Abstract
This report describes an improved protocol to generate stranded, barcoded RNA-seq libraries to capture the whole transcriptome. By optimizing the use of duplex specific nuclease (DSN) to remove ribosomal RNA reads from stranded barcoded libraries, we demonstrate improved efficiency of multiplexed next generation sequencing (NGS). This approach detects expression profiles of all RNA types, including miRNA (microRNA), piRNA (Piwi-interacting RNA), snoRNA (small nucleolar RNA), lincRNA (long non-coding RNA), mtRNA (mitochondrial RNA) and mRNA (messenger RNA) without the use of gel electrophoresis. The improved protocol generates high quality data that can be used to identify differential expression in known and novel coding and non-coding transcripts, splice variants, mitochondrial genes and SNPs (single nucleotide polymorphisms).
Keywords: RNA-seq, transcriptome, duplex-specific nuclease, gene expression1
1. Introduction
Next-generation sequencing (NGS) technologies represent the most advanced molecular tools currently available to interrogate the complexities of the human transcriptome in its entirety [1, 2, 3]. RNA-seq NGS has the capability and capacity to detect all types of gene expression, i.e., the whole transcriptome, including small noncoding RNAs, mRNAs, lincRNAs, novel transcripts, and repetitive elements in a manner capable of identifying splice variants and SNPs [4, 5, 6]. A whole transcriptome analysis requires the reduction of rRNA products for efficient NGS while incorporating barcoded adapters for multiplexing. While current RNA-seq library generation protocols utilize poly(A+) selection, RNase H, Ribozero and random hexamer priming for rRNA reduction [3, 7, 8, 9, 10], here we describe an improved protocol for capturing the strand-specific whole transcriptome of gene expression. This new method utilizes re-association kinetics and duplex specific nuclease (DSN) to preferentially remove highly abundant rRNA species. The use of specially designed barcoded TrueSeq adapters facilitates multiplexing and sequencing of samples on the Illumina HiSeq platform [11, 12].
1.1 Work flow for RNA-seq
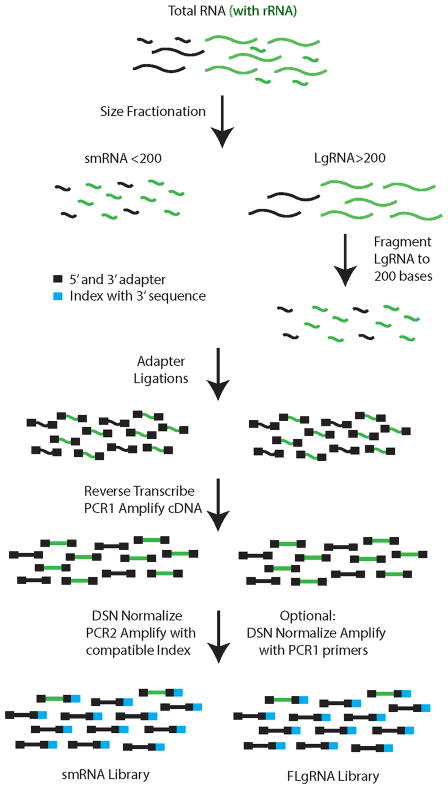
The key steps used to generate barcoded, stranded, whole transcriptome RNA-seq libraries are illustrated in Fig. 1. Beginning with total RNA, samples are fractionated by size to separate RNA species <200 nt (smRNA) from large RNA >200 nt (LgRNA). The LgRNAs are chemically fragmented to an average of 200 nt in length [13]. The ends are prepared and the adapters are sequentially added to the ends to retain strandedness. Following PCR amplification, the libraries are normalized using hybridization kinetics, duplex specific nuclease (DSN), and PCR amplification with the addition of barcoded index sequences for multiplexing samples in a single flow cell lane [11, 12]. This approach allows the capture of all RNA species independent of size or poly(A) status.
Fig. 1.
Library generation workflow. Starting with total RNA, size fractionation into smRNA (<200 nt) and LgRNA (>200 nt) generates whole transcriptome coverage in two fractions. Following fragmentation of LgRNA (FLgRNA), protocols to generate stranded RNA-seq libraries are essentially the same for both fractions. Duplex specific nuclease (DSN) is utilized to remove rRNA sequences while retaining all other transcript library elements. Index sequences are added after DSN treatment, during PCR2, to generate libraries that can be used for multiplexed next generation sequencing with the Illumina platform.
The improved method described in this report is practical for multiplexed RNA-seq on the Illumina HiSeq platform. For Illumina multiplex sequencing, it is critical to plan the index sequence combinations in advance due to limitations with this platform. Not all index sequences are compatible for de-multiplexing and should be carefully reviewed prior to the addition of these sequences [11]. Our approach utilizes DSN normalization yet allows flexibility for addition of the barcodes later, with the option of generating multiple versions of the same library with different barcoded adapter sequences.
2. Materials and methods
2.1 Cell culture
For this study, we used a pair of isogenic epithelial ovarian cancer cell lines (A2780 and A2780-C1R5) [14]. The culture conditions for parental A2780 line (sensitive to the chemotherapy cisplatin) A2780-C1R5 (derived from A2780 after several rounds of cisplatin selection and thus represents acquired drug resistance) have been described previously [14]. Briefly, cells were grown in RPMI 1640 media (ThermoFisher Scientific, Wilmington, MA 01887, cat. no. MT-15-040-CV) supplemented with 10% fetal bovine serum (Atlanta Biological, Norcross, GA 30093, cat. no. S11050) and 2 mM glutamine, 100 units U/mL penicillin and 100 μg/mL Streptomycin (ThermoFisher Scientific, Wilmington, MA 01887, cat. no. MT30-009-CI) at 37°C in 5% CO2 in a Steri-Cycle CO2 incubator (ThermoFisher Scientific, Wilmington, MA 01887, model 370 Series).
2.2 RNA isolation
RNA was isolated from trypsinized cells utilizing a modified version of the Qiagen RNeasy AllPrep method (Qiagen Sciences, Valencia, CA 91355, cat. no. 80004). Pellets were harvested at 4°C, 1,000 rpm (129 g) for 5 min in an Eppendorf fixed angle rotor (Hauppauge, NY 11788, cat. no. F34-6-38) using a Falcon 15 mL conical (ThermoFisher Scientific, Wilmington, MA 01887,, cat. no. 14-959-70C) in an Eppendorf 5810R centrifuge at 4°C. Cells were lysed in 600 μL RNeasy RLT buffer (Qiagen Sciences, Valencia, CA 91355, cat. no. 79216) with 1% BME (vol/vol) by vortexing or raking the tube across a micro-centrifuge tube rack about 20 times. Cell lysis was completed by centrifugation through the Qiashredder spin column (Qiagen Sciences, Valencia, CA 91355, cat. no. 79654) for 2 min at 13,300 rpm (17,000 g) in an AccuSpin Micro17 centrifuge (ThermoFisher Scientific, Wilmington, MA 01887, cat. no. 13-100-675). The flow-thru was collected and passed over an AP-DNA column (10,200 rpm, 10,000 g) for 20 s to remove DNA. Ethanol (900 μL) was added to a 60% final concentration (vol/vol) to the RNA/protein fraction before RNA capture using an RNeasy spin column (centrifugation at 10,200 rpm for 20 s). The protein flow-thru fraction was stored at −80°C for later use. The RNeasy column was washed with Qiagen RWT buffer (Qiagen Sciences, Valencia, CA 91355, cat. no. 1067933) containing ethanol (as recommended by the manufacturer) for 20 s at 10,200 rpm. The RNeasy column was then washed with 500uL RPE buffer by centrifugation for 20 s at 10,200 rpm followed by centrifugation for 5 min at 13,300 rpm. The RNeasy column was also washed with 500μL of 80% (vol/vol) ethanol by centrifugation for 5 min at 13,300 rpm. The column was transferred to a clean collection tube and centrifuged for an additional 5 min at 13,300 rpm to dry the column. Elution of the total RNA fraction into a clean collection tube was accomplished by soaking the column matrix with 50uL DIW (supplied in the kit) for one minute followed by centrifugation for one minute at 13,300 rpm. Total RNA samples were stored at −80°C until further use. RNA integrity was monitored using a Bioanalyzer and the Agilent RNA 6000 Pico assay (Agilent Technologies, Santa Clara, CA, 95051, cat. no. 5067-1513). An RNA integrity number (RIN) greater than 8 is recommended for all samples.
2.3 RNA size fractionation
Size fractionation of total RNA samples around 200 nt was accomplished utilizing a supplemental Qiagen protocol [13]. For optimal results, 1–5 μg total RNA was diluted into 100 μL DIW and combined with 350 μL Qiagen RLT buffer containing 1% BME. Next, 302 μL ethanol was added to 40% (vol/vol) final concentration (0.67 vol. 100% ethanol) and passed the sample over a Qiagen RNeasy Minelute column (Qiagen Sciences, Valencia, CA 91355, cat. no. 74204), which allowed small RNA species (smRNA) less than 200 nt to pass through while retaining RNA larger than 200 nt (LgRNA). Then, ethanol in the smRNA fraction was increased to 60% by adding 375 μL (50% vol/vol) 100% ethanol. Both bound RNA fractions (smRNA, LgRNA) were washed with 500uL RPE buffer (20 s at 10,200 rpm) followed by 500 μL 80% ethanol (vol/vol) for 5 min, 13,300 rpm spins respectively. The samples in the columns were dried by centrifugation in a clean tube for 5 min at 13,300 rpm. RNA was recovered by elution with 34 μL DIW (de-ionized water) for smRNA or 37 μL DIW for LgRNA and centrifuged for 1 min at 13,300 rpm. The RNA concentrations were determined using a Nanodrop 2000 (ThermoFisher Scientific, Wilmington, MA 01887). Bioanalyzer profiles (Agilent RNA 6000 Pico assay) for all RNA fractions are shown in supplemental figure S1.
2.4 RNA fragmentation
The LgRNA fraction was fragmented to an average length of 150 nt prior to the adapter ligation. For optimal results, start with 500–2,000 ng LgRNA in 36 μL DIW. To this, add 4 μL 10X Ambion fragmentation reagent (Life Technologies, Grand Island, NY 14072, cat. no. AM8740) and incubate the sample for 4 min at 70°C. Quench the reaction with 4 μL stop buffer (supplied) and place on ice. Purify the fragmented LgRNA (FLgRNA) using an RNeasy MinElute, 40% (vol/vol) ethanol as described above (sec. 2.3) and elute in 18 μL DIW. The Bioanalyzer profile for FLgRNA is shown in supplemental figure S1D.
2.5 RNA end repair
smRNA or FLgRNA ends were repaired before adapter ligation in the same manner essentially as described [12]. RNA (17 μL) was phosphatase-treated with 1 μL Antarctic phosphatase (New England Biolabs, Ipswich, MA 01938, cat. no. #M0289L), 2 μL 10X reaction buffer and 0.5 μL RNasin (Promega Corporation, Madison, WI 53711, cat. no. N2511) for 30 min at 37°C. Phosphatase was heat-inactivated for 5 min at 65°C. The 5′ end of the RNA was phosphorylated at 37°C for 60 min with 2 μL (20 U) T4 polynucleotide phosphate (New England Biolabs, Ipswich, MA 01938, cat. no. M0201L), 5 μL 10X reaction buffer (supplied), 5 μL 10mM ATP (Epicentre Biotechnologies, Madison, WI 53713, cat. no. RA02825), 0.5 μL RNasin and 17 μL DIW. RNA samples were purified with RNeasy MinElute columns (described previously for smRNA, sec. 2.3) after adding 175 μL RLT (with 1% BME), 338 μL ethanol (60% vol/vol final concentration) and then the end repaired RNA was eluted with 15 μL DIW.
2.6 Library adapter ligations
Samples consisting of 13 μL (5 picomole) end repaired RNA were heat denatured with 2 μL of 10 μM (15 picomole) RS-TS-3′ adapter (Suppl Table S1) for 2 min at 70°C and quenched on ice. While one ice the samples were combined and ligated with the following: 2 μL T4 RNA ligase 2 truncated (New England Biolabs, Ipswich, MA 01938, cat. no. M0242L), 2 μL 10X reaction buffer (supplied), 1.6 μL 100mM MgCl2 and 1 μL RNasin. Samples were incubated overnight at 16°C. Before the 5′ adapter was ligated, an aliquot of RS-TS-5′ adapter (Suppl Table S1) was heat-denatured at 70C for 2 min, quenched on ice, then the RS-TS-5′ adapter was ligated at 20°C for 2 h with 2 μL (40 U) T4 RNA ligase 1 (New England Biolabs, Ipswich, MA 01938, cat. no. M0204L), 1 μL 10X reaction buffer (supplied), 3 μL 10 mM ATP, and 1.5 μL denatured 10 μM RS-TS-5′ adapter. If necessary, ligated RNA was stored at −80°C until it was purified with an RNeasy MinElute column using 98 μL RLT (with 1% BME), 85 μL ethanol (final 40% vol/vol) for FLgRNA or 190 μL ethanol (final 60% vol/vol) for smRNA. RNeasy column purification was accomplished as described previously (sec 2.3) and adapter ligated RNA was eluted with 12 μL DIW. Samples can be stored at −80°C until further use. Both RS-TS-3′ and RS-TS-5′ HPLC purified adapters were purchased from Integrated DNA Technologies (Coralville, IA 52241).
2.7 Reverse transcription
Starting with 10 μL 3′-5′ adapter ligated RNA (sec. 2.6), 2.5 μL RS-TS-PCR1A primer (Suppl Table S1) was added, the mixture was heat denatured for 2 min at 70°C, and then quenched on ice. Reverse transcription was carried out by adding 2.5 μL Superscript RT III/RNaseOut enzyme mix plus 13.5 μL 2X Superscript reaction buffer (Life Technologies, Grand Island, NY 14072, cat. no. 18080-400) and incubating the reaction at 50°C for 60 min. Once the reverse transcription reaction was completed, the RNA was digested with 1 μL/10 U RNase H (Life Technologies, Grand Island, NY 14072, cat. no. AM2293) for 20 min at 37°C followed by PCR1 amplification. The RS-TS-PCR1A HPLC purified primer was purchased from Integrated DNA Technologies (Coralville, IA 52241).
2.8 PCR1 amplification of RNA-seq library
The following reagents were combined with the reverse transcription reaction; 20 μL 5X Phusion HF buffer (New England Biolabs, Ipswich, MA 01938, cat. no. F-530), 2 μL (25 μM) RS-TS-PCR1A primer (Suppl Table S1), 2 μL (25 μM) RS-PCR-2C, 1 μL 100 mM dNTP (25 μM each, dATP, dCTP, dGTP, dTTP) (G-Biosciences, 9800 Page Ave, St. Louis, MO 63132, cat no. 786–460), 2 μL 40 mM Na2EDTA, 1 μL Phusion DNA polymerase (2 U/μL (New England Biolabs, Ipswich, MA 01938, cat. no. M0530S) and 42.5 μL DIW. Libraries were amplified by incubating at 98°C for 30 s, followed by 15 cycles of 98°C for 10 s, 55°C for 30 s, 72°C for 15 s followed by one cycle of 72°C for 10 min. A two chamber MJ PTC-200 Peltier thermal cycler (MJ Research, Waltham, MA 02451) was used for all amplifications. All HPLC purified primers were purchased from Integrated DNA Technologies (Coralville, IA 52241).
2.9 AMPure XP purification
PCR purifications were accomplished with Agencourt AMPure XP magnetic beads (Beckman Coulter, Brea, CA 92821, cat. no. A63880). To purify RNA-seq libraries following PCR1 (amplification 1) reactions, 1.6:1 vol/vol (160 μL) AMPure XP beads were added to the FLgRNA or 2.0:1 vol/vol (200 μL) AMPure XP beads were added to the smRNA samples. The bead mixtures were vortexed briefly and incubated at ambient temperature (20°C) for 5 min in a 1.5 mL microfuge tube. The tube was moved to a Magna-Sep stand (Life Technologies, Grand Island, NY 14072, cat. no. K1585–01) for 5 min to allow magnetic beads to collect on the side of the tube with the bound DNA. The DNA/beads were washed with 700 μL 80% (vol/vol) ethanol as follows: while on the stand, tubes were inverted, then removed, rotated 180°, and inserted back into the magnetic stand and the wash discarded. This step was performed twice. After the second wash solution was discarded and the tubes were briefly centrifuged (with the beads oriented outward), returned to the magnetic stand, and any remaining ethanol was discarded. The beads were dried in the tube with the lid open at 40°C for 6 min in a heating block or at ambient temperature until completely dried. DNA was eluted from the magnetic beads by gently pipetting with 30 μL EB (Qiagen Sciences, Valencia, CA 91355, cat. no. 1014609). The tube was returned to the stand to bind the magnetic beads in order to remove the DNA in EB buffer. The DNA was quantified using the Nanodrop 2000 (ThermoFisher Scientific, Wilmington, MA 01887) and then 0.6 μL 5% (vol/vol) Tween 20 (Promega Corporation, Madison, WI 53711, cat. no. H5152) was added. Size selection using the different ratios of AMPure XP beads is shown in supplemental figure S2.
2.10 rRNA product reduction with duplex specific nuclease (DSN)
We used a modified version of the Illumina protocol to remove rRNA sequences with DSN [12]. To remove rRNA sequences from the RNA-seq libraries, 100 ng PCR1 DNA library, determined by Qubit dsDNA HS assay, (Life Technologies, Grand Island, NY 14072, cat. no. Q32854), was combined with 2 μL 10X DSN20 hybridization buffer (200 mM NaCl, 280 mM Tris-HCl, 220 mM Tris-Base (pH 8.0 automatically), 1.5 μL 5% Tween 20, and brought the final volume to 20 μL with DIW. Samples were denatured in the hybridization mix at 98°C for 2 min and hybridized at 68°C for 5 h. In a separate PCR chamber, 20 μL aliquots were preheated of 2X DSNM reaction buffer (56 mM Tris- HCl, 44 mM Tris Base [pH 8.0 automatically], 40 mM MgCl2) at 68°C. The preheated DSNM buffer was mixed into the hybridization reaction and pre-incubated for 10 min at 68°C. Then, 1 μL (1 U) DSN (Wako Chemicals, Richmond, VA 23237, cat. no. EA002) was added to hybridization/DSNM reaction, mixed well and incubated 10 min at 68°C. Multiple reactions were performed concurrently at 30 s or 1 min intervals, starting when the DSNM buffer was added to the hybridization reaction. Then the reaction was stopped with 42 μL ice cold 2X DSNES stop buffer (100mM Na2EDTA, pH 8, 500 mM NaCl) and transferred to −20°C. The DNA was recovered by AMPure XP purification using 1.6:1 vol/vol (135 μL) AMPure XP beads for FLgRNA libraries and 2.0:1 vol/vol (168 μL) AMPure XP beads for smRNA libraries as described previously (sec. 2.9). Reactions were kept cold prior to purification with AMPure XP beads.
2.11 PCR2 amplification of normalized RNA-seq library with indexed primer
AMPure XP purified DSN normalized RNA-seq libraries were combined with 20 μL 5X Phusion HF buffer (New England Biolabs, Ipswich, MA 01938, cat. no. F-530), 2 μL (25 μM) RS-TS PCR-I-# indexed primer (Suppl Table S1), 2 μL (25 μM) RS-PCR-2C, 1 μL 100 mM dNTP (25 μM each, dATP, dCTP, dGTP, dTTP)(G-Biosciences, St. Louis, MO 63132, cat no. 786–460), 1 μL Phusion DNA polymerase (2 U/μL, New England Biolabs, 240 County Rd, Ipswich, MA 01938, cat. no. M0530S) and 68 μL DIW. Libraries were amplified by incubating at 98°C for 30 s, followed by 17 cycles of 98°C for 10 s, 55°C for 30 s, 72°C for 15 s followed by one cycle of 72°C for 10 min. AMPure XP purification was repeated as described previously (sec. 2.9) except 1.6:1 vol/vol ratios (160 μL) AMPure XP beads were used for both FLgRNA and smRNA libraries to eliminate adapter dimers. The increased size of the smRNA library elements due to the addition of the extended 3′ sequences with barcode dictated this. Bioanalyzer traces illustrating adapter dimer (AD) removal with 1.6:1 Ampure XP ratios following the PCR2 amplification, (after DSN normalization and barcode addition) are shown in supplemental figure S2. Barcode index combinations for multiplexing are limited and should be planned before the PCR2 reaction [11]. Barcode combinations used in this study are shown in supplemental table S2. Libraries can also be amplified after DSN normalization without adding the barcode sequence by using RS-TS-PCR1A primer (instead of the RS-TS PCR-I-# index primer), but a 2.0:1 (vol/vol) ratio of AMPure XP beads was required during purification of smRNA libraries as described for smRNA PCR1 AMPure XP cleanup in section 2.9. All HPLC purified primers were purchased from Integrated DNA Technologies (Coralville, IA 52241).
2.12 Quantitative Real-Time PCR (qRT-PCR) for RNA-seq libraries
The fold change (reduction) of rRNA products from the DSN normalization was quantified with qRT-PCR by the relative quantification method [15]. In the current study, qRT-PCR was performed using a Roche LightCycler 480 system (Roche Applied Science, Indianapolis, IN 46250) with the Roche SYBR Green I master mix (cat. no. 04707516001). rRNA sequence removal was monitored using qRT-PCR by comparing PCR1 amplification (sec. 2.8) libraries and DSN normalized PCR2 amplification (sec. 2.11). The PCR mix consisted of 5 μL diluted RNA-seq library template (0.5 ng/μL), 1 μL 5 μM forward and reverse primers, 4 μL DIW and 10 μL 2X SYBR Master mix. Real Time PCR programming for the Roche 480 run Cycle was as follows: 1-cycle 95°C, 5 min, 50-cycles 95°C for 15 s, 60°C for 15 s, 72°C for 30 s and 1-cycle 95°C for 5 s, 60°C for 60 s, 95°C for 0 s and 40°C hold (for melting curve analysis). For smRNA libraries, primers designed to the U6 gene and 5S rRNA were used monitor DSN efficacy. qRT-PCR on FLgRNA libraries required EEF1A1 primers for the reference gene and either 18S rRNA or 28S rRNA primers sets to validate the DSN normalization. All primer and adapter sequences are shown in supplemental table S1. All qRT-PCR primers (standard salt free purification) were obtained from Fisher Scientific, Pittsburgh, PA 15275.
2.13 qRT-PCR with cDNA from total RNA
In order to determine changes in transcript levels between platinum sensitive and resistant cell lines, standard qRT-PCR was utilized. Starting with 2 μg total RNA derived from cells, combine with 1.5 μL DNase (1.5 U, Promega Corporation, 2800 Woods Hollow Rd, Madison, WI 53711, cat. no. M198A), 3 μL 5X MMLV reverse transcriptase buffer (Promega supplied), 1uL RNasin (Promega cat. no. N251B) and the volume was increased to 15 μL with DIW. The DNase reaction was incubated for 60 min at 37°C and then inactivated with DNase at 80°C for 5 min. cDNA was made by adding 2 μL/1μg random hexamer (Qiagen Sciences, Valencia, CA 91355, cat. no. 79236) and denatured at 70°C for 2 min. Then, the DNase-treated RNA was combined with 5 μL 5X MMLV reverse transcriptase buffer (Promega supplied), 2 μL 10 mM dNTP mix (Promega cat. no. U1511), 1 μL RNasin (Promega cat. no. N251B), 1.6 μL MMLV Reverse Transcriptase (Promega cat. no. M1708), 13.4 μL DIW and incubated at 42°C for 60 min, then the cDNA was denatured at 95°C for 5 min. Prior to qRT-PCR, the cDNA was diluted with 160 μL DIW, and the sample was stored at −20°C. qRT-PCR was carried out as described in sec. 2.12 except that 5 μL of diluted cDNA was used instead of diluted RNA-seq PCR1. All qRT-PCR primers (standard salt free purification) were obtained from Fisher Scientific, Pittsburgh, PA 15275.
2.14 Library quantification for Illumina HiSeq2000 50 base single read (SR50) analysis
Accurate quantification was necessary before combining samples for multiplex sequencing. All RNA-seq libraries were pulse centrifuged in EBT buffer to 10,000 rpm (9,200 g) and mixed gently prior to dilutions. The concentration of the library was determined by Qubit analysis and then an aliquot was diluted to 0.5 ng/μL to run on the Bioanalyzer (BioA) high sensitivity (HS) DNA chip (Agilent Technologies, Santa Clara, CA, 95051, cat. no. 5067–4626). Using the BioA region analysis tool, the BioA nanomolar (nM) concentration was determined based on the dilution factor. The BioA region analysis tool was used to determine the average fragment size (# bp), from which the average molecular weight of the library was calculated based on the equation: . Then the Qubit nM concentration of the library was calculated using the molecular weight (from BioA) and the Qubit ng/μL concentration with equation: . Using the higher nM concentration from both calculations described above, each barcoded library (to be mixed for a single lane) was diluted to 34 nM and combined using equal volumes with compatible index combinations (Suppl Table S2). The final concentration determination for the flow cell lane mixture was done by repeating the Qubit and the BioA HS DNA methods described above (this section). The best concentration estimate was again made by taking the higher value of the two concentration estimates (as just described) and carefully diluting (as just described) the multiplexed library mixture to 17 nM. For the Illumina HiSeq2000 platform, 2 μL of the 17 nM mixture of a 200 base average fragment size was loaded into one flow cell lane. Bioanalyzer (BioA) profiles for smRNA and FLgRNA libraries are shown in supplemental figure S4.
2.15 Sequencing
Sequencing was performed on the Illumina HiSeq2000 Sequencer. Library fragments from Section 2.14 were denatured and bridge-amplified on the HiSeq flow cell (TruSeq SR Cluster Kit v3-cBot-HS) prior to sequencing on the HiSeq2000 Sequencer. A 50-cycle TruSeq v3 SBS kit was used for sequencing the library fragments as well as the index portion of the TruSeq adapters. Manufacture’s instruction was followed to combine compatible barcoded libraries per sequencing lane. The barcode combinations were further crosschecked by Illumina Experiment Manager software. Demultiplexing of sequenced data was performed using CASAVA v1.8.2 fastq files. Each index sequence read was compared to the index sequence specified in the sample sheet. One mismatch was allowed per barcode.
2.16 Read Preparation
Multiple rounds of trimming and cleaning were performed to remove low quality and adapter sequences from the reads. Initial cleaning and trimming was accomplished with Trimmomatic (version 0.22) [16]; with the following parameters: “-phred33 ILLUMINACLIP:adapter.fa:2:30:10 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:17”. The adapter file varied with sample based on the sequencing adapters and use of barcodes. Additional trimming to remove short adapter sequences was done with fastx_clipper (version 0.0.13.2) with the parameters: “–l 17 –v –M 1”. For time consuming later steps, identical sequences within a particular sample were collapsed to make a pseudo-read with a known count.
2.17 Repeat Masking
The collapsed read set was repeat masked using RepeatMasker (version open-3.2 [17]) with the following parameters: -e crossmatch -species human -no_is -frag 300000 -small–xm. The abundance of the collapsed read set was then used to calculate the actual abundance of reads containing a particular class of repeat.
2.18 Read Mapping
Read mapping was performed with tophat2 (version 2.0.6) [19] using the –b2-very-sensitive --read-edit-dist 2 --max-multihits 100 --keep-tmp --library-type fr-secondstrand parameters. Tophat2 relies on bowtie2 (version 2.0.2). The reads were mapped to the human genome (hg19 from UCSC) using the Gencode annotation (version 13; [18]). Reads were additionally mapped to the known bacterial genomes (from NCBI as of Nov 30th, 2012) using bowtie2. Reads mapped with tophat2 were associated with genes using custom perl scripts that allowed no more than 2 unmapped bases to make an association. Reads that mapped to multiple genes were marked as such.
2.19 Identification of novel genes and splice variants
Cufflinks (version 2.0.2) [19]) and cuffmerge were used to identify reads that were consistent with a novel gene or splice form. For small genes/RNAs (less than 200 bps) a minimum of five reads were required. Novelty was assessed against the combined gencode (version 13) and UCSC repeat annotations.
2.20 SNP detection
Single nucleotide polymorphisms were identified using samtools mpileup and VarScan (version 2.3.3) [20, 21]. VarScan was run with the following parameters: “pileup2snp --min-reads2 5 --min-var-freq 0.3”. From these results, a set of high confidence SNPs (with coverage of 20 or more reads) were identified.
3.Results
3.1 RNA preparation
Total RNA was isolated from A2780 tissue culture cells as described in section 2.2 such that all RNA species were retained, as shown in Supplemental Fig. S4. The total RNA (Suppl Fig. S2A) sample was effectively sized fractionated around 200 nt using an RNeasy MinElute column with ethanol manipulations as described in sec. 3.3 to generate smRNA <200 (Suppl Fig. S2B) and LgRNA >200 nt samples (Suppl Fig. S2C). Libraries were prepared directly from the smRNA fraction while the LgRNA sample was chemically fragmented (FLgRNA) as described in sec. 2.4 prior to library preparation. The size distribution profile for FLgRNA is shown in Suppl Fig. S2D.
3.2 Duplex specific nuclease (DSN) removal of rRNA products
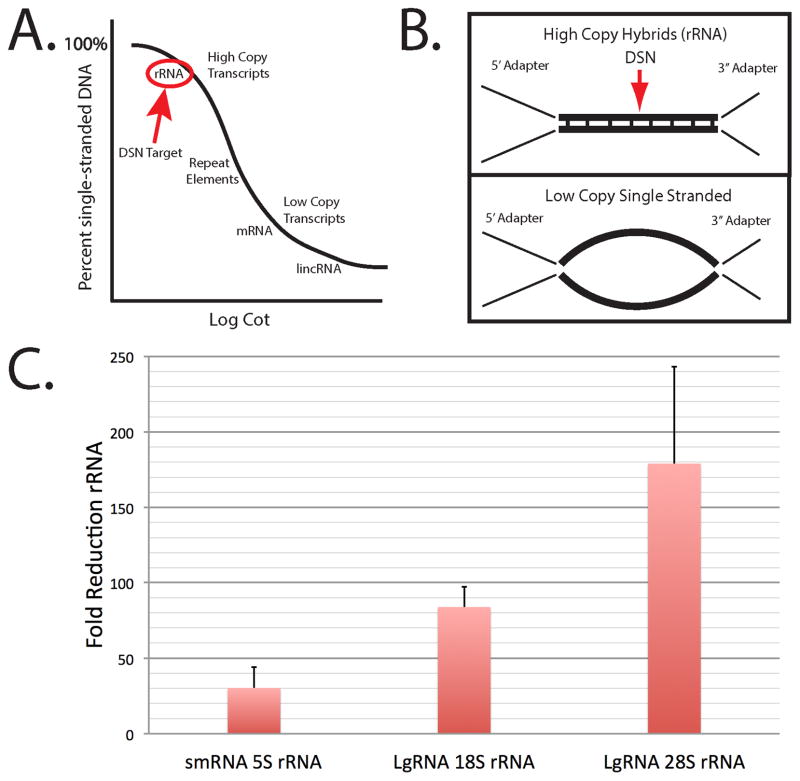
Optimization of the DSN reaction condition was necessary for library preparations. The goal was to allow high copy rRNA products to hybridize during a limited time course and become double stranded DSN targets while retaining single stranded adapter end sequences found on all library components Fig. 2. Optimal temperature and salt concentrations were predicted using GC content. By using a defined salt concentration, we predicted Tm’s (temperature at which the sequence remains half hybridized) as shown in Suppl Table S3. Towards this means we adjusted salt conditions (20 mM NaCl) and added surfactant (0.5% Tween20) during the five hour, 68°C hybridization. These conditions allowed high concentration rRNA sequences to hybridize due to GC content while retaining single stranded low copy transcripts (Fig. 2A, B). Library adapter ends (high copy) remain single stranded due to GC content (see Suppl Table S3). For optimized DSN reactions following the hybridization, we adjusted the MgCl2 concentrations to maximize DSN activity and implemented a more robust reaction stop buffer. The resulting reduction of rRNA products by these optimized DSN conditions was monitored by qRT-PCR, comparing rRNA in the PCR1 (pre-DSN) fraction with PCR2 product following DSN treatment (Fig. 2C). We monitored 5S rRNA (121 nt) in the smRNA libraries and tested 18S rRNA and 28S rRNA reduction in the FLgRNA libraries. The resulting reduction by DSN was 30/80/160-fold for 5S/18S/28S rRNA sequences respectively. The 5.8S rRNA (156 nt) could not be used as a DSN validation target gene in the smRNA libraries since the concentration was too low (Suppl Fig. S5).
Fig. 2.
Duplex specific nuclease (DSN) removal of rRNA sequences from libraries. Panel A, Theoretical hybridization curve based on copy number (concentration), and time (Cot). B) Predicted hybrid structures are illustrated. Here we show the predicted hybridization structures obtained with our optimized DSN conditions based on concentration (copy number), salts (20 mM NaCl), temperature (68°C), GC content (see Suppl Table S3). The arrow indicates the hybridized high-copy sequences (from rRNAs) in the library that are targeted by DSN. C) qRT-PCR results for rRNA reduction in smRNA (small RNA <200 nt) and FLgRNA (fragmented large RNA >200 nt) libraries following the DSN treatments. Fold change in smRNA libraries were determined by comparing the 5S rRNA levels in PCR1 with post-DSN PCR2 products. The reduction of 18S rRNA and 28S rRNA in FLgRNA libraries was determined in the same manner.
We successfully manipulated AMPure XP bead ratios to capture smRNA library components while removing adapter dimers from these preparations (Suppl Fig. S2). The smRNA libraries provided a narrow window for selection since miRNA elements were only 20 bp longer than the 118 bp adapter dimers to be removed following the DSN treatment (Suppl Fig. S3).
3.3 RNA-seq library validation
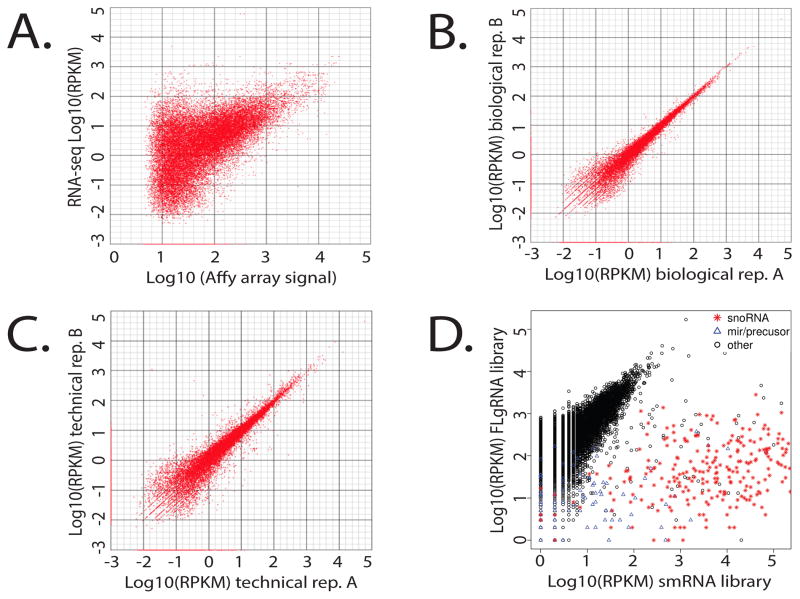
The Illumina single-read, strand specific 50 nt read (SR50) library data was validated using several methods. These libraries were generated from platinum sensitive and resistant A2780 epithelial ovarian cancer cells that had been previously analyzed on the Affymetrix U133+ platform [14]. Comparison of the RNA-seq data to the Affymetrix data yielded a Pearson correlation of r=0.6 (Fig. 3A), which was similar to previous studies [22]. We also observed a correlation of r=0.93 for biological replicates (Fig. 3B), which was similar to the technical replicates, r=0.92 (Fig. 3C). Several genes in the analysis indicated no change in expression levels between the platinum sensitive and resistant cells. These genes were sorted by RPKM values (Reads Per Kilobase per Million reads) into three categories; high RPKM ≥100, mod RPKM =10, and low RPKM<1. We then generated cDNA from these cell lines and performed quantitative qRT-PCR to validate expression levels changes. A strong correlation was observed for both the high and moderate RPKM gene sets, but correlations for the low RPKM set were gene dependent (Suppl Fig. S6).
Fig. 3.
Library Validation: Pearson correlations and scatter plots. A) RNA-seq vs. Affymetrix U133+ (r = 0.6), B) Biological replicates (r = 0.93), C) Technical replicates (r = 0.92). D) Scatter plot comparing FLgRNA (fragmented large RNA >200 nt) library with smRNA (small RNA <200 nt) library demonstrating enrichment of snoRNA and miRNA in the smRNA library.
3.4 RNA-seq data summary
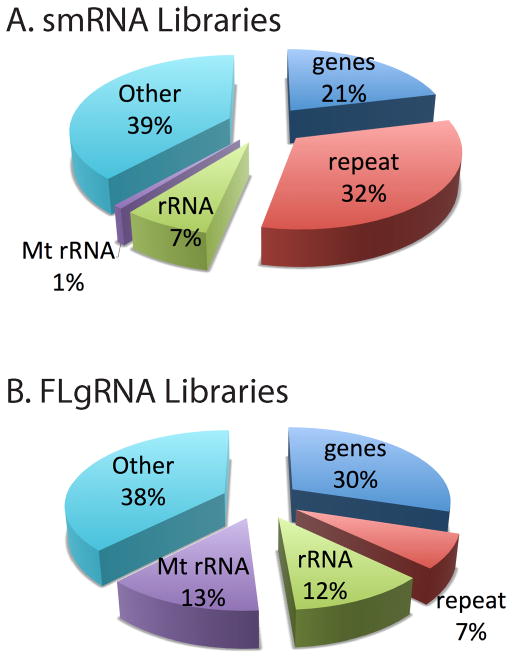
The distribution of mapped reads highlights the major types of genes detected by this RNA-seq protocol. The percentages of different RNA types has been summarized for the small RNA <200 nt (smRNA) libraries in Fig. 4A, and fragmented large RNA (>200 nt) libraries (FLgRNA) is shown in Fig. 4.B. With this protocol, we detected transcripts from approximately 12,000 known genes, 200 unknown genes, 140 snoRNAs, 100 miRNAs, 27 mitochondrial genes, 100 alternatively spliced genes, 1,300 repeat elements, and 500 significant SNPs within coding sequences. Table 1 highlights the whole transcriptome data captured with this protocol.
Fig. 4.
RNA-seq library data summaries. RNA-seq libraries demonstrating whole transcriptome coverage using duplex specific nuclease (DSN) reduction of rRNA reads on size fractionated total RNA from A2780 cell lines. A) This summary of the small RNA library (smRNA <200 nt) mapped reads illustrates the distribution of reads captured. B) Long RNA (>200 nt) was used to generate this summary of fragmented large RNA (FLgRNA) library mapped reads produced with this protocol.
Table 1.
Whole transcriptome data (gene counts) for smRNA and FLgRNA libraries1
| smRNA Librarya | FLgRNA Libraryb | |
|---|---|---|
| Known Genes | 5808 | 11218 |
| Novel Genes | 3 | 204 |
| Splice Variants | 0 | 101 |
| Unique SNPsc | 17 | 420 |
| CDS SNPsc | 28 | 545 |
| snoRNA | 133 | 146 |
| miRNA | 99 | 15 |
| mt genesd | 26 | 27 |
smRNA library, from total RNA<200 nt
FLgRNA library, fragmented from total RNA >200 nt
All SNPs filtered for p values <0.001
mt, mitochondrial genes detected
Summary of transcript analyses. Depth and types of analyses applicable to the sequence data generated from this protocol.
4. Discussion
Numerous methods have been described for the removal of rRNA sequences from RNA-seq library preparations but few have been demonstrated for whole transcriptome. Using the protocol described here, the whole transcriptome is captured from the libraries by using hybridization kinetics and duplex specific nuclease (DSN) to remove rRNA sequences. Protocols that capture small ncRNAs, such as miRNAs, typically require gel electrophoresis with potential cross-contamination, but the method described in this article is an alternative approach applicable to all types of RNA.
Our DSN protocol differs from the Illumina protocol in several key ways. First, the RNA isolation protocol has been optimized with a modified version of the Qiagen AllPrep RNeasy kit with RWT buffer wash to retain miRNAs and utilize the Qiagen RNA fractionation to separate small RNAs from the larger RNAs (Suppl Fig. 1), allowing us to investigate the whole transcriptome. Superscript III (Life Technologies) was used in the reverse transcriptase reaction followed by RNaseH treatment for improved yields. After reverse transcriptase and RNaseH treatment, EDTA was added prior to PCR1 amplification to reduce the MgCl2 concentration for higher fidelity. We have also optimized the DSN protocol as follows: the hybridization conditions are adjusted to select for rRNA sequences without adapter hybridization (Fig. 2). These changes include 20mM NaCl (Illumina uses 500mM), mixed Tris-HCl/Base buffers in place of Hepes for pH, salt control and thermal stability (Hepes is unstable at high temperatures). Tween is added as a surfactant to prevent nucleic acids from sticking to the sides during the hybridization. The MgCl2 is increased to 20mM during the DSN enzyme treatment (DSNM buffer, Sec. 2.10) for optimal activity (again with mixed Tris buffering) and a more robust DSN stop buffer is also applied for the stop reaction (2X DSNES buffer, Sec. 2.10). Following DSN treatment, AMPureXP clean-up conditions are adapted to retain miRNA components in the smRNA libraries (Sec. 2.9, Suppl Fig. 2). Primers and PCR primer annealing temperatures are modified for better thermal stability and specificity (Suppl Fig. 7).
The RNA-seq protocol described here offers several distinct advantages compared to other protocols. The removal of rRNA using DSN is economical and does not require costly repetitive cycles of bead hybridizations or addition of synthetic oligonucleotides to the samples. Our method minimizes the potential for off-target effects (non-rRNA), because DSN activity and dsDNA hybrid stability are both impeded by mismatches in hybrid structures [23]. Gel purifications, which are cumbersome and time consuming, are not required, eliminating potential cross-contaminations. RNA conversion to cDNA is performed from a single-stranded ligation product, preserving the strand identity of the RNA transcripts while also allowing the capture of small RNAs and identification of intergenic ncRNAs frequently located within other transcription units in the opposite orientation [4, 5, 6]. Barcode/index sequence additions for multiplexing options are flexible, allowing one to combine up to four samples in a single flow cell lane in order to produce nearly 70 million reads per sample for sufficient coverage. This provides sufficiently deep sequencing for an in-depth analysis suitable to monitoring expression levels, splicing variants, and SNP analysis. Our DSN normalization technique can be implemented with other sequencing platforms as well as formalin-fixed paraffin-embedded (FFPE) samples [24] and emerging array multiplexing technologies [25].
Drawbacks to this protocol, although minor, are recognized. A five-hour hybridization is required for DSN normalization with a dual chamber (or separate) PCR instrument to pre-heat the DSN reaction buffer before combining with the hybridization reaction (2.10). Salt concentrations are critical. To ensure proper conditions in the DSN hybridization reaction, one must pay close attention at key steps. It is necessary to make the hybridization buffer in order to obtain the proper pH without additional salts (sec. 2.10). For similar reasons, it is also important to use the modified wash protocol described for AMPure XP purification of the PCR1 reaction prior to the DSN hybridization reaction (sec. 2.9). Accurate library concentration estimates are required prior to submission for sequencing (2.14). As with other NGS protocols, tissue cultures, xenografts, or other RNA sources must be free of contamination to prevent loss of valuable flow cell capacity. Considerable pre-planning is necessary to ensure compatibility of barcode index sequences (Suppl Table S2) based on recommendations by Illumina [11]. Primers and adapters frequently must be designed or redesigned (within the constraints of the Illumina platform) based on our criteria standards (Suppl Fig. S7).
Of the approximately 1,400 human miRNAs, only ~25% are expressed at a given time. The vast majority (~75%) display tissue specific silencing by one or both epigenetic mechanisms, e.g. CpG methylation and/or H3K27 methylation [26]. Based on these approximations and the fact that cell lines were analyzed in the current study (less complexity compared to tissues), we would expect to observe expression of about 350 miRNAs. Our bioinformatics analysis, under optimal conditions, detects an average of 414 miRNAs in the A2780 cell lines (if we consider reads with low counts, ≥2). We also detect significant numbers of miRNAs in fragmented total RNA not fractionated by size (data not shown); however, a comparison made with size fractionated smRNA and FLgRNA libraries provides additional information regarding RNA turnover and processing rates by read count ratios in the two libraries (our unpublished observation).
5. Conclusions
We describe a protocol for generating stranded RNA-seq libraries that capture the whole transcriptome. This method utilizes an improved duplex specific nuclease (DSN) strategy for removing rRNA reads from next generation sequencing (NGS). The data generated using this method is of high quality, actually exceeding the accuracy of qRT-PCR, a gold standard in the gene expression field. Our method, as with other RNA-seq protocols, preserves sample integrity, allowing for detection of splicing variants as well as SNPs. Library barcoding is flexible, allowing investigators to take full advantage of the current multiplexing technology available with NGS. All known types of RNA are represented in library samples generated using this protocol, which yield nearly 70,000,000 reads for each of four separate barcoded samples from a single flow cell lane. Finally, this method should be highly amendable to future advances in NGS technology with adjustments made to accommodate adapter sequence hybridization stabilities.
Supplementary Material
Suppl Fig. S1. Bioanalyzer (BioA) profiles for RNA fractions. Panel A shows the BioA tracing for the total RNA before fractionation. B) smRNA (<200 nt) sample. C)BioA profile for LgRNA (>200 nt) and D)BioA tracing for fragmented LgRNA (FLgRNA).
Suppl Fig. S2. Bioanalyzer profiles for the AMPureXP purification strategy. A) 20 bp DNA ladder. B) 20 bp ladder purified with 2.0:1 ratio of AMPureXP beads to aqueous DNA volume. C) 20 bp ladder purified with 1.6:1 ratio of beads to DNA. D) Normalized barcoded smRNA library purified with 2.0:1 ratio of beads with adapter dimers (AD) present. E) Normalized barcoded smRNA library purified with 1.6:1 ratio of beads with adapter dimers (AD) selectively removed.
Suppl Fig. S3. Details of Illumina SR50 (stranded single read 50 base) RNA-seq library construction. Libraries were generated from total RNA following size fractionation in small RNA (smRNA <200 nt) and fragmented large RNA (FLgRNA) from total RNA >200 nt. We implemented a two-step strategy. Duplex specific nuclease (DSN) was first used to remove high copy rRNA products prior to adding index sequences (in PCR2) for multiplexing on the Illumina HiSeq2000 sequencer.
Suppl Fig. S4. Bioanalyzer high sensitivity (BioA HS) DNA profiles for SR50 libraries. A) BioA HS trace for smRNA library. B) BioA HS trace for FLgRNA library.
Suppl Fig. S5. qRT-PCR cycle counts (Cp values) for rRNA sequences found in PCR1 libraries (see Suppl Fig. 3 and sec. 2.8) prior to duplex specific nuclease (DSN) normalization. Final RNA-seq libraries were tested for DSN efficiency and removal of rRNA sequences. U6 snRNA and EEF1A1 are reference genes for smRNA and FLgRNA libraries, respectively. Low Cp values (11–12) for 5S rRNA, 18S rRNA, and 28S rRNA indicate high copy DSN targets; however, 5.8S rRNA Cp = 35 indicates low copy number and not recommended for DSN validation.
Suppl Fig. S6. qRT-PCR validation of RNA-seq data for transcripts with no fold change between A2780 platinum sensitive and resistant cell lines. A)Transcripts demonstrating RNA-seq High RPKM reads(>100) compared to qRT-PCR fold change. B)Moderate (MOD) RPKM reads (=10) in RNA-seq compared to qRT-PCR fold change. C)Low RPKM (<1) RNA-seq reads exhibiting no fold change are compared to qRT-PCR fold change. RPKM is defined as Reads Per Kilobase per Million reads.
Suppl Fig. S7. Primer and adapter design criteria. Primer and adapter design criteria are described in detail with each consideration itemized.
Acknowledgments
We thank Drs. Curt Balch, Ram Podicheti, and Kurt Zimmer for helpful discussion and Dr. Irene Newton and the Center for Genomics and Bioinformatics at Indiana University for the use of their Agilent Bioanalyzer. This work was also funded by the National Cancer Institute Award CA085289, the Integrative Cancer Biology Program CA1113001, P50 CA083639, U54 CA151668, and CA125806 and the Ovarian Cancer Research Fund (PPDIU01.2011). This research is based upon work supported by the National Science Foundation under grant No. ABO-1062432.
Footnotes
Abbreviations: smRNA: small total RNA <200 nt, LgRNA: large total RNA >200 nt, FLgRNA: fragmented LgRNA, DSN: duplex-specific nuclease
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
David F.B. Miller, Email: millerdf@indiana.edu.
Pearlly S. Yan, Email: Pearlly.Yan@osumc.edu.
Aaron Buechlein, Email: abuechle@cgb.indiana.edu.
Benjamin A. Rodriguez, Email: Benjamin.Rodriguez@bcm.edu.
Ayse S. Yilmaz, Email: AyseSelen.Yilmaz@osumc.edu.
Shokhi Goel, Email: shgoel@indiana.edu.
Hai Lin, Email: linhai@umail.iu.edu.
Bridgette Collins-Burow, Email: bcollin1@tulane.edu.
Lyndsay V. Rhodes, Email: lvanhoy@tulane.edu.
Chris Braun, Email: cbbraun@imail.iu.edu.
Sunila Pradeep, Email: SPradeep@mdanderson.org.
Rajesha Rupaimoole, Email: RRupaimoole@mdanderson.org.
Mehmet Dalkilic, Email: dalkilic@indiana.edu.
Anil K. Sood, Email: asood@mdanderson.org.
Matthew E. Burow, Email: mburow@tulane.edu.
Haixu Tang, Email: hatang@indiana.edu.
Tim H. Huang, Email: huangt3@uthscsa.edu.
Yunlong Liu, Email: yunliu@iupui.edu.
Douglas B. Rusch, Email: drusch@cgb.indiana.edu.
Kenneth P. Nephew, Email: knephew@indiana.edu.
References
- 1.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet. 11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 3.Wang L, Si Y, Dedow LK, Shoa Y, Liu P, Brutnell TP. A low-cost library construction protocol and data analysis pipeline for Illumina-based strand-specific multiplex RA-seq. Plos One. 2011;10:1–12. 31–46. doi: 10.1371/journal.pone.0026426. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 5.Cabili MN, Trapnell C, Goff L, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Djebali S, Davis CA, Merkel A, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morlan JD, Qu K, Sinicropi DV. Selective depletion of rRNA enables whole transcriptome profiling of archival fixed tissue. Plos One. 2012;7:1–8. doi: 10.1371/journal.pone.0042882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pease J, Sooknanan R. A rapid, directional RNA-seq library preparation workflow for Illumina sequencing. Nat Methods Applic Note. 2012;9:i–ii. [Google Scholar]
- 9.Zhong S, Jound J, Zheng YU, Chen Y, Liu B, Shoa Y, Xiang JZ, Fei Z, Giovannoi JJ. High-throughput Illumina strand-specific sequencing library preparation. Cold Spring Harb Protoc. 2011;10:940–949. doi: 10.1101/pdb.prot5652. [DOI] [PubMed] [Google Scholar]
- 10.Levin Jz, Yassour M, Adiconis X, Nusbaum C, Thompson DA, Friedman N, Gnirke A, Regev A. Comprehensive comparative analysis of strand-specific RNA sequencing methods. Nat Methods. 2010;7:709–715. doi: 10.1038/nmeth.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Illumina. TruSeq sample preparation best practices and troubleshooting Catalog # FC-930-1019 Part # 15025666 Rev. A. 2011 Jun; http://support.illumina.com/documents/MyIllumina/f517eea6-71ff-477d-a10c-79420260641b/TruSeq_SamplePrep_Best_Practices_Guide_15025666_A.pdf.
- 12.Illumina. Application for duplex-specific thermostable nuclease (DSN) to normalize RNA samples for Illumina sequencing, Part # 15014673 Rev. C. 2010 Jun; http://support.illumina.com/documents/MyIllumina/7836bd3e-3358-4834-b2f7-80f80acb4e3f/DSN_Normalization_SamplePrep_Application_Note_15014673_C.pdf.
- 13.Qiagen . Purification of miRNA from total RNA, cultured cells, soft tissues, or plant tissues using the RNeasy Mini kit and RNeasy MinElute cleanup kit, supplemental Qiagen protocol. Qiagen Sciences; Valencia, CA 91355: protocol RY20 Jan-05. [Google Scholar]
- 14.Li M, Balch C, Montgomery JS, Jeong M, Chung JH, Yan P, Huange TH, Kim S, Nephew KP. Integrated analysis of DNA methylation and gene expression reveals specific signaling pathways associated with platinum resistance in ovarian cancer. BMC Genomics. 2009;2 doi: 10.1186/1755-8794-2-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Lohse M, Bolger AM, Nagel A, Fernie AR, Lunn JE, Stitt M, Usadel B. RobiNA: a user-friendly, integrated software solution for RNA-Seq-based transcriptomics. Nucleic Acids Res. 2012;40:W622–627. doi: 10.1093/nar/gks540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smit A, Hubley R, Green P. Not Title. nd Dec 01; retrieved from http://www.repeatmasker.org/ 2012.
- 18.Harrow J, Frankish A, Gonzalez JM, Tapanari E, et al. Gencode: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, Van Baren MJ, Salzberg SL, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Handsaker B, Wysoker A, Fennel T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller CA, Mardis ER, Ding L, Wilson RK. VarScan 2: somatic mutation and copy number alteration discovery in cancer exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shagin DA, Rebrikov DV, Kozhemyako VB, Altshuler IM, Shcheglov AS, Zhulidov PA, Bogdanova EA, Staroverov DB, Rasskazov VA, Lukyanov S. A novel method for SNP detection using a new duplex-specific nuclease from crab hepatopancreas. Genome Res. 2002;12:1935–1942. doi: 10.1101/gr.547002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sinicropi D, Qu K, Collin F, Crager M, Liu M, Pelham RJ, Pho M, et al. Whole transcriptome RNA-Seq analysis of breast cancer recurrence risk using formalin-fixed paraffin-embedded tumor tissue. Plos One. 2012;7 doi: 10.1371/journal.pone.0040092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prokopec SD, Watson JD, Waggott DM, et al. Systematic evaluation of medium-throughput mRNA abundance platforms. RNA. 2013;19:51–62. doi: 10.1261/rna.034710.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baer C, Claus R, Plass C. Genome-wide regulation of miRNAs in cancer. Cancer Res. 2013;73:473–477. doi: 10.1158/0008-5472.CAN-12-3731. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Suppl Fig. S1. Bioanalyzer (BioA) profiles for RNA fractions. Panel A shows the BioA tracing for the total RNA before fractionation. B) smRNA (<200 nt) sample. C)BioA profile for LgRNA (>200 nt) and D)BioA tracing for fragmented LgRNA (FLgRNA).
Suppl Fig. S2. Bioanalyzer profiles for the AMPureXP purification strategy. A) 20 bp DNA ladder. B) 20 bp ladder purified with 2.0:1 ratio of AMPureXP beads to aqueous DNA volume. C) 20 bp ladder purified with 1.6:1 ratio of beads to DNA. D) Normalized barcoded smRNA library purified with 2.0:1 ratio of beads with adapter dimers (AD) present. E) Normalized barcoded smRNA library purified with 1.6:1 ratio of beads with adapter dimers (AD) selectively removed.
Suppl Fig. S3. Details of Illumina SR50 (stranded single read 50 base) RNA-seq library construction. Libraries were generated from total RNA following size fractionation in small RNA (smRNA <200 nt) and fragmented large RNA (FLgRNA) from total RNA >200 nt. We implemented a two-step strategy. Duplex specific nuclease (DSN) was first used to remove high copy rRNA products prior to adding index sequences (in PCR2) for multiplexing on the Illumina HiSeq2000 sequencer.
Suppl Fig. S4. Bioanalyzer high sensitivity (BioA HS) DNA profiles for SR50 libraries. A) BioA HS trace for smRNA library. B) BioA HS trace for FLgRNA library.
Suppl Fig. S5. qRT-PCR cycle counts (Cp values) for rRNA sequences found in PCR1 libraries (see Suppl Fig. 3 and sec. 2.8) prior to duplex specific nuclease (DSN) normalization. Final RNA-seq libraries were tested for DSN efficiency and removal of rRNA sequences. U6 snRNA and EEF1A1 are reference genes for smRNA and FLgRNA libraries, respectively. Low Cp values (11–12) for 5S rRNA, 18S rRNA, and 28S rRNA indicate high copy DSN targets; however, 5.8S rRNA Cp = 35 indicates low copy number and not recommended for DSN validation.
Suppl Fig. S6. qRT-PCR validation of RNA-seq data for transcripts with no fold change between A2780 platinum sensitive and resistant cell lines. A)Transcripts demonstrating RNA-seq High RPKM reads(>100) compared to qRT-PCR fold change. B)Moderate (MOD) RPKM reads (=10) in RNA-seq compared to qRT-PCR fold change. C)Low RPKM (<1) RNA-seq reads exhibiting no fold change are compared to qRT-PCR fold change. RPKM is defined as Reads Per Kilobase per Million reads.
Suppl Fig. S7. Primer and adapter design criteria. Primer and adapter design criteria are described in detail with each consideration itemized.