Abstract
Prostate cancer (PCa) is one of the most common cancers among men in Western developed countries and its incidence has increased considerably in many other parts of the world, including China. The etiology of PCa is largely unknown but is thought to be multifactorial, where inherited genetics plays an important role. In this article, we first briefly review results from studies of familial aggregation and genetic susceptibility to PCa. We then recap key findings of rare and high-penetrance PCa susceptibility genes from linkage studies in PCa families. We devote a significant portion of this article to summarizing discoveries of common and low-penetrance PCa risk-associated single-nucleotide polymorphisms (SNPs) from genetic association studies in PCa cases and controls, especially those from genome-wide association studies (GWASs). A strong focus of this article is to review the literature on the potential clinical utility of these implicated genetic markers. Most of these published studies described PCa risk estimation using a genetic score derived from multiple risk-associated SNPs and its utility in determining the need for prostate biopsy. Finally, we comment on the newly proposed concept of genetic score; the notion is to treat it as a marker for genetic predisposition, similar to family history, rather than a diagnostic marker to discriminate PCa patients from non-cancer patients. Available evidence to date suggests that genetic score is an objective and better measurement of inherited risk of PCa than family history. Another unique feature of this article is the inclusion of genetic association studies of PCa in Chinese and Japanese populations.
Keywords: biopsy, Chinese, family history, genetic score, heritability, prostate cancer (PCa), prostate-specific antigen (PSA), PSA screen, single-nucleotide polymorphisms (SNPs)
Incidence of prostate cancer (PCa)
Prostate cancer (PCa) is the second most frequently diagnosed cancer and the sixth leading cause of cancer-related death in men, with an estimated 914 000 new cases and 258 000 deaths per year globally in 2008.1,2 Incidence and mortality rates of PCa vary considerably between different countries and races. The highest incidence of PCa is found in Australia and New Zealand, Northern America and Western Europe, with estimated age-standardized incidence rates greater than 80 per 100 000 in 2008. In contrast, the incidence rate is about tenfold lower in Asia, with estimated age-standardized incidence rate at less than 8 per 100 000 in 2008. However, in some Asian countries, the PCa incidence has increased considerably in recent decades. For example, age-standardized incidence rates increased by three- to eightfold between 1973–1977 and 1998–2002, from 5.1 to 15, 0.8 to 6.9 and 6.7 to 21.5 per 100 000 in Hong Kong (China), Shanghai (China) and Hiroshima (Japan), respectively.3 A combination of factors may contribute to the differences and temporal changes in PCa incidence rates, including genetic factors, Western diet, longer life expectancy and increased use of prostate-specific antigen (PSA) tests in these Asian countries.
Family history of PCa
Evidence for an inherited component of PCa was reported as early as 19604 and was later confirmed in many retrospective case–control studies as well as prospective studies. In case–control studies, the proportion of positive family history of PCa among case subjects diagnosed with the disease was compared with control subjects without a diagnosis of PCa through a retrospective questionnaire. In prospective studies, information on family history of PCa is collected first for all study subjects and rate of PCa diagnosis in the follow-up period is compared between subjects with or without family history.
In 2003, two large meta-analyses of family history based on published studies between 1966 and 2002 family history of PCa were published.5,6 Several important observations can be summarized from these two meta-analyses. First, a positive family history was significantly associated with increased risk for PCa, although the estimates of relative risk (RR) vary considerably among studies and even between the two meta-analyses. The RR of positive family history for PCa was estimated to be 2.50 (95% confidence interval (CI): 2.2–2.8) in one meta-analysis5 and 1.93 (95% CI: 1.65–2.26) in another meta-analysis.6 Second, the RR for PCa is higher in men with a positive family history in first-degree relatives (father, brothers and sons) than that in the second-degree relatives; these RRs were 2.22 (95% CI: 2.06–2.40) and 1.88 (95% CI: 1.54–2.30), respectively.6 Third, the RR for PCa is higher in men with affected brothers (RR=2.87; 95% CI: 2.21–3.73) than an affected father (RR=2.12; 95% CI: 1.82–2.51).6 Fourth, the RR for PCa is highest in men whose relatives were diagnosed with the disease before age of 60 years and the RR decreased with age.
After the two meta-analyses in 2003, additional large prospective studies were published. These studies all confirmed positive family history as a risk factor for PCa; however, the estimated RRs were generally lower, and again showed substantial interstudy differences; the RR was 1.31 in the placebo group of the Prostate Cancer Prevention Trial,7 1.47 in the placebo group of the Reduction by Dutasteride of Prostate Cancer Events (REDUCE) trial,8 1.83 in the Health Professionals Follow-up Study9 and 1.91 in the α-tocopherol, β-carotene Cancer Prevention Study.10 Several factors may have contributed to the different risk estimates of family history for PCa among studies. The methods (first-degree relatives or extended families) and survey instruments may have affected the results. Recall bias may contribute to higher estimates of RR in retrospective studies than prospective studies. PCa patients are more likely than controls to share and seek information about PCa among their relatives, and thus are more likely to report a positive family history. In prospective studies, recall bias does not exist because family history information is collected for each subject at baseline. Differential levels of PCa detection among men with or without family history may also contribute to higher estimates of RR, leading to diagnostic bias. Men who have a positive family history are more likely to undergo PCa screening and thus are more likely to be diagnosed with PCa. Diagnostic bias is less of concern in clinical trials (Prostate Cancer Prevention Trial and REDUCE) because all men undergo prostate biopsy regardless of their clinical presentations such as positive family history. Lastly, the short follow-up time of most cohort studies may contribute to lower Odds Ratio (OR) estimates. For example, the study subjects in the REDUCE study were followed only 4 years for the detection of PCa. The effect of family history on PCa risk would presumably be more prominent if subjects are followed for a longer time. The key findings from studies of family history of PCa are summarized in Box 1.
Box 1.

Family history of PCa. PCa, prostate cancer.
Twin studies of PCa
Because both shared genetics and household environment among relatives may contribute to the observed association of positive family history and PCa risk, other types of studies are needed to dissect these two confounding factors. Twin studies provide an excellent study design to disentangle the relative importance of environmental and genetic influences. By comparing the concordance rate of PCa between monozygotic (MZ) twins and dizygotic (DZ) twins, heritability (h2), a measure of the proportion of observable differences in a trait between individuals within a population that is due to genetic differences can be estimated. An h2 of 1 indicates that the trait is completely influenced by genetic factors while an h2 of 0 indicates that the trait is completely influenced by environmental factors.
The first reports of PCa twin studies came from two groups at the same time in 1997. In a cohort of 31 848 twin pairs from the World War II veteran twin study, Page and colleagues11 found that the concordance rate of PCa was significantly higher in MZ twins (27.1%) than in DZ twins (7.1%), with an estimated h2 of 0.57. In a cohort of 10 503 twin pairs from the Swedish Twin Registry, Ahlbom and colleagues12 found PCa concordance rates of 20% and 4% in MZ and DZ twins, respectively, with an estimated h2 of 0.36. Since then, several additional twin studies of PCa were reported, and all found higher concordance rate of PCa in MZ twins than DZ twins.13,14 The most well-known twin study was 44 788 twin pairs from Sweden, Danish and Finnish twin registries.15 The concordance rates of PCa were 21% and 6% in MZ twins and DZ twins, respectively, and the estimated h2 of PCa was 0.42, the highest among all common types of cancer.
Segregation analysis of PCa
Segregation analysis is a study design to further infer the mode of inheritance of a disease, i.e., whether it is polygenic (many genes with each contributing a small effect), monogenic (single gene with a dominant or recessive effect) or non-genetic. Segregation analysis is carried out by first identifying probands (index cases) and then systematically evaluating the disease status of their family members. The mode of inheritance is inferred by comparing concordance of a disease among different degrees of relatives and spouses. Results of segregation analysis provide the rationale and model for linkage analysis for identification of chromosomal locations of major susceptibility genes.
The first segregation analysis of PCa was reported in 1991 from 691 PCa families recruited at Johns Hopkins Hospital and a rare autosomal dominant gene was the best model to explain the clustering of PCa in these families.16 Since then, ∼10 segregation analyses of PCa were published.17,18,19,20,21,22,23,24,25 Although various results were reported in these studies, they generally suggested that the mode of inheritance of PCa is a mix of polygenic inheritance with several rare autosomal dominant or recessive genes. These findings are now corroborated by the discovery of more than 50 common low-penetrance PCa risk-associated single-nucleotide polymorphisms (SNPs) (i.e., polygenic) and several rare high-penetrance PCa susceptibility genes (major gene), as described in detail below.
Rare and high-penetrance PCa susceptibility genes
Linkage studies are used to identify chromosomal regions harboring major PCa susceptibility genes in families with multiple affected patients. Polymorphic markers (microsatellite repeats or SNPs) across the genome are genotyped in family members and their cosegregation (linkage) with the disease is tested using an logarithm (base 10) of odds (LOD) score. An LOD score over 3 is typically considered as evidence that the linkage is statistically significant. After a chromosomal region is identified, fine mapping and sequencing methods are used to pinpoint susceptibility genes.
The first genome-wide linkage analysis of PCa was reported in 1996; chromosome 1q24, named as HPC1, was implicated to harbor a PCa susceptibility gene based on 91 PCa families ascertained at Johns Hopkins Hospital and Sweden.26 A gene in this region, RNASEL, was later identified as a PCa susceptibility gene in February of 2002 using a combination of fine mapping and direct sequencing.27 The first reported PCa susceptibility gene was ELAC2 in February of 2001, identified from positional cloning and mutation screening of 17p, a chromosomal region that was implicated in a genome-wide linkage analysis of several large PCa families from Utah.28 The third PCa susceptibility gene, MSR1, was discovered in October of 2002 after fine mapping and direct sequencing of a linkage region at 8p22–23.29 The molecular mechanisms of these three candidate PCa susceptibility genes are largely unknown and confirmations of these genes in other series of PCa families are inconsistent.
In addition to these three linkage regions, multiple chromosomal regions have been suggested to harbor PCa susceptibility genes from multiple genome-wide linkage analyses in PCa families ascertained in North America, Europe and Australia.30,31,32,33,34,35,36,37,38,39,40,41,42,43,44 Notably, the International Consortium for Prostate Cancer Genetics (ICPCG) was formed in 1996 to coordinate efforts among investigators across the world for identifying PCa susceptibility genes. To date, more than 2000 families with at least two men affected with PCa have been recruited by members of ICPCG. Combined genome-wide linkage analyses by ICPCG has led to the identification of multiple chromosomal regions harboring PCa genes.45,46,47,48,49,50 However, because linkage analysis is susceptible to several confounders of complex diseases such as genetic heterogeneity within and between families, phenocopy (i.e., PCa is caused other than genetic alterations) and incomplete penetrance, they are prone to false-positive and false-negative findings. As a result, few of the reported PCa linkage regions and suggested genes within these regions could be consistently replicated.
A rare success was the recent identification of PCa susceptibility gene, HOXB13.51 To identify PCa susceptibility genes within a previously implicated linkage region at 17q21–22, more than 200 genes in the targeted region were evaluated by next-generation sequencing in 94 probands selected from families linked to the region. Probands from four families were discovered to have a rare but recurrent mutation (G84E) in HOXB13 (rs138213197), a homeobox transcription factor gene that is important in prostate development. All 18 men with PCa and available DNA in these four families carried the mutation. In addition, the carrier rate of the G84E mutation was increased by a factor of approximately 20 in 5083 unrelated PCa patients of European descent, with the mutation found in 72 subjects (1.4%), as compared with one in 1401 control subjects (0.1%) (P=8.5×10−7). The mutation was significantly more common in men with early-onset, familial PCa (3.1%) than in those with late-onset, non-familial PCa (0.6%) (P=2.0×10−6).
Strikingly, within a year since the initial discovery, 10 studies investigating HOXB13 in PCa families and population-based case–control subjects of European descent were published and all confirmed the finding.52,53,54,55,56,57,58,59,60,61 These studies continued to reveal characteristics of the G84E mutation of HOXB13: (i) it is likely a founder mutation that originated in Nordic countries;54,56,59,61 (ii) its carrier frequency was at ∼5% among 2443 PCa families of the ICPCG;56 (iii) mutation carriers transmit the mutation to affected offspring significantly more than the chance of 50%56 (iv) its frequency was significantly higher in PCa patients than unaffected men in the general population, with the RR for PCa estimated between 3.3 and 20.1 among various studies;51,52,53,54,55,56,57,58,59,60,61 (v) its frequency was particularly high among PCa patients with an early age at diagnosis or with a positive family history and was highest if they have both of these characteristics;51,52,53,54,55,56,57,59,60,61 and (vi) the association with pathological features of PCa remains to be demonstrated.
In addition, HOXB13 was also found to increase PCa risk in the Chinese population.62 By sequencing this gene in 96 unrelated PCa patients from the Chinese Consortium for Prostate Cancer Genetics (ChinaPCa), a different mutation in the HOXB13 gene (G135E) was found. Further evaluation of this novel mutation found that it was seen five times in 1422 PCa patients but never in 1536 unaffected controls from the ChinaPCa. The key findings from studies of rare and high-penetrance PCa susceptibility are summarized in Box 2.
Box 2.

Rare and high-penetrance PCa susceptibility genes. PCa, prostate cancer.
Common and low-penetrance risk-associated SNPs
Different from linkage analysis which is designed to identify rare and high-penetrance genes in families with multiple affected members, genetic association studies are designed to identify common and low-penetrance genes by comparing the allele/genotype frequencies of genetic markers (typically SNPs) between cases and controls from the general population. Although genetic association studies remain susceptible to confounders of complex diseases such as heterogeneity, phenocopy and incomplete penetrance, the impact of such confounders on results is relatively smaller compared to linkage analysis.
Among many challenges faced in genetic association studies, the selection of SNPs and criteria for claiming statistically significant SNPs are particularly worth noting. In the late 1990s and early 2000s, genetic association studies of PCa and other complex diseases were hypothesis-driven and relied on SNPs in candidate genes and pathway genes. While such genes may be biologically plausible, sequence variants with functional impact related to PCa development may not necessarily exist in these genes. The other major problem for candidate or pathway approaches was the liberal criteria used for judging statistical significance.63 A P<0.05 or a P-value cutoff based on a Bonferroni correction was typically used for claiming significance because few tests were performed. However, false positives remained a serious problem because multiple such studies were performed and studies showing significant findings were more likely to be published. Confirmation in multiple independent studies is an effective tool to guard against such false positives, but it was not widely adopted during that period. As a result of these challenges, few association findings from candidate and pathway genes have been consistently replicated.
Similar to the candidate gene approach, genetic association studies have also been performed to target chromosomal regions implicated by linkage analysis. Again, few consistent associations were found, likely due to poorly defined linkage regions and multiple testing. One success was the PCa association at 8q24 reported by the deCODE genetics.64 In this seminal study, Amundadottir and colleagues64 first identified a PCa linkage at 8q24 from a genome-wide linkage analysis in 323 PCa families in Iceland. They then genotyped hundreds of genetic markers in a 10-Mb interval at the region among 869 unrelated PCa cases and 596 unaffected controls and identified an association signal. After further fine mapping of this region in additional cases and controls from Iceland, they established a statistical association of PCa risk with markers at 8q24; the strongest PCa associated SNP was rs1447295, with an RR for PCa estimated at 1.72 (P=1.7×10−9). This association was also confirmed in additional case–control populations from Sweden and the United States (both Caucasians and African Americans). Impressively, the PCa association at 8q24 was confirmed in almost all published studies, making it the first and most consistent PCa risk-associated SNP.
With the development of high-throughput and low-cost genotyping arrays, it became feasible to systematically screen hundreds and thousands of SNPs in the genome for their association with disease risk, without a need to limit to specific genes and regions. The identification of a genetic association for age-related macular degeneration by a genome-wide association study (GWAS) in 2005 provided the first empirical evidence that this approach could be successful.65 This agnostic approach is scientifically sound because it requires a simple assumption that can most likely hold: that inherited variants somewhere in the genome may account for genetic susceptibility to a disease. In addition, much more stringent criteria are required to declare statistical significance, in part forced by the large number of tests involved in GWAS. For example, a P-value cutoff of 5×10−8 is typically required for a study-wise type I error of 0.05 to account for ∼1 million tests performed in GWAS (0.05/1 000 000). Furthermore, confirmation in independent study populations is also required for GWAS. As a result, thousands of SNPs have been identified to be consistently associated with risk for many complex diseases since 2005, including PCa (http://www.genome.gov/gwastudies/). Interestingly, many of these SNPs are not located in apparent candidate genes or pathways, and some are not within genes.66 These findings not only provide novel insight to disease etiology but also explain in part the failure of candidate gene association studies.
The first two GWASs of PCa were reported in May 2007.64,68 By systematically evaluating 550 000 SNPs across the genome in 1172 cases and 1157 controls of European origin nested from the Cancer Genetic Markers of Susceptibility, Yeager and colleagues67 confirmed the PCa association with rs1447295 and identified another independent association at 8q24 (rs6983267), ∼70 Kb centromeric to rs1447295. By combining their results with four additional studies with a total of 4296 cases and 4299 controls, the association was highly significant (P=9.42×10−13). The estimated RRs were 1.26 and 1.58 for heterozygous and homozygous carriers, respectively. At the same time, the deCODE genetics reported a PCa GWAS in 1453 PCa patients and 3064 controls from Iceland.68 Again, in addition to confirming the PCa association of rs1447295, they identified yet another new PCa association at 8q24 (rs16901979), ∼360 Kb from rs1447295 and ∼288 Kb from rs6983267. The association was confirmed in three additional Caucasian populations from Spain, the Netherlands and the United States, with a combined P-value of 1.1×10−12. The estimated RR was 1.79. Interestingly, this association was also confirmed in an African-American population (RR=1.34; P=0.005).
Shortly after these two PCa GWASs, multiple PCa associations were reported from GWAS, including two independent loci at 17q12 and 17q24 by Gudmundsson et al.69 in August 2007, one locus at 9q33 by Duggan et al.70 in December 2007, four novel loci at 7p15, 10q11, 10q26 and 11q13 by Thomas et al.71 in March 2008, five novel loci at 3p12, 6q25, 7q21, 19q13 and Xp11 by Eeles et al.72 in March 2008 and two novel loci at 2p15 and Xp11 by Gudmundsson et al.73 in March 2008. In October 2009, more PCa risk-associated SNPs in the genome were reported from PCa GWAS, including additional novel loci at 8q24 by Al Olama et al.74 and by Yeager et al.,75 seven novel loci at 2p21, 2q31, 4q22, 4q24, 8p21, 11p15 and 22q13 by Eeles et al.76 and three new loci at 3q21, 8q24 and 19q13 by Gudmundsson et al.77 In 2011, several additional PCa risk-associated SNPs were reported, including two new loci at 2q37 and 12q13 by Schumacher et al.78 and seven loci by Kote-Jarai et al.79 of the Prostate Cancer Association Group to Investigate Cancer Associated Alterations in the Genome (PRACTICAL) Consortium.
Novel PCa risk associations have also been found in other races and ethnicities. Haiman and colleagues searched 1 047 986 SNPs across the genome in 3425 PCa cases and 3290 controls that were African Americans.80 Only the 8q24 region exceeded genome-wide significance level in the discovery stage. However, by following up suggestive regions across the genome among additional 1842 cases and 3265 controls that were African Americans, they identified a novel PCa association at 17q21. The most significant SNP at the region was rs7210100, with an RR for PCa estimated at 1.51 (P=3.4×10−13). The frequency of the risk allele is ∼5% in men of African descent, but is rare in other populations (<1%). Multiple PCa associations were also found in a Japanese population.81,82 In 2010, by evaluating about half a million SNPs across the genome in 1583 cases and 3386 controls that were Japanese, Takata and colleagues81 found eight independent associations with PCa in the first stage of GWAS, including six previously implicated associations in Caucasians (two at 8q24, 3p12, 8p21, 10q11 and 17q12) and two novel associations at 5p15 and 6q22. In addition, by following additional promising SNPs in the second stage with 3001 more cases and 5415 more controls, they found three more novel PCa associations (2p24, 6p21 and 13q22). In a second report in 2012, Akamatsu and colleagues82 identified three additional PCa associations at 11q12, 10q26, and 3p11 after following more promising SNPs from the same Japanese PCa GWAS in a larger study.
Most recently, a PCa GWAS in a Chinese population was reported.83 By performing a PCa GWAS in 1497 cases and 1008 controls, Xu and colleagues observed one region (8q24) in the genome that exceeded genome-wide significance level. Suggestive evidence for PCa associations was also found for eight loci previously implicated in European populations and four loci previously implicated in the Japanese population (P<0.05). In addition, by following up additional promising SNPs in independent Chinese study subjects (2987 PCa cases and 7926 controls), they found two novel PCa associations at chromosomes 9q31.2 (rs817826, P=5.45×10−14) and 19q13.4 (rs103294, P=5.34×10−16). The rs103294 marker at 19q13.4 is in strong linkage equilibrium with a 6.7-kb germline deletion that removes the first six of seven exons in LILRA3, a gene-regulating inflammatory response.
To date, more than 50 PCa risk-associated SNPs have been consistently associated with PCa risk in Caucasians, African Americans, Japanese and Chinese from GWAS and fine mapping of implicated regions84,85,86,87 (Supplementary Table 1), more than any other type of cancer. Several observations may be summarized from these findings. First, most of these associations can be consistently replicated in independent study populations. Second, few of these associations were in well-known PCa candidate genes and pathways, and many are in inter-genic regions (Supplementary Table 2). This observation reveals our currently limited knowledge on PCa etiology and demonstrates the advantage of a systematic genome-wide approach. Third, while several genetic variants confer PCa risk in multiple races, some PCa associations are race specific. Fourth, most of these variants are common in general populations but confer modest risk to PCa, with RR typically between 1.1 and 1.2, although a few of them have RR over 1.50. However, when these SNPs were considered together, they confer stronger risk to PCa88,89 (see below). The key findings from studies of common and low-penetrance risk-associated SNPs are summarized in Box 3.
Box 3.

Common and low-penetrance risk-associated SNPs. SNP, single-nucleotide polymorphism.
PCa risk assessment using SNPs
The discovery of more than 50 PCa risk-associated SNPs from GWAS in the past 7 years was a major accomplishment in understanding PCa etiology, especially considering that only three risk factors (age, race and family history) were established for PCa prior to GWAS. These findings provide new targets and directions for biological studies to understand the well-observed genetic susceptibility to PCa. These PCa risk-associated SNPs may also have potential utility in the screening, diagnosis, prevention and treatment of PCa.
One of the obvious applications of PCa risk-associated SNPs is for identifying men with higher risk for PCa. In 2008, Zheng and colleagues88 examined the cumulative effect of the first five PCa risk-associated SNPs on PCa risk and demonstrated that increasing number of PCa risk-associated alleles was significantly associated with increasing PCa risk in a Swedish population-based case–control study (P=3.93×10−28 for a trend test). In men who had any five or more of genetic risk factors (five risk genotypes and positive family history), the RR for PCa was 9.46 (P=1.29×10−8), as compared to men without any of the factors. Later in 2009, the same group provided further evidence for the cumulative effect in a new study.89 The methods used in this study differed from the previous one in three major aspects. First, the number of SNPs was increased from five to 14 PCa risk-associated SNPs. Second, in addition to the Swedish case–control population, the study included another nested case–control study population from the Prostate, Lung, Colon and Ovarian (PLCO) Cancer Screening Trial in the United States Third, an absolute risk was calculated for easier interpretation by physicians and patients. Considering men with 11 risk alleles (average in general population) and negative family history as having baseline risk, men who had 14 or more risk alleles and positive family history had ORs of 4.92 and 3.88 for PCa in the Swedish and US study, respectively. Once a man's SNP genotypes and family history are known, his absolute risk for PCa can be readily calculated and easily interpreted. For example, 55-year-old men with a positive family history and 14 or more risk alleles have 52% and 41% risks of being diagnosed with PCa in the next 20 years in the Swedish and US populations, respectively. In comparison, without knowledge of genotype and family history, these men had an average population absolute risk of 13%.
Several additional studies evaluated the cumulative effect of PCa risk-associated SNPs on PCa risk.90,91,92,93,94 Salina et al.90 replicated the cumulative effect of the five SNPs in a US population (P=1.5×10−20). They observed an improved, but not statistically significant, area under the curve (AUC) of a receiver operating characteristic (ROC) in discriminating cases from controls using a model with these five SNPs. The AUCs were 0.63 and 0.66, respectively, for a model with previously known risk factors only (age, serum PSA and family history) and a model adding five SNPs. Kote-Jarai et al.91 evaluated 15 PCa risk-associated SNPs in a large PRACTICAL consortium with 7370 cases and 5742 controls and found a strong cumulative effect of these SNPs on PCa risk. Men in the top 10% of the risk distribution based on these 15 SNPs had a 2.1-fold increased risk relative to general population rates. Lindström et al.93 evaluated 25 PCa risk-associated SNPs in 7509 prostate cancer cases and 7652 controls within the National Cancer Institute Breast and Prostate Cancer Cohort Consortium (BPC3). Compared with men in the lowest tenth percentile of a simple count of risk alleles, men in the highest tenth percentile had more than a fivefold risk of developing PCa. The absolute 10-year risk for 50-year-old men with a family history ranged from 1.6% (tenth percentile of genetic risk) to 6.7% (ninetieth percentile of genetic risk). For men without a family history, the risk ranged from 0.8% (tenth percentile) to 3.4% (ninetieth percentile). They also compared the discriminative performance of three models: (i) age and family history; (ii) age and SNPs and (iii) age, family history and SNPs. They found that model 2 was significantly better than model 1, and model 3 was similar to model 2, suggesting that SNPs had a better discriminative performance than family history, especially in men with age younger than 60 years. Klein et al.94 compared the discriminative performance of SNPs and PSA in 943 men diagnosed with PCa and 2829 matched controls in the Malmo Diet and Cancer cohort from Sweden. They found the AUC of SNPs alone for discriminating PCa was 0.57. However, the AUC was significantly lower than that of PSA alone (0.79).
The cumulative effect of PCa risk-associated SNPs on PCa risk was also seen in Japanese and Chinese populations.95,96 Akamatsu et al.95 evaluated 16 SNPs that were significantly associated with PC risk among three Japanese study populations, including a population of 689 cases and 749 male controls for model generation, and two populations comprising 3294 cases and 6281 controls for model validation. The model with these SNPs had AUCs of 0.68, 0.66 and 0.66, respectively, for discriminating PCa cases and controls in these three populations. They also showed that the AUC of the genetic model was similar regardless of PSA levels. In a Chinese study, Zheng et al.96 evaluated 33 PCa risk-associated SNPs implicated in populations of European descent among 1108 PCa cases and 1525 controls of ChinaPCa. They found that genetic score based on these SNPs was significantly higher for cases than controls (P=5.91×10−20), and was significantly associated with risk of PCa in a dose-dependent manner (P for trend: 4.78×10−18). The AUC of the genetic model in discriminating cases and controls was 0.60. When only SNPs that were implicated in the Chinese population were used, the AUC was slightly increased to 0.62.
Clinical utility for prostate biopsy
Risk assessment is critical to differentiate individual risk for PCa for personalized medicine. PCa risk-associated SNPs could be added to existing predictors to improve performance of risk assessment. While its utility may include targeted PSA screening, chemoprevention, diagnosis and treatment, most studies published to date reported the utility genomic risk assessment in prostate biopsy.95,97,98,99 Currently, age, family history, PSA levels, prostate volumes and other clinical variables are used by urologists to determine the need for prostate biopsy for detection of PCa. However, only about 30%–40% men who meet the indications for biopsy are positive for PCa. To assess whether PCa risk-associated SNPs can be used to better stratify patients' risk for a positive biopsy, Aly et al.97 evaluated 35 implicated PCa risk-associated SNPs in 5241 men who underwent a prostate biopsy in Stockholm, Sweden, during 2005 to 2007. When comparing an existing clinical model based on age, PSA, free-to-total PSA and family history, adding SNPs could significantly reduce the number of biopsy needed for detecting the same number of PCa patients. For example, 22.7% biopsies could be avoided in this cohort at a cost of missing a PCa diagnosis in 3% of patients characterized as having an aggressive disease. In another study, Kader et al.98 compared the performance of 33 PCa risk-associated SNPs with existing clinical parameters in predicting positive prostate biopsy rate in the REDUCE trial. All men in the trial had an initial negative prostate biopsy and underwent study-mandated biopsies at years 2 and 4.100 PCa risk can be estimated and ranked for each patient based on the best clinical model only and the combined clinical and genetic model (with 33 SNPs). For the clinical model only, the positive biopsy rates during the 4-year follow-up were 17%, 23% and 37%, respectively for patients with the lowest (first quartile), intermediate and highest (fourth quartile) risk. In contrast, the positive biopsy rates during the 4-year follow-up were 14%, 22% and 42%, respectively for patients with the lowest, intermediate and highest risk based on the combined clinical and genetic model. Adding SNPs to the best clinical model reclassified PCa risk in 33% of men, and the reclassified risk had a significantly better correlation to biopsy outcomes. Importantly, they found that the benefit of adding the SNPs was greatest among men at intermediate clinical risk (twenty-fifth percentile to seventy-fifth percentile). In addition, the AUC for discriminating prostate biopsy increased from 0.62 using the best clinical model to 0.66 using the combined clinical and genetic model. Similar benefit of adding PCa risk-associated SNPs for predicting biopsy outcomes was also reported in a Japanese study.95
The clinical utility of genetic risk assessment for targeted prostate biopsy was also reported in the Chinese population. PCa risk-associated SNPs in Chinese have been evaluated for SNPs initially discovered in Caucasians101 and Japanese.102 A PCa GWAS in the Chinese population also revealed two novel PCa-risk associated SNPs.83 A comprehensive evaluation of all 53 reported PCa risk-associated SNPs reported by the end of 2012 among 1922 PCa cases and 2175 controls selected from the ChinaPCa found evidence for association in the Chinese population at P<0.05.103 Based on this information, Jiang et al.99 performed a study to assess the clinical utility of these 24 PCa risk-associated SNPs for differentiating PCa risk in a biopsy cohort in Shanghai, China. Among 308 patients that underwent prostate biopsy at Huashan Hospital between April 2011 and August 2012, 141 (45.8%) were diagnosed with PCa. Genetic score calculated based on these 24 SNPs was significantly higher in patients with PCa (median=1.30) than without (median=0.89) (P=3.81×10−6). The difference remained significant after adjusting for age and total PSA (P=0.007). The PCa detection rate increased with increasing genetic score; 26.3%, 43.2% and 60.0% for men with lower (<0.5), average (0.5–1.5) and higher (>1.5) genetic score, respectively (P-trend=0.0003). For patients with moderately elevated PSA levels (1.6–20 ng ml−1), the PCa detection rate was 31.2% overall and were 16.7%, 31.2% and 40.9% for men with lower (<0.5), average (0.5–1.5) and higher (>1.5) genetic score, respectively (P-trend=0.03). For patients with PSA≥20 ng ml−1, however, the PCa detection rates were high (>69%) regardless of genetic score. They concluded that a genetic score based on PCa risk-associated SNPs is an independent predictor of prostate biopsy outcomes in Chinese men and may be helpful to determine the need for prostate biopsy among patients within a ‘gray zone' of PCa risk.
Divergent views on clinical utility of PCa risk-associated SNPs
Despite consistent results in almost all published studies demonstrating that genetic score based on PCa risk-associated SNPs is a strong risk factor for PCa and predicts PCa independent of existing variables such age, family history and PSA levels,88,89,90,91,92,93,94,95,96,97,98,99 there is resistance for the genetic testing and its clinical use among physicians and researchers. Most of the reservations center on the issues related to small improvement of genetic score in AUC for discriminating PCa cases and controls. Other concerns are related to the ability to properly interpret results by physicians and patients, potential anxiety caused by the genetic testing among patients and their family members, cost and time for performing genetic testing, as well as other ethical, legal and social implications.104 While acknowledging that the improvement of AUC was small in absolute terms (for example, it increased from 0.62 to 0.66 after adding 33 SNPs in the REDUCE study98,105), the increase was considerable in relative terms, a 33% improvement ((0.66−0.62)/(0.62−0.50)). The rather large difference between absolute and relative benefit reflects the poor ability of the current clinical approach to predict prostate biopsy outcome. For complex diseases such as PCa, many factors, including PCa risk-associated SNPs, may contribute to its development. The usefulness of a novel predictor should be judged by comparative effectiveness, i.e., whether it improves over existing clinical practice. This is particularly true if a new biomarker is easy to measure, non-invasive and has a low cost. For the 33 PCa risk-associated SNPs, they can be measured in a panel from blood or saliva samples at a cost similar to a PSA test.
More importantly, it is worth noting that genetic score calculated from PCa risk-associated SNPs is a predictive marker. Genetic score is not intended to be used as a diagnostic marker for discriminating PCa patients vs. non-patients. Rather, genetic score should be used as a measurement for genetic susceptibility, similar to family history. When genetic score is used for identifying high-risk subjects for targeted PSA screening, its performance should be compared to age and family history, and AUC is not the most appropriate measurement for assessing its performance. For the purpose of identifying high-risk subjects for targeted biopsy, the performance of genetic score can be evaluated in comparison with other existing clinical variables.
Genetic score should be used to measure genetic susceptibility to PCa, similar to family history
Both family history and genetic score are measures of genetic susceptibility to PCa. An advantage of family history is that the information can be obtained without a laboratory test. However, rather than providing a direct measurement of the patient's inherited risk, family history captures PCa information of their relatives. Consequently, family history is influenced by family size, age and survival status of male relatives, recall ability, family communication and prevalence of the disease in various populations. The fact that brothers are estimated to have exactly the same inherited risk for PCa based on family history highlights its limitation because they actually share only 50% of their genetic makeup on average. Genetic score requires a genetic test and can be calculated based on genotypes of multiple PCa risk-associated SNPs, weighted by their RR to PCa. A genetic score of 1.0 indicates an average risk in the general population. Because genetic score is a continuous variable, it offers better resolution to distinguish genetic risk, for example, about 50%, 8% and 2% of the US population have a genetic risk that is one-, two- and threefold higher than the average risk in the general population based on a genetic score calculated from 33 PCa risk-associated SNPs.98 To comprehensively compare the performance of these two measurements, Sun and colleagues106 performed a head-to-head comparison of these two measurements in five study populations. They found that genetic score outperforms family history in objectiveness, precision and discriminative ability. For objectiveness, they found that the proportion of men with a positive family history of PCa differed considerably among these five study populations, while mean genetic score was similar. For example, among three geographic regions from the REDUCE trial, the proportion of positive family history was significantly different; 4.2%, 10.9% and 22.8% in Eastern Europe, Western Europe and North America, respectively (P<0.001). These large differences were found even though the same protocol was used to obtain family history information. In contrast, the mean genetic score was similar among different geographic regions within the REDUCE study (from 0.95 to 0.97, P=0.88). For the precision of measuring association, they found the RR for PCa differed significantly among studies for family history but was similar for genetic score. For example, the RRs of family history for PCa were 1.20, 1.53 and 1.91 in North America, Eastern Europe and Western Europe, respectively within the REDUCE trial. In contrast, the RR estimates of genetic score for PCa were similar among these five study populations, from 1.69 to 1.82. For discriminative performance, they found the AUC of genetic score (0.58–0.62) for discriminating cases vs. controls was significantly higher than family history (0.51–0.55) in each of these five study populations (P<0.05). This last observation was also found in other studies comparing these two variables.93,97 Based on these pieces of evidence, we would argue that if family history is accepted and used by urologists and primary care physicians to assess an individual's risk for PCa, genetic score should be also be used to improve assessment of inherited risk for PCa.
Another major advantage of genetic score over family history is in populations where PCa incidence was historically low. Positive family history of PCa in these geographic regions and countries such as China is extremely low and almost uninformative. Genetic score, on the other hand, does not depend on PCa incidence and is the only effective measurement of genetic susceptibility to PCa.99 The key findings from studies of PCa risk assessment using SNPs and potential clinical utility are summarized in Box 4.
Box 4.
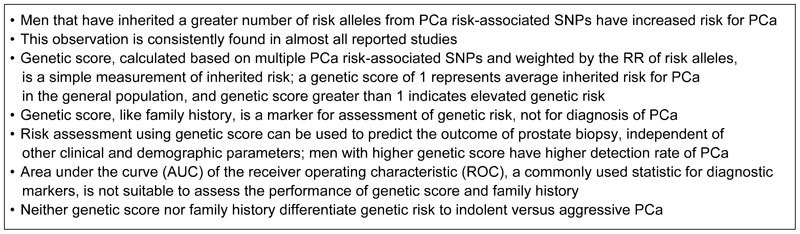
PCa risk assessment using SNPs and potential clinical utility. PCa, prostate cancer; SNP, single-nucleotide polymorphism.
Future directions
Although excellent progress has been made in delineating the genetic basis of PCa, more genetic studies are needed to better understand the genetic susceptibility to PCa, especially aggressive types of PCa. To date, few genetic markers have been identified that can consistently distinguish indolent vs. aggressive PCa risk. Such genetic markers would be extremely important to address the current debate on overdiagnosis and overtreatment of PCa.
Great efforts are currently underway to identify rare and high-penetrance mutations using Exome SNP arrays that contain more than 250 000 known mutations across the genome. In addition, ultimate association studies using whole-genome sequencing are becoming feasible in the near future.57 These new approaches may provide unique opportunity to identify genetic variants that predispose individuals to the risk of aggressive PCa. Finally, one important area that needs special attention is to understand the impact of genetic test results on perception and behavior.
Acknowledgments
This work was partially funded by the National Key Basic Research Program Grant 973 (No.2012CB518301) to JX, the Key Project of the National Natural Science Foundation of China (No.81130047) to JX, intramural grants from Fudan University ‘Thousand Talents Program' and Huashan Hospital to JX and the National Institutes of Health (No.NCI CA129684) to JX.
No competing financial interests.
Footnotes
Supplementary Information accompanies the paper on Asian Journal of Andrology's website (http://www.nature.com/aja).
Supplementary Information
References
- Ferlay J, Shin HR, Bray F, Forman D, Mathers C, et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, et al. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- Cullen J, Elsamanoudi S, Brassell SA, Chen Y, Colombo M, et al. The burden of prostate cancer in Asian nations. J Carcinog. 2012;11:7. doi: 10.4103/1477-3163.94025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolf CM. An investigation of the familial aspects of carcinoma of the prostate. Cancer. 1960;13:739–44. doi: 10.1002/1097-0142(196007/08)13:4<739::aid-cncr2820130414>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Johns LE, Houlston RS. A systematic review and meta-analysis of familial prostate cancer risk. BJU Int. 2003;91:789–94. doi: 10.1046/j.1464-410x.2003.04232.x. [DOI] [PubMed] [Google Scholar]
- Goh CL, Schumacher FR, Easton D, Muir K, Henderson B, et al. Genetic variants associated with predisposition to prostate cancer and potential clinical implications. J Intern Med. 2012;271:353–65. doi: 10.1111/j.1365-2796.2012.02511.x. [DOI] [PubMed] [Google Scholar]
- Thompson IM, Ankerst DP, Chi C, Goodman PJ, Tangen CM, et al. Assessing prostate cancer risk: results from the Prostate Cancer Prevention Trial. J Natl Cancer Inst. 2006;98:529–34. doi: 10.1093/jnci/djj131. [DOI] [PubMed] [Google Scholar]
- Thomas JA, 2nd, Gerber L, Moreira DM, Hamilton RJ, Bañez LL, et al. Prostate cancer risk in men with prostate and breast cancer family history: results from the REDUCE study (R1) J Intern Med. 2012;272:85–92. doi: 10.1111/j.1365-2796.2011.02504.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YC, Page JH, Chen R, Giovannucci E. Family history of prostate and breast cancer and the risk of prostate cancer in the PSA era. Prostate. 2008;68:1582–91. doi: 10.1002/pros.20825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn J, Moslehi R, Weinstein SJ, Snyder K, Virtamo J, et al. Family history of prostate cancer and prostate cancer risk in the Alpha-Tocopherol, Beta-Carotene Cancer Prevention (ATBC) Study. Int J Cancer. 2008;123:1154–9. doi: 10.1002/ijc.23591. [DOI] [PubMed] [Google Scholar]
- Page WF, Braun MM, Partin AW, Caporaso N, Walsh P. Heredity and prostate cancer: a study of World War II veteran twins. Prostate. 1997;33:240–5. doi: 10.1002/(sici)1097-0045(19971201)33:4<240::aid-pros3>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Ahlbom A, Lichtenstein P, Malmström H, Feychting M, Hemminki K, et al. Cancer in twins: genetic and nongenetic familial risk factors. J Natl Cancer Inst. 1997;89:287–93. doi: 10.1093/jnci/89.4.287. [DOI] [PubMed] [Google Scholar]
- Verkasalo PK, Kaprio J, Koskenvuo M, Pukkala E. Genetic predisposition, environment and cancer incidence: a nationwide twin study in Finland, 1976–1995. Int J Cancer. 1999;83:743–9. doi: 10.1002/(sici)1097-0215(19991210)83:6<743::aid-ijc8>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Neale RE, Mineau G, Whiteman DC, Brownbill PA, Murphy MF. Childhood and adult cancer in twins: evidence from the Utah genealogy. Cancer Epidemiol Biomarkers Prev. 2005;14:1236–40. doi: 10.1158/1055-9965.EPI-04-0723. [DOI] [PubMed] [Google Scholar]
- Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, et al. Environmental and heritable factors in the causation of cancer—analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- Carter BS, Steinberg GD, Beaty TH, Childs B, Walsh PC. Familial risk factors for prostate cancer. Cancer Surv. 1991;11:5–13. [PubMed] [Google Scholar]
- Grönberg H, Damber L, Damber JE, Iselius L. Segregation analysis of prostate cancer in Sweden: support for dominant inheritance. Am J Epidemiol. 1997;146:552–7. doi: 10.1093/oxfordjournals.aje.a009313. [DOI] [PubMed] [Google Scholar]
- Schaid DJ, McDonnell SK, Blute ML, Thibodeau SN. Evidence for autosomal dominant inheritance of prostate cancer. Am J Hum Genet. 1998;62:1425–38. doi: 10.1086/301862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhage BA, Baffoe-Bonnie AB, Baglietto L, Smith DS, Bailey-Wilson JE, et al. Autosomal dominant inheritance of prostate cancer: a confirmatory study. Urology. 2001;57:97–101. doi: 10.1016/s0090-4295(00)00891-8. [DOI] [PubMed] [Google Scholar]
- Gong G, Oakley-Girvan I, Wu AH, Kolonel LN, John EM, et al. Segregation analysis of prostate cancer in 1,719 white, African-American and Asian-American families in the United States and Canada. Cancer Causes Control. 2002;13:471–82. doi: 10.1023/a:1015755219674. [DOI] [PubMed] [Google Scholar]
- Baffoe-Bonnie AB, Kiemeney LA, Beaty TH, Bailey-Wilson JE, Schnell AH, et al. Segregation analysis of 389 Icelandic pedigrees with Breast and prostate cancer. Genet Epidemiol. 2002;23:349–63. doi: 10.1002/gepi.10188. [DOI] [PubMed] [Google Scholar]
- Valeri A, Briollais L, Azzouzi R, Fournier G, Mangin P, et al. Segregation analysis of prostate cancer in France: evidence for autosomal dominant inheritance and residual brother-brother dependence. Ann Hum Genet. 2003;67 Pt 2:125–37. doi: 10.1046/j.1469-1809.2003.00022.x. [DOI] [PubMed] [Google Scholar]
- Conlon EM, Goode EL, Gibbs M, Stanford JL, Badzioch M, et al. Oligogenic segregation analysis of hereditary prostate cancer pedigrees: evidence for multiple loci affecting age at onset. Int J Cancer. 2003;105:630–5. doi: 10.1002/ijc.11128. [DOI] [PubMed] [Google Scholar]
- Pakkanen S, Baffoe-Bonnie AB, Matikainen MP, Koivisto PA, Tammela TL, et al. Segregation analysis of 1,546 prostate cancer families in Finland shows recessive inheritance. Hum Genet. 2007;121:257–67. doi: 10.1007/s00439-006-0310-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacInnis RJ, Antoniou AC, Eeles RA, Severi G, Guy M, et al. Prostate cancer segregation analyses using 4390 families from UK and Australian population-based studies. Genet Epidemiol. 2010;34:42–50. doi: 10.1002/gepi.20433. [DOI] [PubMed] [Google Scholar]
- Smith JR, Freije D, Carpten JD, Grönberg H, Xu J, et al. Major susceptibility locus for prostate cancer on chromosome 1 suggested by a genome-wide search. Science. 1996;274:1371–4. doi: 10.1126/science.274.5291.1371. [DOI] [PubMed] [Google Scholar]
- Carpten J, Nupponen N, Isaacs S, Sood R, Robbins C, et al. Germline mutations in the ribonuclease L gene in families showing linkage with HPC1. Nat Genet. 2002;30:181–4. doi: 10.1038/ng823. [DOI] [PubMed] [Google Scholar]
- Tavtigian SV, Simard J, Teng DH, Abtin V, Baumgard M, et al. A candidate prostate cancer susceptibility gene at chromosome 17p. Nat Genet. 2001;27:172–80. doi: 10.1038/84808. [DOI] [PubMed] [Google Scholar]
- Xu J, Zheng SL, Komiya A, Mychaleckyj JC, Isaacs SD, et al. Germline mutations and sequence variants of the macrophage scavenger receptor 1 gene are associated with prostate cancer risk. Nat Genet. 2002;32:321–5. doi: 10.1038/ng994. [DOI] [PubMed] [Google Scholar]
- Xu J, Meyers D, Freije D, Isaacs S, Wiley K, et al. Evidence for a prostate cancer susceptibility locus on the X chromosome. Nat Genet. 1998;20:175–9. doi: 10.1038/2477. [DOI] [PubMed] [Google Scholar]
- Edwards S, Meitz J, Eles R, Evans C, Easton D, et al. Results of a genome-wide linkage analysis in prostate cancer families ascertained through the ACTANE consortium. Prostate. 2003;57:270–9. doi: 10.1002/pros.10301. [DOI] [PubMed] [Google Scholar]
- Schleutker J, Baffoe-Bonnie AB, Gillanders E, Kainu T, Jones MP, et al. Genome-wide scan for linkage in Finnish hereditary prostate cancer (HPC) families identifies novel susceptibility loci at 11q14 and 3p25–26. Prostate. 2003;57:280–9. doi: 10.1002/pros.10302. [DOI] [PubMed] [Google Scholar]
- Wiklund F, Gillanders EM, Albertus JA, Bergh A, Damber JE, et al. Genome-wide scan of Swedish families with hereditary prostate cancer: suggestive evidence of linkage at 5q11.2 and 19p13.3. Prostate. 2003;57:290–7. doi: 10.1002/pros.10303. [DOI] [PubMed] [Google Scholar]
- Lange EM, Gillanders EM, Davis CC, Brown WM, Campbell JK, et al. Genome-wide scan for prostate cancer susceptibility genes using families from the University of Michigan prostate cancer genetics project finds evidence for linkage on chromosome 17 near BRCA1. Prostate. 2003;57:326–34. doi: 10.1002/pros.10307. [DOI] [PubMed] [Google Scholar]
- Friedrichsen DM, Stanford JL, Isaacs SD, Janer M, Chang BL, et al. Identification of a prostate cancer susceptibility locus on chromosome 7q11–21 in Jewish families. Proc Natl Acad Sci USA. 2004;101:1939–44. doi: 10.1073/pnas.0308336100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillanders EM, Xu J, Chang BL, Lange EM, Wiklund F, et al. Combined genome-wide scan for prostate cancer susceptibility genes. J Natl Cancer Inst. 2004;96:1240–7. doi: 10.1093/jnci/djh228. [DOI] [PubMed] [Google Scholar]
- Camp NJ, Farnham JM, Cannon Albright LA. Genomic search for prostate cancer predisposition loci in Utah pedigrees. Prostate. 2005;65:365–74. doi: 10.1002/pros.20287. [DOI] [PubMed] [Google Scholar]
- Chang BL, Isaacs SD, Wiley KE, Gillanders EM, Zheng SL, et al. Genome-wide screen for prostate cancer susceptibility genes in men with clinically significant disease. Prostate. 2005;64:356–61. doi: 10.1002/pros.20249. [DOI] [PubMed] [Google Scholar]
- Slager SL, Zarfas KE, Brown WM, Lange EM, McDonnell SK, et al. Genome-wide linkage scan for prostate cancer aggressiveness loci using families from the University of Michigan Prostate Cancer Genetics Project. Prostate. 2006;66:173–9. doi: 10.1002/pros.20332. [DOI] [PubMed] [Google Scholar]
- Lange EM, Ho LA, Beebe-Dimmer JL, Wang Y, Gillanders EM, et al. Genome-wide linkage scan for prostate cancer susceptibility genes in men with aggressive disease: significant evidence for linkage at chromosome 15q12. Hum Genet. 2006;119:400–7. doi: 10.1007/s00439-006-0149-6. [DOI] [PubMed] [Google Scholar]
- Christensen GB, Camp NJ, Farnham JM, Cannon-Albright LA. Genome-wide linkage analysis for aggressive prostate cancer in Utah high-risk pedigrees. Prostate. 2007;67:605–13. doi: 10.1002/pros.20554. [DOI] [PubMed] [Google Scholar]
- Schaid DJ, Stanford JL, McDonnell SK, Suuriniemi M, McIntosh L, et al. Genome-wide linkage scan of prostate cancer Gleason score and confirmation of chromosome 19q. Hum Genet. 2007;121:729–35. doi: 10.1007/s00439-007-0368-5. [DOI] [PubMed] [Google Scholar]
- Baffoe-Bonnie AB, Kittles RA, Gillanders E, Ou L, George A, et al. Genome-wide linkage of 77 families from the African American Hereditary Prostate Cancer study (AAHPC) Prostate. 2007;67:22–31. doi: 10.1002/pros.20456. [DOI] [PubMed] [Google Scholar]
- Stanford JL, FitzGerald LM, McDonnell SK, Carlson EE, McIntosh LM, et al. Dense genome-wide SNP linkage scan in 301 hereditary prostate cancer families identifies multiple regions with suggestive evidence for linkage. Hum Mol Genet. 2009;18:1839–48. doi: 10.1093/hmg/ddp100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Dimitrov L, Chang BL, Adams TS, Turner AR, et al. A combined genomewide linkage scan of 1,233 families for prostate cancer-susceptibility genes conducted by the international consortium for prostate cancer genetics. Am J Hum Genet. 2005;77:219–29. doi: 10.1086/432377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaid DJ, McDonnell SK, Zarfas KE, Cunningham JM, Hebbring S, et al. Pooled genome linkage scan of aggressive prostate cancer: results from the International Consortium for Prostate Cancer Genetics. Hum Genet. 2006;120:471–85. doi: 10.1007/s00439-006-0219-9. [DOI] [PubMed] [Google Scholar]
- Camp NJ, Cannon-Albright LA, Farnham JM, Baffoe-Bonnie AB, George A, et al. Compelling evidence for a prostate cancer gene at 22q12.3 by the International Consortium for Prostate Cancer Genetics. Hum Mol Genet. 2007;16:1271–8. doi: 10.1093/hmg/ddm075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen GB, Baffoe-Bonnie AB, George A, Powell I, Bailey-Wilson JE, et al. Genome-wide linkage analysis of 1,233 prostate cancer pedigrees from the International Consortium for Prostate Cancer Genetics using novel sumLINK and sumLOD analyses. Prostate. 2010;70:735–44. doi: 10.1002/pros.21106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Cancel-Tassin G, Valeri A, Cussenot O, Lange EM, et al. Chromosomes 4 and 8 implicated in a genome wide SNP linkage scan of 762 prostate cancer families collected by the ICPCG. Prostate. 2012;72:410–26. doi: 10.1002/pros.21443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey-Wilson JE, Childs EJ, Cropp CD, Schaid DJ, Xu J, et al. Analysis of Xq27–28 linkage in the international consortium for prostate cancer genetics (ICPCG) families. BMC Med Genet. 2012;13:46. doi: 10.1186/1471-2350-13-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing CM, Ray AM, Lange EM, Zuhlke KA, Robbins CM, et al. Germline mutations in HOXB13 and prostate-cancer risk. N Engl J Med. 2012;366:141–9. doi: 10.1056/NEJMoa1110000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breyer JP, Avritt TG, McReynolds KM, Dupont WD, Smith JR. Confirmation of the HOXB13 G84E germline mutation in familial prostate cancer. Cancer Epidemiol Biomarkers Prev. 2012;21:1348–53. doi: 10.1158/1055-9965.EPI-12-0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbari MR, Trachtenberg J, Lee J, Tam S, Bristow R, et al. Association between germline HOXB13 G84E mutation and risk of prostate cancer. J Natl Cancer Inst. 2012;104:1260–2. doi: 10.1093/jnci/djs288. [DOI] [PubMed] [Google Scholar]
- Karlsson R, Aly M, Clements M, Zheng L, Adolfsson J, et al. A population-based assessment of germline HOXB13 G84E mutation and prostate cancer risk. Eur Urole-pub ahead of print 20 July 2012. [DOI] [PubMed]
- Schroeck FR, Zuhlke KA, Siddiqui J, Siddiqui R, Cooney KA, et al. Testing for the recurrent HOXB13 G84E germline mutation in men with clinical indications for prostate biopsy. J Urol. 2013;189:849–53. doi: 10.1016/j.juro.2012.09.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Lange EM, Lu L, Zheng SL, Wang Z, et al. HOXB13 is a susceptibility gene for prostate cancer: results from the International Consortium for Prostate Cancer Genetics (ICPCG) Hum Genet. 2013;132:5–14. doi: 10.1007/s00439-012-1229-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudmundsson J, Sulem P, Gudbjartsson DF, Masson G, Agnarsson BA, et al. A study based on whole-genome sequencing yields a rare variant at 8q24 associated with prostate cancer. Nat Genet. 2012;44:1326–9. doi: 10.1038/ng.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stott-Miller M, Karyadi DM, Smith T, Kwon EM, Kolb S, et al. HOXB13 mutations in a population-based, case–control study of prostate cancer. Prostatee-pub ahead of print 5 November 2012; doi:10.1002/pros.22604. [DOI] [PMC free article] [PubMed]
- Laitinen VH, Wahlfors T, Saaristo L, Rantapero T, Pelttari LM, et al. HOXB13 G84E mutation in Finland; population-based analysis of prostate, breast and colorectal cancer risk. Cancer Epidemiol Biomarkers Prev. 2013;22:452–60. doi: 10.1158/1055-9965.EPI-12-1000-T. [DOI] [PubMed] [Google Scholar]
- Kluźniak W, Wokołorczyk D, Kashyap A, Jakubowska A, Gronwald J, et al. The G84E mutation in the HOXB13 gene is associated with an increased risk of prostate cancer in Poland. Prostate. 2013;73:542–8. doi: 10.1002/pros.22594. [DOI] [PubMed] [Google Scholar]
- Chen Z, Greenwood C, Isaacs WB, Foulkes WD, Sun J, et al. The G84E mutation of HOXB13 is associated with increased risk for prostate cancer: results from the REDUCE trial. Carcinogenesise-pub ahead of print 11 March 2013. [DOI] [PMC free article] [PubMed]
- Lin X, Qu L, Chen Z, Xu C, Ye D, et al. A novel Germline mutation in HOXB13 is associated with prostate cancer risk in Chinese men. Prostate. 2013;73:169–75. doi: 10.1002/pros.22552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K. A comprehensive review of genetic association studies. Genet Med. 2002;4:45–61. doi: 10.1097/00125817-200203000-00002. [DOI] [PubMed] [Google Scholar]
- Amundadottir LT, Sulem P, Gudmundsson J, Helgason A, Baker A, et al. A common variant associated with prostate cancer in European and African populations. Nat Genet. 2006;38:652–8. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–9. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci USA. 2009;106:9362–7. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeager M, Orr N, Hayes RB, Jacobs KB, Kraft P, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645–9. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- Gudmundsson J, Sulem P, Manolescu A, Amundadottir LT, Gudbjartsson D, et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet. 2007;39:631–7. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- Gudmundsson J, Sulem P, Steinthorsdottir V, Bergthorsson JT, Thorleifsson G, et al. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat Genet. 2007;39:977–83. doi: 10.1038/ng2062. [DOI] [PubMed] [Google Scholar]
- Duggan D, Zheng SL, Knowlton M, Benitez D, Dimitrov L, et al. Two genome-wide association studies of aggressive prostate cancer implicate putative prostate tumor suppressor gene DAB2IP. J Natl Cancer Inst. 2007;99:1836–44. doi: 10.1093/jnci/djm250. [DOI] [PubMed] [Google Scholar]
- Thomas G, Jacobs KB, Yeager M, Kraft P, Wacholder S, et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008;40:310–5. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- Eeles RA, Kote-Jarai Z, Giles GG, Al Olama AA, Guy M, et al. Multiple newly identified loci associated with prostate cancer susceptibility. Nat Genet. 2008;40:316–21. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- Gudmundsson J, Sulem P, Rafnar T, Bergthorsson JT, Manolescu A, et al. Common sequence variants on 2p15 and Xp11.22 confer susceptibility to prostate cancer. Nat Genet. 2008;40:281–3. doi: 10.1038/ng.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Olama AA, Kote-Jarai Z, Giles GG, Guy M, Morrison J, et al. Multiple loci on 8q24 associated with prostate cancer susceptibility. Nat Genet. 2009;41:1058–60. doi: 10.1038/ng.452. [DOI] [PubMed] [Google Scholar]
- Yeager M, Chatterjee N, Ciampa J, Jacobs KB, Gonzalez-Bosquet J, et al. Identification of a new prostate cancer susceptibility locus on chromosome 8q24. Nat Genet. 2009;41:1055–7. doi: 10.1038/ng.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eeles RA, Kote-Jarai Z, Al Olama AA, Giles GG, Guy M, et al. Identification of seven new prostate cancer susceptibility loci through a genome-wide association study. Nat Genet. 2009;41:1116–21. doi: 10.1038/ng.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudmundsson J, Sulem P, Gudbjartsson DF, Blondal T, Gylfason A, et al. Genome-wide association and replication studies identify four variants associated with prostate cancer susceptibility. Nat Genet. 2009;41:1122–6. doi: 10.1038/ng.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher FR, Berndt SI, Siddiq A, Jacobs KB, Wang Z, et al. Genome-wide association study identifies new prostate cancer susceptibility loci. Hum Mol Genet. 2011;20:3867–75. doi: 10.1093/hmg/ddr295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kote-Jarai Z, Al Olama AA, Leongamornlert D, Tymrakiewicz M, Saunders E, et al. Identification of a novel prostate cancer susceptibility variant in the KLK3 gene transcript. Hum Genet. 2011;129:687–94. doi: 10.1007/s00439-011-0981-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haiman CA, Chen GK, Blot WJ, Strom SS, Berndt SI, et al. Genome-wide association study of prostate cancer in men of African ancestry identifies a susceptibility locus at 17q21. Nat Genet. 2011;43:570–3. doi: 10.1038/ng.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata R, Akamatsu S, Kubo M, Takahashi A, Hosono N, et al. Genome-wide association study identifies five new susceptibility loci for prostate cancer in the Japanese population. Nat Genet. 2010;42:751–4. doi: 10.1038/ng.635. [DOI] [PubMed] [Google Scholar]
- Akamatsu S, Takata R, Haiman CA, Takahashi A, Inoue T, et al. Common variants at 11q12, 10q26 and 3p11.2 are associated with prostate cancer susceptibility in Japanese. Nat Genet. 2012;44:426–9, S1. doi: 10.1038/ng.1104. [DOI] [PubMed] [Google Scholar]
- Xu J, Mo Z, Ye D, Wang M, Liu F, et al. Genome-wide association study in Chinese men identifies two new prostate cancer risk loci at 9q31.2 and 19q13.4. Nat Genet. 2012;44:1231–5. doi: 10.1038/ng.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SL, Stevens VL, Wiklund F, Isaacs SD, Sun J, et al. Two independent prostate cancer risk-associated Loci at 11q13. Cancer Epidemiol Biomarkers Prev. 2009;18:1815–20. doi: 10.1158/1055-9965.EPI-08-0983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Zheng SL, Wiklund F, Isaacs SD, Purcell LD, et al. Evidence for two independent prostate cancer risk-associated loci in the HNF1B gene at 17q12. Nat Genet. 2008;40:1153–5. doi: 10.1038/ng.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu FC, Sun J, Wiklund F, Isaacs SD, Wiley KE, et al. A novel prostate cancer susceptibility locus at 19q13. Cancer Res. 2009;69:2720–3. doi: 10.1158/0008-5472.CAN-08-3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Zheng SL, Wiklund F, Isaacs SD, Li G, et al. Sequence variants at 22q13 are associated with prostate cancer risk. Cancer Res. 2009;69:10–5. doi: 10.1158/0008-5472.CAN-08-3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SL, Sun J, Wiklund F, Smith S, Stattin P, et al. Cumulative association of five genetic variants with prostate cancer. N Engl J Med. 2008;358:910–9. doi: 10.1056/NEJMoa075819. [DOI] [PubMed] [Google Scholar]
- Xu J, Sun J, Kader AK, Lindström S, Wiklund F, et al. Estimation of absolute risk for prostate cancer using genetic markers and family history. Prostate. 2009;69:1565–72. doi: 10.1002/pros.21002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinas CA, Koopmeiners JS, Kwon EM, FitzGerald L, Lin DW, et al. Clinical utility of five genetic variants for predicting prostate cancer risk and mortality. Prostate. 2009;69:363–72. doi: 10.1002/pros.20887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kote-Jarai Z, Easton DF, Stanford JL, Ostrander EA, Schleutker J, et al. Multiple novel prostate cancer predisposition loci confirmed by an international study: the PRACTICAL Consortium. Cancer Epidemiol Biomarkers Prev. 2008;17:2052–61. doi: 10.1158/1055-9965.EPI-08-0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Kader AK, Hsu FC, Kim ST, Zhu Y, et al. Inherited genetic markers discovered to date are able to identify a significant number of men at considerably elevated risk for prostate cancer. Prostate. 2011;71:421–30. doi: 10.1002/pros.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindström S, Schumacher FR, Cox D, Travis RC, Albanes D, et al. Common genetic variants in prostate cancer risk prediction—results from the NCI Breast and Prostate Cancer Cohort Consortium (BPC3) Cancer Epidemiol Biomarkers Prev. 2012;21:437–44. doi: 10.1158/1055-9965.EPI-11-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein RJ, Hallden C, Gupta A, Savage CJ, Dahlin A, et al. Evaluation of multiple risk-associated single nucleotide polymorphisms versus prostate-specific antigen at baseline to predict prostate cancer in unscreened men. Eur Urol. 2012;61:471–7. doi: 10.1016/j.eururo.2011.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akamatsu S, Takahashi A, Takata R, Kubo M, Inoue T, et al. Reproducibility, performance, and clinical utility of a genetic risk prediction model for prostate cancer in Japanese. PLoS One. 2012;7:e46454. doi: 10.1371/journal.pone.0046454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Liu F, Lin X, Wang X, Ding Q, et al. Predictive performance of prostate cancer risk in Chinese men using 33 reported prostate cancer risk-associated SNPs. Prostate. 2012;72:577–83. doi: 10.1002/pros.21462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aly M, Wiklund F, Xu J, Isaacs WB, Eklund M, et al. Polygenic risk score improves prostate cancer risk prediction: results from the Stockholm-1 cohort study. Eur Urol. 2011;60:21–8. doi: 10.1016/j.eururo.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kader AK, Sun J, Reck BH, Newcombe PJ, Kim ST, et al. Potential impact of adding genetic markers to clinical parameters in predicting prostate biopsy outcomes in men following an initial negative biopsy: findings from the REDUCE trial. Eur Urol. 2012;62:953–61. doi: 10.1016/j.eururo.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Liu F, Wang Z, Na R, Zhang L, et al. Prediction of prostate cancer from prostate biopsy in Chinese men using a genetic score derived from 24 prostate cancer risk-associated SNPs. Prostate 2013. In press. [DOI] [PMC free article] [PubMed]
- Andriole GA, Bostwick D, Brawley OW, Gomella LG, Marberger M, et al. The influence of dutasteride on the risk of biopsy-detectable prostate cancer: outcomes of the REduction by DUtasteride of Prostate Cancer Events (REDUCE) study. N Engl J Med. 2010;362:1192–202. doi: 10.1056/NEJMoa0908127. [DOI] [PubMed] [Google Scholar]
- Liu F, Hsing AW, Wang X, Shao Q, Qi J, et al. Systematic confirmation study of reported prostate cancer risk-associated single nucleotide polymorphisms in Chinese men. Cancer Sci. 2011;102:1916–20. doi: 10.1111/j.1349-7006.2011.02036.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Liu F, Hsing AW, Wang X, Shao Q, et al. Replication and cumulative effects of GWAS-identified genetic variations for prostate cancer in Asians: a case–control study in the ChinaPCa consortium. Carcinogenesis. 2012;33:356–60. doi: 10.1093/carcin/bgr279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na R, Liu F, Zhang P, Ye D, Xu C, et al. Evaluation of reported prostate cancer risk-associated SNPs from genome-wide association studies of various racial populations in Chinese men. Prostate 2013. In press. [DOI] [PMC free article] [PubMed]
- Turner AR, Kader AK, Xu J. Utility of genome-wide association study findings: prostate cancer as a translational research paradigm. J Intern Med. 2012;271:344–52. doi: 10.1111/j.1365-2796.2012.02522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kader AK, Xu J. Reply from Authors re: Matthew R. Cooperberg. Will biomarkers save prostate cancer screening. Eur Urol. 2012;62:962–3. doi: 10.1016/j.eururo.2012.06.034. [DOI] [PubMed] [Google Scholar]
- Sun J, Na R, Hsu FC, Zheng SL, Wiklund F, et al. Genetic score is an objective and better measurement of inherited risk of prostate cancer than family history. Eur Urol. 2013;63:585–7. doi: 10.1016/j.eururo.2012.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


