Abstract
Aims: Monothiol glutaredoxins (1-C-Grxs) are small proteins linked to the cellular iron and redox metabolism. Trypanosoma brucei brucei, model organism for human African trypanosomiasis, expresses three 1-C-Grxs. 1-C-Grx1 is a highly abundant mitochondrial protein capable to bind an iron–sulfur cluster (ISC) in vitro using glutathione (GSH) as cofactor. We here report on the functional and structural analysis of 1-C-Grx1 in relation to its ISC-binding properties. Results: An N-terminal extension unique to 1-C-Grx1 from trypanosomatids affects the oligomeric structure and the ISC-binding capacity of the protein. The active-site Cys104 is essential for ISC binding, and the parasite-specific glutathionylspermidine and trypanothione can replace GSH as the ligands of the ISC. Interestingly, trypanothione forms stable protein-free ISC species that in vitro are incorporated into the dithiol T. brucei 2-C-Grx1, but not 1-C-Grx1. Overexpression of the C104S mutant of 1-C-Grx1 impairs disease progression in a mouse model. The structure of the Grx-domain of 1-C-Grx1 was solved by nuclear magnetic resonance spectroscopy. Despite the fact that several residues—which in other 1-C-Grxs are involved in the noncovalent binding of GSH—are conserved, different physicochemical approaches did not reveal any specific interaction between 1-C-Grx1 and free thiol ligands. Innovation: Parasite Grxs are able to coordinate an ISC formed with trypanothione, suggesting a new mechanism of ISC binding and a novel function for the parasite-specific dithiol. The first 3D structure and in vivo relevance of a 1-C-Grx from a pathogenic protozoan are reported. Conclusion: T. brucei 1-C-Grx1 is indispensable for mammalian parasitism and utilizes a new mechanism for ISC binding. Antioxid. Redox Signal. 19, 665–682.
Introduction
Glutaredoxins (Grxs) are 10–12-kDa proteins that occur in most living organisms and belong to the thioredoxin-fold superfamily (64). Based on sequence features, Grxs are clustered into six classes (69), with I and II being the most populated ones. Class I proteins comprise single-domain Grxs with either a dithiol (CP/G/S/FYC, 2-C-Grxs) or a monothiol (CP/SYS, 1-C-Grxs) active site. Class II proteins are 1-C-Grxs with a CGFS motif present as single- or multidomain proteins (69). Despite sharing several residues and a common fold, both classes evolved distinct functionalities. 2-C-Grxs employ glutathione (GSH) to reduce protein–GSH mixed disulfides (deglutathionylation) as well as inter- or intramolecular disulfides (48). In most cases, the latter type of reaction involves both active-site cysteines, whereas protein deglutathionylation proceeds via a monothiol mechanism requiring only the first cysteine (26). In addition, after the seminal article of Lillig et al. (49), several 2-C-Grxs were shown to coordinate a [2Fe-2S] iron–sulfur cluster (ISC) (8, 12, 22, 70). As 2-C-Grxs are monomeric proteins, coordination of the ISC by the N-terminal active cysteines of each monomer leads to dimerization through the cluster. The remaining two coordination positions are occupied by GSH molecules (19, 22, 38, 39, 69, 70).
Innovation.
In eukaryotes, the biogenesis of the iron–sulfur cluster (ISC) and many ISC-proteins—several of them being essential for life—is a process confined to the mitochondria, where monothiol glutaredoxins (1-C-Grxs) are key components of the machinery. This study discloses a novel ISC-binding mechanism, several distinctive structural features, and the in vivo indispensability of the mitochondrial 1-C-Grx1 from a biomedically relevant protozoa. The finding that parasite-specific low-molecular-mass thiols act as ISC ligands provides evidence for a new link between the thiol-redox and iron metabolism in these organisms. Some of these peculiarities may be exploited for specific drug design.
In contrast, most class II 1-C-Grxs studied so far lack a disulfide reductase activity, but serve as structural domains for ISC binding and/or ISC-dependent protein–protein interactions (31, 35, 41, 42, 44, 47, 59). ISC ligation by 1-C-Grxs also requires the active-site cysteine and GSH, and several structures of 1-C-Grxs in their apo- and holoform have shed light onto the molecular determinants and the mechanism of the ISC assembly (25, 36, 39, 46, 50). Monodomain 1-C-Grxs are central components of the highly conserved biosynthetic pathway of the iron–sulfur proteins (4, 13, 23, 41, 53, 54, 67, 74, 76), a process that in eukaryotes is mainly compartmentalized in the mitochondria. Multidomain 1-C-Grxs, such as Grx3 and 4 in yeast, were shown to be key elements in intracellular iron trafficking and the cytosolic ISC assembly machinery (35, 42, 55). The physiological relevance of these proteins in iron trafficking, ISC biogenesis/mobilization, and heme biosynthesis was demonstrated by genetic approaches for several organisms (4, 13, 23, 27, 53, 54, 67, 74, 76).
Protozoan parasites of the genus Trypanosoma and Leishmania are responsible for devastating diseases of humans and livestock in the subtropical regions. Subspecies of the Trypanosoma brucei complex occur in sub-Saharan Africa and cause human sleeping sickness (T. brucei rhodesiense and T. brucei gambiense) and Nagana disease in cattle (T. brucei brucei). All trypanosomatids evolved a unique thiol-redox system based on trypanothione [T(SH)2], trypanothione reductase, and the thiol–disulfide oxidoreductase tryparedoxin (43). Trypanothione is synthesized from GSH and spermidine with mono-glutathionylspermidine (Gsp) as an intermediate. The trypanothione system replaces the ubiquitous GSH/GSH reductase and thioredoxin/thioredoxin reductase systems and is indispensable for parasite survival (reviewed in 43). Two 2-C-Grxs (2-C-Grx1 and 2) (12) and three 1-C-Grxs occur in both the mammalian bloodstream and the insect procyclic form of T. brucei (17, 24). The parasite 1-C-Grx1 and 1-C-Grx2 are single-domain mitochondrial proteins, while 1-C-Grx3 contains an additional N-terminal thioredoxin-like domain (17, Comini, unpublished). 1-C-Grx1 is an abundant protein capable of assembling in vitro a [2Fe-2S] cluster using GSH as ligand (17). Complementation studies in Saccharomyces cerevisiae deficient in the mitochondrial Grx5 demonstrate that only 1-C-Grx1 modestly rescued the mutant phenotype (24). The lack of functional conservation of T. brucei 1-C-Grx1 in comparison with various prokaryotic and eukaryotic 1-C-Grxs (53) suggests a significant structural and/or biochemical specificity of the parasite protein. The indispensability of 1-C-Grx1 for infective T. brucei was inferred from the refractoriness to obtain RNA interference and single-knockout cell lines of this parasite stage (17).
Here we report on the identification of a trypanosomatid-specific N-terminal extension affecting the oligomeric structure of 1-C-Grx1 and the assembly of an ISC using parasite-specific low-molecular-mass (LMM) thiols as ligands via a previously unreported mechanism. Elucidation of the nuclear magnetic resonance (NMR) structure of the isolated Grx domain of 1-C-Grx1 revealed atomic details that account for the peculiar behavior of the trypanosomal protein. Importantly, animal infection experiments with T. brucei overexpressing an active-site mutant of 1-C-Grx1 highlighted a critical role of the mitochondrial protein for parasite infectivity.
Results
The role of a unique N-terminal stretch of 1-C-Grx1 in protein oligomerization and ISC binding
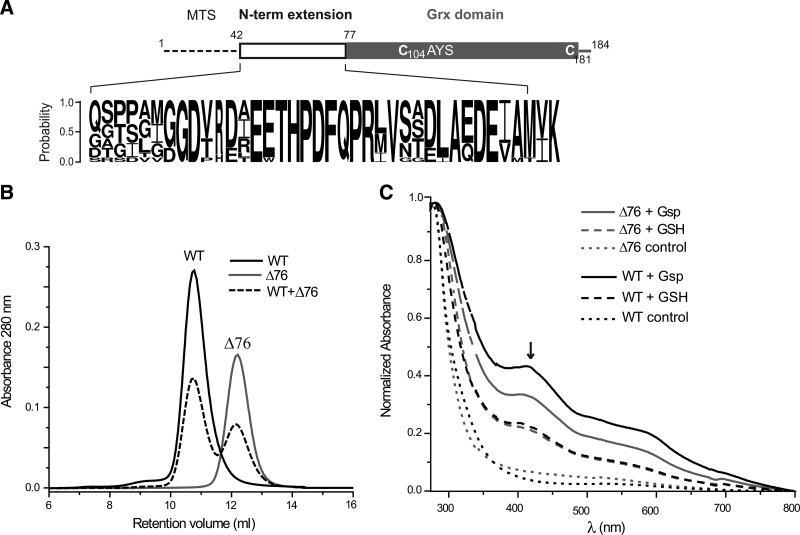
The genomes of trypanosomatids encode three putative 1-C-Grxs. 1-C-Grx1 and 1-C-Grx2 are single-domain proteins containing an N-terminal mitochondrial localization peptide, while 1-C-Grx3 is a Trx/1-C-Grx hybrid lacking any evident targeting signal (15). Apo-1-C-Grx1 differs from the clearly monomeric 1-C-Grx2 and 1-C-Grx3 in eluting from the gel chromatography column with a mass intermediate between that expected for a monomer or a noncovalent homodimer (17, 24). To disclose the structural elements responsible for this oligomeric conformation, comparative sequence and mutational analyses were performed. A multiple sequence alignment highlighted an intriguing difference between Tb1-C-Grx1 and other 1-C-Grxs, namely, a 35-residue-long stretch (residues 42–76) located between the putative mitochondrial targeting signal (residues 1–41) and the Grx domain (residues 77–184) (Fig. 1A) (15). Database searches with PSI-BLAST using the 35-mer peptide of Tb1-C-Grx1 as a probe exclusively retrieved sequences from trypanosomatid homologs (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/ars), pointing to a remarkable uniqueness of this sequence. Bioinformatic analyses predicted a nonregular structure for the first half of this peptide, whereas the second half likely folds into a helical structure (Supplementary Fig. S1). Despite a moderate sequence conservation of this region between 1-C-Grx1 from T. brucei, T. cruzi, and Leishmania major (mean sequence identity of 38–42%), it contains a highly conserved motif [E55(E/W)THPDFQPR64] (numbering according to T. brucei 1-C-Grx1, Fig. 1A and Supplementary Fig. S1). Based on this finding, a mutant of Tb1-C-Grx1 lacking the first 76 residues (mitochondrial targeting sequence and N-terminal extension; (Δ76 Tb1-C-Grx1) was generated. Size-exclusion chromatography (SEC, Fig. 1B) revealed that the mature wild-type (WT) protein (residues 42–184; WT Tb1-C-Grx1) eluted with a retention volume nearly corresponding to that expected for a dimeric protein species (apparent molecular mass of 28.6±2.2 kDa; theoretical mass of the subunit 16.2 kDa), whereas (Δ76 Tb1-C-Grx1 eluted at an apparent molecular mass of 16.1±0.2 kDa, which is close to the theoretical mass of the monomer (12.3 kDa). Gel filtration analysis of an equimolar mixture of the recombinant mature and truncated proteins did not yield species with intermediate molecular masses (Fig. 1B).
FIG. 1.
Structural organization, gel chromatography, and ISC formation of WT and Δ76 Tb1-C-Grx1. (A) Schematic representation of Tb1-C-Grx1 with the putative MTS, the N-terminal extension, and the Grx domain with the active-site CAYS (Cys104) as well as Cys181. The conservation in the N-terminal extension of 1-C-Grx1s from different trypanosomatids is shown as logo (see Supplementary Fig. S1 for details). (B) SEC of mature (WT), truncated (Δ76), and a mixture of both proteins. (C) UV–visible spectra of ISC reconstitution mixtures for 50 μM WT (black lines) and Δ76 (gray lines) Tb1-C-Grx1 in the presence of 150 μM GSH (dashed) or Gsp (solid). The cysteine desulfurase was omitted in control reactions (dotted lines). The spectra were normalized for the absorbance at 280 nm. The black arrow indicates the characteristic absorbance at 420 nm of the holocomplex. 1-C-Grx, monothiol glutaredoxin; Grx, glutaredoxin; GSH, glutathione; Gsp, glutathionylspermidine; ISC, iron–sulfur cluster; SEC, size-exclusion chromatography; WT, wild type; Tb, Trypanosoma brucei; MTS, mitochondrial targeting sequence.
As shown previously, Tb1-C-Grx1 does not catalyze a wide variety of reactions that are carried out by classical Grxs (24), but is capable to assemble an ISC in both the test tube and when produced in a heterologous organism such as Escherichia coli (17). To determine if the N-terminal extension contributes to ISC binding, WT and Δ76 Tb1-C-Grx1 were subjected to the ISC reconstitution assays in the presence of GSH or Gsp as LMM thiol. Both proteins revealed absorption peaks at 320 and 420 nm (Fig. 1C), which are characteristics of an 1-C-Grx-bound ISC (8, 17, 18, 30, 49, 52, 62, 63, 70). However, the lability of the holo-(Δ76 Tb1-C-Grx1, when freed from the reconstitution components, precluded SEC analysis. Taken together, the N-terminal region of Tb1-C-Grx1 does not appear to be critical for ISC binding under in vitro conditions, but may contribute to stabilize the holocomplex. Therefore, a potential involvement of the N-terminal tail in LMM thiol binding was investigated by NMR spectroscopy (see below).
The active-site cysteine is indispensable for ISC binding
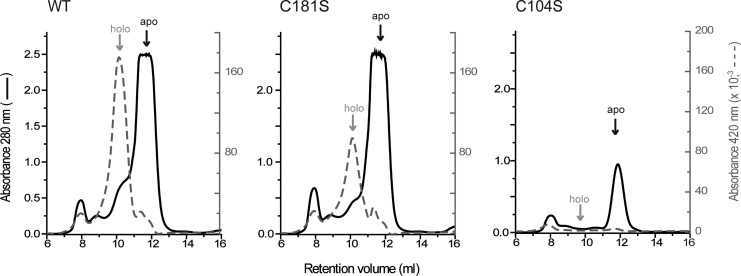
All Grxs capable to bind an ISC use the N-terminal active-site cysteine and GSH as thiol ligands for metal coordination (8, 70). Tb1-C-Grx1 contains, in addition to the active-site Cys104, a nonconserved Cys181 located three residues from the C-terminus (Fig. 1A). To verify the predicted role of the active-site cysteine in ISC binding, C104S and C181S mutants of His-tagged full-length Tb1-C-Grx1 were generated by site-directed mutagenesis and expressed in E. coli. These mutations did neither affect the oligomeric state (Supplementary Fig. S2) nor the far-UV circular-dichroism (CD) spectra (not shown) of the recombinant proteins. The first evidence for Cys104 being responsible for ISC binding was the brownish color of the Ni2+-affinity chromatography column loaded with the bacterial lysate expressing the WT and C181S, but not C104S 1-C-Grx1 (Supplementary Fig. S2). This observation was confirmed by subjecting freshly purified His-tagged Tb1-C-Grx1 isoforms to SEC. A small fraction of the WT and C181S proteins eluted in a peak absorbing at both 280 nm and 420 nm (Fig. 2). These species eluted at apparent molecular masses of 44–52 kDa, suggesting a dimeric complex in comparison to the apoform with an observed mass of ∼30 kDa (see below). Moreover, the peak assigned to the holoprotein disappeared when treated with EDTA (Supplementary Fig. S2). As expected, the C104S mutant runs as a single species that lacked absorption at 420 nm (Fig. 2).
FIG. 2.
ISC binding by different Tb1-C-Grx1 species overexpressed in Escherichia coli. His-tagged WT, C181S, and C104S Tb1-C-Grx1 eluted from the Ni2+- affinity chromatography column were subjected to SEC with online detection at 280 nm (black solid line) and 420 nm (gray dashed line). The WT and C181S proteins displayed a minor peak or shoulder, preceding the apoform (black arrow), with an absorbance at 420 nm (gray arrow). This peak corresponds to the holoprotein (holo) and is absent in the elution profile of the C104S mutant. The peak eluting at ∼8 ml corresponds to the exclusion volume (proteins with molecular masses≥75 kDa).
Parasite-specific thiols can replace GSH as ISC ligand
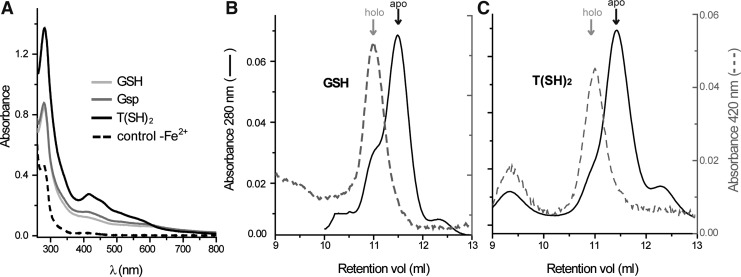
T(SH)2 is the main LMM thiol of trypanosomes (43). Recently, we reported a potential novel function for this dithiol, namely, as a ligand of the ISC bound to dithiol Tb2-C-Grx1 (12, 15). Here, we extended this analysis to verify the capacity of Tb1-C-Grx1 to use parasite-specific LMM thiols for ISC binding. Tag-free Tb1-C-Grx1 incorporates ISCs with Gsp or T(SH)2 as nonprotein thiol ligands that display absorption spectra similar to those obtained with GSH (Fig. 3A; 17). SEC of holo-Tb1-C-Grx1 reconstituted with GSH or T(SH)2 revealed two peaks. In both cases, the main fraction represented the free protein (apo), eluting with a mass of ∼30 kDa and lacking absorption at 420 nm. A minor protein fraction of a higher apparent molecular mass (∼40 kDa) exhibited absorbance at 420 nm (Fig. 3B, C). As described above for holo-Tb1-C-Grx1 isolated from the recombinant E. coli cells (Fig. 2), the apparent molecular mass of the holoproteins (∼40 kDa) was significantly lower than that expected for a globular tetramer (58–68 kDa), but in accordance with a dimeric conformation.
FIG. 3.
In vitro ISC binding by Tb1-C-Grx1. (A) Tag-free Tb1-C-Grx1 WT (50 μM) was subjected to the ISC reconstitution assay in the presence of 1 mM GSH, 1 mM Gsp, or 500 μM T(SH)2 (1 mM thiol). Control assays lacked Fe2+. Identical results were obtained for His-tagged proteins (not shown). (B, C) About 100 μg protein from the ISC reconstitution mixtures with GSH (B) or T(SH)2 (C) was subjected to SEC with online detection at 280 nm (solid black line) and 420 nm (dashed gray line). In both cases, apo-Tb1-C-Grx1 showed the expected retention volume and no absorbance at 420 nm, while the peak containing the chromophore (“holo”) eluted at lower retention volume. T(SH)2, trypanothione.
LMM thiols show negligible interaction with apo-1-C-Grx1
We next investigated whether LMM thiols bind noncovalently to Tb1-C-Grx1 as an initial requirement for ISC assembly. Isothermal titration calorimetry of Tb1-C-Grx1 with GSH did not reveal any interaction under the conditions assayed (50 μM protein titrated with up to 500 μM GSH in 20 mM Tris–HCl buffer, pH 7.8, at 25°C; not shown); neither the protein interacted with GSH immobilized on a sepharose matrix (not shown). Also, monitoring Trp fluorescence (the protein contains a single Trp at position 142 close to the active-site motif; see Supplementary Fig. S3) and the near-UV CD spectra of the protein treated with GSH up to 10-fold molar excess showed only minor spectral changes at high thiol:protein ratios (Supplementary Fig. S3). After sequence-specific NMR assignment of (Δ76 1-C-Grx1 (see below), the 15N-heteronuclear single-quantum coherence (HSQC) spectra of this protein were recorded during titration with GSH or T(SH)2. In contrast to the results reported for other Grxs using the same methodology (37, 58), no significant chemical-shift perturbations were observed for (Δ76 1-C-Grx1 up to a thiol:protein molar ratio of 100:1 and 50:1 for GSH and T(SH)2, respectively (Supplementary Fig. S3). To verify or exclude a role of the N-terminal extension in ligand binding, an identical experiment was conducted with the mature full-length protein. Only minor shifts in few peaks of the 15N-HSQC spectra of WT Tb1-C-Grx1 were observed upon titration with both GSH and T(SH)2. For both ligands, the perturbations were rather small with<10 peaks showing a combined 15N,1H chemical-shift changes larger than 0.02 ppm, with the largest shift being 0.13 ppm. Further, the observed shifts do not reach a saturation plateau even at the highest thiol:protein ratio tested [125:1 and 100:1 for GSH and T(SH)2, respectively] (Supplementary Fig. S3). Although it is not possible to provide a quantitative estimation of the dissociation constant for this interaction, the data clearly show that GSH and T(SH)2 display a very low affinity (≥10 mM) toward WT Tb1-C-Grx1. Despite that the backbone NMR assignment is not available for the WT protein, most of the peaks undergoing perturbations during the titrations could be tentatively assigned by comparison with the HSQC spectrum of Δ76 Tb1-C-Grx1 (see below). The majority of these peaks correspond to residues nearby the putative GSH-binding pocket (Cys104, Ser107, Trp142, Gly157, Ile160, and Thr162) with the exception of Thr180, which precedes the C-terminal cysteine located on the opposite face of the protein. Thus, Tb1-C-Grx1 diverges from other ISC-binding Grxs in lacking any significant interaction with the free thiols, at least in vitro (37, 58).
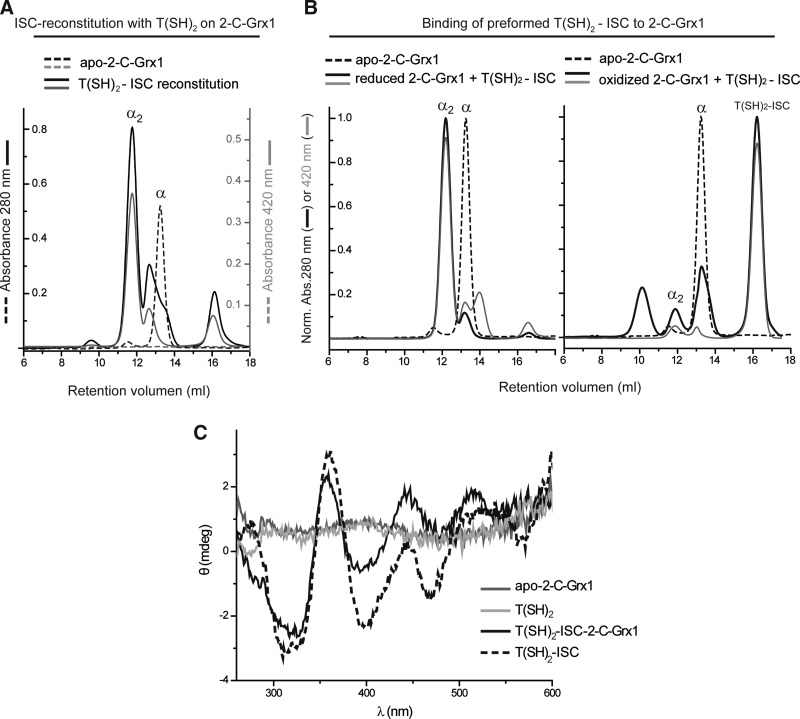
Tb2-C-Grx1, but not 1-C-Grx1, incorporates a preformed trypanothione–ISC complex
Recently, we have shown that a free T(SH)2-ISC complex can be incorporated into the dithiolic Tb2-C-Grx1, converting the monomeric protein into an ISC-linked dimer (12). Here we studied if Tb1-C-Grx1 is also capable of accepting preformed ISC complexes. The T(SH)2-ISC complex was produced (Fig. 4A). Interestingly, the protein-free complex was stable for several hours (not shown) even after purification from the reaction mixture by SEC under aerobic conditions (Fig. 4B). It displayed the typical UV–visible spectra of ISCs bound to proteins or the recently reported [2Fe-2S]-(GSH)4 complex (Fig. 4C) (65). The CD spectrum (Fig. 4D) differed from those reported for ISC-proteins (4, 36, 49, 70), but was almost identical to that of holo-Tb2-C-Grx1 assembled with T(SH)2 as ligand (Fig. 5A, C). Incubation of WT or Δ76 apo-Tb1-C-Grx1 with the T(SH)2-ISC complex followed by gel filtration chromatography did not reveal any holo-Tb1-C-Grx1 (not shown). Under similar conditions, reduced, but not oxidized, Tb2-C-Grx1 resulted in a dimeric holoprotein (Fig. 5B). These results highlight a remarkable difference in ISC binding between these parasite 1-C-Grx and 2-C-Grx, which may reflect distinct cellular roles (ISC transfer vs. ISC redox regulator; 4, 12, 31, 36, 49) and/or the requirement of other components of the ISC assembly machinery to allow ISC binding to 1-C-Grx1. In line with this proposal, the heterologous expression of recombinant His-tagged 1-C-Grx1 yielded the holoprotein in higher quantities and a more stable form than the in vitro reconstruction system.
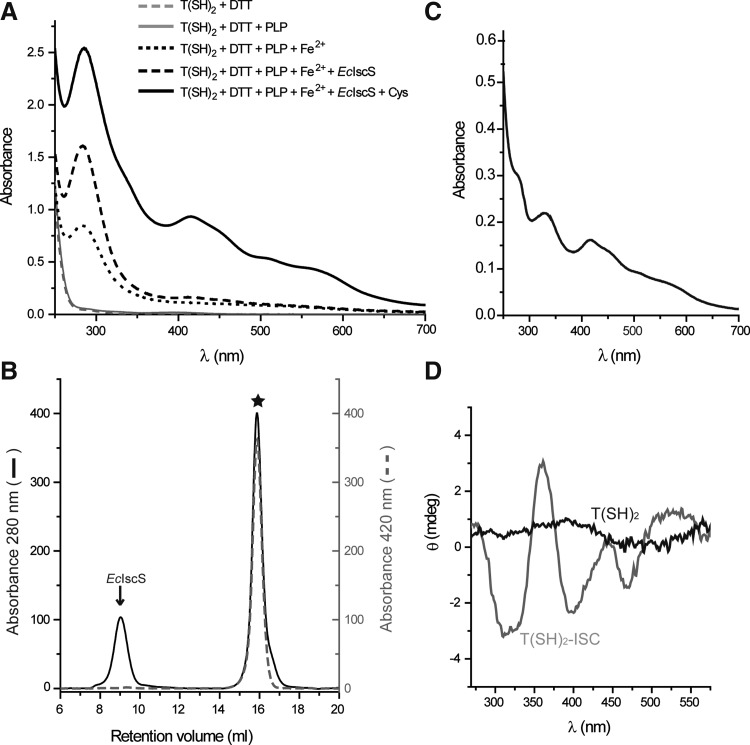
FIG. 4.
ISC formation on trypanothione. (A) One mM T(SH)2 was incubated with the individual components of the reconstitution assay (5 mM dithiothreitol, 10 μM pyridoxal-5′-phosphate, 500 μM Fe2+, 5 μM EcIscS, and 500 μM cysteine) as described under the Material and Methods section, but in the absence of Grxs. The UV–visible spectrum of each mixture was recorded after centrifugation. (B) The ISC reconstitution mixture containing all components (black solid line in A) was subjected to gel chromatography. The major peak (star) elutes with a retention time corresponding to an LMM component. EcIscS indicates the elution peak of E. coli cysteine desulfurase. (Abs. 280 nm: black solid line, Abs. 420 nm: gray dashed line). (C) The UV–visible spectrum of the peak fraction from B (star) containing the T(SH)2-ISC complex. (D) The CD Spectra of the T(SH)2-ISC complex (gray line) and free T(SH)2 (black line). CD, circular dichroism; LMM, low molecular mass.
FIG. 5.
The parasite 2-C-Grx1 binds preformed T(SH)2-ISC complexes. (A) About 100 μM Tb2-C-Grx1 was subjected to a reconstitution mixture with 1 mM T(SH)2, followed by gel chromatography with online detection at 280 nm (black lines) and 420 nm (gray lines). The ISC reconstitution mixture is shown in solid lines, while the reduced free 2-C-Grx1 (apo) is shown in dashed lines. The peaks corresponding to the apo- (monomeric) and holo- (dimeric) 2-C-Grx1 are indicated as α and α2, respectively. (B) A protein-free T(SH)2-ISC complex produced and isolated by SEC as described in Figure 4 was incubated with reduced (left) or oxidized (right) Tb2-C-Grx1 in buffer A at room temperature for 30 min before subjecting the mixture to SEC. Incubation of reduced Tb2-C-Grx1 with the preformed T(SH)2-ISC complex yielded mainly a dimeric protein species (solid black line) that absorbed at 420 nm (solid gray line), whereas oxidized 2-C-Grx1 did not bind the free T(SH)2-ISC complex. For comparison, the chromatogram of apo-2-C-Grx1 is included (black dashed lines), and all profiles are shown normalized. (C) The CD spectra of holo-Tb2-C-Grx1 reconstituted with T(SH)2-ISC (α2 in A; solid black line), apo-2-C-Grx1 (solid gray line), free T(SH)2 (solid light gray line), and isolated T(SH)2-ISC complex (black dotted line). 2-C-Grx, dithiol glutaredoxin.
NMR structure of the Grx domain of 1-C-Grx1
To obtain high-resolution structural information on Tb1C-Grx1, we focused on the truncated protein, which encompasses the full sequence of a classical Grx domain. This strategy was chosen not only because it can be challenging to obtain precise structures of proteins larger than 30 kDa, but also because the mature WT protein showed significant degradation under the standard conditions required for the NMR analysis. For the NMR analysis, Δ76 Tb1-C-Grx1 was expressed in an isotopically labeled form. The protein showed well-dispersed 15N- and 13C-HSQC spectra indicative of a folded polypeptide (Supplementary Fig. S4). The good quality of the acquired NMR spectra allowed the achievement of nearly complete resonance assignments and the calculation of a well-defined protein structure as documented in Supplementary Table S1 and Figure 6. The solution structure of Δ76 Tb1-C-Grx1 (Fig. 6A) is characterized by a core of four β-strands (β1, residues 91–95; β2, residues 122–125; β3, residues 147–150; and β4, residues 153–156) surrounded by five α-helices (α1, residues 79–88; α2, residues 105–116; α3, residues 130–138; α4, residues 158–167; and α5, residues 169–177). A classical β-bulge is present at the end of β4 that, as consequence, is significantly twisted. Helices α1 and α3 are located at one side of the β-sheet plane and are oriented almost orthogonally to each other. Helices α2, α4, and α5 are located on the opposite side, where α2 and α4 are essentially parallel. Helices α4 and α5 are almost continuous in the sequence, but are structurally tilted by 90°, most likely facilitated by the conserved Gly168 interspaced between the helices. All the helices are amphipathic with hydrophobic side chains pointing toward the core of the protein, thus stabilized by an extensive network of hydrophobic interactions mostly conserved in 1-C-Grxs (25). The solvent-exposed Cys104 is located N-terminal to α2, at the end of a loop that is present only in class II 1-C-Grxs [reviewed in Ref. (15)]. Two hydrogen bonds, the one between Met103-NH and Lys96-O’ and the one between Lys96-NH and the side chain of Ser107, play a role in determining the geometry of the CAYS active site and of the preceding loop. Lys96 is fully conserved in all 1-C-Grxs and is involved in GSH binding (see below). The orientation of its side chain is determined by van der Waal's contacts with surrounding Val126, Leu127, and Ile145 and the positive charge points toward the thiol group of Cys104, probably contributing to the decrease of its pKa (71a). Pro146, spatially close to Cys104, adopts a cis-conformation, as deduced from the presence of strong sequential dαα and the absence of sequential dαδ nuclear Overhauser effect (NOEs) between Ile145 and Pro146 (75) and from the chemical shifts of the Cβ and Cγ resonances for this proline (72).
FIG. 6.
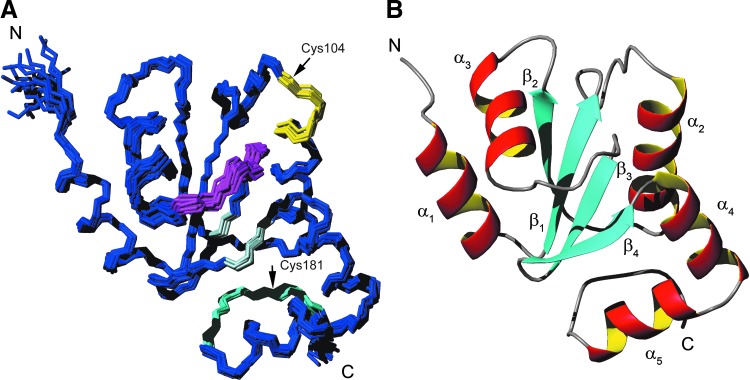
Nuclear magnetic resonance structure of Δ76 Tb1-C-Grx1. (A) Superposition of the backbone of the 20 conformers with the best CYANA target function, refined with Amber. Active-site residues (104–107, yellow), cis-proline-containing loop (residues 141–146, magenta), residues involved in the formation of the conserved β-bulge (148, 155, and 156, light blue), and residues from C-terminus (178–182, cyan) are highlighted. The positions of Cys104 and Cys181 are indicated with black arrows. (B) Ribbon representation showing secondary-structure elements. The program MOLMOL was used to prepare these figures To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
Most of the conserved residues on the surface of the protein map on the same region (15), adjacent to the active site and corresponding to the binding pocket of GSH described for other Grxs (11, 36, 39, 50, 58). Among these, the residues corresponding to Lys96, Cys104, Thr144, and Asp159 have been shown to be crucial for both GSH binding and ISC assembly in 1-C-Grxs (Fig. 7A) (62). A comparison with the structure of the ISC-bound dimeric form of E. coli monothiol Grx4 (EcGrx4; PDB ID 2WIC) (36) shows that most of these residues have the correct spatial orientation for the interaction with noncovalently bound GSH. However, a significant difference can be observed for the loop containing Thr144 and Ile145, which in Δ76 Tb1-C-Grx1 appears to be closer to Asp159 (Fig. 7A). As a consequence, Thr144 occludes a cavity hosting the γ-Glu moiety of GSH in EcGrx4, and the side chain of Ile145 protrudes from the pocket (Fig. 7A, B); hence, both residues preclude the binding of the thiol ligand to Tb1-C-Grx1 as opposed to EcGrx4 (Fig. 7C). In Tb1-C-Grx1, this loop seems to have some structural plasticity as the 15N-HSQC peaks of Ser140, Trp142, Thr144, and Ile145 appear broader than the average, suggesting a conformational exchange in this region (Supplementary Fig. S4). Notably, also Asp159, located at the beginning of α4 and spatially close to Thr144, shows a very broad resonance. The electrostatic potential around the putative GSH-binding pocket is positive due to the presence of conserved basic residues, whereas on the opposite side of the polypeptide, a negatively charged region is present that is not generally conserved in 1-C-Grxs and may be a surface for protein–protein interactions (Supplementary Fig. S5) (15).
FIG. 7.
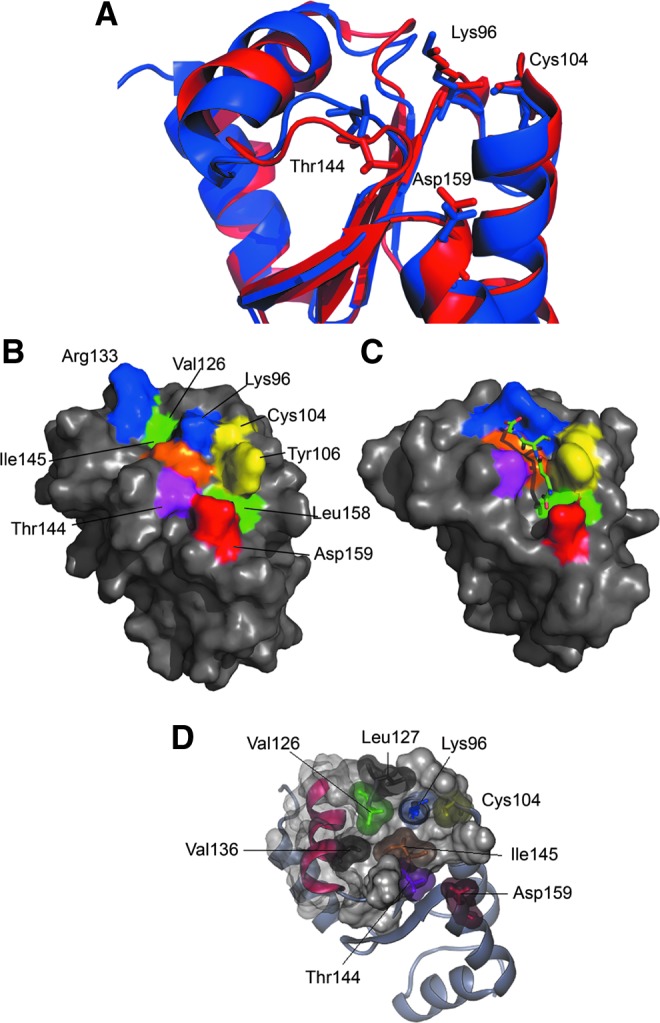
Structural features of the Tb1-C-Grx1 active-site region. (A) Superposition of the protein portion encompassing the putative GSH-binding pocket in apo-Δ76 Tb1-C-Grx1 (red, this work) and E. coli Grx4 in an ISC-bound dimeric form (blue; PDB-ID 2WIC; a single monomer is shown, and the ISC and GSH molecules are omitted). (B) Conserved residues predicted to be important for the binding of GSH are mapped on the surface of Δ76 Tb1-C-Grx1 with different colors: Lys96 and Arg133 in blue; Cys104 and Tyr106 in yellow; Thr144 in magenta; Ile145 in orange; and Asp159 in red; the hydrophobic residues Val126 and Leu158 in the pocket are shown in green. (C) The corresponding residues in E. coli Grx4 (PDB ID 2WIC) are represented with the same colors; the noncovalently bound GSH molecule is shown as sticks. The program PyMol was used for preparing the pictures. (D) Residues Ile145, Val 136, Val126, and Leu127, forming the hydrophobic network discussed in the text, are represented by their van der Waal's surfaces. Other residues important for GSH binding are highlighted with the same color used in panel (B) To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
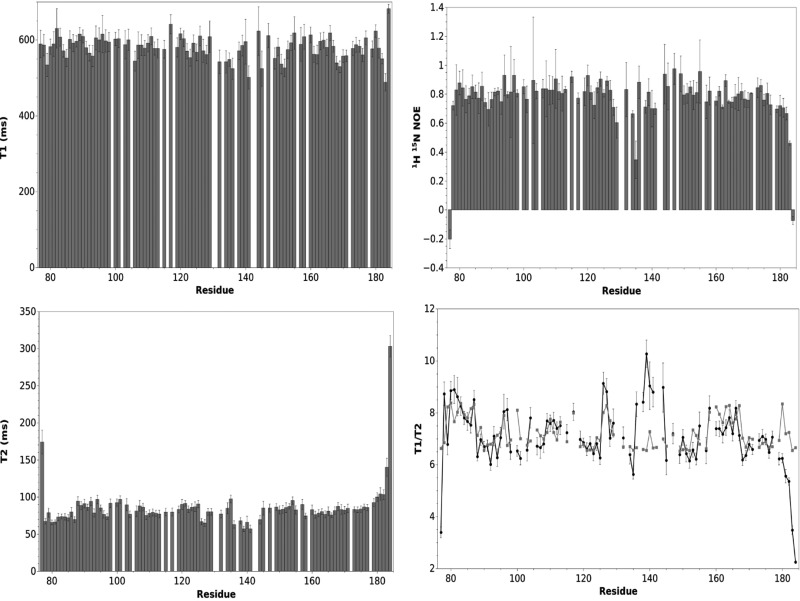
The backbone dynamics of Grx1 were investigated by 15N relaxation measurements. The correlation time for protein tumbling (τc) was estimated from the T1/T2 ratios (5), yielding a value of 7.9±0.6 ns, fully consistent with the protein being a monomer; the value calculated by the HYDRONMR program (7) from the monomeric structure was 7.7 ns. 15N{1H}-NOEs (Fig. 8) indicate that the protein is well ordered, with limited dynamics in the sub-nanosecond time scale, except for very few residues at the N- and C-terminus and within helix α3 (Fig. 6A), which is known to be strongly distorted in several proteins of the thioredoxin-fold family (64).
FIG. 8.
Backbone dynamics of Δ76 Tb1-C-Grx1. Backbone amide 15N longitudinal (T1) and transverse (T2) relaxation time and 15N{1H}-NOE measured at 600 MHz and 298 K. Experimental (black circle) and HYDRONMR (gray square) T1/T2 are compared. T1 and T2 were obtained by fitting cross peak volumes, measured as a function of the relaxation delay, to a single exponential decay using the NmrPipe software package (20). Spectra were recorded with 9 (10 [twice], 50, 100, 200 [twice], 400, 700, 1000 [twice], 1300, and 1600 ms) and 8 delay times (16.31 [twice], 32.64, 48.96 [twice], 65.28, 97.92 [twice], 146.89, 179.52, and 228.5 ms) for T1 and T2 measurements, respectively. NOE values were calculated as the ratio of peak volumes in spectra recorded with and without saturation. NOE, nuclear Overhauser effect.
Residues 178–182, following α5 (Fig. 6A) do not have a regular secondary structure. They have average 15N{1H}-NOE values slightly lower than the average for the entire protein (0.70 vs. 0.80) (Fig. 8), but their conformation is well defined. This segment is kept in its position by a hydrogen bond between Arg182-NH and Leu117-O’ and stabilized by hydrophobic interactions involving Ile179, Leu174, Ile150, and Val91. At the end of this segment, Cys181, the only other cysteine of the protein, is located at a distance of 26 Å from Cys104. Thus, a significant rearrangement and/or local unfolding would be required to allow formation of an intramolecular disulfide suggested previously to occur under mild oxidizing conditions (51). Notably, also in EcGrx4, formation of a disulfide bridge between two cysteines spatially distant in the available structure has been proposed (25).
HYDRONMR provides a method to calculate the T1/T2 ratio when the structure of the protein is available (7), and deviations between experimental and calculated values can be related to chemical exchange or fast internal motion. The method is particularly valuable in discriminating between anisotropy and chemical exchange without the need of relaxation measurements at different magnetic fields. The plot of experimental and calculated T1/T2 values for Δ76 Tb1-C-Grx1 (Fig. 8) clearly shows that the experimental values are significantly higher than those calculated with HYDRONMR for residues 136–144, corresponding to the C-terminus of α3 and the entire cis-proline-containing loop. Thus, this portion of the protein probably undergoes a conformational exchange. Other residues suggested by this method to undergo motion in the μs–ms time scale are Lys96, Cys104, Val126, and Leu127. Interestingly, the first two have been proposed to be essential for the binding of GSH, while Val126 forms part of the GSH pocket and is in contact with Val136, located on α3.
Overexpression of C104S 1-C-Grx1 and, to a lesser extent, of WT 1-C-Grx1 impairs the infectivity of African trypanosomes
Previous attempts to silence the expression of 1-C-Grx1 in the infective stage of T. brucei by RNA interference and gene knockout approaches were unsuccessful (17). The alleles encoding 1-C-Grx1 could be replaced in the insect form of the parasite, only if an inducible ectopic copy of the gene was coexpressed (17). Altogether, these results were strong indirect evidence for the indispensability of 1-C-Grx1 for parasite viability. This was further supported by a recent genome-wide RNA interference analysis published by Alsford et al. (1). To assess the relevance of 1-C-Grx1 for parasite survival under in vitro and in vivo conditions, experiments were conducted with a T. brucei cell line overexpressing C104S, which conserves the in-solution conformation of the WT isoform, but is unable to bind an ISC (Fig. 2 and Supplementary Fig. S2).
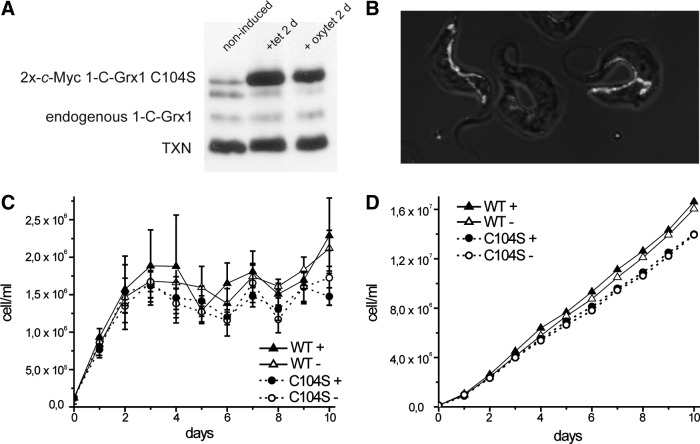
Cell lines of bloodstream parasites harboring a tetracycline (tet)-inducible ectopic copy of the C104S 1-C-Grx1 gene fused to a C-terminal cMyc2 sequence were generated, and three independent clones were phenotypically characterized. Incubation of the cells with tet or oxytetracycline (oxytet) induced the expression of C104S 1-C-Grx1-cMyc2 to a level 7–10-fold higher than that of the authentic protein (Fig. 9A and Supplementary Fig. S5). These levels are comparable to those achieved for the overexpression of the WT protein with the same host cell/vector system (17). For all clones, maximum expression of the C104S mutant occurred already 24 h upon tet/oxytet addition and remained constant for at least 28 days of continued induction (not shown). Immunofluorescence microscopy with polyclonal anti-1-C-Grx1 (Fig. 9B and Supplementary Fig. S5) or monoclonal anti-cMyc (Supplementary Fig. S5) antibodies confirmed the correct localization of the mutant protein in the single mitochondrion of the parasite. As previously observed for parasites overexpressing WT 1-C-Grx1 cMyc2 (17), bloodstream trypanosomes overexpressing the C104S mutant protein proliferated at similar rates as the corresponding noninduced cells (Fig. 9C, D; Supplementary Fig. S5).
FIG. 9.
Phenotypic characterization of bloodstream trypanosomes overexpressing an ectopic copy of C104S Tb1-C-Grx1-cMyc2. (A) Total cell extracts from 5×106 cells induced during 48 h with 1 μg/ml tet or 10 μg/ml oxytet were separated on an SDS-15% PAGE and proteins of interest revealed by Western blot. The endogenous (∼16 kDa) and 2x-c-Myc-tagged C104S (∼18 kDa) Tb1-CGrx1 were detected using guinea pig serum α-Tb1-C-Grx1. Detection of tryparedoxin (TXN) with rabbit serum α-T. brucei tryparedoxin served as a loading control (see Supplementary Fig. S5). (B) Merge image showing the mitochondrial localization of Tb1-C-Grx1 C104S in bloodstream T. brucei. Parasites induced for 48 h with 10 μg/ml oxytet were treated with Mitotracker® (mitochondrial marker), fixed, and incubated with purified guinea pig serum α-Tb1-C-Grx1 (see the Materials and Methods section). The image depicts the superimposition of both staining on a bright-field image (original pictures are shown in Supplementary Fig. S5). (C) Representative proliferation of T. brucei 514–1313 (WT) and C104S (clone 3, Supplementary Fig. S5). Cells were inoculated at 1×105 cells/ml in the culture medium with (+) or without (−) 10 μg/ml oxytet. Every 24 h, parasites were counted and diluted to the initial cell density in a fresh medium. Shown are the mean values and standard deviations from four independent experiments. (D) The cumulative cell density of the data plotted in (A). oxytet, oxytetracycline; tet, tetracycline; SDS-PAGE, sodium dodecyl sulfate–polyacrylamide gel electrophoresis.
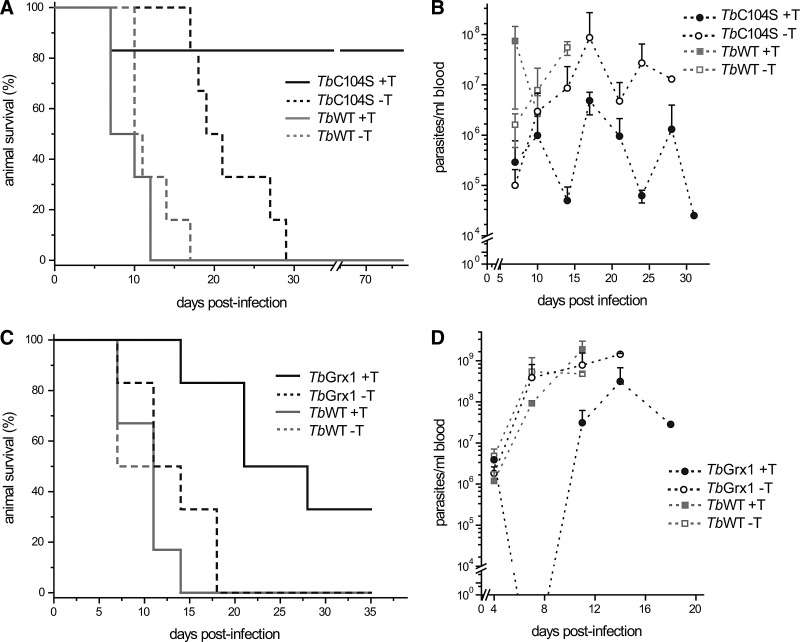
Because in vitro culture conditions not necessarily reflect the in vivo challenges the pathogenic organisms face in their mammalian hosts, the relevance of 1-C-Grx1 for parasite survival was addressed using a mouse infection model. Groups of six mice fed (+T) or not (−T) with oxytet in the drinking water were infected with the parental (TbWT) and the transgenic cell lines with tet-inducible overexpression of WT 1-C-Grx1-cMyc2 (TbGrx1; see reference 17 for details) and C104S 1-C-Grx1-cMyc2 (TbC104S, see Material and Methods for details). Disease development was evaluated by visual inspection of animal health status, parasitemia, and survival. As depicted in the Kaplan–Meier survival plots of Figure 10A, all mice infected with TbWT parasites died between days 11 and 16 postinfection regardless of the presence (TbWT+T) or absence (TbWT−T) of oxytet in the drinking water, which rules out any influence of the antibiotic in disease progression. The estimated medium survival time (50th percentile, T50) of animals infected with TbC104S−T was 19 days, 9 days longer than the control groups TbWT independently of the addition of oxytet. The slightly lower proliferation rate of noninduced transgenic C104S parasites compared to the parental cell line may account for this difference in infection onset (Fig. 9C, D and Supplementary Fig. S5). Nevertheless, all animals from this group had succumbed at day 28. In contrast, five out of six (83%) of the animals infected with TbC104S+T survived for at least 2 months (Fig. 10A). At this time point, oxytet treatment was interrupted and animal health monitored for another 2 weeks. Parasites remained undetectable in the blood, and no disease symptoms were recorded. In line with the survival profile, the parasitemia in animals infected with TbC104S+T was one to three orders of magnitude lower than that observed in the control groups (TbWT+T or−T, and TbC104S−T; Fig. 10B) and, interestingly, displayed a cyclical pattern of increase and decrease (Fig. 10B), resembling the parasitemia waves observed in the chronic stage of the infection.
FIG. 10.
Survival and parasitemia analysis of mice infected with WT, 1-C-Grx1 C104S-, and 1-C-Grx1-overexpressing T. brucei. Mature female BALB/cJ mice (n=6) were infected intraperitoneally with 104 bloodstream parasites from the isogenic cell line 514–1313 (TbWT) and transgenic cell lines for tet-inducible expression of C-myc-tagged WT (TbGrx1) and C104S mutant of Tb1-C-Grx104S (TbC104S). Groups of mice were given (+) or not (−) with 1 mg/ml oxytet in drinking water to induce in vivo the expression of the trans-genes. The parasite level in blood (parasitemia) was evaluated regularly in animals from all the groups by light microscopy. The results are depicted as Kaplan–Meier survival (A, C) and parasitemia (B, D) plots for the experiments involving the TbWT strain and the cell line TbC104S (A, B) and TbGrx1 (C, D).
An identical infection experiment was carried out with parasites overexpressing WT 1-C-Grx1-cMyc2 (TbGrx1). Again, the survival plot of mice from control groups infected with Tb-WT parasites was similar not influenced by oxytet administration (Fig. 10C). Mice from the TbGrx1-T group showed a slightly extended survival (18 days for total animal death) with respect to both control groups (TbWT+/−T: 11 days for total animal death). Nevertheless, this difference was not statistically significant (p<0.01 by log-rank test). In contrast, parasites induced in vivo to express WT 1-C-Grx1-cMyc2 (TbGrx1+T) were less infective resulting in a 33% overall survival rate after 35 days. At this experimental end point, no parasites were detected in the blood of the two living animals, which, in addition, did not present any morbidity signs. The medium survival time for this group (T50=27 days) was at least twice compared to the control groups (TbWT+/−T and TbGrx1−T; T50=10–13 days). Parasitemia developed very similarly for animals infected with TbWT dosed or not with oxytet and TbGrx1−T. In contrast, the parasite density in most animals from the TbGrx1+T group (Fig. 10D) was significantly lower and in agreement with the survival profile of this group (Fig. 10C). Interestingly, animals infected with TbGrx1+T displayed a significantly lower overall survival (33%) compared to mice infected with TbC104S+T (animal survival rate of 83%), indicating that parasites induced to express a 1-C-Grx1 lacking its active-site Cys are less competent to withstand the in vivo growth conditions.
Discussion
Proteins belonging to the thioredoxin-fold superfamily have evolved different functionalities by subtle modifications of their common architecture. In this respect, 1-C-Grxs represent a novel and ubiquitous protein subfamily with a thioredoxin-like fold where most of its members lack the disulfide oxidoreductase activity, are capable to coordinate an ISC and participate in intracellular iron homeostasis (3, 31, 69). African trypanosomes are equipped with three 1-C-Grxs (17, 24). 1-C-Grx1 probably plays an indispensable role in mitochondrial ISC biogenesis of the parasite (17), but, strikingly, only modestly rescued the function of the ortholog protein in a yeast mutant (24). This contrasted with the data for other 1-C-Grxs that readily counteract the deleterious phenotype of S. cerevisiae devoid of mitochondrial Grx5 (53). The present study was conducted to get a deeper insight into the biochemical and structural determinants for the specificity of the trypanosomal protein, as well as to assess its biological relevance for infection.
In silico analysis revealed that the 35-residue-long N-terminal extension preceding the Grx domain of Tb1-C-Grx1 is a structural feature exclusive to 1-C-Grx1 from trypanosomatids that confers conformational flexibility to the protein (see below) and results in a more stable ISC assembly. In this respect, titration experiments with LMM thiols indicate that this element may influence the conformation of residues located in the contiguous Grx domain. Additional roles for the N-terminal extension, such as mediating ISC delivery and/or interaction with putative partner proteins of the trypanosomatid ISC machinery, cannot be excluded and will be explored in future studies.
ISC reconstitution in vitro using tag-free or His-tagged WT protein with GSH, Gsp, or T(SH)2 resulted in holospecies with apparent masses of about 40 kDa, slightly higher than that of the apoprotein (∼30 kDa). The strikingly higher apparent mass (∼50 kDa) of the holoform of His-tagged Tb1-C-Grx1 (WT or C181S mutant) produced in vivo by E. coli is not yet fully understood, but may be a consequence of a conformational change or a different ISC stoichiometry generated by the bacterial ISC biosynthetic machinery. This potential ability of holo-Tb1-C-Grx1 to adopt different conformations is in agreement with a recent structural analysis of apo-/holo-EcGrx4 (36). The authors of this study even hypothesized that the conformational changes required for GSH-mediated ISC binding/release would be induced in vivo by acceptor proteins or chaperones. Moreover, it is worth to note that similar variations on gel filtration behavior were recently reported for apo and holo-EcGrx4 (77), indicating an intrinsic conformational plasticity of these proteins. Nevertheless, the precise nature and stoichiometry of the different holospecies of Tb1-C-Grx1 deserves further investigation.
Coordination of the ISC by Tb1-C-Grx1 is similar to that reported for other 1-C-Grxs. It involves the active-site cysteine and an LMM thiol, which—as shown here—can be either GSH or the parasite-specific Gsp and T(SH)2. The analysis of the 3D structures of Grxs with the ISC bound revealed that GSH is noncovalently linked to a groove with several conserved residues (see below) (15, 36, 39, 58). An interaction of the free thiol with the apoprotein may occur before ISC binding or during ISC assembly, an issue not addressed so far for any 1-C-Grx. Here we demonstrate that Tb1-C-Grx1 does interact in vitro with free GSH or T(SH)2, although with extremely low affinity (Kd≥10 mM). Taking into account that the ex vivo intracellular concentrations of GSH and T(SH)2 in T. brucei have been reported to fluctuate below 0.4 mM (2, 21), it seems very unlikely that this interaction will be predominant under physiological conditions. However, we cannot exclude the possibility that Tb1-C-Grx1 binds the thiol ligands upon ISC assembly and/or in the presence of additional cofactors such as chaperones that are recognized as important components of the cellular ISC biosynthetic machinery (54). This may also explain the higher yield and stability of the holo-Tb1-C-Grx1 produced in vivo by E. coli compared to the reconstituted protein.
In trypanosomatids, T(SH)2 replaces GSH in the maintenance of redox homeostasis and xenobiotic detoxification (10, 43) and other cellular functions requiring a LMM thiol cofactor. Recently, GSH has been proposed to be the major chelating agent of the labile iron pool (34) and to be absolutely indispensable for intracellular iron homeostasis and cytosolic ISC maturation (45). Thus, our finding that a stable T(SH)2-ISC complex can be formed in vitro poses a new biological role for T(SH)2 in the iron–sulfur metabolism of trypanosomatids, either as a ligand of ISC proteins or as an intracellular ISC carrier, as recently proposed for GSH (65). As shown here, under in vitro conditions, T(SH)2 was capable to fulfill very efficiently these functions: it acted as a thiol ligand of holo-Tb1-C-Grx1 and holo-Tb2-C-Grx1, and as a carrier for the assembly of the ISC-T(SH)2 complex into Tb2-C-Grx1, but not Tb1-C-Grx1. This remarkable difference in the capacity and affinity of the 2-C-Grx and 1-C-Grx (note also that in vitro reconstituted or in vivo synthesized holo-1-C-Grx1 was rather unstable compared to 2-C-Grx1) for ISC assembly is not surprising and can be ascribed to the distinct functions the ISC and these proteins play in the cell. For instance, 1-C-Grxs are considered not to be the primary scaffolds for ISC assembly, but important components mediating the transfer of ISC to target proteins (31, 54), and such a function would be disfavored if the ISC is tightly bound to the protein. On the other hand, 2-C-Grxs are multipurpose oxidoreductases, for some of which the ISC has been shown to negatively regulate the enzyme activity and to be released upon oxidative treatment (12, 49).
The first 3D structure of a 1-C-Grx from a pathogenic protozoa with an unusual thiol-redox system is presented. The construct used comprises the full Grx domain but lacks the N-terminal region that is absent in orthologs from other organisms. 15N-relaxation data confirmed that the truncated protein is monomeric even at the high concentration used for NMR. The global fold of the Grx domain of Tb1-C-Grx1 is remarkably similar, both in terms of length and orientation of secondary structural elements, not only to other 1-C-Grxs of known structure but also to more distantly related 2-C-Grxs, the latter proteins principally lacking the long, folded loop preceding the active site. Other structural features observed, conserved in related Grxs, and proposed to have a role in shaping the GSH pocket (25) are a cis-proline close to the active site and a β-bulge distorting the β4.
A significant structural difference between Tb1-C-Grx1 and other 1-C-Grxs is the conformation of the loop (Ser140 to Pro146) containing the cis-proline that significantly distorts the putative GSH pocket as observed by a comparative analysis with the structure of holo-EcGrx4 (Fig. 7A). As a matter of fact, we have shown that the interaction of Δ76 Tb1-C-Grx1, and also of the full-length protein, with GSH and T(SH)2 is negligible. It is worth to note that Ile145, which sterically prevents cofactor binding at the active site pocket, is described as an important element that modulates the activities of Trx-fold proteins (i.e., Trx, Grx, DsbC, and DsbG) (66). Whether in 1-C-Grxs this element plays a similar role and/or is modulated by ISC binding remains to be investigated. Notably, the different conformation of the loop observed in Tb1-C-Grx1 is imposed by a steric interaction between the side chains of Ile145 and Val136 (Fig. 7D). Despite that the presence of bulky, hydrophobic residues in these positions is a conserved feature in 1-C-Grxs, very small changes in the structure of the C-terminal end of α3, where Val136 is located, can have a significant effect on the conformation of the loop. In the other available structures of 1-C-Grxs, the side chains of residues corresponding to Val136 have a slightly different orientation, allowing the loop to adopt the observed conformation. Interestingly, in Tb1-C-Grx1, Val136 is part of a hydrophobic network connecting not only Ile145 but also Val126, Leu127, and Met103 (before the active-site Cys104). Therefore, subtle structural changes at the C-terminal end of α3 could have a significant effect on the shape of the GSH-binding pocket (Fig. 7D). The possibility of a conformational change modulating the shape of this pocket is supported by the dynamics in the μs–ms time scale observed for the cis-Pro-containing loop. Interestingly, differences in backbone dynamics between reduced and oxidized E. coli Grx1 have been suggested to play an important role in the binding of GSH (40), and more recently, a correlation between catalytic turnover and conformational fluctuations has been revealed for human Grx1 (37). The structure of Tb1-C-Grx1 shows that α3 is tightly connected to the N-terminus through different interactions involving residues located in these regions of the protein. Not surprisingly, NMR titration experiments performed on WT Tb1-C-Grx1 indicate that GSH and T(SH)2 interact with the protein, albeit with very low affinity, supporting the notion that the loop might undergo subtle conformational rearrangements in the presence of the N-terminal extension.
The indispensability of 1-C-Grx1 for the infective stage of African trypanosomes has previously been inferred from the refractoriness of trypanosomes for gene-specific knockout and knockdown (17) and recently confirmed by a genome-scale RNAi study (1). A clue about the biological role of this protein in bloodstream T. brucei was further obtained in vitro by phenotypic characterization of a cell line overexpressing WT 1-C-Grx1. While a 5-fold to 10-fold overexpression of 1-C-Grx1 did not affect cell growth (note that a similar outcome was obtained for parasites overexpressing the C104S mutant), this condition impaired the parasite's capacity to cope with stimuli that disrupt iron and redox homeostasis (17). These stimuli are a part of the host innate immune defense mechanisms aimed at controlling T. brucei colonization [for a review, see Refs. (56, 71)]. Specifically, the restriction of iron availability at a systemic level is a well-documented mechanism triggered by trypanosomal infection in the host that is accompanied by the upregulation of several genes, such as hemooxygenase 1, ferroportin, and ferritin, which regulate the iron levels (57, 60, 73). Thus, the importance of iron at the interface host–parasite and the putative role of 1-C-Grx1 in iron metabolism prompted us to investigate the relevance of this protein on a physiological context, namely, a murine infection model, and using transgenic trypanosomes that allow for the inducible overexpression of both WT and a C104S mutant of 1-C-Grx1. In both cases, parasites induced to overexpress in vivo the two proteins displayed an impaired capacity to establish and maintain animal infection as compared to the control groups (i.e., mice infected with nonoxytet-induced cell lines and WT parasites). However, parasite survival was significantly more affected in trypanosomes induced to overexpress the mutant C104S rather than the WT form of 1-C-Grx1, as evidenced from the overall lower parasite levels and increased mouse survival rate of the TbC104S T+group.
Our data do not allow concluding on the precise function played by 1-C-Grx1 in the parasite mitochondrion—for example, as a component of the ISC assembly/transfer multiprotein complex, single ISC-delivering unit, or enzymatic entity—but provides clear evidence that its role depends on its active-site Cys104, as shown for the yeast ortholog (6). The deleterious effect caused by overexpression of 1-C-Grx1 C104S may lie on a dominant-negative interaction of the apo-mutant with the endogenous WT protein and/or with its protein partners, which may affect the metabolic output of the pathway—for example, reduced ISC biogenesis with accumulation of ISC precursors and apoforms of ISC proteins—as reported for a yeast Grx5 mutant (6, 67). On the other hand, the in vivo growth defect displayed by trypanosomes with a nonphysiological higher level of exogenous WT 1-C-Grx1 may be a consequence of an enhanced synthesis of ISC proteins, an increased sequestration of ISC in 1-C-Grx1 and/or formation of nonproductive heterocomplexes with components of the biosynthetic machinery, resulting in an unbalanced metabolic output.
Based on the data reported here, the proposed lack of functional conservation between Tb1-C-Grx1 and the orthologs from different organisms (24) can now be ascribed to the biochemical and structural specialization of the trypanosomatid protein. The use of parasite-specific thiols as ISC ligands, the ISC-binding mechanisms together with other structural features discussed above are important molecular determinants for the specificity of this protein (17). Moreover, although the role of 1-C-Grx1 remains yet elusive, our results are conclusive with regard to the critical function of the mitochondrial protein in facilitating host infection and disease progression, and specifically in demonstrating Cys104 as a major modulator of protein activity in vitro and in vivo. Our future efforts will be directed at gaining further insights into the participation of 1-C-Grx1 in parasite's iron metabolism, the role of the second nonconserved Cys181, and the structure of the full-length protein.
Materials and Methods
Reagents
Trypanothione [T(SH)2] was produced as described in Comini et al. (14). Chemicals, molecular biology and cell culture reagents and kits, antibodies, chromatographic columns, and services are detailed in Supplementary Data.
DNA constructs
The E. coli expression plasmids pET-trx1b/WT Tb1-C-Grx1 and pET-trx1b/Δ76 Tb1-C-Grx1 were constructed as described in Supplementary Data and used to generate the tag-free mature (residues Gln42-Leu184; Tb1-C-Grx1 WT) and truncated (residues Met77-Leu184; Δ76 Tb1-C-Grx1) forms of T. brucei brucei 1-C-Grx1. The pET-trx1b vector was kindly provided by Gunther Stier (EMBL). The pQE30-Tb1-C-Grx1 plasmid (24) was used to express recombinant His-tagged Tb1-C-Grx1 WT. Single cysteine mutants C104S and C181S were generated using this vector and the QuickChange® Site-Directed mutagenesis Kit from Stratagene and/or by PCR with specific oligonucleotides (see Supplementary Table S2). The vector pHD1700-Tb1-C-Grx1-c-Myc2 (17), which allows for the tet-inducible expression of ectopic genes in T. brucei, served as a template to produce the C104S mutant as described in Supplemental Information.
Expression and purification of recombinant proteins
Recombinant proteins were heterologously expressed in E. coli cells essentially as described previously (8, 12, 17, 24) (for details, see Supplementary Data). In all cases, the first purification step was an Ni2+-affinity chromatography (HisTrap®; General Electric Healthcare Life Sciences [GE]). To obtain tag-free Tb1-C-Grx1 WT or Δ76, the fusion protein was expressed in E. coli BL21 (DE3) cells, purified by HisTrap, treated with 3C-type tobacco etch virus protease to cleave the N-terminal His-tagged E. coli Trx, and repurified on HisTrap. Finally, the proteins were subjected to SEC on a HiLoad Superdex 75 prep-grade column (GE). Recombinant His-tagged versions of Tb1-C-Grx1 (WT, C104S, and C181S) contain an MRGSH6GS stretch upstream of the initial Gln42, while tag-free WT and Δ76 Tb1-C-Grx1 possess an N-terminal extension of GAMG and GA, respectively, derived from the cloning strategy. Recombinant T. brucei 2-C-Grx1 was generated as described previously (12).15N and 15N/13C uniformly labeled Δ76 Tb1-C-Grx1 was produced in E. coli BL21 (DE3) cells grown in an M9 minimal medium containing [13C]glucose and/or 15NH4Cl as a sole carbon and nitrogen source, respectively. The labeled protein was purified as described above. The final SEC step was omitted, and the buffer exchanged to 50 mM sodium phosphate buffer, pH 7.0, 150 mM NaCl, and 10 mM dithiothreitol (DTT), in H2O/D2O (90:10% v/v) using a HiTrap desalting column (GE). Finally, Δ76 Tb1-C-Grx1 was concentrated up to 1 mM by ultrafiltration before subjecting to an NMR analysis (see below).
Analytical techniques
All recombinant proteins were subjected to absorption and fluorescence spectroscopy analysis, sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), analytical SEC, and mass spectrometry to confirm the purity and quality (for details, see Supplementary Data). The protein concentration was determined by the bicinchoninic acid assay (Sigma-Aldrich) using bovine serum albumin as a standard or quantified by absorption at 280 nm using the corresponding molar absorption coefficient (ɛ) determined according to Pace et al. (61): 12,190 M−1 cm−1 for tag-free WT and Δ76 Tb1-C-Grx1, 9970 M−1 cm−1 for His-tagged versions of Tb1-C-Grx1, 11,460 M−1 cm−1 for Tb2-C-Grx1, and 39,700 M−1 cm−1 for E. coli cysteine desulfurase (EcIscS, see below). The spectra were recorded in a dual-beam Varian Cary 50 spectrophotometer and Varian Cary Eclipse spectrofluorimeter (Agilent Technologies) with a thermostatized multicell holder. A J-810 spectropolarimeter (Jasco, Inc.), equipped with a temperature-controlled cell holder, was used for CD measurements. Analytical SEC was performed in a Superdex 75 10/300 GL column (GE) coupled to ÄKTA-FPLC system (GE) with online UV–visible detection. Standard globular proteins (6.5–75 kDa; GE kit) were used for the calibration of the columns.
NMR spectroscopy
The NMR spectra for assignment and structure calculation were acquired at 298 K on 900, 800, and 500 Avance Bruker spectrometers equipped with triple-resonance cryoprobes. The NMR experiments used for these purposes are summarized in Supplementary Table S3. Data were processed and the spectra were analyzed using TOPSPIN 1.3 (Bruker BioSpin GmbH) and CARA 1.9 (http://cara.nmr.ch), respectively. The backbone dynamic properties of Δ76 Tb1-C-Grx1 and titration assays with LMM thiols were measured by 15N-HSQC using a Bruker DMX 600 MHz spectrometer with a room temperature (RT) probe. For the titration experiments, stock solutions of GSH and T(SH)2 were prepared in 50 mM sodium phosphate buffer, pH 7.0, 150 mM NaCl, and 10 mM DTT in H2O/D2O (90:10% v/v) and added to the NMR tube containing the full-length or the truncated 1-C-Grx1 (protein concentration 0.2–0.4 mM) to obtain different thiol:protein ratios. The combined amide chemical-shift perturbation (Δδ) was calculated as follows: Δδ=[((ΔδH)2+(ΔδN/5)2)/2]1/2. Distance constraints for structure determination were obtained from the 15N-edited and 13C-edited 3D NOESY-HSQC spectra. Backbone dihedral angle constraints were derived from 15N,13C’,13Cα,13Cβ, and Hα chemical shifts using TALOS+(72). An automated method, based on the ATNOS/CANDID algorithms (32, 33), was used for NOESY peak picking and assignment, implemented in the UNIO’10 package (28), combined with the torsion angle dynamics for structure calculation using CYANA 2.1 (29). A cis-conformation of the peptide bond preceding Pro146 was imposed for structure calculation. The 20 conformers with the lowest residual target function values were subjected to refinement by restrained molecular dynamics in explicit water, with the AMBER package through the AMPS–NMR interface (9). The relaxation properties were predicted by hydrodynamic calculations with the software HYDRONMR (7) using a standard value of 3.3Å for the atomic radius element. The resonance assignment and the structural coordinates of Δ76 Tb1-C-Grx1 have been deposited at the BioMagResBank (www.bmrb.wisc.edu) with the accession code 18,485 and the Protein Data Bank (www.pdb.org) with entry code 2ltk, respectively.
Isolation, reconstitution, and characterization of holoprotein species
To isolate the holoform of His-tagged Tb1-C-Grx1 directly from recombinant E. coli, 100 μM FeCl3 was added to the culture medium before induction, and all buffers used for purification were extensively degassed and bubbled with argon. The protein eluted from the HisTrap column was stored under argon-saturated atmosphere and protected from light. The fresh eluates were analyzed by UV–visible spectroscopy and SEC. In vitro ISC reconstitution of holoproteins was done with tag-free or His-tagged proteins as reported (8, 17), with minor modifications. Briefly, 50–100 μM protein was incubated under an argon atmosphere with 5 mM DTT, 10 μM pyridoxal-5′-phosphate, and 0.5–1 mM LMM thiol [GSH, Gsp, or T(SH)2] in 50 mM sodium phosphate, pH 7.8, and 300 mM NaCl, for 10 min at RT. Then, 100–500 μM Fe(NH4)2(SO4)2 and an identical concentration of cysteine plus 2.5–5 μM EcIscS were added. The reaction was allowed to proceed at RT under a constant argon flush in closed polypropylene tubes. After 1-h incubation, the tubes were centrifuged 5 min at ≥14,000 g, and the supernatant was subjected to UV–visible spectroscopy, CD spectroscopy, or analytical SEC.
Parasite culture and transgenic cell lines
A bloodstream T. brucei (strain Lister 427) cell line 514–1313, suitable for the tet-inducible expression systems, was cultivated in an HMI-9 medium supplemented with 10% v/v fetal bovine serum and antibiotics (0.2 μg/ml phleomycin, 2.5 μg/ml gentamycin, and 5 μg/ml hygromycin B) (17). The preparation of cell lines with a tet-inducible ectopic copy of Tb1-C-Grx1 WT was described in an earlier publication (17). To generate cells with controlled expression of Tb1-C-Grx1C104S, 3×107 trypanosomes from the parental cell line 514–1313 were electroporated with 10–100 μg of NotI-linearized pHD1700-Tb1-C-Grx1C104S, using an ECM 630 (BTX) electroporator and a 2-mm-gap electroporation cuvette. The electroshock settings and hygromycin-dependent selection of transfectans were performed as described previously (17). The expression of the ectopic genes was induced by adding 1–10 μg/ml tet or oxytet to the culture medium and confirmed by Western blot (see below).
Western blot and immunofluorescence analysis
Western blots were performed on total cell extracts from quantified parasites that were previously separated under reducing conditions on an SDS/12%–15% PAGE. Guinea pig and rabbit polyclonal antibodies against Tb1-C-Grx1 (17) and T. brucei tryparedoxin (TXN, 16), respectively, and monoclonal antibody anti-c-Myc (clone 9E10; Roche) were used at dilutions of 1:500, 1:2,000–5,000, and 1:400, respectively. The corresponding horseradish peroxidase-conjugated goat anti-guinea pig IgG, anti-rabbit IgG, and anti-mouse IgG secondary antibodies were diluted at a 1:10,000 dilution. Reactive bands were detected by chemiluminescence using ECL plus (GE Healthcare) and quantified by a densitometric analysis using the program ImageJ (NCBI; see Supplementary Data for details). For staining of mitochondria, parasites were harvested by centrifugation at 2000 g for 10 min at RT, incubated for 25 min at 37°C in 10 ml of fresh medium containing 0.25 μM MitoTracker® RedCMXRos, washed twice with phosphate-buffered saline (PBS), and fixed in 4% (w/v) paraformaldehyde for 18 min at RT. For subsequent indirect immunofluorescence analysis, the fixed cells were handled essentially as described in (17) and then incubated 1 h with purified guinea pig anti-Tb1-C-Grx1 (1:500) and mouse monoclonal anti-c-Myc (1:250). Alexa-Fluor488-labeled goat anti-guinea pig IgG and anti-mouse IgG at a dilution 1:1,000 were used as secondary antibodies. Nucleic acids were stained with TOPRO (Invitrogen). The parasites were visualized with the spectral confocal microscope Leica TCS SP5.
Phenotypic analysis of infective T. brucei overexpressing 1-C-Grx1 C104S
The growth phenotype of bloodstream T. brucei stably transfected with pHD1700-Tb1-C-Grx1C104S-c-Myc2 was evaluated for the three different clones. Batch and continuous culture experiments were initiated by inoculating the parasites at 104 or 105 cells/ml in a fresh culture medium containing or not 1 μg/ml tet or 10 μg/ml oxytet. For batch cultures, the cell density was assessed, and oxytet was replaced on a daily basis. For a continuous culture, cells were counted every 24 and 48 h and then diluted to the initial cell density in a fresh medium. The expression of Tb1-C-Grx1C104S was verified in different cultures and time points by Western blot.
Animal infection model
Groups of 6–8-week-old female mice from the strain BALB/cJ were housed on individual ventilated cages with negative pressure (Sealsafe rack, Tecniplast) in controlled environmental conditions with food and water administered ad libitum. The experimental procedures complied with the national law and international FELASA guidelines for the laboratory animal's protocols and were approved by the Institut Pasteur de Montevideo Animal Care Committee. The infectivity of the bloodstream T. brucei cell lines 1-C-Grx1-c-Myc2 (TbGrx1), 1-C-Grx1-C104S-c-Myc2 (TbC104S), and WT (parental cell line 514–1313, TbWT) was evaluated in mice fed or not with 1 mg/ml oxytet in drinking water. Fresh oxytet was given to animals starting at day −3 (before infection) and replaced every 48 h until indicated. These experimental conditions proved successful to induce in vivo the expression of a tet-inducible reporter gene, namely, a modified green fluorescence protein, exogenously incorporated to bloodstream T. brucei (Sardi and Comini, unpublished). Six mice per group were infected with a single intraperitoneal injection of 104 infective parasites suspended in 0.3 ml PBS. Animals' health status and survival were monitored in a daily basis, searching for signs of discomfort or pain. The parasite levels in mice were determined regularly in blood samples taken from the submandibular sein as described earlier (68). Mice showing an impaired health status and/or with a parasite load ≥108 cells/ml of blood were euthanized. Data are depicted as Kaplan–Meier survival plots and parasitemia plots. The log-rank test was used for statistical comparison of survival curves.
Supplementary Material
Abbreviations Used
- 1-C-Grx
monothiol glutaredoxin
- 2-C-Grx
dithiol glutaredoxin
- CD
circular dichroism
- DTDPy
4,4′-dithiodipyridine
- DTNB
5,5′-dithio-bis-2-nitrobenzoic acid
- DTT
dithiothreitol
- EDTA
ethylenediaminetetraacetic acid
- Grx
glutaredoxin
- GSH
glutathione
- Gsp
glutathionylspermidine
- HSQC
heteronuclear single-quantum coherence
- IPTG
isopropyl-β-D-thiogalactopyranoside
- ISC
iron–sulfur cluster
- LMM
low molecular mass
- MTS
mitochondrial targeting sequence
- NMR
nuclear magnetic resonance
- NOE
nuclear Overhauser effect
- oxytet
oxytetracycline
- PBS
phosphate-buffered saline
- PCR
polymerase chain reaction
- PLP
pyridoxal-5′-phosphate
- PMSF
phenylmethylsulfonyl fluoride
- RT
room temperature
- SDS-PAGE
sodium dodecyl sulfate–polyacrylamide gel electrophoresis
- SEC
size-exclusion chromatography
- T(SH)2
trypanothione
- Tb
Trypanosoma brucei
- tet
tetracycline
- TEV
tobacco etch virus
- TLCK
Nα-tosyl-l-lysine chloromethyl ketone
- WT
wild type
Acknowledgments
This work was supported by a grant from the Agencia Nacional de Investigación e Innovación (ANII; Innova Uruguay, Agreement No. DCI-ALA/2007/19.040 between Uruguay and the European Commission) to MAC. BM and MAC acknowledge the financial support from ANII (PhD and short-term-visit fellowships) and PEDECIBA. Luciana Fleitas (Lab. Redox Biology of Trypanosomes), Magdalena Portela (Analytical Biochemistry and Proteomic Unit, Institut Pasteur de Montevideo), Gabriel Fernández (Transgenic and Animal Experimentation Unit, Institut Pasteur de Montevideo), and Lorenzo Gesiot (Padova University) are acknowledged for technical assistance in biochemistry, mass spectrometry, animal manipulation, and NMR experiments, respectively. Natalie Dirdjaja (Biochemie Zentrum Heidelberg) and Jochen Rettig (present address University of Bern, Swiss) are gratefully acknowledged for assistance during preparation of trypanothione and DNA constructs, respectively. Financial support by the Access to Research Infrastructures activity in the 7th Framework Programme of the EC/Project number: 261863, Bio-NMR is gratefully acknowledged for providing access to NMR spectrometers; Dr. Frank Löhr and the staff of the Center for Biomolecular Magnetic Resonance at the J.-W. Goethe University in Frankfurt for onsite assistance and the WeNMR project (European FP7 e-Infrastructure grant, contract no. 261572, www.wenmr.eu), for the use of Web portals and computing facilities.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Alsford S. Turner DJ. Obado SO. Sanchez-Flores A. Glover L. Berriman M. Hertz-Fowler C. Horn D. High-throughput phenotyping using parallel sequencing of RNA interference targets in the African trypanosome. Genome Res. 2011;21:915–924. doi: 10.1101/gr.115089.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ariyanayagam MR. Fairlamb AH. Ovothiol and trypanothione as antioxidants in trypanosomatids. Mol Biochem Parasitol. 2001;115:189–198. doi: 10.1016/s0166-6851(01)00285-7. [DOI] [PubMed] [Google Scholar]
- 3.Atkinson HJ. Babbitt PC. An atlas of the thioredoxin fold class reveals the complexity of function-enabling adaptations. PLoS Comput Biol. 2009;5:e1000541. doi: 10.1371/journal.pcbi.1000541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bandyopadhyay S. Gama F. Molina-Navarro MM. Gualberto JM. Claxton R. Naik SG. Huynh BH. Herrero E. Jacquot JP. Johnson MK. Rouhier N. Chloroplast monothiol glutaredoxins as scaffold proteins for the assembly and delivery of [2Fe-2S] clusters. EMBO J. 2008;27:1122–1133. doi: 10.1038/emboj.2008.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barbato G. Ikura M. Kay LE. Pastor RW. Bax A. Backbone dynamics of calmodulin studied by 15N relaxation using inverse detected two-dimensional NMR spectroscopy: the central helix is flexible. Biochemistry. 1992;31:5269–5278. doi: 10.1021/bi00138a005. [DOI] [PubMed] [Google Scholar]
- 6.Bellí G. Polaina J. Tamarit J. De La Torre MA. Rodríguez-Manzaneque MT. Ros J. Herrero E. Structure-function analysis of yeast Grx5 monothiol glutaredoxin defines essential amino acids for the function of the protein. J Biol Chem. 2002;277:37590–37596. doi: 10.1074/jbc.M201688200. [DOI] [PubMed] [Google Scholar]
- 7.Bernadò P. de la Torre JG. Pons M. Interpretation of 15N NMR relaxation data of globular protein using hydrodynamic calculations with HYDRONMR. J Biomol NMR. 2002;23:139–150. doi: 10.1023/a:1016359412284. [DOI] [PubMed] [Google Scholar]
- 8.Berndt C. Hudemann C. Hanschmann E-M. Axelsson R. Holmgren A. Lillig CH. How does iron-sulfur cluster coordination regulate the activity of human glutaredoxin 2? Antioxid Redox Signal. 2007;9:151–157. doi: 10.1089/ars.2007.9.151. [DOI] [PubMed] [Google Scholar]
- 9.Bertini I. Case DA. Ferella L. Giachetti A. Rosato A. A Grid-enabled web portal for NMR structure refinement with AMBER. Bioinformatics. 2011;27:2384–2390. doi: 10.1093/bioinformatics/btr415. [DOI] [PubMed] [Google Scholar]
- 10.Bocedi A. Dawood KF. Fabrini R. Federici G. Gradoni L. Pedersen JZ. Ricci G. Trypanothione efficiently intercepts nitric oxide as a harmless iron complex in trypanosomatid parasites. FASEB J. 2010;24:1035–1042. doi: 10.1096/fj.09-146407. [DOI] [PubMed] [Google Scholar]
- 11.Bushweller JH. Billeter M. Holmgren A. Wüthrich K. The nuclear magnetic resonance s1lution structure of the mixed disulfide between Escherichia coli glutaredoxin(C14S) and glutathione. J Mol Biol. 1994;235:1585–1597. doi: 10.1006/jmbi.1994.1108. [DOI] [PubMed] [Google Scholar]
- 12.Ceylan S. Seidel V. Ziebart N. Berndt C. Dirdjaja N. Krauth-Siegel RL. The dithiol gl1utaredoxins of african trypanosomes have distinct roles and are closely linked to the unique trypanothione metabolism. J Biol Chem. 2010;285:35224–35237. doi: 10.1074/jbc.M110.165860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng N-H. AtGRX4, an Arabidopsis chloroplastic monothiol glutaredoxin, is able to suppress yeast grx5 mutant phenotypes and respond to oxidative stress. FEBS Lett. 2008;582:848–854. doi: 10.1016/j.febslet.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 14.Comini MA. Dirdjaja N. Kaschel M. Krauth-Siegel RL. Preparative enzymatic synthesis of trypanothione and trypanothione analogues. Int J Parasitol. 2009;39:1059–1062. doi: 10.1016/j.ijpara.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 15.Comini MA. Krauth-Siegel RL. Bellanda M. Mono- and dithiol glutaredoxins in the trypanothione based redox metabolism of pathogenic trypanosomes. Antioxid Redox Signal. 2013;19:708–722. doi: 10.1089/ars.2012.4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Comini MA. Krauth-Siegel RL. Flohé L. Depletion of the thioredoxin homologue trparedoxin impairs antioxidative defence in African trypanosomes. Biochem J. 2007;402:43–49. doi: 10.1042/BJ20061341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Comini MA. Rettig J. Dirdjaja N. Hanschmann E-M. Berndt C. Krauth-Siegel RL. Monothiol glutaredoxin-1 is an essential iron-sulfur protein in the mitochondrion of African trypanosomes. J Biol Chem. 2008;283:27785–27798. doi: 10.1074/jbc.M802010200. [DOI] [PubMed] [Google Scholar]
- 18.Couturier J. Koh CS. Zaffagnini M. Winger AM. Gualberto JM. Corbier C. Decottignies P. Jacquot J-P. Lemaire SD. Didierjean C. Rouhier N. Structure-function relationship of the chloroplastic glutaredoxin S12 with an atypical WCSYS active site. J Biol Chem. 2009;284:9299–9310. doi: 10.1074/jbc.M807998200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Couturier J. Stroher E. Albetel A-N. Roret T. Muthuramalingam M. Tarrago L. Seidel T. Tsan P. Jacquot J-P. Johnson MK. Dietz K-J. Didierjean C. Rouhier N. Arabidopsis chloroplastic glutaredoxin C5 as a model to explore the molecular determinants for iron-sulfur cluster binding into glutaredoxins. J Biol Chem. 2011;286:27515–27527. doi: 10.1074/jbc.M111.228726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delaglio F. Grzesiek S. Vuister GW. Zhu G. Pfeifer J. Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 21.Fairlamb AH. Henderson GB. Bacchi CJ. Cerami A. In vivo effects of difluoromethylornithine on trypanothione and polyamine levels in bloodstream forms of Trypanosoma brucei. Mol Biochem Parasitol. 1987;24:185–191. doi: 10.1016/0166-6851(87)90105-8. [DOI] [PubMed] [Google Scholar]
- 22.Feng Y. Zhong N. Rouhier N. Hase T. Kusunoki M. Jacquot J-P. Jin C. Xia B. Structural insight into poplar glutaredoxin C1 with a bridging iron-sulfur cluster at the active site. Biochemistry. 2006;45:7998–8008. doi: 10.1021/bi060444t. [DOI] [PubMed] [Google Scholar]
- 23.Fernandes AP. Fladvad M. Berndt C. Andrésen C. Lillig CH. Neubauer P. Sunnerhagen M. Holmgren A. Vlamis-Gardikas A. A novel monothiol glutaredoxin (Grx4) from Escherichia coli can serve as a substrate for thioredoxin reductase. J Biol Chem. 2005;280:24544–24552. doi: 10.1074/jbc.M500678200. [DOI] [PubMed] [Google Scholar]
- 24.Filser M. Comini MA. Molina-Navarro MM. Dirdjaja N. Herrero E. Krauth-Siegel RL. Cloning, functional analysis, and mitochondrial localization of Trypanosoma brucei monothiol glutaredoxin-1. Biol Chem. 2008;389:21–32. doi: 10.1515/BC.2007.147. [DOI] [PubMed] [Google Scholar]
- 25.Fladvad M. Bellanda M. Fernandes AP. Mammi S. Vlamis-Gardikas A. Holmgren A. Sunnerhagen M. Molecular mapping of functionalities in the solution structure of reduced Grx4, a monothiol glutaredoxin from Escherichia coli. J Biol Chem. 2005;280:24553–24561. doi: 10.1074/jbc.M500679200. [DOI] [PubMed] [Google Scholar]
- 26.Gallogly MM. Starke DW. Mieyal JJ. Mechanistic and kinetic details of catalysis of thiol-disulfide exchange by glutaredoxins and potential mechanisms of regulation. Antioxid Redox Signal. 2009;11:1059–1081. doi: 10.1089/ars.2008.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gerdes SY. Scholle MD. Campbell JW. Balázsi G. Ravasz E. Daugherty MD. Somera AL. 7yrpides NC. Anderson I. Gelfand MS. Bhattacharya A. Kapatral V. D'Souza M. Baev MV. Grechkin Y. Mseeh F. Fonstein MY. Overbeek R. Barabási A-L. Oltvai ZN. Osterman AL. Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J Bacteriol. 2003;185:5673–5684. doi: 10.1128/JB.185.19.5673-5684.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guerry P. Herrmann T. Comprehensive automation for NMR structure determination of proteins. Methods Mol Biol. 2012;831:429–451. doi: 10.1007/978-1-61779-480-3_22. [DOI] [PubMed] [Google Scholar]
- 29.Güntert P. Mumenthaler C. Wüthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J Mol Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 30.Haunhorst P. Berndt C. Eitner S. Godoy JR. Lillig CH. Characterization of the human monothiol glutaredoxin 3 (PICOT) as iron-sulfur protein. Biochem Biophys Res Commun. 2010;394:372–376. doi: 10.1016/j.bbrc.2010.03.016. [DOI] [PubMed] [Google Scholar]
- 31.Herrero E. de la Torre-Ruiz MA. Monothiol glutaredoxins: a common domain for multiple functions. Cell Mol Life Sci. 2007;64:1518–1530. doi: 10.1007/s00018-007-6554-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herrmann T. Güntert P. Wüthrich K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J Mol Biol. 2002;319:209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- 33.Herrmann T. Güntert P. Wüthrich K. Protein NMR structure determination with automated NOE-identification in the NOESY spectra using the new software ATNOS. J Biomol NMR. 2002;24:171–189. doi: 10.1023/a:1021614115432. [DOI] [PubMed] [Google Scholar]
- 34.Hider RC. Kong XL. Glutathione: a key component of the cytoplasmic labile iron pool. Biometals. 2011;24:1179–1187. doi: 10.1007/s10534-011-9476-8. [DOI] [PubMed] [Google Scholar]
- 35.Hoffmann B. Uzarska MA. Berndt C. Godoy JR. Haunhorst P. Lillig CH. Lill R. Mühlenhoff U. The multidomain thioredoxin-monothiol glutaredoxins represent a distinct functional group. Antioxid Redox Signal. 2011;15:19–30. doi: 10.1089/ars.2010.3811. [DOI] [PubMed] [Google Scholar]
- 36.Iwema T. Picciocchi A. Traore DAK. Ferrer J-L. Chauvat F. Jacquamet L. Structural basis for delivery of the intact [Fe2S2] cluster by monothiol glutaredoxin. Biochemistry. 2009;48:6041–6043. doi: 10.1021/bi900440m. [DOI] [PubMed] [Google Scholar]
- 37.Jensen KS. Winther JR. Teilum K. Millisecond dynamics in glutaredoxin during catalytic turnover is dependent on substrate binding and absent in the resting states. J Am Chem Soc. 2011;133:3034–3042. doi: 10.1021/ja1096539. [DOI] [PubMed] [Google Scholar]
- 38.Johansson C. Kavanagh KL. Gileadi O. Oppermann U. Reversible sequestration of active site cysteines in a 2Fe-2S-bridged dimer provides a mechanism for glutaredoxin 2 regulation in human mitochondria. J Biol Chem. 2007;282:3077–3082. doi: 10.1074/jbc.M608179200. [DOI] [PubMed] [Google Scholar]
- 39.Johansson C. Roos AK. Montano SJ. Sengupta R. Filippakopoulos P. Guo K. von Delft F. 9olmgren A. Oppermann U. Kavanagh KL. The crystal structure of human GLRX5: iron-sulfur cluster co-ordination, tetrameric assembly and monomer activity. Biochem J. 2011;433:303–311. doi: 10.1042/BJ20101286. [DOI] [PubMed] [Google Scholar]
- 40.Kelley JJ., 3rd Caputo TM. Eaton SF. Laue TM. Bushweller JH. Comparison of backbone dynamics of reduced and oxidized Escherichia coli glutaredoxin-1 using 15N NMR relaxation measurements. Biochemistry. 1997;36:5029–5044. doi: 10.1021/bi962181g. [DOI] [PubMed] [Google Scholar]
- 41.Kim K-D. Chung W-H. Kim H-J. Lee K-C. Roe J-H. Monothiol glutaredoxin Grx5 interacts with Fe-S scaffold proteins Isa1 and Isa2 and supports Fe-S assembly and DNA integrity in mitochondria of fission yeast. Biochem Biophys Res Commun. 2010;392:467–472. doi: 10.1016/j.bbrc.2010.01.051. [DOI] [PubMed] [Google Scholar]
- 42.Kim KD. Kim HJ. Lee KC. Roe JH. Multi-domain CGFS-type glutaredoxin Grx4 regulates iron homeostasis via direct interaction with a repressor Fep1 in fission yeast. Biochem Biophys Res Commun. 2011;408:609–614. doi: 10.1016/j.bbrc.2011.04.069. [DOI] [PubMed] [Google Scholar]
- 43.Krauth-Siegel RL. Comini MA. Redox control in trypanosomatids, parasitic protozoa with trypanothione-based thiol metabolism. Biochim Biophys Acta. 2008;1780:1236–1248. doi: 10.1016/j.bbagen.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 44.Kumánovics A. Chen OS. Li L. Bagley D. Adkins EM. Lin H. Dingra NN. Outten CE. Keller G. Winge D. Ward DM. Kaplan J. Identification of FRA1 and FRA2 as genes involved in regulating the yeast iron regulon in response to decreased mitochondrial iron-sulfur cluster synthesis. J Biol Chem. 2008;283:10276–10286. doi: 10.1074/jbc.M801160200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kumar C. Igbaria A. D'Autreaux B. Planson A-G. Junot C. Godat E. Bachhawat AK. Delaunay-Moisan A. Toledano MB. Glutathione revisited: a vital function in iron metabolism and ancillary role in thiol-redox control. EMBO J. 2011;30:2044–2056. doi: 10.1038/emboj.2011.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li L. Cheng N. Hirschi KD. Wang X. Structure of Arabidopsis chloroplastic monothiol glutaredoxin AtGRXcp. Acta Crystallogr D Biol Crystallogr. 2010;66:725–732. doi: 10.1107/S0907444910013119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li H. Mapolelo DT. Dingra NN. Naik SG. Lees NS. Hoffman BM. Riggs-Gelasco PJ. Huynh BH. Johnson MK. Outten CE. The yeast iron regulatory proteins Grx3/4 and Fra2 form heterodimeric complexes containing a [2Fe-2S] cluster with cysteinyl and histidyl ligation. Biochemistry. 2009;48:9569–9581. doi: 10.1021/bi901182w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lillig CH. Berndt C. Holmgren A. Glutaredoxin systems. Biochim Biophys Acta. 2008;1780:1304–1317. doi: 10.1016/j.bbagen.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 49.Lillig CH. Berndt C. Vergnolle O. Lönn ME. Hudemann C. Bill E. Holmgren A. Characterization of human glutaredoxin 2 as iron-sulfur protein: a possible role as redox sensor. Proc Natl Acad Sci U S A. 2005;102:8168–8173. doi: 10.1073/pnas.0500735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luo M. Jiang Y-L. Ma X-X. Tang Y-J. He Y-X. Yu J. Zhang R-G. Chen Y. Zhou C-Z. Structural and biochemical characterization of yeast monothiol glutaredoxin Grx6. J Mol Biol. 2010;398:614–622. doi: 10.1016/j.jmb.2010.03.029. [DOI] [PubMed] [Google Scholar]
- 51.Melchers J. Dirdjaja N. Ruppert T. Krauth-Siegel RL. Glutathionylation of trypanosomal thiol redox proteins. J Biol Chem. 2007;282:8678–8694. doi: 10.1074/jbc.M608140200. [DOI] [PubMed] [Google Scholar]
- 52.Mesecke N. Mittler S. Eckers E. Herrmann JM. Deponte M. Two novel monothiol glutaredoxins from Saccharomyces cerevisiae provide further insight into iron-sulfur cluster binding, oligomerization, and enzymatic activity of glutaredoxins. Biochemistry. 2008;47:1452–1463. doi: 10.1021/bi7017865. [DOI] [PubMed] [Google Scholar]
- 53.Molina-Navarro MM. Casas C. Piedrafita L. Bellí G. Herrero E. Prokaryotic and eukaryotic monothiol glutaredoxins are able to perform the functions of Grx5 in the biogenesis of Fe/S clusters in yeast mitochondria. FEBS Lett. 2006;580:2273–2280. doi: 10.1016/j.febslet.2006.03.037. [DOI] [PubMed] [Google Scholar]
- 54.Mühlenhoff U. Gerber J. Richhardt N. Lill R. Components involved in assembly and dislocation of iron-sulfur clusters on the scaffold protein Isu1p. EMBO J. 2003;22:4815–4825. doi: 10.1093/emboj/cdg446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mühlenhoff U. Molik S. Godoy JR. Uzarska MA. Richter N. Seubert A. Zhang Y. Stubbe J. Pierrel F. Herrero E. Lillig CH. Lill R. Cytosolic monothiol glutaredoxins function in intracellular iron sensing and trafficking via their bound iron-sulfur cluster. Cell Metab. 2010;12:373–385. doi: 10.1016/j.cmet.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Namangala B. Contribution of innate immune responses towards resistance to African trypanosome infections. Scand J Immunol. 2012;75:5–15. doi: 10.1111/j.1365-3083.2011.02619.x. [DOI] [PubMed] [Google Scholar]
- 57.Nishimura K. Nakaya H. Nakagawa H. Matsuo S. Ohnishi Y. Yamasaki S. Effect of Trypanosoma brucei brucei on erythropoiesis in infected rats. J Parasitol. 2011;97:88–93. doi: 10.1645/GE-2522.1. [DOI] [PubMed] [Google Scholar]
- 58.Noguera V. Walker O. Rouhier N. Jacquot J-P. Krimm I. Lancelin J-M. NMR reveals a novel glutaredoxin-glutaredoxin interaction interface. J Mol Biol. 2005;353:629–641. doi: 10.1016/j.jmb.2005.08.035. [DOI] [PubMed] [Google Scholar]
- 59.Oh JG. Jeong D. Cha H. Kim JM. Lifirsu E. Kim J. Yang DK. Park CS. Kho C. Park S. Yoo YJ. Kim DH. Kim J. Hajjar RJ. Park WJ. PICOT increases cardiac contractility by inhibiting PKCζ activity. J Mol Cell Cardiol. 2012;53:53–63. doi: 10.1016/j.yjmcc.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 60.Omotainse SO. Anosa VO. Erythrocyte response to Trypanosoma brucei in experimentally infected dogs. Rev Elev Med Vet Pays Trop. 1992;45:279–283. [PubMed] [Google Scholar]
- 61.Pace CN. Vajdos F. Fee L. Grimsley G. Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Picciocchi A. Saguez C. Boussac A. Cassier-Chauvat C. Chauvat F. CGFS-type monothiol glutaredoxins from the cyanobacterium Synechocystis PCC6803 and other evolutionary distant model organisms possess a glutathione-ligated [2Fe-2S] cluster. Biochemistry. 2007;46:15018–15026. doi: 10.1021/bi7013272. [DOI] [PubMed] [Google Scholar]
- 63.Qi W. Cowan JA. Mechanism of glutaredoxin-ISU [2Fe-2S] cluster exchange. Chem Commun (Camb) 2011;47:4989–4991. doi: 10.1039/c0cc05079b. [DOI] [PubMed] [Google Scholar]
- 64.Qi Y. Grishin NV. Structural classification of thioredoxin-like fold proteins. Proteins. 2005;58:376–388. doi: 10.1002/prot.20329. [DOI] [PubMed] [Google Scholar]
- 65.Qi W. Li J. Chain CY. Pasquevich GA. Pasquevich AF. Cowan JA. Glutathione complexed fe-s centers. J Am Chem Soc. 2012;134:10745–10748. doi: 10.1021/ja302186j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ren G. Stephan D. Xu Z. Zheng Y. Tang D. Harrison RS. Kurz M. Jarrott R. Shouldice SR. Hiniker A. Martin JL. Heras B. Bardwell JCA. Properties of the thioredoxin fold superfamily are modulated by a single amino acid residue. J Biol Chem. 2009;284:10150–10159. doi: 10.1074/jbc.M809509200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rodríguez-Manzaneque MT. Tamarit J. Bellí G. Ros J. Herrero E. Grx5 is a mitochondrial glutaredoxin required for the activity of iron/sulfur enzymes. Mol Biol Cell. 2002;13:1109–11721. doi: 10.1091/mbc.01-10-0517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Roldán A. Comini MA. Crispo M. Krauth-Siegel RL. Lipoamide dehydrogenase is essential for both bloodstream and procyclic Trypanosoma brucei. Mol Microbiol. 2011;81:623–639. doi: 10.1111/j.1365-2958.2011.07721.x. [DOI] [PubMed] [Google Scholar]
- 69.Rouhier N. Couturier J. Johnson MK. Jacquot J-P. Glutaredoxins: roles in iron homeostasis. Trends Biochem Sci. 2010;35:43–52. doi: 10.1016/j.tibs.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rouhier N. Unno H. Bandyopadhyay S. Masip L. Kim S-K. Hirasawa M. Gualberto JM. Lattard V. Kusunoki M. Knaff DB. Georgiou G. Hase T. Johnson MK. Jacquot J-P. Functional, structural, and spectroscopic characterization of a glutathione-ligated [2Fe-2S] cluster in poplar glutaredoxin C1. Proc Natl Acad Sci U S A. 2007;104:7379–7384. doi: 10.1073/pnas.0702268104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Salmon D. Vanwalleghem G. Morias Y. Denoeud J. Krumbholz C. Lhommé F. Bachmaier S. Kador M. Gossmann J. Dias FB. De Muylder G. Uzureau P. Magez S. Moser M. De Baetselier P. Van Den Abbeele J. Beschin A. Boshart M. Pays E. Adenylate cyclases of Trypanosoma brucei inhibit the innate immune response of the host. Science. 2012;337:463–466. doi: 10.1126/science.1222753. [DOI] [PubMed] [Google Scholar]
- 71a.Sardi F. Manta B. Portillo-Ledesma S. Knoops B. Comini MA. Ferrer-Sueta G. Determination of acidity and nucleophilicity in thiols by reaction with monobromobimane and fluorescence detection. Anal Biochem. 2013;435:74–82. doi: 10.1016/j.ab.2012.12.017. [DOI] [PubMed] [Google Scholar]
- 72.Shen Y. Bax A. Prediction of Xaa-Pro peptide bond conformation from sequence and chemical shifts. J Biomol NMR. 2010;46:199–204. doi: 10.1007/s10858-009-9395-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stijlemans B. Vankrunkelsven A. Brys L. Raes G. Magez S. De Baetselier P. Scrutinizing the mechanisms underlying the induction of anemia of inflammation through GPI-mediated modulation of macrophage activation in a model of African trypanosomiasis. Microbes Infect. 2010;12:389–399. doi: 10.1016/j.micinf.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 74.Wingert RA. Galloway JL. Barut B. Foott H. Fraenkel P. Axe JL. Weber GJ. Dooley K. Davidson AJ. Schmid B. Schmidt B. Paw BH. Shaw GC. Kingsley P. Palis J. Schubert H. Chen O. Kaplan J. Zon LI. Deficiency of glutaredoxin 5 reveals Fe-S clusters are required for vertebrate haem synthesis. Nature. 2005;436:1035–1039. doi: 10.1038/nature03887. [DOI] [PubMed] [Google Scholar]
- 75.Wüthrich K. NMR of Proteins and Nucleic Acids. New York: John Wiley & Sons; 1986. [Google Scholar]
- 76.Ye H. Jeong SY. Ghosh MC. Kovtunovych G. Silvestri L. Ortillo D. Uchida N. Tisdale J. Camaschella C. Rouault TA. Glutaredoxin 5 deficiency causes sideroblastic anemia by specifically impairing heme biosynthesis and depleting cytosolic iron in human erythroblasts. J Clin Invest. 2010;120:1749–1761. doi: 10.1172/JCI40372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yeung N. Gold B. Liu NL. Prathapam R. Sterling HJ. Willams ER. Butland G. The E. coli monothiol glutaredoxin GrxD forms homodimeric and heterodimeric FeS cluster containing complexes. Biochemistry. 2011;50:8957–8969. doi: 10.1021/bi2008883. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.