUP techniques recover spermatozoa with reduced levels of SDF.5, 9, 25 Regarding the DGC method, some studies have reported that sperm preparation techniques involving centrifugation are associated with iatrogenic reactive oxygen species generation and with an increase in sperm DNA damage.5, 26 In the case of the SUP approach, most studies agree that this technique allows the efficient elimination of spermatozoa with high levels of DNA damage.4, 5, 6 Nevertheless, none of these studies makes any distinction between the types of DNA damage present in the spermatozoa, either before or after sample processing. Therefore, the results presented in this study are novel. The identification of the type of damage present in a processed sample may be important for the assessment of its fertility potential. Although the significance of the different types of DNA damage in sperm has not been fully elucidated, double-strand DNA breaks (DSBs) are considered more deleterious than single-strand DNA breaks (SSBs). DSBs have been described as a cause of chromosomal abnormalities and they are not as easily repaired as SSBs.27, 28
Although the implications of using DNA damaged spermatozoa for ART are not clear, the results of several prior studies are cause for concern. Sperm DNA damage has been shown to negatively impact the ICSI embryo development, pregnancy rates and offspring health.29, 30, 31 Given that certain DNA damage in the sperm may be repaired by the oocyte, the most moderate hypothesis would state that a subclinical level of both single- and double-strand DNA damage could be repaired by the oocyte in the male pronucleus stage.32, 33, 34 However, in cases in which DNA lesions remain unrepaired in the male pronuclei, certain genetic abnormalities could potentially be transmitted to the offspring or directly prevent the embryo from undergoing normal cleavage.35, 36, 37
DSBs are considered critical lesions in the formation of chromosomal aberrations.38, 39 Unlike the SSB repair processes, which rely on the presence of an undamaged strand opposite the lesion, most DSB repair processes possess a mutagenic potential. Because no template is available for repair synthesis, all of the repair mechanisms (single-strand annealing or non-homologous end-joining), apart from the conservative homologous recombination repair, which may be employed to restore the original sequence at the DSB site, will necessarily produce deletions and point mutations at the repair site.38, 40 Hence, the presence of a high level of dsDD in sperm is likely to overwhelm the oocyte's repair capacity and possibly allow the transmission of genetic abnormalities to the offspring. The presence of extensive ssDD may require as well additional repair activity from the oocyte and, in certain cases, compromise the development of the embryo.
Genetic integrity is particularly important in selecting sperm for intracytoplasmic sperm injection. Although functionally competent spermatozoa are not a prerequisite for intracytoplasmic sperm injection, the use of DNA-damaged spermatozoa for this technique could have adverse consequences that may appear during the post-implantation development of the offspring, rather than before.41 Clearly, information concerning the amount and type of DNA damage removed after sperm selection and the amount and type of remaining DNA damage could be critical in understanding the outcome of ART procedures and preventing undesirable consequences. Additionally, the methodology proposed here could also be used in cases in which sperm selection is reinforced using magnetic-activated cell sorting. These fractions could easily be assessed, especially to detect ssDD and dsDD, to analyse the effectiveness of the antibodies in removing highly damaged DNA sperm using Annexin V as a target.42
The data obtained in the present study highlight the limitations of the use of sperm motility as an indicator of general sperm quality and underscore the need to examine the sperm processing protocols used in each laboratory to determine their ability to eliminate DNA damage and prevent the potential transmission of genetic mutations via ART. It is also of great importance to characterize the various types of damage present in spermatozoa in terms of their origin, magnitude and impact on fertility.
In summary, the results of the present study suggest that both the DGC and the SUP methods are efficient in recovering highly motile spermatozoa and in eliminating highly damaged spermatozoa and sperm containing dsDD. However, the SUP approach is not as effective as the DGC technique in decreasing the quantity of ssDD remaining after selection. The assessment of DNA integrity in combination with semen analysis provides a better characterisation of the semen sample and may be used as a tool for improving ART outcomes.
Author contributions
ME participated in the design of the study, carried out the DNA fragmentation analyses, collected the data and drafted the manuscript. MI and IG carried out the conventional semen parameters analysis and collected the data. JS performed the statistical analysis. AG participated in the design of the study and supervised data collection. JG participated in the design of the study, supervised the data analysis and helped to draft the manuscript.
Acknowledgments
The authors would like to thank the donors participating in this study for providing the samples. Financial support for this study was provided by the Ministerio de Ciencia y Tecnología, Spain (BFU2007-66340/BFI; CGL 2005-02898/BOS).
The authors declare no competing financial interests.
References
- Boomsma CM, Heineman MJ, Cohlen BJ, Farquhar C. Semen preparation techniques for intrauterine insemination. Cochrane Database Syst Rev. 2007;4:CD004507. doi: 10.1002/14651858.CD004507.pub3. [DOI] [PubMed] [Google Scholar]
- Centola GM, Herko R, Andolina E, Weisensel S. Comparison of sperm preparation methods: effect on recovery, motility, motion parameters and hyperactivation. Fertil Steril. 1998;70:1173–5. doi: 10.1016/s0015-0282(98)00352-5. [DOI] [PubMed] [Google Scholar]
- Mortimer D. Sperm preparation methods. J Androl. 2000;21:357–66. [PubMed] [Google Scholar]
- Spano M, Cordelli E, Leter G, Lombardo F, Lenzi A, et al. Nuclear chromatin variations in human spermatozoa undergoing swim-up and cryopreservation evaluated by the flow cytometric sperm chromatin structure assay. Mol Hum Reprod. 1999;5:29–37. doi: 10.1093/molehr/5.1.29. [DOI] [PubMed] [Google Scholar]
- Zini A, Finelli A, Phang D, Jarvi K. Influence of semen processing technique on human sperm DNA integrity. Urology. 2000;56:1081–4. doi: 10.1016/s0090-4295(00)00770-6. [DOI] [PubMed] [Google Scholar]
- Younglai EV, Holt D, Brown P, Jurisicova A, Casper RF. Sperm swim-up techniques and DNA fragmentation. Hum Reprod. 2001;16:1950–3. doi: 10.1093/humrep/16.9.1950. [DOI] [PubMed] [Google Scholar]
- Meseguer M, Santiso R, Garrido N, García-Herrero S, Remohí J, et al. Effect of sperm DNA fragmentation on pregnancy outcome depends on oocyte quality. Fertil Steril. 2011;95:124–8. doi: 10.1016/j.fertnstert.2010.05.055. [DOI] [PubMed] [Google Scholar]
- Stevanato J, Bertolla RP, Barradas V, Spaine DM, Cedenho AP, et al. Semen processing by density gradient centrifugation does not improve sperm apoptotic deoxyribonucleic acid fragmentation rates. Fertil Steril. 2008;90:889–90. doi: 10.1016/j.fertnstert.2007.01.059. [DOI] [PubMed] [Google Scholar]
- Sakkas D, Manicardi GC, Tomlinson M, Mandrioli M, Bizzaro D, et al. The use of two density gradient centrifugation techniques and the swim-up method to separate spermatozoa with chromatin and nuclear DNA anomalies. Hum Reprod. 2000;15:1112–26. doi: 10.1093/humrep/15.5.1112. [DOI] [PubMed] [Google Scholar]
- Enciso M, Sarasa J, Agarwal A, Fernández JL, Gosálvez J. A two-tailed Comet assay for assessing DNA damage in spermatozoa. Reprod Biomed Online. 2009;18:609–16. doi: 10.1016/s1472-6483(10)60003-x. [DOI] [PubMed] [Google Scholar]
- Fernández JL, Muriel L, Goyanes V, Segrelles E, Gosálvez J, et al. Simple determination of human sperm DNA fragmentation with an improved sperm chromatin dispersion test. Fertil Steril. 2005;84:833–42. doi: 10.1016/j.fertnstert.2004.11.089. [DOI] [PubMed] [Google Scholar]
- WHO . Cambridge; Cambridge University Press; 2010. WHO Laboratory Manual for the Examination of the Human Semen and Semen Cervical Mucus Interaction. [Google Scholar]
- Gosálvez J, Cortés-Gutierrez E, Nuñez R, Fernandez JL, Caballero P, et al. A dynamic assessment of sperm DNA fragmentation versus sperm viability in proven fertile human donors. Fertil Steril. 2009;92:1915–9. doi: 10.1016/j.fertnstert.2008.08.136. [DOI] [PubMed] [Google Scholar]
- Gosálvez J, de la Torre J, López-Fernández C, Pérez-Gutiérrez L, Ortega L, et al. DNA fragmentation dynamics in fresh versus frozen thawed 1 plus gradient isolated human spermatozoa. Syst Biol Reprod Med. 2010;56:27–36. doi: 10.3109/19396360903515430. [DOI] [PubMed] [Google Scholar]
- Enciso M, Muriel M, Fernández JL, Goyanes V, Segrelles E, et al. Infertile men with varicocele show a high relative proportion of sperm cells with intense nuclear damage level, evidenced by the SCD test. J Androl. 2006;27:106–11. doi: 10.2164/jandrol.05115. [DOI] [PubMed] [Google Scholar]
- Fernández JL, Muriel L, Rivero MT, Goyanes V, Vázquez R, et al. The sperm chromatin dispersion test: a simple method for the determination of sperm DNA fragmentation. J Androl. 2003;24:59–66. [PubMed] [Google Scholar]
- Schmid TE, Kamischke A, Bollwein H, Nieschlag E, Brinkworth MH. Genetic damage in oligozoospermic patients detected by fish, inverse restriction site mutation assay, sperm chromatin structure assay and the comet assay. Hum Reprod. 2003;18:1474–80. doi: 10.1093/humrep/deg259. [DOI] [PubMed] [Google Scholar]
- Sakkas D, Manicardi GC, Tomlinson M, Mandrioli M, Bizzaro D, et al. The use of two density gradient centrifugation techniques and the swim-up method to separate spermatozoa with chromatin and nuclear DNA anomalies. Hum Reprod. 2000;15:1112–6. doi: 10.1093/humrep/15.5.1112. [DOI] [PubMed] [Google Scholar]
- Hammadeh ME, Kühnen A, Amer AS, Rosenbaum P, Schmidt W. Comparison of sperm preparation methods: effect on chromatin and morphology recovery rates and their consequences on the clinical outcome after in vitro fertilization embryo transfer. Int J Androl. 2001;24:360–8. doi: 10.1046/j.1365-2605.2001.0317a.x. [DOI] [PubMed] [Google Scholar]
- Allamaneni SS, Agarwal A, Rama S, Ranganathan P, Sharma RK. Comparative study on density gradients and swim-up preparation techniques utilizing neat and cryopreserved spermatozoa. Asian J Androl. 2005;7:86–92. doi: 10.1111/j.1745-7262.2005.00008.x. [DOI] [PubMed] [Google Scholar]
- Jackson RE, Bormann CL, Hassun PA, Rocha AM, Motta EL, et al. Effects of semen storage and separation techniques on sperm DNA fragmentation. Fertil Steril. 2010;94:2626–30. doi: 10.1016/j.fertnstert.2010.04.049. [DOI] [PubMed] [Google Scholar]
- Ricci G, Perticarari S, Boscolo R, Montico M, Guaschino S, et al. Semen preparation methods and sperm apoptosis: swim-up versus gradient-density centrifugation technique. Fertil Steril. 2009;91:632–8. doi: 10.1016/j.fertnstert.2007.11.068. [DOI] [PubMed] [Google Scholar]
- Zini A, Mak V, Phang D, Jarvi K. Potential adverse effect of semen processing on human sperm deoxyribonucleic acid integrity. Fertil Steril. 1999;72:496–9. doi: 10.1016/s0015-0282(99)00295-2. [DOI] [PubMed] [Google Scholar]
- Stevanato J, Bertolla RP, Barradas V, Spaine DM, Cedenho AP, et al. Semen processing by density gradient centrifugation does not improve sperm apoptotic deoxyribonucleic acid fragmentation rates. Fertil Steril. 2008;90:889–90. doi: 10.1016/j.fertnstert.2007.01.059. [DOI] [PubMed] [Google Scholar]
- Ricci G, Boscolo R, Martinelli M, Perticarari S. Methods for defecting sperm apoptosis. Fertil Steril. 2009;92:19–20. doi: 10.1016/j.fertnstert.2008.12.109. [DOI] [PubMed] [Google Scholar]
- Twigg J, Irvine DS, Houston P, Fulton N, Michael L, et al. Iatrogenic DNA damage induced in human spermatozoa during sperm preparation: protective significance of seminal plasma. Mol Hum Reprod. 1998;4:439–45. doi: 10.1093/molehr/4.5.439. [DOI] [PubMed] [Google Scholar]
- Collins AR, Dobson VL, Duinská M, Kennedy G, Ttina R. The comet assay: what can it really tell us. Mutat Res. 1997;375:183–93. doi: 10.1016/s0027-5107(97)00013-4. [DOI] [PubMed] [Google Scholar]
- Speit G, Dennog C, Lampl L. Biological significance of DNA damage induced by hyperbaric oxygen. Mutagenesis. 1998;13:85–7. doi: 10.1093/mutage/13.1.85. [DOI] [PubMed] [Google Scholar]
- Ahmadi A, Ng SC. Fertilizing ability of DNA-damaged spermatozoa. J Exp Zool. 1999;284:696–704. doi: 10.1002/(sici)1097-010x(19991101)284:6<696::aid-jez11>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Fernández-González R, Moreira PN, Pérez-Crespo M, Sánchez-Martín M, Ramírez MA, et al. Long-term effects of mouse intracytoplasmic sperm injection with DNA-fragmented sperm on health and behavior of adult offspring. Biol Reprod. 2008;78:761–72. doi: 10.1095/biolreprod.107.065623. [DOI] [PubMed] [Google Scholar]
- Zini A, Boman J, Belzile E, Ciampi A. Sperm DNA damage is associated with an increased risk of pregnancy loss after IVF and ICSI: systematic review and meta-analysis. Hum Reprod. 2008;23:2663–8. doi: 10.1093/humrep/den321. [DOI] [PubMed] [Google Scholar]
- Brandriff B, Pedersen RA. Repair of the ultraviolet-irradiated male genome in fertilized mouse eggs. Science. 1981;211:1431–3. doi: 10.1126/science.7466400. [DOI] [PubMed] [Google Scholar]
- Ashwood-Smith MJ, Edwards RG. DNA repair by oocytes. Mol Hum Reprod. 1996;2:46–51. doi: 10.1093/molehr/2.1.46. [DOI] [PubMed] [Google Scholar]
- Lehman CW, Jeong-Yu S, Trautman JK, Carroll D. Repair of heteroduplex DNA in Xenopus laevis oocytes. Genetics. 1994;138:459–70. doi: 10.1093/genetics/138.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavitrano M, Camaioni A, Fazio VM, Dolci S, Farace MG, et al. Sperm cells as vectors for introducing foreign DNA into eggs: genetic transformation of mice. Cell. 1989;57:717–23. doi: 10.1016/0092-8674(89)90787-3. [DOI] [PubMed] [Google Scholar]
- Perry AC, Wakayama T, Kishikawa H, Kasai T, Okabe M, et al. Mammalian transgenesis by intracytoplasmic sperm injection. Science. 1999;284:1180–3. doi: 10.1126/science.284.5417.1180. [DOI] [PubMed] [Google Scholar]
- Bonduelle M, Aytoz A, van Assche E, Devroey P, Liebaers I, et al. Incidence of chromosomal aberrations in children born after assisted reproduction through Intracytoplasmatic sperm injection. Hum Reprod. 1998;13:781–2. doi: 10.1093/humrep/13.4.781. [DOI] [PubMed] [Google Scholar]
- Pfeiffer P, Goedecke W, Obe G. Mechanisms of DNA double-strand break repair and their potential to induce chromosomal aberrations. Mutagenesis. 2000;15:289–302. doi: 10.1093/mutage/15.4.289. [DOI] [PubMed] [Google Scholar]
- Marchetti F, Essers J, Kanaar R, Wyrobek AJ. Disruption of maternal DNA repair increases sperm-derived chromosomal aberrations. Proc Natl Acad Sci. 2007;104:17725–9. doi: 10.1073/pnas.0705257104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer P. The mutagenic potential of DNA double-strand break repair. Toxicol Lett. 1998;96–97:119–29. doi: 10.1016/s0378-4274(98)00058-7. [DOI] [PubMed] [Google Scholar]
- Bonduelle M, Liebaers I, Deketelaere V, Derde MP, Camus M, et al. Neonatal data on a cohort of 2889 infants born after ICSI (1991–1999) and of 2995 infants born after IVF (1983–1999) Hum Reprod. 2002;17:671–94. doi: 10.1093/humrep/17.3.671. [DOI] [PubMed] [Google Scholar]
- Said TM, Grunewald S, Paasch U, Rasch M. Agarwal et al Effects of magnetic activated cell sorting on sperm motility and cryosurvival rates. Fertil Steril. 2005;83:1442–6. doi: 10.1016/j.fertnstert.2004.11.052. [DOI] [PubMed] [Google Scholar]


