Abstract
Streptococcus suis is an important zoonotic agent causing severe diseases in pigs and humans. To date, 33 serotypes of S . suis have been identified based on antigenic differences in the capsular polysaccharide. The capsular polysaccharide synthesis (cps) locus encodes proteins/enzymes that are responsible for capsular production and variation in the capsule structures are the basis of S . suis serotyping. Multiplex and/or simplex PCR assays have been developed for 15 serotypes based on serotype-specific genes in the cps gene cluster. In this study, we developed a set of multiplex PCR (mPCR) assays to identify the 33 currently known S . suis serotypes. To identify serotype-specific genes for mPCR, the entire genomes of reference strains for the 33 serotypes were sequenced using whole genome high-throughput sequencing, and the cps gene clusters from these strains were identified and compared. We developed a set of 4 mPCR assays based on the polysaccharide polymerase gene wzy, one of the serotype-specific genes. The assays can identify all serotypes except for two pairs of serotypes: 1 and 14, and 2 and 1/2, which have no serotype-specific genes between them. The first assay identifies 12 serotypes (serotypes 1 to 10, 1/2, and 14) that are the most frequently isolated from diseased pigs and patients; the second identifies 10 serotypes (serotypes 11 to 21 except 14); the third identifies the remaining 11 serotypes (serotypes 22 to 31, and 33); and the fourth identifies a new cps cluster of S . suis discovered in this study in 16 isolates that agglutinated with antisera for serotypes 29 and 21. The multiplex PCR assays developed in this study provide a rapid and specific method for molecular serotyping of S . suis .
Introduction
Streptococcus suis is one of the most important swine pathogens worldwide, responsible for cases of septicemia with sudden death, meningitis, arthritis, endocarditis, and pneumonia amongst other diseases [1], and is considered a major problem in the swine industry [2]. It is also an emerging zoonotic agent. Humans can be infected when in close contact with pigs or pork products through skin wounds, or through consumption of raw pork [3-5]. S . suis human infections commonly lead to meningitis [6]. Septic shock, endocarditis, cellulitis, peritonitis, rhabdomyolysis, arthritis, spondylodiscitis, pneumonia, uveitis, and endophthalmitis can also occur [7]. During the past few years, the number of human S . suis infections reported worldwide has increased significantly, with most cases reported in Asia [8-10].
Presently, 33 serotypes (type 1 through 31, 33, and 1/2) of S . suis have been identified [11]. Former S . suis serotypes 32 and 34 have been reclassified as Streptococcus orisratti [11]. Although there is no clear association between specific serotypes and a given pathological condition, strains isolated from diseased pigs primarily belong to serotype 2 in most countries, followed by serotypes 3, 4, 5, 7, 8, and 1/2 in Asian countries [12-14]. In some European countries, serotype 9 is also frequently recovered from diseased animals, followed by serotype 1 and 14 [15-17]. However, in Canada, serotypes 2, 3, and 1/2 are the three most prevalent serotypes followed by serotypes 4, 7, and 8 [18,19]. Serotype 2 is the most prevalent serotype isolated from human cases in many countries [20], but serotypes 1, 4, 5, 14, 16, and 24 have also been reported [21-25].
Serotyping is one of the most important diagnostic tools for S . suis and remains a valuable method to understand the epidemiology of a particular outbreak or to monitor serotype prevalence, as well as to guide vaccine development. Currently, S . suis serotypes are routinely identified by the agglutination or co-agglutination tests using serotype-specific antisera [26]. Although these techniques are relatively simple, producing antisera is laborious, time-consuming, and expensive. In addition, auto-agglutinating strains cannot be serotyped using antisera. The S . suis serotypes are determined by the antigenicity of the capsule [27-29]. Production of the capsule is encoded by capsular polysaccharide synthesis genes located at the cps locus [30,31]. Molecular serotyping by PCR amplification of serotype specific cps genes does not require antisera and is an attractive alternative to the current agglutination and co-agglutination tests. Several simplex PCR and multiplex PCR (mPCR) assays to identify S . suis serotypes 1, 14, 2, 1/2, 3, 4, 5, 7, 8, 9, 10, 16, 19, 23, and 25 have been reported [32-38]. However, there are 18 serotypes of S . suis that cannot be identified using the PCR assay available.
In the present study, we sequenced the genomes of the 33 S . suis serotype reference strains (1 to 31, 33, and 1/2) as well as one field isolate, using Illumina sequencing to obtain sequences of the cps gene clusters to identify serotype-specific genes. We developed a set of 4 mPCR assays, based on the serotype-specific polysaccharide polymerase gene wzy, for molecular serotyping of S . suis .
Material and Methods
Bacterial strains
Reference strains for 33 S . suis serotypes, 1 to 31, 33, and 1/2 from the S . suis strain collection at the University of Montréal, Montreal, Canada [39] and one field isolate from a healthy pig (see below) were used for genome sequencing. One serotype 14 clinical strain isolated from a patient [40], and 83 S . suis field strains isolated between 2011 and 2012 from clinically healthy pigs in slaughter houses in Beijing, Jiangsu province, and Sichuan province were used for evaluation of PCR typing. All isolates were serotyped using the agglutination test (serum provided by Statens Serum Institute, Copenhagen, Denmark). The strains were grown overnight on Columbia blood base agar plates (Guangzhou Detgerm Microbiological Science, P. R. China) at 37° C and a single colony was inoculated in 5 ml of Todd-Hewitt broth (THB, Oxoid Ltd., London, UK) and incubated for 8 h at 37° C with agitation (100 rpm). Streptococcus pneumoniae strains ATCC700657, ATCC700670, ATCC700676, ATCC700902, ATCC700906, ATCC49619, Streptococcus bovis ATCC33317, and Streptococcus pyogenes ATCC700294 were from our laboratory collection. Klebsiella pneumoniae 46117-3, Streptococcus pyogenes 32003, Streptococcus sanguis 32214, Enterococcus faecalis 32221, Streptococcus oralis 32231, Streptococcus lutetiensis 033, Streptococcus thermophilus 20174, Streptococcus mutans 10387, Streptococcus agalactiae 10465, and Streptococcus acidominimus 21026 were purchased from the China Center of Industrial Culture Collection. Streptococcus orisratti strains originally classified as the reference strains for S . suis serotypes 32 (strain EA1172.91) and 34 (strain 92-2742) were also from the S . suis strain collection at the University of Montréal, Montreal, Canada [11].
Whole genome sequencing and identification of the cps locus
Genomic DNA of bacterial strains was isolated and purified with the Wizard Genomic DNA Purification kit (Promega, Madison, MI). Genomic DNA was sequenced by Solexa sequencing after constructing a paired-end (PE) library with an average insert length of 500 bp to 2,000 bp. The reads were 100 bp in length generated with Illumina Solexa GA IIx (Illumina, San Diego, CA) and assembled into scaffolds using the program SOAP de novo (Release 2.04, http://soap.genomics.org.cn/soapdenovo.html). Each cps locus sequence was identified from the draft sequence based on the S . suis cps locus characteristics previously reported [30,31,41]. The Artemis program (www.sanger.ac.uk) was used to identify cps open reading frames (ORFs) and annotations [42]. BLAST and PSI-BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) were used to search several databases [43] including the GenBank (www.ncbi.nlm.nih.gov/GenBank), the Clusters of Orthologous Groups (COG; www.ncbi.nlm.nih.gov/COG/), and Pfam (pfam.sanger.ac.uk) protein motif databases [44,45]. cps genes were named according to the nomenclature for the S . suis serotype 2 cps locus [31]. The cps genes for a serotype were named with the serotype number followed by a letter from A to Z, in order, e.g., Cps11N means the Nth ORF from serotype 11. Only ORFs A to D are genetically highly similar across different serotypes.
The TMHMM v2.0 analysis program (http://www.cbs.dtu.dk/services/TMHMM/) was used to identify potential transmembrane segments from the amino acid sequences.
Identification of serotype-specific genes in the cps loci
The local BLAST program BLAST+ applications (downloaded from ftp://ftp.ncbi.nlm.nih.gov/blast/executables/LATEST) were performed on a Microsoft Windows platform (ftp://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/LATEST/user_manual.pdf). The genome sequences of the 33 serotype reference strains plus 18 genome sequences already deposited in the GenBank database were used to build a local database. Each cps gene was compared to the local database using the BLASTn program with default parameters. The E-value cut off for a significant match was set at 10−10 [46]. Serotype-specific genes were identified when the BLAST results showed no similarity to sequences of other serotype strains.
Sequence alignment and comparisons were performed using the ClustalW program [47]. Phylogenetic trees for the wzy gene of the 33 S . suis reference strains and other Streptococcus spp. were generated using the neighbor-joining method using the program MEGA 5.0 [48].
Primer design
Using the Primer-BLAST program (http://www.ncbi.nlm.nih.gov/tools/primer-blast/), primers were designed to have similar physical characteristics in order to allow simultaneous amplification in the same conditions and multiplex reactions. The lengths of the primers were between 20 and 23 bp, their melting temperatures were between 47.91 and 50.94 °C, and the expected amplicon sizes ranged between 153 and 1,006 bp. The primers based on the conserved region of thrA, a housekeeping gene, were designed to serve as internal controls [49]. The GenBank accession numbers of the genes used for primer design for the mPCR are shown in Table 1. The primers were synthesized by Sangon Biotech (Shanghai) and dissolved in TE buffer (10 mM Tris-Cl, 1 mM EDTA [pH 8.0]) to obtain 20 µM stock solutions.
Table 1. Serotype specific primers used in this study.
| Serotypes | Sequences (5′–3′)d | Targeted Genes | GenBank accession no. | Multiplex PCR assaye | PCR product size (bp) |
|---|---|---|---|---|---|
| 1 & 14a | Forward: 450-TCTTATAACAGGCGTCAAAACA-471 | cps1I | JX986790 | 1st | 153 |
| Reverse: 602-ATCGGTATAAAAGCAAGACACA-581 | JX986804 | ||||
| 2 & 1/2a | Forward: 544-TTCGTATTAACTTACTTGGCGT-565 | cps2I | KC537364 | 1st | 363 |
| Reverse:906-TAAATCCCCATATGCCAAATCC-885 | KC537384 | ||||
| 3a | Forward: 442-ACATCCATTGCAGGAGTAGT-461 | cps3L | KC537365 | 1st | 210 |
| Reverse:651-TGCAGTTCCAAAATTCTTCGT-631 | |||||
| 4a | Forward: 399-TGATATTGGCTATCTTTTGGGG-420 | cps4K | KC537366 | 1st | 542 |
| Reverse: 940-TTCCCCCTTCAAATAAACTCTG-919 | |||||
| 5a | Forward: 583-AGGTATGTCTTCTTATTCGCAG-604 | cps5L | KC537367 | 1st | 428 |
| Reverse: 1010-ATAATCCCTCCTGATACTAGGC-989 | |||||
| 6a | Forward: 141-TGGTGTCTTTCTACCTGCAA-160 | cps6I | KC537368 | 1st | 705 |
| Reverse:845-TCACCAAGATACGTGAACCA-826 | |||||
| 7a | Forward: 364-AAAATTCGTTCCATTGTAGGTG-385 | cps7L | KC537369 | 1st | 609 |
| Reverse: 972-TGAAGTTGAAGCTGGTGATAAA-951 | |||||
| 8a | Forward: 130-ATCGCTTCAAATAAGGTAGGAG-151 | cps8K | JX986797 | 1st | 268 |
| Reverse: 397-TGTAGGCCGTAATATCAACAAA-376 | |||||
| 9a | Forward:2-TGAAAGTAGGTATATCTCAGCA-23 | cps9J | KC537370 | 1st | 809 |
| Reverse: 810-AAAGAATTGAATCCCACCTGAG-789 | |||||
| 10a | Forward:25-CTATCACTACCACGGAATGC-44 | cps10M | JX986799 | 1st | 303 |
| Reverse:327-TAACCGTCCGTCTAGAATGT-308 | |||||
| 11a | Forward: 46-ATTGTTACGATTTGGGCGAT-65 | cps11N | KC537371 | 2nd | 512 |
| Reverse:557-GAACCCCATGTAGTTATGGC-538 | |||||
| 12a | Forward:1295- CATGGGAACTGTACAGGATAAG-1316 | cps12J | KC537372 | 2nd | 171 |
| Reverse: 1465-CCACCTTACTACCTGTTTTACC-1444 | |||||
| 13a | Forward: 30-GCTTGTAGCGAATTTTGGTATT-51 | cps13L | JX961643 | 2nd | 741 |
| Reverse:770-CCATTAGATGTATTTGCTCCCA-749 | |||||
| 15a | Forward: 565-ACCTACTCAAGAACATCCTTTC-586 | cps15K | JX961644 | 2nd | 458 |
| Reverse: 1022-GTAACTAAAACAGCAAACGTCA-1001 | |||||
| 16a | Forward: 551-ATCAACAAACATTTTCGAGGAC-572 | cps16I | KC537373 | 2nd | 223 |
| Reverse: 773-GCTGAATAATAGATTCGTCCTGT-751 | |||||
| 17a | Forward: 37-TTGCCGTATAAGGTCTTAGTTG-58 | cps17O | KC537374 | 2nd | 380 |
| Reverse: 416-ATCTGACGGTAAATGTTCTCTG-395 | |||||
| 18a | Forward: 689-ATAGGCTGTACTTTGATAACCG-710 | cps18N | KC537375 | 2nd | 310 |
| Reverse:998-AGCCTATCGCTCAAAAACTTAT-977 | |||||
| 19a | Forward:589-ATTATTATAGGGCAAAGCAGGG-610 | cps19L | KC537376 | 2nd | 674 |
| Reverse:1262-ATCGTACACAACAAAACGATTC-1241 | |||||
| 20a | Forward: 236-TAATCGTTGCCTTTGAGCAT-255 | cps20I | KC537377 | 2nd | 938 |
| Reverse: 1173-CGCTATATAAGGAAACCTCGG-1153 | |||||
| 21a | Forward: 218-TGGCAGACTTCTTTTCTCAC-237 | cps21P | KC537378 | 2nd | 858 |
| Reverse: 1075-CCTGTAGCGCCTCATAAAAC-1056 | |||||
| 22a | Forward: 183-AGGATCGGTAAGTTTAGGTACA-204 | cps22K | KC537379 | 3rd | 158 |
| Reverse:340-ATGCAGTAAAACACGAAAACAA-319 | |||||
| 23a | Forward: 250-TATTATAGTCCGATGCAAGCAG-271 | cps23J | JX986802 | 3rd | 461 |
| Reverse:710-ATGAGAACGAAACGGAATAGTT-689 | |||||
| 24a | Forward: 736-GATAGCAATGTAATCCAATCGC-757 | cps24M | KC537380 | 3rd | 204 |
| Reverse:939-GTAGGTTCCCCTAGTAAGAAGT-918 | |||||
| 25a | Forward: 477-ATTGAGTCCTTTTACTGGTAGC-498 | cps25M | JX986803 | 3rd | 390 |
| Reverse:866-TACTGAGCTACATAATCCCACA-845 | |||||
| 26a | Forward:663-CAAAATTCCTGGATTAACGCTT-684 | cps26P | KC537381 | 3rd | 315 |
| Reverse:977-CGATCTGAGGACTTATCAAGAA-956 | |||||
| 27a | Forward: 354-GTGGTTTTGGAGGATATTTTCG-375 | cps27K | JX961652 | 3rd | 530 |
| Reverse:883-ATTGAGATAAACTACTCCGTGC-862 | |||||
| 28a | Forward: 38-GGGCACTTGTTTTACTTCCT-57 | cps28L | JX961653 | 3rd | 896 |
| Reverse:933-GCCAAGTAATACCCTACCTG-914 | |||||
| 29a | Forward: 314-AAAGTGCCTATTCTGGGATTTT-335 | cps29L | JX961654 | 3rd (4th) | 263 |
| Reverse:576-TAAAGGCAACTTCCACATTGTA-555 | |||||
| 30a | Forward: 581-TTGGGCTTGTAAATAGTGAGAG-602 | cps30I | KC537382 | 3rd | 625 |
| Reverse:1205-CGATTAGATAAGCGCATTTGTT-1184 | |||||
| 31a | Forward: 19-CATATGTTTTCGTGGGGAGT-38 | cps31L | JX961656 | 3rd | 1006 |
| Reverse:1024-GTGATGAAAACATCGTTGGTAG-1003 | |||||
| 33a | Forward: 353-GAGTTGCGACCTATTATTCTCA-374 | cps33K | KC537383 | 3rd | 731 |
| Reverse:1083-GAATTGAACAACGACTGCAATA-1062 | |||||
| 21b | Forward: 13-TTGATAACAGGAGCAAACTCAT-34 | cps21H | KC537385 | 4th | 455 |
| Reverse: 467-TTACCATAAATCATCGGTGGTC-446 | |||||
| 21b | Forward: 78-AGTAGAAAGAGGGTACAAGGTT-99 | cps21I | KC537385 | 4th | 311 |
| Reverse: 388-CAGGTATGTTCCGTTTAGAACT-367 | |||||
| Allc | Forward: 1180-GAAAATATGAAGAGCCATGTCG-1201 | thrA | CP000837 | All 4 | 120 |
| Reverse: 1299-GACAACGAACATAACAGAAACTTC-1276 |
a Primers used in the multiplex PCR reactions.
b Primers used for screening the distribution of the cpsH and cpsI among 19 isolates which were positive for serotype 29 by mPCR.
c The thrA gene refer to S . suis GZ1 (GenBank NO. CP000837).
d The numbers flanking the oligonucleotide primers represent the positions in the target genes.
e Primers included in one or more of the four mPCR assays.
mPCR and detection of mPCR products
The different mPCR assays contained the same reagents except for primers. mPCR was performed using 2×Taq PCR Master Mix containing Taq DNA polymerase: 0.05 units/µl; MgCl2: 4 mM; dNTP: 4 mM; and buffer (Biomed, Beijing, China). The reaction mixture (20 µl) for each PCR consisted of 10 µl 2×Taq PCR Master Mix, and 0.2 µM of each primer. The PCR program for the mPCR reactions was as follows: 94° C for 5 min, followed by 30 cycles: 94° C for 30 s, 58° C for 40 s, and 72° C for 50 s; with a final extension of 72° C for 5 min in a thermocycler (Senso, Germany). The PCR products were analyzed with gel electrophoresis using 2% agarose gels and 0.5×TBE buffer in an electrophoresis chamber (32 cm between electrodes). The running time was 40 min at the voltage of 160 V and the current of 66 mA. PCR products were DNA sequenced.
To evaluate the sensitivity of the mPCR assays, S . suis reference strains were growth to an OD600 of 0.6 in broth culture which was roughly equivalent to 1×108 colony forming unit (CFU)/ml. This culture was diluted down in 10-fold serial dilutions, approximately from 1×108 CFU/ml to 10 CFU/ml. One ml of each dilution was used for DNA preparation using the Wizard Genomic DNA Purification kit (Promega, Madison, MI). At same time each dilution was plated out for CFU quantification to determine the actual number of cells used for DNA preparation. The amount of template used was based on the actual CFU count to work out the minimum number of CFUs required for the mPCR assays. This method assumed full recovery of genomic DNA during DNA preparation.
Results
Identification of the target genes for the mPCR assays
Comparison of all 33 cps gene clusters showed that the first four genes in the cps cluster were conserved in all reference strains while the central or last parts of the cps gene clusters contained the serotype-specific genes. One to 10 serotype-specific genes were identified for each serotype. However, no serotype-specific genes were found to distinguish between serotypes 1 and 14 or between serotypes 2 and 1/2. As previously shown [41], the cps gene clusters of these two pairs of serotypes are highly similar.
The function of most cps genes was predicted based on similarities to proteins found in searching the databases described in the M & M. However, database searches with Cps11N, Cps13L, Cps17O, Cps18N, Cps22K, Cps24M, Cps26P, and Cps28L failed to identify any significant similarity with any other proteins in the GenBank. Hydrophobicity analysis showed that they are all very hydrophobic proteins and that they have 9 to 13 predicted transmembrane segments, which is a typical topology for Wzy, a protein that polymerizes polysaccharide repeat units [50]. Accordingly, these genes were named as wzy.
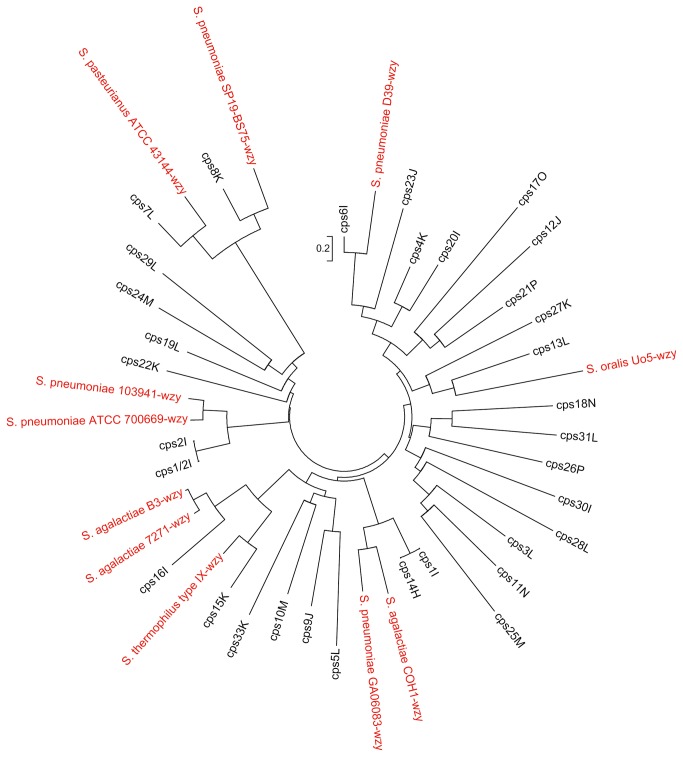
The serotype-specific genes of each serotype and their predicted functions are shown in Table S1. Note that there are some cps gene name discrepancies between the Wang et al. [41] and Okura et al. [51] reports. In our study, the cps gene names are the same as in the Okura et al. report. cps1I and cps1J were named as cps1H and cps1I respectively in the Wang et al. report (GenBank NO. JF273644). cps5H, cps5I, cps5J, cps5K, cps5L, cps5M, cps5N, and cps5O were named as cps5I, cps5J, cps5K, cps5L, cps5M, cps5N, cps5O, and cps5P, respectively, in the Wang et al. report (GenBank NO. JF273648). The serotype-specific genes encode glycosyltransferase, acetyltransferase, phosphotransferase, polysaccharide polymerase (Wzy), or flippase (Wzx). Of these serotype-specific genes, only wzy exists in all of the serotypes. Thus, with the exception of the cps1I/cps14H pair and the cps2I/cps1/2I pair, there is high sequence divergence in the wzy genes of different serotypes in S . suis (Figure 1). Therefore, the wzy gene was chosen as the target gene to develop the PCR assays for molecular serotyping.
Figure 1. The tree was constructed using the neighbor-joining algorithm based on wzy genes of the thirty-three reference strains of S . suis and other Streptococcus spp strains (in red color).
Bar, sequence dissimilarity.
Development and evaluation of the mPCR assays
First, we designed serotype-specific PCR primers based on the wzy gene and performed simplex PCRs to determine the specificity of each primer pair using template DNA extracted from the 33 reference strains. Each pair of primers amplified the predicted PCR product specifically from the DNA samples of the corresponding serotype, which was confirmed by DNA sequencing of the PCR products.
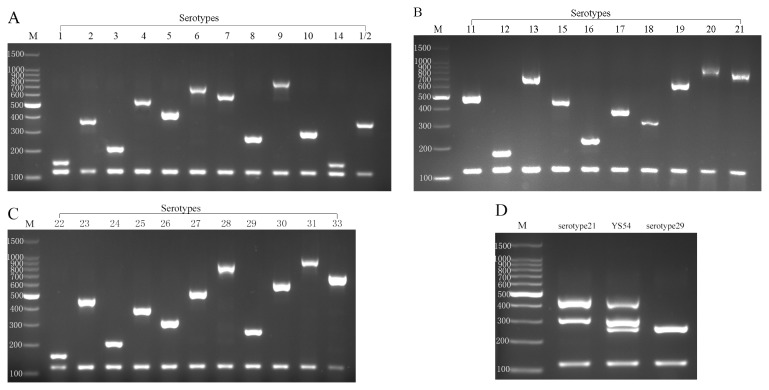
Three mPCR assays were then designed based on the simplex PCR assays above. A primer pair that amplifies a 120 bp fragment from the thrA gene was added to each mPCR as an internal control since thrA is present in all strains. Assay 1 was designed to identify the most common serotypes isolated from human and swine infections (serotypes 1 to 10, 1/2, and 14); assay 2, to identify serotypes 11 to 21 (except 14); and assay 3, to identify serotypes 22 to 33 (except 32). DNA samples prepared from the 33 reference strains were analyzed using the three mPCR assays. For each DNA sample two bands were produced, one of which was the internal control, as expected, while the other was the serotype-specific wzy gene. As anticipated, the mPCR assays could not differentiate serotype 1 from serotype 14, or serotype 2 from serotype 1/2. Non-specific amplification bands were not observed in any of the samples tested. The amplicon sizes allowed good separation on 2% agarose gels, where each PCR product could be unambiguously identified by size (Figure 2). The specificity of the mPCR assays was tested using 19 other Streptococcus spp. strains and one Klebsiella pneumoniae strain. No cross-amplification products were detected from these strains (results not shown).
Figure 2. Multiplex PCR (assay 1, 2, 3) products of S . suis reference strains representing all 33 serotypes(A,B,C).
Multiplex PCR (assay 4) products of S . suis isolate YS54 and serotype 21 and 29 reference strains(D). PCR products were electrophoresed on a 2% (wt/vol) agarose gel, stained with goldenview, and photographed under UV light. Serotypes are indicated above the lanes. Lane M: 100-bp DNA ladder markers (Biomed, Beijing, China), the sizes (bp) are indicated on the left.
The detection limit of the mPCR assays for all except two serotypes (serotype 9 and 20) was 104 CFU. For serotypes 9 and 20, it was 105 CFU.
Molecular serotyping results determined by the mPCR assays were compared with those obtained with the sero-agglutination test using 84 S . suis isolates (Table 2). There was complete consistency between the two techniques for 68 strains. However, 16 strains showed agglutination with both serotypes 29 and 21 antisera but were identified as serotype 29 by mPCR. This discrepancy is discussed further below.
Table 2. Typing obtained with 84 isolates of S . suis using the multiplex PCR assays and the agglutination test with serotype-specific antisera.
| Serotype using antiserum | No. of isolates tested | Serotype using multiplex PCR |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1&14 | 1/2& 2 | 5 | 7 | 8 | 9 | 10 | 11 | 12 | 15 | 16 | 21 | 22 | 24 | 29 | 30 | 31 | ||
| 1 | 1 | 1 | ||||||||||||||||
| 14 | 1 | 1 | ||||||||||||||||
| 2 | 8 | 8 | ||||||||||||||||
| 1/2 | 3 | 3 | ||||||||||||||||
| 5 | 8 | 8 | ||||||||||||||||
| 7 | 3 | 3 | ||||||||||||||||
| 8 | 2 | 2 | ||||||||||||||||
| 9 | 1 | 1 | ||||||||||||||||
| 10 | 3 | 3 | ||||||||||||||||
| 11 | 9 | 9 | ||||||||||||||||
| 12 | 8 | 8 | ||||||||||||||||
| 15 | 1 | 1 | ||||||||||||||||
| 16 | 1 | 1 | ||||||||||||||||
| 21 | 1 | 1 | ||||||||||||||||
| 22 | 1 | 1 | ||||||||||||||||
| 24 | 4 | 4 | ||||||||||||||||
| 29 | 3 | 3 | ||||||||||||||||
| 30 | 8 | 8 | ||||||||||||||||
| 31 | 2 | 2 | ||||||||||||||||
| 29/21* | 16 | 16 | ||||||||||||||||
agglutinating with both serotypes 29 and 21 antisera and serotype 29 positive in the third multiplex PCR assay.
Development of an mPCR assay for typing strains agglutinated with both serotypes 29 and 21 antisera
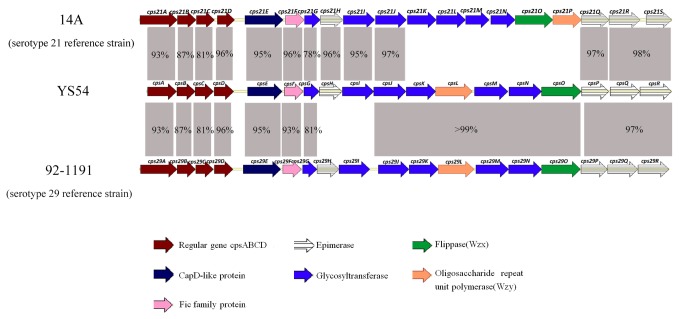
As described above, 16 isolates agglutinated with both serotype 29 and 21 antisera but were only positive for serotype 29 using mPCR. To reveal the genetic basis of the discrepancy, we sequenced the genome of one of these 16 isolates (YS54). The cps gene cluster of YS54 was compared with those of S . suis serotype 21 and 29 reference strains, 14A and 92-1191 respectively.
The sizes of the cps gene clusters in YS54 (GenBank accession number KC537387), 14A (serotype 21 reference strain, GenBank accession number KC537385), and 92-1191 (serotype 29 reference strain, GenBank accession number KC537386) were 20,579 bp, 20,263 bp, and 20,135 bp, respectively. Differences between the cps gene clusters of these three strains are shown in Figure 3. The cps genes of YS54 are highly similar to serotype 29 strain 92-1191 except for cpsH and cpsI. The cpsH and cpsI of YS54 were highly similar to those of serotype 21 strain 14A, while the cpsH and cpsI of strain 14A shared no similarity with strain 91-1191. CPS29H showed 53% identity to the nucleoside-diphosphate-sugar epimerase of Clostridium clariflavum (GenBank accession number YP_005048548). CPS29I showed 44% identity to the glycosyltransferase of Enterococcus faecium (GenBank accession number EJV43441). CPS21H shared 59% identity with the UDP-sugar epimerase of Amphibacillus xylanus (GenBank accession number YP_006845901). CPS21I shared 55% identity with the group 1 glycosyltransferase of Acetivibrio cellulolyticus (GenBank accession number ZP_09465963). Therefore the cps gene cluster of YS54 is novel.
Figure 3. Comparisons of the cps loci of isolate YS54 and serotype 21 and 29 reference strains.
The numbers between the cps loci present identities of the cps genes.
To identify this novel cps gene cluster by PCR, we designed a fourth mPCR assay containing three pairs of primers targeting cps29L, cps21H, cps21I, as well as the internal control thrA. The 16 isolates with cross agglutination and three serotype 29 isolates were tested. The three serotype 29 isolates were identified as serotype 29, yielding the same amplification pattern as the serotype 29 reference strain, while the 16 isolates with cross agglutination showed the same amplification pattern as strain YS54 (Figure 2).
Discussion
In this study we developed four mPCR assays as a molecular serotyping scheme for S . suis . The scheme encompasses all S . suis serotypes that are differentiated by serotype-specific genes. The mPCR assays can supplement or supercede earlier methods developed for only 15 S . suis serotypes (1, 14, 2, 1/2, 3, 4, 5, 7, 8, 9, 10, 16, 19, 23, and 25) [32-38]. Tien et al. recently reported that the reference strains of serotypes 20, 22, 26, and 33 do not belong to S . suis [52]. However reclassification of these S . suis serotypes has not been widely accepted. In addition, strains belonging to these serotypes are still isolated from diseased pigs [19]. As a consequence, we decided to include all of these serotypes in our mPCR assays.
In this study, we used whole genome sequencing to obtain the full cps gene cluster from all serotypes to identify serotype-specific genes. We compared our cps sequences with the sequence data published recently by Okura et al. [51], where cps1, cps1/2, cps6, cps12-15, cps17, cps18, cps21, cps22, cps24, cps26-29, cps31, and cps33 were 100% identical; while cps11, cps20, and cps30 had 1 bp differences. We also compared our sequences with the sequences reported by Wang et al. [41], where cps3, cps4, cps5, cps8, cps23, and cps25 were 100% identical; while cps7, cps9, cps10, cps16, and cps19 had 3 bp, 26 bp, 2 bp, 57 bp and 1 bp differences, respectively [41]. Since the strains used were the same for the 8 serotypes with discrepant sequence, the differences may be the result of mutations during subculture or sequencing errors. Additionally our cps2 is 100% identical to the cps of S735, a serotype 2 strain [53].
The choice of gene targets for serotype specificity was an important consideration in developing the PCR serotyping assays. The targets used previously for identification of certain serotypes by PCR were based on various serotype-specific genes [32,33,35-38]; whereas our mPCR assay was developed based on the serotype-specific wzy genes from all of the serotypes. The formation of capsular polysaccharides in S . suis was proposed to be similar to several other Streptococcus species synthesized by the Wzy-dependent pathway where repeat units are built on the inner face of the cytoplasmic membrane, transported to the outer face of the membrane with Wzx flippase, and polymerized with Wzy polymerase [41,50,54]. Wzy-dependent polymers usually contain various sugars and glycosidic linkages. The specificity of the Wzy polymerase determines the linkage it catalyzes between sugars on the growing chain and the next repeat unit [55]. As shown in Figure 1, the wzy genes in S . suis serotypes share low identity with other Streptococcus spp. (e.g. cps2I/cps1/2I share 64% identity with 84% coverage with the wzy of Streptococcus pneumoniae strain 103941, and cps7L shares 64% identity with 67% coverage with the wzy of Streptococcus pasteurianus ATCC 43144). The wzy genes from the different serotypes except those between serotype 1 and 14 and serotype 2 and 1/2 share very little DNA sequence identity. Therefore, the wzy gene is ideally suited as a target for molecular serotyping in S . suis . The sequence divergence eliminates non-specific amplification from other serotypes or other species.
We developed the mPCR serotyping assays based on conventional PCR because it is widely available and more affordable than real-time PCR; in particular, it is more readily deployable in developing countries where most of the S . suis infections occur. Conventional PCR also allowed more targets (up to 12 targets in our assays) to be included in one mPCR assay than real-time PCR, which depends on the number of colors (up to six) that a real-time PCR machine is able to detect. Since it was not possible to formulate one mPCR assay to include all serotype specific gene targets, we have developed four mPCR assays to detect these serotypes. The disadvantage of multiple assays is the increased workload. To alleviate this, we designed the first mPCR assay to identify the most common serotypes recovered from clinical samples [12-19]. This assay should be performed first. If the strain is not identifiable by this assay, the 3 other assays can then be used. This strategy reduces workload with minimal delay in reporting typing results.
As previously reported the cps gene clusters of serotype 1 and serotype 14, and of serotype 2 and serotype 1/2 are very similar with the nucleotide sequence of the wzy genes being nearly identical in these two pairs of serotypes. Therefore, the mPCR assays developed in this study cannot discriminate these two pairs of serotypes. Differentiation of serotype 1 and 14, and serotype 2 and 1/2 will require the use of serotype specific antisera. Okura et al. suggested the antigenic differences between serotypes 1 and 14 may be attributed to point mutations in cpsG and cpsE in the two serotypes [51]. Thus differentiation of these 2 pairs of serotypes using mutational changes may be feasible.
Sixteen strains agglutinated with both serotype 29 and 21 antisera and can only be differentiated using the fourth mPCR assay. The serotype 21/29 strains were recovered in different years and, more importantly, in different parts of China and are probably unrelated epidemiologically. These strains potentially belong to a new serotype. Indeed, serotype 1/2 was recognized as a serotype due to cross reaction with both serotype 1 and serotype 2 antisera [27]. It is important to note that cross-reacting strains (between 2 or even 3 serotypes) are frequently observed when serotyping high numbers of strains (M. Gottschalk, unpublished observations, International Reference Laboratory for S . suis serotyping). Therefore, testing such strains with the mPCR assays may identify new cps gene clusters.
We show that cps29H and cps29I were replaced by cps21H and cps21I in these 16 isolates. Even though the predicted functions of cpsH and cpsI are glycosyltransferase and epimerase respectively in both serotype 21 and serotype 29, their amino acids were very different. The cpsH and cpsI in these cross-reaction strains must transfer the same glycosyl group as that in CPS21 leading to the formation of the shared CPS epitope(s) with CPS21. This may be an explanation of the cross-reaction with both antisera 21 and 29 in these strains. A similar situation can be found between a serotype Ib of group B streptococcus and serotype 35B of S. pneumoniae [56,57]. We recommend that strains positive for serotype 29 in the third mPCR assay should be tested using the fourth mPCR assay to determine whether the isolates belong to this new cps gene cluster type.
The detection limit of the mPCR assays was in the range of 104 CFU to 105 CFU, which appeared to be low. However, some other mPCR based methods also reported similar level of sensitivity. The detection limit of an mPCR for serotyping Neisseria mengingitidis was 1 ng of purified DNA which is equivalent to 4×105 genomes [58]. The range of detection limits of an mPCR assay for identification and differentiation of Campylobacter species was between 2.5×105 CFU and 2.5×1010 using unpurified DNA template prepared using the boiling method [59]. We tested the sensitivity of our method using purified DNA prepared from known number of cells. We assumed that all cells used for the DNA preparation were fully recovered as genomic DNA. Since some loss of DNA may have occurred during purification, the actual sensitivity might be higher.
Field strains used in this study were recovered from tonsils of clinically healthy pigs. Strains originating from carrier animals may explain why the distribution of the serotypes in this study varied from the most common serotypes previously described. The mPCR test developed here may be used to survey a large collection of strains from both diseased and healthy animals from different geographical regions to determine the distribution of different S . suis serotypes.
In conclusion, the mPCR based molecular serotyping method we developed for S . suis is a relatively systematic typing tool with which all except two pairs of serotypes of S . suis can be identified. It provides a fast and cost-effective way to determine the serotype of an isolate of the currently recognized serotypes. The set of 4 mPCR assays developed in this study was tested using bacterial isolates only. Future studies will aim to develop this mPCR-based typing method to directly detect and serotype S . suis from clinical samples.
Supporting Information
The serotype-specific genes of all serotypes.
(DOCX)
Funding Statement
This study was supported by grant (2013ZX10004221, 2013ZX10004216-001-002 and 812111251) from Minister of Science and Technology, People's Republic of China. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Wertheim HF, Nghia HD, Taylor W, Schultsz C (2009) Streptococcus suis: an emerging human pathogen. Clin Infect Dis 48: 617-625. doi:10.1086/596763. PubMed: 19191650. [DOI] [PubMed] [Google Scholar]
- 2. Gottschalk M (2012) Streptococcocis, In: Straw BE, Zimmerman JJ, D’Allaire S, Taylor DJ. Diseases of Swine. Ames, IA, USA: Blackwell Publishing; pp. 841-855. [Google Scholar]
- 3. Kay R, Cheng AF, Tse CY (1995) Streptococcus suis infection in Hong Kong. QJM 88: 39-47. PubMed: 7894987. [PubMed] [Google Scholar]
- 4. Yu HJ, Liu XC, Wang SW, Liu LG, Zu RQ et al. (2005) Matched case-control study for risk factors of human Streptococcus suis infection in Sichuan Province, China. Zhonghua Liu Xing Bing Xue Za Zhi 26: 636-639. PubMed: 16471206. [PubMed] [Google Scholar]
- 5. Takeuchi D, Kerdsin A, Pienpringam A, Loetthong P, Samerchea S et al. (2012) Population-based study of Streptococcus suis infection in humans in Phayao Province in northern Thailand. PLOS ONE 7: e31265. doi:10.1371/journal.pone.0031265. PubMed: 22363601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Arends JP, Zanen HC (1988) Meningitis caused by Streptococcus suis in humans. Rev Infect Dis 10: 131-137. doi:10.1093/clinids/10.1.131. PubMed: 3353625. [DOI] [PubMed] [Google Scholar]
- 7. Huang YT, Teng LJ, Ho SW, Hsueh PR (2005) Streptococcus suis infection. J Microbiol Immunol Infect 38: 306-313. PubMed: 16211137. [PubMed] [Google Scholar]
- 8. Hui AC, Ng KC, Tong PY, Mok V, Chow KM et al. (2005) Bacterial meningitis in Hong Kong: 10-years’ experience. Clin Neurol Neurosurg 107: 366-370. doi:10.1016/j.clineuro.2004.10.006. PubMed: 16023529. [DOI] [PubMed] [Google Scholar]
- 9. Yu H, Jing H, Chen Z, Zheng H, Zhu X et al. (2006) Human Streptococcus suis outbreak, Sichuan, China. Emerg Infect Dis 12: 914-920. doi:10.3201/eid1206.051194. PubMed: 16707046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mai NT, Hoa NT, Nga TV, Linh le D, Chau TT et al. (2008) Streptococcus suis meningitis in adults in Vietnam. Clin Infect Dis 46: 659-667. doi:10.1086/527385. PubMed: 19413493. [DOI] [PubMed] [Google Scholar]
- 11. Hill JE, Gottschalk M, Brousseau R, Harel J, Hemmingsen SM et al. (2005) Biochemical analysis, cpn60 and 16S rDNA sequence data indicate that Streptococcus suis serotypes 32 and 34, isolated from pigs, are Streptococcus orisratti . Vet Microbiol 107: 63-69. doi:10.1016/j.vetmic.2005.01.003. PubMed: 15795078. [DOI] [PubMed] [Google Scholar]
- 12. Kataoka Y, Sugimoto C, Nakazawa M, Morozumi T, Kashiwazaki M (1993) The epidemiological studies of Streptococcus suis infections in Japan from 1987 to 1991. J Vet Med Sci 55: 623-626. doi:10.1292/jvms.55.623. PubMed: 8399744. [DOI] [PubMed] [Google Scholar]
- 13. Wei Z, Li R, Zhang A, He H, Hua Y et al. (2009) Characterization of Streptococcus suis isolates from the diseased pigs in China between 2003 and 2007. Vet Microbiol 137: 196-201. doi:10.1016/j.vetmic.2008.12.015. PubMed: 19185432. [DOI] [PubMed] [Google Scholar]
- 14. Kim D, Han K, Oh Y, Kim CH, Kang I et al. (2010) Distribution of capsular serotypes and virulence markers of Streptococcus suis isolated from pigs with polyserositis in Korea. Can J Vet Res 74: 314-316. PubMed: 21197232. [PMC free article] [PubMed] [Google Scholar]
- 15. Aarestrup FM, Jorsal SE, Jensen NE (1998) Serological characterization and antimicrobial susceptibility of Streptococcus suis isolates from diagnostic samples in Denmark during 1995 and 1996. Vet Microbiol 60: 59-66. doi:10.1016/S0378-1135(98)00147-3. PubMed: 9595627. [DOI] [PubMed] [Google Scholar]
- 16. Wisselink HJ, Smith HE, Stockhofe-Zurwieden N, Peperkamp K, Vecht U (2000) Distribution of capsular types and production of muramidase-released protein (MRP) and extracellular factor (EF) of Streptococcus suis strains isolated from diseased pigs in seven European countries. Vet Microbiol 74: 237-248. doi:10.1016/S0378-1135(00)00188-7. PubMed: 10808092. [DOI] [PubMed] [Google Scholar]
- 17. Schultsz C, Jansen E, Keijzers W, Rothkamp A, Duim B et al. (2012) Differences in the population structure of invasive Streptococcus suis strains isolated from pigs and from humans in The Netherlands. PLOS ONE 7: e33854. doi:10.1371/journal.pone.0033854. PubMed: 22563452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Messier S, Lacouture S, Gottschalk M (2008) Distribution of Streptococcus suis capsular types from 2001 to 2007. Can Vet J 49: 461-462. PubMed: 18512456. [PMC free article] [PubMed] [Google Scholar]
- 19. Gottschalk M, Lacouture S, Bonifait L, Roy D, Fittipaldi N et al. (2013) Characterization of Streptococcus suis isolates recovered between 2008 and 2011 from diseased pigs in Quebec, Canada. Vet Microbiol 162: 819-825. doi:10.1016/j.vetmic.2012.10.028. PubMed: 23177911. [DOI] [PubMed] [Google Scholar]
- 20. Gottschalk M, Xu J, Calzas C, Segura M (2010) Streptococcus suis: a new emerging or an old neglected zoonotic pathogen? Future Microbiol 5: 371-391. doi:10.2217/fmb.10.2. PubMed: 20210549. [DOI] [PubMed] [Google Scholar]
- 21. Kerdsin A, Oishi K, Sripakdee S, Boonkerd N, Polwichai P et al. (2009) Clonal dissemination of human isolates of Streptococcus suis serotype 14 in Thailand. J Med Microbiol 58: 1508-1513. doi:10.1099/jmm.0.013656-0. PubMed: 19661209. [DOI] [PubMed] [Google Scholar]
- 22. Nghia HD, Hoa NT, Linh le D, Campbell J, Diep TS et al. (2008) Human case of Streptococcus suis serotype 16 infection. Emerg Infect Dis 14: 155-157. doi:10.3201/eid1401.070534. PubMed: 18258097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kerdsin A, Dejsirilert S, Sawanpanyalert P, Boonnark A, Noithachang W et al. (2011) Sepsis and spontaneous bacterial peritonitis in Thailand. Lancet 378: 960. doi:10.1016/S0140-6736(11)60923-9. PubMed: 21890062. [DOI] [PubMed] [Google Scholar]
- 24. Gottschalk M, Segura M, Xu J (2007) Streptococcus suis infections in humans: the Chinese experience and the situation in North America. Anim Health Res Rev 8: 29-45. doi:10.1017/S1466252307001247. PubMed: 17692141. [DOI] [PubMed] [Google Scholar]
- 25. Vilaichone RK, Vilaichone W, Nunthapisud P, Wilde H (2002) Streptococcus suis infection in Thailand. J Med Assoc Thai 85 Suppl 1: S109-S117. PubMed: 12188400. [PubMed] [Google Scholar]
- 26. Higgins R, Gottschalk M (1990) An update on Streptococcus suis identification. J Vet Diagn Invest 2: 249-252. doi:10.1177/104063879000200324. PubMed: 2094457. [DOI] [PubMed] [Google Scholar]
- 27. Demoor CE (1963) Septicaemic Infections in Pigs, Caused by Haemolytic Streptococci of New Lancefield Groups. Designated R S And T Antonie Van Leeuwenhoek 29: 272-280. doi:10.1007/BF02046069. [DOI] [PubMed] [Google Scholar]
- 28. Elliott SD (1966) Streptococcal infection in young pigs. I. An immunochemical study of the causative agent (PM streptococcus). J Hyg (Lond) 64: 205-212. doi:10.1017/S0022172400040468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Windsor RS, Elliott SD (1975) Streptococcal infection in young pigs. IV. An outbreak of streptococcal meningitis in weaned pigs. J Hyg (Lond) 75: 69-78. doi:10.1017/S0022172400047070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Smith HE, Damman M, van der Velde J, Wagenaar F, Wisselink HJ et al. (1999) Identification and characterization of the cps locus of Streptococcus suis serotype 2: the capsule protects against phagocytosis and is an important virulence factor. Infect Immun 67: 1750-1756. PubMed: 10085014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Smith HE, de Vries R, van’t Slot R, Smits MA (2000) The cps locus of Streptococcus suis serotype 2: genetic determinant for the synthesis of sialic acid. Microb Pathog 29: 127-134. doi:10.1006/mpat.2000.0372. PubMed: 10906268. [DOI] [PubMed] [Google Scholar]
- 32. Smith HE, van Bruijnsvoort L, Buijs H, Wisselink HJ, Smits MA (1999) Rapid PCR test for Streptococcus suis serotype 7. FEMS Microbiol Lett 178: 265-270. doi:10.1111/j.1574-6968.1999.tb08686.x. PubMed: 10499276. [DOI] [PubMed] [Google Scholar]
- 33. Smith HE, Veenbergen V, van der Velde J, Damman M, Wisselink HJ et al. (1999) The cps genes of Streptococcus suis serotypes 1, 2, and 9: development of rapid serotype-specific PCR assays. J Clin Microbiol 37: 3146-3152. PubMed: 10488168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang K, Fan W, Wisselink H, Lu C (2011) The cps locus of Streptococcus suis serotype 16: Development of a serotype-specific PCR assay. Vet Microbiol 153: 403-406. doi:10.1016/j.vetmic.2011.05.050. PubMed: 21715110. [DOI] [PubMed] [Google Scholar]
- 35. Wang K, Sun X, Lu C (2012) Development of Rapid Serotype-Specific PCR Assays for Eight Serotypes of Streptococcus suis . J Clin Microbiol 50: 3329-3334. doi:10.1128/JCM.01584-12. PubMed: 22875885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wisselink HJ, Joosten JJ, Smith HE (2002) Multiplex PCR assays for simultaneous detection of six major serotypes and two virulence-associated phenotypes of Streptococcus suis in tonsillar specimens from pigs. J Clin Microbiol 40: 2922-2929. doi:10.1128/JCM.40.8.2922-2929.2002. PubMed: 12149353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Marois C, Bougeard S, Gottschalk M, Kobisch M (2004) Multiplex PCR Assay for Detection of Streptococcus suis Species and Serotypes 2 and 1/2 in Tonsils of Live and Dead Pigs. J Clin Microbiol 42: 3169-3175. doi:10.1128/JCM.42.7.3169-3175.2004. PubMed: 15243078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Anusak K, Dejsirilert S, Akeda Y, Sekizaki T, Hamada S et al. (2012) Fifteen Streptococcus suis serotypes identified by multiplex PCR. J Med Microbiol, 61: 1669–72. PubMed: 22918870. [DOI] [PubMed] [Google Scholar]
- 39. Harel J, Higgins R, Gottschalk M, Bigras-Poulin M (1994) Genomic relatedness among reference strains of different Streptococcus suis serotypes. Can J Vet Res 58: 259-262. PubMed: 7534206. [PMC free article] [PubMed] [Google Scholar]
- 40. Ye C, Bai X, Zhang J, Jing H, Zheng H et al. (2008) Spread of Streptococcus suis sequence type 7, China. Emerg Infect Dis 14: 787-791. doi:10.3201/eid1405.070437. PubMed: 18439362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang K, Fan W, Cai L, Huang B, Lu C (2011) Genetic analysis of the capsular polysaccharide synthesis locus in 15 Streptococcus suis serotypes. FEMS Microbiol Lett 324: 117-124. doi:10.1111/j.1574-6968.2011.02394.x. PubMed: 22092812. [DOI] [PubMed] [Google Scholar]
- 42. Mural RJ (2000) ARTEMIS: a tool for displaying and annotating DNA sequence. Brief Bioinform 1: 199-200. doi:10.1093/bib/1.2.199. PubMed: 11465031. [DOI] [PubMed] [Google Scholar]
- 43. Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389-3402. doi:10.1093/nar/25.17.3389. PubMed: 9254694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tatusov RL, Natale DA, Garkavtsev IV, Tatusova TA, Shankavaram UT et al. (2001) The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res 29: 22-28. doi:10.1093/nar/29.4.e22. PubMed: 11125040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L et al. (2002) The Pfam protein families database. Nucleic Acids Res 30: 276-280. doi:10.1093/nar/30.1.276. PubMed: 11752314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Prosdocimi F, Faria-Campos AC, Peixoto FC, Pena SD, Ortega JM et al. (2002) Clustering of Schistosoma mansoni mRNA sequences and analysis of the most transcribed genes: implications in metabolism and biology of different developmental stages. Mem Inst Oswaldo Cruz 97 Suppl 1: 61-69. doi:10.1590/S0074-02762002000100009. PubMed: 12426597. [DOI] [PubMed] [Google Scholar]
- 47. Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673-4680. doi:10.1093/nar/22.22.4673. PubMed: 7984417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tamura K, Peterson D, Peterson N, Stecher G, Nei M et al. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731-2739. doi:10.1093/molbev/msr121. PubMed: 21546353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. King SJ, Leigh JA, Heath PJ, Luque I, Tarradas C et al. (2002) Development of a multilocus sequence typing scheme for the pig pathogen Streptococcus suis: identification of virulent clones and potential capsular serotype exchange. J Clin Microbiol 40: 3671-3680. doi:10.1128/JCM.40.10.3671-3680.2002. PubMed: 12354864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bentley SD, Aanensen DM, Mavroidi A, Saunders D, Rabbinowitsch E et al. (2006) Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLOS Genet 2: e31. doi:10.1371/journal.pgen.0020031. PubMed: 16532061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Okura M, Takamatsu D, Maruyama F, Nozawa T, Nakagawa I et al. (2013) Genetic Analysis of Capsular Polysaccharide Synthesis Gene Clusters from All Serotypes of Streptococcus suis: Potential Mechanisms for Generation of Capsular Variation. Appl Environ Microbiol 79: 2796-2806. doi:10.1128/AEM.03742-12. PubMed: 23416996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. le Tien, Nishibori T, Nishitani Y, Nomoto R, Osawa R (2013) Reappraisal of the taxonomy of Streptococcus suis serotypes 20, 22, 26, and 33 based on DNA-DNA homology and sodA and recN phylogenies. Vet Microbiol 162: 842-849. doi:10.1016/j.vetmic.2012.11.001. PubMed: 23245487. [DOI] [PubMed] [Google Scholar]
- 53. Boyle B, Vaillancourt K, Bonifait L, Charette SJ, Gottschalk M et al. (2012) Genome sequence of the swine pathogen Streptococcus suis serotype 2 strain S735. J Bacteriol 194: 6343-6344. doi:10.1128/JB.01559-12. PubMed: 23105076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yother J (2011) Capsules of Streptococcus pneumoniae and other bacteria: paradigms for polysaccharide biosynthesis and regulation. Annu Rev Microbiol 65: 563-581. doi:10.1146/annurev.micro.62.081307.162944. PubMed: 21721938. [DOI] [PubMed] [Google Scholar]
- 55. Chaffin DO, Beres SB, Yim HH, Rubens CE (2000) The serotype of type Ia and III group B streptococci is determined by the polymerase gene within the polycistronic capsule operon. J Bacteriol 182: 4466-4477. doi:10.1128/JB.182.16.4466-4477.2000. PubMed: 10913080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cieslewicz MJ, Chaffin D, Glusman G, Kasper D, Madan A et al. (2005) Structural and genetic diversity of group B streptococcus capsular polysaccharides. Infect Immun 73: 3096-3103. doi:10.1128/IAI.73.5.3096-3103.2005. PubMed: 15845517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mavroidi A, Aanensen DM, Godoy D, Skovsted IC, Kaltoft MS et al. (2007) Genetic relatedness of the Streptococcus pneumoniae capsular biosynthetic loci. J Bacteriol 189: 7841-7855. doi:10.1128/JB.00836-07. PubMed: 17766424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhu H, Wang Q, Wen L, Xu J, Shao Z et al. (2012) Development of a multiplex PCR assay for detection and genogrouping of Neisseria meningitidis . J Clin Microbiol 50: 46-51. doi:10.1128/JCM.00918-11. PubMed: 22090406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wang G, Clark CG, Taylor TM, Pucknell C, Barton C et al. (2002) Colony multiplex PCR assay for identification and differentiation of Campylobacter jejuni, C. coli, C. lari, C. upsaliensis, and C. fetus subsp. fetus . J Clin Microbiol 40: 4744-4747. doi:10.1128/JCM.40.12.4744-4747.2002. PubMed: 12454184. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The serotype-specific genes of all serotypes.
(DOCX)