Abstract
Many bacteria are naturally competent, able to bind and take up DNA from their extracellular environment. This DNA can serve as a significant source of nutrients, in addition to providing genetic material for recombination. The regulation of competence in several model organisms highlights the importance of this nutritional function, although it has often been overlooked. Natural competence is induced by starvation in Haemophilus influenzae, the model for competence regulation in the gamma-proteobacteria. This induction depends on the activation of the global metabolic regulator CRP, which occurs upon depletion of phosphotransferase sugars. In this work, we show that the depletion of purine nucleotides under competence-inducing conditions activates the CRP-dependent competence-specific regulator Sxy. Depletion of extra- or intra-cellular purine nucleotides activates Sxy translation, while high levels inhibit it. This is modulated by the stem structure formed by sxy mRNA. The exact mechanism by which the nucleotide depletion signal is transduced is unclear, but it does not involve direct binding of purine intermediates to the sxy stem, and does not require Hfq or competence proteins. Similar regulation occurs in the relatives of H. influenzae, Actinobacillus pneumoniae and A. suis, confirming the importance of processes enabling competent bacteria to exploit the abundant DNA in their environments.
Introduction
Natural competence is the ability of bacteria to actively take up DNA from their environment. Since this DNA can recombine with the chromosome and change the cell's genotype (transformation), natural transformation is a major mechanism of genetic exchange, shaping bacterial genomes and spreading alleles that increase bacterial survival and virulence (Domingues et al., 2012; Livermore, 2012). However, most DNA taken up by competent cells is degraded, providing nucleotides, elements (C, N, P) and energy, but no genetic information (Pifer and Smith, 1985; Stewart and Carlson, 1986). This suggests that DNA uptake could make a significant contribution to cellular metabolism. However, although nutritional signals affect competence development in most model systems, their roles are often thought to be indirect (Solomon and Grossman, 1996; Macfadyen, 2000; Finkel and Kolter, 2001; Palchevskiy and Finkel, 2006; Bosse et al., 2009; Johnsborg and Havarstein, 2009; Kristensen et al., 2012).
Nutritional signals play a prominent role in competence of Haemophilus influenzae, the model for gamma-proteobacteria. The traditional method for competence induction abruptly deprives cells of both nucleotides and the carbon/energy resources needed for nucleotide synthesis (Maughan et al., 2008). On the molecular level, when phosphotransferase (PTS) sugars are unavailable, a rise in intracellular cAMP levels activates CRP (cAMP Receptor Protein) to induce transcription of sugar utilization genes (containing the canonical CRP-N binding site) and of the competence activator sxy (Dorocicz et al., 1993; Macfadyen, 2000; Redfield et al., 2005). Sxy and CRP together then activate transcription of the 25 genes in the CRP-S regulon (containing the non-canonical CRP-S binding site), whose products enable the cell to take up DNA (Redfield et al., 2005; Cameron and Redfield, 2008; Sinha et al., 2012).
Though the PTS sugar depletion cascade that controls cAMP and CRP is well understood, the signals and mechanisms that control Sxy translation are less clear. In addition to transcriptional control by CRP, Sxy is subject to translational control by an mRNA stem structure (Cameron et al., 2008). Competence development requires the unfolding of this stem, and mutants in which the stem is weakened are hypercompetent (Cameron et al., 2008). In Vibrio cholerae and its relatives, where the regulation of competence and of Sxy has also been extensively studied, a similar stem in the sxy homologue tfoX (Yamamoto et al., 2010) transduces a nutritional signal, the availability of the N-acetylglucosamine polymer chitin (Meibom et al., 2005; Pollack-Berti et al., 2010; Yamamoto et al., 2010; 2011; Antonova and Hammer, 2011; Suckow et al., 2011; Antonova et al., 2012; Blokesch, 2012; Lo Scrudato and Blokesch, 2012; Seitz and Blokesch, 2012). In response to chitin, a small Hfq-regulated RNA, tfoR, modulates tfoX translation by base pairing with part of the stem (Yamamoto et al., 2011).
Although the regulatory inputs controlling H. influenzae sxy translation are not known, depleted nucleotide pools have been implicated in competence induction. The starvation medium that maximally induces competence (MIV) lacks nucleotides, and microarray studies have confirmed that the abrupt transfer from rich medium to MIV sharply induces purine biosynthesis genes (Herriott et al., 1970; Redfield et al., 2005). In addition, Macfadyen et al. showed that the addition of extracellular purine nucleotides represses natural competence (Macfadyen et al., 2001). The biggest repressing effect was seen with the nucleotides AMP and GMP and the nucleoside guanosine, while adenosine had a smaller effect and bases had little; this difference was attributed to differences in the transport process (Macfadyen et al., 2001).
Our understanding of H. influenzae's purine metabolism and regulation derives from parallels with Escherichia coli and is shown schematically in Fig. 1. Extracellular nucleotides are converted to their respective nucleosides in the periplasm, and these are transported into the cytosol by NupC. In the cytosol, DeoD converts purine nucleosides to their respective bases; these can in turn be converted back into nucleotides by the HPRT and APRT phosphoribosyl transferases (Gots and Benson, 1973). Engodenous purine biosynthesis involves multiple steps to convert PRPP to the intermediate ribonucleoside monophosphate IMP, and is repressed by the master regulator PurR when extracellular nucleotides are available (Fig. 1). Aside from purine biosynthesis genes, PurR regulates genes for purine transport, salvage and interconversion pathways, and also downregulates the genes in pyrimidine biosynthetic and transport pathways (Ravcheev et al., 2002; Zhang et al., 2008; Cho et al., 2011). In the presence of guanine or hypoxanthine, E. coli PurR binds to a 16 bp palindromic sequence located within the promoter region of pur regulon genes (Cho et al., 2011). In H. influenzae, similar binding sites are found upstream of pur gene homologues (Mironov et al., 1999; Ravcheev et al., 2002). PurR's role in natural competence is not known, though Macfadyen et al. hypothesized that it might repress one or more competence genes to prevent DNA uptake when purines are abundant (Macfadyen et al., 2001). In this work we investigate the role of nucleotide availability on competence regulation and find that purine availability instead controls competence by controlling Sxy translation. We also show that nucleotide availability regulates competence in Actinobacillus pneumoniae and A. suis, relatives of H. influenzae, confirming the importance of such regulation.
Fig. 1.
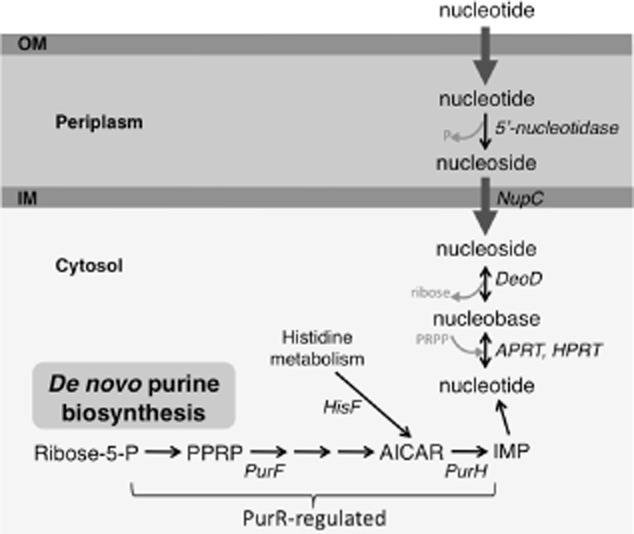
Purine nucleotide transport and metabolism in H. influenzae. Both extracellular purine pools and endogeneous purine biosynthesis affect purine nucleotide levels in the cytoplasm. Extracellular nucleotides are transported across the outer membrane and immediately converted to nucleosides by a 5′-nucleotidase. These nucleosides are then transported across the inner membrane by NupC. Once inside the cytosol, nucleosides are converted to bases and nucleotides. De novo purine biosynthesis uses ribose-5-P to generate the purine intermediate IMP in multiple steps requiring PurR-regulated enzymes. IMP can be converted to the purine nucleotides AMP or GMP. PRPP, phosphoribosyl pyrophosphate; APRT, adenine phosphoribosyltransferase; HPRT, hypoxanthine-guanine phosphoribosyltransferase; AICAR, 5-Aminoimidazole-4-carboxamide ribonucleotide.
Results
Extracellular purine nucleotides reduce Sxy translation
Macfadyen et al. showed that the addition of purine nucleotides reduced competence 100-fold (Macfadyen et al., 2001). They also showed that AMP addition reduced expression of two competence genes, suggesting that it acts on competence induction rather than later stages of competence. We confirmed this by showing that AMP addition only repressed competence when added as or shortly after cells were transferred to MIV, and that it had no effect when added at later time points when cells are already competent (Fig. 2). Macfadyen et al. also showed that AMP addition does not change the PTS/cAMP response to MIV (Macfadyen et al., 2001), so we hypothesized that it might affect the competence-specific activator Sxy. Western blotting showed that Sxy protein levels were severely reduced by the addition of AMP (Fig. 3A, bottom graph), suggesting that competence is low in the presence of purine nucleotides because Sxy levels are low. In contrast, qPCR showed that sxy mRNA was unchanged by the addition of AMP (Fig. 3A, top graph), suggesting that purine nucleotides specifically regulate Sxy translation.
Fig. 2.
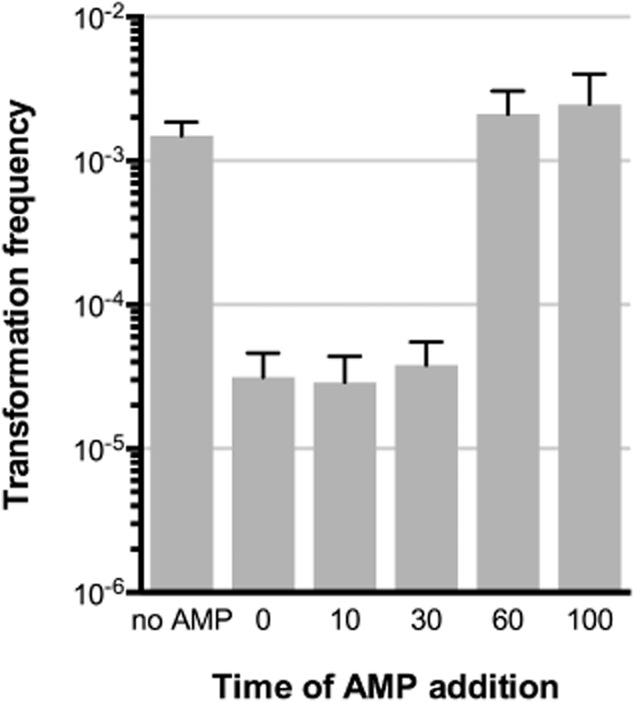
Effect of the timing of AMP addition to the inhibition of competence. sBHI-grown cells were transferred to MIV with 1 mM AMP added at different times during the MIV 100 min incubation. Transformation frequency was measured after 100 min. Each bar represents the mean of three biological replicates ± standard deviations.
Fig. 3.
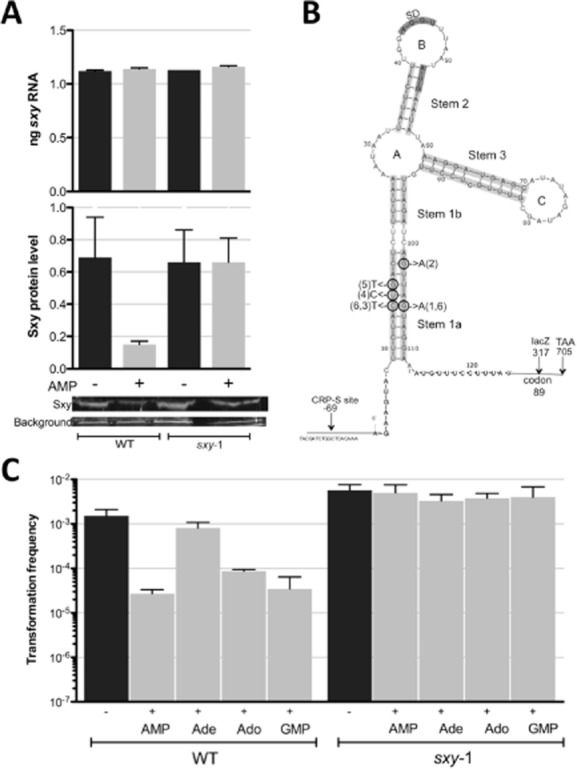
Purines repress Sxy expression unless the mRNA stem is weakened.
A. qPCR (top) and western blot (bottom) analysis of sxy RNA and protein expression in the presence (grey bars) and absence (black bars) of 1 mM AMP after 100 min in MIV (see Experimental procedures for details). Error bars represent the mean of three biological replicates. Sxy protein levels are relative to the unidentified protein used for internal standardization in Cameron et al. (2008). A representative Western blot below the bottom graph shows Sxy protein and the standardization protein. WT, Wild type.
B. Secondary structure of sxy mRNA. The stem structure, the position of individual mutations and other relevant sequence features are shown.
C. sBHI-grown cells were transferred at OD600 0.2 to MIV with (grey bars) or without (black bars) 1 mM purine source for 100 min before measuring transformation frequency (Ade = adenine, Ado = adenosine). Each bar represents the mean of three biological replicates ± standard deviations. WT, Wild type.
To determine whether the based-paired stem of sxy mRNA is the sensor that responds to purine nucleotides, we measured RNA and protein levels in the hypercompetent mutant sxy-1. In this mutant, a G-A transition at position 106 destabilizes the mRNA stem (Fig. 3B) and increases sxy translatability; analysis of compensatory mutations confirmed the essential role of base pairing (Redfield, 1991; Cameron et al., 2008). In contrast to wild type, sxy-1 cells maintained high levels of Sxy protein in MIV + AMP (Fig. 3A, bottom graph), and competence was unaffected by the addition of AMP or other purine sources (adenine, adenosine or GMP) (Fig. 3C). Together these results provide further evidence that purine nucleotides directly affect Sxy translation. Competence was also independent of AMP in the other available stem sxy-2-5 mutants (Fig. 4), whose mutations fall elsewhere in the sxy mRNA stem, but have the same destabilizing effect (Fig. 3B) (Cameron et al., 2008). The sxy-6 mutant, whose compensatory mutations restore base pairing in the stem and thus increase its stability (Cameron et al., 2008), served as a negative control (Figs 3B and 4). The murE749 hypercompetent strain (Ma and Redfield, 2000) provided another important control because the mutation that makes it hypercompetent is in murE itself and not in the sxy stem. The sensitivity of murE749 to AMP (Fig. 4) confirmed the specificity of the stem mutants' resistance to AMP. The insensitivity of mutants with destabilized stems to AMP is thus not simply attributable to higher levels of Sxy protein, and suggests that the sxy mRNA stem plays an important role in sensing and/or responding to nucleotide availability.
Fig. 4.
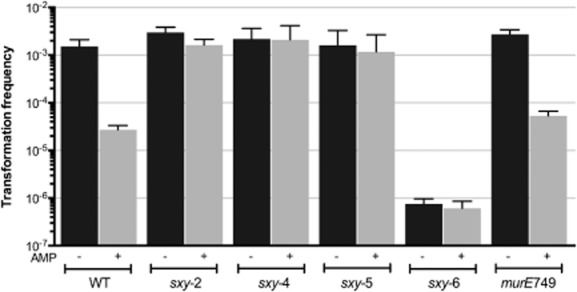
Competence is insensitive to purine addition in stem loop mutants. sBHI-grown cells were transferred at OD600 0.2 to MIV with (grey bars) or without (black bars) 1 mM AMP for 100 min before measuring transformation frequency. Each bar represents the mean of three biological replicates ± standard deviations. WT, Wild type.
PurR's role in purine nucleotide repression of competence is indirect
Macfadyen et al. found candidate PurR binding sites in the promoters of several CRP-S genes, suggesting that they may be repressed under conditions when PurR is active (Macfadyen et al., 2001). We therefore checked for this additional level of competence regulation by purine nucleotides.
The first step was to search for candidate PurR binding sites in the CRP-S promoters of competence gene. Using the E. coli PurR motif as a starting point, Ravcheev et al. developed species-specific scoring matrices for H. influenzae and several other gamma-proteobacteria (Ravcheev et al., 2002). We used their H. influenzae matrix to score the H. influenzae genome. Promoters with high-scoring sites include expected purine biosynthesis genes, as well as cytoplasmic and periplasmic phosphatases (Table 1). The only competence gene was rec2 (score 5.31), whose product is required for translocation of ssDNA across the inner membrane (Barouki and Smith, 1985). The regions upstream of other H. influenzae CRP-S genes did not contain candidate PurR sites (highest score: 4.19, upstream of the sxy gene). Though the candidate rec2 site had not been detected in previous studies (Mironov et al., 1999; Ravcheev et al., 2002), it is conserved in 20 sequenced H. influenzae genomes. However, such sites were not found upstream of rec2 homologues in other Pasteurellaceae or gamma-proteobacteria (E. coli, V. cholerae), and recent ChIP and gene expression studies of E. coli did not identify rec2 as a PurR target (Cho et al., 2011).
Table 1.
Putative PurR binding sites in H. influenzae.
| Score | Sequence | ORF | Position | Transcript | Process |
|---|---|---|---|---|---|
| 6.00 | ACGCAAACGTTTGCTT | HI0887 | −63 | purH-purD-glyA | purine biosynthesis |
| 5.97 | AAGCAAACGTTTGCTA | HI1726 | −77 | hemH (purC) | purine biosynthesis |
| 5.85 | AAGCAAACGTTTGCGA | HI1429 | −63 | purM-purN | purine biosynthesis |
| 5.75 | AAGCAAACGTTTGCAT | HI0752 | 1 | purL | purine biosynthesis |
| 5.74 | AAGAAAACGTTTGCGT | HI1206 | −82 | cvpA-purF | purine biosynthesis |
| 5.70 | AGGCAAACGTTTGCTA | HI1615 | −75 | purE-purK | purine biosynthesis |
| 5.68 | AAGTAAACGTTTGCGT | HI0125 | −83 | HI0125 | unknown |
| 5.31 | ATGCAAACGGTTGCTT | HI0061 | −70 | rec2 | natural competence |
| 5.27 | TAGCAAACGCTTTCTT | HI0609 | −45 | folD | 1-carbon metabolism |
| 5.16 | TTGCAAACGGTTGCTT | HI1736 | −95 | HI1736 | unknown |
| 5.11 | TGGCAAACGATTGCTA | HI0495 | −85 | aphA | phosphatase, periplasmic |
| 4.88 | AAGTAAACGATTGTGT | HI0464 | −34 | rpiA-serA | purine biosynthesis |
| 4.84 | AGGAAAACGTTTCCGT | HI0638 | −26 | hflD-purB | purine biosynthesis |
| 4.59 | ATGCAAACGATTACTC | HI0667 | −76 | glpX | phosphatase,cytoplasmic |
| 4.54 | ATAAAATCGTTTGCTA | HI1153 | −41 | hpt | purine biosynthesis |
The PurR-binding motif of Ravcheev et al. (2002) was used to score the H. influenzae Rd KW20 genome (see Experimental procedures). The sequences for ORFs on the reverse strand have been reverse complemented. The rec2 site is shaded and bold. ORF indicates the gene nearest to the site. Position indicates the distance of the start codon from the distal end of the site (total site length = 16 bp). Transcript indicates the predicted genes regulated by the PurR site. Process indicates annotated metabolic processes encoded by the transcript. All 20 sequenced H. influenzae genomes had putative sites upstream of rec2, 18 with the same score as Rd KW20, and two strains (22.4–21 and PittEE) with better sites (site: ATGCAAACGTTTGCTT, score: 5.75).
To directly test whether PurR represses H. influenzae rec2, we measured expression of a rec2::lacZ fusion in the presence and absence of PurR. We constructed an insertion deletion mutant of purR, replacing amino acids 5 to 299 of PurR with a kanamycin-resistance cassette, ensuring that both the DNA-binding and catalytic activities were disrupted. In this strain, purine biosynthesis genes are expected to be constitutively expressed at all growth stages. As negative and positive controls, we also tested expression of lacZ fusions to the CRP-S gene comA and to the PurR-regulated gene purH. As shown in Fig. 5A and B, deletion of PurR did not change rec2 or comA expression, when CRP/Sxy expression was low (after overnight culture or in early log phase), intermediate (late log phase), or high (upon transfer to MIV). As expected, expression of rec2 and comA remained low in cells transferred to AMP-supplemented MIV, and did not change upon deletion of purR. In contrast, purH was strongly induced by the absence of PurR under all conditions tested (Fig. 5C). A careful time-course monitoring rec2 expression throughout growth in sBHI also showed no effect of purR deletion (data not shown). These results confirm that PurR does not repress rec2.
Fig. 5.
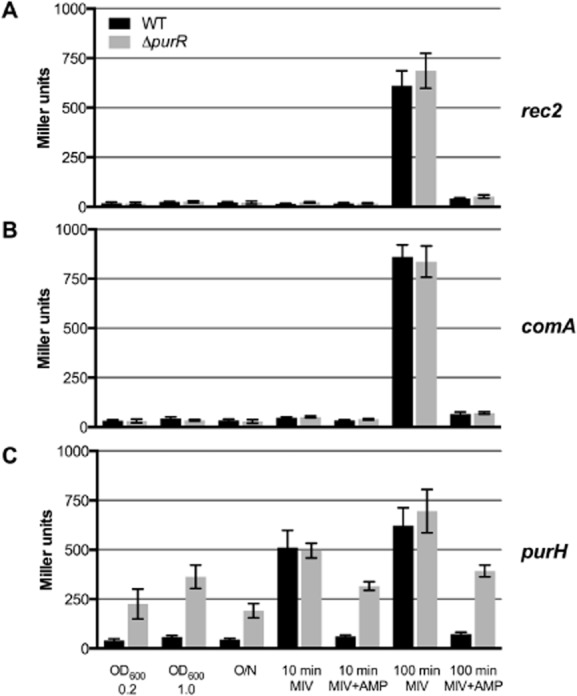
rec2 expression is unchanged by the absence of PurR. Expression of the H. influenzae chromosomal rec2::lacZ (A), comA::lacZ (B) and purH::lacZ (C) fusions in the presence (black, WT: wild type) or absence (grey) of PurR. β-Galactosidase activity of the fusions under different conditions is shown as Miller units. Each bars represents the mean of three biological replicates ± standard deviations.
One further test was to assess competence of the purR− strain in the presence of purines: if purine nucleotides' repressing effect on competence is mediated through PurR, the effect should disappear in the purR− strain. As shown in Fig. 6A, the addition of AMP still reduced competence in the purR− strain. These results in combination with those of Fig. 5 confirm that purine repression of competence is not mediated by PurR's repression of competence genes.
Fig. 6.
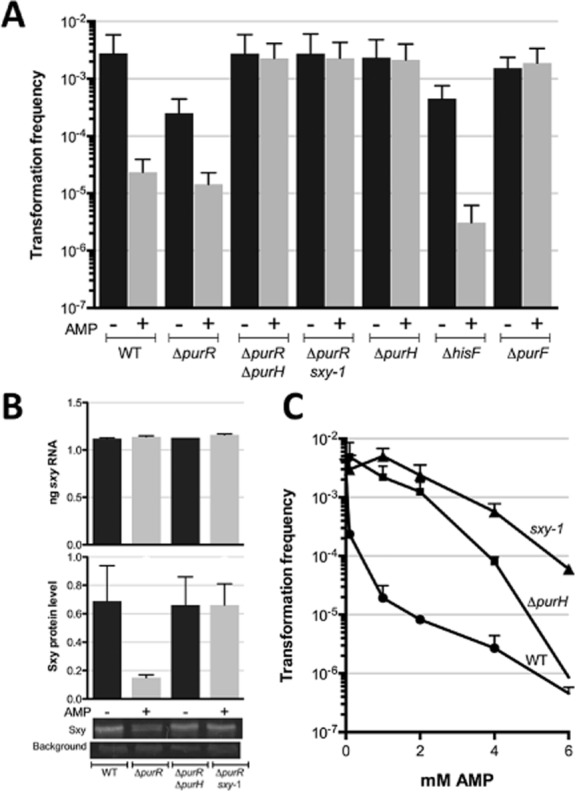
Natural competence in the presence and absence of AMP. sBHI-grown cells were transferred to MIV with (grey bars) or without (black bars) 1 mM AMP (A), or with increasing concentrations of AMP (C) for 100 min before measuring transformation frequency. Each bar or point represents the mean of three biological replicates ± standard deviations. Circles: WT (wild type), squares: ΔpurH, triangles: sxy-1.
B. qPCR (top) and western blot (bottom) analysis of sxy RNA and protein expression in the presence (grey bars) and absence (black bars) of 1 mM AMP after 100 min in MIV (see Experimental procedures for details). Error bars represent the mean of three biological replicates. Sxy protein levels are relative to the unidentified protein used for internal standardization in Cameron et al. (2008). A representative Western blot below the bottom graph shows Sxy protein and the standardization protein.
Increased intracellular purine nucleotides repress sxy expression by blocking translation
Figure 6A shows that not only was competence not raised in cells lacking purR, it was lowered ∼ 10-fold; this was not due to differences in growth of the mutant strain (Fig. S2). Cells lacking PurR are expected to have elevated intracellular purine nucleotide pools due to constitutive expression of purine biosynthesis genes, so we hypothesized that the reduced competence of the purR− strain might have the same cause as that of wild-type cells treated with extracellular purine nucleotides.
To test whether elevated purine pools cause the competence defect of the purR− strain, we eliminated endogeneous purine biosynthesis in this strain by further knocking out purH, whose product converts AICAR into IMP, the last common precursor of AMP and GMP (Fig. 1). In this strain, all purine biosynthesis genes are constitutively expressed but purines cannot be synthesized. As shown in Fig. 6A, the purH− mutation restored competence of the purR− strain to wild-type levels, confirming our hypothesis. Next, we measured Sxy protein levels and found them to be significantly reduced in the purR− strain, and similar to wild type in the purR− purH− strain (Fig. 6B). As expected, sxy mRNA levels were similar in all strains (Fig. 6B). Finally, because mutations that weaken the sxy mRNA stem overcome the inhibitory effect of high extracellular purines, we tested whether they could also overcome the competence defect of the purR− strain. As shown in Fig. 6A, the competence defect of the purR− strain was abolished by the sxy-1 mutation. Collectively these results confirm that increased purine nucleotide pools, whether extracellular or intracellular, reduce competence by blocking Sxy translation.
Because the removal of endogeneous purine biosynthesis overcame the repression of competence by high intracellular purine pools (purR− purH− strain), we tested its effect on the repression of competence by extracellular AMP. As shown in Fig. 6A, addition of the purH− mutation made the purR− strain insensitive to 1 mM AMP. Surprisingly, a purR+ purH− strain was also insensitive to AMP (Fig. 6A), suggesting that the repression of competence by AMP is overcome by the accumulation of intermediates in the purine biosynthesis pathway upstream of PurH. In Salmonella enterica, purH− cells have increased PPRP pools due to feedback inhibition of purF and other upstream purine biosynthesis genes by AICAR (Allen et al., 2002; Bazurto and Downs, 2011). Since PRPP is needed to convert bases to nucleotides, it is possible that cells lacking purH are better able to convert extracellular purines. To test whether the phenotype of the purH− strain is attributed to increased PRPP pools, we created a knockout in purF. PurF carries out the first step in the purine biosynthetic pathway, the conversion of PPRP (phosphoribosyl pyrophosphate) to ribosylamine-5P (Fig. 1). We also created a knockout of hisF, a cyclase enzyme required for the last step of histidine metabolism that also produces AICAR (Fig. 1). We found that competence was still inhibited by AMP in cells lacking hisF, while cells lacking purF, like those lacking purH, were insensitive to extracellular AMP addition (Fig. 6A). This confirms that increased intracellular PRPP pools enable competent cells to overcome the inhibitory effects of extracellular purine nucleotides. Because the inhibition of competence by purine nucleotides is concentration-dependent (Macfadyen et al., 2001), we tested whether AMP at higher concentrations would overcome the higher PRPP pools of purH− cells and inhibit competence. As shown in Fig. 6C, competence was inhibited by AMP concentrations as low as 0.1 mM in wild-type cells, whereas comparable inhibition was only seen upwards of 4 mM AMP in purH− cells. In contrast, inhibition of the sxy-1 strain occurred only at concentrations high enough to significantly affect viability (Figs 6C and S1).
Purine regulation of Sxy translation is indirect
The above results suggest that an increase in intracellular purine pools limits Sxy translation and competence. The simplest model is that cytosolic purine intermediates directly bind to the sxy mRNA stem, and stabilize it to repress translation. Testing whether the Sxy stem itself responds to purines is complicated by additional levels of regulation present in H. influenzae. Thus we instead measured the expression of a translational sxy::lacZ fusion in the presence and absence of purine sources in E. coli. In this fusion, the lacZ::kan cassette is fused to codon 89 of the H. influenzae sxy coding sequence, leaving the stem intact (Fig. 3B) (Cameron et al., 2008). As positive and negative controls, we used plasmid-encoded versions of the transcriptional lacZ fusions to H. influenzae purH and comA described above. We tested expression of all fusions both in rich and minimal media. As expected, the expression of purH was lowered by AMP addition in both media (Fig. 7). The difference in expression between these media likely reflects inhibition of purH expression by abundant purines in the Luria–Bertani (LB) medium. Also as expected, the expression of the negative control comA was the same in both media and was unchanged by AMP addition (Fig. 7). The expression from the sxy stem was high [two- to fivefold higher than that seen under competence-inducing conditions in H. influenzae (Cameron et al., 2008)], but was not changed by AMP even at higher concentrations, or by GMP, adenine or adenosine (Fig. 7). This suggests that exogeneous purine nucleotides do not limit Sxy translation in E. coli.
Fig. 7.
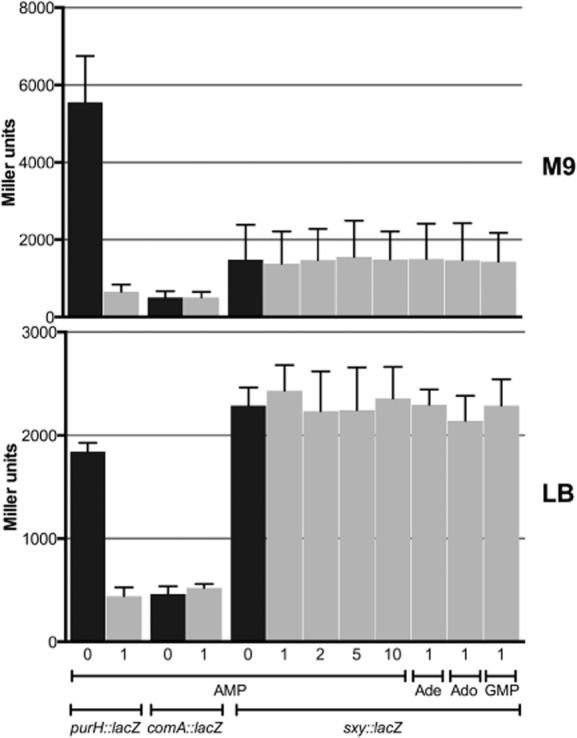
The Sxy stem loop does not directly respond to purines. The expression of plasmid-encoded fusions in E. coli was measured in the presence (grey bars) or absence (black bars) of purine sources, after 30 min in M9 minimal medium (top graph) or LB (bottom graph). The β-galactosidase activity from the translational sxy::lacZ fusion, and transcriptional comA::lacZ and purH::lacZ fusions is shown as Miller units. The concentrations of AMP are given below each bar in mM; adenine (Ade) and adenosine (Ado) were used at 1 mM. Each bar represents the mean of three biological replicates ± standard deviations.
Purine regulation of Sxy translation does not require Hfq
We next tested whether Sxy translation was regulated by a small regulatory or antisense RNA, which would respond (directly or indirectly) to purine levels. Translation of the V. cholerae sxy homologue is activated by the global RNA regulator Hfq and the small RNA tfoR (Yamamoto et al., 2011). Moreover, Cameron et al. showed that such regulation is possible in H. influenzae, as a ssDNA oligo designed to be complementary to part of H. influenzae sxy mRNA competed with stem formation in vitro and increased translation (Cameron et al., 2008).
To test whether Hfq is involved in regulation of sxy expression in H. influenzae, we examined the effect on competence of a deletion of hfq. Competence of the hfq− strain was reduced about 10-fold, a defect that was overcome by the sxy-1 stem mutation (Fig. 8), suggesting that Hfq may participate in the unfolding of the stem to enable translation. However, competence was still strongly repressed by AMP in the hfq− strain by the same factor as wild-type cells (Fig. 8), indicating that if Hfq participates in sxy regulation, it does not do so by sensing purine pools.
Fig. 8.
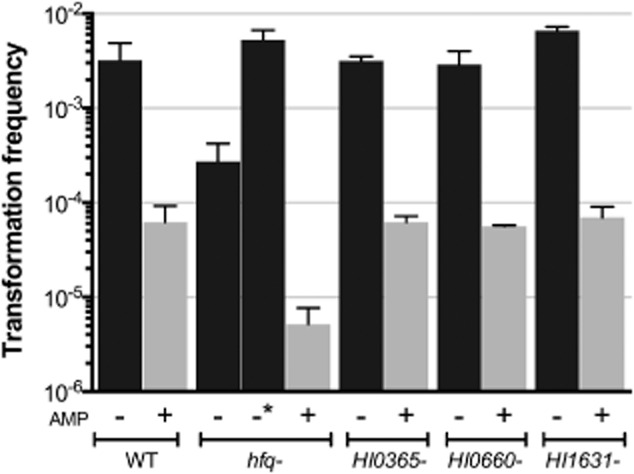
AMP inhibits competence in mutants of hfq, HI0365, HI0660 and HI1631. sBHI-grown cells were transferred at OD600 0.2 to MIV with (grey bars) or without (black bars) 1 mM AMP for 100 min before measuring transformation frequency. Each bars represents the mean of at least two biological replicates ± standard deviations. WT, Wild type; *hfq− sxy-1.
Purine regulation of Sxy translation does not require HI0365, HI0660 or HI1631
We recently characterized the competence phenotypes of every CRP-S gene (Sinha et al., 2012). Several have no role in the DNA uptake process and lack any characterized functions, so we tested whether any of these might affect competence regulation in the presence of purine nucleotides through feedback regulation of Sxy translation. As shown in Fig. 8 (right bars), the competence of knockouts of HI0365, HI0660 and HI1631 was still repressed by AMP, confirming that their products do not affect purine regulation of competence.
The impact of purine nucleotides on competence is not limited to H. influenzae
We examined whether high purine pools also prevent competence in relatives of H. influenzae, by testing the effect of AMP addition in two other naturally competent Pasteurellacean species, Actinobacillus pleuropneumoniae and Actinobacillus suis (Redfield et al., 2006; Maughan et al., 2008). Natural competence is known to be induced by starvation and dependent on Sxy/CRP in A. pleuropneumoniae (Bosse et al., 2009), but has not been studied in A. suis.
We first demonstrated that A. suis is naturally competent in the starvation medium used to induce competence in other Pasteurellaceae, and found that this is entirely dependent on Sxy (Fig. 9). In addition, like other Pasteurellaceae, the A. suis H91-0380 genome contains homologues of essential competence gene, with conserved CRP-S sites in their promoters (Table 2). As shown in Fig. 9, competence was reduced by addition of AMP in both species, suggesting that the mechanisms by which AMP regulates competence are conserved. Since A. pleuropneumoniae and A. suis are both distant relatives of H. influenzae, we conclude that purine nucleotide repression of Sxy translation is likely to be an ancestral property of the Pasteurellaceae.
Fig. 9.
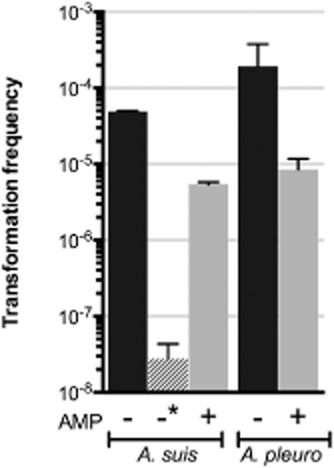
AMP inhibits competence in Actinobacillus pleuropneumoniae (A. pleuro) HS143 and A. suis ATCC15557. sBHI-grown cells were transferred at OD600 0.2 to MIV with (grey bars) or without (black bars) 0.1 mM AMP for 100 min before measuring transformation frequency. Each bar represents the mean of three biological replicates ± standard deviations. *A. suis Δsxy. The hashed bar represents the limit of detection of the sxy− strain.
Table 2.
Competence genes homologues and their CRP-S site in the sequenced strain of A. suis H91-0380
| Score | Sequence | Position | Transcript | Start | End |
|---|---|---|---|---|---|
| 32.0 | TTTTGCGATCCTGATCGAAAAA | −87 | comNOPQ | 1786332 | 1788485 |
| 31.3 | TTTTGCGATCTGGATCGCAAAC | −83 | rec2 | 675504 | 677798 |
| 30.9 | TTTTGCGATCTTGATCGAAAAT | −165 | HI0365/pilF2 | 82751 | 84535 |
| 30.6 | TTTTTCGATCTATATCGCAAAA | −76 | comF | 2430643 | 2431320 |
| 30.4 | TTTTGCGATCAAGATCGAGAAA | −99 | comE1 | (2267009) | (2267374) |
| 30.2 | TTTTGCGATCCTGATCGAGAAA | −87 | comM | 2145444 | 2146973 |
| 29.8 | TTTTTCGATGAAGATCGCAAAA | −133 | comABCDE | (1349840) | (1353261) |
| 29.7 | TTTTGTGATCTCAATCGAAAAA | −181 | dprA | 2097055 | 2098209 |
| 28.0 | TTTTTCGACGCAGATCGCAAAA | −78 | radC | (1640152) | (1640826) |
| 27.1 | ATTCTCGATCCGGATCGCAAAA | −85 | pilABCD | (1114637) | (1118340) |
| 18.4 | TTTCAGGTAGAATGTCGCACAA | −57 | ssb | 986417 | 986953 |
| 16.4 | TTGTTGCATCATTTCGCCAAAA | −96 | HI0569/60 | 2413794 | 2413813 |
The sequences for ORFs on the reverse strand have been reverse complemented. Position indicates the distance of the start codon from the distal end of the site. Transcript indicates the predicted genes regulated by the CRP-S site. Start and End indicate the coordinates of the transcript.
Discussion
Twenty years ago, we hypothesized that competent cells take up DNA primarily for its nucleotides, and predicted that competence genes would be induced by nucleotide limitation (Redfield, 1993). In this work, we show that extracellular purine nucleotides specifically reduce expression of the competence regulator Sxy, by inhibiting its translation. This reduction does not involve Hfq-dependent small RNA processes or three competence-regulated proteins of unknown function. Expression of a sxy::lacZ translational fusion was unaffected by extracellular purine intermediates in E. coli. Moreover, the sxy stem structure bears no resemblance to known purine riboswitches and preliminary analyses have shown that sxy mRNA does not bind purine compounds in a riboswitch assay. This suggests an alternative model of regulation, where one or more purine-binding proteins sense purine levels in the cytoplasm and activate or repress regulators of Sxy translation. Further work is required to determine if such regulators exist and how they act.
We have shown that nucleotide abundance also regulates competence in relatives of H. influenzae that inhabit similar niches in different hosts: H. influenzae colonizes the human nasopharynx and middle ear, while A. pleuropneumoniae and A. suis colonize the upper respiratory tracts of pigs. DNA is abundant in both niches, as it is in many mucosal environments, and can serve as a rich nutrient source. The importance of DNA as a nutrient and its involvement in competence regulation has also been demonstrated in other families of the gamma-proteobacteria. For example, Finkel and co-workers showed that E. coli could grow by using DNA as the sole source of carbon and energy, and that this required homologues of H. influenzae's competence genes (Finkel and Kolter, 2001; Palchevskiy and Finkel, 2006). We have shown that these homologues are required for DNA uptake by E. coli, though conditions that naturally induce this process are not known (Sinha and Redfield, 2012). The transfer to starvation conditions does not induce E. coli sxy expression, suggesting that other signals are likely required (Sinha et al., 2009; Sinha and Redfield, 2012). Another example of nucleotide abundance regulating competence comes from V. cholerae, whose competence is inhibited by cytidine (Antonova et al., 2012). This was attributed to disrupted interactions between CRP and the nucleoside-scavenging cytidine repressor CytR. H. influenzae lacks a homologue of CytR, so the mechanisms of nucleotide inhibition are different in both organisms. Though Macfadyen et al. found no effect of pyrimidine nucleotides on H. influenzae competence (Macfadyen et al., 2001), it is possible that an inhibitory effect exists in vivo. H. influenzae cannot synthesize pyrimidines de novo and relies instead on salvage from the medium of pyrimidines or precursors. Pyrimidine pools thus never get depleted under our standard laboratory conditions, because they are abundant in rich medium (sBHI) and because the MIV medium used to induce competence contains the pyrimidine precursor citrulline.
All Pasteurellaceae sxy genes are preceded by long intergenic regions that could form into stem structures like those of H. influenzae sxy and V. cholerae tfoX, and contribute to regulation. At present, most transcriptional start sites have not yet been mapped, which prevents predictions of stem structures and more detailed studies of regulation. Like the V. cholerae tfoX stem, the H. influenzae sxy stem may integrate multiple signals to regulate natural competence: aside from purine availability, it might also respond to separate signals through Hfq. Further work is required to confirm whether Hfq regulates sxy expression, as well as to identify the conditions under which regulation occurs and the small RNA(s) involved. A search of the genome for small regulatory RNAs using RNAPredator (Eggenhofer et al., 2011) or the reverse search mode of TargetRNA (http://snowwhite.wellesley.edu/targetRNA/index_2.html) found some sequences with complementarity to sxy mRNA, but these were often short or showed modest identity. In contrast, performing the same search with the V. cholerae tfoX sequence identified tfoR within the top matches (and a search with tfoR identified tfoX). The best chance of identifying comparable small RNAs in H. influenzae would be to screen genomic libraries for clones that change sxy expression as was done to identify tfoR (Yamamoto et al., 2011).
This work dissects the regulation of a biological process to inform an understanding of its evolutionary function. DNA is abundant in the human nasopharynx where H. influenzae lives, and we have demonstrated the importance of intracellular nucleotide depletion for competence induction, through the competence regulator Sxy. Because sxy transcription requires CRP (Cameron et al., 2008), the nucleotide depletion and sugar depletion signals are tightly linked. Further competence-inducing signals may exist as in other bacteria, and each clarifies the conditions under which DNA uptake is favourable.
Experimental procedures
Bacterial strains, plasmids and culture conditions
The strains and plasmids used in this study are listed in Table 3. Escherichia coli was cultured at 30°C or 37°C in LB medium, with ampicillin (100 μg ml−1) and spectinomycin (40 μg ml−1) when required. H. influenzae was cultured at 37°C in brain heart infusion (BHI) medium supplemented with NAD (2 mg ml−1) and hemin (10 mg ml−1) (sBHI), with novobiocin (2.5 μg ml−1), spectinomycin (20 μg ml−1), chloramphenicol (2 μg ml−1) or kanamycin (7 μg ml−1) when required. Actinobacillus pleuropneumoniae was cultured at 37°C on BHI agar supplemented with 10% Levinthal's Base with or without 50 μg ml−1 kanamycin, or in BHI broth supplemented with 100 μg ml−1 β-nicotinamide adenine dinucleotide (NAD). A. suis was obtained from Janet MacInnes and cultured at 37°C in plain BHI medium, with or without 25 μg ml−1 nalidixic acid.
Table 3.
Bacterial strains and plasmids
| Name | Organism | Genotype | References/notes |
|---|---|---|---|
| Bacterial strains | |||
| HS143 | A. pleuropneumoniae | WT | Bosse et al. (2009) |
| 4074 ΔsodC | A. pleuropneumoniae | sodC::kan | Bosse et al. (2009); donor DNA for transformation assays |
| ATCC 15557 | A. suis | WT | Wetmore et al. (1963) |
| ATCC 15557 Δsxy | A. suis | sxy::spc | This study |
| SO4 nalR | A. suis | nalR | Janet MacInnes; donor DNA for transformation assays |
| DH5α | E. coli | F80lacZ Δ(lacIZYA-argF) endA1 | Taylor et al. (1993); used for routine cloning |
| SW102 | E. coli | F- mcrA Δ(mrr-hsdRMS-mcrBC) Φ80dlacZΔM15 ΔlacX74 deoR recA1 endA1 araD139 Δ(ara, leu)7649 rpsL nupG [λcI857 (cro-bioA) <> tet] ΔgalK | Warming et al. (2005); used for recombineering |
| KW20 | H. influenzae | WT | Alexander and Leidy (1951) |
| MAP7 | H. influenzae | strR kanR novR nalR spcR vioR stvR | Barcak et al. (1991); donor DNA for transformation assays |
| ΔpurR | H. influenzae | KW20 purR::kan | This study |
| ΔpurR2 | H. influenzae | KW20 purR::spc | This study |
| ΔpurH | H. influenzae | KW20 purH::spc | This study |
| ΔpurR purH | H. influenzae | KW20 purR::kan purH::spc | This study |
| rec2::lacZ | H. influenzae | KW20 rec2::lacZ-cm (operon fusion) | Gwinn et al. (1997) |
| comA::lacZ | H. influenzae | KW20 comA::lacZ-cm (operon fusion) | Gwinn et al. (1997) |
| purH::lacZ | H. influenzae | KW20 purH::lacZ-kan (operon fusion) | This study |
| ΔpurR rec2:lacZ | H. influenzae | KW20 rec2::lacZ-cm (operon fusion) purR::kan | This study |
| ΔpurR comA::lacZ | H. influenzae | KW20 comA::lacZ-cm (operon fusion) purR::kan | This study |
| ΔpurR purH::lacZ | H. influenzae | KW20 purH::lacZ-kan (operon fusion) purR::spc | This study |
| sxy-1 | H. influenzae | KW20 sxyG106A | Redfield (1991) |
| sxy-2 | H. influenzae | KW20 sxyG102A | Cameron et al. (2008) |
| sxy-4 | H. influenzae | KW20 sxyT15C | Cameron et al. (2008) |
| sxy-5 | H. influenzae | KW20 sxyG16T | Cameron et al. (2008) |
| sxy-6 | H. influenzae | KW20 sxyC14T,G106A | Cameron et al. (2008) |
| murE749 | H. influenzae | KW20 murEG13033A | Ma and Redfield (2000) |
| Δhfq | H. influenzae | KW20 hfq::spc | This study |
| Δhfq sxy-1 | H. influenzae | KW20 hfq::spc sxyG106A | This study |
| ΔHI0365 | H. influenzae | KW20 HI0365::spc | Sinha et al. (2012) |
| ΔHI0660 | H. influenzae | KW20 HI0660::spc | Sinha et al. (2012) |
| ΔHI1631 | H. influenzae | KW20 HI1631::spc | Sinha et al. (2012) |
| rpoB* | H. influenzae | KW20 rpoB* | This study |
| Plasmids | |||
| pcomA::lacZ | / | comA::lacZ-kan in pGEMT-Easy (operon fusion) | This study |
| ppurH::lacZ | / | purH::lacZ-kan in pGEMT-Easy (operon fusion) | This study |
| pLBSF2 | / | sxy89::lacZ-kan in pGEM (protein fusion) | Cameron et al. (2008) |
Unless otherwise stated, AMP, adenine, adenosine, GMP and AICAR were used at final concentrations of 1 mM for H. influenzae and E. coli, and 0.1 mM for A. pleuropneumoniae and A. suis. These concentrations do not affect cell viability (Fig. S1).
Strain construction and cloning
The sequences of primers used are given in Table S1. All strains were verified by PCR and sequencing. Mutations were transferred to the chromosome of A. suis or H. influenzae by natural transformation of MIV-competent cells (see below).
KW20 ΔpurR/KW20 ΔpurR sxy-1: A 3 kb region flanking and including purR was PCR-amplified and cloned into pGEMT-Easy (Promega) to generate pGpurR. Inverse PCR on pGpurR amplified a fragment lacking sequences between the first 32 bp and last 132 bp of purR, with KpnI and EcoRI restriction enzymes sites added to the forward and reverse primers respectively. The PCR fragment was digested, and ligated to a KpnI/EcoRI-digested kanR cassette from pUC4K (Taylor and Rose, 1988). Positive clones (pGpurR::kan) transformed into DH5α were isolated on medium containing ampicillin and kanamycin. The insert was PCR amplified using the universal primers M13F and M13R (Promega) and used for transformation into H. influenzae KW20 and sxy-1 cells. A second ΔpurR strain (ΔpurR2, purR::spc) was constructed in KW20 in the same way, but with spcR from pRSM2832 (Tracy et al., 2008) replacing kanR, and using blunt-end cloning instead of KpnI/EcoRI.
KW20 ΔpurH: The strategy used to construct ΔpurR was followed, with the following differences: the region flanking and including purH in pGpurH was 4 kb, and the amplified inverse PCR fragment lacked sequences between the first 229 bp and last 442 bp of purH and contained HindIII and EcoRI sites. This was ligated to a HindIII/EcoRI-digested spcR cassette from pRSM2832 (Tracy et al., 2008) to generate pGpurH::spc.
KW20 ΔpurR purH: The insert from pGpurH::spc was used to transform competent cells of ΔpurR::kan. Double transformants were selected on medium containing kanamycin and spectinomycin.
KW20 Δhfq/KW20 Δhfq sxy-1: The hfq::spec strain was constructed by recombineering using the procedure described in Sinha et al. (2012), replacing the sequence between the first and last seven codons of the hfq coding sequence with a spcR cassette pRSM2832 (Tracy et al., 2008). This mutation was transferred to KW20 and to sxy-1 cells.
ppurH::lacZ and pcomA::lacZ: A 4 kb region flanking and including purH was PCR-amplified and ligated into pGEM-Easy (Promega) to generate plasmid pGpurH. This plasmid was linearized with ClaI, blunt-ended using Klenow and ligated to a blunt lacZ-kan cassette excised from pLKZ83 with BamHI (Barcak et al., 1991). The same strategy was followed with pGcomA-E (Sinha et al., 2012) which was linearized with BglII and directly ligated to the BamHI-digested lacZ cassette. Clones in the forward orientation were selected on medium containing ampicillin, kanamycin and Xgal. The purH::lacZ insert and the comA::lacZ and rec2::lacZ fusions of Gwinn et al. (1997) were also transferred to KW20 and to ΔpurR by natural transformation.
ATCC 15557 Δsxy: This sxy::spec derivative was constructed by recombineering using the procedure described in Sinha et al. (2012), replacing the sequence between the first and last seven codons of the sxy coding sequence in strain ATCC 15557 with a spcR cassette pRSM2832 (Tracy et al., 2008).
Transformation assays
Haemophilus influenzae cells were made competent by transfer of sBHI-grown cells to MIV medium at OD600 0.2–0.25, and incubation for 100 min at 37°C as described previously (Poje and Redfield, 2003). When testing the effect of purine repression, AMP and other purine sources were added to MIV with the cells.
To measure transformation, competent cells were incubated with 1 μg of MAP7 chromosomal DNA in 1 ml of MIV culture for 15 min at 37°C, after which the free DNA was degraded by incubation with DNase I (10 μg ml−1) for 5 min. Cells were then diluted and plated on sBHI agar with and without novobiocin. Transformation frequencies were calculated by dividing the number of novobiocin-resistant colony-forming units (cfu) by the total number of cfu. Transformation in rich medium was measured in the same way, after adding 1 μg DNA to 1 ml culture aliquots.
Search for PurR binding sites
Candidate PurR binding sites were identified in the H. influenzae Rd KW20 genome using the species-specific position weight matrix of Ravcheev et al. (2002). Scoring used custom R scripts, including the SeqInR add-on package to manipulate sequences (Charif and Lobry, 2007). Searches using E. coli PurR motifs (Mironov et al., 1999; Cho et al., 2011) on the H. influenzae genome found the same sites. A cutoff score of 4.5 captured all of the PurR-regulated genes identified by Ravcheev et al. (2002). An additional requirement of location between −100 and +20 of annotated start codons eliminated false-positive sites outside of promoter regions. blast was used to identify rec2 homologues in other sequenced Haemophilus influenzae, Pasteurellaceae and selected gamma-proteobacteria, and the highest scoring PurR site from −200 to +20 of the start was determined.
Search for CRP-S homologues in A. suis
Competence genes in the A. suis H91-0380 genome sequence were first identified by blast using A. pleuropneumoniae L20 homologues (Bosse et al., 2009). Next, their upstream regions were searched for candidate CRP-S binding sites, using a scoring matrix produced using the method of Schneider et al. (1986) from the set of Pasteurellaceaen CRP-S sites identified by Cameron and Redfield (2006).
β-Galactosidase assays
Haemophilus influenzae strains containing chromosomal lacZ fusions were grown in sBHI or transferred to MIV as described above. E. coli strains containing plasmid-encoded lacZ fusions were grown in LB or in M9 with 0.4% glucose. Cultures were incubated with or without an added purine source and sampled at regular time intervals. After sampling, cells were immediately assayed for OD600 and pelleted by centrifugation, supernatants were removed and cell pellets were frozen at −20°C for later assays of β-galactosidase activity following the protocol of Miller (1992).
Quantitative measurements of sxy expression
Real-time qPCR was used to measure sxy transcript levels, while Western blotting was used to measure Sxy protein levels. All procedures were carried out as described in Cameron et al. (2008), in which the primers and antibody used are also described. For each sample, sxy RNA levels were adjusted using murG levels, while Sxy protein levels were adjusted using an unidentified protein of constant abundance (Cameron et al., 2008) (‘background’ in Fig. 3).
Acknowledgments
This work was supported by an operating grant from the Canadian Institutes for Health Research to R. J. R. We thank Janet MacInnes for providing the Actinobacillus suis strains and for providing expertise on working with this organism.
Supporting information
Additional supporting information may be found in the online version of this article at the publisher's web-site.
References
- Alexander HE, Leidy G. Determination of inherited traits of H. influenzae by desoxyribonucleic acid fractions isolated from type-specific cells. J Exp Med. 1951;93:345–359. doi: 10.1084/jem.93.4.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen S, Zilles JL, Downs DM. Metabolic flux in both the purine mononucleotide and histidine biosynthetic pathways can influence synthesis of the hydroxymethyl pyrimidine moiety of thiamine in Salmonella enterica. J Bacteriol. 2002;184:6130–6137. doi: 10.1128/JB.184.22.6130-6137.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonova ES, Hammer BK. Quorum-sensing autoinducer molecules produced by members of a multispecies biofilm promote horizontal gene transfer to Vibrio cholerae. FEMS Microbiol Lett. 2011;322:68–76. doi: 10.1111/j.1574-6968.2011.02328.x. [DOI] [PubMed] [Google Scholar]
- Antonova ES, Bernardy EE, Hammer BK. Natural competence in Vibrio cholerae is controlled by a nucleoside scavenging response that requires CytR-dependent anti-activation. Mol Microbiol. 2012;86:1215–1231. doi: 10.1111/mmi.12054. [DOI] [PubMed] [Google Scholar]
- Barcak GJ, Chandler MS, Redfield RJ, Tomb JF. Genetic systems in Haemophilus influenzae. Methods Enzymol. 1991;204:321–342. doi: 10.1016/0076-6879(91)04016-h. [DOI] [PubMed] [Google Scholar]
- Barouki R, Smith HO. Reexamination of phenotypic defects in rec-1 and rec-2 mutants of Haemophilus influenzae Rd. J Bacteriol. 1985;163:629–634. doi: 10.1128/jb.163.2.629-634.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazurto JV, Downs DM. Plasticity in the purine-thiamine metabolic network of Salmonella. Genetics. 2011;187:623–631. doi: 10.1534/genetics.110.124362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blokesch M. Chitin colonization, chitin degradation and chitin-induced natural competence of Vibrio cholerae are subject to catabolite repression. Environ Microbiol. 2012;14:1898–1912. doi: 10.1111/j.1462-2920.2011.02689.x. [DOI] [PubMed] [Google Scholar]
- Bosse JT, Sinha S, Schippers T, Kroll JS, Redfield RJ, Langford PR. Natural competence in strains of Actinobacillus pleuropneumoniae. FEMS Microbiol Lett. 2009;298:124–130. doi: 10.1111/j.1574-6968.2009.01706.x. [DOI] [PubMed] [Google Scholar]
- Cameron AD, Redfield RJ. Non-canonical CRP sites control competence regulons in Escherichia coli and many other gamma-proteobacteria. Nucleic Acids Res. 2006;34:6001–6014. doi: 10.1093/nar/gkl734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron AD, Redfield RJ. CRP binding and transcription activation at CRP-S sites. J Mol Biol. 2008;383:313–323. doi: 10.1016/j.jmb.2008.08.027. [DOI] [PubMed] [Google Scholar]
- Cameron AD, Volar M, Bannister LA, Redfield RJ. RNA secondary structure regulates the translation of sxy and competence development in Haemophilus influenzae. Nucleic Acids Res. 2008;36:10–20. doi: 10.1093/nar/gkm915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charif D, Lobry JR. Seqin{R} 1.0-2: A Contributed Package to the {R} Project for Statistical Computing Devoted to Biological Sequences Etrieval and Analysis. in Structural Approaches to Sequence Evolution: Molecules, Networks, Populations. New York: Springer Verlag; 2007. [Google Scholar]
- Cho BK, Federowicz SA, Embree M, Park YS, Kim D, Palsson BO. The PurR regulon in Escherichia coli K-12 MG1655. Nucleic Acids Res. 2011;39:6456–6464. doi: 10.1093/nar/gkr307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingues S, Harms K, Fricke WF, Johnsen PJ, da Silva GJ, Nielsen KM. Natural transformation facilitates transfer of transposons, integrons and gene cassettes between bacterial species. PLoS Pathog. 2012;8:e1002837. doi: 10.1371/journal.ppat.1002837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorocicz IR, Williams PM, Redfield RJ. The Haemophilus influenzae adenylate cyclase gene: cloning, sequence, and essential role in competence. J Bacteriol. 1993;175:7142–7149. doi: 10.1128/jb.175.22.7142-7149.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggenhofer F, Tafer H, Stadler PF, Hofacker IL. RNApredator: fast accessibility-based prediction of sRNA targets. Nucleic Acids Res. 2011;39:W149–W154. doi: 10.1093/nar/gkr467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel SE, Kolter R. DNA as a nutrient: novel role for bacterial competence gene homologs. J Bacteriol. 2001;183:6288–6293. doi: 10.1128/JB.183.21.6288-6293.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gots JS, Benson CE. Genetic control of bacterial purine phosphoribosyltransferases and an approach to gene enrichment. Adv Exp Med Biol. 1973;41:33–39. doi: 10.1007/978-1-4684-3294-7_5. [DOI] [PubMed] [Google Scholar]
- Gwinn ML, Stellwagen AE, Craig NL, Tomb JF, Smith HO. In vitro Tn7 mutagenesis of Haemophilus influenzae Rd and characterization of the role of atpA in transformation. J Bacteriol. 1997;179:7315–7320. doi: 10.1128/jb.179.23.7315-7320.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herriott RM, Meyer EM, Vogt M. Defined nongrowth media for stage II development of competence in Haemophilus influenzae. J Bacteriol. 1970;101:517–524. doi: 10.1128/jb.101.2.517-524.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsborg O, Havarstein LS. Regulation of natural genetic transformation and acquisition of transforming DNA in Streptococcus pneumoniae. FEMS Microbiol Rev. 2009;33:627–642. doi: 10.1111/j.1574-6976.2009.00167.x. [DOI] [PubMed] [Google Scholar]
- Kristensen BM, Sinha S, Boyce JD, Bojesen AM, Mell JC, Redfield RJ. Natural transformation of Gallibacterium anatis. Appl Environ Microbiol. 2012;78:4914–4922. doi: 10.1128/AEM.00412-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livermore DM. Fourteen years in resistance. Int J Antimicrob Agents. 2012;39:283–294. doi: 10.1016/j.ijantimicag.2011.12.012. [DOI] [PubMed] [Google Scholar]
- Lo Scrudato M, Blokesch M. The regulatory network of natural competence and transformation of Vibrio cholerae. PLoS Genet. 2012;8:e1002778. doi: 10.1371/journal.pgen.1002778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C, Redfield RJ. Point mutations in a peptidoglycan biosynthesis gene cause competence induction in Haemophilus influenzae. J Bacteriol. 2000;182:3323–3330. doi: 10.1128/jb.182.12.3323-3330.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfadyen LP. Regulation of competence development in Haemophilus influenzae. J Theor Biol. 2000;207:349–359. doi: 10.1006/jtbi.2000.2179. [DOI] [PubMed] [Google Scholar]
- Macfadyen LP, Chen D, Vo HC, Liao D, Sinotte R, Redfield RJ. Competence development by Haemophilus influenzae is regulated by the availability of nucleic acid precursors. Mol Microbiol. 2001;40:700–707. doi: 10.1046/j.1365-2958.2001.02419.x. [DOI] [PubMed] [Google Scholar]
- Maughan H, Sinha S, Wilson L, Redfield RJ. Pasteurellaceae: Biology, Genomics and Molecular Aspects. Norfolk, UK: Caister Academic Press; 2008. [Google Scholar]
- Meibom KL, Blokesch M, Dolganov NA, Wu CY, Schoolnik GK. Chitin induces natural competence in Vibrio cholerae. Science. 2005;310:1824–1827. doi: 10.1126/science.1120096. [DOI] [PubMed] [Google Scholar]
- Miller JH. A Short Course in Bacterial Genetics: A Laboratory Manual and Handbook for Escherichia coli and Related Bacteria. Plainview, NY: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- Mironov AA, Koonin EV, Roytberg MA, Gelfand MS. Computer analysis of transcription regulatory patterns in completely sequenced bacterial genomes. Nucleic Acids Res. 1999;27:2981–2989. doi: 10.1093/nar/27.14.2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palchevskiy V, Finkel SE. Escherichia coli competence gene homologs are essential for competitive fitness and the use of DNA as a nutrient. J Bacteriol. 2006;188:3902–3910. doi: 10.1128/JB.01974-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pifer ML, Smith HO. Processing of donor DNA during Haemophilus influenzae transformation: analysis using a model plasmid system. Proc Natl Acad Sci USA. 1985;82:3731–3735. doi: 10.1073/pnas.82.11.3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poje G, Redfield RJ. Transformation of Haemophilus influenzae. Methods Mol Med. 2003;71:57–70. doi: 10.1385/1-59259-321-6:57. [DOI] [PubMed] [Google Scholar]
- Pollack-Berti A, Wollenberg MS, Ruby EG. Natural transformation of Vibrio fischeri requires tfoX and tfoY. Environ Microbiol. 2010;12:2302–2311. doi: 10.1111/j.1462-2920.2010.02250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravcheev DA, Gel'fand MS, Mironov AA, Rakhmaninova AB. Purine regulon of gamma-proteobacteria: a detailed description. Genetika. 2002;38:1203–1214. [PubMed] [Google Scholar]
- Redfield RJ. sxy-1, a Haemophilus influenzae mutation causing greatly enhanced spontaneous competence. J Bacteriol. 1991;173:5612–5618. doi: 10.1128/jb.173.18.5612-5618.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redfield RJ. Genes for breakfast: the have-your-cake-and-eat-it-too of bacterial transformation. J Hered. 1993;84:400–404. doi: 10.1093/oxfordjournals.jhered.a111361. [DOI] [PubMed] [Google Scholar]
- Redfield RJ, Cameron AD, Qian Q, Hinds J, Ali TR, Kroll JS, Langford PR. A novel CRP-dependent regulon controls expression of competence genes in Haemophilus influenzae. J Mol Biol. 2005;347:735–747. doi: 10.1016/j.jmb.2005.01.012. [DOI] [PubMed] [Google Scholar]
- Redfield RJ, Findlay WA, Bosse J, Kroll JS, Cameron AD, Nash JH. Evolution of competence and DNA uptake specificity in the Pasteurellaceae. BMC Evol Biol. 2006;6:82. doi: 10.1186/1471-2148-6-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider TD, Stormo GD, Gold L, Ehrenfeucht A. Information content of binding sites on nucleotide sequences. J Mol Biol. 1986;188:415–431. doi: 10.1016/0022-2836(86)90165-8. [DOI] [PubMed] [Google Scholar]
- Seitz P, Blokesch M. Cues and regulatory pathways involved in natural competence and transformation in pathogenic and environmental Gram-negative bacteria. FEMS Microbiol Rev. 2012;37:336–363. doi: 10.1111/j.1574-6976.2012.00353.x. [DOI] [PubMed] [Google Scholar]
- Sinha S, Redfield RJ. Natural DNA uptake by Escherichia coli. PLoS ONE. 2012;7:e35620. doi: 10.1371/journal.pone.0035620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S, Cameron ADS, Redfield RJ. Sxy induces a CRP-S regulon in Escherichia coli. J Bacteriol. 2009;191:5180–5195. doi: 10.1128/JB.00476-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S, Mell JC, Redfield RJ. Seventeen Sxy-dependent cyclic AMP receptor protein site-regulated genes are needed for natural transformation in Haemophilus influenzae. J Bacteriol. 2012;194:5245–5254. doi: 10.1128/JB.00671-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon JM, Grossman AD. Who's competent and when: regulation of natural genetic competence in bacteria. Trends Genet. 1996;12:150–155. doi: 10.1016/0168-9525(96)10014-7. [DOI] [PubMed] [Google Scholar]
- Stewart GJ, Carlson CA. The biology of natural transformation. Annu Rev Microbiol. 1986;40:211–235. doi: 10.1146/annurev.mi.40.100186.001235. [DOI] [PubMed] [Google Scholar]
- Suckow G, Seitz P, Blokesch M. Quorum sensing contributes to natural transformation of Vibrio cholerae in a species-specific manner. J Bacteriol. 2011;193:4914–4924. doi: 10.1128/JB.05396-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor LA, Rose RE. A correction in the nucleotide sequence of the Tn903 kanamycin resistance determinant in pUC4K. Nucleic Acids Res. 1988;16:7762. doi: 10.1093/nar/16.1.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RG, Walker DC, McInnes RR. E. coli host strains significantly affect the quality of small scale plasmid DNA preparations used for sequencing. Nucleic Acids Res. 1993;21:1677–1678. doi: 10.1093/nar/21.7.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tracy E, Ye F, Baker BD, Munson RS., Jr Construction of non-polar mutants in Haemophilus influenzae using FLP recombinase technology. BMC Mol Biol. 2008;9:101. doi: 10.1186/1471-2199-9-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 2005;33:e36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetmore PW, Thiel JF, Herman YF, Harr JR. Comparison of selected Actinobacillus species with a hemolytic variety of Actinobacillus from irradiated swine. J Infect Dis. 1963;113:186–194. doi: 10.1093/infdis/113.3.186. [DOI] [PubMed] [Google Scholar]
- Yamamoto S, Morita M, Izumiya H, Watanabe H. Chitin disaccharide (GlcNAc)2 induces natural competence in Vibrio cholerae through transcriptional and translational activation of a positive regulatory gene tfoXVC. Gene. 2010;457:42–49. doi: 10.1016/j.gene.2010.03.003. [DOI] [PubMed] [Google Scholar]
- Yamamoto S, Izumiya H, Mitobe J, Morita M, Arakawa E, Ohnishi M, Watanabe H. Identification of a chitin-induced small RNA that regulates translation of the tfoX gene, encoding a positive regulator of natural competence in Vibrio cholerae. J Bacteriol. 2011;193:1953–1965. doi: 10.1128/JB.01340-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Morar M, Ealick SE. Structural biology of the purine biosynthetic pathway. Cell Mol Life Sci. 2008;65:3699–3724. doi: 10.1007/s00018-008-8295-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


