Abstract
Significance: Glutaredoxins are ubiquitous small thiol proteins of the thioredoxin-fold superfamily. Two major groups are distinguished based on their active sites: the dithiol (2-C-Grxs) and the monothiol (1-C-Grxs) glutaredoxins with a CXXC and a CXXS active site motif, respectively. Glutaredoxins are involved in cellular redox and/or iron sulfur metabolism. Usually their functions are closely linked to the glutathione system. Trypanosomatids, the causative agents of several tropical diseases, rely on trypanothione as principal low molecular mass thiol, and their glutaredoxins readily react with the unique bis(glutathionyl) spermidine conjugate. Recent Advances: Two 2-C-Grxs and three 1-C-Grxs have been identified in pathogenic trypanosomatids. The 2-C-Grxs catalyze the reduction of glutathione disulfide by trypanothione and display reductase activity towards protein disulfides, as well as protein-glutathione mixed disulfides. In vitro, all three 1-C-Grxs as well as the cytosolic 2-C-Grx of Trypanosoma brucei can complex an iron–sulfur cluster. Recently the structure of the 1-C-Grx1 has been solved by NMR spectroscopy. The structure is very similar to those of other 1-C-Grxs, with some differences in the loop containing the conserved cis-Pro and the surface charge distribution. Critical Issues: Although four of the five trypanosomal glutaredoxins proved to coordinate an iron–sulfur cluster in vitro, the physiological role of the mitochondrial and cytosolic proteins, respectively, has only started to be unraveled. Future Directions: The use of trypanothione by the glutaredoxins has established a novel role for this parasite-specific dithiol. Future work should reveal if these differences can be exploited for the development of novel antiparasitic drugs. Antioxid. Redox Signal. 19, 708–722.
Trypanosomatids and Their Unique Thiol Redox Metabolism
Trypanosomatids are protozoan parasites of the order Kinetoplastida, which represents one of the most ancient eukaryotic lineages that diverged after acquisition of a mitochondrion. Members of this family are responsible for a variety of tropical diseases. Trypanosoma brucei rhodesiense and T. b. gambiense are the causative agents of African sleeping sickness, T. cruzi is the etiologic agent of American Chagas' disease, and distinct species of Leishmania are responsible for oriental sore and black fever. T. b. brucei, T. congolense, T. equiperdum, and T. vivax cause animal trypanosomiasis. The parasites present a complex life cycle with stages in specific arthropods and the mammalian host(s). Trypanosomatids display a number of morphological and metabolic peculiarities (50). For instance, the parasite thiol redox homeostasis is maintained by the unique trypanothione/trypanothione reductase couple, which substitutes for the glutathione/glutathione reductase and the thioredoxin/thioredoxin reductase systems of the host (24, 39, 44, 45). Trypanothione [T(SH)2; Fig. 1A] is synthesized from glutathione and spermidine with monoglutathionylspermidine (Gsp) as intermediate (44). The dithiol T(SH)2 constitutes the main low molecular mass thiol of trypanosomatids and acts as redox cofactor in a plethora of cellular functions (39, 44, 45). The transfer of reducing equivalents from T(SH)2 to protein targets can be mediated by thioredoxin- (Trx), tryparedoxin- (TXN) and glutaredoxin-type oxidoreductases (12, 20, 63, 77; Fig. 1B). Because of its extremely low concentration in T. brucei and T. cruzi (66, 79), the physiological significance of the parasite Trxs remains, however, elusive. TXN, a parasite-specific distant relative of the Trx protein family, represents the main multipurpose oxidoreductase (15, 52). The parasite glutaredoxins (Grxs) probably have rather specialized functions and are the subject of this review. T(SH)2 has also been shown to complex to electrophiles such as heavy metals and drugs or to ligate endobiotics (i.e., methylglyoxal) or nitro–iron complexes (7, for recent reviews see 44, 45). On the other hand, iron is an essential element that trypanosomatids take up from the extracellular milieu by mechanisms that differ between species (reviewed in 55, 83). Inside the parasites, the metal is incorporated into proteins as direct cofactor (e.g., ribonucleotide reductase, alternative oxidase, the four superoxide dismutases; 21) or in form of an iron–sulfur complex (e.g., aconitase, fumarate hydratases), wherein it fulfills important redox or structural functions (55, 83). Iron is also required for hemoproteins. However, pathogenic trypanosomatids are auxotrophic for heme and obtain the cofactor via uptake of host heme-binding proteins or precursors (e.g., haptoglobin–hemoglobin complex, coproporphobilinogen III; reviewed in 55, 75).
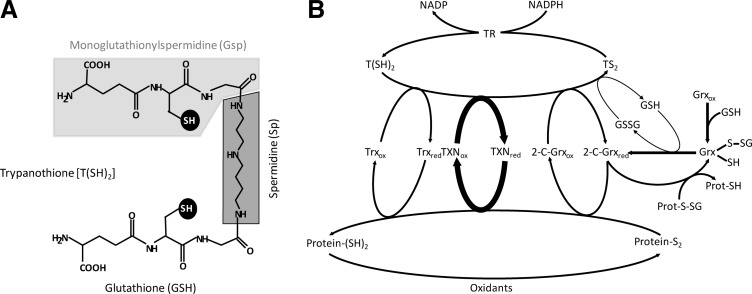
FIG. 1.
The trypanothione-based redox metabolism of trypanosomatids. (A) The main low molecular mass thiol is trypanothione [T(SH)2]. It is synthesized from two molecules of glutathione (GSH, light gray background) and one spermidine (Sp, dark gray background) with mono(glutathionyl)spermidine (Gsp; gray backgrounds) as intermediate. The two thiol groups acting as redox cofactors or ligands are highlighted on a black background. (B) The NADPH-dependent flavoenzyme trypanothione reductase (TR) catalyzes the reduction of trypanothione disulfide (TS2) to T(SH)2 and, thus, is responsible for maintaining the cellular thiol redox homeostasis. T(SH)2 is the direct reducing agent for the parasite thioredoxin (Trx), tryparedoxin (TXN), and dithiol glutaredoxin (2-C-Grx) type oxidoreductases, as well as for glutathione disulfide (GSSG). In vitro all three oxidoreductases catalyze the reduction of intra and/or intermolecular protein disulfides (Protein-S2) to the respective reduced forms (Protein-(SH)2); however TXN is by far the most efficient multipurpose oxidoreductase of the parasites. Trx displays an extremely low cellular concentration and its physiological role, if any, is not known. The parasite Grxs also catalyze the reduction of protein disulfides and specifically, of glutathione–protein mixed disulfides (Prot-S-SG), as well as of GSSG by T(SH)2. The subindex “red” or “ox” denotes the dithiol (reduced) and disulfide (oxidized) form of the proteins.
Glutaredoxins Form a Large and Diverse Protein Family
Glutaredoxins are ubiquitous small thiol proteins of the Trx-fold superfamily (68). According to the number of cysteines in the active site, glutaredoxins were initially classified as dithiolic (CXXC motif; 2-C-Grx) and monothiolic (CXXS motif; 1-C-Grx) proteins. However, a recent and thorough phylogenetic analysis identified six distinct classes of glutaredoxins, where the first two are the most widespread ones, present in most prokaryotic and eukaryotic organisms (74). Class I contains CPYC-type 2-C-Grxs and both mono- and dithiol closely related variants with a CPYS, CGYC, CPFC, or CSY[C/S] motif. Class II proteins are 1-C-Grxs with a conserved CGFS active site and can be subdivided into two groups: proteins with a single 1-C-Grx domain and those composed of an N-terminal Trx-like domain that is followed by one or more 1-C-Grx domains (33; 74; Fig. 2). The latter proteins occur almost exclusively in eukaryotes and represent a distinct functional group (35).
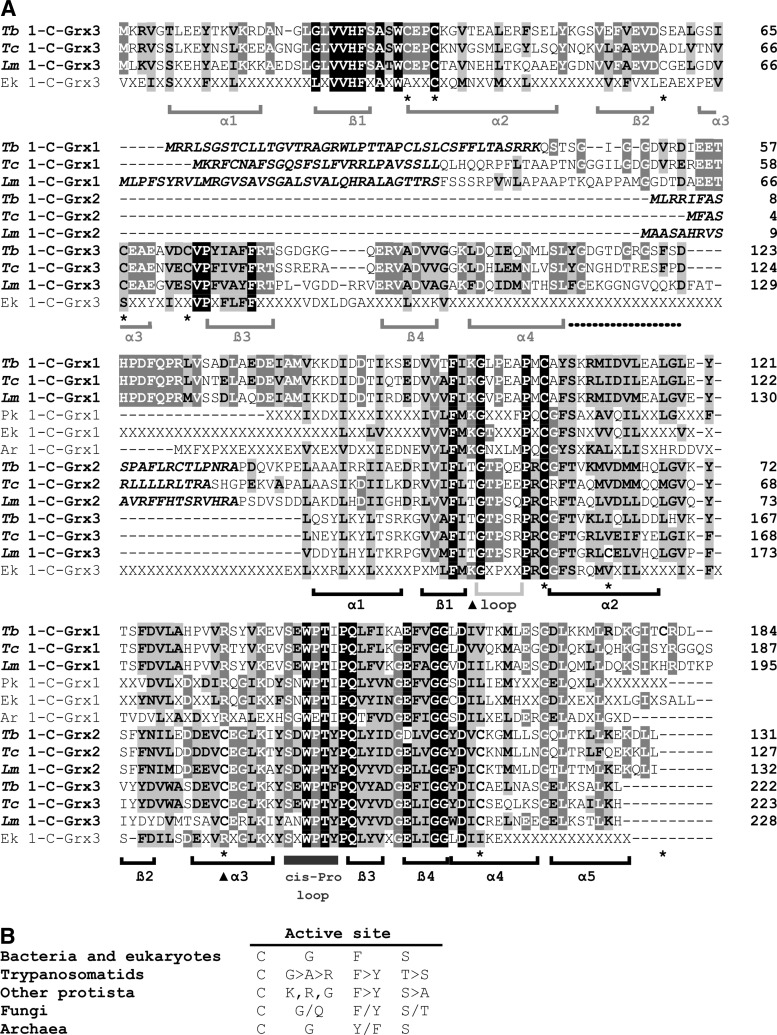
FIG. 2.
Sequence analysis of monothiol glutaredoxins. (A) 1-C-Grxs from trypanosomatids (Tb, Trypanosoma brucei; Tc, Trypanosoma cruzi; Lm, Leishmania major) were aligned with the consensus sequences obtained for prokaryote (Pk), eukaryote (Ek), and archeabacteria (Ar) homologues using the ClustalW algorithm (85) and manually adjusted as necessary. Accession numbers are AJ619696, AM489503 and AM489504 for T. brucei 1-C-Grx1, 2 and 3, respectively; XP_807837, XP_803206 and XP_813048 for T. cruzi 1-C-Grx1, 2 and 3, respectively; NP_047037, CAJ01951 and XP_843232 for L. major 1-C-Grx1, 2 and 3, respectively. Residues given in italic indicate the predicted mitochondrial targeting sequence of Kinetoplastida 1-C-Grx1 and 2. Residues shown in black on gray and white on gray represent residues that are similar and identical, respectively, in at least 40% of the aligned sequences. Residues that are strictly conserved in all sequences analyzed are shown white on black. Cysteine residues in the 1-C-Grxs from trypanosomatids are indicated with an asterisk at the bottom of the alignment. The arrow heads mark basic residues suggested to be involved in glutathione binding in classical Grxs (28, 40). The consensus sequences for the three phylogenetic domains were obtained by alignment of characterized or putative 1-C-Grxs from representative organisms. The sequences used for prokaryotes were from Agrobacterium tumefaciens (α-protobacterium, Acc. Nr. AAK87621.1), Neisseria gonorrheae (β-protobacterium, YP_207507.1), Myxococcus xanthus (δ-protobacterium, ABF89434.1), Escherichia coli (γ-protobacterium, 1YKA) and Synechococcus elongatus (cyanobacterium, BAD78595.1); for archaeabacteria: Haloarcula marismortui (AAV46243.1), Natronomonas pharaonis (YP_326686.1), Haloquadratum walsbyi (CAJ51793.1), and Halobacterium sp. (AAG18993.1); for eukaryotes: Gallus gallus (NP_001008472.1 and XP_421826.1), Danio rerio (AAH59659.1 and NP_001005950.1), Homo sapiens (NP_057501.2 and AAH05289.1), Caenorhabditis elegans (CAB11547.1 and NP_001023756.1), Apis mellifera (XP_625213.1 and XP_392870.1), Tetraodon negroviridis (CAG00128.1 and CAG02746.1), Saccharomyces cerevisiae (Q02784, Q03835 and P32642), Arabidopsis thaliana (AY157988), Tribolium castaneum (XP_975383.1 and XP_972466.1), Porphyra purpurea (P51384), Bos taurus (XP_582303.1 and AAX46537.1), and Xenopus tropicalis (AAH75374.1 and NP_001017209.1). The consensus sequences show only residues that were common to more than 75% of the proteins analyzed. X represents any amino acid. The secondary structure motifs below the alignment refer to the α-helical (α) and ß-sheet (ß) regions in the NMR structure of T. brucei 1-C-Grx1 (black) as well as the homology model of the Trx domain of T. brucei 1-C-Grx3 (gray). The dotted line marks the linker region of T. brucei 1-C-Grx3 and the light and dark gray bar indicates the insertion preceding the active site and the cys-Pro loop. (B) The consensus active site motif of 1-C-Grxs from representatives of the prokaryotic and eukaryotic domains and the family of Trypanosomatidae was obtained from the sequences listed above. The following sequences were used for other protista: Cryptosporidium parvum (CAD98438), Tetrahymena thermophila (XP 001016225, XP 001032143 and XP 001008985), Paramecium tetraurelia (CAK57552, CAK90692 and CAK55785), and Plasmodium falciparum (CAG25239 and CAD50844, for Glp2 and Glp3, respectively); fungi: Encephalitozoon cuniculi (NP 597481), Cocidioides immitis (XP 001244791), and Mortierella alpina (CAB 56513); Archaea: Haloarcula morismortui (AAV46243), Natromonas pharaonis (YP326686), and Haloquadratum walsbyi (CAJ 51793).
The Grx system was first described in the 1980s as “thioltransferase” (3) and as the donor of reducing equivalents for ribonucleotide reductase in an Escherichia coli strain lacking Trx 1 (33). Grxs are usually monomeric proteins that occur in different subcellular compartments (51, 53, 65). There they act as thiol/disulfide oxidoreductases catalyzing the (de)glutathionylation of proteins as well as the reduction of dehydroascorbate and protein disulfides (5, 76). Novel roles have recently emerged for some 2-C-Grxs such as human Grx2 (42, 46), poplar GrxC1 (25), and T. brucei 2-C-Grx1 (12) (see below) based on their capability to bind iron–sulfur clusters (ISCs).
1-C-Grxs were first identified in yeast and subsequently in different organisms (reviewed in 33). With few exceptions, such as yeast Grx 6 (54, 59) and trypanosomal 1-C-Grx1 (16, 27, Fleitas, Manta, and Comini, unpublished), 1-C-Grxs are monomeric proteins in the apo-state (13, 18, 25, 28, 47, 59, 69, 82). Similarly to 2-C-Grxs, distinct isoforms of 1-C-Grxs are found in the cytosol, nucleus, mitochondria as well as plant chloroplast (reviewed in 86). Except for yeast Grx6 and Grx7 (59, 60), most 1-C-Grxs lack or have negligible classical disulfide reductase activity (18, 26, 27, 43, 69, 82). Instead, the capability to coordinate ISCs appears to be a common feature for 1-C-Grxs (16, 40, 42, 43, 54, 67, 89) with yeast Grx7 being, so far, the only known exception (59, 60). The latter feature determines an evolutionary conserved and indispensable role of 1-C-Grxs in the biogenesis and assembly of iron–sulfur proteins (11, 61) and other cell-specific regulatory functions such as the (in)activation of nuclear transcription factors (35, 62, 86).
In trypanosomatids, five Grxs have been identified which are members of class I (T. brucei 2-C-Grx1 and 2) and II (T. brucei 1-C-Grx1, 2 and 3) proteins. The parasite Grxs have a variety of distinctive biochemical, structural and biological features, and are functionally linked to the trypanothione system (12, 16, 27, 56) (Table 1). These proteins are subject of this review.
Table 1.
Small Proteins of the Thioredoxin Protein Family Identified in African Trypanosomes
| |
T. brucei protein |
||||||
|---|---|---|---|---|---|---|---|
| Properties | 2-C-Grx1a | 2-C-Grx2a | TXNb | Trxc | 1-C-Grx1d | 1-C-Grx2d | 1-C-Grx3d |
| Gene Accession N° | Tb11.47.0012 | Tb927.1.1770 | Tb927.3.3780 | Tb09.160.2020 | AJ619696 | AM489503 | AM489504 |
| Subunit molecular mass (kDa) | 10,576 | 12,152 | 15,891 | 12,135 | 15,931 | 12,595 | 24,624 |
| Oligomeric structure (apo/holo) | Monomer/Dimeric ISC | Monomer | Monomer | Monomer | Dimer / Dimeric ISC | Monomer / Dimeric ISC | Monomer / Dimeric ISC |
| 3D-structure (PDB code) | NDe | ND | 1O73 | 1R26 | 2LTK | ND | ND |
| Active site motif | CPYC | CQFC | CPPC | CGPC | CAYS | CGFT | CGFT |
| Putative in vivo reducing agent | T(SH)2 | T(SH)2 | T(SH)2 | T(SH)2 | T(SH)2/TXN | ND | ND |
| Subcellular localization | cytosol | mitochondrial | cytosol | cytosolf | mitochondrial | mitochondrial | cytosolf |
| Putative function | Protein deglutationylation Reduction of GSSG by T(SH)2 Iron sulfur metabolism |
Protein disulfide reduction Protein deglutationylation |
Protein disulfide reduction | Protein disulfide reduction | ISC metabolism | unknown | unknown |
| Indispensability under culture conditions | no | yes | yes | no | yes | ND | ND |
| Total cellular concentration (μM)g | about 2 | 0.1–0.4 | 80–90 | ≪1 | 5–30 | <0.5 | 2–6 |
Data taken from references a12; b15, 53; c29, 70, 78, 79; d16, 27, 55a, 58. eND, not determined, fsuggested localization based on the absence of a putative targeting signal, g in the mammalian bloodstream form. The light gray shading refers to biophysical and structural properties. The dark gray shading denotes biochemical and biological functions of the proteins.
Trypanosomatids Possess Two Distantly Related Dithiol Glutaredoxins
The genome sequences of trypanosomes revealed two single copy genes for 2-C-Grxs. One gene encodes a protein with an overall sequence identity of 40% compared to human Grx2c but with the active site motif (CPYC) of human Grx1. Consequently, the protein has been named 2-C-Grx1. The protein encoded by the second gene in T. brucei, 2-C-Grx2, possesses an unusual CQFC motif and shares only 25% of its residues with 2-C-Grx1. The 2-C-Grx2 sequences are highly conserved in trypanosomatids, while counterparts in organisms outside the order of Kinetoplastida have not been found. Remarkably, despite an overall sequence identity of 80% among the 2-C-Grx2-type proteins from different trypanosomatids, the Leishmania representatives have a serine residue instead of the second active site cysteine and thus are, by definition, 1-C-Grxs. Alignment of the parasite proteins with structurally characterized 2-C-Grxs from other organisms revealed that residues reported to form a binding groove for GSH on the protein surface are conserved (12). However, a Lys that precedes the active site motif and other charged or polar residues found to interact with the glycine carboxylate of GSH are replaced. These substitutions may support the interaction of the parasite 2-C-Grxs with T(SH)2 where the glycine does not carry a negative charge but forms an amide bond with the spermidine. However, it should be stressed that in E. coli Grx2, which has the highest HED reductase activity of the three bacterial dithiol Grxs (E. coli 2-C-Grx 1 to 3), the Lys is replaced by a Tyr. This supports previous reports that the characteristic γ-Glu-Cys moiety is more important for GSH recognition than the glycine carboxylate (23, 81). As will be outlined in this review, the trypanosomal 2-C-Grxs are involved in reactions with both GSH and/or T(SH)2. Thus, amino acid exchanges that would result in a mutually exclusive specificity for T(SH)2 or GSH cannot be expected.
The role of both 2-C-Grxs has been studied in African trypanosomes (12), and recombinant T. cruzi 2-C-Grx2 has been partially characterized (56). Neither 2-C-Grx1 nor 2-C-Grx2 of T. brucei possesses an obvious targeting signal. Immuno-fluorescence analyses revealed 2-C-Grx1 mainly in the cytosol in both the mammalian bloodstream form and the procyclic insect form of T. brucei (12). In contrast, 2-C-Grx2 occurs in the single mitochondrion of the parasite. Upon fractionated digitonin lysis of procyclic parasites, 2-C-Grx2 co-eluted with cytochrome c, suggesting its presence in the intermembrane space. This localization would correspond to that described for a subfraction of Grx1 in rat hearts (65).
Interplay between the 2-C-Grxs and the trypanothione system
The mode of action of 2-C-Grxs is generally closely linked to the glutathione system. The dithiol form of 2-C-Grxs is regenerated by the spontaneous reaction of the intramolecular disulfide of the protein with two molecules of GSH, and the resulting GSSG is then reduced by NADPH and glutathione reductase (GR). Trypanosomatids lack GR, while containing about 300 μM of each free GSH and T(SH)2 (for reviews see 44, 45). Both thiols spontaneously reduce the parasite 2-C-Grxs. However, the apparent second order rate constants differ significantly, being 100 M−1s−1 and 105 M−1sec−1 for the reaction of GSH and T(SH)2, respectively (12). Thus, T(SH)2 is about 1000-fold more efficient compared to GSH and should be the main physiological reducing agent of the parasite 2-C-Grxs.
In trypanosomatids, GSH is regenerated from GSSG by direct thiol/disulfide exchange with T(SH)2. However, this spontaneous reaction is probably not sufficient at lowered T(SH)2 concentrations as they may occur under severe oxidative stress conditions. To mimic this situation, the reaction of 9 μM T(SH)2 with 20 μM GSSG—conditions at which the spontaneous reaction is negligible—was measured. 2-C-Grx1 and 2-C-Grx2 catalyzed the reaction with 106 U/mg and 1.8 U/mg, respectively (12), showing that 2-C-Grx1 is significantly more efficient compared to 2-C-Grx2 both from T. brucei (12) as well as T. cruzi (56). Thus, in trypanosomes the cytosolic 2-C-Grx1 may substitute for GR providing kinetic control for keeping GSH in the reduced state (Fig. 1B).
Oxidoreductase activity of the parasite 2-C-Grxs
The T. brucei 2-C-Grxs reduce the mixed disulfide between GSH and either 2-mercaptoethanol (HED assay) or cysteine residues of bovine serum albumin (BSA) and T. brucei peroxiredoxin. The kcat/Km values towards the glutathionylated model proteins were about 1×105 M−1 s−1 and 1×104 M−1 s−1, respectively, values very similar to those reported for human Grx1 and Grx2 (30, 41). In contrast, T. brucei TXN catalyzes the deglutathionylation of BSA-SSG with a much lower catalytic efficiency of only 4×102 M−1 s−1. Thus, protein deglutathionylation may indeed be a specific in vivo role of the parasite 2-C-Grxs (Table 1).
The trypanosomal 2-C-Grxs also catalyze the reduction of protein disulfides such as insulin disulfide, used as in vitro model substrate. Although this reaction was first shown to be catalyzed by Trxs (37), many Grxs also act as protein disulfide reductase, although mostly with lower efficiency. With T(SH)2 as reducing agent, E. coli Grx1 (used as a control) displayed the highest activity, followed by T. brucei 2-C-Grx2, whereas 2-C-Grx1 had only low activity (12). When comparing the reduction of insulin disulfide by DTE and T(SH)2, both parasite proteins had the highest activity with T(SH)2, whereas E. coli Grx1 exerted identical activity with both dithiols. The superior reducing capacity of T(SH)2 towards the parasite Grxs suggests some specific interactions. However, the fact that E. coli Grx1 equally well accepted DTE and T(SH)2 strongly suggests that individual glutaredoxins do not display a strict thiol specificity. Taken together, the parasite 2-C-Grxs may act as physiological reductases for both protein disulfides and protein-glutathione mixed disulfides (Figs. 1b and 3). In the case of the 2-C-Grx2—especially in the insect stage of the parasite with its fully elaborated mitochondrion—these types of substrates may originate from reactive oxygen species leaking out from the respiratory chain (Fig. 3). In this respect, mammalian Grx1 and Grx2 have been suggested to regulate the local thiol redox homeostasis in the intermembrane space and matrix, respectively (65).
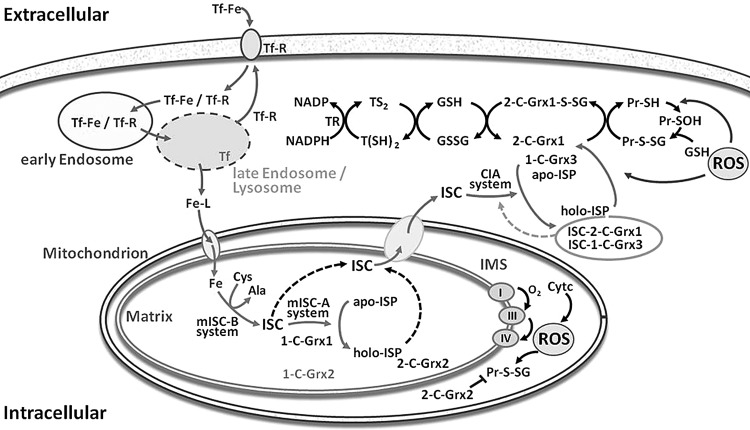
FIG. 3.
Putative roles of the glutaredoxins in the thiol and iron metabolism of African trypanosomes. African trypanosomes obtain iron by the uptake of iron-loaded transferrin (Tf-Fe) from the host blood via receptor-mediated (Tf-R) endocytosis. In the late endosome, Fe is released and exported into the cytosol. Here it circulates in complex with low molecular mass ligands (Fe–L) and can be translocated into the mitochondrial matrix. As shown for yeast and mammalian cells as well as partially for trypanosomes, the mitochondrion is the primary site for the biosynthesis of ISC that requires two specialized machineries. The mitochondrial Iron Sulfur Cluster Biogenesis (mISC-B) system synthesizes the ISC from iron and cysteine (as sulfur donor), and the Iron Sulfur Cluster Assembly (mISC-A) system then transfers the pre-formed ISC from scaffold proteins to acceptor iron sulfur proteins (ISP). T. brucei 1-C-Grx1 is an indispensable (iron sulfur) protein, probably participating in the mISC-A system. The parasite 1-C-Grx2 is a low abundant and functionally nonredundant orthologue of 1-C-Grx1. How ISC is exported from the mitochondrion is not yet known. In the cytosol, 1-C-Grx3 and 2-C-Grx1 may be part of the cytosolic iron assembly (CIA) machinery that transforms apo-iron sulfur proteins (apo-ISP) into the respective holo-proteins (holo-ISP). Alternatively, ISC formation on 2-C-Grx1 may result in an inactive form that, in the presence of high levels of reactive oxygen species (ROS), may be converted again in the free protein (for further abbreviations, see legend of Fig. 1). 2-C-Grx2 in the—intermembrane space (IMS) and/or matrix of the—mitochondrion plays probably a crucial role for the reduction of protein disulfides and/or glutathionylated proteins (Pr-S-SG) produced by reactive oxygen species (ROS) originating from the respiratory chain and cytochrome c (cyt c) activity present only in the insect stage of the parasite. The scheme is based on data from reference 55 and citations therein.
As described above, in contrast to the 2-C-Grx2 proteins from T. brucei and T. cruzi, the Leishmania orthologues display a serine instead of the second active site cysteine. One may speculate that the proteins have distinct functions in the individual parasites, but this is not very likely. A serine replacing the second cysteine may also stabilize the thiolate of the nucleophilic first cysteine. By precluding formation of an intramolecular disulfide, this substitution may even result in a protein species with higher activity in reactions that follow a monothiol mechanism such as protein deglutathionylation (19, 30, 41). The precise role of the second active site cysteine residue in the catalytic mechanism of 2-C-Grxs is, however, still an open question. Replacement of the cysteine by a serine can result in increased (19, 30, 41) or decreased (19) activities, depending on the individual protein. As shown recently, the GrxC5, localized in chloroplasts of Arabidopsis thaliana, exclusively relies on a monothiol mechanism to reduce insulin disulfide (17), a reaction regarded to follow a dithiol mechanism (9). On the other hand, in yeast Grx8 both active site cysteines are involved in the (low) activity towards glutathione-mixed disulfides and a refined catalytic model for both mono- and dithiol Grxs has been suggested (22).
Functional comparison of the parasite 2-C-Grxs with thioredoxin and tryparedoxin
Besides the two 2-C-Grxs described above, trypanosomatids encode two other types of dithiol oxidoreductases, namely Trx(s) and the parasite-specific TXN(s). The physiological role of Trx in both African trypanosomes (79) and T. cruzi (66) is not yet known. Silencing of the gene by RNA interference in procyclic T. brucei and the deletion of both alleles in infective cells did not affect the proliferation of the parasites, at least under culture conditions (79). TXNs have a molecular mass of about 16,000 Da and are characterized by a CPPC active site motif. They are mainly cytosolic proteins (84). In vitro, TXN transfers the reducing equivalents from T(SH)2 onto ribonucleotide reductase (20), methionine sulfoxide reductase (2), as well as 2-Cys-peroxiredoxins (8, 63) and non-selenium glutathione peroxidase-type enzymes (34, 77). Thus, this parasite-specific oxidoreductase exerts many functions that in other organisms are fulfilled by Trxs and/or Grxs. Recombinant T. brucei 2-C-Grx1 can replace TXN in the transfer of electrons onto ribonucleotide reductase, although the reaction is less efficient. In vitro, TXN is a more potent protein disulfide reductase compared to the 2-C-Grxs, while its activity towards glutathionylated proteins is very low (12). Thus, protein (de)glutathionylation is probably a physiological function of the parasite 2-C-Grxs that is not accomplished by TXN.
Pathogenic Trypanosomes Encode Three Monothiol Glutaredoxins
The genomes of T. brucei, T. cruzi, and L. major (http://tritrypdb.org/tritrypdb/) encode three homologues of 1-C-Grxs (Fig. 2A). The proteins are encoded by single copy genes located on different chromosomes. 1-C-Grx1 and 1-C-Grx2 are single domain proteins, while in 1-C-Grx3 the monothiol glutaredoxin domain is C-terminal of a Trx domain. As expected, the primary structures of the individual 1-C-Grxs are more similar among the different parasites (mean identity of about 65%) than those of the three proteins within the same organism (mean identity of 32% for all three 1-C-Grxs from T. brucei, T. cruzi, or L. major). Interestingly, the Grx domain of 1-C-Grx3 is more closely related to the primary structure of 1-C-Grx2 than to that of 1-C-Grx1 (mean identity of 39% and 26%, respectively). This may suggest that 1-C-Grx3 has originated by a fusion event between a 1-C-Grx2 ancestor and a Trx. While most bacterial and eukaryotic 1-C-Grxs have a conserved CGFS motif (33, 74), the proteins from trypanosomatids present a rather variable active site (Fig. 2B). To figure out if this is a peculiarity of the parasites or extends to other organisms, a PSI-BLAST search using T. brucei 1-C-Grxs sequences against the NCBI nonredundant protein database was performed. Homologous sequences from more than 200 organisms were identified. The few proteins with an active site motif divergent from the consensus CGFS sequence were found in protista, fungi, and members of the Archaea (Fig. 2B). Another interesting result from this search was that of all eukaryotic Trx/1-C-Grx chimera identified, only the trypanosomal 1-C-Grx3 contains a Trx-like domain with a dithiol active site (Fig. 2A).
Expression of the 1-C-Grxs in African trypanosomes
So far, the proteins from T. brucei represent the best studied 1-C-Grxs from a protozoan organism. All three proteins are expressed in both the mammalian infective as well as the insect stage of T. brucei, with the highest levels found in stationary phase and starving parasites (16). The single domain proteins 1-C-Grx1 and 1-C-Grx2 reside in the mitochondrion in both life stages of T. brucei (16, 27, Comini: unpublished data). The subcellular localization of 1-C-Grx3 has not been studied but is likely to be cytosolic due to the absence of recognizable targeting signals. The total cellular concentration of 1-C-Grx1 in logarithmically growing bloodstream T. brucei is about 5 μM (16). This indicates an unusually high mitochondrial concentration of about 200 μM when taking into account that the rudimentary organelle of this parasite form has been reported to account for only 2.3% of the total cell volume (64). The concentration of 1-C-Grx2 is at least two orders of magnitude lower than that of 1-C-Grx1 (16).
Oligomeric state and redox properties of the T. brucei 1-C-Grxs
Recombinant T. brucei 1-C-Grx1 exists as a noncovalent homodimer in both the apo-form and with bound iron–sulfur cluster, whereas the apo-forms of 1-C-Grx2 and 1-C-Grx3 are monomeric proteins (16, 27). An N-terminal region of about ∼37 residues, exclusive of the kinetoplastid 1-C-Grx1s, has recently been identified to be responsible for dimerization of the T. brucei and T. cruzi proteins (55a). In vitro, T. brucei 1-C-Grx1 lacks any general thiol-disulfide oxidoreductase activity (27) but is susceptible to oxidation by GSSG and Gsp disulfide (16, 58). In addition to the active site Cys104, T. brucei 1-C-Grx1 contains a second cysteine, namely Cys181, located at the fourth position from the C-terminus (Fig. 2A). In contrast to the specificity displayed by E. coli Grx4 and S. cerevisiae Grx5, Cys181—not Cys104—forms a mixed disulfide with GSH and Gsp (58), which can trigger the formation of an intramolecular Cys104–Cys181 disulfide bridge (16, 58). In vitro, both oxidation states are reduced by TXN or the T(SH)2/TR system (27, 58). The physiological significance of the oxidized forms of 1-C-Grx1 as well as the potential reducing agent in vivo remains to be elucidated since Cys181 is not conserved. Interestingly, preliminary data indicate that also the active site cysteine in the Grx domain of T. brucei 1-C-Grx2 and 1-C-Grx3 is refractory to thiolation by GSH and Gsp while other cysteine residues get specifically modified (Melchers J. PhD Thesis, Heidelberg University, 2008). Future work should reveal if this is a peculiarity of the trypanosomal 1-C-Grxs and/or may play a regulatory role in protein function.
The Potential Role of Glutaredoxins in the Parasite Iron–Sulfur Metabolism
Features of the iron–sulfur complexes coordinated by 2-C-Grx1 and 1-C-Grxs
The distinct physiological function(s) of the glutaredoxins in T. brucei are not yet known. Remarkably, in vitro all three 1-C-Grxs as well as 2-C-Grx1 are capable of binding an iron–sulfur complex (ISC). Upon purification from recombinant E. coli, the proteins were obtained as light brown solutions with absorption maxima at 320 nm and 420 nm, in addition to the protein peak at 280 nm (12, 16), indicative of a protein/iron–sulfur complex (6, 75). Size exclusion chromatography of these species revealed that binding of the ISC led to the dimerization of the monomeric apo-forms of 2-C-Grx1, 1-C-Grx2, and 1-C-Grx3 (12, Manta and Comini, unpublished; Fig. 4). In the case of 1-C-Grx1, ISC-binding increased the hydrodynamical radius of the homodimeric protein (16, 55a). Other 1-C-Grxs shown to form dimeric apo-proteins are S. cerevisiae Grx6 and Grx7 (59). Binding of an ISC to yeast Grx6 results in tetramerization of the protein (59). The [2Fe–2S] nature of the ISC has been determined for 1-C-Grx1 (16). As reported for several ISC-glutaredoxin complexes (6, 25, 40, 42, 43, 54, 67, 75), most probably two GSH molecules act as the low molecular mass ligands of the ISC in the holo-form of the trypanosomal proteins extracted from E. coli. In vitro reconstitution of the iron–sulfur proteins in the presence of Gsp or T(SH)2 revealed that these parasite specific thiols can easily substitute for GSH as the nonprotein ligand(s). Strikingly, the dithiol T(SH)2 was capable to ligate the chromophor also to the monomeric form of 2-C-Grx1 which probably represents an intermediate during formation of the holo-protein (12, 55a).
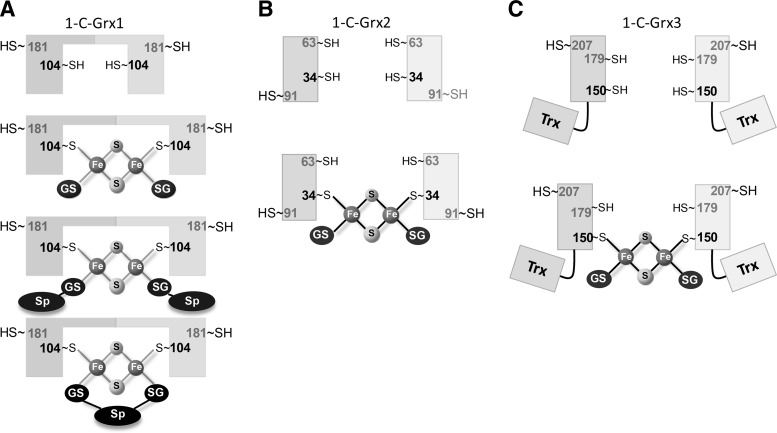
FIG. 4.
Working models for iron–sulfur cluster assembly into 1-C-Grxs from trypanosomatids. (A) 1-C-Grx1 is a homodimeric protein in free form as well as after binding an ISC complex. ISC coordination involves the active site Cys104 (bold) from each subunit and two additional sulfhydryl groups provided by either two molecules of GSH (-SG) or Gsp (–SG-Sp) or by one trypanothione [(-SG)2-Sp] molecule. (B) and (C) Apo 1-C-Grx2 and 1-C-Grx3 are monomeric proteins that dimerize upon binding of an ISC. Depicted are the putative complexes involving the active site Cys34 and Cys150 (bold), respectively, and two molecules of GSH as nonprotein ligands.
A cysteine in the first position and the absence of a proline in the second position of the active site motif, as well as the noncovalent binding of GSH are conserved features among glutaredoxins capable of coordinating an ISC (6, 25, 40, 42, 43, 54, 67, 75). For cluster coordination, T. brucei 1-C-Grx1 also relies on its active site cysteine but the protein is not definite with respect to the nature of the thiol ligand and the mechanism of ISC binding (55a). T. brucei 2-C-Grx1 was the first authentic 2-C-Grx with the canonical CPYC active site shown to incorporate an ISC (12). These data suggest that additional molecular features determine the productive binding of an ISC to the trypanosomal Grxs. The protein–ISC complexes formed by T. brucei 2-C-Grx1 and 1-C-Grx1 are labile to air and/or H2O2 exposure (12, 16). The yield of holo-complexes obtained upon in vitro reconstitution is dependent on the low molecular mass thiol coordinated, being highest with T(SH)2 followed by Gsp and finally GSH (12, 55a). This may suggest that in the parasite T(SH)2 serves as the ligand resulting in a comparably stable ISC. Interestingly, T(SH)2, but not the monothiols GSH and Gsp, can form a protein-free iron-containing complex which subsequently can be incorporated into 2-C-Grx1 (12, 55a).
Glutaredoxins and regulation of the iron–sulfur metabolism
The mitochondria of eukaryotic cells are responsible for the de novo synthesis of ISCs that are either assembled to iron–sulfur proteins or transferred to the cytosol for further incorporation into target proteins via a specialized assembly system (48). This system is operative in African trypanosomes and the depletion of a cysteine desulfurase and a primary ISC-scaffold protein has been shown to be deleterious for the insect stage of the parasite (80). 1-C-Grxs have been initially recognized as key components of the mitochondrial machinery probably being involved in the chaperone-assisted transfer of ISCs to acceptor proteins (Fig. 3) (11, 73, 87, 88). Complementation studies using a yeast mutant defective in mitochondrial Grx5, which is unable to maturate iron–sulfur proteins and accumulates toxic levels of intracellular iron, demonstrated the evolutionary conserved function for 1-C-Grxs from different eukaryotic and prokaryotic organisms (11, 13, 61, 87). Intriguingly, respective experiments with the three T. brucei 1-C-Grxs revealed that only 1-C-Grx1 modestly rescued the mutant phenotype (27). The striking lack of functional conservation is indicative of a significant structural and/or functional divergence of the parasite proteins. Any genetic manipulation of T. brucei aimed at abrogating or downregulating 1-C-Grx1 failed, which strongly suggests an essential role of the protein in the mitochondrion of the parasite and the incapacity of 1-C-Grx2 to take over this function (16). A regulatory function for 1-C-Grx1 in the mitochondrial iron/redox metabolism can also be envisaged. Bloodstream T. brucei takes up iron via a transferrin receptor (Fig. 3). Cells overexpressing (>10-fold) an ectopic copy of 1-C-Grx1 are more affected in vitro upon iron deprivation as well as oxidative stress and cannot sustain infection in the mammalian host when compared to wild-type parasites (16, 55a).
In yeast, the cytosolic Trx/1-C-Grx hybrids Grx3 and Grx4 are involved in intracellular iron sensing and trafficking. The mechanism includes the ISC-mediated interaction of Grx3 and Grx4 with transcription factors that, depending on the iron levels, regulate operons controlling iron usage or uptake (35, 62). It seems unlikely that the parasite 1-C-Grx3 plays a similar role, owing to the fact that transcription factors are almost absent in trypanosomes and gene expression is mainly controlled at post-transcriptional level (14). However, 1-C-Grx3 and 2-C-Grx1 may indeed play a role in the cytosolic ISC assembly machinery, for example, as ISC scaffold protein and/or by mediating ISC transfer to apo-proteins (Fig. 3). Alternatively, as proposed for the mitochondrial human Grx2 (49), the ISC formation may serve as a redox sensor for the regulation of 1-C-Grx3 and 2-C-Grx1. Under conditions with lowered free thiol levels, the complex may be broken down releasing the catalytically active monomeric protein species.
Structural Basis for Monothiol Glutaredoxin Function
The Trx fold, a four-stranded ß-sheet flanked by three α-helices, is amongst the most conserved structural motifs found in proteins that are not necessarily related in function (57; Fig. 5). Modifications of this core fold lead to remarkable structural diversity that allows for classification of its members into eleven distinct evolutionary families (68). A network-based computational approach, used to create similarity-based maps of the Trx fold class, has demonstrated that Grxs are not a cohesive superfamily and that 1-C-Grxs are distinctly related with other classes within the superfamily (4). Two key signatures distinguish 1-C-Grxs with a CGFS active site (class II) from classical 2-C-Grxs and other 1-C-Grxs (class I, see above). These are an insertion of five residues forming a loop that precedes the active site, as well as a highly conserved WP motif within another loop that contains also the conserved cis-Pro that is part of the putative binding site for GSH (Fig. 2A). Mesecke et al. (59) already suggested that these structural elements may be relevant in determining the redox activity of 1-C-Grxs. Remarkably, 1-C-Grxs that contain these structural elements lack general oxidoreductase activity (e.g., yeast Grx5, E. coli Grx4; Plasmodium falciparum Glp1, T. brucei 1-C-Grx1) (26, 27, 69, 72). In contrast, yeast Grx6, classified as 1-C-Grx, for its CSYS active site, but lacking this loop and the conserved WP motif, maintains the ability to reduce the mixed disulfide of glutathionylated ß-mercaptoethanol (54, 59). Mutational analysis of 1-C-Grxs should reveal whether these structural motifs are sufficient to determine the lack of redox activity in certain protein subgroups.
FIG. 5.
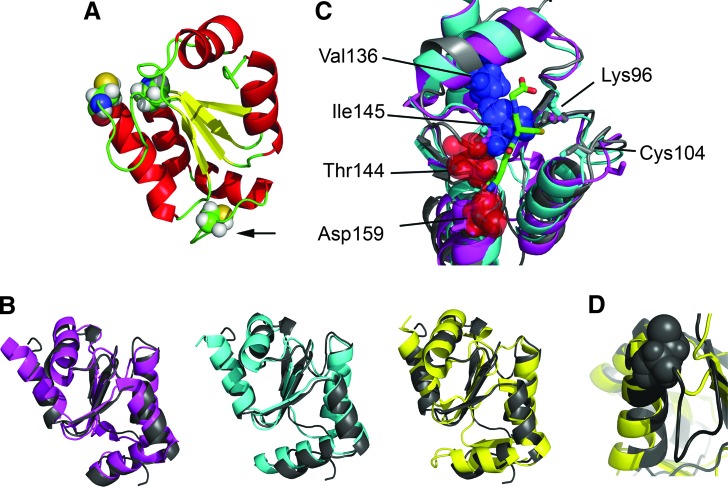
Three-dimensional structure of T. brucei 1-C-Grx1 in comparison with other 1-C-Grxs. (A) The structure of T. brucei 1-C-Grx1 (residues 76–184) was determined by multidimensional NMR spectroscopy (PDB ID 2LTK). The active site Cys104, the conserved cis-Pro146 facing the active site (upper left part) and the only other cysteine residue (Cys181, marked by an arrow) are shown as spheres. (B) Overlay of the solution structure of T. brucei 1-C-Grx1 (gray) with the crystal structures of 1-C-Grxs: S. cerevisiae Grx5 (magenta, PDB-id 3GX8, backbone-RMSD: 1.5 Å), E. coli Grx4 (cyan, PDB-id 2WCI, backbone-RMSD: 1.5 Å) and S. cerevisiae Grx6 (yellow, PDB-id 3L4N, backbone-RMSD: 1.7 Å). The abbreviations for the proteins correspond to those in the respective publication. (C) Superposition of the structures of T. brucei 1-C-Grx1 (gray; PDB-ID 2LTK), E. coli Grx4 in ISC-bound dimeric form (cyan; PDB-ID 2WIC; a single monomer is shown with the noncovalently bound GSH; the ISC is omitted), and S. cerevisiae Grx5 (magenta, PDB-ID 3GX8). Residues shown to be critical for GSH binding and ISC ligation are depicted as sticks (corresponding to Lys96, Cys104, Thr144, and Asp159 in T. brucei 1-C-Grx); the side chains of Val136 and Thr144, Ile145, and Asp159 in T. brucei 1-C-Grx1 are indicated with spheres; the glutathione moiety from the structure of E. coli holo-Grx4 is shown in green. It is evident that the change in the conformation of the loop containing the cis-Pro146 observed in T. brucei 1-C-Grx1 significantly distorts the GSH pocket and precludes the binding of glutathione as determined in E. coli Grx4. (D) Enlargement of the active site region of T. brucei 1-C-Grx1 (gray) and S. cerevisiae Grx6 (yellow) showing the presence and absence of the loop that precedes the active site in the corresponding protein. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
Structural analysis of a truncated version of T. brucei 1-C-Grx1 as well as 1-C-Grx3
Very recently, we achieved the complete NMR assignment of a truncated version of T. brucei 1-C-Grx1 (residues 76–184, protein lacking the mitochondrial targeting signal and the stretch responsible for dimerization) and the structure was solved (PDB 2LTK; [55a]) based on the semi-automated analysis of NOE-derived interproton distance (32). The overall fold comprises of a four-stranded ß-sheet, flanked by five α-helices (Fig. 5A) and thus is very similar to those of other 1-C-Grxs (Fig. 5B), as well as classical Grxs (not shown). As expected, the active-site Cys104 is exposed to the solvent and the side chain of Lys96 is in proximity of the thiol group, probably contributing to decrease its pKa-value (76a). Most of the residues shown to be involved in GSH binding and ISC assembly in other 1-C-Grxs (e.g., Lys96, Cys104, and Asp 159 of T. brucei 1-C-Grx1) (10, 42, 54, 67) are conserved in T. brucei 1-C-Grx1 (Figs. 2A and 5C) and show a spatial orientation similar to that observed in the dimeric structure of E. coli Grx4 coordinating a 2Fe-2S cluster which presents a noncovalently bound GSH for each monomer (PDB 2WCI; 40; Fig 5C). A remarkable exception is Thr144. The residue is part of a loop whose conformation differs from that of the respective residues in the crystal structures of yeast Grx5 (PDB 3GX8; Wang et al. unpublished) and E. coli Grx4. In the latter structures, the conserved loop, containing the monothiol WPT motif, adopts a very similar conformation stabilized by a network of hydrogen bonds. Intriguingly, T. brucei 1-C-Grx1 is not capable of establishing this H-bond mediated anchoring of the loop, mainly because Phe149 replaces an otherwise conserved tyrosine or tryptophan residue (Tyr108 in yeast Grx5, Trp 75 in E. coli Grx4) that contributes with its side chain to fix the conformation of the structural element. The different conformation of the loop in T. brucei 1-C-Grx1, which significantly distorts the GSH binding pocket, is dictated by steric interactions between Ile145 and Val136. Experimental evidence indicates that this loop presents structural plasticity and we have proposed that the backbone dynamics of this region may modulate the binding of the glutathione moiety and/or influence the catalytic properties of the protein (55a). Interestingly, Ile145 precedes the conserved cis-Pro which has been proposed to be crucial for determining the redox properties of different Trx fold proteins (71). In addition, Tb 1-C-Grx1, as well as other kinetoplastid's 1-C-Grxs, present a 5-residues insertion upstream the active site (see Fig. 2A) that lacks secondary structure (Fig. 5D) and has been proposed to contribute to the almost negligible redox activity of 1-C-Grxs from other organisms (see above; 59).
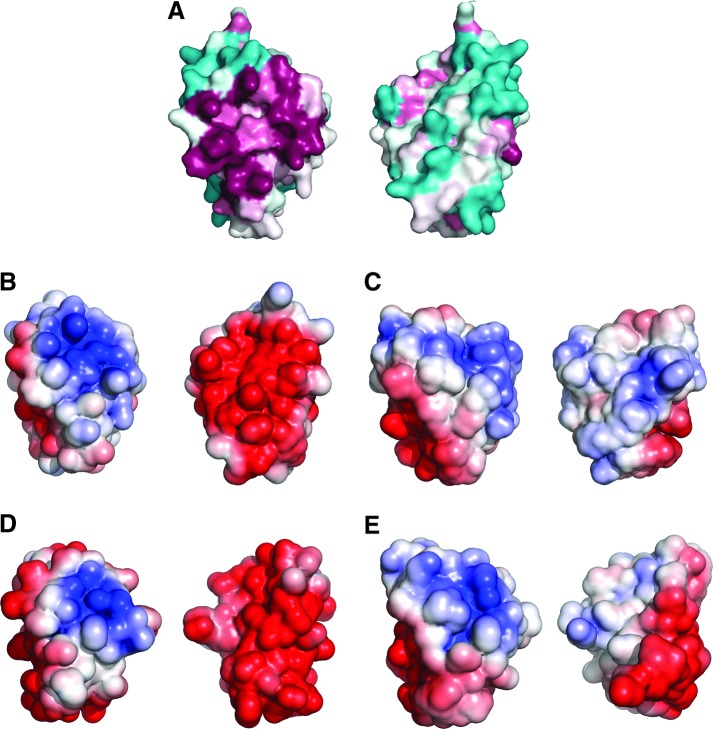
Analysis of the electrostatic surface of T. brucei 1-C-Grx1 discloses two main areas that are characterized by a concentration of identical charges: a conserved, positively charged surface patch that comprises most of the putative GSH binding residues and a negatively charged region on the opposite side of the molecules (Fig. 6). The latter region of negative electrostatic potential is also observed in yeast Grx5 where it is even more extended (PDB 3GX8). In contrast, in E. coli Grx4 the same region is occupied by a hydrophobic patch (28, 40). The functional role of these regions has yet not been addressed, and one possibility is that they serve as specific protein–protein interaction areas.
FIG. 6.
Molecular surface of T. brucei 1-C-Grx1 and 3. (A) Surface amino acid conservation in T. brucei 1-C-Grx1 calculated with the Consurf server using the 150 unique 1-C-Grx sequences with the lowest E-value from PSI-BLAST. Conserved and variable residues are depicted in purple and cyan, respectively. Electrostatic potential mapped onto the molecular surface of (B) T. brucei 1-C-Grx1, (C) T. brucei C-terminal domain of Grx3, (D) S. cerevisiae Grx5, and (E) E. coli Grx4; red and blue denote negatively and positively charged residues, respectively. Each image shows two views of the same protein rotated by z-180°. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
In comparison to the other types of parasite 1-C-Grxs, the trypanosomatid 1-C-Grx1s show a 4–10 residues long C-terminal extension. In the case of T. brucei 1-C-Grx1, these residues form a relatively rigid segment (as probed by 15N relaxation experiments), without regular secondary structure. At the fourth position form the C-terminus Cys181 is located (Figs. 2A and 5A) which in vitro can form an intramolecular disulfide with the active-site Cys104 (58). In the available structure the two residues are positioned at a distance far beyond that required for a disulfide bridge, and only a substantial conformational change or a local unfolding would allow formation of this covalent link. Interestingly, a structural rearrangement has also been hypothesized for E. coli Grx4 to explain the reaction between two cysteine residues distantly located in the solved structure (28). The segment comprising residues 180–182 adopts an extended conformation with parallel backbone NH and CO bonds oriented toward the exterior of the protein. Under oxidative conditions, this could promote a transient association of GSSG via antiparallel β-strand backbone hydrogen bonds with subsequent formation of a cross-strand disulfide between the cysteine of GSH and Cys181. Cross-strand disulfides are highly strained (38) and this intermediate complex could in turn trigger the conformational transition required for the formation of the intramolecular disulfide. The conformation of Cys181, together with the distortion of the GSH pocket described above, might explain why this cysteine, and not Cys104, is glutathionylated in Tb 1-C-Grx1.
So far, there is no structural information available for multi-domain 1-C-Grxs. As described above, T. brucei 1-C-Grx3 is a Trx/1-C-Grx hybrid protein for which, due to the high structural conservation of this class of proteins, the backbone conformation of both domains can be easily predicted in silico. The recently achieved backbone assignment for both domains of T. brucei 1-C-Grx3 and the measurement of a set of residual dipolar couplings (RDCs) allowed us to validate the models in term of type and orientation of secondary structure elements (unpublished). The two domains of 1-C-Grx3 are linked by eleven residues (residues 112–122, Fig. 2A), which are not conserved and therefore cannot be modeled. Interestingly, our 15N relaxation measurements and proteolysis experiments (unpublished) show that this linker is not particularly flexible compared with the rest of the protein. Accordingly, RDCs measurements support a defined relative orientation of the two domains. Ongoing studies aim at determining the relative spatial arrangement of the two domains and their interaction interface. This should shed light on the role of the N-terminal Trx domain that at least in yeast has been shown to be essential for the function of multi-domain Trx/1-C-Grxs whose physiological role cannot be replaced by single domain 1-C-Grxs (35).
Summary and Outlook
Trypanosomatids are equipped with two dithiol and three monothiol glutaredoxins. The specialization of the trypanosomal proteins in using T(SH)2 as cofactor for redox reactions and/or ISC binding constitutes a remarkable evolutionary adaptation of Grxs to the unique thiol redox metabolism of these parasites. The ability of 2-C-Grx1 to catalyze the reduction of GSSG by T(SH)2 has probably evolved to keep glutathione in the reduced state–especially under oxidative stress conditions–in organisms that lack a GR. Both 2-C-Grxs reduce protein disulfides as well as GSH-protein mixed disulfides. While the former type of reaction, at least in vitro, is even more efficiently catalyzed by TXN, protein deglutathionylation seems to be a specific function of the parasite 2-C-Grxs.
Interestingly, four of the five T. brucei Grxs can coordinate an ISC in vitro. Putative roles in ISC and/or iron metabolism can be expected for the single domain 1-C-Grxs, considering that this is an almost strictly conserved function for this protein subgroup (33, 74). The precise role of 1-C-Grx1 in the parasite mitochondrion remains unknown. The protein is indispensable for the mammalian infective form and not functionally redundant with 1-C-Grx2. Recent progresses in the biochemical and structural characterization of the parasite 1-C-Grxs have displayed distinct properties such as T(SH)2 as preferred thiol ligand, homodimeric structure, electrostatic surface, and the inferred mechanism for ISC binding. These features are likely responsible for the specificity of the trypanosomal proteins and, hence, their inability to substitute Grx5 in yeast mitochondria. 2-C-Grx1 and probably also 1-C-Grx3 are cytosolic proteins. In vitro, 2-C-Grx1 coordinates an ISC, but the significance of this reaction in vivo is not yet known. As suggested for human Grx2, formation of the complex may convert the protein into a catalytically inactive form unless the level of low molecular mass thiols is dramatically lowered such as under severe oxidative stress conditions. Alternatively, 2-C-Grx1 may play a role in the maturation of cytosolic ISC proteins. Future work, including oxidative challenge, alteration of iron homeostasis, and in vivo studies, on 2-C-Grx1-depleted or -deficient parasites should reveal the physiological functions of the protein. 2-C-Grx2 is the only parasite Grx that does not bind an ISC in vitro. It may play an important role in the mitochondrial redox metabolism, especially of procyclic parasites with their fully elaborated organelle. The identification that the 2-C-Grx2 orthologues from all Leishmania species have a single active site cysteine poses additional challenges for the functional role(s) and classification of (the parasite) glutaredoxins.
Abbreviations Used
- 1-C-Grx
monothiol glutaredoxin
- 2-C-Grx
dithiol glutaredoxin
- 3D
tridimensional
- Ar
archeabacteria
- CIA
cytosolic iron assembly
- cyt c
cytochrome c
- DTE
dithioerythritol
- Ek
eukaryote
- Fe
iron
- Fe–L
iron-low molecular mass ligand complexes
- Grx
glutaredoxin
- GSH
glutathione
- Gsp
mono glutathionylspermidine
- GSSG
glutathione disulfide
- IMS
intermembrane space
- ISC
iron sulfur cluster
- ISP
iron sulfur proteins
- mISC-A
mitochondrial iron sulfur cluster assembly machinery
- mISC-B
mitochondrial iron sulfur cluster biogenesis
- ND
not determined
- NMR
nuclear magnetic resonance
- PDB
protein data bank
- Pk
prokaryote
- Pr-S-SG
glutathionylated protein
- RDCs
residual dipolar couplings
- RMSD
root-mean-square deviation
- ROS
reactive oxygen species
- Spd
spermidine
- Tf
transferrin
- TfR
transferrin receptor
- TR
trypanothione reductase
- Trx
thioredoxin
- TS2
trypanothione disulfide
- T(SH)2
trypanothione
- TXN
tryparedoxin
Acknowledgments
Financial support by the Access to Research Infrastructures activity in the 7th Framework Programme of the EC (Project number: 261863, Bio-NMR) for conducting the research is gratefully acknowledged by Massimo Bellanda. Marcelo Comini acknowledges support from the Agencia Nacional de Investigación e Innovación (Grant Innova Uruguay, agreement No. DCI–ALA/2007/19.040 between Uruguay and the European Commission).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Alphey MS. Gabrielsen M. Micossi E. Leonard GA. McSweeney SM. Ravelli RB. Tetaud E. Fairlamb AH. Bond CS. Hunter WN. Tryparedoxins from Crithidia fasciculata and Trypanosoma brucei: photoreduction of the redox disulfide using synchrotron radiation and evidence for a conformational switch implicated in function. J Biol Chem. 2003;278:25919–25925. doi: 10.1074/jbc.M301526200. [DOI] [PubMed] [Google Scholar]
- 2.Arias DG. Cabeza MS. Erben ED. Carranza PG. Lujan HD. Tellez Inon MT. Iglesias AA. Guerrero SA. Functional characterization of methionine sulfoxide reductase A from Trypanosoma spp. Free Radic Biol Med. 2011;50:37–46. doi: 10.1016/j.freeradbiomed.2010.10.695. [DOI] [PubMed] [Google Scholar]
- 3.Askelöf P. Axelsson K. Eriksson S. Mannervik B. Mechanism of action of enzymes catalyzing thiol-disulfide interchange. Thioltransferases rather than transhydrogenases. FEBS Lett. 1974;38:263–267. doi: 10.1016/0014-5793(74)80068-2. [DOI] [PubMed] [Google Scholar]
- 4.Atkinson H. Babbitt PC. An atlas of the thioredoxin fold class reveals the complexity of function-enabling adaptations. PLoS Comput. Biol. 2009;5:e1000541. doi: 10.1371/journal.pcbi.1000541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beer SM. Taylor ER. Brown SE. Dahm CC. Costa NJ. Runswick MJ. Murphy MP. Glutaredoxin 2 catalyzes the reversible oxidation and glutathionylation of mitochondrial membrane thiol proteins: Implications for mitochondrial redox regulation and antioxidant defense. J Biol Chem. 2004;279:47939–47951. doi: 10.1074/jbc.M408011200. [DOI] [PubMed] [Google Scholar]
- 6.Berndt C. Hudemann C. Hanschmann EM. Axelsson R. Holmgren A. Lillig CH. How does iron–sulfur cluster coordination regulate the activity of human glutaredoxin 2? Antioxid Redox Signal. 2007;9:151–157. doi: 10.1089/ars.2007.9.151. [DOI] [PubMed] [Google Scholar]
- 7.Bocedi A. Dawood KF. Fabrini R. Federici G. Gradoni L. Pedersen JZ. Ricci G. Trypanothione efficiently intercepts nitric oxide as a harmless iron complex in trypanosomatid parasites. FASEB J. 2010;24:1035–1042. doi: 10.1096/fj.09-146407. [DOI] [PubMed] [Google Scholar]
- 8.Budde H. Flohé L. Hecht HJ. Hofmann B. Stehr M. Wissing J. Lunsdorf H. Kinetics and redox-sensitive oligomerisation reveal negative subunit cooperativity in tryparedoxin peroxidase of Trypanosoma brucei brucei. Biol Chem. 2003;384:619–633. doi: 10.1515/BC.2003.069. [DOI] [PubMed] [Google Scholar]
- 9.Bushweller JH. Aslund F. Wüthrich K. Holmgren A. Structural and functional characterization of the mutant Escherichia coli glutaredoxin (C14→S) and its mixed disulfide with glutathione. Biochemistry. 1992;31:9288–9293. doi: 10.1021/bi00153a023. [DOI] [PubMed] [Google Scholar]
- 10.Bushweller JH. Billeter M. Holmgren A. Wüthrich K. The nuclear magnetic resonance solution structure of the mixed disulfide between Escherichia coli glutaredoxin (C14S) and glutathione. J Mol Biol. 1994;235:1585–1597. doi: 10.1006/jmbi.1994.1108. [DOI] [PubMed] [Google Scholar]
- 11.Camaschella C. Campanella A. De Falco L. Boschetto L. Merlini R. Silvestri L. Levi S. Iolascon A. The human counterpart of zebrafish shiraz shows sideroblastic-like microcytic anemia and iron overload. Blood. 2007;110:1353–1358. doi: 10.1182/blood-2007-02-072520. [DOI] [PubMed] [Google Scholar]
- 12.Ceylan S. Seidel V. Ziebart N. Berndt C. Dirdjaja N. Krauth-Siegel RL. The dithiol glutaredoxins of African trypanosomes have distinct roles and are closely linked to the unique trypanothione metabolism. J Biol Chem. 2010;285:35224–35237. doi: 10.1074/jbc.M110.165860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng NH. Liu JZ. Brock A. Nelson RS. Hirschi KD. AtGRXcp, an Arabidopsis chloroplastic glutaredoxin, is critical for protection against protein oxidative damage. J Biol Chem. 2006;281:26280–26288. doi: 10.1074/jbc.M601354200. [DOI] [PubMed] [Google Scholar]
- 14.Clayton C. Shapira M. Post-transcriptional regulation of gene expression in trypanosomes and leishmanias. Mol Biochem Parasitol. 2007;156:93–101. doi: 10.1016/j.molbiopara.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 15.Comini MA. Krauth-Siegel RL. Flohé L. Depletion of the thioredoxin homologue tryparedoxin impairs antioxidative defense in African trypanosomes. Biochem J. 2007;402:43–49. doi: 10.1042/BJ20061341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Comini MA. Rettig J. Dirdjaja N. Hanschmann EM. Berndt C. Krauth-Siegel RL. Monothiol glutaredoxin-1 is an essential iron-sulfur protein in the mitochondrion of African trypanosomes. J Biol Chem. 2008;283:27785–27798. doi: 10.1074/jbc.M802010200. [DOI] [PubMed] [Google Scholar]
- 17.Couturier J. Ströher E. Albetel AN. Roret T. Muthuramalingam M. Tarrago L. Seidel T. Tsan P. Jacquot JP. Johnson MK. Dietz KJ. Didierjean C. Rouhier N. Arabidopsis chloroplastic glutaredoxin C5 as a model to explore molecular determinants for iron-sulfur cluster binding into glutaredoxins. J Biol Chem. 2011;286:27515–27527. doi: 10.1074/jbc.M111.228726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deponte M. Becker K. Rahlfs S. Plasmodium falciparum glutaredoxin-like proteins. Biol Chem. 2005;386:33–40. doi: 10.1515/BC.2005.005. [DOI] [PubMed] [Google Scholar]
- 19.Discola KF. de Oliveira MA. Rosa Cussiol JR. Monteiro G. Bárcena JA. Porras P. Padilla CA. Guimarães BG. Netto LE. Structural aspects of the distinct biochemical properties of glutaredoxin 1 and glutaredoxin 2 from Saccharomyces cerevisiae. J Mol Biol. 2009;385:889–901. doi: 10.1016/j.jmb.2008.10.055. [DOI] [PubMed] [Google Scholar]
- 20.Dormeyer M. Reckenfelderbaumer N. Lüdemann H. Krauth-Siegel RL. Trypanothione-dependent synthesis of deoxyribonucleotides by Trypanosoma brucei ribonucleotide reductase. J Biol Chem. 2001;276:10602–10606. doi: 10.1074/jbc.M010352200. [DOI] [PubMed] [Google Scholar]
- 21.Dufernez F. Yernaux C. Gerbod D. Noël C. Chauvenet M. Wintjens R. Edgcomb VP. Capron M. Opperdoes FR. Viscogliosi E. The presence of four iron-containing superoxide dismutase isozymes in trypanosomatidae: characterization, subcellular localization, and phylogenetic origin in Trypanosoma brucei. Free Radic Biol Med. 2006;40:210–225. doi: 10.1016/j.freeradbiomed.2005.06.021. [DOI] [PubMed] [Google Scholar]
- 22.Eckers E. Bien M. Stroobant V. Herrmann JM. Deponte M. Biochemical characterization of dithiol glutaredoxin 8 from Saccharomyces cerevisiae: The catalytic redox mechanism redux. Biochemistry. 2009;48:1410–1423. doi: 10.1021/bi801859b. [DOI] [PubMed] [Google Scholar]
- 23.Elgán TH. Berndt KD. Quantifying Escherichia coli glutaredoxin-3 substrate specificity using ligand-induced stability. J Biol Chem. 2008;283:32839–32847. doi: 10.1074/jbc.M804019200. [DOI] [PubMed] [Google Scholar]
- 24.Fairlamb AH. Cerami A. Metabolism and functions of trypanothione in the Kinetoplastida. Annu Rev Microbiol. 1992;46:695–729. doi: 10.1146/annurev.mi.46.100192.003403. [DOI] [PubMed] [Google Scholar]
- 25.Feng Y. Zhong N. Rouhier N. Hase T. Kusunoki M. Jacquot JP. Jin C. Xia B. Structural insight into poplar glutaredoxin C1 with a bridging iron-sulfur cluster at the active site. Biochemistry. 2006;45:7998–8008. doi: 10.1021/bi060444t. [DOI] [PubMed] [Google Scholar]
- 26.Fernandes AP. Fladvad M. Berndt C. Andrésen C. Lillig CH. Neubauer P. Sunnerhagen M. Holmgren A. Vlamis-Gardikas A. A novel monothiol glutaredoxin (Grx4) from Escherichia coli can serve as a substrate for thioredoxin reductase. J Biol Chem. 2005;280:24544–24552. doi: 10.1074/jbc.M500678200. [DOI] [PubMed] [Google Scholar]
- 27.Filser M. Comini MA. Molina-Navarro MM. Dirdjaja N. Herrero E. Krauth-Siegel RL. Cloning, functional analysis, and mitochondrial localization of Trypanosoma brucei monothiol glutaredoxin-1. Biol Chem. 2008;389:21–32. doi: 10.1515/BC.2007.147. [DOI] [PubMed] [Google Scholar]
- 28.Fladvad M. Bellanda M. Fernandes AP. Mammi S. Vlamis-Gardikas A. Holmgren A. Sunnerhagen M. Molecular mapping of functionalities in the solution structure of reduced Grx4, a monothiol glutaredoxin from Escherichia coli. J Biol Chem. 2005;280:24553–24561. doi: 10.1074/jbc.M500679200. [DOI] [PubMed] [Google Scholar]
- 29.Friemann R. Schmidt H. Ramaswamy S. Forstner M. Krauth-Siegel RL. Eklund H. Structure of thioredoxin from Trypanosoma brucei brucei. FEBS Lett. 2003;554:301–205. doi: 10.1016/s0014-5793(03)01173-6. [DOI] [PubMed] [Google Scholar]
- 30.Gallogly MM. Starke DW. Leonberg AK. Ospina SM. Mieyal JJ. Kinetic and mechanistic characterization and versatile catalytic properties of mammalian glutaredoxin 2: Implications for intracellular roles. Biochemistry. 2008;47:11144–11157. doi: 10.1021/bi800966v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gerdes SY. Scholle MD. Campbell JW. Balázsi G. Ravasz E. Daugherty MD. Somera AL. Kyrpides NC. Anderson I. Gelfand MS. Bhattacharya A. Kapatral V. D'Souza M. Baev MV. Grechkin Y. Mseeh F. Fonstein MY. Overbeek R. Barabási AL. Oltvai ZN. Osterman AL. Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J Bacteriol. 2003;185:5673–5684. doi: 10.1128/JB.185.19.5673-5684.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guerry P. Herrmann T. Comprehensive automation for NMR structure determination of proteins. Methods Mol Biol. 2012;831:429–451. doi: 10.1007/978-1-61779-480-3_22. [DOI] [PubMed] [Google Scholar]
- 33.Herrero E. de la Torre-Ruiz MA. Monothiol glutaredoxins: A common domain for multiple functions. Cell Mol Life Sci. 2007;64:1518–1530. doi: 10.1007/s00018-007-6554-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hillebrand H. Schmidt A. Krauth-Siegel RL. A second class of peroxidases linked to the trypanothione metabolism. J Biol Chem. 2003;278:6809–6815. doi: 10.1074/jbc.M210392200. [DOI] [PubMed] [Google Scholar]
- 35.Hoffmann B. Uzarska MA. Berndt C. Godoy JR. Haunhorst P. Lillig CH. Lill R. Mühlenhoff U. The multidomain thioredoxin-monothiol glutaredoxins represent a distinct functional group. Antioxid Redox Signal. 2011;15:19–30. doi: 10.1089/ars.2010.3811. [DOI] [PubMed] [Google Scholar]
- 36.Holmgren A. Hydrogen donor system for Escherichia coli ribonucleoside-diphosphate reductase dependent upon glutathione. Proc Natl Acad Sci USA. 1976;73:2275–2279. doi: 10.1073/pnas.73.7.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holmgren A. Thioredoxin catalyzes the reduction of insulin disulfides by dithiothreitol and dihydrolipoamide. J Biol Chem. 1979;254:9627–9632. [PubMed] [Google Scholar]
- 38.Indu S. Kochat V. Thakurela S. Ramakrishnan C. Varadarajan R. Conformational analysis and design of cross-strand disulfides in antiparallel β-sheets. Proteins. 2011;79:244–260. doi: 10.1002/prot.22878. [DOI] [PubMed] [Google Scholar]
- 39.Irigoín F. Cibils L. Comini MA. Wilkinson SR. Flohé L. Radi R. Insights into the redox biology of Trypanosoma cruzi: Trypanothione metabolism and oxidant detoxification. Free Radic Biol Med. 2008;45:733–742. doi: 10.1016/j.freeradbiomed.2008.05.028. [DOI] [PubMed] [Google Scholar]
- 40.Iwema T. Picciocchi A. Traore DA. Ferrer JL. Chauvat F. Jacquamet L. Structural basis for delivery of the intact [Fe2S2] cluster by monothiol glutaredoxin. Biochemistry. 2009;48:6041–6043. doi: 10.1021/bi900440m. [DOI] [PubMed] [Google Scholar]
- 41.Johansson C. Lillig CH. Holmgren A. Human mitochondrial glutaredoxin reduces S-glutathionylated proteins with high affinity accepting electrons from either glutathione or thioredoxin reductase. J Biol Chem. 2004;279:7537–7543. doi: 10.1074/jbc.M312719200. [DOI] [PubMed] [Google Scholar]
- 42.Johansson C. Kavanagh KL. Gileadi O. Oppermann U. Reversible sequestration of active site cysteines in a 2Fe-2S-bridged dimer provides a mechanism for glutaredoxin 2 regulation in human mitochondria. J Biol Chem. 2007;282:3077–3082. doi: 10.1074/jbc.M608179200. [DOI] [PubMed] [Google Scholar]
- 43.Johansson C. Roos AK. Montano SJ. Sengupta R. Filippakopoulos P. Guo K. von Delft F. Holmgren A. Oppermann U. Kavanagh KL. The crystal structure of human GLRX5: iron–sulfur cluster co-ordination, tetrameric assembly and monomer activity. Biochem J. 2011;433:303–311. doi: 10.1042/BJ20101286. [DOI] [PubMed] [Google Scholar]
- 44.Krauth-Siegel RL. Comini MA. Redox control in trypanosomatids, parasitic protozoa with trypanothione-based thiol metabolism. Biochim Biophys Acta. 2008;1780:1236–1248. doi: 10.1016/j.bbagen.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 45.Krauth-Siegel RL. Leroux AE. Low-molecular-mass antioxidants in parasites. Antioxid Redox Signal. 2012;17:583–607. doi: 10.1089/ars.2011.4392. [DOI] [PubMed] [Google Scholar]
- 46.Lee DW. Kaur D. Chinta SJ. Rajagopalan S. Andersen JK. A disruption in iron sulfur center biogenesis via inhibition of mitochondrial dithiol glutaredoxin 2 may contribute to mitochondrial and cellular iron dysregulation in mammalian glutathione-depleted dopaminergic cells: Implications for Parkinson's disease. Antioxid Redox Signal. 2009;11:2083–2094. doi: 10.1089/ars.2009.2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li L. Cheng N. Hirschi KD. Wang X. Structure of Arabidopsis chloroplastic monothiol glutaredoxin AtGRXcp. Acta Crystallogr D Biol Crystallogr. 2010;66:725–732. doi: 10.1107/S0907444910013119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lill R. Mühlenhoff U. Maturation of iron-sulfur proteins in eukaryotes: Mechanisms, connected processes, and diseases. Annu Rev Biochem. 2008;77:669–700. doi: 10.1146/annurev.biochem.76.052705.162653. [DOI] [PubMed] [Google Scholar]
- 49.Lillig CH. Berndt C. Vergnolle O. Lönn ME. Hudemann C. Bill E. Holmgren A. Characterization of human glutaredoxin 2 as iron-sulfur protein: A possible role as redox sensor. Proc Natl Acad Sci USA. 2005;102:8168–8173. doi: 10.1073/pnas.0500735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lopes AH. Souto-Padrón T. Dias FA. Gomes MT. Rodrigues GC. Zimmermann LT. Alves e Silva TL. Vermelho AB. Trypanosomatids: Odd organisms, devastating diseases. Open Parasitol J. 2010;4:30–59. [Google Scholar]
- 51.Lönn ME. Hudemann C. Berndt C. Cherkasov V. Capani F. Holmgren A. Lillig CH. Expression pattern of human glutaredoxin 2 isoforms: Identification and characterization of two testis/cancer cell-specific isoforms. Antioxid Redox Signal. 2008;10:547–557. doi: 10.1089/ars.2007.1821. [DOI] [PubMed] [Google Scholar]
- 52.Lüdemann H. Dormeyer M. Sticherling C. Stallmann D. Follmann H. Krauth-Siegel RL. Trypanosoma brucei tryparedoxin, a thioredoxin-like protein in African trypanosomes. FEBS Lett. 1998;431:381–385. doi: 10.1016/s0014-5793(98)00793-5. [DOI] [PubMed] [Google Scholar]
- 53.Lundberg M. Johansson C. Chandra J. Enoksson M. Jacobsson G. Ljung J. Johansson M. Holmgren A. Cloning and expression of a novel human glutaredoxin (Grx2) with mitochondrial and nuclear isoforms. J Biol Chem. 2001;276:26269–26275. doi: 10.1074/jbc.M011605200. [DOI] [PubMed] [Google Scholar]
- 54.Luo M. Jiang YL. Ma XX. Tang YJ. He YX. Yu J. Zhang RG. Chen Y. Zhou CZ. Structural and biochemical characterization of yeast monothiol glutaredoxin Grx6. J Mol Biol. 2010;398:614–622. doi: 10.1016/j.jmb.2010.03.029. [DOI] [PubMed] [Google Scholar]
- 55.Manta B. Fleitas L. Comini MA. Iron metabolism in pathogenic trypanosomes. In: Rajica Arora S., editor. Iron Metabolism. Croatia: INTECH; 2012. pp. 1–40. [Google Scholar]
- 55a.Manta B. Pavan C. Sturlese M. Medeiros A. Crispo M. Berndt C. Krauth-Siegel RL. Bellanda M. Comini MA. Iron-sulfur cluster binding by mitochondrial monothiol glutaredoxin-1 of Trypanosoma brucei: Molecular basis of iron-sulfur cluster coordination and relevance for parasite infectivity. Antioxid Redox Signal. 2013;19:665–682. doi: 10.1089/ars.2012.4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marquez VE. Arias DG. Piattoni CV. Robello C. Iglesias AA. Guerrero SA. Cloning, expression, and characterization of a dithiol glutaredoxin from Trypanosoma cruzi. Antioxid Redox Signal. 2010;12:787–792. doi: 10.1089/ars.2009.2907. [DOI] [PubMed] [Google Scholar]
- 57.Martin JL. Thioredoxin: A fold for all reasons. Structure. 1995;3:245–250. doi: 10.1016/s0969-2126(01)00154-x. [DOI] [PubMed] [Google Scholar]
- 58.Melchers J. Dirdjaja N. Ruppert T. Krauth-Siegel RL. Glutathionylation of trypanosomal thiol redox proteins. J Biol Chem. 2007;282:8678–8694. doi: 10.1074/jbc.M608140200. [DOI] [PubMed] [Google Scholar]
- 59.Mesecke N. Mittler S. Eckers E. Herrmann JM. Deponte M. Two novel monothiol glutaredoxins from Saccharomyces cerevisiae provide further insight into iron-sulfur cluster binding, oligomerization, and enzymatic activity of glutaredoxins. Biochemistry. 2008;47:1452–1463. doi: 10.1021/bi7017865. [DOI] [PubMed] [Google Scholar]
- 60.Mesecke N. Spang A. Deponte M. Herrmann JM. A novel group of glutaredoxins in the cis-Golgi critical for oxidative stress resistance. Mol Biol Cell. 2008;19:2673–2680. doi: 10.1091/mbc.E07-09-0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Molina-Navarro MM. Casas C. Piedrafita L. Bellí G. Herrero E. Prokaryotic and eukaryotic monothiol glutaredoxins are able to perform the functions of Grx5 in the biogenesis of Fe/S clusters in yeast mitochondria. FEBS Lett. 2006;580:2273–2280. doi: 10.1016/j.febslet.2006.03.037. [DOI] [PubMed] [Google Scholar]
- 62.Muhlenhoff U. Molik S. Godoy JR. Uzarsk MA. Richter N. Seubert A. Zhang Y. Stubbe J. Pierrel F. Herrero E. Lillig CH. Lill R. Cytosolic monothiol glutaredoxins function in intracellular iron sensing and trafficking via their bound iron–sulfur cluster. Cell Metab. 2010;12:373–385. doi: 10.1016/j.cmet.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nogoceke E. Gommel DU. Kiess M. Kalisz HM. Flohé L. A unique cascade of oxidoreductases catalyses trypanothione-mediated peroxide metabolism in Crithidia fasciculata. Biol Chem. 1997;378:827–836. doi: 10.1515/bchm.1997.378.8.827. [DOI] [PubMed] [Google Scholar]
- 64.Nolan DP. Voorheis HP. The mitochondrion in bloodstream forms of Trypanosoma brucei is energized by the electrogenic pumping of protons catalysed by the F1F0-ATPase. Eur J Biochem. 1992;209:207–216. doi: 10.1111/j.1432-1033.1992.tb17278.x. [DOI] [PubMed] [Google Scholar]
- 65.Pai HV. Starke DW. Lesnefsky EJ. Hoppel CL. Mieyal JJ. What is the functional significance of the unique location of glutaredoxin 1 (GRx1) in the intermembrane space of mitochondria? Antioxid Redox Signal. 2007;9:2027–2033. doi: 10.1089/ars.2007.1642. [DOI] [PubMed] [Google Scholar]
- 66.Piattoni CV. Blancato VS. Miglietta H. Iglesias AA. Guerrero SA. On the occurrence of thioredoxin in Trypanosoma cruzi. Acta Trop. 2006;97:151–160. doi: 10.1016/j.actatropica.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 67.Picciocchi A. Saguez C. Boussac A. Cassier-Chauvat C. Chauvat F. CGFS-type monothiol glutaredoxins from the cyanobacterium Synechocystis PCC6803 and other evolutionary distant model organisms possess a glutathione-ligated [2Fe-2S] cluster. Biochemistry. 2007;46:15018–15026. doi: 10.1021/bi7013272. [DOI] [PubMed] [Google Scholar]
- 68.Qi Y. Grishin NV. Structural classification of thioredoxin‐like fold proteins. Proteins. 2005;58:376–388. doi: 10.1002/prot.20329. [DOI] [PubMed] [Google Scholar]
- 69.Rahlfs S. Fischer M. Becker K. Plasmodium falciparum possesses a classical glutaredoxin and a second, glutaredoxin-like protein with a PICOT homology domain. J Biol Chem. 2001;276:37133–37140. doi: 10.1074/jbc.M105524200. [DOI] [PubMed] [Google Scholar]
- 70.Reckenfelderbäumer N. Lüdemann H. Schmidt H. Steverding D. Krauth-Siegel RL. Identification and functional characterization of thioredoxin from Trypanosoma brucei brucei. J Biol Chem. 2000;275:7547–7552. doi: 10.1074/jbc.275.11.7547. [DOI] [PubMed] [Google Scholar]
- 71.Ren G. Stephan D. Xu Z. Zheng Y. Tang D. Harrison RS. Kurz M. Jarrott R. Shouldice SR. Hiniker A. Martin JL. Heras B. Bardwell JC. Properties of the thioredoxin fold superfamily are modulated by a single amino acid residue. J Biol Chem. 2009;284:10150–10159. doi: 10.1074/jbc.M809509200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rodríguez-Manzaneque MT. Ros J. Cabiscol E. Sorribas A. Herrero E. Grx5 glutaredoxin plays a central role in protection against protein oxidative damage in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:8180–8190. doi: 10.1128/mcb.19.12.8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rodríguez-Manzaneque MT. Tamarit J. Bellí G. Ros J. Herrero E. Grx5 is a mitochondrial glutaredoxin required for the activity of iron/sulfur enzymes. Mol Biol Cell. 2002;13:1109–1121. doi: 10.1091/mbc.01-10-0517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rouhier N. Couturier J. Johnson MK. Jacquot J-P. Glutaredoxins: Roles in iron homeostasis. Trends Biochem Sci. 2010;35:43–52. doi: 10.1016/j.tibs.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rouhier N. Unno H. Bandyopadhyay S. Masip L. Kim SK. Hirasawa M. Gualberto JM. Lattard V. Kusunoki M. Knaff DB. Georgiou G. Hase T. Johnson MK. Jacquot JP. Functional, structural, and spectroscopic characterization of a glutathione-ligated [2Fe-2S] cluster in poplar glutaredoxin C1. Proc Natl Acad Sci USA. 2007;104:7379–7384. doi: 10.1073/pnas.0702268104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ruoppolo M. Lundström-Ljung J. Talamo F. Pucci P. Marino G. Effect of glutaredoxin and protein disulfide isomerase on the glutathione-dependent folding of ribonuclease A. Biochemistry. 1997;36:12259–12267. doi: 10.1021/bi970851s. [DOI] [PubMed] [Google Scholar]
- 76a.Sardi F. Manta B. Portillo-Ledesma S. Knoops B. Comini MA. Ferrer-Sueta G. Determination of acidity and nucleophilicity in thiols by reaction with monobromobimane and fluorescence detection. Anal Biochem. 2013 doi: 10.1016/j.ab.2012.12.017. [In Press]. [DOI] [PubMed] [Google Scholar]
- 77.Schlecker T. Schmidt A. Dirdjaja N. Voncken F. Clayton C. Krauth-Siegel RL. Substrate specificity, localization, and essential role of the glutathione peroxidase-type tryparedoxin peroxidases in Trypanosoma brucei. J Biol Chem. 2005;280:14385–14394. doi: 10.1074/jbc.M413338200. [DOI] [PubMed] [Google Scholar]
- 78.Schmidt H. Krauth-Siegel RL. Functional and physicochemical characterization of the thioredoxin system in Trypanosoma brucei. J Biol Chem. 2003;278:46329–46336. doi: 10.1074/jbc.M305338200. [DOI] [PubMed] [Google Scholar]
- 79.Schmidt A. Clayton CE. Krauth-Siegel RL. Silencing of the thioredoxin gene in Trypanosoma brucei brucei. Mol Biochem Parasitol. 2002;125:207–210. doi: 10.1016/s0166-6851(02)00215-3. [DOI] [PubMed] [Google Scholar]
- 80.Smíd O. Horáková E. Vilímová V. Hrdy I. Cammack R. Horváth A. Lukes J. Tachezy J. Knock-downs of iron-sulfur cluster assembly proteins IscS and IscU down-regulate the active mitochondrion of procyclic Trypanosoma brucei. J Biol Chem. 2006;281:28679–28686. doi: 10.1074/jbc.M513781200. [DOI] [PubMed] [Google Scholar]
- 81.Srinivasan U. Mieyal PA. Mieyal JJ. pH profiles indicative of rate-limiting nucleophilic displacement in thioltransferase catalysis. Biochemistry. 1997;36:3199–3206. doi: 10.1021/bi962017t. [DOI] [PubMed] [Google Scholar]
- 82.Tamarit J. Belli G. Cabiscol E. Herrero E. Ros J. Biochemical characterization of yeast mitochondrial Grx5 monothiol glutaredoxin. J Biol Chem. 2003;278:25745–25751. doi: 10.1074/jbc.M303477200. [DOI] [PubMed] [Google Scholar]
- 83.Taylor MC. Kelly JM. Iron metabolism in trypanosomatids, and its crucial role in infection. Parasitology. 2010;137:899–917. doi: 10.1017/S0031182009991880. [DOI] [PubMed] [Google Scholar]
- 84.Tetaud E. Giroud C. Prescott AR. Parkin DW. Baltz D. Biteau N. Baltz T. Fairlamb AH. Molecular characterisation of mitochondrial and cytosolic trypanothione-dependent tryparedoxin peroxidases in Trypanosoma brucei. Mol Biochem Parasitol. 2001;116:171–183. doi: 10.1016/s0166-6851(01)00320-6. [DOI] [PubMed] [Google Scholar]
- 85.Thompson JD. Higgins DG. Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vilella F. Alves R. Rodríguez-Manzaneque MT. Bellí G. Swaminathan S. Sunnerhagen P. Herrero E. Evolution and cellular function of monothiol glutaredoxins: involvement in iron–sulfur cluster assembly. Comp Funct Genomics. 2004;5:328–341. doi: 10.1002/cfg.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wingert RA. Galloway JL. Barut B. Foott H. Fraenkel P. Axe JL. Weber GJ. Dooley K. Davidson AJ. Schmid B. Paw BH. Shaw GC. Kingsley P. Palis J. Schubert H. Chen O. Kaplan J. Zon LI. Tübingen 2000 Screen Consortium. Deficiency of glutaredoxin 5 reveals Fe–S clusters are required for vertebrate haem synthesis. Nature. 2005;436:1035–1039. doi: 10.1038/nature03887. [DOI] [PubMed] [Google Scholar]
- 88.Ye H. Jeong SY. Ghosh MC. Kovtunovych G. Silvestri L. Ortillo D. Uchida N. Tisdale J. Camaschella C. Rouault TA. Glutaredoxin 5 deficiency causes sideroblastic anemia by specifically impairing heme biosynthesis and depleting cytosolic iron in human erythroblasts. J Clin Invest. 2010;120:1749–1761. doi: 10.1172/JCI40372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yeung N. Gold B. Liu NL. Prathapam R. Sterling HJ. Willams ER. Butland G. The E. coli monothiol glutaredoxin GrxD forms homodimeric and heterodimeric FeS cluster containing complexes. Biochemistry. 2011;50:8957–8969. doi: 10.1021/bi2008883. [DOI] [PMC free article] [PubMed] [Google Scholar]