Abstract
A crisis continues to brew within the pharmaceutical research and development (R&D) enterprise: productivity continues declining as costs rise, despite ongoing, often dramatic scientific and technical advances. To reverse this trend, we offer various suggestions for both the expansion and broader adoption of modeling and simulation (M&S) methods. We suggest strategies and scenarios intended to enable new M&S use cases that directly engage R&D knowledge generation and build actionable mechanistic insight, thereby opening the door to enhanced productivity. What M&S requirements must be satisfied to access and open the door, and begin reversing the productivity decline? Can current methods and tools fulfill the requirements, or are new methods necessary? We draw on the relevant, recent literature to provide and explore answers. In so doing, we identify essential, key roles for agent-based and other methods. We assemble a list of requirements necessary for M&S to meet the diverse needs distilled from a collection of research, review, and opinion articles. We argue that to realize its full potential, M&S should be actualized within a larger information technology framework—a dynamic knowledge repository—wherein models of various types execute, evolve, and increase in accuracy over time. We offer some details of the issues that must be addressed for such a repository to accrue the capabilities needed to reverse the productivity decline. © 2013 Wiley Periodicals, Inc.
INTRODUCTION
Pharmaceutical research and development (R&D) is in the midst of a productivity decline. Unified, transdisciplinary, in silico modeling and simulation (M&S) methods are viewed broadly as a promising countermeasure. Agent-based (AB) modeling is a phrase used currently to identify relatively young modeling methods that utilize software agents. We review and present evidence that AB methods will be essential contributors to successful M&S countermeasures.
TRI-FOCUS: AGENT-BASED M&S, R&D, AND EXPLANATORY MECHANISMS
The development of agent modeling tools and the availability of increasingly detailed, varied, and abundant data coupled with advances in computation have made possible a growing number of agent-based modeling and simulation (AB M&S) applications across a variety of non-biomedical domains and disciplines. To illustrate, we identify early1–15 and more recent16–32 use cases. Macal and North33 provide additional examples and a tutorial on AB M&S methods. Within the biomedical domain, AB M&S is a relatively new approach for studying systems composed of interacting components, some of which can be autonomous. The methods are used primarily to gain insight into mechanisms responsible for phenomena of living systems. We cite early34–40 and more recent41–56 examples of such applications. Amigoni and Schiaffonati57, An et al.58, and Edelman et al.59 provide biomedically-focused reviews. Several applications exemplify the expanding variety of applications relevant to the pharmaceutical sciences.46,60–72
Pharmaceutical science stakeholders agree that there is a crisis within the broad pharmaceutical R&D domain (private and public): productivity continues to decline, even in the face of dramatic scientific and technical advances accompanied by a data deluge, especially at the molecular level. A flurry of recent reviews and commentaries73–102 discuss the problems from several different perspectives and offer strategies and scenarios for how M&S methods can and are being used to enhance productivity. The different perspectives include pharmacometrics [including translational and physiologically based pharmacokinetics–pharmacodynamics (PBPK), and disease progression M&S], quantitative and systems pharmacology, and model-based drug development. Given the apparent fit between designed uses of AB M&S methods and the multifarious problems cited in the reviews that can benefit from computational M&S methods, it is surprising that only two of the citations mention AB M&S methods. No examples are presented within those citations of any current use of AB M&S methods within pharmaceutical companies. Does that situation signal ripe opportunity for AB M&S methods to add new value beyond that delivered using the established methods? That question, although provocative, is premature because one should not try to force-fit a particular M&S method to a particular set of problems. We should first evaluate where and how the cited domain experts envision M&S methods being used to reverse the productivity trend. We can then select those M&S methods that enable those uses.
The cited experts anticipate shifting focus from analysis of data to discovery and challenge of explanatory mechanisms. They see M&S becoming integral and essential to envisioned discovery and development processes that are dramatically more productive in part because of improvements in use of M&S methods across all R&D activities. Yet they also make clear that the established practice is to select one or more modeling tools, drawn primarily from those already in use,75 to address the particular problem at hand. The practice is the same independent of stage in the R&D process. Various scientists, often separated in time and space across R&D processes, integrate the derived information. Anticipating that having M&S integrated across R&D activities can improve productivity, what new demands are placed on M&S methods? We show why multifaceted, networked M&S use cases require drawing from an expanded computational modeling method and tool repertoire. Can current methods and tools fulfill the requirements? No. We justify that answer and discuss why AB and more advanced M&S methods are essential. It becomes clear that an analog (see Box 1) based knowledge repository will be needed. We identify M&S method requirements, and that brings into focus additional M&S use cases that we believe the repository's framework will likely need to enable.
BOX 1.
DEFINITIONS
Actor: an entity identifiable by an observer as a cause of an effect; an entity that participates in a process (plays a part); in computer science it is a mathematical model of computation that treats actors as the universal primitives: in response to a received message, an actor can make local decisions, create more actors, send messages, and determine how to respond to the next message received.
Agent: a software object that can schedule its own events (within an analog, it is quasi-autonomous); it senses and is part of its environment; it pursues and can revise an agenda within a larger script; some of its attributes and actions may be designed to represent biological counterparts, whereas others will deal with issues of software execution.
Agent-based: something formulated with or built up from agents; in this context, it identifies a simulation model in which quasi-autonomous agents are key components. Terms that are often synonymous within the M&S literature include individual-based and multi-agent.
Framework: a carefully crafted assemblage of tools, devices (some software, some hardware, and occasionally even some wet-ware), usage protocols, good practices, etc., governed by a set of component interoperability standards. Upon satisfying use cases and listed requirements, we expect the framework to be an extensible, distributed, open, and loosely coupled (yet unified from the user's perspective).
Analog: a software device that has (some) aspects and attributes that are similar to those of its R&D referent yet can exist and operate in isolation and in the absence of its R&D counterpart; in biomedical M&S, a model implemented in software that, when executed, produces phenomena that mimic one or more attributes measured or observed during referent wet-lab experiments. In this context, most analogs will be suitable for experimentation.
In silico experiment: it is precisely analogous to a wet-lab experiment. An analog is a hypothesis: the mechanism produced upon execution by interacting components will result in phenomena, often at different scales, that are similar, or not, to prespecified wet-lab phenomena. Measurement of features during execution enables testing the hypothesis. That activity is an in silico experiment.
Analog based knowledge repository: easily accessible, organized framework feature. Its content is an up-to-date instantiation of all relevant mechanistic knowledge in an accessible, easily understood, observable, and interactive form. It contains annotated records of analogs (current and falsified), their mechanisms, how they were composed plus the rationale, along with records of in silico experiments. All use cases (see ‘Paramount use cases’ subsection) can be achieved by employing the analog based knowledge repository. We envision domain experts making go/no-go decisions after interactive exploration of many scenarios (simulations) within the repository.
MODELING, SIMULATION, AND THE PRODUCTIVITY CRISIS
Ideally, one should begin any M&S project without bias for any particular model types or methods.103 Selecting M&S methods or tools in advance of specifying uses cases can constrain and even bias thinking about explanatory mechanisms. Hence, the first task is to clearly identify near and longer term expectations. Among the questions to be answered are these: what are the problems? What questions need to be answered? What new knowledge is sought and how will it be used? What decisions must be made? When are the deadlines? What resources are available? etc. Within pharmaceutical R&D, these questions are complicated by the nested, networked, multi-year, evolving nature of the overall enterprise. Scientific insight achieved at an early stage may be critical downstream (e.g., during dose ranging studies or clinical trials), and early planning for that prospect may influence selection of M&S methods in important ways. Yao et al.102 illustrate the variety of relationships and common computational methods that support them (see Figure S1 in Supporting Information).
Consider a model and method judged appropriate for a particular discovery or early development problem. The insight sought is intended to be useful for downstream as well as for current decision-making. Can the model, method, and information be easily utilized as needed within a tight time window at a downstream go/no-go decision juncture? If doing so proves problematic because of differences in models, methods, and/or tools, or perhaps because of inadequate records of analogs and (more notably) their in silico experiments, etc., uncertainties and delays increase unnecessarily along with the risk of a flawed decision. The situation is complicated further by a larger, pressing, overarching need: reverse the declining trend in productivity.
The cited experts make the case that M&S methods are essential to simultaneously increasing productivity of therapeutic drug development, and facilitating the recursive cycle of new knowledge buildup aimed at improved quality care. Accepting that position, we view the enterprise as a networked, experiment-intensive system that begins small (significantly left on the Systems Information spectrum in Figure 1). As R&D progresses, the system evolves and expands. It becomes a large, distributed, multiscale, multi-aspect M&S challenge. For a particular aspect of the system, its location on the Systems Information spectrum in Figure 1 inches to the right. In what variety of ways do the cited experts envision M&S methods being used? Answers are provided in Supporting Information-Use Cases; a sample of answers is provided in Box 3. We propose a set of seven paramount use cases that are derived from and subsume those particular use cases. We then present five requirements, which we argue must be satisfied to enable achieving those paramount use cases efficiently. We discuss M&S methods that will enable a unified M&S approach to satisfy those requirements. To our knowledge, such an analysis has not been done. We argue that such an examination is necessary and essential to discover strategies capable of restoring and enhancing productivity.
FIGURE 1.
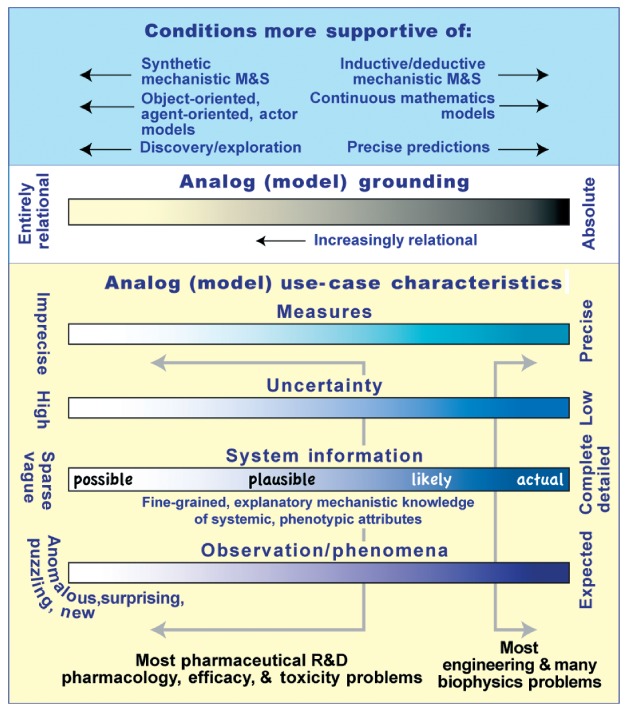
A particular analog use case can be characterized by an approximate location on each of the four lower spectra. Spectrum location along with specifics about how and for what the simulation model will be used within the larger R&D context, determine which M&S methods are most appropriate for a given use case. Within a pharmaceutical R&D context, a use case includes details of the specific, biology-focused wet-lab experiment that analog execution is intended to model in some way. Depending on use case location on the spectra, an analog can be located anywhere along a spectrum of software devices (models) ranging from synthetic (all components designed to be plugged together and are thus replaceable) to purely inductive models. System Information includes current conceptual knowledge about the mechanisms on which wet-lab experiments focused. As the R&D process advances, evidence will shrink the space of possible mechanisms. The result will be a set of plausible analog mechanisms supported by validation evidence. Later, that set too will shrink. The result will be a smaller set of likely mechanisms (those that have survived several falsification experiments). An R&D project's product can be successful without being able to designate key mechanisms as either actual or likely. Actual mechanisms are typically known for engineered systems, but are typically lacking in therapeutics. Grounding is discussed in Box 2; additional information is provided as Supporting Information. Conditions on the far right of the bottom four spectra are supportive of models (typically, continuous equations) that rely exclusively on absolute grounding. Hunt at al.104 make the case that when left of center on one or more of the bottom three spectra, models should rely more on relational grounding. When on the far left, early stage, purely qualitative M&S is still useful and productive: it facilitates goal-oriented research efforts by clarifying (unifying) current thinking about referent phenomena. Such models would typically be coarse grain and use relational grounding. Spectra colors were selected arbitrarily.
BOX 3.
CLUSTERED EXAMPLES OF SPECIFIC M&S USE CASES
These examples (selected randomly) were drawn from those listed in Supporting Information-Use Cases. They illustrate the diversity and scope of particular envisioned73–102 M&S use cases.
Knowledge integration
Quantitatively address questions concerning the functional relationship between prognostic factors, dosage, and outcomes.86,93,96
Integrate the basic components to describe and understand the complex interplay between the pharmacology of drug action and (patho-) physiological systems.85,93
Management of uncertainty
Account for uncertainty in the underlying assumptions and thus resulting prediction of drug effects.76,91
Decision support
Document decision-making from discovery through development to regulatory filing and improve answering postfiling questions and approval, as well as life cycle management of the asset.96
Provide a decision-making tool for selecting effective and safe doses, optimizing study sample sizes, evaluating alternative trial designs and making rational go/no-go decisions based on the probability of achieving predefined study goals.75,78,79,91,96,97
Preclinical and clinical development
Facilitate design and/or selection of lead compounds, selection of the first-in-human dose, early clinical trial design, and proof-of-concept studies of experimental drugs and drug combinations.74,75,82,83,85,102
Simulate outcomes of alternative study designs before the experimental investigation commences—incorporating different doses and/or different patients and computing the probability of a successful trial given the characterized patient population and proposed treatment regimens.75,78,86
Predict the toxic potential of chemicals and human adverse effect, and generate hypotheses about the putative molecular mechanisms of chemical-induced injury.94,99
Drug-disease modeling
Use to understand the relationship between drug effect and the natural progression of the underlying disease.76,85,91,93
Describe macrophysiological processes within a particular disease state and use to understand likely modulation of those processes with specific interventions.73
Drug–drug interaction, special populations
Guide investigations in pediatric, the elderly and other special populations.76,84,88–91,95 Identify and assess complex drug-drug interactions early in drug development so that clinical studies could be planned or prioritized to assess the risk.79
Prediction
Allow for extrapolation of the PK properties across species and compounds.85,86,89,95
Ultimately predict human PK from in silico, in vitro, and physicochemical data.83
INTEGRATIVE MANAGEMENT OF KNOWLEDGE AND UNCERTAINTY
Knowledge integration is an essential, paramount use case. For this discussion, we limit attention to knowledge relevant to the primary R&D project focus: the particular disease, morbidity, or health issue; the biologic or chemical entity treatment intervention; the pharmacological, toxicological, and clinical outcomes that are consequences of treatments; and the variety of mechanisms (even when vague) that are offered to explain those phenomena. We ignore other important categories of knowledge that can impact go/no-go decisions, but the approach and framework described below can be expanded to include them. Examples of those categories include regulatory science; chemistry, manufacturing, and control; human resources; accessible contract services; etc.
The cited reviews discuss roles of various computational models and methods in knowledge integration. However, we explain below that little knowledge actually resides in those models. Much of the mechanistic insight resides within mental models, and that presents a problem: mental model differences, similarities, and inconsistencies are difficult, and often impossible to ascertain. Mental models are subject to their own forms of error introduction and propagation. Increasing reliance on synthetic analogs (defined below under Mechanism-Based Approach) presents advantages because they can evolve into executable representations of what we know (or think we know) about biological systems. Those representations are called executable biological knowledge embodiments64,65,104 and dynamic knowledge representations.34,58,60,61,105–109 Such executions are suitable for knowledge discovery.58,108,109 Knowledge embodiment is made feasible because synthetic analogs provide concrete instances of that knowledge rather than analytic descriptions of conceptual representations. When an analog is executed, it demonstrates when, how, and where our knowledge matches or fails to match referent system details, which enables and facilitates knowledge discovery.58,108,109
For current knowledge and beliefs to be useful (especially in a social context like shared model usage, validation, and falsification), it must be embedded in an analog and visible to the user (which is often not the case now). Analogs help build schemata for knowledge (and ignorance) representation, which can provide a mechanism for the curation and maintenance of the embedded knowledge. Users must be able to readily identify the knowledge and be able to discuss it, rely on it, dispute it, and falsify it (or not), all while reflecting on how that knowledge impacts the project. To achieve these capabilities, most, if not all, modeling activities and all mechanism representations will need to take place within a framework of the type described in Box 1 and discussed below. The R&D project related content of that framework will become part of an interactive, analog based knowledge repository.
It should be noted that knowledge is not embedded in the any of the variety of pharmacometric or systems pharmacology models identified in the reviews.73–102 Model refers specifically to the equations used to describe the referent aspects, dynamics, and features75,79,84–90,93,95 as implemented in software. As documented by An105,107 and Hunt et al.64,104 embedding knowledge in pharmacometric and ordinary differential equation (ODE) models is challenging, if not problematic. Equations are typically used to describe patterns in data. The associated conceptual models do reflect knowledge, but it exists separate from the equations. Humans interpreting the I/O of the cited equations use prose and sketches to provide the semantic grounding for those models in terms of both the idealized, conceptual model and knowledge of the referent system. That semantic grounding is done manually, and is separate from the equations and their software implementations. Hence, neither knowledge nor semantics is embedded in those equations. However, the equation models are typically encapsulated within ‘ready to use’ software tools and packages. Bouzom et al.7575 lists and describes the more frequently used general and constrained tools and packages. The builders, especially of the constrained, domain-focused tools and packages, invest considerable effort in collecting, organizing, and enabling use of domain knowledge, information, and data related to a variety of model use cases. So doing considerably lowers the barrier to model parameterization and scenario exploration. The preceding points are expanded upon in Supporting Information using two concrete examples (Box 2).
BOX 2.
ANALOG GROUNDING ISSUES
Grounding issues must be addressed to satisfy the listed requirements. Key issues discussed in detail by Hunt et al.104 are summarized in Supporting Information. The units, dimensions, and/or objects to which a variable or model constituent refers establish groundings. Absolute grounding: variables, parameters, and input–output (I/O) are in real-world units. Relational grounding: variables, parameters, and I/O are in units defined by other system components. Relational grounding requires a separate analog-to-referent mapping model. To satisfy requirements, a spectrum of model classes, methods, and groundings (illustrated in Figure 1) will be needed: absolute grounding occupies one extreme; relational grounding occupies the other.
Absolute grounding provides simple, interpretive mappings between simulation output, parameter values, and referent data. However, complex issues must be addressed each time one of the following occurs: expand the model to include additional phenomena; combining models; and/or model context changes. Expansions are challenging, even infeasible, when center-left in Figure 1. Reusability is hindered in part because the conflated semi-mechanistic, equation-based model and the model-to-referent mapping model have different use cases.
Analogs must evolve (become more complicated) as R&D advances and new mechanistic insight accumulate. That evolution will require changing, adding, and removing component linkages within analogs. We recommend keeping most component groundings relational. So doing facilitates component replacement, limiting any one component formulation solely to its coupling with the others. Any component can be replaced at will as long as the minimal I/O interface requirements are met. Starting with relationally grounded analog components allows the modeler to iterate progressively from qualitative to quantitative validation.
Acknowledging and Reducing Uncertainty
We focus on uncertainties directly associated with the mechanistic theories on which the project is based. At the start of a typical project, uncertainty is pervasive. Consequently, projects typically begin considerably left in Figure 1 Uncertainty and System Information spectra. Uncertainty sources are varied and plentiful. They need to be identified, annotated, and updated. As the project advances successfully, we inch to the right on both the System Information and Uncertainty spectra. Productivity-enhancing M&S methods must facilitate and increase the pace of that movement in real time. We can infer that new M&S methods will be needed that are capable of generating new knowledge without requiring new wet-lab experiments. We can also infer that they must also be capable of explorartion and advanced selection of strategies to reduce uncertainties systematically in real time in parallel with advancing wet-lab experiments. Preference must be given to M&S methods that can do both.
Reliance on computational models that simply abstract away uncertainties is counterproductive: so doing does yield simple, easily managed models, which can be useful for particular use cases, but it also adds a new source of uncertainty. The core source of uncertainty is the experimental evidence on which the project's mechanistic theories are based (in the context of all other available knowledge and insight), and we recognize that current theory at a particular project stage is just one drawn from a space of possible or plausible mechanistic theories.
Studies conducted by Amgen110 and Bayer HealthCare111 found that published, preclinical, scientific findings that were important to their R&D efforts could be confirmed in only 11–25% of cases. A rule-of-thumb among early-stage venture capital firms is that at least 50% of published studies, even those in top-tier academic journals, cannot be repeated.112 Consequently, there is significant risk that the evidence supporting the conceptual mechanistic models on which the project is based is flawed (even if the theory proves reasonably correct). Even when experiments are repeated, variability of results can be considerable. Rarely can measurements made on cell culture models be mapped quantitatively 1:1 to comparable measures made on animal models. Similarly, animal model phenomena rarely map 1:1 to human counterparts. Hence, there is considerable uncertainty about how results from different experiments can or should be mapped to—and thus influence—the conceptual mechanistic models on which project scientists are relying. Such uncertainty and mapping dilemmas present problems for those using conventional inductive, equation-based models (typically ODEs) of the type discussed in the cited reviews. Modelers understand the issues: when using such methods, they would prefer to be on the far right of Figure 1. The conventional strategy is to request more data. So doing postpones mechanistic modeling. Unfortunately, it may be necessary for the project to advance beyond the next few go/no-go decision points before the requested data begins to come available. While waiting for the required data, the accepted strategy for dealing with uncertainty issues is to abstract them away: if we make particular simplifying assumptions, we can create an idealized, hypothetical mechanistic scenario that, if fully validated, will place us on the right side of Figure 1 where inductive, predictive models can be reliable. In doing so, model-grounding issues, discussed below, are typically ignored. As the project advances, there is no straightforward way to ‘add back’ the various uncertainties abstracted away, even if they could be measured, or undo the simplifying assumptions.
Analogs are particularly useful in managing uncertainty because they can be used to simulate possible and plausible mechanisms when center-left on the three lower Figure 1 spectra (although our focus here is on uncertainty, other elements that are often also abstracted away, such as biological or experimental details, may also be handled more concretely by analogs). An analog's mechanism is a consequence of components interacting during execution. Monte Carlo variations in component specifications, rules specifying interactions, and parameterizations can cause phenomena generated during each execution to be unique. That process simulates the non-deterministic nature of biological phenomena. That variety can also represent uncertainty about generative mechanisms, experimental variability, and intra- and interindividual variability. The challenge then becomes to follow a disciplined protocol, as done in Sheikh-Bahaei and Hunt7171, Hunt et al.109, and Lam and Hunt113, to identify and apply constraints in conjunction with in silico experimentation (see Box 1) to systematically shrink the space of acceptable Monte Carlo variations. Having multiple, equally satisfactory analogs of the same referent is an acknowledgment and representation of uncertainties that can shrink but not vanish.
Avoiding Information Loss, ‘Warts and All’, is Essential
A product of modeling efforts cited in the reviews, irrespective of model type, is derived measures that are recorded for use by others. Examples include mean predicted ED50 (±SD), clearance, half-life, volume of distribution, etc. Examples described in Supporting Information-Use Cases73–102 illustrate that derived measures acquired during an early R&D stage can influence the perspective taken or focal aspect of interest at a later stage. However, derived measures are lossy (information is lost). Features present in the raw experimental observations are lost. Contextual information, including experiment method details, assay information, etc., may also be lost. Information loss is a concern: later, it may prove critical. Similar to issues discussed above, it becomes increasingly infeasible to recover lost information. Today, preventing such information loss can be challenging: proper measures are required to preserve, manage, and transfer information. During a project's lifetime (and thereafter), it requires attention to those preservation details across different functional groups through each development stage, from preclinical to postmarketing. The risk of critical information loss increases for conceptual model descriptions enriched with quantitative, mechanistic, pharmacological, physiological, and systems biology details that do not readily reduce to simple, mathematical or statistical descriptions.
Despite the above concerns, we stress that derived measures along with highly abstracted models are essential: researchers need them. They provide a much needed bridge from particular and specific concrete modeling to the more powerful generalized models from which researchers will develop the theory for therapy controlled normal-to-morbid or diseased-to-normal transitions. For this reason, the framework must facilitate access to, and the analog based knowledge repository must house, curate, and facilitate access to analogs, metadata, and much of the experimental observations from which the metadata were derived.
Consider measures derived from multiple different models separated in time, combined and extended beyond their original model use cases, and how they may influence downstream conceptual models used for go/no-go decision-making. Conceptual models can be expected to push the go/no-go decision in one direction. A quite different direction may result if all the original models, including those that at an earlier time were judged ‘failures’, could be re-run together to directly inform decision makers when that decision is required. An analog based knowledge repository fulfilling the requirements presented below will guard against information loss and knowledge distortion. Having intuitive, easily understood analogs will reduce dependency on, and usefulness of, lossy, derived measures to inform domain experts' conceptual models.
EFFECTIVE DECISION-MAKING SUPPORT REQUIRES A UNIFIED FRAMEWORK
Today, domain experts rely primarily on their own conceptual models—informed, of course, by computational models—to make go/no-go decisions. Improved productivity can be achieved by changing that relationship: domain experts make go/no-go decisions after using simulations to explore many scenarios within a computational framework built and equipped specifically for scenario exploration in addition to enabling the multiple M&S methods needed to satisfy the requirements below. It will be the ‘framework’ within which all model use cases occur. We envision the framework becoming an analog based knowledge repository: an up-to-date instantiation of all accumulated, new, and proprietary mechanistic knowledge in an accessible, easily understood, observable, and interactive form. As a consequence, R&D project team members will have reduced dependency on the difficult to challenge conceptual mechanistic models of current, past, and no longer available domain experts.
Current experiment records and/or protocols capture and preserve some current, underlying, mechanistic conceptualizations, however, that contextual information is typically decoupled from data and can become unavailable in subsequent phases of development. A good practice will be that information is documented and provided as part of annotation within the framework. It will not be ancillary, as in OpenABM.115 In order to effectively incorporate data, especially wet-lab data across discovery and development stages for future use, automated capabilities will be needed that enable and facilitate metadata annotations, which may include biological, anatomical, and physiological details across biological scales. Of course, the framework will also provide methods for storage, curation, composition, and execution of analogs representing what is known about domain-specific referent systems. Its core constituents (specific items) will be data, semantic relationships, workflow actions, and computational components. There will also be a variety of derived constituents including data sets, semantic networks (e.g., XML ontologies), workflows, plus analogs and their components. Under Resources, we list currently available tools that can be used to organize and complete all of the preceding tasks.
As an example of a knowledge repository decision support use case, consider a task to estimate the likelihood of success for a clinical trial given a compound and cohort sample specifications. The following may seem futuristic, yet tools for enabling all capabilities (listed under Web Resources) are in use today within different domains. The project's inter-disciplinary team, including regulatory scientists, statisticians, programmers, regulatory staff, etc., populate the knowledge repository with experimental protocols (Workflows under Resources), experimental data sets, analog mechanisms (Executable models under Resources) representing the cohort and compounds, and maps between the terms (objects, methods, variables, parameters, graphs, etc.) used in all these elements. Software agents and actors within the repository simplify the process. A user specifies a set of objectives or criteria for a successful (or unacceptable) clinical trial outcome and assembles at least one analog that might plausibly generate data satisfying the objectives. Repository agents facilitate the composition by periodically checking the consistency and completeness of the evolving analog. The user then executes the analog according to study design workflow(s). Execution results will exhibit the systemic causation and variability generated by fine-grain, networked events embedded in analog components. Repository agents will compare and contrast the results to the objectives and present the user with similarity scores validating or falsifying analogs. Repository agents may also present a suite of alternatively composed analogs consistent with the data, ontologies, and workflows previously installed. Similarity scores provide a rudimentary estimate of the likelihood of success for the clinical trial. Further execution and similarity scores of alternative analogs provide refinement of and confidence (or the lack thereof) in those estimates. Note that because we are left of center in Figure 1, the approach avoids assuming the existence of a perfectly accurate analog. Any analog that achieves all face validation targets and satisfies the various similarity measures will be considered valid (until falsified). We envision all of the preceding being completed within hours.
As a technical note, an analog may contain components based on different formalisms. Some may be graph theoretic. Some may use ODEs. Others will be (or will use) agents. The preceding discussions illustrate that referring to the modeling activities as being AB is misleading. Agent-directed or agent-oriented are more accurate descriptions.114
FROM PARTICULAR TO PARAMOUNT M&S USE CASES
We worked through each of the cited reviews and commentaries73–102 and identified more than ninety general and specific M&S use cases. Although each article presented its assessment from a particular perspective, there was, as might be expected, considerable overlap in specific use cases. That overlap guided and facilitated clustering them into seven categories that span all pharmaceutical R&D activities. Given use cases, we sought general requirement statements that would subsume two or more uses within each category. They are listed in the subsequent section.
Paramount Use Cases
The following are called paramount M&S use cases because they subsume the particular use cases in Supporting Information-Use Cases clustered under the following seven categories.
Knowledge Integration
Given multiple organizational perspectives (disease state, prognostic factors, drug characteristics, cohort variability, disease progression, drug effects, timelines, budgets, etc.) and multiple data sets (qualitative and quantitative; some possibly incommensurate) from various experiments and experimental models, solve for a collection of alternative, composable, explanatory mechanisms parameterized by, and validated against, available data. Here, ‘solve’ is defined as applying constraints within an iterative protocol (see e.g., Tang and Hunt116, Park et al.117, Engelberg et al.118) to shrink a set of possible mechanisms into a much smaller set of plausible (supported early by qualitative validations) and incrementally more likely mechanisms (supported by quantitative validation).
Use collected mechanisms to assemble concrete, consistent, comprehensive, clinically relevant ‘stories’ about the co-evolution, during and following treatment, of subject (multiscale), the condition being treated, and treatment as well as (absorption, distribution, metabolism, and elimination) ADME plus response monitoring in the case of a drug. In this context, a story is the narrative created during simulation, and a simulation results from executing an in silico experiment. There are several reasons why a good simulation story is important.119,120 Use assembled stories to discover and make explicit potential conflicts, voids, and ambiguities within an analog based knowledge repository, thereby providing immediate predictive and analytical use, while bringing into focus paths for repository improvement.
Management of Uncertainty
Present for in silico experimentation, plausible mechanisms exhibiting variability, both composite and singular, that are qualitatively and quantitatively similar to that seen (or expected) in laboratory experiments or clinical trials, including matching changes in variability across different cohorts and study designs. Use simulations to estimate probability of achieving clinical target efficacy outcomes, and when feasible, estimate probability of specified, undesired effects.
Decision Support
Present contextualized assemblies of plausible mechanisms parameterized to provide an evidence-based justification for componentized estimates of likelihood of success at critical stages within the development process. Use the assemblies and their stories to estimate efficacy and safety windows, including confidence intervals, and to demonstrate sample sizes within appropriate population cohorts needed for constraining study outcomes to within those windows.
Preclinical and Clinical Development
Preclinical and clinical development along with postmarketing uses are covered by requirements listed above and below.
Drug-disease Modeling
Solve for plausible mechanism composites satisfying multiple long-term aspects of diseases, drug effects and fates within cohorts, placebo effect, and disease modifying interventions. Use those mechanisms to predict compound behavior (PK) within contexts of interest, including hypothetical mechanisms for lack of adherence, dropout, and multiple drug interaction.
Drug–drug Interaction, Special Populations
Solve for plausible mechanism composites of compound interactions of interest, including time-dependent inhibition, induction, and competition between a parent compound and its metabolites. Solve for plausible mechanism composites for various cohorts (animals, children, and adults) based on classifications of compound behavior in each cohort. Hypothesize and build mechanism translation maps between analog cohorts (e.g., between adults and children). Use plausible translation maps to design clinical trials for human cohorts.
Prediction
Solve for plausible mechanism composites by multi-objective search within constraints defined by fine-grained classifications of cohorts (particular attributes at multiple levels and scales) and compounds (e.g., particular physicochemical properties), including their formulation, across model types (in vitro to in vivo, animal to human). Use the plausible solutions to predict the outcomes of precise and particular regimens on individually characterized cohorts, both across (translation) and within model types.
REQUIREMENTS
To enable all paramount use cases, the framework must enable, and analog systems must meet, these five requirements.
-
An analog's components and spaces will be concrete (enabling knowledge embodiment), wherein its details will be directly defined by its use cases. Analog components will be somewhat modular, in schedule as well as state. So doing helps accomplish the following framework activities.
It enables defining and annotating component- and module-to-biological counterpart mappings, making them explicit, intuitive, and easily understood. It enables experimentation on concrete analogs. It is essential for wet-lab R&D scientists and decision makers to easily follow, interpret, and comment, unassisted, on simulation details. For that, the embodied knowledge needs to be easily accessible, which requires transparency in representation and execution—form and function.
It enables making modules quasi-autonomous and thus more biomimetic.65,104 Analog components composing virtual cells, organs, animal models, and ultimately individuals, must exhibit some level of autonomous behavior in order to improve similarity with biological counterpart phenotypes, while increasing their explicative and predictive utility.
Components can be adapted easily to represent different past and future experiment designs and protocols.
It is straightforward to change mechanistic detail (granularity, resolution) to simulate additional attributes or experiments. It facilitates scaling (translation, morphing) among in silico experimental systems to represent transitions from in vitro to animal models, from animal model to human cohorts, and from normal health to morbid.
It enables reusing analogs and components along with their embedded knowledge, for study of new chemical entities (or biologics) and new intervention scenarios under similar or different morbidity constraints or epigenetic influences.
It facilitates verification through unit testing, where each component can be tested in isolation as well as in the composed analog context.
It facilitates versioning, where each component can evolve independent of other components.
It enables building trust in surviving analogs by accumulating direct in silico-to-wet-lab validation evidence, where measures taken during in silico experiments are mapped quantitatively to counterpart measures taken during wet-lab experiments. So doing reduces reliance on derived measures, which can mask information loss, assumptions, and uncertainty removal. Many of those validation exercises will rely extensively on pharmacometrics and conventional modeling methods.
It facilitates archiving analog and mechanism evolution along with in silico experiment successes and failures within the framework. The latter is particularly important. When an analog or in silico experiment fails in some way, we acquire new knowledge, e.g., a feature of an analog mechanism thought to have a particular in vitro biological counterpart, does not. However, in a different context, that mechanism or some variant may prove useful.
-
Components and spaces can be assembled easily to simulate current, past, and future laboratory or clinical experiments. Generating many alternative, plausible, testable (through in silico experimentation) components for each function/structure and then selecting against those that fail, is needed to insure generation of new knowledge. So doing helps accomplish two framework activities.
It becomes increasingly easy to construct (plug together) and explore alternative mechanistic hypotheses and intervention scenarios. It facilitates contrasting their predictions during simulation. So doing can help avoid dictating ‘best methods’ prematurely. The latter keeps the door open for yet-to-be-stated M&S use cases while increasing opportunities for serendipitous insight and/or discovery.
It becomes increasingly easy to construct multiscale, multiresolution, multi-attribute analogs (eventually individualized virtual patients) composed of heterogeneous (form, function, methods, formalisms, etc.) components.
Simulation experiments are feasible in the presence and absence of chemical entity objects (hereafter, CE-objects). They are also feasible in the presence of multiple CE-objects. Components within analogs can recognize different CE-objects and adjust their response accordingly.
Coarse grain (from the perspective of biological organization) phenomena will derive mostly from local component interactions at a finer grain (local includes a living entity's immediate environment). When required, finer grain mechanisms can respond to coarser grain phenomena.
-
Semi-automated modeling methods are needed to more rapidly complete three critical activities:
Conduct in silico experimentation to explore and shrink spaces of competing mechanistic hypotheses plus alternative mechanism instantiations and parameterizations.
Use cross-model validation methods (based on quantitative similarity measures) to discover parsimonious options to increase and decrease component and analog granularity when milestones change and when new questions require new use cases, which necessitates changing targeted attributes and/or shifting attention to new aspects and phenomena.
Discover testable hypotheses about how changes in clinical, field, or laboratory measures may be linked mechanistically to observed or observable changes in particular biological level phenomena, especially molecular-level phenomena.
MECHANISM-BASED APPROACH
New projects are typically initiated based on evidence (even if limited) supporting conceptual mechanistic models (morbidity or disease progression, cause-effect, pharmacology, clinical outcomes, etc.) that often include hypothesized molecular targets. There are large gaps in the mechanistic knowledge landscape that must be filled strategically to facilitate making ‘correct’ go/no-go decisions. The mechanistic landscape spans cell cultures, model organisms, and humans. It also spans pharmacology, toxicology, disposition, metabolism, and more. Today, no one member of the R&D enterprise has a comprehensive ‘view’ of that landscape, yet it is clear from the cited experts that full knowledge of the current state of that landscape and what can be predicted from it is needed at each go/no-go decision juncture. Knowledge of—or use of one or more features on—that mechanism landscape is common to all identified model use cases.
The ability to navigate, use, and leverage current mechanistic insight is a common feature of all paramount use cases. Enabling those use cases requires transitioning from models that are separate and distinct computational (or diagrammatic) descriptions of conceptual representations (e.g., the systems biology, PBPK, and pharmacodynamic models discussed in the cited reviews) to concrete instances of that knowledge. Doing so is feasible using synthetic analogs of mechanisms.65 We create analogs by combining (plugging together) specific elements, often varied and diverse, so as to form a coherent biomimetic whole. The analog is synthetic because it is a software mechanism constructed from extant, autonomous components (in this case, executable software components) whose existence and purpose are independent of the model or mechanistic landscape that they comprise. The expectation is that, upon execution, the interacting elements and components—the mechanism undergoing simulation—will exhibit event sequences and outcomes that are measurably similar to counterpart mechanisms within the corresponding wet-lab experiments that will be used to validate the analog. See Yan et al.119, Lam and Hunt113, Tang and Hunt116, Engelberg et al.118, and Park et al.118, for validated examples of synthetic analogs and their mechanisms, all designed for use center-left on the three lower Figure 1 spectra.
Exploratory Iteration
One final, critical capability is needed in order to enable the remaining paramount use cases. It must be straightforward to repeat any simulation experiment using a different compound, set of compounds, or no compound. So doing can be accomplished most easily by building analogs using object-oriented software methods and using different mobile objects to map to different compounds of interest. We require that objects representing drugs can be added (or not) and, when ‘inactive’, their presence will not interfere with any already validated mechanism. Examples of that approach are provided in Sheikh-Bahaei and Hunt71, Lam and Hunt113, Park et al.118, Yan et al.121, and Sheikh-Bahaei et al.122 In those reports, one CE-object maps to a very small amount of referent compound in a small aliquot of measurable material taken from a referent wet-lab experiment, such as culture media, blood, tissue, etc. In time, these CE-objects will carry information that distinguishes one from another, such as structure specifications, particular physicochemical properties, affinities for biological substrates, etc. It follows that any analog component that might feasibly interact with a CE-object must be able to ‘read’ structure specifications and particular physicochemical properties, and use that information to adjust (or not) its interactions with that object.
To enable the latter, analog components will need to be ‘intelligent’ (able to use artificial intelligence methods): they will be programmed with the results of many earlier validation and falsification experiments and can—absent user intervention—arrive at a customized parameterization that determines how they will interact with a new CE-object. To illustrate, imagine that analogs of each of a battery of in vitro model systems have achieved degrees of validation for ten compounds. One analog's referent is an in vitro system used to characterize metabolic profiles and predict human metabolic clearance. Focus on that analog: upon validation, for each mechanistic event, it retains its parameterizations for all ten CE-objects. The project team needs an answer to this question. Given three, competing, new chemical entities, are their expected metabolic clearance values within a target range? Each analog component, for each unique mechanistic event, can use available framework tools, follow a provided protocol, and construct a predictive map from the space of selected structure specifications and particular physicochemical properties to the space of validated parameterizations. We extend the mapping to the three new compounds and arrive at unique analog parameterizations for each CE-object counterpart. We then conduct analog clearance experiments using each of the three CE-objects. We use the results to predict wet-lab clearance measures. Sheikh-Bahaei and Hunt71 described a prototype example of such a protocol. With a knowledge base of only ten compounds, we would have only limited trust in those predictions. However, we would expect to have more trust after doing the same using a knowledge base of fifty compounds. At that stage we could begin characterizing families of compounds as well as particular molecules. As more information is embedded, the knowledge repository increasingly facilitates the work of the domain expert.
The same basic approach can be used to explore expected outcomes of in silico toxicology experiments, experiments in analogs of animal disease models, and even analogs of normal and special human cohorts. There would be no technical barriers to simulation experiments that explore possible drug–drug interactions.
Beyond Paramount Use Cases
Figure 1 helps to bring into focus important challenges faced by any new, high risk, high gain pharmaceutical R&D project. It can be characterized as being left of center on the bottom three spectra. A task is to acquire just enough new information and knowledge (move right in Figure 1) to make better-informed, ‘good’ go/no-go decisions sooner within budget constraints and in the face of considerable uncertainty. The expectation is that simulations will use available knowledge to provide essential information and/or guidance moving forward. To do so, M&S methods must be engaged at the start of the project. Otherwise there will be considerable risk that M&S efforts will lag behind wet-lab efforts. The logical beginning scenario would be to pull together components from models successfully supporting more mature projects, modify them as appropriate, and assemble them to begin being synergistic with wet-lab experiments. Even when the new effort is far left in Figure 1 spectra, speculative and qualitative analogs can be constructed to help researchers think clearly, explicitly, and concretely about the referent and design specific, well-focused wet-lab experiments. That scenario is an example of model use cases beyond those that directly support product development and approval. It also speaks to requirements, covered by those above, which have apparently not yet been considered.
A related, also essential use case will occur once a new product has been approved: revisit the process in silico (from start to finish) to explore alternative R&D, knowledge acquisition paths that would have been more time, cost, and/or knowledge effective. To do so will require simulating phases of the R&D process, including various wet-lab experiments performed or not. Such exploratory simulation may seem futuristic, but it is easy to believe that, with such capabilities, the field will have achieved dramatic productivity gains and that is the objective. These additional M&S use cases illustrate that future development of simulation models must take place within a common framework, if the simulations are expected to make a lasting contribution to the knowledge base. Simulated R&D activities will be analogs of past or future, real or considered R&D activities. None of the analogs used, however, will be fully detailed or fully validated. Because we will be center-left on the bottom three Figure 1 spectra, we know that they will (always) be flawed in ways yet to be determined. Those flaws will be hidden by built-in uncertainties.
Simulations that support and add plausible detail to early conceptual models, such as hybrids of systems biology, PBPK, and pharmacodynamic models, will be useful. Simulation models that can also falsify aspects of those conceptual models, earlier rather than later, will be especially valuable because hypothesis falsification, not validation, generates the new knowledge that will be needed for making go/no-go decisions.123,124 Enabling mechanism falsification in addition to mechanism validation is another, important use case not specifically identified in the cited reviews. Falsification of an analog requires that its mechanism be concrete and particular.65 Plausible conceptual mechanisms, especially those that are more complicated, can in theory be falsified using wet-lab experimentation, but doing so can be challenging and time- and resource-intensive.
Analog Based Knowledge Repository and AB Models
Representing within a knowledge repository the variety of data structures, experimental observations, and documented biological phenomena that may influence go/no-go decisions during the R&D projects is beyond the scope of any one model of computation (MoC). A successful analog based knowledge repository will require the co-existence of multiple, occasionally inconsistent, yet equally valid, models of the same referent(s), as has been done within other domains.125 Inconsistency robustness126 is important to the aspect-oriented nature of scientific M&S.65,104,127 The management of inconsistent models is part of the strategy for integrating MoCs into a coherent knowledge repository. The choice of underlying MoC for any particular activity is driven by the focal aspects or model (analog or component) use cases. Variety in aspects is better realized by variety in MoCs. For example, a dataflow MoC can co-exist with a discrete event analog of a mouse cancer model, the former representing a functional relationship between high volume population-oriented variables (like concentration in a PBPK model) and the latter representing mechanistic relationships between unique, biomimetic components. For the foreseeable future, such a repository will not, itself, be AB, but it will be partially agent-directed.115 Agents such as Experiment Agents used in our work118,128–130 follow protocols and perform tasks exactly analogous to those human modelers perform: set up and execute various models (analogs, modules, etc.) and use pharmacometric methods. Drawing on work from Lee and colleagues125,131,132, we anticipate no restrictions being placed on MoCs, except where required for managed composition, execution, and analysis as has been done with Ptolemy II125 for M&S distributed computing scenarios. Depending on use case requirements, analogs may be realized by any given MoC, including continuous-time systems, ABMs, ODEs, systems and process networks, Stream X-Machines, etc.
Conclusion
An eruption of recent articles, including those cited,73–102 is focusing attention on the declining productivity crisis stressing pharmaceutical R&D. The hundred plus authors of the more than thirty cited reviews and commentaries argue from different perspectives (pharmacometrics, quantitative and systems pharmacology, model-based drug development, etc.) for expanded use of M&S methods as the primary means to reverse the downward productivity trend. They describe more than ninety particular M&S use cases. The Box 3 includes particular examples. They were merged into paramount use cases listed above. We made the case that facile realization of those use cases will require use of an analog based knowledge repository that fulfills five requirements. AB M&S methods will play important roles along with those methods already in use, but additional capabilities and methods are required. Importantly, there are no technology gaps. The five requirements can be satisfied by the tools listed under Resources, which are in use within other domains.
Reversing the productivity decline requires rethinking how M&S methods can and should be used within the larger R&D process. It requires shifting focus from analysis of data to discovery of explanatory mechanisms. The latter, conditioned paramount use cases, requires implementing the requirements listed above. As illustrated in Figure 1, and as explained in Hunt et al.104 and Supporting Information, so doing requires increased reliance on relational grounding and diminished reliance on absolute grounding. It also requires being clear about how computational models are being used and continuously exploring how barriers to participation in M&S activities can be lowered.
Acknowledgments
This work was supported in part by the CDH Research Foundation, the Alternatives Research and Development Foundation, and the EPA (G10C20235). The funding bodies had no role in study design; in the collection, analysis, and interpretation of data; in the writing of the manuscript; or in the decision to submit the manuscript for publication. We gratefully acknowledge commentary and suggestions provided by Brenden Peterson, Rada Savic, and Andre J. Jackson.
WEB RESOURCES
Listed below are available, open-source tools of the type that can be used now to begin satisfying the listed interactive Knowledge Repository requirements
Databases
PostgreSQL (relational): http://www.postgresql.org/
ZOBD: http://zodb.org/
NoSQL: http://nosql-database.org/
RDF/SPARQL: http://www.w3.org/TR/rdf-sparql-query/
Data [de]composition
HDF: http://www.hdfgroup.org/HDF5/
Semantics
Protoge: http://protege.stanford.edu/
Ontopia: http://www.ontopia.net/
Jena: https://jena.apache.org/
Pellet: http://clarkparsia.com/pellet/ (complete)
Racer: http://www.sts.tu-harburg.de/∼r.f.moeller/racer/ (complete)
Hermit: http://hermit-reasoner.com/
Executable models
Code
Eclipse: http://eclipse.org/
Netbeans: http://netbeans.org/
Graphical programming
Ptolemy II: http://ptolemy.eecs.berkeley.edu/ptolemyII/index.htm
NetLogo: http://ccl.northwestern.edu/netlogo/
ArgoUML: http://argouml.tigris.org/
Modeling infrastructure
JAMES II: http://wwwmosi.informatik.uni-rostock.de/mosi/projects/cosa/james-ii
CC3D: http://www.compucell3d.org/
MASON: http://cs.gmu.edu/∼eclab/projects/mason/
Actor model133
Erlang: http://www.erlang.org/
Scala: http://www.scala-lang.org/node/242
Workflow
Kepler: http://kepler-project.org/
Taverna: http://www.taverna.org.uk/
Galaxy: http://galaxyproject.org/
Triana: http://www.trianacode.org/
Graphs
NetworkX: http://networkx.lanl.gov/
Supplementary material
REFERENCES
- 1.Arthur WB, Durlauf SN, Lane DA. The Economy As An EvolvingComplex System II, Santa Fe Institute Series. Santa Fe, NM: Westview Press; 1997. [Google Scholar]
- 2.Axelrod R. The Complexity of Cooperation: Agent-Based Models ofCompetition and Collaboration. Priceton, NJ: Princeton University Press; 1997. [Google Scholar]
- 3.Bankes SC. Agent-based modeling: a revolution? Proc Natl Acad Sci USA. 2002;99:7199–7200. doi: 10.1073/pnas.072081299. doi: 10.1073/pnas.072081299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonabeau E. Agent-based modeling: methods and techniques for simulating human systems. Proc Natl Acad Sci USA. 2002;99(suppl 3):7280–7287. doi: 10.1073/pnas.082080899. doi: 10.1073/pnas.082080899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davidsson P. Multi agent based simulation: beyond social simulation. In: Moss S, Davidsson P, editors. Multi Agent Based Simulation. LectureNotes in Computer Science. Vol. 1979. Berlin: Springer; 2000. pp. 97–107. doi: 10.1007/3-540-44561-7_7. [Google Scholar]
- 6.DeAngelis DL, Cox DK, Coutant CC. Cannibalism and size dispersal in young-of-the-year largemouth bass: experiment and model. Ecol Model. 1979;8:133–148. [Google Scholar]
- 7.DeAngelis DL, Mooij WM. Individual-based modeling of ecological and evolutionary processes. Annu Rev Ecol Evol Syst. 2005;36:147–168. doi: 10.1146/annurev.ecolsys.36.102003.152644. [Google Scholar]
- 8.Epstein JM, Axtell R. Growing Artificial Societies: Social Science from the Bottom Up. Washington, DC: Brookings Institution Press and The MIT Press; 1996. [Google Scholar]
- 9.Grimm V. Ten years of individual-based modelling in ecology: what have we learned and what could we learn in the future? Ecol Model. 1999;115:129–148. doi: 10.1016/S0304-3800(98)00188-4. [Google Scholar]
- 10.Judson OP. The rise of the individual-based model in ecology. Trends Ecol Evol. 1994;9:9–14. doi: 10.1016/0169-5347(94)90225-9. doi: 10.1016/ 0169-5347(94)90225-9. [DOI] [PubMed] [Google Scholar]
- 11.Kephart JO. Software agents and the route to the information economy. Proc Natl Acad Sci USA. 2002;99(suppl 3):7207–7213. doi: 10.1073/pnas.082080499. doi: 10.1073/pnas.082080499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.LeBaron B. Short-memory traders and their impact on group learning in financial markets. Proc Natl Acad Sci USA. 2002;99(suppl 3):7201–7206. doi: 10.1073/pnas.072079699. doi: 10.1073/pnas.072079699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parunak HVD, Savitt R, Riolo RL. Agent-based modeling vs. equation-based modeling: a case study and users' guide. In: Sichman JS,, Conte R, Gilbert N, editors. Multi-Agent Systems and Agent BasedSimulation Lecture Notes in Computer Science. Vol. 1534. Berlin: Springer; 1998. pp. 10–25. doi: 10.1007/10692956_2. [Google Scholar]
- 14.Rose KA, Cowan JH. Predicting fish population dynamics: compensation and the importance of site-specific considerations. Environ Sci Policy. 2000;3(suppl 1):433–443. doi: 10.1016/S1462-9011(00)00054-X. [Google Scholar]
- 15.Schelling TC. Micromotives and Macrobehavior. New York, NY: W.W. Norton & Company, Inc; 1978. [Google Scholar]
- 16.Auchincloss AH, Riolo RL, Brown DG, Cook J, Diez Roux AV. An agent-based model of income inequalities in diet in the context of residential segregation. Am J Prev Med. 2011;40:303–311. doi: 10.1016/j.amepre.2010.10.033. doi: 10.1016/j.amepre.2010.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bobashev GV, Goedecke DM, Yu F, Epstein JM. A hybrid epidemic model: combining the advantages of agent-based and equation-based approaches. In: Henderson SG, Biller B, Hsieh MH, Shortle J, Tew JD, Barton RR, editors. Proceedings of the 2007 Winter Simulation Conference IEEE, New York. 2007. pp. 1511–1516. doi: 10.1109/WSC.2007.4419767. [Google Scholar]
- 18.Bousquet F, Le Page C. Multi-agent simulations and ecosystem management: a review. Ecol Model. 2004;176:313–332. doi: 10.1016/j.ecolmodel.2004.01.011. [Google Scholar]
- 19.Caplat P, Anand M, Bauch C. Symmetric competition causes population oscillations in an individual-based model of forest dynamics. Ecol Model. 2008;211:491–500. doi: 10.1016/j.ecolmodel.2007.10.002. [Google Scholar]
- 20.Castella J-C, Verburg PH. Combination of process-oriented and pattern-oriented models of land-use change in a mountain area of Vietnam. Ecol Model. 2007;202:410–420. doi: 10.1016/j.ecolmodel.2006.11.011. [Google Scholar]
- 21.Charles S, Subtil F, Kielbassa J, Pont D. An individual-based model to describe a bullhead population dynamics including temperature variations. Ecol Model. 2008;215:377–392. doi: 10.1016/j.ecolmodel.2008.04.005. [Google Scholar]
- 22.Faugeras B, Maury O. Modeling fish population movements: from an individual-based representation to an advection–diffusion equation. J Theor Biol. 2007;247:837–848. doi: 10.1016/j.jtbi.2007.04.012. doi: 10.1016/j.jtbi.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 23.Garcia R. Uses of agent-based modeling in innovation/new product development research. J Prod Innovat Manage. 2005;22:380–398. doi: 10.1111/j.1540-5885.2005.00136.x. [Google Scholar]
- 24.Garcia R, Jager W. From the special issue editors: agent-based modeling of innovation diffusion. J Prod Innovat Manage. 2011;28:148–151. doi: 10.1111/j.1540-5885.2011.00788.x. [Google Scholar]
- 25.Grimm V, Railsback SF. Individual-based Modeling and Ecology. Princeton, NJ: Princeton University Press; 2005. [Google Scholar]
- 26.Grosman PD, Jaeger JAG, Biron PM, Dussault C, Oullet J-P. Trade-off between road avoidance and attraction by roadside salt pools in moose: An agent-based model to assess measures for reducing moose-vehicle collisions. Ecol Model. 2011;222:1423–1435. doi: 10.1016/j.ecolmodel.2011.01.022. [Google Scholar]
- 27.López-Sánchez M, Noria X, Rodríguez JA, Gilbert N. Multi-agent based simulation of news digital markets. Int Jf Comput Sci Appl. 2005;2:7–14. [Google Scholar]
- 28.Kohler TA, Gumerman GJ, Reynolds RG. Simulating ancient societies. Sci Am. 2005;293:76–82. doi: 10.1038/scientificamerican0705-76. 84. doi: 10.1038/scientificamerican0705-76. [DOI] [PubMed] [Google Scholar]
- 29.Pan X, Han CS, Dauber K, Law KH. A multi-agent based framework for the simulation of human and social behaviors during emergency evacuations. AI Soc. 2007;22:113–132. doi: 10.1186/1471-2105-6-228. [Google Scholar]
- 30.Sato H. Simulation of the vegetation structure and function in a Malaysian tropical rain forest using the individual-based dynamic vegetation model SEIB-DGVM. Forest Ecol Manage. 2009;257:2277–2286. doi: 10.1016/j.foreco.2009.03.002. [Google Scholar]
- 31.Tang W, Bennett DA. Agent-based modeling of animal movement: a review. Geography Compass. 2010;4:682–700. doi: 10.1111/j.1749-8198.2010.00337.x. [Google Scholar]
- 32.Rand R, Rust RT. Agent-based modeling in marketing: Guidelines for rigor. Int J Res Market. 2011;28:181–193. doi: 10.1016/j.ijresmar.2011.04.002. [Google Scholar]
- 33.Macal CM, North MJ. Tutorial on agent-based modelling and simulation. J Simul. 2010;4:151–162. doi: 10.1057/jos.2010.3. [Google Scholar]
- 34.An G. Agent-based computer simulation and sirs: building a bridge between basic science and clinical trials. Shock. 2001;16:266–273. doi: 10.1097/00024382-200116040-00006. [DOI] [PubMed] [Google Scholar]
- 35.Casal A, Sumen C, Reddy TE, Alber MS, Lee PP. Agent-based modeling of the context dependency in T cell recognition. J Theor Biol. 2005;236:376–391. doi: 10.1016/j.jtbi.2005.03.019. doi: 10.1016/j.jtbi.2005.03.019. [DOI] [PubMed] [Google Scholar]
- 36.Kreft JU, Booth G, Wimpenny JWT. Bacsim, a simulator for individual-based modelling of bacterial colony growth. Microbiology. 1998;144:3275–3287. doi: 10.1099/00221287-144-12-3275. doi: 10.1099/00221287-144-12-3275. [DOI] [PubMed] [Google Scholar]
- 37.Mansury Y, Kimura M, Lobo J, Deisboeck TS. Emerging patterns in tumor systems: simulating the dynamics of multicellular clusters with an agent-based spatial agglomeration model. J Theor Biol. 2002;219:343–370. doi: 10.1006/jtbi.2002.3131. doi: 10.1006/jtbi.2002.3131. [DOI] [PubMed] [Google Scholar]
- 38.Segovia-Juarez JL, Ganguli S, Kirschner D. Identifying control mechanisms of granuloma formation during M. tuberculosis infection using an agent-based model. J Theor Biol. 2004;231:357–376. doi: 10.1016/j.jtbi.2004.06.031. doi: 10.1016/j.jtbi.2004.06.031. [DOI] [PubMed] [Google Scholar]
- 39.Thorne BC, Bailey AM, Peirce SM. Combining experiments with multi-cell agent-based modeling to study biological tissue patterning. Brief Bioinform. 2007;8:245–257. doi: 10.1093/bib/bbm024. doi: 10.1093/bib/bbm024. [DOI] [PubMed] [Google Scholar]
- 40.Walker DC, Southgate J, Hill G, Holcombe M, Hose DR, Wood SM, Mac Neil S, Smallwood RH. The epitheliome: agent-based modelling of the social behaviour of cells. Biosystems. 2004;76:89–100. doi: 10.1016/j.biosystems.2004.05.025. doi: 10.1016/j.biosystems.2004.05.025. [DOI] [PubMed] [Google Scholar]
- 41.Artzy-Randrup Y, Rorick MM, Day K, Chen D, Dobson AP, Pascual M. Population structuring of multi-copy, antigen-encoding genes in Plasmodium falciparum. eLife. 2012;1:e00093. doi: 10.7554/eLife.00093. doi: 10.7554/ eLife.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bailey AM, Lawrence MB, Shang H, Katz AJ, Peirce SM. Agent-based model of therapeutic adipose-derived stromal cell trafficking during ischemia predicts ability to roll on P-selectin. PLoS Comput Biol. 2009;5:e1000294. doi: 10.1371/journal.pcbi.1000294. doi: 10.1371/journal.pcbi.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bogle G, Dunbar PR. Agent-based simulation of T-cell activation and proliferation within a lymph node. Immunol Cell Biol. 2010;88:172–179. doi: 10.1038/icb.2009.78. doi: 10.1038/icb.2009.78. [DOI] [PubMed] [Google Scholar]
- 44.Fallahi-Sichani M, El-Kebir M, Marino S, Kirschner DE, Linderman JJ. Multiscale computational modeling reveals a critical role for TNF-α receptor 1 dynamics in tuberculosis granuloma formation. J Immunol. 2011;186:3472–3483. doi: 10.4049/jimmunol.1003299. doi: 10.4049/jimmunol.1003299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Joyce KE, Laurienti PJ, Hayasaka S. Complexity in a brain-inspired agent-based model. Neural Networks. 2012;33:275–290. doi: 10.1016/j.neunet.2012.05.012. doi: 10.1016/j.neunet.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim PS, Lee PP. Modeling protective anti-tumor immunity via preventative cancer vaccines using a hybrid agent-based and delay differential equation approach. PLoS Comput Biol. 2012;8:e1002742. doi: 10.1371/journal.pcbi.1002742. doi: 10.1371/journal.pcbi.1002742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Klann MT, Lapin A, Reuss M. Agent-based simulation of reactions in the crowded and structured intracellular environment: Influence of mobility and location of the reactants. BMC Syst Biol. 2011;5:71. doi: 10.1186/1752-0509-5-71. doi: 10.1186/1752-0509-5-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li NY, Verdolini K, Clermont G, Mi Q, Rubinstein EN, Hebda PA, Vodovotz Y. A patient-specific in silico model of inflammation and healing tested in acute vocal fold injury. PLoS ONE. 2008;3:e2789. doi: 10.1371/journal.pone.0002789. doi: 10.1371/journal.pone.0002789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marino S, El-Kebir M, Kirschner D. A hybrid multi-compartment model of granuloma formation and T cell priming in Tuberculosis. J Theor Biol. 2011;280:50–62. doi: 10.1016/j.jtbi.2011.03.022. doi: 10.1016/j.jtbi.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martínez IV, Gómez EJ, Hernando ME, Villares R, Mellado M. Agent-based model of macrophage action on endocrine pancreas. Int J Data Mining Bioinform. 2012;6:355–368. doi: 10.1504/ijdmb.2012.049293. doi: 10.1504/IJDMB.2012.049293. [DOI] [PubMed] [Google Scholar]
- 51.Mina P, di Bernardo M, Savery NJ, Tsaneva-Atanasova K. Modelling emergence of oscillations in communicating bacteria: a structured approach from one to many cells. J R Soc Interface. 2013;10:20120612. doi: 10.1098/rsif.2012.0612. doi: 10.1098/rsif.2012.0612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pogson M, Holcombe M, Smallwood R, Qwarnstrom E. Introducing spatial information into predictive NF-kappaB modeling—an agent-based approach. PLoS ONE. 2008;3:e2367. doi: 10.1371/journal.pone.0002367. doi: 10.1371/journal.pone.0002367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smallwood R. Computational modeling of epithelial tissues. Wiley Interdiscip Rev Syst Biol Med. 2009;1:191–201. doi: 10.1002/wsbm.18. doi: 10.1002/wsbm.18. [DOI] [PubMed] [Google Scholar]
- 54.Stern JR, Christley S, Zaborina O, Alverdy JC, An G. Integration of TGF-β- and EGFR-based signaling pathways using an agent-based model of epithelial restitution. Wound Repair Regen. 2012;20:862–871. doi: 10.1111/j.1524-475X.2012.00852.x. doi: 10.1111/j.1524-475X.2012.00852.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tang J, Enderling H, Becker-Weimann S, Pham C, Polyzos A, Chen CY, Costes SV. Phenotypic transition maps of 3D breast acini obtained by imaging-guided agent-based modeling. Integr Biol. 2011;3:408–421. doi: 10.1039/c0ib00092b. doi: 10.1039/C0IB00092B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang H, Vaksman Z, Litwin DB, Shi P, Kaplan HB, Igoshin OA. The mechanistic basis of Myxococcus xanthus rippling behavior and its physiological role during predation. PLoS Comput Biol. 2012;8:e1002715. doi: 10.1371/journal.pcbi.1002715. doi: 10.1371/journal.pcbi.1002715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Amigoni F, Schiaffonati V. Multiagent-based simulation in biology—a critical analysis. Model-Based Reason Sci Technol Med Stud Comput Intell. 2007;64:179–191. [Google Scholar]
- 58.An G, Mi Q, Dutta-Moscato J, Vodovotz Y. Agent-based models in translational systems biology. Wiley Interdiscip Rev Syst Biol Med. 2009;1:159–171. doi: 10.1002/wsbm.45. doi: 10.1002/wsbm.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Edelman LB, Eddy JA, Price ND. In silico models of cancer. Wiley Interdiscip Rev Syst Biol Med. 2010;2:438–459. doi: 10.1002/wsbm.75. doi: 10.1002/wsbm.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.An G. Insilico experiments of existing and hypothetical cytokine-directed clinical trials using agent-based modeling. Crit Care Med. 2004;32:2050–2060. doi: 10.1097/01.ccm.0000139707.13729.7d. doi: 10.1097/01.CCM.0000139707.13729.7D. [DOI] [PubMed] [Google Scholar]
- 61.An G, Bartels J, Vodovotz Y. In silico augmentation of the drug development pipeline: examples from the study of acute inflammation. Drug Dev Res. 2011;72:187–200. doi: 10.1002/ddr.20415. doi: 10.1002/ddr.20415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Alberts S, Keenan MK, D'Souza RM, An G. Data-parallel techniques for simulating a mega-scale agent-based model of systemic inflammatory response syndrome on graphics processing units. Simulation. 2012;88:895–907. doi: 10.1177/0037549711425180. [Google Scholar]
- 63.Fallahi-Sichani M, Flynn JL, Linderman JJ, Kirschner DE. Differential risk of tuberculosis reactivation among anti-TNF therapies is due to drug binding kinetics and permeability. J Immunol. 2012;188:3169–3178. doi: 10.4049/jimmunol.1103298. doi: 10.4049/jimmunol.1103298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hunt CA, Ropella GEP. Moving beyond in silico tools to in silico science in support of drug development research. Drug Dev Res. 2011;72:153–161. doi: 10.1002/ddr.20412. [Google Scholar]
- 65.Hunt CA, Ropella GE, Lam TN, Tang J, Kim SH, Engelberg JA, Sheikh-Bahaei S. At the biological modeling and simulation frontier. Pharmaceut Res. 2009;26:2369–2400. doi: 10.1007/s11095-009-9958-3. doi: 10.1007/s11095-009-9958-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim SH, Jackson AJ, Hur R, Hunt CA. Individualized, discrete event, simulations provide insight into inter- and intra-subject variability of extended-release, drug products. Theor Biol Med Model. 2012;9:39. doi: 10.1186/1742-4682-9-39. doi: 10.1186/1742-4682-9-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lam TN, Hunt CA. Discovering plausible mechanistic details of hepatic drug interactions. Drug Metab Dispos. 2009;37:237–246. doi: 10.1124/dmd.108.023820. doi: 10.1124/dmd.108.023820. [DOI] [PubMed] [Google Scholar]
- 68.Marino S, Linderman JJ, Kirschner DE. A multifaceted approach to modeling the immune response in tuberculosis. Wiley Interdiscip Rev Syst Biol Med. 2011;3:479–489. doi: 10.1002/wsbm.131. doi: 10.1002/wsbm.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Park S, Ropella GE, Kim SH, Roberts MS, Hunt CA. Computational strategies unravel and trace how liver disease changes hepatic drug disposition. J Pharmacol Exp Ther. 2009;328:294–305. doi: 10.1124/jpet.108.142497. doi: 10.1124/jpet.108.142497. [DOI] [PubMed] [Google Scholar]
- 70.Pepper JW. Drugs that target pathogen public goods are robust against evolved drug resistance. Evol Appl. 2012;5:757–761. doi: 10.1111/j.1752-4571.2012.00254.x. doi: 10.1111/j.1752-4571.2012.00254.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sheikh-Bahaei S, Hunt CA. Enabling clearance predictions to emerge from in silico actions of quasi-autonomous hepatocyte components. Drug Metab Dispos. 2011;39:1910–1920. doi: 10.1124/dmd.111.038703. doi: 10.1124/dmd.111.038703. [DOI] [PubMed] [Google Scholar]
- 72.Sun X, Zhang L, Tan H, Bao J, Strouthos C, Zhou X. Multi-scale agent-based brain cancer modeling and prediction of TKI treatment response: incorporating EGFR signaling pathway and angiogenesis. BMC Bioinform. 2012;13:218. doi: 10.1186/1471-2105-13-218. doi: 10.1186/1471-2105-13-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Allerheiligen SR. Next-generation model-based drug discovery and development: quantitative and systems pharmacology. Clin Pharmacol Ther. 2010;88:135–137. doi: 10.1038/clpt.2010.81. doi: 10.1038/clpt.2010.81. [DOI] [PubMed] [Google Scholar]
- 74.Baneyx G, Fukushima Y, Parrott N. Use of physiologically based pharmacokinetic modeling for assessment of drug-drug interactions. Future Med Chem. 2012;4:681–693. doi: 10.4155/fmc.12.13. doi: 10.4155/fmc.12.13. [DOI] [PubMed] [Google Scholar]
- 75.Bouzom F, Ball K, Perdaems N, Walther B. Physiologically based pharmacokinetic (PBPK) modelling tools: how to fit with our needs? Biopharm Drug Dispos. 2012;33:55–71. doi: 10.1002/bdd.1767. doi: 10.1002/bdd.1767. [DOI] [PubMed] [Google Scholar]
- 76.Edginton AN, Joshi G. Have physiologically-based pharmacokinetic models delivered? Expert Opin Drug Metab Toxicol. 2011;7:929–934. doi: 10.1517/17425255.2011.585968. doi: 10.1517/17425255.2011.585968. [DOI] [PubMed] [Google Scholar]
- 77.Gobburu JV, Lesko LJ. Quantitative disease, drug, and trial models. Annu Rev Pharmacol Toxicol. 2009;49:291–301. doi: 10.1146/annurev.pharmtox.011008.145613. doi: 10.1146/annurev.pharmtox.011008.145613. [DOI] [PubMed] [Google Scholar]
- 78.Grasela TH, Slusser R. Improving productivity with model-based drug development: an enterprise perspective. Clin Pharmacol Ther. 2010;88:263–268. doi: 10.1038/clpt.2010.117. doi: 10.1038/clpt.2010.117. [DOI] [PubMed] [Google Scholar]
- 79.Grillo JA, Zhao P, Bullock J, Booth BP, Lu M, Robie-Suh K, Berglund EG, Pang KS, Rahman A, Zhang L, et al. Utility of a physiologically-based pharmacokinetic (PBPK) modeling approach to quantitatively predict a complex drug-drug-disease interaction scenario for rivaroxaban during the drug review process: implications for clinical practice. Biopharm Drug Dispos. 2012;33:99–110. doi: 10.1002/bdd.1771. doi: 10.1002/bdd.1771. [DOI] [PubMed] [Google Scholar]
- 80.Huisinga W, Telgmann R, Wulkow M. The virtual laboratory approach to pharmacokinetics: design principles and concepts. Drug Discov Today. 2006;11:800–805. doi: 10.1016/j.drudis.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 81.Iyengar R, Zhao S, Chung SW, Mager DE, Gallo JM. Merging systems biology with pharmacodynamics. Sci Transl Med. 2012;4:126ps7. doi: 10.1126/scitranslmed.3003563. doi: 10.1126/scitranslmed.3003563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jiang W, Kim S, Zhang X, Lionberger RA, Davit BM, Conner DP, Yu LX. The role of predictive biopharmaceutical modeling and simulation in drug development and regulatory evaluation. Int J Pharm. 2011;418:151–160. doi: 10.1016/j.ijpharm.2011.07.024. doi: 10.1016/j.ijpharm.2011.07.024. [DOI] [PubMed] [Google Scholar]
- 83.Jones HM, Dickins M, Youdim K, Gosset JR, Attkins NJ, Hay TL, Gurrell IK, Logan YR, Bungay PJ, Jones BC, Gardner IB. Application of PBPK modelling in drug discovery and development at Pfizer. Xenobiotica. 2012;42:94–106. doi: 10.3109/00498254.2011.627477. doi: 10.3109/00498254.2011.627477. [DOI] [PubMed] [Google Scholar]
- 84.Khalil F, Láer S. Physiologically based pharmacokinetic modeling: methodology, applications, and limitations with a focus on its role in pediatric drug development. J Biomed Biotechnol. 2011;2011:907461. doi: 10.1155/2011/907461. doi: 10.1155/2011/907461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mager DE, Jusko WJ. Development of translational pharmacokinetic-pharmacodynamic models. Clin Pharmacol Ther. 2008;83:909–912. doi: 10.1038/clpt.2008.52. doi: 10.1038/clpt.2008.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nucci G, Gomeni R, Poggesi I. Model-based approaches to increase efficiency of drug development in schizophrenia: a can't miss opportunity. Expert Opin Drug Discovry. 2009;4:837–856. doi: 10.1517/17460440903036073. doi: 10.1517/17460440903036073. [DOI] [PubMed] [Google Scholar]
- 87.Prokop A, Michelson S. Systems Biology in Biotech and Pharma. Springer Briefs in Pharmaceutical Science and Drug Development. Vol. 2. New York: Springer; 2012. Development: multiscale CSB—simulation tools; pp. 87–102. doi: 10.1007/978-94-007-2849-3_8. [Google Scholar]
- 88.Rostami-Hodjegan A. Physiologically based pharmacokinetics joined with in vitroin vivo extrapolation of ADME: a marriage under the arch of systems pharmacology. Clin Pharmacol Ther. 2012;92:50–61. doi: 10.1038/clpt.2012.65. doi: 10.1038/clpt.2012.65. [DOI] [PubMed] [Google Scholar]
- 89.Rowland M, Peck C, Tucker G. Physiologically-based pharmacokinetics in drug development and regulatory science. Ann Rev Pharmacol Toxicol. 2011;51:45–73. doi: 10.1146/annurev-pharmtox-010510-100540. doi: 10.1146/annurev-pharmtox-010510-100540. [DOI] [PubMed] [Google Scholar]
- 90.Sinha V, Kimko HHC. Recent developments in physiologically based pharmacokinetic modeling. Clin Trial Simul AAPS Adv Pharm Sci Series. 2011;1:483–499. doi:1.0.1007/978-1-4419-7415-0_21. [Google Scholar]
- 91.Suryawanshi S, Zhang L, Pfister M, Meibohm B. The current role of model-based drug development. Expert Opin Drug Discovry. 2010;5:311–321. doi: 10.1517/17460441003713470. doi: 10.1517/17460441003713470. [DOI] [PubMed] [Google Scholar]
- 92.Zhang L, Pfister M, Meibohm B. Concepts and challenges in quantitative pharmacology and model-based drug development. AAPS J. 2008;10:552–559. doi: 10.1208/s12248-008-9062-3. doi: 10.1208/s12248-008-9062-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang L, Sinha V, Forgue ST, Callies S, Ni L, Peck R, Allerheiligen SR. Model-based drug development: the road to quantitative pharmacology. J Pharmacokinet Pharmacodyn. 2006;33:369–393. doi: 10.1007/s10928-006-9010-8. [DOI] [PubMed] [Google Scholar]
- 94.Jack J, Wambaugh J, Shah I. Systems toxicology from genes to organs. Methods Mol Biol (Clifton, NJ) 2013;930:375–397. doi: 10.1007/978-1-62703-059-5_17. doi: 10.1007/978-1-62703-059-5_17. [DOI] [PubMed] [Google Scholar]
- 95.Kuepfer L, Lippert J, Eissing T. Multiscale mechanistic modeling in pharmaceutical research and development. Adv Exp Med Biol. 2012;736:543–561. doi: 10.1007/978-1-4419-7210-1_32. doi: 10.1007/978-1-4419-7210-1_32. [DOI] [PubMed] [Google Scholar]
- 96.Wetherington JD, Pfister M, Banfield C, Stone JA, Krishna R, Allerheiligen S, Grasela DM. Model-based drug development: strengths, weaknesses, opportunities, and threats for broad application of pharmacometrics in drug development. J Clin Pharmacol. 2010;50(9 suppl):31S–46S. doi: 10.1177/0091270010377629. doi: 10.1177/0091270010377629. [DOI] [PubMed] [Google Scholar]
- 97.van der Graaf PH, Benson N. Systems pharmacology: bridging systems biology and pharmacokinetics-pharmacodynamics (PKPD) in drug discovery and development. Pharmaceut Res. 2011;28:1460–1464. doi: 10.1007/s11095-011-0467-9. doi: 10.1007/s11095-011-0467-9. [DOI] [PubMed] [Google Scholar]
- 98.Morgan P, Van Der Graaf PH, Arrowsmith J, Feltner DE, Drummond KS, Wegner CD, Street SD. Can the flow of medicines be improved? Fundamental pharmacokinetic and pharmacological principles toward improving Phase II survival. Drug Discov Today. 2012;17:419–424. doi: 10.1016/j.drudis.2011.12.020. doi: 10.1016/j.drudis.2011.12.020. [DOI] [PubMed] [Google Scholar]
- 99.Valerio LG. In silico toxicology for the pharmaceutical sciences. Toxicol Appl Pharmacol. 2009;241:356–370. doi: 10.1016/j.taap.2009.08.022. doi: 10.1016/j.taap.2009.08.022. [DOI] [PubMed] [Google Scholar]
- 100.Vicini P. Multiscale modeling in drug discovery and development: future opportunities and present challenges. Clin Pharmacol Ther. 2010;88:126–129. doi: 10.1038/clpt.2010.87. doi: 10.1038/clpt.2010.87. [DOI] [PubMed] [Google Scholar]
- 101.van der Graaf PH. CPT: Pharmacometrics Syst Pharmacol. 2012;1:e8. doi: 10.1038/psp.2012.8. doi: 10.1038/psp.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yao L, Evans JA, Rzhetsky A. Novel opportunities for computational biology and sociology in drug discovery. Trends Biotechnol. 2010;28:161–170. doi: 10.1016/j.tibtech.2010.01.004. doi: 10.1016/j.tibtech.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shannon RE. 1998;1:7–14. Introduction to the art and science of simulation. In Simulation Conference Proceedings, 1998. Winter. [Google Scholar]
- 104.Hunt CA, Ropella GEP, Lam TN, Gewitz AD. Relational grounding facilitates development of scientifically useful multiscale models. Theor Biol Med Model. 2011;8:35. doi: 10.1186/1742-4682-8-35. doi: 10.1186/1742-4682-8-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.An G. Introduction of an agent-based multi-scale modular architecture for dynamic knowledge representation of acute inflammation. Theor Biol Med Model. 2008;5:11. doi: 10.1186/1742-4682-5-11. doi: 10.1186/1742-4682-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.An G. Dynamic knowledge representation using agent-based modeling: ontology instantiation and verification of conceptual models. Methods Mol Biol (Clifton, NJ) 2009;500:445–468. doi: 10.1007/978-1-59745-525-1_15. doi: 10.1007/978-1-59745-525-1_15. [DOI] [PubMed] [Google Scholar]
- 107.An GC. Translational systems biology using an agent-based approach for dynamic knowledge representation: an evolutionary paradigm for biomedical research. Wound Repair Regen. 2010;18:8–12. doi: 10.1111/j.1524-475X.2009.00568.x. doi: 10.1111/j.1524-475X.2009.00568.x. [DOI] [PubMed] [Google Scholar]
- 108.Ropella GEP, Hunt CA. Foundations for Extended Life Cycle Biological Models. CiteSeerX10.1.1.1.2664 2003. Available at: http://biosystems.ucsf.edu/publications/Ropella_UCITR_2003.pdf (accessed January 3, 2013)
- 109.Hunt CA, Ropella GE, Yan L, Hung DY, Roberts MS. Physiologically based synthetic models of hepatic disposition. J Pharmacokinet Pharmacodyn. 2006;33:737–772. doi: 10.1007/s10928-006-9031-3. [DOI] [PubMed] [Google Scholar]
- 110.Begley CG, Ellis LM. Drug development: raise standards for preclinical cancer research. Nature. 2012;483:531–533. doi: 10.1038/483531a. doi: 10.1038/483531a. [DOI] [PubMed] [Google Scholar]
- 111.Prinz F, Schlange T, Asadullah K. Believe it or not: how much can we rely on published data on potential drug targets? Nat Rev Drug Discov. 2011;10:712. doi: 10.1038/nrd3439-c1. doi: 10.1038/nrd3439-c1. [DOI] [PubMed] [Google Scholar]
- 112.Osherovich L. Hedging against academic risk. Sci Business eXchange. 2011;4 (15). doi:10.1038/scibx. 2011.416. [Google Scholar]
- 113.Lam TN, Hunt CA. Mechanistic insight from in silico pharmacokinetic experiments: roles of P-glycoprotein, Cyp3A4 enzymes, and microenvironments. J Pharmacol Exp Ther. 2010;332((2)):398–412. doi: 10.1124/jpet.109.160739. doi: 10.1124/jpet.109.160739. [DOI] [PubMed] [Google Scholar]
- 114.OpenABM. Available at: http://www.openabm.org/standards. (Accessed January 5, 2013)
- 115.Yilmaz L, Óren T. Agent-directed Simulation and Systems Engineering. 2009. Wiley Series in Systems Engineering and Management. Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim, ISBN: 978-3-527-40781-1. [Google Scholar]
- 116.Tang J, Hunt CA. Identifying the rules of engagement enabling leukocyte rolling, activation, and adhesion. PLoS Comput Biol. 2010;6:e1000681. doi: 10.1371/journal.pcbi.1000681. doi: 10.1371/journal.pcbi.1000681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Park S, Kim SH, Ropella GEP, Roberts MS, Hunt CA. Tracing multiscale mechanisms of drug disposition in normal and diseased livers. J Pharmacol Exp Ther. 2010;334:124–136. doi: 10.1124/jpet.110.168526. doi: 10.1124/jpet.110.168526. [DOI] [PubMed] [Google Scholar]
- 118.Engelberg JA, Datta A, Mostov KE, Hunt CA. MDCK cystogenesis driven by cell stabilization within computational analogues. PLoS Comput Biol. 2011;7:e1002030. doi: 10.1371/journal.pcbi.1002030. doi: 10.1371/journal.pcbi.1002030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bigelow JH, Davis PK. (2002). Implications of multi-resolution modeling (MRM) and exploratory analysis for validation. Proceedings of Foundations, 2002 V&V Workshop. http://citeseerx.ist.psu.edu/viewdoc/download?doi:10.1.1.118.9430&rep=rep1&type=pdf. (Accessed February 17, 2013)
- 120.Danos V, Feret J, Fontana W, Harmer R, Krivine J. Rule-based modelling of cellular signalling. CONCUR 2007—concurrency theory. Lect Notes Comput Sci. 2007;4703:17–41. doi: 10.1007/978-3-540-74407-8_3. [Google Scholar]
- 121.Yan L, Sheihk-Bahaei S, Park S, Ropella GEP, Hunt CA. Predictions of hepatic disposition properties using a mechanistically realistic, physiologically based model. Drug Metab Dispos. 2008;36:759–768. doi: 10.1124/dmd.107.019067. doi: 10.1124/dmd.107.019067. [DOI] [PubMed] [Google Scholar]
- 122.Sheikh-Bahaei S, Maher JJ, Hunt CA. Computational experiments reveal plausible mechanisms for changing patterns of hepatic zonation of xenobiotic clearance and hepatotoxicity. J Theor Biol. 2010;265:718–733. doi: 10.1016/j.jtbi.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Popper K. The Growth of Scientific Knowledge. 2002. Conjectures and refutations. . London, UK: Routledge. [Google Scholar]
- 124.Popper K. The Logic of Scientific Discovery. London, UK: Routledge; 2002. The Logic of Scientific Discovery. ISBN 0-203-99462-0. [Google Scholar]
- 125.Hylands C, Lee EA, Liu J, Liu X, Neuendorffer S, Xiong Y, Zhao Y, Zheng H. Overview of the Ptolemy project, Technical Report UCB/ERL M01/11, University of California, Berkeley, 2001. Available at: http://ptolemy.berkeley.edu/publications/papers/03/overview/overview03.pdf. (Accessed January 5, 2013)
- 126.Hewitt C. 2008. Formalizing common sense for scalable inconsistency-robust information integration using Direct Logic (TM) reasoning and the Actor Model. arXiv:0812.4852 v82.
- 127.Yilmaz Y, Hunt CA. Advanced concepts and generative simulation formalisms for creative discovery systems engineering. In: Tolk A, Jain LC, editors. Intelligent-Based Systems Engineering ISRL 10. Germany: Springer Berlin; 2011. pp. 233–258. doi: 10.1007/978-3-642-17931-0_9 (Chapter 9) [Google Scholar]
- 128.Kim SH, Park S, Ropella GEP, Hunt CA. Agent-directed tracing of multi-scale drug disposition events within normal and diseased In Silico Livers. Int J Agent Technol Syst. 2010;2:1–17. doi: 10.4018/jats/2010070101. [Google Scholar]
- 129.Ropella GEP, Hunt CA. Cloud computing and validation of expandable in silico livers. BMC Syst Biol. 2010;4:168. doi: 10.1186/1752-0509-4-168. doi: 10.1186/1752-0509-4-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ropella GEP, Kennedy RC, Hunt CA. Falsifying an enzyme induction mechanism within a validated, multiscale liver model. Int J Agent Technol Syst. 2012;4:1–14. doi: 10.4018/jats.2012070101. [Google Scholar]
- 131.Goderis A, Brooks C, Altintas I, Lee EA, Goble C. Heterogeneous composition of models of computation. Future Gener Comput Syst. 2009;25:552–560. 2009. doi: 10.1016/j.future.2008.06.014. [Google Scholar]
- 132.Eidson JE, Lee EA, Matic S, Seshia SA, Zou J. Distributed real-time software for cyber-physical systems. Proc IEEE. 2011;100:45–59. doi: 10.1109/JPROC.2011.2161237. (Accessed January 5, 2013) [Google Scholar]
- 133.Hewitt C. Actor model of computation: scalable robust information systems. 2010;arXiv:1008.1459. v28. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


