Abstract
RNA editing is a post-transcriptional process, which results in base substitution modifications to RNA. It is an important process in generating protein diversity through amino acid substitution and the modulation of splicing events. Previous studies have suggested a link between gene-specific reductions in adenosine to inosine RNA editing and aging in the human brain. Here we demonstrate that changes in RNA editing observed in humans with age are not observed during aging in healthy rats. Furthermore, we identify a conserved editing site in rats, in Cog3. We propose that either age-related changes in RNA editing are specific to primates or humans, or that they are the manifestation of disease pathology. Since rodents are often used as model organisms for studying aging, these findings demonstrate the importance of understanding species-specific differences in RNA biology during aging.
Electronic supplementary material
The online version of this article (doi:10.1007/s10522-013-9433-8) contains supplementary material, which is available to authorized users.
Keywords: RNA editing, RNA-seq, Cerebral cortex, Brain aging
Introduction
Adenosine to inosine (A-to-I) RNA editing is a post-transcriptional process that alters the sequences of RNA molecules. The adenosine deaminases ADAR and ADARB1 convert specific adenosine residues on RNA to inosine bases. During translation, sequencing, and splicing, inosine is recognized as guanosine. Therefore, A-to-I RNA editing has important implications in altering specific amino acids, miRNA targeting, and in the modulation of alternative splicing (Nishikura 2010).
Targets of A-to-I RNA editing are often genes involved in neurotransmission in the central nervous system (CNS), although editing is known to occur in other tissues. Changes in A-to-I RNA editing have been implicated in the development of cancer inside and outside the CNS (Cenci et al. 2008; Galeano et al. 2010; Paz et al. 2007; Shah et al. 2009), and in various neurodegenerative diseases, such as dyschromatosis symmetrica hereditaria (DSH), amyotrophic lateral sclerosis (ALS), Alzheimer’s disease, and Huntington’s disease. In addition, the process has implications in epilepsy, depression, schizophrenia and is associated with a greater risk of suicide (Akbarian et al. 1995; Farajollahi and Maas 2010; Maas et al. 2006). More recently it was identified that single nucleotide polymorphisms in ADARB1 and ADARB2 are associated with extreme longevity in humans (Sebastiani et al. 2009). Furthermore, it was recently observed that editing of the p53-inducible Cyfip2 (Cytoplasmic FMR1 interacting protein 2) significantly declines with age in human cerebral cortex. However, editing of Gabra3 (Gamma-aminobutyric acid A receptor subunit alpha 3) was found not to decline with age in humans. This suggests that RNA editing efficiency may be linked to aging in a gene-specific manner (Nicholas et al. 2010).
Humans are known to have far more targets of A-to-I RNA editing than rodents, due to the spread of Alu repeats during human evolution (Neeman et al. 2006). However, there are several known targets of A-to-I RNA editing that are conserved from rodents to humans (Levanon et al. 2005). The brown rat, Rattus norvegicus, is a well-established model species for studying aging and has been used in neurological research for many years. The brown rat is held as a model for mammalian behavioral and neurodegenerative studies (Jacob 1999; Wood et al. 2013). Comparative approaches are essential to fully understand aging; hence it is crucial to establish differences between humans and rodents to assess their importance to inform about human aging. Functional alterations, through RNA editing or other means, may also contribute to species differences in aging. Since studies linking RNA editing to aging have only been reported in humans, this study aimed to investigate whether the effect of age on RNA editing efficacy is conserved in the brown rat.
Methods
Rat tissues used in this study were supplied from a previous experiment (Merry et al. 2008). All animal husbandry procedures undertaken in this study were carried out in accordance with the provisions of the United Kingdom Animals (Scientific Procedures) Act 1986. Male BN rats (SubstrainBN/SsNOlaHSD) were obtained from Harlan UK at 21–28 days of age and maintained under barrier conditions on a 12 h light: 12 h dark cycle (08:00–20:00). The health status of the rats was monitored at regular intervals through the screening of sentinel animals. All rats were fed ad libitum and sacrificed at 6, 12, and 28 months of age. None of the animals exhibited any signs of pathology when sacrificed. Each age group had six rats, from which brain samples were taken, flash frozen, and stored at −80 °C.
RNA was extracted from cerebral cortex of rats using the RNeasy lipid tissue kit (Qiagen). The quality of the extracted RNA was assessed using the Agilent 2100 Bioanalyzer; all RNA integrity numbers (RINs) were above 8.0, indicating that the RNA had minimal degradation. For RNA extraction from brain tissue, RIN > 8 represents a high quality threshold (Bettscheider et al. 2011). The samples were pooled in pairs (leaving 3 samples per age group). Ribosomal RNA was removed from the pooled samples using the Eukaryote Ribominus Kit (Invitrogen) and confirmed with the Agilent 2100 Bioanalyzer.
RNA-seq data was generated by SOLiD sequencing (Applied Biosystems) from these samples in a previous study (Wood et al. 2013). The RNA-seq results from the SOLiD system were output as color space FASTA and quality files. These were converted into FASTQ format using a Python script from Galaxy (http://main.g2.bx.psu.edu/). The FASTQ files were mapped to the Ensembl release 65 rat reference genome (RGSC 3.4 assembly, May 2010 gene build) using Bowtie (Langmead et al. 2009) and settings appropriate to SOLiD data. For each sample, ~33.6 million reads were generated (range, 29.5–39.8 million reads). On average 16.7 million reads per sample were mapped to the reference genome (range, 13.8–21.4 million reads, ~50 % of reads generated were mapped). All data have been submitted to GEO under the accession GSE34272 (Wood et al. 2013). A mismatch analysis was performed on the aligned reads using Bambino in order to generate candidate editing sites (Edmonson et al. 2011). These candidates were selected using custom Python scripts for A-G mismatches within exons in genes on the positive strand, and T-C mismatches on the negative strand. Results were narrowed by selecting for non-synonymous mismatches and for those with a high number of read counts. Selected candidate targets were reverse transcribed using M-MLV reverse transcriptase (Invitrogen) and amplified by PCR using rTaq (TaKaRa). Editing levels were then analyzed by Sanger sequencing using the 3730 DNA Analyzer (Applied Biosystems) and quantifications were calculated by peak height analysis (Eggington et al. 2011).
Results
Cerebral cortex from 6, 12, and 28 month old rats were studied by RNA-seq to identify transcriptomic differences that occur during aging (Wood et al. 2013). These data were used to identify mismatches between the sequenced reads and the genome. Using Bambino (Edmonson et al. 2011), we identified candidate A-to-I RNA editing sites in the brown rat and used Sanger sequencing to validate these from RNA extracted from the cerebral cortex of the same rats. This analysis led to the identification of a new editing site in rats in the conserved oligomeric Golgi complex subunit 3 (Cog3) at position Chr15:61477456 (rn5 rat genome build). This modification results in a codon change from AUU to IUU, and an amino acid change from isoleucine to valine. Editing of this site has previously been reported in humans (Shah et al. 2009), and more recently in mice (Danecek et al. 2012), demonstrating conservation of the editing site between humans and rodents.
Using Sanger sequencing, editing levels of known conserved targets of A-to-I RNA editing were analyzed from the cerebral cortex of rats used in the RNA-seq: Gabra3, Cyfip2, Kcna1, Flna, and Blcap (which has three editing sites, known as Y/C, Q/R, and K/R), and the newly identified site in Cog3, using primers given in Table 1. The RNA editing enzymes, ADAR and ADARB1, have different specificities for base sequence and structure of their target sites (Riedmann et al. 2008). Therefore, different targets of RNA editing may be edited by both enzymes, or preferentially by just one. These differences have previously been hypothesized as a possible cause of the gene-specific RNA editing changes observed during aging in humans (Nicholas et al. 2010). The RNA editing sites studied here are targeted to different degrees by ADAR and ADARB1. For example, while Cyfip2 and Flna are targeted primarily by ADARB1 (Riedmann et al. 2008), Gabra3 is targeted by both ADARs (Ohlson et al. 2007), and Blcap is targeted mostly by ADAR (Riedmann et al. 2008). Therefore, it should be possible to identify whether any protein-specific age-related changes in RNA editing occur in the RNA editing targets that were studied.
Table 1.
Oligonucleotide sequences used for PCR amplification and sequencing of the RNA editing targets, Gabra3, Cyfip2, Kcna1, Flna, Cog3, and Blcap
| Gene name | Oligonucleotide sequences |
|---|---|
| Gabra3 | Fwd 5′-TGTCACAAGTTTCTTTCTGGCTTA-3′ |
| Rev 5′-TACCTTCTTGCCTTCCCAAG-3′ | |
| Cyfip2 | Fwd 5′-ATGGGCTTTGGCCTCTATCT-3′ |
| Rev 5′-ATGTTGTACTGGGGGCTGAT-3′ | |
| Kcna1 | Fwd 5′-ATGAGGGAGTTAGGGCTGCT-3′ |
| Rev 5′-GATCAGTTGCGGTGCAGTTA-3′ | |
| Flna | Fwd 5′-AAAGGATGGCTCTTGTGGTG-3′ |
| Rev 5′-CTATGCACCTTGGCATCAAT-3′ | |
| Cog3 | Fwd 5′-CTTCACGGGATGTTGTATCC-3′ |
| Rev 5′-TGAACTCCTCCAGTGGCTCT-3′ | |
| Blcap | Fwd 5′-AGCTCCTGGAGAGAGAGTCG-3′ |
| Rev 5′-AGCAAGTAGAAGCCCATGAA-3′ |
The mean lifespan of the rat strain used for this study was 28.06 ± 0.72 months (n = 102), determined from a previous study (Merry et al. 2008). Therefore the ages of the rats used in this study, aged 6, 12, and 28 months represent 21.4, 42.7, and 99.8 % of the mean lifespan, respectively. We compared the editing levels of six human cerebral cortex samples from 76 to 80 year old individuals (mean 77.7 years) from a previous study (Nicholas et al. 2010) to the levels of editing present in 28 month old rats. The 76–80 year old human samples analyzed represented 99.5 % of the mean human lifespan, assuming a human life expectancy of 78.0 years (United States Census Bureau 2012). These samples were compared based on their similar sample size, ages, and percentage of mean lifespan. RNA editing levels of Gabra3 remain similar in 28-month-old rats (93.2 % ± 1.5) and 76–80 year old humans (91.5 % ± 2.9). Editing levels of Cyfip2 also remain high in 28-month-old rats (88.0 % ± 1.3). However, editing of Cyfip2 declines to 70.9 % ± 5.7 in 76–80 year old humans. Thus, age-matched 28-month-old rats maintain high levels of Cyfip2 editing, compared to 76–80 year old humans.
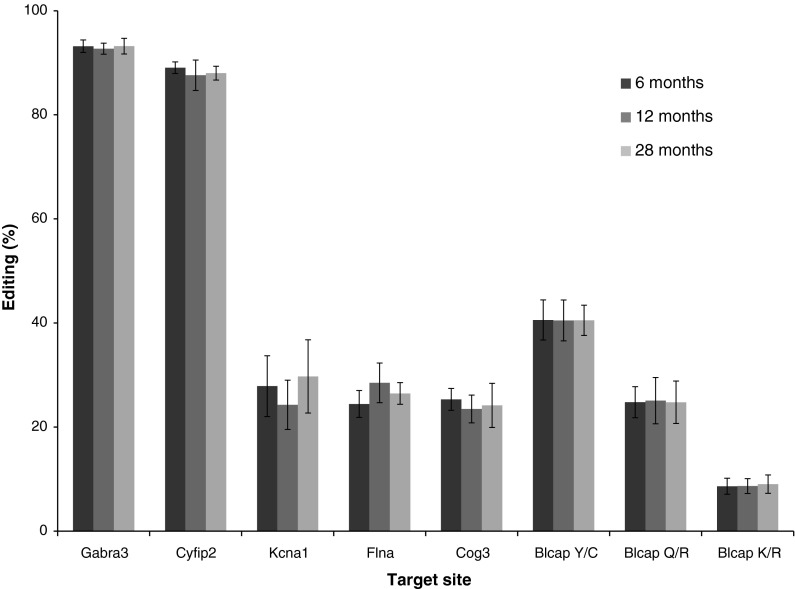
Although we observe differences in RNA editing between humans and rats, we do not identify significant changes in RNA editing levels of Gabra3, Cyfip2, Kcna1, Flna, Blcap or Cog3 with age in rats by one-way ANOVA (Fig. 1; Table S1) or collectively by two-way ANOVA, F(2,128) = 0.430, p = 0.652. Importantly, no change was identified for Cyfip2 (r2 = 0.021), in contrast to humans (Nicholas et al. 2010). Therefore, RNA editing of these targets in the brown rat is maintained throughout the average adult lifespan.
Fig. 1.
RNA editing percentages of the conserved target sites within Gabra3, Cyfip2, Kcna1, Flna, Cog3, and the Y/C, Q/R, and K/R sites of Blcap, were quantified from cerebral cortex of rats aged 6, 12, and 28 months. Quantification was determined by Sanger sequencing and peak height analysis. Error bars represent one standard deviation from the mean
Discussion
We report a new editing site in the brown rat in Cog3 which causes an amino acid change from isoleucine to valine. This site has recently been identified in humans and mice, demonstrating its evolutionary conservation (Danecek et al. 2012; Shah et al. 2009). Cog3 is part of a protein complex in the Golgi apparatus that localises to the membrane, and is required for correct protein glycosylation (Shestakova et al. 2006). It has not yet been identified whether editing of Cog3 can modulate its functionality, although its conservation between rodents and humans implies that it could have a functional role.
Conserved targets of RNA editing were studied in the brown rat, in order to investigate whether an age-related decline of RNA editing is conserved from humans through to the brown rat. Editing levels of Gabra3, Cyfip2, Kcna1, Flna, Blcap, and Cog3 were analyzed in 6-, 12-, and 28-month-old rats. We demonstrate that there are no significant differences in editing levels between RNA editing targets at any age in rat cerebral cortex. Unlike in elderly humans (Nicholas et al. 2010), 28-month-old rats show no deterioration of RNA editing in the targets tested in this study. Thus, within the average lifespan of the rat, we see no evidence for changes in RNA editing with age. We cannot, however, rule out the possibility that RNA editing levels may decrease in rats after the age of 28 months. In this study, only male rats were analyzed, so sex specific effects cannot be ruled out. However, no differences in RNA editing have been reported to date between sexes (Zhu et al. 2012), and no differences in gender were observed during aging in humans (Nicholas et al. 2010).
At younger ages, humans and rats appear to show no differences in RNA editing of Cyfip2 and Gabra3. In elderly humans it was identified that editing of Cyfip2 was reduced showing a significant decline with age (Nicholas et al. 2010). In contrast, we identify that editing levels of Cyfip2 are maintained in the rat across the mean lifespan, from 6- to 28-months of age. This suggests that the gene-specific link between RNA editing of Cyfip2 and aging is not conserved between humans and rats. Due to the increased lifespan within the primate lineage, and since RNA editing is known to be highly prevalent in the primate lineage (Eisenberg et al. 2005), our hypothesis is that this gene-specific link to aging may be confined to primates.
Alternatively, the decrease in RNA editing in Cyfip2 found in humans could be due to disease pathology. Many human patients used in the previous study died from heart failure (Nicholas et al. 2010). In contrast, all the rats used in our study were sacrificed when healthy. Although there has been no report of an association between changes in RNA editing and heart failure, it remains a possibility that this could have affected RNA editing, whether directly or indirectly.
Rodent models are frequently used in biogerontological studies, and although many aspects of aging are shared between rodents and humans, there are many important differences in their physiology, susceptibility to diseases, and their population demographics (Demetrius 2005). Furthermore, it has recently been shown that gene expression changes in response to inflammation are considerably different between mice and humans (Seok et al. 2013). This demonstrates that not only at the physiological level, but also at the underlying genetic level, the responses of rodent models to biological processes can be very different to those in humans. Since rodents are often used as model organisms for studying aging, it is vital to understand how biological processes, such as RNA editing, are differentially affected during aging, depending on the species.
This study demonstrates that the differences in RNA editing between rodents and humans are an important consideration when using rodent models to study aging. In contrary to previous findings in humans, we report that A-to-I RNA editing of evolutionarily conserved targets does not decline with age in rats. Furthermore, we identify that the gene-specific link of RNA editing to aging, which was identified in humans, is not conserved through to the brown rat. We interpret that age-related changes observed in RNA editing could be specific to primates or humans, or that they may be a result of pathology, as opposed to aging per se.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
The authors would like to thank Patricia Murray for guidance, Erez Levanon for critical reading of the manuscript, and the BBSRC (BBD5265291 and BBH0084971) for financially supporting this work.
Disclosure
The authors declare no conflict of interest.
References
- Akbarian S, Smith MA, Jones EG. Editing for an AMPA receptor subunit RNA in prefrontal cortex and striatum in Alzheimer’s disease, Huntington’s disease and schizophrenia. Brain Res. 1995;699:297–304. doi: 10.1016/0006-8993(95)00922-D. [DOI] [PubMed] [Google Scholar]
- Bettscheider M, Murgatroyd C, Spengler D. Simultaneous DNA and RNA isolation from brain punches for epigenetics. BMC Res Notes. 2011;4:314. doi: 10.1186/1756-0500-4-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenci C, Barzotti R, Galeano F, et al. Down-regulation of RNA editing in pediatric astrocytomas: ADAR2 editing activity inhibits cell migration and proliferation. J Biol Chem. 2008;283:7251–7260. doi: 10.1074/jbc.M708316200. [DOI] [PubMed] [Google Scholar]
- Danecek P, Nellåker C, McIntyre RE, et al. High levels of RNA-editing site conservation amongst 15 laboratory mouse strains. Genome Biol. 2012;13:26. doi: 10.1186/gb-2012-13-4-r26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demetrius L. Of mice and men. EMBO Rep. 2005;6:S39–S44. doi: 10.1038/sj.embor.7400422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmonson MN, Zhang J, Yan C, et al. Bambino: a variant detector and alignment viewer for next-generation sequencing data in the SAM/BAM format. Bioinformatics. 2011;27:865–866. doi: 10.1093/bioinformatics/btr032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggington JM, Greene T, Bass BL. Predicting sites of ADAR editing in double-stranded RNA. Nat Commun. 2011;2:319. doi: 10.1038/ncomms1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg E, Nemzer S, Kinar Y, et al. Is abundant A-to-I RNA editing primate-specific? Trends Genet. 2005;21:77–81. doi: 10.1016/j.tig.2004.12.005. [DOI] [PubMed] [Google Scholar]
- Farajollahi S, Maas S. Molecular diversity through RNA editing: a balancing act. Trends Genet. 2010;26:221–230. doi: 10.1016/j.tig.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galeano F, Leroy A, Rossetti C, et al. Human BLCAP transcript: new editing events in normal and cancerous tissues. Int J Cancer. 2010;127:127–137. doi: 10.1002/ijc.25022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob HJ. Functional genomics and rat models. Genome Res. 1999;9:1013–1016. doi: 10.1101/gr.9.11.1013. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levanon EY, Hallegger M, Kinar Y, et al. Evolutionarily conserved human targets of adenosine to inosine RNA editing. Nucleic Acids Res. 2005;33:1162–1168. doi: 10.1093/nar/gki239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas S, Kawahara Y, Tamburro KM, Nishikura K. A-to-I RNA editing and human disease. RNA Biol. 2006;3:1–9. doi: 10.4161/rna.3.1.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merry BJ, Kirk AJ, Goyns MH. Dietary lipoic acid supplementation can mimic or block the effect of dietary restriction on life span. Mech Ageing Dev. 2008;129:341–348. doi: 10.1016/j.mad.2008.04.004. [DOI] [PubMed] [Google Scholar]
- Neeman Y, Levanon EY, Jantsch MF, Eisenberg E. RNA editing level in the mouse is determined by the genomic repeat repertoire. RNA. 2006;12:1802–1809. doi: 10.1261/rna.165106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas A, De Magalhaes JP, Kraytsberg Y, et al. Age-related gene-specific changes of A-to-I mRNA editing in the human brain. Mech Ageing Dev. 2010;131:445–447. doi: 10.1016/j.mad.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu Rev Biochem. 2010;79:321–349. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlson J, Pedersen JS, Haussler D, Ohman M. Editing modifies the GABA(A) receptor subunit alpha3. RNA. 2007;13:698–703. doi: 10.1261/rna.349107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz N, Levanon EY, Amariglio N, et al. Altered adenosine-to-inosine RNA editing in human cancer. Genome Res. 2007;17:1586–1595. doi: 10.1101/gr.6493107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedmann EM, Schopoff S, Hartner JC, Jantsch MF. Specificity of ADAR-mediated RNA editing in newly identified targets. RNA. 2008;14:1110–1118. doi: 10.1261/rna.923308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastiani P, Montano M, Puca A, et al. RNA editing genes associated with extreme old age in humans and with lifespan in C. elegans. PLoS ONE. 2009;4:e8210. doi: 10.1371/journal.pone.0008210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seok J, Warren HS, Cuenca AG, et al. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci USA. 2013 doi: 10.1073/pnas.1222878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah SP, Morin RD, Khattra J, et al. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature. 2009;461:809–813. doi: 10.1038/nature08489. [DOI] [PubMed] [Google Scholar]
- Shestakova A, Zolov S, Lupashin V. COG complex-mediated recycling of Golgi glycosyltransferases is essential for normal protein glycosylation. Traffic. 2006;7:191–204. doi: 10.1111/j.1600-0854.2005.00376.x. [DOI] [PubMed] [Google Scholar]
- United States Census Bureau (2012) Life expectancy by sex, age, and race: 2008. http://www.census.gov/compendia/statab/2012/tables/12s0104.pdf. Accessed 9 May 2013
- Wood SH, Craig T, Li Y, et al. Whole transcriptome sequencing of the aging rat brain reveals dynamic RNA changes in the dark matter of the genome. Age. 2013;35:763–776. doi: 10.1007/s11357-012-9410-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Urban DJ, Blashka J, et al. Quantitative analysis of focused A-to-I RNA editing sites by ultra-high-throughput sequencing in psychiatric disorders. PLoS ONE. 2012;7:e43227. doi: 10.1371/journal.pone.0043227. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.