Abstract
Bacillus subtilis is a soil-dwelling Gram-positive bacterial species that has been extensively studied as a model of biofilm formation and stress-induced cellular differentiation. The tetrameric protein, SinR, has been identified as a master regulator for biofilm formation and linked to the regulation of the early transition states during cellular stress response, such as motility and biofilm-linked biosynthetic genes. SinR is a 111-residue protein that is active as a dimer of dimers, composed of two distinct domains, a DNA-binding helix-turn-helix N-terminus domain and a C-terminal multimerization domain. In order for biofilm formation to proceed, the antagonist, SinI, must inactivate SinR. This interaction results in a dramatic structural rearrangement of both proteins. Here we report the full-length backbone and side chain chemical shift values in addition to the experimentally derived secondary structure predictions as the first step towards directly studying the complex interaction dynamics between SinR and SinI.
Keywords: NMR, SinR, Bacillus subtilis, Biofilm regulation
Biological context
Bacteria have a plethora of defense mechanisms in their arsenal to help them cope with their rapidly changing, and often harsh, microenvironments. The most robust and dependable method at their disposal is the formation of a biofilm. Generally, a biofilm can be described as a complex community of cells that have adhered to a surface and “glued” themselves together in an extracellular polymeric matrix of DNA, polysaccharides, and protein. In recent years, biofilms have taken center stage in microbial research, because they are capable of being both a great tool and, simultaneously, our worst enemy (Hall-Stoodley et al. 2004). Clinically, biofilms are responsible for the rapidly increasing population of antibiotic resistant infections caused by such familiar species as methicillin-resistant Staphylococcus aureus (MRSA) and multi-drug resistant Acinetobacter baumannii. From an industrial point of view, biofilms are a costly hazard that can disrupt production, utilities, and slow shipping schedules (Fux et al. 2005). Yet, biofilms also have the potential to be a tremendous aid as a bioremediation tool (i.e. water purification and treatment, hazardous waste degradation) and as an alternative fuel source (Logan 2009; Singh et al. 2006).
Over the last few years, the non-pathogenic, soil-dwelling bacterial species, Bacillus subtilis, has become a well-studied model organism for biofilm formation in Gram-positive species. With the exception of the response regulator, Spo0A, the most important regulatory protein in B. subtilis biofilm formation is the master repressor, SinR (Kearns et al. 2005). SinR regulates the expression of two vital biofilm operons, the eps and the tapA-sipW-tasA operons, which control the biosynthesis of biofilm matrix components (Chu et al. 2008). In order to initiate biofilm formation, the bacteria have to sequester SinR using two key components: SinI and SlrR. SinI is a major antagonist for SinR and binds to the C-terminal domain, disrupting the multimerization event required for activity; while, SlrR controls the function of SinR through an epigenetic switch (Chai et al. 2009, 2010b). In low concentrations, the interaction between SinR and SlrR has a similar result to the SinR–SinI interaction, but as the concentration of SlrR increases, the SlrR–SinR complex further promotes biofilm formation by repressing a series of motility and autolysin genes that prevent chain formation, thus re-purposing SinR to aid in biofilm formation (Chai et al. 2010a).
SinR is a 111-residue protein (13 kDa) that forms a functional tetramer with two distinct domains: the N-terminal DNA binding domain and the C-terminal multimerization domain. The N-terminal domain (residues 1–69) is a well-conserved cro/C1-type helix-turn-helix (HTH) domain. The C-terminal domain (residues 74–111) is a unique helical bundle domain that is the site of both dimerization and tetramerization, and the region targeted by SinI, which interacts with the C-terminus to break up the SinR tetramer forming a heterodimer (Kearns et al. 2005). For SinI to disarm SinR, a series of energy intensive hurdles must be overcome. SinI must first dissociate from its own homodimer, disrupt the SinR homotetramer interface, and then trigger the dissociation of the SinR homodimer to form the extremely stable SinI:SinR heterodimer (Scott et al. 1999). As a first step toward understand the complex dynamics of this mechanism and to improve upon the existing structural models for this protein, we report the backbone and side chain chemical shift assignments of the full length SinR protein and their resulting secondary structure prediction.
Methods and experiments
The plasmid containing the full length SinR (SinRFL) from B. subtilis used in this study was provided by Prof. Richard Losick at Harvard University. The N-terminal domain (SinRN, residues 1–69) vector was created using the QuikChange® II site-directed mutagenesis kit (Agilent Technologies) to introduce a stop codon after E69; while, the C-terminal domain (SinRC, residues 69–111) was extracted from the SinR gene using PCR and cloned into pET-28a (EMD Millipore) with a thrombin-cleavable N-terminal His6-affinity tag. All three expression constructs were transformed into E. coli BL21(DE3) cells (EMD Millipore) for expression. The proteins were uniformly label with 13C/15N using M9T media supplemented with ammonium chloride (15N) and/or D-glucose-13C6 at 34 °C. Protein expression was induced with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) when the cultures reached an OD600 of −0.7. The cells were harvested by centrifugation 6–8 h post-induction at 11,000×g and stored at −80 °C. In addition to the uniformly labeled cultures, SinRC was cultured in a selective-labeling media (Griffey et al. 1985) in order to incorporate [13C6, 15N2] lysine and [13C6, 15N] leucine (SinRC-Sp).
SinRFL was resuspended and sonicated in Buffer A (10 mM Tris Base, 300 mM NaCl, 1 mM EDTA, 0.3 mM DTT, 10 mM MgCl2, and 0.02 % sodium azide at pH 8.0) prior to clarification through centrifugation at 20,000×g for 25 min. The lysate supernatant was decanted and passed over SP-Sepharose HP (GE Healthcare) and eluted using a gradient from 300 mM to 1 M NaCl (Buffer A with the addition of 700 mM NaCl). Following elution, SinRFL was dialyzed back into Buffer A and then passed over Heparin-Agarose Type I (Sigma-Aldrich), where the protein was eluted using Buffer B (50 mM Tris Base, 800 mM NaCl, 1 mM EDTA, 0.3 mM DTT, 10 mM MgCl2, and 0.02 % sodium azide at pH 8.5). SinRN was purified as described above using the following Buffer C (10 mM Tris Base, 10 mM NaCl, 1 mM EDTA, 0.02 % sodium azide at pH 7.9) and an elution gradient from 10 mM to 500 mM NaCl for both resins in place of Buffers A and B.
SinRC and SinRC-Sp were resuspended in Buffer D (50 mM Tris Base, 300 mM NaCl, 0.02 % sodium azide at pH 7.9), sonicated, and passed over Ni–NTA Agarose (QIAGEN) following clarification via centrifugation as stated above. The protein was eluted from the resin using a gradient from 0 mM to 500 mM imidazole (Buffer D with 500 mM imidazole). After pooling the eluted protein, the tag was immediately cleaved from the dilute protein using thrombin (EMD Millipore) and incubated at room temperature overnight to completely remove the affinity tag. The cleavage reaction was terminated using a 10 min incubation with AEBSF (Acros Organics) and passed over Phenyl-Sepharose (Sigma-Aldrich) to remove the tag and thrombin. Prior to NMR experimentation, all three proteins were extensively dialyzed into 20 mM MES, 200 mM NaCl, 0.02 % sodium azide at pH 6.0.
All NMR experiments were performed at 310 K on either a Varian Inova 600 MHz or a Bruker Avance 700 MHz, both equipped with cryoprobes. Backbone chemical shifts were assigned in a sequential manner from the following experiments: 2D [15N–1H] HSQC, 2D [15N–1H] TROSY-HSQC, HNCO, HN(CA)CO, CBCA(CO)NH, HNCACB, and C(CO)NH (15 and 18 ms). Sidechain proton chemical shifts were assigned using the following experiments: HBHA(CO)NH, H(CCO)NH, 15N-TOCSY-HSQC (50 and 75 ms), and an HCCH-TOCSY. The SinRC-Sp chemical shifts were assigned using the following: 2D [15N-1H] HSQC, CBCANH, HNCACB, 15N-TOCSY-HSQC (50 and 75 ms), and an HCCH-TOCSY. Aromatic assignments were made from a 13C-aromatic NOESY (80 ms). Data was processed using NMRPipe (Delaglio et al. 1995) and analyzed using NMRView (Johnson and Blevins 1994). Dihedral angles and secondary structure predictions were calculated using the program TALOS+ (Shen et al. 2009) and Cα chemical shift index deviations (Wishart and Sykes 1994).
Assignments and data deposition
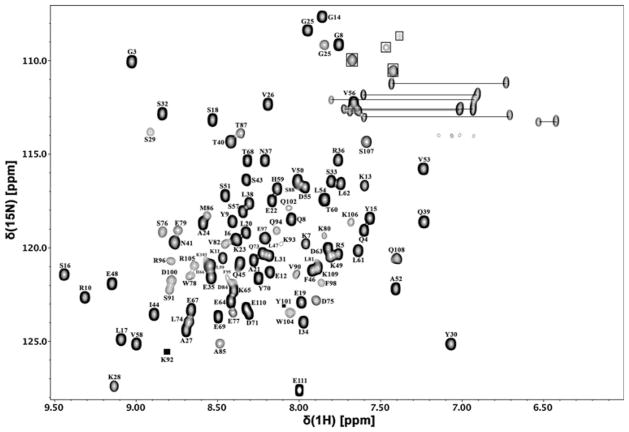
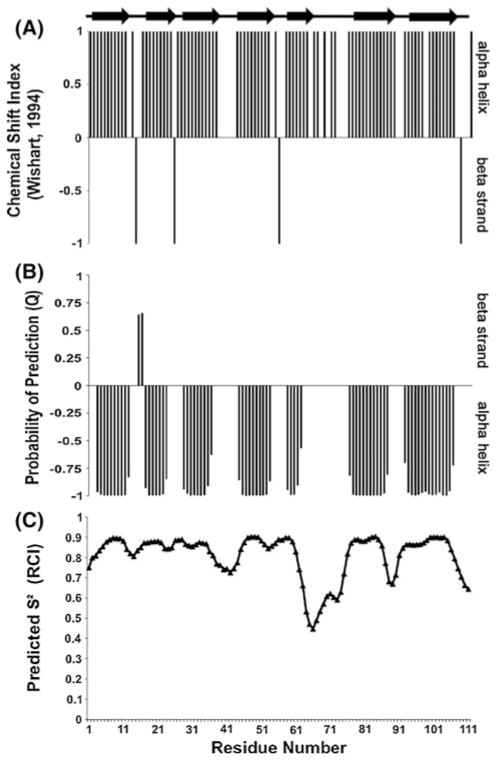
Overall, 99 % of the assignable backbone amide residueshave been identified with the exception of second residue as shown in the 2D [1H–15N] TROSY-HSQC spectrum (Fig. 1) using full length SinR in combination with the individual domains (SinRN, SinRC, SinRC-Sp) to aid in deconvoluting peak overlap. Nearly complete total carbon (98 %) and proton (97 %) assignments have also been obtained. Using a combination of TALOS + and the Wishart Cα chemical shift index prediction method (Fig. 2), the second structure of both domains was found to fit the cro/C-1 HTH DNA-binding domain (5 helix bundle) and the unique SinI binding domain that were observed in the X-ray crystal models by the Wilkinson group (Colledge et al. 2011; Scott et al. 1999). The chemical shift assignments have been deposited in the BioMagResBank (http://www.bmrb.wisc.edu) underthe accession number 19008.
Fig. 1.
2D [1H–15N] TROSY-HSQC spectrum of 450 μM SinR protein from Bacillus subtilis in 20 mM MES, 200 mM NaCl, and 0.02 % sodium azide at pH 6.0 and 310 K using a 700 MHz spectrometer (solid boxes represent peaks that are low intensity, open boxes highlight arginine Hε–Nε chemical shifts)
Fig. 2.
a Chemical shift index (CSI) plot for SinR Cα chemical shifts using the Wishart–Sykes identification method where dense positive regions indicate helices and dense negative regions represent β-strands; b TALOS+ secondary structure predictions where a dense negative region reflects an α-helix and a dense negative region is a β-strand; c TALOS+ predicted S2 order parameter values based on the random coil index (RCI) for residue
Acknowledgments
SDS, ALO, and JC performed and analyzed the experimental work. This research was supported by NIH grant RO1-GM055769 (JC) and the V Foundation for Cancer Research.
Footnotes
The authors declare no competing financial interest.
Contributor Information
Sean D. Stowe, Department of Molecular and Structural Biochemistry, North Carolina State University, Raleigh, NC 27695-7622, USA
Andrew L. Olson, Department of Molecular and Structural Biochemistry, North Carolina State University, Raleigh, NC 27695-7622, USA
Richard Losick, Department of Molecular and Cellular Biology, Harvard University, Cambridge, MA 02138, USA.
John Cavanagh, Email: john_cavanagh@ncsu.edu, Department of Molecular and Structural Biochemistry, North Carolina State University, Raleigh, NC 27695-7622, USA.
References
- Chai Y, Kolter R, Losick R. Paralogous antirepressors acting on the master regulator for biofilm formation in Bacillus subtilis. Mol Microbiol. 2009;74(4):876–887. doi: 10.1111/j.1365-2958.2009.06900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y, Kolter R, Losick R. Reversal of an epigenetic switch governing cell chaining in Bacillus subtilis by protein instability. Mol Microbiol. 2010a;78(1):218–229. doi: 10.1111/j.1365-2958.2010.07335.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y, Norman T, Kolter R, Losick R. An epigenetic switch governing daughter cell separation in Bacillus subtilis. Genes Dev. 2010b;24(8):754–765. doi: 10.1101/gad.1915010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu F, Kearns DB, McLoon A, Chai Y, Kolter R, Losick R. A novel regulatory protein governing biofilm formation in Bacillus subtilis. Mol Microbiol. 2008;68(5):1117–1127. doi: 10.1111/j.1365-2958.2008.06201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colledge VL, Fogg MJ, Levdikov VM, Leech A, Dodson EJ, Wilkinson AJ. Structure and Organisation of SinR, the master regulator of biofilm formation in Bacillus subtilis. J Mol Biol. 2011;411(3):597–613. doi: 10.1016/j.jmb.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister G, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6(3):277–293. doi: 10.1007/bf00197809. [DOI] [PubMed] [Google Scholar]
- Fux CA, Costerton JW, Stewart PS, Stoodley P. Survival strategies of infectious biofilms. Trends Microbiol. 2005;13(1):34–40. doi: 10.1016/j.tim.2004.11.010. [DOI] [PubMed] [Google Scholar]
- Griffey RH, Redfield AG, Loomis RE, Dahlquist FW. Nuclear magnetic resonance observation and dynamics of specific amide protons in T4 lysozyme. Biochemistry. 1985;24(4):817–822. doi: 10.1021/bi00325a001. [DOI] [PubMed] [Google Scholar]
- Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Micro. 2004;2(2):95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- Johnson B, Blevins R. NMR View: a computer program for the visualization and analysis of NMR data. J Biomol NMR. 1994;4(5):603–614. doi: 10.1007/bf00404272. [DOI] [PubMed] [Google Scholar]
- Kearns DB, Chu F, Branda SS, Kolter R, Losick R. A master regulator for biofilm formation by Bacillus subtilis. Mol Microbiol. 2005;55(3):739–749. doi: 10.1111/j.1365-2958.2004.04440.x. [DOI] [PubMed] [Google Scholar]
- Logan BE. Exoelectrogenic bacteria that power microbial fuel cells. Nat Rev Micro. 2009;7(5):375–381. doi: 10.1038/nrmicro2113. [DOI] [PubMed] [Google Scholar]
- Scott DJ, Leejeerajumnean S, Brannigan JA, Lewis RJ, Wilkinson AJ, Hoggett JG. Quaternary re-arrangement analysed by spectral enhancement: the interaction of a sporulation repressor with its antagonist. J Mol Biol. 1999;293(5):997–1004. doi: 10.1006/jmbi.1999.3221. [DOI] [PubMed] [Google Scholar]
- Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44(4):213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Paul D, Jain RK. Biofilms: implications in bioremediation. Trends Microbiol. 2006;14(9):389–397. doi: 10.1016/j.tim.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Wishart D, Sykes B. The 13C chemical-shift index: a simple method for the identification of protein secondary structure using 13C chemical-shift data. J Biomol NMR. 1994;4(2):171–180. doi: 10.1007/bf00175245. [DOI] [PubMed] [Google Scholar]