Abstract
Objectives
The goal of this study was to characterize the regulation of heme and non-heme iron in human failing hearts.
Background
Iron is an essential molecule for cellular physiology, but in excess it facilitates oxidative stress. Mitochondria are the key regulators of iron homeostasis through heme and iron-sulfur cluster synthesis. Since mitochondrial function is depressed in failing hearts and iron accumulation can lead to oxidative stress, we hypothesized that iron regulation may also be impaired in heart failure (HF).
Methods
We measured mitochondrial and cytosolic heme and non-heme iron levels in failing human hearts retrieved during cardiac transplant surgery. In addition, we examined the expression of genes regulating cellular iron homeostasis, heme biosynthetic pathway, and microRNAs that may potentially target iron regulatory networks.
Results
While cytosolic non-heme iron levels were reduced in HF, mitochondrial iron content was maintained. Moreover, we observed a significant increase in heme levels in failing hearts, with corresponding feedback inhibition of the heme synthetic enzymes and no change in heme degradation. The rate-limiting enzyme in heme synthesis, δ-aminolevulinic acid synthase 2 (ALAS2), was significantly upregulated in HF. Overexpression of ALAS2 in H9c2 cardiac myoblasts resulted in increased heme levels, and hypoxia and erythropoietin treatment increased heme production through upregulation of ALAS2. Finally, increased heme levels in cardiac myoblasts were associated with excess production of reactive oxygen species and cell death, suggesting a maladaptive role for increased heme in HF.
Conclusions
Despite global mitochondrial dysfunction, heme levels are maintained above baseline in human failing hearts.
Keywords: heme, ALAS2, mitochondria, iron, heart failure
INTRODUCTION
Heart failure (HF) rates have soared over the last decade, with almost 6 million Americans affected and the prevalence of the disease rising steadily (1). Molecular mechanisms of HF are complex (2), but mitochondrial dysfunction is an early and common finding in hypertrophied and failing hearts (3). Virtually all aspects of mitochondrial physiology are deranged in HF (4), including mitochondrial number and biogenesis (5), energetic capacity (6), and production of reactive oxygen species (ROS) (7) . In addition to generating ATP, mitochondria play a major role in the regulation of iron balance and synthesis of heme and iron-sulfur (Fe/S) clusters (8, 9); however these processes have not been systematically examined in failing hearts.
Indirect evidence points at a link between cellular and mitochondrial iron regulation and HF. Iron is an essential molecule that functions as a cofactor in mitochondrial cytochromes, antioxidant enzymes and more, and its deficiency is associated with cardiomyopathy (10). On the other hand, the redox properties of iron make it an ideal catalyst for the production of toxic hydroxyl radical (OH·) by the Fenton reaction (11), and cardiac dysfunction is a prominent feature of iron overload diseases, such as hemochromatosis (12). Elevated iron levels were noted in the hearts of mice overexpressing the α subunit of the Gq protein, a model of cardiomyopathy (13); however, cellular distribution of iron has not been assessed. Recently, we have shown that accumulation of iron specifically in the mitochondria through a reduction in the levels of ATP-binding cassette transporter B8 (ABCB8), a protein involved in mitochondrial iron export, leads to the development of cardiomyopathy in mice (14). Similarly, aggregation of iron inside the mitochondria has been observed in the hearts of Friedreich’s ataxia patients (15), who develop progressive and lethal cardiac dysfunction (16). Thus, maintenance of iron balance inside the heart appears to be critical for its function, but it remains unknown how iron regulation is altered in failing human hearts.
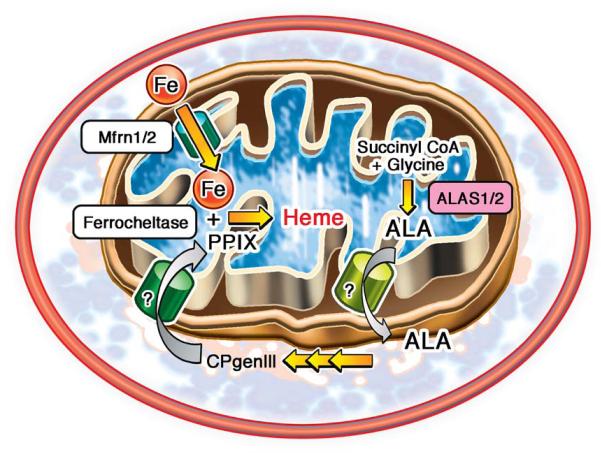
Iron enters the mitochondria through an inner-membrane transporter mitoferrin 2 (Mfrn2) (17, 18), and can either be stored in a complex with mitoferritin (FtMt), or used in heme and Fe/S cluster biosynthetic pathways (19). Heme production begins in the mitochondria with condensation of glycine and succinyl CoA to form δ-aminolevulinic acid (ALA) by the rate-limiting enzyme ALA synthase (ALAS) (9). The next five conversions are carried out in the cytosol. Finally, the synthesis is completed in the mitochondria with insertion of an iron atom into protoporphyrin IX (PPIX) to form heme by ferrochelatase (9) (Figure 1). In the heart, heme functions as a catalytic or structural subunit of mitochondrial electron transport chain (ETC) complexes, myoglobin, antioxidant enzymes and components of the cytochrome p450 (20). Moreover, heme can be broken down by heme oxygenases (HMOX) into elemental iron, carbon monoxide and cardioprotective antioxidant biliverdin (21). Despite the essential role heme plays in the heart, this molecule remains understudied outside of the erythropoietic system.
Figure 1.
Schematic representation of cellular heme synthesis pathway
Fe/S cluster assembly requires over 20 enzymes and scaffolding proteins, and takes place primarily in the mitochondria (8). Fe/S clusters are incorporated into mitochondrial, cytoplasmic and nuclear proteins involved in oxidative phosphorylation, DNA repair, purine metabolism and heme production (22, 23). Disruption of Fe/S cluster synthesis may also lead to cardiomyopathy, as noted in patients with FRDA (24) and mice with deletion of ABCB8 (14), although these conditions are also associated with mitochondrial iron accumulation and ROS.
Given the critical role mitochondria play both in iron homeostasis and in HF, we aimed to characterize the changes in cytosolic and mitochondrial heme and non-heme iron regulation in human failing hearts. Our results demonstrate that both cytosolic and mitochondrial heme levels are increased in failing hearts, with feedback inhibition of the heme synthetic enzymes, except for ALAS2, which is increased in heart failure and whose expression was previously reported to be restricted to hematopoietic system. We also show that in H9c2 cardiac myoblasts, ALAS2 expression and heme levels are regulated by hypoxia and erythropoietin (EPO), the two pathways that are often altered in failing hearts. Furthermore, we demonstrate that increased heme levels are associated with elevated oxidative stress and loss of viability in cultured cardiomyoblasts.
METHODS
Human samples
Tissue samples were obtained from the tissue bank at Feinberg Cardiovascular Research Institute (Northwestern University) and consisted of samples from non-failing (n=10) and failing ischemic (n=10) human hearts. Failing ischemic tissues were obtained from the explanted hearts of cardiac transplant recipients. Non-failing heart tissue samples were obtained from unmatched organ donors whose hearts were unsuitable for transplantation but had no known cardiac disease. Explanted hearts were immediately placed in cold cardioplegic solution and subsequently frozen in liquid nitrogen. Protocols for tissue procurement were approved by the Institutional Review Board of the Northwestern University. Informed consent was obtained from all transplant patients and from the families of organ donors before tissue collection.
Cell Culture
H9c2 cardiac myoblasts were purchased from ATCC and kept in complete DMEM medium (ATCC) supplemented with 10% fetal bovine serum (Invitrogen) and 1% penicillin-streptomycin (P/S). For hypoxic experiments, cells were maintained in a hypoxic chamber at 37°C, 5%CO2 in the presence of 1% O2 for up to 8 days. Medium was replaced every two days and cells were collected under hypoxia prior to the analysis. For pharmacologic treatments, cells were grown to 80-90% confluence and incubated with 10 μM hemin (Sigma-Aldrich) and 0.6mg/mL erythropoietin (Sigma-Aldrich) for 48 hours in complete medium.
Mitochondrial fractionation
For tissue samples, we used the Mitochondria Isolation Kit for Tissue (Pierce) according to the manufacturer’s dounce homogenization protocol for hard tissue. Homogenized tissue was treated with reagents in the presence of EDTA-free protease inhibitors (Protease Arrest, G-Biosciences). To maintain the integrity of the mitochondria, samples were kept on ice during the isolation process. Following differential centrifugation at 4°C, the supernatant (cytosolic fraction) was collected, and the remaining pellet containing the mitochondria-enriched fraction was dissolved in 1% Triton X-100 (Sigma-Aldrich) in Tris buffered saline (Cellgro).
Non-heme iron measurement
Mitochondrial and cytosolic non-heme iron was quantified using a commercial Iron Assay Kit (BioVision). Briefly, tissue lysate was mixed with acidic solution to release protein-bound iron, followed by a reduction of iron to its ferrous form and incubation with Ferene S compound to produce a colored complex. The absorbance was then measured on Spectra Max Plus microplate reader at 593 nm and normalized to protein concentration of each sample.
Heme iron measurement
For determination of total heme levels, ~5 mg of frozen tissue was homogenized in 1% Triton-X100 in TBS and centrifuged at 5,000 × g for 10 minutes to remove debris. For determination of mitochondrial and cytosolic heme levels, mitochondrial fraction was isolated using Mitochondrial Isolation Kit for Tissue (Pierce) according to the manufacturer’s protocol. Protein concentration of cytosolic or mitochondrial lysate was quantified by BCA assay (Pierce, IL) and heme was quantified as described (25). Briefly, equal amounts of protein were mixed with 2 M oxalic acid, heated to 95°C for 30 minutes to release iron from heme and generate fluorescent protoporphyrin IX. Samples were then centrifuged for 10 min at 1,000 × g at 4°C to remove debris; the fluorescence of the supernatant was assessed at 405 nm / 600 nm on Spectra Max Gemini fluorescence microplate reader and normalized to protein concentration of each sample. For determination of unsaturated protoporphyrin IX levels, incubation and heating with oxalic acid steps were omitted. Instead, samples were diluted in PBS, followed by fluorescence measurement and normalization to protein content.
Knockdown and overexpression of ALAS2
For knockdown experiments, ALAS2 and control non-targeting siRNA (Thermo Scientific) were transfected into H9c2 cells using Dharmafect I reagent (Thermo Scientific) for 48 hours according to the manufacturer’s protocol. For hypoxia experiments, transfections were repeated every 48 hours to maintain low levels of ALAS2 expression throughout the time course of the study. Knockdown efficiency was confirmed by qRT-PCR and Western blot analyses.
ALAS2 overexpression was achieved through lentiviral transduction. Lentiviral particles coding for ALAS2-GFP fusion protein or GFP-only control were transduced into H9c2 cells at equal multiplicity of infection for 48 hours and overexpression was confirmed by Western blotting.
Quantitative Real-Time PCR
RNA was isolated with RNA STAT-60 (TEL-TEST, Inc), reverse-transcribed with a Random Hexamer (Applied Biosystems), and amplified on a 7500 Fast Real-Time PCR system with SYBR Green PCR Master Mix (Applied Biosystems). Primers were designed using Primer3 (v. 0.4.0) software to target sequences spanning an exon-intron-exon boundary and their specificity was confirmed by running a dissociation curve. mRNA levels were calculated by the comparative threshold cycle method and normalized to β-actin gene.
MitoDNA/genomic DNA
Total DNA was isolated using the DNeasy Blood & Tissue Kit (Qiagen) and the ratio of mitochondrially-encoded COX I to the genomic 18S gene was determined by quantitative RT-PCR using SYBR Green PCR Master Mix (Applied Biosystems).
Western Blot
~ 5mg of tissue was homogenized in RIPA buffer (Thermo Scientific) in the presence of Protease Arrest protease inhibitors (G-Biosciences), centrifuged at 5,000 × g for 15 minutes to remove debris, and protein concentration of the supernatant determined by BCA assay. Fifteen-30 μg of protein were resolved on SDS-PAGE gels and transferred to nitrocellulose membranes (Invitrogen). The membranes were probed with antibodies against ferritin light and heavy chains (Sigma-Aldrich), HMOX1 (Abcam), ALAS1/2 (Abcam), ferrochelatase (Proteintech), phosphor-JAK2 (Tyr1007/1008), JAK2 (Millipore), NF-E2 (Proteintech), GAPDH (Santa Cruz) and tubulin (Abcam). HRP-conjugated donkey anti-rabbit and donkey anti-mouse were used as secondary antibodies (Santa Cruz) and visualized by Pierce SuperSignal Chemiluminescent Substrates.
Complex IV activity
Complex IV activity was determined using Complex IV Human Enzyme Activity Microplate Assay Kit (Abcam) according to the manufacturer’s protocol. Briefly, complex IV was immobilized in the wells of a 96-well plate by immunocapturing and the specific activity was determined colorimetrically as oxidation of cytochrome c, the substrate of complex IV, by measuring absorbance change at 550 nm for 2 hours at 30°C at 5-min intervals.
Mitochondrial ROS Quantification
MitoSox Red (Invitrogen) was used to assess mitochondrial O2• production (26). Cells were visualized by microscopy and ROS levels were quantified by ImageJ software. Four fields per each sample were obtained and averaged. Nuclei were counterstained with Hoescht 33342 dye (Invitrogen) and subtracted from the total Mitosox fluorescence to exclude the signal from localization of dye into the nucleus. An accurate overlay between Hoescht and nuclear MitoSox dye was achieved by adjusting microscopy settings prior to data collection, with representative single-channel and overlay images presented in Figure S1A. In addition, total MitoSox fluorescence without nuclear signal subtraction was quantified by ImageJ.
Cell Death Analysis
Following ALAS2 overexpression or hemin treatment, cells were double-labeled with propidium iodine (PI, Sigma-Aldrich, USA) and Alexa Fluor® 350-conjugated Annexin V (Molecular Probes, NY), and analyzed by flow cytometry in a FacsCanto flow cytometer (BD Bioscience). The data are presented as the sum of apoptotic and necrotic cells normalized to the control.
MicroRNA Experiments
Taqman microRNA assays (Applied Biosystems) were used to quantify miR144, 145, 148a, 148b and 152 levels according to the manufacturer protocol. Levels of these microRNAs in failing hearts were quantified by qRT-PCR and normalized to the expression of small RNA U6. To overexpress miR145 we used Pre-miR miRNA Precursors, and to downregulate miR145 we used Anti-miR miRNA Inhibitors (Ambion). Scrambled Pre-miR and anti-scrambled Anti-miR were used in control experiments, respectively.
Statistical Analysis
Data are expressed as mean ± SEM. Statistical significance was assessed with the unpaired Student’s t-test; a P value of less than 0.05 was considered statistically significant.
RESULTS
Heme is increased in failing hearts
Mitochondria are the primary site of heme and iron-sulfur cluster synthesis, and their function is severely impaired in heart failure (Figure 1). In this study, we characterized changes in cellular and mitochondrial iron homeostasis in failing human hearts. We used left ventricular myocardial tissue from ten patients with terminal heart failure obtained during the heart transplant surgery and ten non-myopathic hearts as controls. Consistent with mitochondrial dysfunction invariably present in HF (27, 28), the mitochondrial DNA copy number and the activity of mitochondrial complex IV were reduced in failing hearts compared to the controls (Figure S1B,C) , thus providing molecular confirmation of the pathology report.
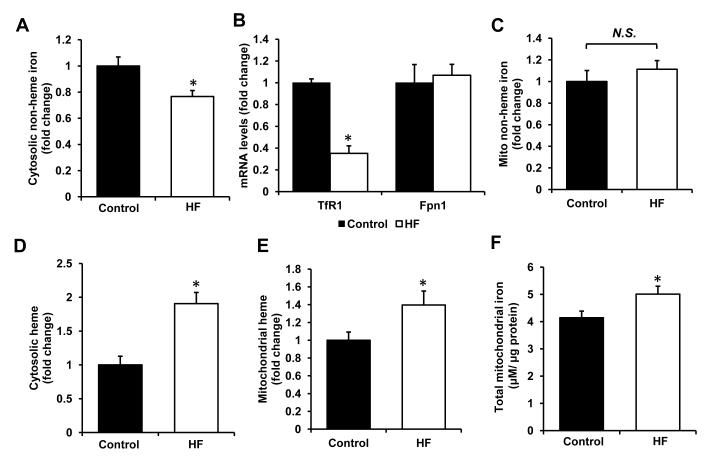
We next measured the levels of non-heme and heme iron in these hearts. There was a significant decrease in cytosolic levels of non-heme iron (Figure 2A) and a trend towards a decrease in the levels of cytosolic iron storage protein ferritin (heavy and light chains, Figure S2A,B). Moreover, mRNA levels of transferrin receptor 1 (TfR1), a protein facilitating cellular iron import, were greatly reduced, while the expression of ferroportin 1 (Fpn1), an iron exporter remained stable (Figure 2B), suggesting that the observed deficiency may be due to a reduction in iron import in cardiomyocytes. However, mitochondrial non-heme iron content was unaltered in failing hearts (Figure 2C), suggesting that the delivery of iron to the mitochondria remains intact in HF.
Figure 2. Heme and non-heme iron regulation in HF.
(A) Cytosolic non-heme iron levels in control and failing hearts normalized to the cytosolic protein concentration (n=10). (B) mRNA expression of TfR1, an iron importer, and Fpn1, an iron exporter, in HF and control groups (n=10). (C) Non-heme iron levels in the mitochondria of control and failing hearts normalized to mitochondrial protein content (n=10). Cytosolic (D) and mitochondrial (E) heme levels in control and failing hearts normalized to the protein concentration of cytosolic and mitochondrial fractions, respectively (n=10). (F) Total mitochondrial iron levels, obtained by addition of non-heme and heme iron content (n=10). Data are presented as mean ± SEM. * p<0.05 vs. control.
Heme levels were significantly increased in the cytosolic (Figure 2D) and mitochondrial (Figure 2E) fractions in failing hearts, as determined by the increase in fluorescence of protoporphyrin IX (PPIX) ring, the last synthetic intermediate in the heme synthesis pathway, (25). The observed increase was not due to accumulation of unsaturated PPIX, as the levels of iron-free PPIX were comparable between the two groups (Figure S2C). Thus, even though mitochondrial biogenesis and energetic function are depressed in HF, mitochondrial iron levels (Figure 2F) (obtained by the addition of heme and non-heme mitochondrial iron contents) and heme content in this organelle are increased in the failing hearts.
ALAS2 expression is induced in HF
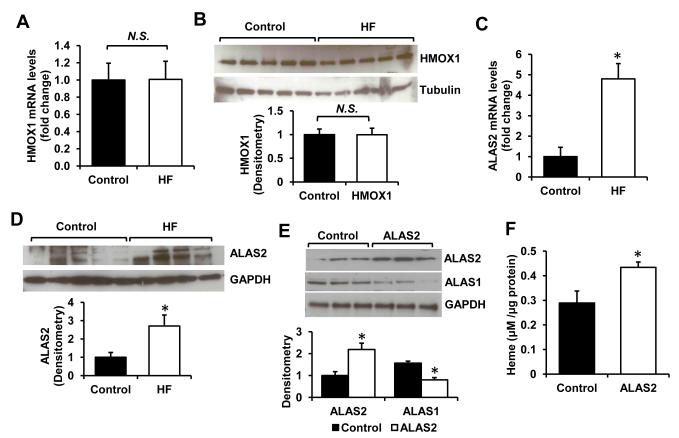
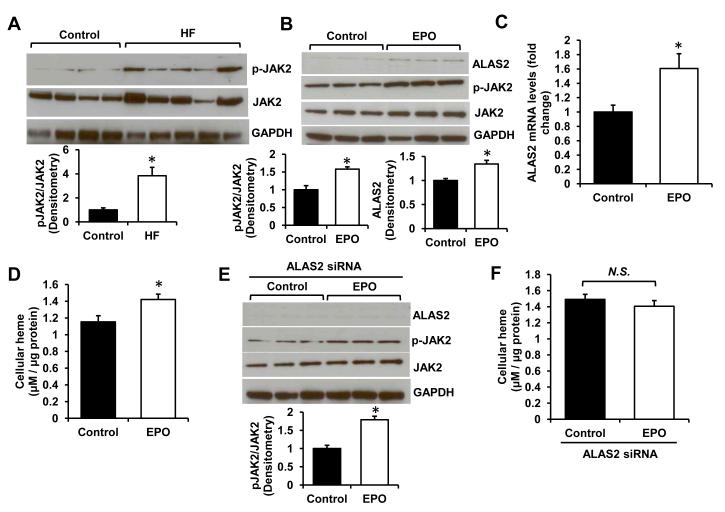
To understand the mechanism of increased heme levels in HF, we assessed the expression of genes directly involved in breakdown or synthesis of heme. The levels of heme oxygenase 1 and 2 (HMOX1/2), the enzymes catalyzing degradation of heme, were not altered (Figure 3A,B, Figure S3A). These results indicate that the increase in heme levels in HF is not likely to be caused by impaired degradation of heme. Many of the enzymes involved in heme synthesis, including ferrochelatase responsible for iron insertion in PPIX, were actually downregulated in the failing hearts (Figure S3B,C), suggestive of a feedback inhibition on the pathway by high cellular levels of heme as have been previously reported (9). However, mRNA and protein levels of ALAS2, a rate-limiting enzyme catalyzing the first committed step in heme synthesis (29), were significantly upregulated in the failing hearts (Figure 3C,D), providing a potential mechanism for heme accumulation in HF.
Figure 3. ALAS2 is upregulated in failing hearts.
mRNA (A) and protein (B) levels of HMOX1 in control and failing hearts (n=5-6). mRNA (C) and protein levels (D) of ALAS2 in control and failing hearts (n=4-6). (E) Westen blot analysis of ALAS1/2 proteins in H9c2 with lentiviral overexpression of ALAS2 enzyme (n=3). Densitometry analyses are presented below the Western blots. (F) Heme levels in H9c2 cells transduced with ALAS2 or control lentiviral vector (n=6). Data are presented as mean ± SEM. * p<0.05 vs. control.
The upregulation of ALAS2 in HF is surprising, since the expression of this isoform was previously reported to be restricted to a hematopoietic cell lineage, and it is not known whether upregulation of ALAS2 in the heart can affect heme levels. To test this, we conducted in vitro experiments in H9c2 cardiac myoblasts. Lentivirus-driven overexpression of human ALAS2 in H9c2 cells resulted in a ~2-fold increase in its protein levels (Figure 3E). Consistent with our findings in human hearts, heme content was elevated in ALAS2-overexpressing cells (Figure 3F). Moreover, mRNA and protein levels of ALAS1, the non-erthroid isoform of the enzyme, were decreased with ALAS2 overexpression (Figure 3E and S3D), consistent with the feedback inhibition by high intracellular heme content (30).
Regulation of ALAS2 in cardiac cells
Mechanisms for regulation of ALAS2 in hematopoietic cells have been extensively characterized, but it is unknown how ALAS2 is regulated in the heart. The major inducer of ALAS2 in developing erythrocytes is GATA1 transcription factor. However, qRT-PCR analysis revealed no expression of GATA1 in human hearts (qRT-PCR cycle 34.85 ± 0.32). In contrast, protein levels of NF-E2, an erythroid transcription factor that has putative binding sites in the promoter of ALAS2 (31), were significantly higher in the HF group compared to control (Figure S4A,B).
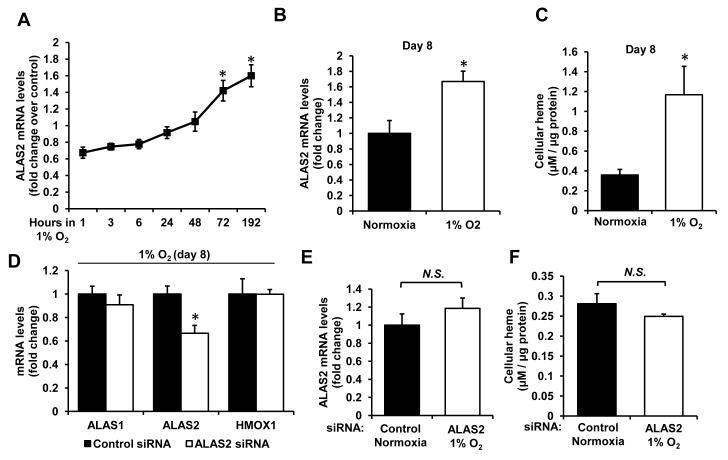
Since ischemia is common in failing hearts and ALAS2 was shown to be regulated by hypoxia-inducible factor 1α in erythroid lineage (32), we assessed regulation of ALAS2 by low oxygen tension by subjecting H9c2 cells to 1% O2 levels. We found ALAS2 expression to be significantly induced by chronic (8-day), but not acute exposure to hypoxia (Figure 4A,B), with a corresponding increase in cellular heme content at day 8 (Figure 4C). Treatment of cells with ALAS2 siRNA during hypoxic incubation effectively ablated an increase in ALAS2 mRNA levels, bringing it down to the normoxic levels (Figure 4D,E), and also prevented the increase in cellular heme (Figure 4F). These data suggest that long-standing ischemia of failing hearts may potentially be responsible for the induction of ALAS2, and provide further evidence that upregulation of ALAS2 in HF may mediate the increase in heme levels.
Figure 4. Regulation of ALAS2 by hypoxia in cardiac myoblasts.
(A) Time-course of ALAS2 mRNA expression in H9c2 cardiac myoblasts subjected to hypoxia (n=6). (B) ALAS2 mRNA levels after 8 days in hypoxia or normoxia (n=6). (C) Cellular heme content after 8 days in hypoxia or normoxia (n=6). (D) mRNA levels of heme synthesis/degradation enzymes in hypoxic cardiac myoblasts with or without siRNA-mediated ALAS2 knockdown (n=6). (E) Comparison of ALAS2 mRNA levels in control normoxic and ALAS2 siRNA-treated hypoxic cells after 8 days (n=6). (F) Heme levels in control normoxic and ALAS2 siRNA hypoxic cells after 8 days (n=6). Data are presented as mean ± SEM. * p<0.05 vs. control.
Finally, ALAS2 is known to be positively regulated by erythropoietin (EPO) in erythroblasts (33) and EPO levels are often elevated in HF patients due to the underlying anemia (34). EPO receptor was previously shown to be expressed in the human heart (35), suggesting that cardiomyocytes may respond to EPO signaling. Western blot analysis of JAK2 kinase, the downstream effector of EPO signaling (36), revealed elevated phosphorylation of this protein consistent with activation of EPO receptor (Figure 5A). Incubation of H9c2 myoblasts with EPO also significantly increased p-JAK2 levels (Figure 5B) and was associated with induction of ALAS2 enzyme on mRNA and protein levels (Figure 5B,C). Heme content was increased in EPO-treated cells (Figure 5D), while suppression of ALAS2 upregulation by EPO using siRNA reversed this increase (Figure 5E,F). Thus, similar to erythropoetic cells, cardiac ALAS2 is responsive to EPO stimulation, which cases an increase in heme levels and may potentially contribute to cardiac heme loading in HF, particularly in the setting of underlying anemia. In summary, we found ALAS2 to be positively regulated by hypoxia and EPO in cardiac cells.
Figure 5. ALAS2 is regulated by erythropoietin.
(A) Western blot analysis of p-JAK2 and JAK2 protein in failing and control hearts (n=4-5). (B) ALAS2 and pJAK2/JAK2 protein levels in H9c2 cells treated with 0.6mg/mL EPO (n=3). (C) qRT-PCR analysis of ALAS2 mRNA expression in H9c2 cells with EPO or vehicle control treatment (n=6). (D) Cellular heme content with EPO treatment in cardiac myoblasts (n=6). (E) Western blot analysis of ALAS2 and pJAK2/JAK2 in ALAS2 siRNA-treated cells incubated with EPO or vehicle control (n=3). (F) Cellular heme content in EPO- or vehicle-treated cells with ALAS2 siRNA knockdown (n=6). Data are presented as mean ± SEM. * p<0.05 vs. control.
Heme and oxidative stress
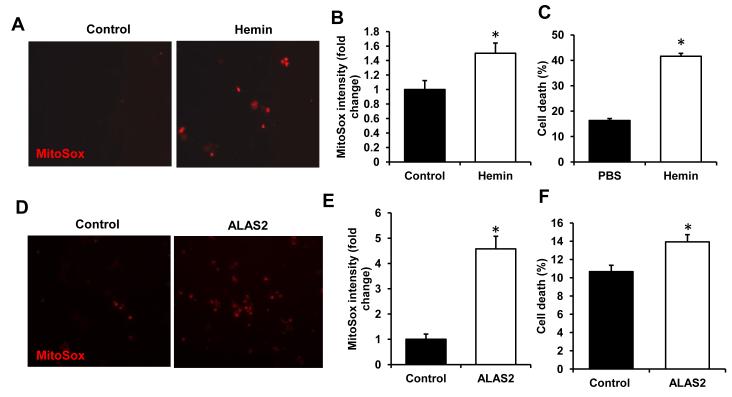
Heart failure is characterized by increased oxidative stress, which significantly disrupts myocardial architecture and signaling (37). Free heme has been suggested to exert toxic effects through oxidation of phospholipids, mtDNA damage, and activation of inflammatory response (38). To determine if excess levels of heme contribute to oxidative stress in cardiac myoblasts, we incubated H9c2 cells with hemin and measured mitochondrial ROS production and cell death using using MitoSox and propidium iodine staining, respectively. Addition of hemin led to a significant increase in ROS levels (Figure 6A,B, Figure S4C) and was associated with the loss of cell viability (Figure 6C). Overexpression of ALAS2 in H9c2 also induced an increase in ROS (Figure 6D,E, Figure S4D) and was associated with increased cell death (Figure 6F). These data suggest that elevated heme production in HF may be maladaptive through enhancing oxidative stress in these hearts.
Figure 6. Heme and ALAS2 overexpression increase oxidative stress.
(A) Representative images of H9c2 incubated with the vehicle control or 10μM hemin for 6 hours and stained with mitochondria-specific ROS-sensitive dye MitoSox. (B) Image J analysis of MitoSox-stained H9c2 following the treatment with vehicle or hemin (n=4-5, 4 fields per sample). (C) Cell death assessed by propidium iodine/Annexin V double-labeling with hemin treatment as in A (n=6). (D) Representative MitoSox images of H9c2 with or without ALAS2 overexpression. (E) Quantification of MitoSox staining with ALAS2 overexpression (n=4-5, 4 fields per sample). (F) Cell death with ALAS2 overexpression assessed as in C (n=6). Data are presented as mean ± SEM. * p<0.05 vs. control.
MiR-145 does not regulate iron homeostasis in HF
MicroRNA are small non-coding RNA molecules that regulate expression of multiple genes by targeting their 3′ untranslated regions (3′UTR) (39). We and others have shown that HF is associated with reduced levels of TfR1, the protein responsible for iron uptake by cardiomyocytes (40). To determine if suppression of TfR1 in HF is mediated by a microRNA, we screened the 3′UTR of TfR1 for putative miR target sequences and the expression of these microRNAs was determined using qRT-PCR (Figure S5A). A significant upregulation of miR-145 was noted in the failing hearts (Figure S5A).
To determine if miR-145 is responsible for the changes in iron homeostasis observed earlier, we overexpressed (Figure S5B) and downregulated (Figure S5C) this miR in H9c2 cardiac myoblasts, followed by the measurement of heme and non-heme iron levels. However, neither of the interventions led to significant changes in heme and non-heme iron content (Figure S5D-G). To determine if miR-145 expression was induced secondarily to the elevated heme levels, we treated H9c2 cells with ALA, the rate-limiting intermediate in heme synthesis pathway, and observed a significant increase in heme levels (Figure S5H), but no change in miR145 expression (Figure S5I). Taken together, these results show that although miR-145 is significantly upregulated in the failing hearts, it does not mediate changes in heme and non-heme iron homeostasis observed in HF and is not regulated by cellular heme levels.
DISCUSSION
The goal of this study was to characterize changes in iron homeostasis occurring in failing human hearts. In spite of mitochondrial dysfunction characteristic of HF, mitochondria-dependent process of heme synthesis was significantly upregulated in failing hearts. We also observed induction of ALAS2, a rate-limiting enzyme in heme synthesis that was previously reported to be expressed exclusively in hematopoietic cells. Moreover, mitochondrial iron levels were maintained at normal levels and were sufficient to support heme synthesis, as we detected very low levels of unsaturated PPIX in the HF group.
It is presently unclear whether the increase in heme levels in HF is an adaptive or maladaptive process. Heme degradation by HMOX1 confers cardiac protection through generation of an antioxidant biliverdin and an anti-inflammatory molecule carbon monoxide (CO) (29). Indeed, induction of HMOX1 was found to be beneficial in HF (41, 42). However, free heme is toxic to the cell as it catalyzes oxidation and breakdown of proteins and DNA, intercalates into lipid bilayers, and damages membrane-bound organelles (38). Similarly, we found that treatment of cardiac myoblasts with hemin significantly elevated ROS generation in the mitochondria and led to a loss in cell viability. Similarly, overexpression of ALAS2, which increases heme content in H9c2 cells, increased both ROS and cell death. On contrary, the levels of HMOX1 were unaltered in the HF samples we examined, suggesting that HMOX1-dependent antioxidation is unlikely to protect the hearts from heme toxicity. Based on the in vitro data, we speculate that the induction of heme synthesis in these hearts may be maladaptive through an increase in oxidative stress. Although our data suggests that the use of iron supplementation and stimulants of erythropoietin in anemic heart failure patients, which yielded controversial results (43), may negatively affect cardiomyocytes through induction of ALAS2 and heme synthesis, potential benefits of iron supplementation, as demonstrated before (44), on other organs and hematopoietic system cannot be excluded. It is important to note, however, that our data only provide an association between induction of ALAS2, elevated cardiac heme levels and heart failure. Generation of a cardiac-specific ALAS2-overexpressing mouse model will allow testing this hypothesis directly.
While our experimental design does not identify the mechanism for increased heme synthesis and ALAS2 induction, we hypothesize that chronic ischemia and EPO may play a role. Two isoforms of ALAS enzyme have been identified. ALAS1 is a ubiquitously expressed housekeeping gene and is negatively regulated by heme (45, 46). On contrary, ALAS2 expression is traditionally studied in the context of erythrocyte maturation, where it is induced to stimulate hemoglobinization (47, 48). Unlike other components of heme synthesis pathway, including ALAS1, ALAS2 is not inhibited by high levels of heme, and is positively regulated by hypoxia (32, 49). Our studies show that ALAS2 is expressed in the heart and is significantly upregulated in HF, providing the first evidence of a heart-specific function for this “non-cardiac” isoform of ALAS. We confirm that, similar to erythroid cells, ALAS2 is regulated by hypoxia in cardiac myoblasts, and that the increase in heme observed in cells subjected to chronic hypoxia is mediated by ALAS2 induction, as knockdown of this protein has also abolished an increase in heme. Moreover, we found that, similar to developing erythrocytes, EPO stimulation induced ALAS2 expression and subsequently elevated heme levels. The significance of these findings in the context of failing hearts remains to be determined. Despite elevated EPO levels, HF patients were found to be resistant to EPO signaling in promoting hematopoiesis (34), but it remains unknown whether the sensitivity of cardiomyocytes to EPO stimulation is also altered.
In addition to showing an increase in heme content in HF, we find that miR-145 is strongly induced in the failing human hearts, but it does not mediate the changes in heme that were observed in HF. MiR-145 is known to play a role in smooth muscle differentiation (50, 51) and also exhibits tumor suppressor activity (52, 53). To the best of our knowledge, this is the first report of miR145 upregulation in heart failure and, naturally, it would be of interest to see if this induction has any functional consequences.
We show that mitochondrial iron levels are increased in the failing hearts due to an increase in heme content, suggesting that mitochondrial iron homeostasis is maintained in spite of severe mitochondrial dysfunction. We would like to note, however, that preservation of mitochondrial iron content does not necessarily imply that all iron-dependent processes remain unaffected. Specifically, it remains to be determined how HF affects the process of Fe/S cluster assembly in the mitochondria and their incorporation into proteins. Fe/S clusters are essential for maintenance of normal energy homeostasis, as they are required for the function of several subunits of electron transport chain (ETC) complexes and the tricarboxylic cycle enzyme aconitase. In fact, we and others report reduced activity of ETC complexes and aconitase in HF, suggesting that Fe/S cluster biosynthesis may be disrupted in failing hearts (54).
Finally, the potential limitations of our studies include the lack of data on demographics, underlying cause of heart failure, and/or medical therapy received by patients prior to the transplant surgery and retrieval of the heart. Likely, our samples come from a heterogeneous patient population. The effects of medications, methods for preservation, and ischemic time for both control and failing hearts are important factors that may have impacted the reported findings. Finally, the mechanisms of cellular iron regulation are complex and multiple. Thus, unmeasured confounders may potentially contribute to the presented findings. Despite these limitations we nevertheless observe a consistent and robust increase in cardiac heme levels in failing hearts. Thus, we can speculate that induction of heme synthesis pathway represents a common and important pathophysiologic finding in HF.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to all members of Feinberg Cardiovascular Research Institute for insightful comments and support. Marina Bayeva is supported by the American Heart Association (AHA) Midwest Affiliate Predoctoral Fellowship (10PRE4430021). Hossein Ardehali is supported by the National Institutes of Health Grants (K02 HL107448, R01 HL104181, and 1P01 HL108795).
List of Abbreviations
- ALA
δ-aminolevulinic acid
- ALAS2
ALA synthase 2
- ABCB10
ATP-binding cassette B10
- ETC
electron transport chain
- EPO
erythropoietin
- Fe/S
iron-sulfur
- HF
heart failure
- HMOX1
heme oxygenase 1
- Mfrn2
mitoferrin 2
- FtMt
mitochondrial ferritin
- PPIX
protoporphyrin IX
- ROS
reactive oxygen species
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no conflicts of interest and have no relationships to disclose
REFERENCES
- 1.Lloyd-Jones D, Adams RJ, Brown TM, Carnethon M, et al. Heart disease and stroke statistics--2010 update: a report from the American Heart Association. Circulation. 2010;121:e46–e215. doi: 10.1161/CIRCULATIONAHA.109.192667. [DOI] [PubMed] [Google Scholar]
- 2.Mudd JO, Kass DA. Tackling heart failure in the twenty-first century. Nature. 2008;451:919–928. doi: 10.1038/nature06798. [DOI] [PubMed] [Google Scholar]
- 3.Karamanlidis G, Bautista-Hernandez V, Fynn-Thompson F, Del Nido P, et al. Impaired mitochondrial biogenesis precedes heart failure in right ventricular hypertrophy in congenital heart disease. Circ Heart Fail. 2011;4:707–713. doi: 10.1161/CIRCHEARTFAILURE.111.961474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bayeva M, Gheorghiade M, Ardehali H. Mitochondria as Therapeutic Target in Heart Failure. J Am Coll Cardiol. 2012 doi: 10.1016/j.jacc.2012.08.1021. doi: 10.1016/j.jacc.2012.08.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sihag S, Cresci S, Li AY, Sucharov CC, et al. PGC-1alpha and ERRalpha target gene downregulation is a signature of the failing human heart. J Mol Cell Cardiol. 2009;46:201–212. doi: 10.1016/j.yjmcc.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ventura-Clapier R, Garnier A, Veksler V, Joubert F. Bioenergetics of the failing heart. Biochim Biophys Acta. 2011;1813:1360–1372. doi: 10.1016/j.bbamcr.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 7.Tsutsui H, Kinugawa S, Matsushima S. Oxidative stress and mitochondrial DNA damage in heart failure. Circ J. 2008;72(Suppl A):A31–37. doi: 10.1253/circj.cj-08-0014. [DOI] [PubMed] [Google Scholar]
- 8.Lill R, Hoffmann B, Molik S, Pierik AJ, et al. The role of mitochondria in cellular iron-sulfur protein biogenesis and iron metabolism. Biochim Biophys Acta. 2012 doi: 10.1016/j.bbamcr.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 9.Khan AA, Quigley JG. Control of intracellular heme levels: heme transporters and heme oxygenases. Biochim Biophys Acta. 2011;1813:668–682. doi: 10.1016/j.bbamcr.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hegde N, Rich MW, Gayomali C. The cardiomyopathy of iron deficiency. Tex Heart Inst J. 2006;33:340–344. [PMC free article] [PubMed] [Google Scholar]
- 11.Aisen P, Enns C, Wessling-Resnick M. Chemistry and biology of eukaryotic iron metabolism. Int J Biochem Cell Biol. 2001;33:940–959. doi: 10.1016/s1357-2725(01)00063-2. [DOI] [PubMed] [Google Scholar]
- 12.Kohgo Y, Ikuta K, Ohtake T, Torimoto Y, et al. Body iron metabolism and pathophysiology of iron overload. Int J Hematol. 2008;88:7–15. doi: 10.1007/s12185-008-0120-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elas M, Bielanska J, Pustelny K, Plonka PM, et al. Detection of mitochondrial dysfunction by EPR technique in mouse model of dilated cardiomyopathy. Free Radic Biol Med. 2008;45:321–328. doi: 10.1016/j.freeradbiomed.2008.04.016. [DOI] [PubMed] [Google Scholar]
- 14.Ichikawa Y, Bayeva M, Ghanefar M, Potini V, et al. Disruption of ATP-binding cassette B8 in mice leads to cardiomyopathy through a decrease in mitochondrial iron export. Proc Natl Acad Sci U S A. 2012;109:4152–4157. doi: 10.1073/pnas.1119338109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Michael S, Petrocine SV, Qian J, Lamarche JB, et al. Iron and iron-responsive proteins in the cardiomyopathy of Friedreich’s ataxia. Cerebellum. 2006;5:257–267. doi: 10.1080/14734220600913246. [DOI] [PubMed] [Google Scholar]
- 16.Payne RM. The Heart in Friedreich’s Ataxia: Basic Findings and Clinical Implications. Prog Pediatr Cardiol. 2011;31:103–109. doi: 10.1016/j.ppedcard.2011.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shaw GC, Cope JJ, Li L, Corson K, et al. Mitoferrin is essential for erythroid iron assimilation. Nature. 2006;440:96–100. doi: 10.1038/nature04512. [DOI] [PubMed] [Google Scholar]
- 18.Paradkar PN, Zumbrennen KB, Paw BH, Ward DM, et al. Regulation of mitochondrial iron import through differential turnover of mitoferrin 1 and mitoferrin 2. Mol Cell Biol. 2009;29:1007–1016. doi: 10.1128/MCB.01685-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang J, Pantopoulos K. Regulation of cellular iron metabolism. Biochem J. 2011;434:365–381. doi: 10.1042/BJ20101825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Furuyama K, Kaneko K, Vargas PD. Heme as a magnificent molecule with multiple missions: heme determines its own fate and governs cellular homeostasis. Tohoku J Exp Med. 2007;213:1–16. doi: 10.1620/tjem.213.1. [DOI] [PubMed] [Google Scholar]
- 21.Wu ML, Ho YC, Lin CY, Yet SF. Heme oxygenase-1 in inflammation and cardiovascular disease. Am J Cardiovasc Dis. 2011;1:150–158. [PMC free article] [PubMed] [Google Scholar]
- 22.Rouault TA, Tong WH. Iron-sulphur cluster biogenesis and mitochondrial iron homeostasis. Nat Rev Mol Cell Biol. 2005;6:345–351. doi: 10.1038/nrm1620. [DOI] [PubMed] [Google Scholar]
- 23.Hausmann A, Samans B, Lill R, Muhlenhoff U. Cellular and mitochondrial remodeling upon defects in iron-sulfur protein biogenesis. J Biol Chem. 2008;283:8318–8330. doi: 10.1074/jbc.M705570200. [DOI] [PubMed] [Google Scholar]
- 24.Napier I, Ponka P, Richardson DR. Iron trafficking in the mitochondrion: novel pathways revealed by disease. Blood. 2005;105:1867–1874. doi: 10.1182/blood-2004-10-3856. [DOI] [PubMed] [Google Scholar]
- 25.Ward JH, Jordan I, Kushner JP, Kaplan J. Heme regulation of HeLa cell transferrin receptor number. J Biol Chem. 1984;259:13235–13240. [PubMed] [Google Scholar]
- 26.Gordon LI, Burke MA, Singh AT, Prachand S, et al. Blockade of the erbB2 receptor induces cardiomyocyte death through mitochondrial and reactive oxygen species-dependent pathways. J Biol Chem. 2009;284:2080–2087. doi: 10.1074/jbc.M804570200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ide T, Tsutsui H, Hayashidani S, Kang D, et al. Mitochondrial DNA damage and dysfunction associated with oxidative stress in failing hearts after myocardial infarction. Circ Res. 2001;88:529–535. doi: 10.1161/01.res.88.5.529. [DOI] [PubMed] [Google Scholar]
- 28.Marin-Garcia J, Goldenthal MJ, Pierpont ME, Ananthakrishnan R. Impaired mitochondrial function in idiopathic dilated cardiomyopathy: biochemical and molecular analysis. J Card Fail. 1995;1:285–291. doi: 10.1016/1071-9164(95)90003-9. [DOI] [PubMed] [Google Scholar]
- 29.Ryter SW, Tyrrell RM. The heme synthesis and degradation pathways: role in oxidant sensitivity. Heme oxygenase has both pro- and antioxidant properties. Free Radic Biol Med. 2000;28:289–309. doi: 10.1016/s0891-5849(99)00223-3. [DOI] [PubMed] [Google Scholar]
- 30.Gotoh S, Nakamura T, Kataoka T, Taketani S. Egr-1 regulates the transcriptional repression of mouse delta-aminolevulinic acid synthase 1 by heme. Gene. 2011;472:28–36. doi: 10.1016/j.gene.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 31.Kramer MF, Gunaratne P, Ferreira GC. Transcriptional regulation of the murine erythroid-specific 5-aminolevulinate synthase gene. Gene. 2000;247:153–166. doi: 10.1016/s0378-1119(00)00103-7. [DOI] [PubMed] [Google Scholar]
- 32.Zhang FL, Shen GM, Liu XL, Wang F, et al. Hypoxic induction of human erythroid-specific delta-aminolevulinate synthase mediated by hypoxia-inducible factor 1. Biochemistry. 2011;50:1194–1202. doi: 10.1021/bi101585c. [DOI] [PubMed] [Google Scholar]
- 33.Sadlon TJ, Dell’Oso T, Surinya KH, May BK. Regulation of erythroid 5-aminolevulinate synthase expression during erythropoiesis. Int J Biochem Cell Biol. 1999;31:1153–1167. doi: 10.1016/s1357-2725(99)00073-4. [DOI] [PubMed] [Google Scholar]
- 34.Okonko DO, Marley SB, Anker SD, Poole-Wilson PA, et al. Suppression of erythropoiesis in patients with chronic heart failure and anaemia of unknown origin: evidence of an immune basis. Int J Cardiol. 2011 doi: 10.1016/j.ijcard.2011.11.081. [DOI] [PubMed] [Google Scholar]
- 35.Depping R, Kawakami K, Ocker H, Wagner JM, et al. Expression of the erythropoietin receptor in human heart. J Thorac Cardiovasc Surg. 2005;130:877–878. doi: 10.1016/j.jtcvs.2004.12.041. [DOI] [PubMed] [Google Scholar]
- 36.Witthuhn BA, Quelle FW, Silvennoinen O, Yi T, et al. JAK2 associates with the erythropoietin receptor and is tyrosine phosphorylated and activated following stimulation with erythropoietin. Cell. 1993;74:227–236. doi: 10.1016/0092-8674(93)90414-l. [DOI] [PubMed] [Google Scholar]
- 37.Tsutsui H, Kinugawa S, Matsushima S. Oxidative stress and heart failure. Am J Physiol Heart Circ Physiol. 2011;301:H2181–2190. doi: 10.1152/ajpheart.00554.2011. [DOI] [PubMed] [Google Scholar]
- 38.Kumar S, Bandyopadhyay U. Free heme toxicity and its detoxification systems in human. Toxicol Lett. 2005;157:175–188. doi: 10.1016/j.toxlet.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 39.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 40.Maeder MT, Khammy O, dos Remedios C, Kaye DM. Myocardial and systemic iron depletion in heart failure implications for anemia accompanying heart failure. J Am Coll Cardiol. 2011;58:474–480. doi: 10.1016/j.jacc.2011.01.059. [DOI] [PubMed] [Google Scholar]
- 41.Lakkisto P, Siren JM, Kyto V, Forsten H, et al. Heme oxygenase-1 induction protects the heart and modulates cellular and extracellular remodelling after myocardial infarction in rats. Exp Biol Med (Maywood) 2011;236:1437–1448. doi: 10.1258/ebm.2011.011148. [DOI] [PubMed] [Google Scholar]
- 42.Wu ML, Ho YC, Yet SF. A central role of heme oxygenase-1 in cardiovascular protection. Antioxid Redox Signal. 2011;15:1835–1846. doi: 10.1089/ars.2010.3726. [DOI] [PubMed] [Google Scholar]
- 43.van Veldhuisen DJ, Anker SD, Ponikowski P, Macdougall IC. Anemia and iron deficiency in heart failure: mechanisms and therapeutic approaches. Nat Rev Cardiol. 2011;8:485–493. doi: 10.1038/nrcardio.2011.77. [DOI] [PubMed] [Google Scholar]
- 44.Anker SD, Comin Colet J, Filippatos G, Willenheimer R, et al. Ferric carboxymaltose in patients with heart failure and iron deficiency. N Engl J Med. 2009;361:2436–2448. doi: 10.1056/NEJMoa0908355. [DOI] [PubMed] [Google Scholar]
- 45.Yoshino K, Munakata H, Kuge O, Ito A, et al. Haeme-regulated degradation of delta-aminolevulinate synthase 1 in rat liver mitochondria. J Biochem. 2007;142:453–458. doi: 10.1093/jb/mvm159. [DOI] [PubMed] [Google Scholar]
- 46.Zheng J, Shan Y, Lambrecht RW, Donohue SE, et al. Differential regulation of human ALAS1 mRNA and protein levels by heme and cobalt protoporphyrin. Mol Cell Biochem. 2008;319:153–161. doi: 10.1007/s11010-008-9888-0. [DOI] [PubMed] [Google Scholar]
- 47.May BK, Dogra SC, Sadlon TJ, Bhasker CR, et al. Molecular regulation of heme biosynthesis in higher vertebrates. Prog Nucleic Acid Res Mol Biol. 1995;51:1–51. doi: 10.1016/s0079-6603(08)60875-2. [DOI] [PubMed] [Google Scholar]
- 48.Cox TC, Sadlon TJ, Schwarz QP, Matthews CS, et al. The major splice variant of human 5-aminolevulinate synthase-2 contributes significantly to erythroid heme biosynthesis. Int J Biochem Cell Biol. 2004;36:281–295. doi: 10.1016/s1357-2725(03)00246-2. [DOI] [PubMed] [Google Scholar]
- 49.Kaneko K, Furuyama K, Aburatani H, Shibahara S. Hypoxia induces erythroid-specific 5-aminolevulinate synthase expression in human erythroid cells through transforming growth factor-beta signaling. FEBS J. 2009;276:1370–1382. doi: 10.1111/j.1742-4658.2009.06878.x. [DOI] [PubMed] [Google Scholar]
- 50.Yamaguchi S, Yamahara K, Homma K, Suzuki S, et al. The role of microRNA-145 in human embryonic stem cell differentiation into vascular cells. Atherosclerosis. 2011;219:468–474. doi: 10.1016/j.atherosclerosis.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 51.Rangrez AY, Massy ZA, Metzinger-Le Meuth V, Metzinger L. miR-143 and miR-145: molecular keys to switch the phenotype of vascular smooth muscle cells. Circ Cardiovasc Genet. 2011;4:197–205. doi: 10.1161/CIRCGENETICS.110.958702. [DOI] [PubMed] [Google Scholar]
- 52.Chen X, Gong J, Zeng H, Chen N, et al. MicroRNA145 targets BNIP3 and suppresses prostate cancer progression. Cancer Res. 2010;70:2728–2738. doi: 10.1158/0008-5472.CAN-09-3718. [DOI] [PubMed] [Google Scholar]
- 53.La Rocca G, Badin M, Shi B, Xu SQ, et al. Mechanism of growth inhibition by MicroRNA 145: the role of the IGF-I receptor signaling pathway. J Cell Physiol. 2009;220:485–491. doi: 10.1002/jcp.21796. [DOI] [PubMed] [Google Scholar]
- 54.Qanud K, Mamdani M, Pepe M, Khairallah RJ, et al. Reverse changes in cardiac substrate oxidation in dogs recovering from heart failure. Am J Physiol Heart Circ Physiol. 2008;295:H2098–2105. doi: 10.1152/ajpheart.00471.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.