Abstract
Background
The HLA (human leukocyte antigen) molecules that present pathogen-derived epitopes to T cells are highly diverse. Correspondingly, many pathogens such as HIV evolve epitope variants in order to evade immune recognition. In contrast, another persistent human pathogen, Mycobacterium tuberculosis, has highly conserved epitope sequences. This raises the question whether there is also a difference in the ability of these pathogens’ epitopes to bind diverse HLA alleles, referred to as an epitope’s binding promiscuity. To address this question, we compared the in silico HLA binding promiscuity of T cell epitopes from pathogens with distinct infection strategies and outcomes of human exposure.
Methods
We used computer algorithms to predict the binding affinity of experimentally-verified microbial epitope peptides to diverse HLA-DR, HLA-A and HLA-B alleles. We then analyzed binding promiscuity of epitopes derived from HIV and M. tuberculosis. We also analyzed promiscuity of epitopes from Streptococcus pyogenes, which is known to exhibit epitope diversity, and epitopes of Bacillus anthracis and Clostridium tetani toxins, as these bacteria do not depend on human hosts for their survival or replication, and their toxin antigens are highly immunogenic human vaccines.
Results
We found that B. anthracis and C. tetani epitopes were the most promiscuous of the group that we analyzed. However, there was no consistent difference or trend in promiscuity in epitopes contained in HIV, M. tuberculosis, and S. pyogenes.
Conclusions
Our results show that human pathogens with distinct immune evasion strategies and epitope diversities exhibit equivalent levels of T cell epitope promiscuity. These results indicate that differences in epitope promiscuity do not account for the observed differences in epitope variation and conservation.
Introduction
MHC (major histocompatibility complex) molecules recognize and bind epitopes derived from foreign and self proteins in order to initiate and maintain adaptive immune responses. The HLA (human leukocyte antigen) alleles that encode MHC molecules are extremely diverse: over 2000 HLA class I, and over 600 HLA class II alleles have been identified [1]. In a model of host–pathogen coevolution, this diversity is maintained through selection for individuals with heterozygous HLA alleles and/or for individuals with rare HLA alleles [2]. Individuals heterozygous for HLA alleles produce a greater diversity of HLA molecules, and therefore may develop an immune response against a greater breadth of pathogen epitopes [3,4]. In addition, pathogens are more likely to develop adaptations in response to the most common HLA alleles in a population, and therefore individuals with rare alleles may have a selective advantage [5]. A common adaptation used by pathogens to avoid immune recognition is antigenic variation, in which epitope variants that decrease the likelihood of HLA binding are selected [6]. Human immunodeficiency virus (HIV) is an extreme example; new variants appear at each successive generation, which eventually leads to immune evasion and disease progression, and has frustrated vaccine development efforts [7]. In this way, hosts and their pathogens can be locked in a never-ending “arms race” to diversify their HLA allele or epitope sequences, respectively. This is known as antagonistic coevolution, or the Red Queen model [5,6].
We recently reported that Mycobacterium tuberculosis, a highly successful persistent human pathogen, has highly conserved epitope sequences [8]. This apparently conflicts with the general model of a host–pathogen coevolution, and emphasizes that M. tuberculosis employs unique approaches to achieve success as a pathogen.
It has also been shown in a variety of pathogens that, even though the adaptive immune response is highly specific, individual HLA class II-restricted peptides [9-13] and HLA class I-restricted peptides [14-18] may bind many different HLA alleles; a trait termed epitope promiscuity. Given the extensive human HLA allele diversity and varied pathogen epitope diversity, we were interested in determining whether the extent of epitope promiscuity varies in pathogens with distinct ecological niches and interactions with human hosts.
We compared epitope promiscuity in M. tuberculosis and HIV, since these human-specific pathogens vary in their epitope diversity [7,19] yet both persist in the face of antigen-specific T cell responses. For contrast, we examined Streptococcus pyogenes, which undergoes antigenic variation [20], but is not able to persist within a host [21], and we examined three vaccine antigens from Bacillus anthracis and Clostridium tetani.
We used the computer algorithms NetMHCpan-2.0 and NetMHCIIpan-2.0 to predict the binding affinity of experimentally determined microbial epitopes to HLA-DR, HLA-A, and HLA-B alleles [22,23]. We then analyzed epitope promiscuity across the majority of human HLA alleles, across the most common HLA alleles found in different geographic regions, and across alleles grouped by similar characteristics. We find that epitopes from B. anthracis and C. tetani are consistently the most promiscuous, and that there is no consistent pattern of promiscuity between M. tuberculosis, HIV and S. pyogenes. Therefore we find similar levels of epitope promiscuity in very different pathogens, and we discuss the impact of promiscuity on host–pathogen coevolution.
Methods
Programs and databases
NetMHCpan-2.0
The NetMHCpan-2.0 algorithm (http://www.cbs.dtu.dk/services/NetMHCpan-2.0/) uses an artificial neural network (ANN) to predict the binding affinity of peptide-MHC (HLA)-I molecule interactions [22,23]. Peptide sequence and HLA sequence information were used as input and experimentally determined affinity data from the IEDB database covering 34 HLA-A and 32 HLA-B alleles were used as output to train the ANN. This method currently gives affinity estimates for peptides with 886 HLA-A alleles and 1,412 HLA-B alleles.
NetMHCIIpan-2.0
The NetMHCIIpan-2.0 algorithm (http://www.cbs.dtu.dk/services/NetMHCIIpan/) uses an ANN to predict the binding affinity of peptide-MHC (HLA)-II molecule interactions [22,23]. Peptide sequence and HLA sequence information were used as input and experimentally determined affinity data from the IEDB database covering 24 HLA-DR alleles were used as output to train the ANN. This method currently provides binding affinity data for 654 HLA-DR alleles.
IEDB
The IEDB (Immune Epitope Database, http://www.immuneepitope.org/) contains peptidic and non-peptidic antibody and T cell epitope data for humans and other animal species. The database contains HLA binding data from diverse antigenic sources, including allergens and multiple human pathogens. It also provides tools for epitope prediction and analysis.
HIV Molecular Immunology Database
The HIV Molecular Immunology Database (http://www.hiv.lanl.gov/content/immunology/index.html) contains data on HIV T cell epitopes, and provides access to tools for epitope prediction and analysis.
dbMHC
The dbMHC (http://www.ncbi.nlm.nih.gov/projects/gv/mhc/main.fcgi?cmd=init) contains DNA seqeuence and other data related to the human MHC, including anthropology resources regarding the distribution and frequency of specific HLA alleles in multiple populations.
Alleles and epitopes used in the study
Epitopes were obtained from the IEDB for all bacterial species, and the HIV Molecular Immunology Database for HIV epitopes. All HLA alleles included in netMHCpan and netMHCIIpan were analyzed. The most common alleles in geographic regions were determined using the IEDB population coverage epitope analysis. Alleles were grouped by supertype as described [24]. The list of all alleles, alleles grouped by population region, alleles grouped by supertype, and all epitopes used in this study are provided in Tables S1, S2, S3, and S4.
Analysis of promiscuity
Binding prediction values were obtained from NetMHCIIpan-2.0 for HLA-DR alleles, and NetMHCpan-2.0 for HLA-A and HLA-B alleles. The two methods give the output binding scores as 1 – log(aff), where aff is the IC50 value in nM. Thus the scores range from 0 to 1, with higher scores indicating higher affinity. We used a receiver operating characteristic (ROC) curve to obtain a threshold prediction value to specify which epitope/alleles combinations were predicted to interact. To construct the ROC curves we ran NetMHCpan-2.0 and NetMHCIIpan-2.0 against their published validation datasets. We plotted the true positive rate (TPR) against the false positive rate (FPR) at different thresholds of binding from 0 to 1. We chose a FPR of 0.05, which corresponded to thresholds of 0.29 for netMHCpan and 0.585 for netMHCIIpan, and TPRs of 0.89 for HLA class I and 0.24 for HLA class II.
We defined promiscuity as the percent of allotypes each epitope was predicted to bind to at each HLA locus. To analyze overall promiscuity of epitopes from each pathogen species, we calculated the mean promiscuity of each group, and compared them with a one-way ANOVA and Tukey post-test using GraphPad Prism 5. We only included epitopes predicted to bind at least one allele in the analyses. To complement this we performed kernel density estimation using reflection for boundary support [25] to estimate the probability density of epitope promiscuity using MATLAB version 7.13.0.564. This was based on a normal kernel function. The density was evaluated at 101 equally spaced points in the interval [0,100]. The probability distribution of points lying outside the relevant region of [0,100], specifically those in the intervals [-100,0] and [100, 200], was reflected onto the distribution between 0 and 100 to arrive at the complete probability distribution. A bandwidth parameter of 15 was used for smoothing the curves. Distributions shifted to the right of the graph indicate higher promiscuity. Epitope promiscuity was analyzed in this way across all HLA alleles, as well as across HLA alleles grouped by population region and by supertype. Analysis of HLA allele promiscuity was carried out in the same way, with promiscuity defined as the percent of epitopes each allotype was predicted to bind each epitope at each HLA locus.
Results
Validation of the netMHCpan and netMHCIIpan methods
The accuracy of the netMHCpan and netMHCIIpan prediction methods has been extensively validated by several previous studies. One study compared experimentally determined peptide-MHC binding affinities with affinities predicted by three different algorithm prediction methods [26]. The authors found a positive correlation between predicted and experimentally determined affinities, and found that this correlation was strongest for the netMHCpan method. Another study evaluated the prediction methods for a larger dataset including alleles that were not included in the set used to train the programs, using a comparison with experimental data from the Immune Epitope Database (IEDB), and showed that the netMHCpan ANN method was top ranking for both affinity and ligand data [27]. In this analysis the authors removed peptides used in program training as well as peptides in the IEDB that had been identified using netMHCpan methods, in order to remove any bias in favor of the netMHCpan methods. An additional study conducted similar, independent evaluations of MHC class I [28] and MHC class II [29] prediction methods, and also found that netMHCpan and netMHCIIpan were consistently the most accurate method when compared with experimental results.
We conducted our own validation studies of the netMHCpan and netMHCIIpan methods by running them against experimental validation datasets. We constructed receiver operating characteristic (ROC) curves for both the methods and confirmed that both methods performed significantly better than random guessing (see Methods). Based on the ROC curve we chose threshold levels for netMHCpan and netMHCIIpan such that the false positive rate (FPR) of detecting epitope binding was 0.05 (Figure S1). This corresponded to a true positive rate (TPR) of 0.89 for HLA class I and 0.24 for HLA class II.
HLA alleles and microbe-derived epitopes
We analyzed all HLA-A and HLA-B, and HLA-DR alleles covered by netMHCpan and netMHCIIpan. We analyzed microbe-derived epitopes in four groups: Bacillus anthracis and Clostridium tetani, Streptococcus pyogenes, Mycobacterium tuberculosis complex, and Human immunodeficiency virus (HIV). B. anthracis and C. tetani were grouped together in all analyses because we found no significant differences between them in mean epitope promiscuity across all alleles (data not shown). These groups were selected for their biological diversity as indicated above, and because epitope information was readily available for each in the IEDB. We included all epitopes available for each pathogen in each database at the time of analysis. Supplementary Tables 1 and 2 provide lists of the alleles and epitopes included in this study.
Promiscuity across all HLA-DR, -A, -B alleles and epitopes
To compare promiscuity between groups, we first determined the percent of alleles that each epitope was predicted to bind and computed mean promiscuity of epitopes from each group. This revealed that epitopes in B. anthracis and C. tetani were most promiscuous across all human HLA-DR, HLA-A and HLA-B alleles (Figure 1), though there was no significant difference in mean promiscuity between B. anthracis and C. tetani and HIV across HLA-DR alleles (Figure 1A). In addition we plotted distributions of the probability density of epitope binding to increasing percentages of HLA alleles using kernel density estimation (see Methods). Each distribution shifted to the right for B. anthracis and C. tetani, indicating that these epitopes were more likely to bind a greater breadth of HLA alleles, which was consistent with the mean promiscuity analysis (Figure 1). HIV epitopes were more promiscuous than M. tuberculosis and S. pyogenes epitopes across HLA-DR alleles (Figure 1A). M. tuberculosis epitopes were more promiscuous than HIV and S. pyogenes epitopes across HLA-A alleles by both analyses, and there was no difference between S. pyogenes and HIV (Figure 1B). HIV epitopes were less promiscuous than all groups across HLA-B alleles, and there was no significant difference between M. tuberculosis and S. pyogenes (Figure 1C). We found that using higher or lower threshold values did not alter the results (data not shown). Therefore, while B. anthracis and C. tetani epitopes were most promiscuous, there was no consistent trend in epitope promiscuity between the other pathogens that extended to multiple HLA loci.
Figure 1. T cell epitope promiscuity across HLA-DR, -A, -B alleles.
Kernel density estimates of epitope promiscuity and graphs of mean epitope promiscuity (see Methods) of B. anthracis and C. tetani (BA & CT; blue), S. pyogenes and (SP; purple), HIV (green) and M. tuberculosis complex (MTBC; orange) across HLA-DR (A), HLA-A (B) and HLA-B (C) alleles. Significant differences in mean promiscuity between groups are indicated with black bars (Tukey’s post-test, p < 0.05). Error bars represent the standard error of the mean.
Although we were primarily interested in epitope promiscuity, the study could also have been framed in terms of the ability of HLA molecules to bind multiple epitopes [10]. Therefore we performed the above analyses for predicted HLA allele promiscuity. We found nearly identical trends to those we found for epitope promiscuity using mean promiscuity analysis (Figure S2). Kernel density estimations were also similar (Figure S2). Thus, given the available data we cannot clearly distinguish the effect of epitope promiscuity from HLA allele promiscuity. However, for simplicity, we chose to focus only on epitope promiscuity in all subsequent analyses.
Promiscuity across the most common HLA-DR, -A, -B alleles in each geographic region
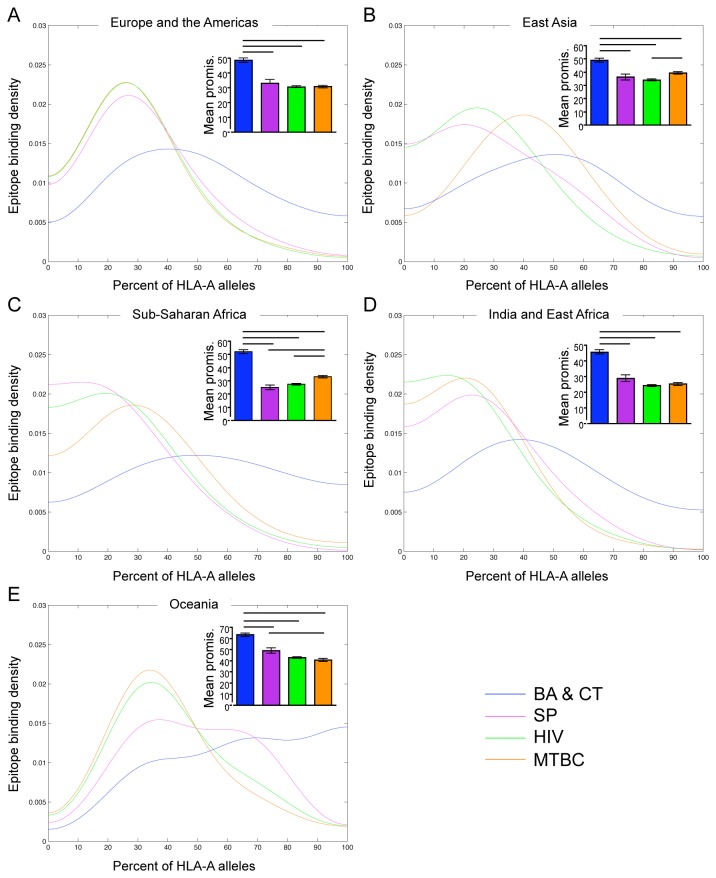
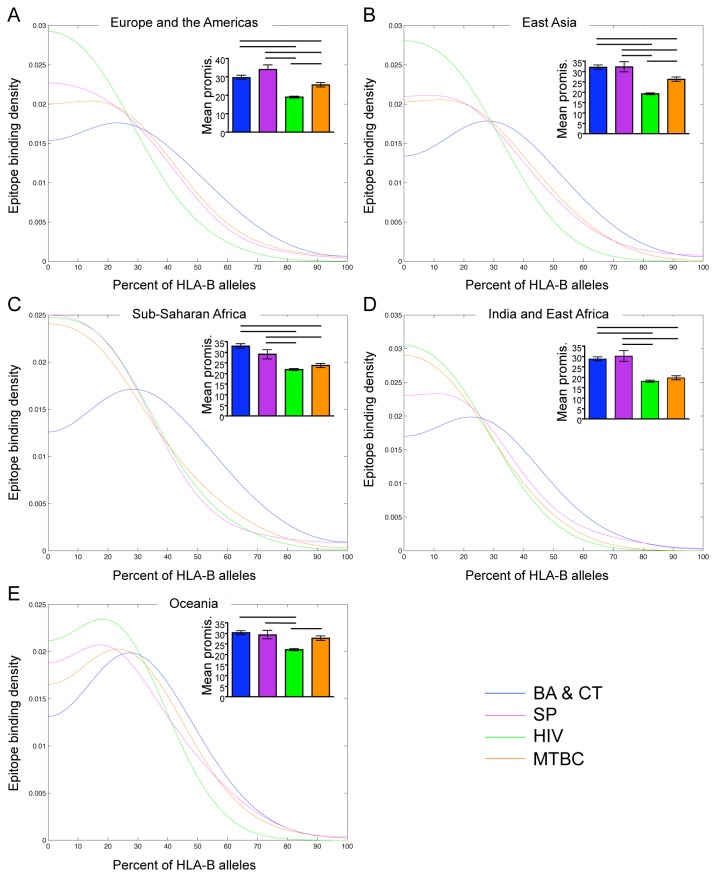
The majority of the alleles used in the above analysis are found at very low frequencies in the human population. Thus we were interested in whether patterns would be more distinct, or if new patterns would emerge, if we focused on only the alleles most prevalent in specific geographic regions. We grouped populations into Europe and the Americas, East Asia, Sub-Saharan Africa, India and East Africa, and Oceania (Table S3) and analyzed the most prevalent alleles in these regions that comprised 60% of the population. We chose these groups because they follow human migration patterns [30] and, correspondingly, HLA alleles are grouped this way in the dbMHC database. Interestingly, the spread and divergence of pathogens like M. tuberculosis also follow these migration patterns [31].
We found that patterns of promiscuity within geographic regions (Figures 2-4) were similar, though not identical, to those found across all alleles (Figure 1). B. anthracis and C. tetani epitopes were most promiscuous across HLA-DR alleles by both analyses in Sub-Saharan African and India and East Africa (Figure 2C-D). S. pyogenes epitopes were least promiscuous in all regions, although the differences in mean promiscuity were not statistically significant for all pair wise comparisons (Figure 2A–E). There was no striking difference in epitope promiscuity across HLA-DR alleles between M. tuberculosis and HIV in all regions except Oceania, where HIV epitopes were more promiscuous (Figure 2A–E). We found that B. anthracis and C. tetani epitopes were most promiscuous across HLA-A alleles in all geographic regions (Figure 3A–E). The other three groups had similar levels of promiscuity in all regions. M. tuberculosis epitopes were more promiscuous in East Asia and Sub-Saharan Africa (Figure 3B,C), and S. pyogenes epitopes were more promiscuous in Oceania (Figure 3E). B. anthracis and C. tetani and S. pyogenes epitopes were most promiscuous across HLA-B alleles in all regions, though this trend was not distinct for S. pyogenes by the Kernel density estimation (Figure 4A–E). M. tuberculosis epitopes were more promiscuous than HIV epitopes in Europe and the Americas, East Asia, and Oceania (Figure 4A,B,E), however there was no striking difference between M. tuberculosis and HIV in Sub-Saharan Africa and India and East Africa (Figure 4C,D).
Figure 2. T cell epitope promiscuity across the most common HLA-DR alleles in geographic regions.
Kernel density estimates of epitope promiscuity and graphs of mean epitope promiscuity (see Methods) of B. anthracis and C. tetani (BA & CT; blue), S. pyogenes and (SP; purple), HIV (green) and M. tuberculosis complex (MTBC; orange) across the most common HLA-DR alleles in Europe and the Americas (A), East Asia (B), Sub-Saharan Africa (C), India and East Africa (D) and Oceania (E). Significant differences in mean promiscuity between groups are indicated with black bars (Tukey’s post-test, p < 0.05). Error bars represent the standard error of the mean.
Figure 3. T cell epitope promiscuity across the most common HLA-A alleles in geographic regions.
Kernel density estimates of epitope promiscuity and graphs of mean epitope promiscuity (see Methods) of B. anthracis and C. tetani (BA & CT; blue), S. pyogenes and (SP; purple), HIV (green) and M. tuberculosis complex (MTBC; orange) across the most common HLA-A alleles in Europe and the Americas (A), East Asia (B), Sub-Saharan Africa (C), India and East Africa (D) and Oceania (E). Significant differences in mean promiscuity between groups are indicated with black bars (Tukey’s post-test, p < 0.05). Error bars represent the standard error of the mean.
Figure 4. T cell epitope promiscuity across the most common HLA-B alleles in geographic regions.
Kernel density estimates of epitope promiscuity and graphs of mean epitope promiscuity (see Methods) of B. anthracis and C. tetani (BA & CT; blue), S. pyogenes (SP; purple), HIV (green) and M. tuberculosis complex (MTBC; orange) across the most common HLA-B alleles in Europe and the Americas (A), East Asia (B), Sub-Saharan Africa (C), India and East Africa (D) and Oceania (E). Significant differences in mean promiscuity between groups are indicated with black bars (Tukey’s post-test, p < 0.05). Error bars represent the standard error of the mean.
Promiscuity within allele supertypes
Despite being highly polymorphic, HLA alleles can be organized into groups that retain similar characteristics with overlapping peptide-binding repertoires, known as supertypes [24]. Supertypes were not specific to population region, as we found a diverse collection of supertypes in each of the allele groups used in the population analysis (data not shown). Thus, we looked at whether there would be trends in epitope promiscuity within groups of alleles with similar characteristics. Supertypes have been best defined for HLA class I alleles [24], therefore for this analysis we focused on HLA-A and HLA-B (Table S4). Using mean promiscuity analysis, we found that B. anthracis and C. tetani were most promiscuous within three of four HLA-A supertypes, and that there was no trend between S. pyogenes, HIV and M. tuberculosis (Figure S3). There was also no striking trend between any of the pathogen groups within HLA-B supertypes (Figure S3).
Discussion
In this study we compared T cell epitope promiscuity between ecologically diverse pathogens with different antigenic diversities. Epitope promiscuity has been shown both experimentally and computationally for peptides derived from different pathogens, including the human papillomavirus [16], HIV [11,15,17,18], and M. tuberculosis [14,32], among others [9,10,12,13]. However, to our knowledge, this is the first study that directly compares promiscuity in multiple pathogens. We found that B. anthracis and C. tetani were the most promiscuous epitope group in the majority of the analyses, and we found no trend in epitope promiscuity between HIV, M. tuberculosis and S. pyogenes. These latter pathogens have extremely different infection strategies and life cycles; thus if there were marked differences in promiscuity between species they should have been detected by our analyses. Importantly, there is no established standard for epitope promiscuity that allows determination whether groups of epitopes are highly promiscuous or not.
The impact of epitope promiscuity on the outcomes of host–pathogen interactions remains to be determined. Promiscuity could benefit the host by increasing the number of antigenic peptides that HLA molecules can present to T cells. This large pool of peptides could also allow for selection of immunodominant peptides that elicit the strongest T cell responses [33]. If this is the case, vaccines containing promiscuous epitopes should be especially effective; and this has been suggested by several studies [34-38]. Our results also showed that epitopes derived from successful vaccine targets, B. anthracis and C. tetani epitopes, were the most promiscuous. Alternatively, peptides recognized by many HLA molecules in a population could predispose to immune exhaustion and transmission of pathogens pre-adapted to the immune response. In this context, promiscuity would be harmful to the host. A recent study of HIV found that HLA molecules associated with slow disease progression also had the lowest levels of promiscuity [39]. The authors propose that carrying HLA molecules with promiscuous binding repertoires makes an individual “functionally homozygous” at the HLA locus, and therefore decreases the heterozygote advantage of having greater HLA allele diversity.
An unexpected finding in the present study is that conserved M. tuberculosis epitopes and variable HIV epitopes have similar breadths of HLA binding. Pathogens evolve variable epitopes because they are less likely to be recognized by one or more of the diverse HLA alleles [5,6]. Therefore, one explanation for the evolution of hyperconserved epitopes is that M. tuberculosis actually benefits from recognition [8]. If promiscuity enhances the host’s ability to recognize pathogens, then we might have expected M. tuberculosis epitopes to be more promiscuous than HIV or S. pyogenes epitopes. As this was not the case, we consider the idea that promiscuity does not globally enhance immune recognition because it diminishes the heterozygote advantage [39]. Whether pathogens benefit from recognition or aim to avoid recognition, they are evolving to exploit the host, presumably to the detriment of the host. Thus, regardless of infection strategy, antagonistic coevolution could emerge: the host benefits from generating new HLA allele combinations that the pathogen has not yet adapted to, and the pathogen benefits from evolving a way to diminish the new HLA diversity. In this manner M. tuberculosis, HIV, and S. pyogenes could have similar extents of promiscuity because each benefits in a similar manner from promiscuous epitopes.
Interestingly, although immune evasion through antigenic variation is the main strategy used by HIV, it has recently been shown that a subset of HIV epitopes is selectively hyperconserved [40]. Similarly, while the majority of identified M. tuberculosis epitopes are highly conserved, a small subset of M. tuberculosis epitopes is variable [8]. It has been proposed that recognition of specific subsets of epitopes may benefit the host or the pathogens during specific stages of the infection. Thus, it is tempting to speculate that epitope promiscuity is a characteristic associated with specific subsets of epitopes – e.g. the epitopes promoting pathogen virulence. Testing this hypothesis by focusing our analysis on these sub-groups would be the next step to determine whether promiscuity depends on the conservation of epitopes or on the virulence cycle of the pathogen.
The advantage of using algorithm-based methods was that we were able to generate data for almost any epitope-HLA combination. In this study we were primarily interested in the potential for different epitopes to bind a diverse array of HLA alleles, and any inconsistencies between the algorithm predictions and biological reality will have applied equally to each pathogen group. In vitro binding assays can be used to complement computational methods by directly testing the ability of epitopes to compete with other peptides known to bind HLA molecules, however there are also limits to computer-based and in vitro systems. Peptides that bind most strongly to HLA molecules do not necessarily elicit the strongest T cell responses [9,41], and thus these methods are not infallible for identifying immunodominant peptides. Techniques that facilitate further discovery of the determinants of peptide epitope generation and recognition, together with the kinetics of epitope generation and presentation during the course of infection [42], will have a major impact on understanding the impact of promiscuity on infection.
Conclusions
We used computer-based algorithms to compare the ability of epitopes from a variety of pathogens to bind multiple of the diverse HLA alleles. We found similar levels of epitope promiscuity in HIV, M. tuberculosis and S. pyogenes, despite differences in biology and epitope diversity. We propose a model where promiscuity benefits a broad range of successful human pathogens because it decreases the functional diversity of HLA alleles in the human population. Studies that examine the binding repertoire of epitopes presented at different stages of human infection will be important to further our understanding of how promiscuity influences host–pathogen interaction, and how it could be incorporated into vaccine design or disease management.
Supporting Information
ROC curves were generated by running NetMHCpan-2.0 and NetMHCIIpan-2.0 against their published validation datasets. The true positive rate (TPR) is plotted against the false positive rate (FPR) at different thresholds of binding from 0 to 1. For subsequent analyses we chose a FPR of 0.05, which corresponded to thresholds of 0.29 for netMHCpan and 0.585 for netMHCIIpan, and TPRs of 0.89 for HLA class I and 0.24 for HLA class II.
(TIF)
Kernel density estimates of epitope promiscuity and graphs of mean HLA allele promiscuity (see Methods) of B. anthracis and C. tetani (BA & CT; blue), S. pyogenes and (SP; purple), HIV (green) and M. tuberculosis complex (MTBC; orange) across HLA-DR (A), HLA-A (B) and HLA-B (C) alleles. Differences in mean promiscuity are indicated with black bars (Tukey’s post-test, p < 0.05). Error bars represent the standard error of the mean.
(TIF)
Graphs of mean epitope promiscuity (see Methods) of B. anthracis and C. tetani (BA and CT), S. pyogenes and (SP), HIV and M. tuberculosis complex (MTBC) across alleles within HLA-A supertypes (A01, A02, A03, A24) and within HLA-B supertypes (B07, B08, B27, B44, B58, B62). For simplicity, Tukey post-tests are not shown because no trend was found between the groups.
(TIF)
The first column refers to the IEDB reference number for bacterial epitopes, and amino acid sequence for HIV epitopes. The second column refers to the microbe group from which the epitope was derived. The third column refers to the protein from which the epitope was derived. The fourth column refers to the class of HLA alleles that the epitopes have been shown to bind, and the class in which they were analyzed in this study.
(PDF)
(PDF)
(PDF)
(PDF)
Funding Statement
This work was supported in part by National Institutes of Health grant R01 AI090928 (JDE), the Belgian American Education Foundation (RC), and the Potts Memorial Foundation (RC). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Robinson J, Mistry K, McWilliam H, Lopez R, Parham P et al. (2011) The IMGT/HLA database. Nucleic Acids Res 39: D1171-D1176. doi:10.1093/nar/gkq998. PubMed: 21071412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Apanius V, Penn D, Slev PR, Ruff LR, Potts WK (1997) The nature of selection on the major histocompatibility complex. Crit Rev Immunol 17: 179-224. doi:10.1615/CritRevImmunol.v17.i2.40. PubMed: 9094452. [DOI] [PubMed] [Google Scholar]
- 3. Dean M, Carrington M, O’Brien SJ (2002) Balanced polymorphism selected by genetic versus infectious human disease. Annu Rev Genomics Hum Genet 3: 263-292. doi:10.1146/annurev.genom.3.022502.103149. PubMed: 12142357. [DOI] [PubMed] [Google Scholar]
- 4. Penn DJ, Damjanovich K, Potts WK (2002) MHC heterozygosity confers a selective advantage against multiple-strain infections. Proc Natl Acad Sci U S A 99: 11260-11264. doi:10.1073/pnas.162006499. PubMed: 12177415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kubinak JL, Ruff JS, Hyzer CW, Slev PR, Potts WK (2012) Experimental viral evolution to specific host MHC genotypes reveals fitness and virulence trade-offs in alternative MHC types. Proc Natl Acad Sci U S A 109: 3422-3427. doi:10.1073/pnas.1112633109. PubMed: 22323587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Deitsch KW, Lukehart SA, Stringer JR (2009) Common strategies for antigenic variation by bacterial, fungal and protozoan pathogens. Nat Rev Microbiol 7: 493-503. doi:10.1038/nrmicro2145. PubMed: 19503065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Johnson WE, Desrosiers RC (2002) Viral persistence: HIV’s strategies of immune system evasion. Annu Rev Med 53: 499-518. doi:10.1146/annurev.med.53.082901.104053. PubMed: 11818487. [DOI] [PubMed] [Google Scholar]
- 8. Comas I, Chakravartti J, Small PM, Galagan J, Niemann S et al. (2010) Human T cell epitopes of Mycobacterium tuberculosis are evolutionarily hyperconserved. Nat Genet 42: 498-503. doi:10.1038/ng.590. PubMed: 20495566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. O’Sullivan D, Arrhenius T, Sidney J, Del Guercio MF, Albertson M et al. (1991) On the interaction of promiscuous antigenic peptides with different DR alleles. Identification of common structural motifs. J Immunol 147: 2663-2669. PubMed: 1717570. [PubMed] [Google Scholar]
- 10. Panina-Bordignon P, Tan A, Termijtelen A, Demotz S, Corradin G et al. (1989) Universally immunogenic T cell epitopes: promiscuous binding to human MHC class II and promiscuous recognition by T cells. Eur J Immunol 19: 2237-2242. doi:10.1002/eji.1830191209. PubMed: 2481588. [DOI] [PubMed] [Google Scholar]
- 11. Kaufmann DE, Bailey PM, Sidney J, Wagner B, Norris PJ et al. (2004) Comprehensive analysis of human immunodeficiency virus type 1-specific CD4 responses reveals marked immunodominance of gag and nef and the presence of broadly recognized peptides. J Virol 78: 4463-4477. doi:10.1128/JVI.78.9.4463-4477.2004. PubMed: 15078927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Krüger S, Schroers R, Rooney CM, Gahn B, Chen SY (2003) Identification of a naturally processed HLA-DR-restricted T-helper epitope in Epstein-Barr virus nuclear antigen type 1. J Immunother 26: 212-221. doi:10.1097/00002371-200305000-00005. PubMed: 12806275. [DOI] [PubMed] [Google Scholar]
- 13. Doolan DL, Southwood S, Chesnut R, Appella E, Gomez E et al. (2000) HLA-DR-promiscuous T cell epitopes from Plasmodium falciparum pre-erythrocytic-stage antigens restricted by multiple HLA class II alleles. J Immunol 165: 1123-1137. PubMed: 10878392. [DOI] [PubMed] [Google Scholar]
- 14. Axelsson-Robertson R, Weichold F, Sizemore D, Wulf M, Skeiky YA et al. (2010) Extensive major histocompatibility complex class I binding promiscuity for Mycobacterium tuberculosis TB10.4 peptides and immune dominance of human leucocyte antigen (HLA)-B*0702 and HLA-B*0801 alleles in TB10.4 CD8 T-cell responses. Immunology 129: 496-505. doi:10.1111/j.1365-2567.2009.03201.x. PubMed: 20002212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Frahm N, Yusim K, Suscovich TJ, Adams S, Sidney J et al. (2007) Extensive HLA class I allele promiscuity among viral CTL epitopes. Eur J Immunol 37: 2419-2433. doi:10.1002/eji.200737365. PubMed: 17705138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nakagawa M, Kim KH, Gillam TM, Moscicki AB (2007) HLA class I binding promiscuity of the CD8 T-cell epitopes of human papillomavirus type 16 E6 protein. J Virol 81: 1412-1423. doi:10.1128/JVI.01768-06. PubMed: 17108051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sabbaj S, Bansal A, Ritter GD, Perkins C, Edwards BH et al. (2003) Cross-reactive CD8+ T cell epitopes identified in US adolescent minorities. J Acquir Immune Defic Syndr 33: 426-438. doi:10.1097/00126334-200308010-00003. PubMed: 12869831. [DOI] [PubMed] [Google Scholar]
- 18. Masemola AM, Mashishi TN, Khoury G, Bredell H, Paximadis M et al. (2004) Novel and promiscuous CTL epitopes in conserved regions of Gag targeted by individuals with early subtype C HIV type 1 infection from southern Africa. J Immunol 173: 4607-4617. PubMed: 15383595. [DOI] [PubMed] [Google Scholar]
- 19. Malim MH, Emerman M (2001) HIV-1 sequence variation: drift, shift, and attenuation. Cell 104: 469-472. doi:10.1016/S0092-8674(01)00234-3. PubMed: 11239404. [DOI] [PubMed] [Google Scholar]
- 20. Fischetti VA, Jones KF, Hollingshead SK, Scott JR (1988) Structure, function, and genetics of streptococcal M protein. Rev Infect Dis 10 Suppl 2: S356-S359. doi:10.1093/cid/10.Supplement_2.S356. PubMed: 3055203. [DOI] [PubMed] [Google Scholar]
- 21. Cunningham MW (2000) Pathogenesis of group A streptococcal infections. Clin Microbiol Rev 13: 470-511. doi:10.1128/CMR.13.3.470-511.2000. PubMed: 10885988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hoof I, Peters B, Sidney J, Pedersen LE, Sette A et al. (2009) NetMHCpan, a method for MHC class I binding prediction beyond humans. Immunogenetics 61: 1-13. doi:10.1007/s00251-008-0341-z. PubMed: 19002680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nielsen M, Justesen S, Lund O, Lundegaard C, Buus S (2010) NetMHCIIpan-2.0 - Improved pan-specific HLA-DR predictions using a novel concurrent alignment and weight optimization training procedure. Immunome Res 6: 9. doi:10.1186/1745-7580-6-9. PubMed: 21073747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sidney J, Peters B, Frahm N, Brander C, Sette A (2008) HLA class I supertypes: a revised and updated classification. BMC Immunol 9: 1. doi:10.1186/1471-2172-9-1. PubMed: 18211710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jones MC (1993) Simple Boundary Correction for Kernel Density-Estimation. Statist Comput 3: 135-146. doi:10.1007/BF00147776. [Google Scholar]
- 26. Peters B, Bui HH, Frankild S, Nielson M, Lundegaard C et al. (2006) A community resource benchmarking predictions of peptide binding to MHC-I molecules. PLOS Comput Biol 2: e65. doi:10.1371/journal.pcbi.0020065. PubMed: 16789818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang H, Lundegaard C, Nielsen M (2009) Pan-specific MHC class I predictors: a benchmark of HLA class I pan-specific prediction methods. Bioinformatics 25: 83-89. doi:10.1093/bioinformatics/btn579. PubMed: 18996943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lin HH, Ray S, Tongchusak S, Reinherz EL, Brusic V (2008) Evaluation of MHC class I peptide binding prediction servers: applications for vaccine research. BMC Immunol 9: 8. doi:10.1186/1471-2172-9-8. PubMed: 18366636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lin HH, Zhang GL, Tongchusak S, Reinherz EL, Brusic V (2008) Evaluation of MHC-II peptide binding prediction servers: applications for vaccine research. BMC Bioinformatics 9 Suppl 12: S22. doi:10.1186/1471-2105-9-S7-P22. PubMed: 19091022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Goebel T (2007) Anthropology. The missing years for modern humans. Science 315: 194-196. doi:10.1126/science.1137564. PubMed: 17218514. [DOI] [PubMed] [Google Scholar]
- 31. Hershberg R, Lipatov M, Small PM, Sheffer H, Niemann S et al. (2008) High functional diversity in Mycobacterium tuberculosis driven by genetic drift and human demography. PLOS Biol 6: e311. doi:10.1371/journal.pbio.0060311. PubMed: 19090620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Weichold FF, Mueller S, Kortsik C, Hitzler WE, Wulf MJ et al. (2007) Impact of MHC class I alleles on the M. tuberculosis antigen-specific CD8+ T-cell response in patients with pulmonary tuberculosis. Genes Immun 8: 334-343. doi:10.1038/sj.gene.6364392. PubMed: 17429413. [DOI] [PubMed] [Google Scholar]
- 33. Eisen HN, Hou XH, Shen C, Wang K, Tanguturi VK et al. (2012) Promiscuous binding of extracellular peptides to cell surface class I MHC protein. Proc Natl Acad Sci U S A 109: 4580-4585. doi:10.1073/pnas.1201586109. PubMed: 22403068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tan PT, Heiny AT, Miotto O, Salmon J, Marques ET et al. (2010) Conservation and diversity of influenza A H1N1 HLA-restricted T cell epitope candidates for epitope-based vaccines. PLOS ONE 5: e8754. doi:10.1371/journal.pone.0008754. PubMed: 20090904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ribeiro SP, Rosa DS, Fonseca SG, Mairena EC, Postól E et al. (2010) A vaccine encoding conserved promiscuous HIV CD4 epitopes induces broad T cell responses in mice transgenic to multiple common HLA class II molecules. PLOS ONE 5: e11072. doi:10.1371/journal.pone.0011072. PubMed: 20552033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lamonaca V, Missale G, Urbani S, Pilli M, Boni C et al. (1999) Conserved hepatitis C virus sequences are highly immunogenic for CD4(+) T cells: implications for vaccine development. Hepatology 30: 1088-1098. doi:10.1002/hep.510300435. PubMed: 10498664. [DOI] [PubMed] [Google Scholar]
- 37. Fonseca SG, Coutinho-Silva A, Fonseca LA, Segurado AC, Moraes SL et al. (2006) Identification of novel consensus CD4 T-cell epitopes from clade B HIV-1 whole genome that are frequently recognized by HIV-1 infected patients. AIDS 20: 2263-2273. doi:10.1097/01.aids.0000253353.48331.5f. PubMed: 17117012. [DOI] [PubMed] [Google Scholar]
- 38. Bryson S, Julien JP, Hynes RC, Pai EF (2009) Crystallographic definition of the epitope promiscuity of the broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2F5: vaccine design implications. J Virol 83: 11862-11875. doi:10.1128/JVI.01604-09. PubMed: 19740978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rao X, Hoof I, Costa AI, van Baarle D, Keşmir C (2011) HLA class I allele promiscuity revisited. Immunogenetics 63: 691-701. doi:10.1007/s00251-011-0552-6. PubMed: 21695550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sanjuán R, Nebot MR, Peris JB, Alcamí J (2013) Immune Activation Promotes Evolutionary Conservation of T-Cell Epitopes in HIV-1. PLOS Biol 11: e1001523 PubMed: 23565057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Morris GP, Allen PM (2012) How the TCR balances sensitivity and specificity for the recognition of self and pathogens. Nat Immunol 13: 121-128. doi:10.1038/ni.2190. PubMed: 22261968. [DOI] [PubMed] [Google Scholar]
- 42. Croft NP, Smith SA, Wong YC, Tan CT, Dudek NL et al. (2013) Kinetics of Antigen Expression and Epitope Presentation during Virus Infection. PLOS Pathog 9: e1003129 PubMed: 23382674. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ROC curves were generated by running NetMHCpan-2.0 and NetMHCIIpan-2.0 against their published validation datasets. The true positive rate (TPR) is plotted against the false positive rate (FPR) at different thresholds of binding from 0 to 1. For subsequent analyses we chose a FPR of 0.05, which corresponded to thresholds of 0.29 for netMHCpan and 0.585 for netMHCIIpan, and TPRs of 0.89 for HLA class I and 0.24 for HLA class II.
(TIF)
Kernel density estimates of epitope promiscuity and graphs of mean HLA allele promiscuity (see Methods) of B. anthracis and C. tetani (BA & CT; blue), S. pyogenes and (SP; purple), HIV (green) and M. tuberculosis complex (MTBC; orange) across HLA-DR (A), HLA-A (B) and HLA-B (C) alleles. Differences in mean promiscuity are indicated with black bars (Tukey’s post-test, p < 0.05). Error bars represent the standard error of the mean.
(TIF)
Graphs of mean epitope promiscuity (see Methods) of B. anthracis and C. tetani (BA and CT), S. pyogenes and (SP), HIV and M. tuberculosis complex (MTBC) across alleles within HLA-A supertypes (A01, A02, A03, A24) and within HLA-B supertypes (B07, B08, B27, B44, B58, B62). For simplicity, Tukey post-tests are not shown because no trend was found between the groups.
(TIF)
The first column refers to the IEDB reference number for bacterial epitopes, and amino acid sequence for HIV epitopes. The second column refers to the microbe group from which the epitope was derived. The third column refers to the protein from which the epitope was derived. The fourth column refers to the class of HLA alleles that the epitopes have been shown to bind, and the class in which they were analyzed in this study.
(PDF)
(PDF)
(PDF)
(PDF)