Abstract
The cells of origin for cancer are the cells within tissues that serve as the target for transformation. Understanding the nature of these cells will benefit disease prevention, diagnosis and prognosis. During the past decade, much progress has been made in understanding the cellular origin for prostate cancer. This review aims to summarize the previous findings, describe the most recent results and discuss some controversies and unresolved issues in this field.
Keywords: cells of origin, prostate cancer, prostate stem cells, lineage hierarchy, lineage tracing, tumor initiation
INTRODUCTION
The cells of origin for cancer denote the cells within tissues that serve as the target for transformation during tumor initiation.1 They are defined differently from the cancer stem cells that represent the cellular subfractions that can regenerate the tumors.2 Characterizing the identity of cells of origin for cancer has been a research area under extensive interrogation. This is mainly due to the enthusiasm to test the hypothesis that distinct cellular origins for cancers determine the histological and molecular features of cancers and account for the heterogeneous nature of certain malignancies. Understanding the nature of these cells will facilitate early diagnosis and accurate prognosis, and may also lead to novel preventive strategies for high-risk patients.
MAJOR APPROACHES FOR STUDYING THE CELLS OF ORIGIN FOR CANCER
Historically, the identities of cellular origins for cancers were assumed based on their histological appearances. Now we know, based on our current knowledge, that random and static observations from tumor specimens can be misleading. Any assumptions made should be validated using more direct genetic approaches. A related, but more elegant hypothesis is that tumors display gene expression profiles that most resemble those of their cells of origin rather than those of other lineages in the same tissue. With this approach, it has been suggested that the mammary gland luminal epithelial progenitors are the cells of origin for the basal-like breast cancer.3 More impressively, brain tumors with identical histological appearances can be divided into distinct groups based on their gene expression profiles that reflect those of their cellular origins.4,5 These studies suggest that the pathological appearance of tumors does not always match their molecular profiles. However, conclusions from these types of studies are still indirect and need further validation using direct genetic studies.
A relatively more direct approach to study the cellular origin for cancer is to establish cell lines with distinct lineage characteristics from human or rodent tissues, transform the cultured cells and determine whether the resulting tumors display different pathological appearance. For example, Ince et al.6 established two types of mammary gland epithelial cell lines that displayed a myoepithelial and a luminal cell phenotype, respectively. When transformed by the same combination of oncogenes, the two types of cells formed tumors with distinct phenotypic appearances and malignant potentials. This study provides direct evidence that the cellular origin for cancer can dictate the tumor phenotype and aggressiveness. However, it should be noted that the phenotypic characteristics of the in vitro cultured cells may not always reflect their origins in vivo.
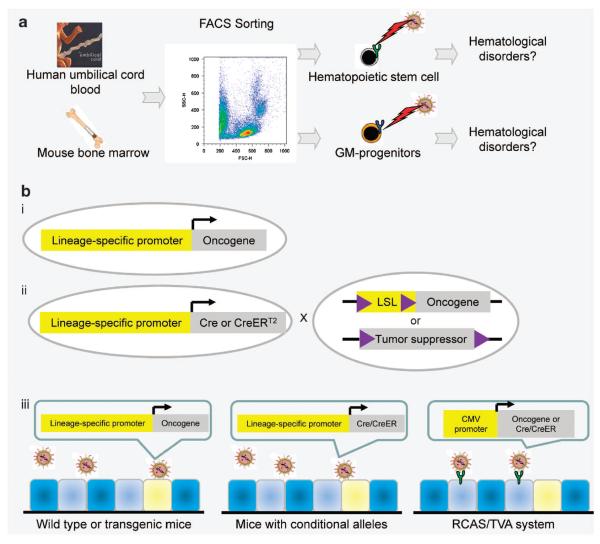
During the past decade, maturation of technologies such as fluorescence-activated cell sorting and the availability of genetically engineered mouse models have made it possible to investigate the cellular origins for cancers using more direct approaches. Figure 1 illustrates the two most popular direct approaches for studying cells of origin for cancer. The first approach is a transplantation assay-based method that was pioneered by studies in the hematopoietic system.7 As shown in Figure 1a, individual hematopoietic lineages are isolated by flow cytometry based on their surface antigen expression profiles. Isolated cells are transduced by either a retrovirus or lentivirus to elicit various oncogenic signaling. Engineered cells are then transplanted into host mice to determine their susceptibility to transformation. With this approach, dozens of studies have concluded that both hematopoietic stem cells and more committed progenitors such as the granulocyte–macrophage progenitors can serve as the cells of origin for hematological disorders in different oncogenic contexts.8–11 Subsequently, this approach has been successfully adapted to studying cells of origin for solid tumors, such as breast12 and prostate cancer,13 as described in detail below. One caveat to this approach is that the expression level of specific oncogenes may influence the susceptibility of different target cell populations.
Figure 1.
Two major direct approaches for studying the cellular origin for cancer. (a) Different hematopoietic cell lineages are isolated by flow cytometry based on their surface antigen expression profiles, and transduced by retrovirus expressing various oncogenic signals. Infected cells are transplanted to determine the susceptibilities of individual cell lineages toward transformation. GM progenitors: granulocyte–macrophage progenitors. (b) Oncogenic signaling is introduced into specific cell lineages in vivo and in situ by lineage-specific promoters (i), or by dual regulation of lineage-specific promoters and the tamoxifen responding CreERT2 (ii), or by various virus infection models (iii). LSL: loxP-flanked transcriptional stop signal. RCAS: replication-competent ASLV long terminal repeat with a Splice acceptor.
The second direct approach to study cellular origin for cancer is to introduce oncogenic signals into specific cell lineages in vivo and in situ in genetically engineered mouse models (Figure 1b). This is often achieved by spatiotemporal activation of oncogenic signaling by lineage-specific promoters. The oncogenic signaling may be initiated upon activation of a lineage-specific promoter (Figure 1b (i)), or by dual regulation of specific promoters and a tamoxifen-responding CreER or a tetracyclin-responding element (Figure 1b (ii)).14 Alternatively, oncogenic signaling can also be specifically introduced into individual lineages by virus infection such as the RCAS/TVA system, and so on (Figure 1b (iii)).15
A typical example is a study from Lapouge et al.,16 in which the authors showed that K-ras activation in hair follicle bulge stem cells leads to squamous skin carcinoma. The same approach has been utilized in many other elegant studies to investigate cellular origin for hematological disorders,17 brain cancers18 and so on. For some organ systems, simplified approaches are available that do not require multiple genetically engineered mouse models and tedious mouse breeding. For examples, epithelial lineages in the lung can be targeted by inhalation of an aerosol of adenovirus,19 whereas mammary gland epithelial lineages can be targeted by intraductal injection of retrovirus, lentivirus or avian virus.20 Although this is the most direct and physiologically relevant approach, it is not always feasible because of the lack of proper animal models. In addition, tumor initiation from a cell lineage with stem cell activity does not necessarily conclude that it is the only cellular origin for cancer. Parallel studies targeting individual downstream lineages are essential to determine whether the stem cell lineage or the differentiated lineages, or both, are susceptible to transformation induced by the introduced oncogenic signaling. Finally, expression of an oncogene driven by lineage-specific promoters or a viral long terminal repeat may be not equivalent to activation of an endogenous oncogene and the level of oncogene expression may influence the susceptibility of the cells of origin.21
PROSTATE EPITHELIAL LINEAGE HIERARCHY AND THE CELLULAR ORIGIN FOR PROSTATE CANCER
Prostate cancer is the second leading cause for cancer-related death in males in the western countries.22 Currently, a major therapeutic challenge for prostate cancer is to distinguish indolent cancers from aggressive ones that need immediate therapeutic intervention. The malignant potential of the prostate cancer may be determined by the transforming capacities of the oncogenic events associated with the disease. Alternatively, it is also hypothesized that the cellular origin for prostate cancer dictates the aggressiveness of the disease. Therefore, much effort has been made in interrogating the identity of the cells of origin for prostate cancer.
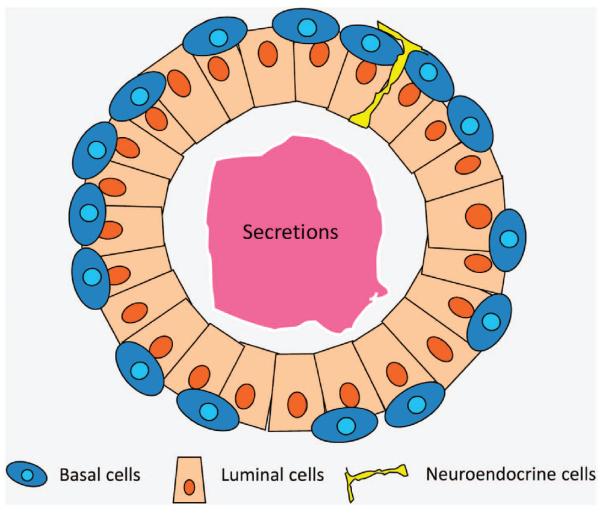
There are three types of epithelial cells in the adult prostate (Figure 2).23 Luminal cells constitute the majority of the prostate epithelia and carry out the secretory function. Most luminal cells depend on androgen signaling for their survival. The low-molecular-weight cytokeratins CK8/18 are typical lineage markers for the luminal cells. The androgen receptor and a homeobox gene Nkx3.1 are also preferentially, but not exclusively, expressed in the luminal cells.24,25 Basal cells are aligned between the basement membrane and luminal cells. They express the high-molecular-weight cytokeratins CK5/14 and a p53 superfamily member p63.26 One classical view of the function of the basal cells is that they serve as a barrier to protect luminal cells from oncogenic insults.27 In addition, classic studies by Isaacs et al.29 demonstrated that rodent prostate epithelia can undergo extensive turnover in response to fluctuating serum testosterone levels, suggesting the existence of a stem cell population. As basal cells do not rely on androgen for their survival, it was hypothesized that prostate stem cells reside in the basal cell layer. Neuroendocrine cells are the third type of epithelial cells and constitute only a very small fraction of cells within prostatic acini.30 They express synaptophysin and chromagranin A, but their functions are still not clear. Additionally, a population of `transit-amplifying' or `intermediate' prostate epithelial cells has been documented in the study of human prostate biology.28,31 These cells express markers of both the basal and luminal cells. Although these cells are very rare in vivo, basal cells often display the intermediate cell phenotype when cultured in vitro.32–34 Because of a lack of proper in vivo experimental approaches for studying neuroendocrine cells and intermediate cells, this review will focus mainly on basal and luminal cell lineages.
Figure 2.
A simplified cartoon image illustrating glandular structures of adult murine prostate.
Historically, luminal cells were thought to be the cellular origin for prostate cancer because prostate cancers are mostly composed of cells that display a luminal cell phenotype. In fact, loss of the expression of the basal cell antigens CK5/14 and p63 has served as a diagnostic criterion for prostate cancer.35 On the other hand, prostate cancer often recurs after antihormonal therapy and becomes castration-resistant. As androgen independence is a major feature of basal cells, it has also been suggested that the cellular origin for prostate cancer is basal cells. In support of this hypothesis, Min et al.36 showed that primary established human basal prostate epithelial PrEC cells can be transformed by oncogenic signaling and can form tumors that are composed of cells displaying a luminal cell phenotype. Finally, intermediate cells have also been observed in human prostate cancer tissues in some reports.37,38 Prostate stem cell antigen, a marker for late intermediate epithelial cells, is also upregulated in human prostate cancers.39 Thus, it has also been suggested that the intermediate epithelial cells may be the cells of origin for prostate cancer. The intermediate epithelial cells have not been definitively defined in vivo under physiological conditions in adult mice, but have been frequently observed during carcinogenesis in various mouse models.40,41 During the past 10 years, more direct approaches have been utilized to investigate prostate epithelial lineage hierarchy as well as the identity of the cellular origin for prostate cancer. Interestingly, those studies have generated some seemingly controversial conclusions.
Previously, we managed to separate individual prostate epithelial cell lineages using flow cytometry, hoping to define the prostate epithelial lineage hierarchy and to determine the cellular origin for prostate cancer using the first approach illustrated in Figure 1a. We established a dissociated prostate cell regeneration assay (Figure 3a)42 based on a classic tissue fragment recombination assay.43 Briefly, dissociated single prostate epithelial cells are combined with embryonically derived urogenital sinus mesenchymal cells (UGSM) and grafted under renal capsules of immunodeficient male host mice. UGSM cells are capable of stimulating proliferation and differentiation of prostate stem cells to regenerate glandular structures de novo. We and others showed using this assay that human and rodent basal cells are capable of regenerating glandular structures that contain all three major epithelial lineages in the prostate.13,44–47 In contrast, basal cell-depleted lineages do not possess this capacity. These studies directly demonstrated the long-existing hypothesis that prostate basal cells possess the stem cell capacity for multi-lineage differentiation.29 In addition, when FACS- (fluorescence-activated cell sorting) sorted cell lineages were infected with lentiviruses that express various oncogenes, only basal cells were able to generate prostatic intraepithelial neoplasia lesions or adenocarcinoma (Figure 3b).13,48,49 Of note, Goldstein et al.48 showed that when FACS-sorted primary human prostate basal cells were transformed, they formed adenocarcinomas that were composed solely of luminal cells. These studies demonstrate that prostate stem cells reside in the basal cell lineage, and that basal cells are an efficient target for transformation in the prostate regeneration assay.
Figure 3.
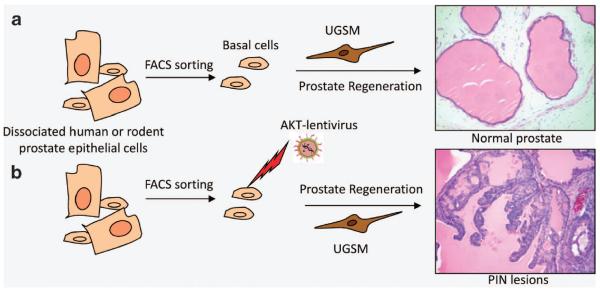
Studies using a dissociated prostate cell regeneration method demonstrate that basal cells possess stem cell activity and are an efficient target for transformation. (a) The prostate regeneration assay. Basal cells are FACS sorted from dissociated human or rodent prostate epithelial cells based on their surface antigen expression profiles. Isolated basal cells are combined with urogenital sinus mesenchymal cells (UGSM) and transplanted under kidney capsules of immunodeficient male hosts. They are capable of forming glandular structures that are reminiscent of normal adult prostates and contain all three prostate epithelial lineages. Other cell lineages do not possess such capacities. (b) When FACS-sorted basal cells are infected with lentivirus expressing a constitutively activated AKT, they can form glandular structures containing prostatic intraepithelial neoplasia (PIN lesions). Other lineages do not generate outgrowths under this condition.
Wang et al.50 employed a lineage-tracing approach to address the same questions (Figure 4). They utilized an Nkx3.1-CreERT2 mouse strain that expresses the tamoxifen-responding CreERT2 under the endogenous promoter of Nkx3.1. In intact mice, Nkx3.1 is mainly expressed by luminal cells but is also expressed in a few basal cells.25 However, using a fluorescence reporter line, Wang et al.29 showed that in castrated Nkx3.1-CreERT2 mice, Nkx3.1 is only expressed in a small fraction of luminal cells, which were termed as the castration-resistant Nkx3.1-expressing (CARN) cells (Figure 4a). They induced extensive epithelial turnover by a classic castration-androgen replacement protocol and showed that CARN cells can generate both basal cells and luminal cells. In addition, when phosphatase and tensin homolog (Pten) is specifically knocked out in CARN cells, castrated mice developed adenocarcinoma upon androgen replacement (Figure 4b). Collectively, this study shows that the CARN luminal cells also possess stem cell activity and can serve as the cellular origin for prostate cancer.
Figure 4.
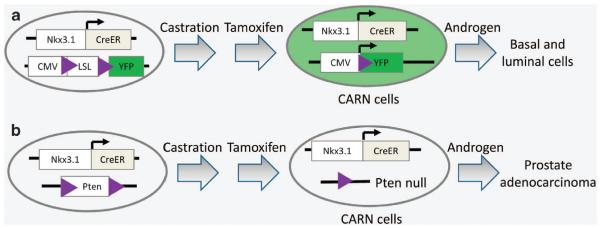
CARN cells represent a distinct stem cell population that can act as the cells of origin for prostate cancer. (a) CARN cells are luminal cells that express Nkx3.1 in castrated mice. They can generate both basal cells and luminal cells in response to androgen stimulation. (b) Pten deletion in CARN cells in castrated mice leads to tumor formation upon androgen replacement.
DETERMINING LINEAGE HIERARCHY AND THE CELLULAR ORIGIN FOR PROSTATE CANCER USING LINEAGE TRACING AND LINEAGE-SPECIFIC GENE TARGETING
The studies described above appeared to be controversial. However, these different conclusions most likely reflect the differences and limitations of the experimental models utilized in those studies. In the prostate regeneration assay, prostate epithelial cells are dissociated into single cells and removed from their normal environmental cues. They are combined with the UGSM cells that possess strong inductive capacity. UGSM cells can reprogram epithelial cells of other tissue origins at various developmental stages into prostate epithelial-like cells.51–53 Therefore, it is unknown whether the biological readout of the assay reflects an obligate or a facultative function of prostate stem cells. In addition, prostate luminal cells by nature do not proliferate and form grafts in this assay. Therefore, the observation that luminal cells cannot be transformed in this assay does not rule out that they cannot be transformed in vivo and in situ.
On the other hand, despite a luminal phenotype, the origin of the CARN cells is unknown. It is possible that basal cells can adapt a CARN cell phenotype in castrated mice. In addition, Nkx3.1 is a transcription factor that has a role in prostate homeostasis24 and has been shown to affect the dynamics of prostate regeneration.54 The Nkx3.1-CreERT2 mouse model was designed so that one allele of Nkx3.1 is functionally null.50 It remains unknown how this may affect stem cell function and lineage maintenance during the induced epithelial turnover. Finally, tumor initiation from the CARN cells occurred in castrated mice. Although it has been reported that castrated males can develop prostate cancer in some rare cases, this may not be similar to mechanisms of cancer initiation in intact adults.
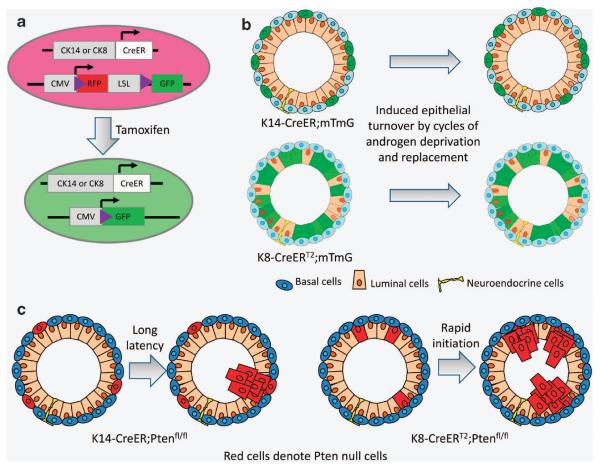
We employed a different experimental approach to further interrogate the lineage hierarchy and cellular origin of prostate cancer. As shown in Figure 5a, prostate basal cells and luminal cells were fluorescently marked individually in adult mice using K14-CreER and K8-CreERT2.40 After induced epithelial turnover, we found that prostate basal cells only generate basal cells, while luminal cells only generate luminal cells (Figure 5b). A previous study using a PSA-CreERT2 model also showed that the luminal cell lineage is self-maintained.55 Our finding is also in agreement with recent studies in the mammary gland showing that the adult myoepithelial and luminal epithelial lineages are also independently sustained.56–58 These studies, together with observations from some other tissue systems,59 highlight a distinct model for adult epithelial maintenance, in which adult epithelial tissues are maintained by unipotent progenitors or self-duplication of epithelial cells.
Figure 5.
Adult murine prostate basal cells and luminal cells are independently self-sustained lineages that both can serve as the cells of origin for prostate cancer. (a) Schematic illustration of the strategies to fluorescently label prostate basal and luminal cells separately. CK14-CreER and CK8-CreERT2 mice are bred with an mTmG fluorescence reporter line. The mTmG reporter mice express red fluorescence protein (RFP) that can be replaced by green fluorescence protein (GFP) upon Cre-LoxP-mediated homologous recombination. Upon tamoxifen induction, GFP is specifically and permanently expressed in the CK14- or CK8-expressing cells and their progenies. (b) Lineage tracing shows that basal cells and luminal cells are independently self-sustained. (c) Both basal cells and luminal cells can serve as cells of origin for prostate cancer in the context of Pten deletion. However, disease initiated from basal cells had a prolonged latency and required the trans-differentiation of basal cells to luminal cells.
We also knocked out the tumor suppressor Pten specifically in basal and luminal cells in intact adult mice using K14-CreER and K8-CreERT2, respectively (Figure 5c).40 Experimental mice were allowed to age to determine whether tumors would form. Our study revealed that prostate luminal cells are very sensitive to the mitogenic signaling induced by Pten deletion. K8-CreERT2;Ptenfl/fl mice developed prostatic intraepithelial neoplasia lesions within 1 month after Pten deletion. In contrast, basal cells are more indolent to oncogenic insults such as loss of Pten or simultaneous loss of Pten and p53. Nevertheless, those mice developed prostatic intraepithelial neoplasia lesions after a long latency. We think this is because basal cells trans-differentiated into luminal cells in the context of these oncogenic signals. As soon as the trans-differentiation took place, the derived luminal cells proliferated in response to the pre-existing oncogenic insults. This study demonstrates that both prostate basal cells and luminal cells can serve as the cellular origin for prostate cancer.
BASAL-LUMINAL DIFFERENTIATION: A CRITICAL PATHOLOGICAL EVENT?
Prostate basal cells are less well-differentiated and proliferate more frequently.60 Therefore, they are more prone to accumulating genetic alterations than luminal cells and may act as a preferred cellular origin for cancer. In addition, prostate basal cells are aligned between luminal cells and the basement membrane and have been proposed to serve as a natural barrier to protect luminal cells from various insults.27 They are exposed to a more `hostile' environment than luminal cells because they are in direct contact with surrounding reactive stroma that generates various cancer-promoting cytokines.61 Therefore, basal cells may have evolved protective molecular circuitries against oncogenic insults through cell-autonomous or non-autonomous mechanisms. In agreement with this, our studies showed that prostate luminal cells are more responsive to mitogenic signaling induced by Pten deletion, while basal cells in situ are more resistant to transformation.
Our study suggests that differentiation of basal cells into luminal cells may have a critical role in prostate cancer initiation. Prostate cancer is unique in that the disease is strictly age-dependent. Seldom do men under 35 develop prostate cancer. Our study showed that K14-Pten mice developed prostate cancer with a long latency: after deletion of Pten in K14-expressing basal cells, it took at least 3 months for K14-Pten mice to develop prostate cancer, which is equivalent to approximately 10 years of human life. This suggests that basal–luminal differentiation is an extremely lengthy and inefficient biological process under certain genetic contexts, which may serve as one explanation as to why prostate cancer is an age-related disease.
Additional evidence suggests a potential role of a deregulated epithelial differentiation program in prostate cancer initiation. A series of TMPRSS2-Ets fusion proteins generated through chromosome translocation have been identified in over 54% of prostate cancers.62 Surprisingly, Ets fusion proteins per se are not sufficient to initiate prostate cancer in several transgenic mouse models.63–66 Of note, two of those studies reported that overexpression of Erg and Etv1 in the prostate leads to a reduction of basal cells within the prostate.65,66 It is tempting to hypothesize that one of the functions of the Ets fusion protein is to drive prostate basal cells to differentiate into prostate luminal cells, and that the cellular origin for Ets fusion protein positive prostate cancer is likely to be prostate basal cells. If this is true, then suppressing the Ets fusion protein-mediated signals that promote basal–luminal differentiation may provide a promising approach to prevent prostate cancer.
Basal cells may also serve as the cellular origin for metastatic prostate cancer. Basal cells display a more mesenchymal phenotype and express genes relevant to epithelial–mesenchymal transition at a higher level than luminal cells. For example, the miR-200 family that positively regulates E-cadherin is expressed at a lower level in basal cells as compared with luminal cells.67–69 Epithelial–mesenchymal transition is a phenotype that is often associated with anchorage-independent survival and higher migratory capability.70 Therefore, basal cells may be more competent to intravasate, survive in the circulating system, extravasate and then differentiate into transformation-competent luminal cells that colonize at distal sites. Although there is no direct evidence to support this hypothesis, it has been shown previously that untransformed murine mammary gland epithelial cells can seed in the lung.71 In addition, our previous studies have demonstrated that basal cells have the capability to survive and differentiate into multiple cell lineages in the transplantation-based prostate regeneration assay.13,49
SUMMARY, CAVEATS AND DISCUSSIONS
Our work demonstrated that adult murine prostate basal cells and luminal cells are independently self-sustained. They both can serve as the cellular origin for prostate cancer. However, basal cells are relatively resistant to oncogenic signals and need to undergo basal–luminal differentiation to initiate cancer40 (Figure 6). These findings are not contradictory to the previous studies showing that basal cells possess multipotent stem cell activity in the prostate regeneration assay.13,44,45 As discussed earlier, the experimental condition of the prostate regeneration assay awakens the plasticity of basal cells, which represent a facultative function that basal cells do not, or rarely, carry under physiological conditions in adult mice. Neither is our work in conflict with previous studies showing that basal cells are efficient targets for transformation in the prostate regeneration assay.48,49 This is because basal cells are capable of differentiating into luminal cells much more efficiently in the prostate regeneration assay. In summary, the prostate regeneration model may reveal some capabilities of adult prostate basal cells under pathological conditions, such as during tumor metastasis (Figure 6b), as described earlier.
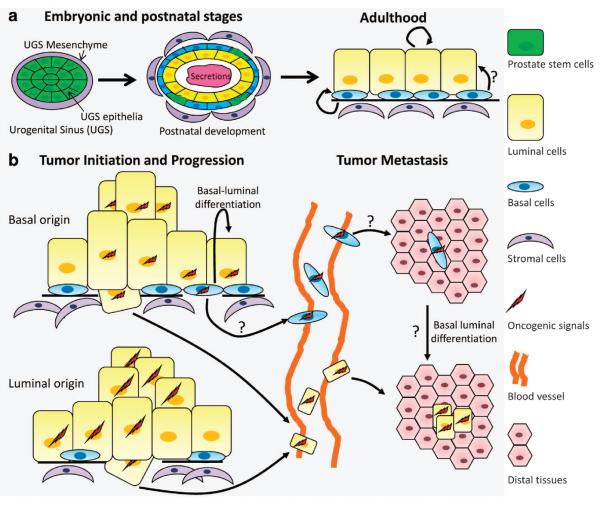
Figure 6.
A model for the prostate lineage hierarchy and the cellular origin for prostate cancer. (a) Prostate epithelial lineage hierarchy. During development, prostate stem cells that express both CK5 and CK8 undergo lineage commitment to generate both basal and luminal cells. Some basal cells retain the stem cell capacity for multi-lineage differentiation at the post-natal developmental stage. In adulthood, prostate basal and luminal cell lineages are mainly self-sustaining. (b) Cellular origin for prostate cancer. Both prostate basal and luminal cells may serve as the cellular origins for primary as well as metastatic prostate cancer. Basal–luminal differentiation may have a critical role in the initiation and progression of prostate cancer with a basal cell origin.
It should be noted that our studies showed that it is in the adulthood that the prostate basal and luminal cell lineages are independently sustained. It has been well-accepted that the prostate epithelia are developed from a stem cell population that expresses both CK5 and CK8 at the embryonic stage (Figure 6a).72 A very recent lineage-tracing study revealed that some murine prostate basal cells retain multipotent capacities at the postnatal developmental stage.73 This developmental stage-dependent switch of the mechanisms for tissue maintenance has also been demonstrated in the mammary gland.56,58
One limitation of our experimental model is that we cannot label prostate basal cells at a high efficiency. The labeling efficiency is the highest in the lateral lobe, and is only about 20%. Similar observations have been made in the mammary gland that myoepithelial cells are relatively refractory to fluorescent marking.56 The molecular basis of this resistance is not known. It is possible that the CK14 promoter in the transgenic mouse model does not work potently in the basal cells. Or alternatively, basal cells express higher levels of ATP-binding cassette transporters so that they can efflux tamoxifen more efficiently.74 With this limitation, we cannot rule out completely that rare bipotent or even multipotent stem cells may exist and can generate all three epithelial lineages in the prostate. However, in the complementary lineage tracing of the luminal lineage, we found no evidence that other lineages contribute significantly to the maintenance of the luminal lineage. Therefore, our study showed convincingly that prostate basal and luminal cell lineages are mainly maintained independently. In summary, even if they exist, multipotent stem cells may only marginally contribute to adult epithelial maintenance. The other interesting finding in our study is that Pten knockout can induce trans-differentiation of basal cells into luminal cells. This is based on the observation that basal to luminal transition is not observed in wild-type mice but is seen in the Pten-null mice. However, in light of the caveats described herein, we cannot exclude the possibility that Pten was deleted in rare bipotent progenitors, which initiate cancer upon differentiating into luminal cells.
The other caveat is that different lineage-specific promoters may mark different subpopulations of the cells within a given lineage. For examples, although lineage tracing using the promoters of CK5 and CK14 showed that myoepithelial cells in adult mouse mammary gland are unipotent,56 a recent study showed that some Axin2-expressing myoepithelial cells in adult mice display bipotent activities during cycles of lactation and involution.58 It will be interesting to see whether similar conclusions derived from our studies will be obtained using mouse models expressing the CreER transgene driven by the promoters of other lineage markers.
Finally, despite the microscopic similarities between their glandular structures, there are fundamental differences between human and rodent prostates. Natural seasonal fluctuations in androgen level may differ between human and rodents, which could have an impact on the biology of the prostate and affect incidence of prostate cancer.75 Human prostate is a lobular organ, while mouse prostate displays a tubular structure. There are more stromal cells in human prostates than in mouse prostates. Human prostate basal cells form a continuous layer. Adjacent basal cells are always in direct contact with each other and form tight junctions. In contrast, the basal cell layer in rodents is punctuated and not continuous.27 Prostate basal cells proliferate more than luminal cells in human prostate, whereas luminal cell proliferation is more frequently observed in rodents.60 Human and rodent prostate cells also differ in terms of the expression of lineage markers such as PSA, CD44 and CD57, and so on.76 Collectively, while studies from rodents provide important insights into molecular and cellular mechanisms underpinning disease initiation and progression in humans, it should be kept in mind that exceptions and differences exist between species.
IMPLICATIONS AND FUTURE DIRECTIONS
Breast cancer has been successfully subclassified into at least five different types based on their gene expression profiles.77 This finding supports the hypothesis that the identity of the cellular origin for cancers determines their histopathological appearances and molecular profiles. Gene expression profiles of large cohorts of human prostate cancer specimens have also been performed.78,79 However, no distinct subtypes of tumors were able to be classified based on their gene expression profiles. This could be accounted for by the fact that prostate cancer is so heterogeneous that sampled specimens fail to reflect the overall genomic compositions and gene expression profiles of tumors, as has been shown in a recent study on renal cell carcinoma.80 On the other hand, unlike breast cancer, treatment-naïve prostate cancer is quite homogeneous in terms of the expression of frequently used lineage markers. Therefore, it is possible that prostate cancers may not be distinguished from each other as distinctively compared to what has been achieved with breast cancers. As an alternative, it has been proposed that prostate cancers be subclassified based on the associated oncogenic signaling, such as the presence of Ets fusion events.81
Although we showed that disease initiation from prostate basal cells have a longer latency in the K14-Pten model, the majority of the tumors are composed of cells that display a luminal cell phenotype. There are no dramatic differences in histological appearance and responses to androgen deprivation between the cancerous lesions initiated from basal and luminal cells. Proliferation and apoptosis indices are also comparable. Therefore, our observations do not support the hypothesis that cellular origin can substantially affect histopathology and malignant potentials of prostate cancers at least in the context of loss of function of Pten and p53. Molecular profiling analyses of these tumors will provide more insights.
Nevertheless, our study raised several additional interesting questions. What is the essential molecular signaling that can induce basal–luminal differentiation? Our recent study showed that Notch signaling is capable of inducing differentiation of basal progenitors, but is not sufficient to drive the basal–luminal differentiation.82 Additionally, the longer latency of the K14-Pten model offers a platform where researchers can study the etiology of cancer initiation from prostate basal cells. It will be interesting to investigate how other oncogenic or inflammatory signaling can accelerate disease initiation in this model. For example, will the expression of Ets fusion proteins in the basal cells of the K14-Pten model accelerate disease initiation by promoting basal–luminal differentiation? These studies should provide novel insights into the molecular mechanisms underpinning prostate cancer initiation.
ACKNOWLEDGEMENTS
I apologize to those authors whose work could not be cited because of space limitation, and thank Drs Michael Ittmann and Jeffrey Rosen for critical reading of the manuscript. This work is supported by NIH R00CA125937 (LX), R01DK092202 (LX) and U01CA141497 (MMI).
Footnotes
CONFLICT OF INTEREST The author declares no conflict of interest.
REFERENCES
- 1.Visvader JE. Cells of origin in cancer. Nature. 2011;469:314–322. doi: 10.1038/nature09781. [DOI] [PubMed] [Google Scholar]
- 2.Clevers H. The cancer stem cell: premises, promises and challenges. Nat Med. 2011;17:313–319. doi: 10.1038/nm.2304. [DOI] [PubMed] [Google Scholar]
- 3.Lim E, Vaillant F, Wu D, Forrest NC, Pal B, Hart AH, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15:907–913. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- 4.Sharma MK, Mansur DB, Reifenberger G, Perry A, Leonard JR, Aldape KD, et al. Distinct genetic signatures among pilocytic astrocytomas relate to their brain region origin. Cancer Res. 2007;67:890–900. doi: 10.1158/0008-5472.CAN-06-0973. [DOI] [PubMed] [Google Scholar]
- 5.Gibson P, Tong Y, Robinson G, Thompson MC, Currle DS, Eden C, et al. Subtypes of medulloblastoma have distinct developmental origins. Nature. 2010;468:1095–1099. doi: 10.1038/nature09587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ince TA, Richardson AL, Bell GW, Saitoh M, Godar S, Karnoub AE, et al. Transformation of different human breast epithelial cell types leads to distinct tumor phenotypes. Cancer Cell. 2007;12:160–170. doi: 10.1016/j.ccr.2007.06.013. [DOI] [PubMed] [Google Scholar]
- 7.Shizuru JA, Negrin RS, Weissman IL. Hematopoietic stem and progenitor cells: clinical and preclinical regeneration of the hematolymphoid system. Annu Rev Med. 2005;56:509–538. doi: 10.1146/annurev.med.54.101601.152334. [DOI] [PubMed] [Google Scholar]
- 8.Huntly BJ, Shigematsu H, Deguchi K, Lee BH, Mizuno S, Duclos N, et al. MOZ-TIF2, but not BCR-ABL, confers properties of leukemic stem cells to committed murine hematopoietic progenitors. Cancer Cell. 2004;6:587–596. doi: 10.1016/j.ccr.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 9.Krivtsov AV, Twomey D, Feng Z, Stubbs MC, Wang Y, Faber J, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442:818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- 10.Cozzio A, Passegue E, Ayton PM, Karsunky H, Cleary ML, Weissman IL. Similar MLL-associated leukemias arising from self-renewing stem cells and short-lived myeloid progenitors. Genes Dev. 2003;17:3029–3035. doi: 10.1101/gad.1143403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barabe F, Kennedy JA, Hope KJ, Dick JE. Modeling the initiation and progression of human acute leukemia in mice. Science. 2007;316:600–604. doi: 10.1126/science.1139851. [DOI] [PubMed] [Google Scholar]
- 12.Welm AL, Kim S, Welm BE, Bishop JM. MET and MYC cooperate in mammary tumorigenesis. Proc Natl Acad Sci USA. 2005;102:4324–4329. doi: 10.1073/pnas.0500470102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xin L, Lawson DA, Witte ON. The Sca-1 cell surface marker enriches for a prostate-regenerating cell subpopulation that can initiate prostate tumorigenesis. Proc Natl Acad Sci USA. 2005;102:6942–6947. doi: 10.1073/pnas.0502320102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuhbandner S, Brummer S, Metzger D, Chambon P, Hofmann F, Feil R. Temporally controlled somatic mutagenesis in smooth muscle. Genesis. 2000;28:15–22. doi: 10.1002/1526-968x(200009)28:1<15::aid-gene20>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 15.von Werder A, Seidler B, Schmid RM, Schneider G, Saur D. Production of avian retroviruses and tissue-specific somatic retroviral gene transfer in vivo using the RCAS/TVA system. Nat Protoc. 2012;7:1167–1183. doi: 10.1038/nprot.2012.060. [DOI] [PubMed] [Google Scholar]
- 16.Lapouge G, Youssef KK, Vokaer B, Achouri Y, Michaux C, Sotiropoulou PA, et al. Identifying the cellular origin of squamous skin tumors. Proc Natl Acad Sci USA. 2011;108:7431–7436. doi: 10.1073/pnas.1012720108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Passegue E, Jamieson CH, Ailles LE, Weissman IL. Normal and leukemic hematopoiesis: are leukemias a stem cell disorder or a reacquisition of stem cell characteristics? Proc Natl Acad Sci USA. 2003;100(Suppl 1):11842–11849. doi: 10.1073/pnas.2034201100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang ZJ, Ellis T, Markant SL, Read TA, Kessler JD, Bourboulas M, et al. Medulloblastoma can be initiated by deletion of Patched in lineage-restricted progenitors or stem cells. Cancer Cell. 2008;14:135–145. doi: 10.1016/j.ccr.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sutherland KD, Proost N, Brouns I, Adriaensen D, Song JY, Berns A. Cell of origin of small cell lung cancer: inactivation of Trp53 and Rb1 in distinct cell types of adult mouse lung. Cancer Cell. 2011;19:754–764. doi: 10.1016/j.ccr.2011.04.019. [DOI] [PubMed] [Google Scholar]
- 20.Du Z, Li Y. RCAS-TVA in the mammary gland: an in vivo oncogene screen and a high fidelity model for breast transformation? Cell Cycle. 2007;6:823–826. doi: 10.4161/cc.6.7.4074. [DOI] [PubMed] [Google Scholar]
- 21.Baker CM, Verstuyf A, Jensen KB, Watt FM. Differential sensitivity of epidermal cell subpopulations to beta-catenin-induced ectopic hair follicle formation. Dev Biol. 2010;343:40–50. doi: 10.1016/j.ydbio.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 23.Abate-Shen C, Shen MM. Molecular genetics of prostate cancer. Genes Dev. 2000;14:2410–2434. doi: 10.1101/gad.819500. [DOI] [PubMed] [Google Scholar]
- 24.Bhatia-Gaur R, Donjacour AA, Sciavolino PJ, Kim M, Desai N, Young P, et al. Roles for Nkx3.1 in prostate development and cancer. Genes Dev. 1999;13:966–977. doi: 10.1101/gad.13.8.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bethel CR, Faith D, Li X, Guan B, Hicks JL, Lan F, et al. Decreased NKX3.1 protein expression in focal prostatic atrophy, prostatic intraepithelial neoplasia, and adenocarcinoma: association with gleason score and chromosome 8p deletion. Cancer Res. 2006;66:10683–10690. doi: 10.1158/0008-5472.CAN-06-0963. [DOI] [PubMed] [Google Scholar]
- 26.Signoretti S, Waltregny D, Dilks J, Isaac B, Lin D, Garraway L, et al. P63 is a prostate basal cell marker and is required for prostate development. Am J Pathol. 2000;157:1769–1775. doi: 10.1016/S0002-9440(10)64814-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.El-Alfy M, Pelletier G, Hermo LS, Labrie F. Unique features of the basal cells of human prostate epithelium. Microsc Res Technol. 2000;51:436–446. doi: 10.1002/1097-0029(20001201)51:5<436::AID-JEMT6>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 28.De Marzo AM, Meeker AK, Epstein JI, Coffey DS. Prostate stem cell compartments: expression of the cell cycle inhibitor p27Kip1 in normal, hyperplastic, and neoplastic cells. Am J Pathol. 1998;153:911–919. doi: 10.1016/S0002-9440(10)65632-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Isaacs JT. Control of cell proliferation and cell death in the normal and neoplastic prostate: a stem cell model. In: Rodgers CH, Coffey DS, Cunha G, Grayhack JT, Hinman F Jr, Horton R, editors. Benigh Prostatic Hyperplasia. US Department of Health and Human Services; Washington, DC: 1987. pp. 85–94. NIH Publication No. 87-2881. [Google Scholar]
- 30.Vashchenko N, Abrahamsson PA. Neuroendocrine differentiation in prostate cancer: implications for new treatment modalities. Eur Urol. 2005;47:147–155. doi: 10.1016/j.eururo.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 31.Litvinov IV, Vander Griend DJ, Xu Y, Antony L, Dalrymple SL, Isaacs JT. Low-calcium serum-free defined medium selects for growth of normal prostatic epithelial stem cells. Cancer Res. 2006;66:8598–8607. doi: 10.1158/0008-5472.CAN-06-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hudson DL, Guy AT, Fry P, O'Hare MJ, Watt FM, Masters JR. Epithelial cell differentiation pathways in the human prostate: identification of intermediate phenotypes by keratin expression. J Histochem Cytochem. 2001;49:271–278. doi: 10.1177/002215540104900214. [DOI] [PubMed] [Google Scholar]
- 33.Peehl DM. Primary cell cultures as models of prostate cancer development. Endocr Relat Cancer. 2005;12:19–47. doi: 10.1677/erc.1.00795. [DOI] [PubMed] [Google Scholar]
- 34.van Leenders G, Dijkman H, Hulsbergen-van de Kaa C, Ruiter D, Schalken J. Demonstration of intermediate cells during human prostate epithelial differentiation in situ and in vitro using triple-staining confocal scanning microscopy. Lab Invest. 2000;80:1251–1258. doi: 10.1038/labinvest.3780133. [DOI] [PubMed] [Google Scholar]
- 35.Humphrey PA. Diagnosis of adenocarcinoma in prostate needle biopsy tissue. J Clin Pathol. 2007;60:35–42. doi: 10.1136/jcp.2005.036442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Min J, Zaslavsky A, Fedele G, McLaughlin SK, Reczek EE, De Raedt T, et al. An oncogene-tumor suppressor cascade drives metastatic prostate cancer by coordinately activating Ras and nuclear factor-kappaB. Nat Med. 2010;16:286–294. doi: 10.1038/nm.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Verhagen AP, Ramaekers FC, Aalders TW, Schaafsma HE, Debruyne FM, Schalken JA. Colocalization of basal and luminal cell-type cytokeratins in human prostate cancer. Cancer Res. 1992;52:6182–6187. [PubMed] [Google Scholar]
- 38.van Leenders GJ, Gage WR, Hicks JL, van Balken B, Aalders TW, Schalken JA, et al. Intermediate cells in human prostate epithelium are enriched in proliferative inflammatory atrophy. Am J Pathol. 2003;162:1529–1537. doi: 10.1016/S0002-9440(10)64286-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tran CP, Lin C, Yamashiro J, Reiter RE. Prostate stem cell antigen is a marker of late intermediate prostate epithelial cells. Mol Cancer Res. 2002;1:113–121. [PubMed] [Google Scholar]
- 40.Choi N, Zhang B, Zhang L, Ittmann M, Xin L. Adult murine prostate basal and luminal cells are self-sustained lineages that can both serve as targets for prostate cancer initiation. Cancer Cell. 2012;21:253–265. doi: 10.1016/j.ccr.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang S, Garcia AJ, Wu M, Lawson DA, Witte ON, Wu H. Pten deletion leads to the expansion of a prostatic stem/progenitor cell subpopulation and tumor initiation. Proc Natl Acad Sci USA. 2006;103:1480–1485. doi: 10.1073/pnas.0510652103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xin L, Ide H, Kim Y, Dubey P, Witte ON. In vivo regeneration of murine prostate from dissociated cell populations of postnatal epithelia and urogenital sinus mesenchyme. Proc Natl Acad Sci USA. 2003;100(Suppl 1):11896–11903. doi: 10.1073/pnas.1734139100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chung LW, Cunha GR. Stromal–epithelial interactions: II. Regulation of prostatic growth by embryonic urogenital sinus mesenchyme. Prostate. 1983;4:503–511. doi: 10.1002/pros.2990040509. [DOI] [PubMed] [Google Scholar]
- 44.Goldstein AS, Lawson DA, Cheng D, Sun W, Garraway IP, Witte ON. Trop2 identifies a subpopulation of murine and human prostate basal cells with stem cell characteristics. Proc Natl Acad Sci USA. 2008;105:20882–20887. doi: 10.1073/pnas.0811411106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lawson DA, Xin L, Lukacs RU, Cheng D, Witte ON. Isolation and functional characterization of murine prostate stem cells. Proc Natl Acad Sci USA. 2007;104:181–186. doi: 10.1073/pnas.0609684104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leong KG, Wang BE, Johnson L, Gao WQ. Generation of a prostate from a single adult stem cell. Nature. 2008;456:804–808. doi: 10.1038/nature07427. [DOI] [PubMed] [Google Scholar]
- 47.Burger PE, Xiong X, Coetzee S, Salm SN, Moscatelli D, Goto K, et al. Sca-1 expression identifies stem cells in the proximal region of prostatic ducts with high capacity to reconstitute prostatic tissue. Proc Natl Acad Sci USA. 2005;102:7180–7185. doi: 10.1073/pnas.0502761102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goldstein AS, Huang J, Guo C, Garraway IP, Witte ON. Identification of a cell of origin for human prostate cancer. Science. 2010;329:568–571. doi: 10.1126/science.1189992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lawson DA, Zong Y, Memarzadeh S, Xin L, Huang J, Witte ON. Basal epithelial stem cells are efficient targets for prostate cancer initiation. Proc Natl Acad Sci USA. 2010;107:2610–2615. doi: 10.1073/pnas.0913873107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang X, Kruithof-de Julio M, Economides KD, Walker D, Yu H, Halili MV, et al. A luminal epithelial stem cell that is a cell of origin for prostate cancer. Nature. 2009;461:495–500. doi: 10.1038/nature08361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taylor RA, Cowin PA, Cunha GR, Pera M, Trounson AO, Pedersen J, et al. Formation of human prostate tissue from embryonic stem cells. Nat Methods. 2006;3:179–181. doi: 10.1038/nmeth855. [DOI] [PubMed] [Google Scholar]
- 52.Neubauer BL, Chung LW, McCormick KA, Taguchi O, Thompson TC, Cunha GR. Epithelial–mesenchymal interactions in prostatic development. II. Biochemical observations of prostatic induction by urogenital sinus mesenchyme in epithelium of the adult rodent urinary bladder. J Cell Biol. 1983;96:1671–1676. doi: 10.1083/jcb.96.6.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Taylor RA, Wang H, Wilkinson SE, Richards MG, Britt KL, Vaillant F, et al. Lineage enforcement by inductive mesenchyme on adult epithelial stem cells across developmental germ layers. Stem Cells. 2009;27:3032–3042. doi: 10.1002/stem.244. [DOI] [PubMed] [Google Scholar]
- 54.Magee JA, Abdulkadir SA, Milbrandt J. Haploinsufficiency at the Nkx3.1 locus. A paradigm for stochastic, dosage-sensitive gene regulation during tumor initiation. Cancer Cell. 2003;3:273–283. doi: 10.1016/s1535-6108(03)00047-3. [DOI] [PubMed] [Google Scholar]
- 55.Liu J, Pascal LE, Isharwal S, Metzger D, Ramos Garcia R, Pilch J, et al. Regenerated luminal epithelial cells are derived from preexisting luminal epithelial cells in adult mouse prostate. Mol Endocrinol. 2011;25:1849–1857. doi: 10.1210/me.2011-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Van Keymeulen A, Rocha AS, Ousset M, Beck B, Bouvencourt G, Rock J, et al. Distinct stem cells contribute to mammary gland development and maintenance. Nature. 2011;479:189–193. doi: 10.1038/nature10573. [DOI] [PubMed] [Google Scholar]
- 57.de Visser KE, Ciampricotti M, Michalak E, Wei-Min Tan D, Speksnijder EN, Hau CS, et al. Developmental stage-specific contribution of LGR5+ cells to basal and luminal epithelial lineages in the postnatal mammary gland. J Pathol. 2012;228:300–309. doi: 10.1002/path.4096. [DOI] [PubMed] [Google Scholar]
- 58.van Amerongen R, Bowman AN, Nusse R. Developmental stage and time dictate the fate of Wnt/beta-catenin-responsive stem cells in the mammary gland. Cell stem cell. 2012;11:387–400. doi: 10.1016/j.stem.2012.05.023. [DOI] [PubMed] [Google Scholar]
- 59.Doupe DP, Alcolea MP, Roshan A, Zhang G, Klein AM, Simons BD, et al. A single progenitor population switches behavior to maintain and repair esophageal epithelium. Science. 2012;337:1091–1093. doi: 10.1126/science.1218835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bonkhoff H, Stein U, Remberger K. The proliferative function of basal cells in the normal and hyperplastic human prostate. Prostate. 1994;24:114–118. doi: 10.1002/pros.2990240303. [DOI] [PubMed] [Google Scholar]
- 61.Tuxhorn JA, Ayala GE, Rowley DR. Reactive stroma in prostate cancer progression. J Urol. 2001;166:2472–2483. [PubMed] [Google Scholar]
- 62.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 63.King JC, Xu J, Wongvipat J, Hieronymus H, Carver BS, Leung DH, et al. Cooperativity of TMPRSS2-ERG with PI3-kinase pathway activation in prostate oncogenesis. Nat Genet. 2009;41:524–526. doi: 10.1038/ng.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Carver BS, Tran J, Gopalan A, Chen Z, Shaikh S, Carracedo A, et al. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet. 2009;41:619–624. doi: 10.1038/ng.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zong Y, Xin L, Goldstein AS, Lawson DA, Teitell MA, Witte ON. ETS family transcription factors collaborate with alternative signaling pathways to induce carcinoma from adult murine prostate cells. Proc Natl Acad Sci USA. 2009;106:12465–12470. doi: 10.1073/pnas.0905931106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Klezovitch O, Risk M, Coleman I, Lucas JM, Null M, True LD, et al. A causal role for ERG in neoplastic transformation of prostate epithelium. Proc Natl Acad Sci USA. 2008;105:2105–2110. doi: 10.1073/pnas.0711711105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang L, Zhao W, Valdez JM, Creighton CJ, Xin L. Low-density Taqman miRNA array reveals miRNAs differentially expressed in prostatic stem cells and luminal cells. Prostate. 2009;70:297–304. doi: 10.1002/pros.21064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 69.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Podsypanina K, Du YC, Jechlinger M, Beverly LJ, Hambardzumyan D, Varmus H. Seeding and propagation of untransformed mouse mammary cells in the lung. Science. 2008;321:1841–1844. doi: 10.1126/science.1161621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang Y, Hayward S, Cao M, Thayer K, Cunha G. Cell differentiation lineage in the prostate. Differentiation. 2001;68:270–279. doi: 10.1046/j.1432-0436.2001.680414.x. [DOI] [PubMed] [Google Scholar]
- 73.Ousset M, Van Keymeulen A, Bouvencourt G, Sharma N, Achouri Y, Simons BD, et al. Multipotent and unipotent progenitors contribute to prostate postnatal development. Nature cell biology. doi: 10.1038/ncb2600. (e-pub ahead of print 16 October 2012) [DOI] [PubMed] [Google Scholar]
- 74.Pascal LE, Oudes AJ, Petersen TW, Goo YA, Walashek LS, True LD, et al. Molecular and cellular characterization of ABCG2 in the prostate. BMC Urol. 2007;7:6. doi: 10.1186/1471-2490-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Moeller H, Goecke B, Herter F. Seasonal and diurnal changes of prostatic androgen receptor and circulating testosterone in young mature rats. Res Exp Med (Berl) 1988;188:451–462. doi: 10.1007/BF01852003. [DOI] [PubMed] [Google Scholar]
- 76.Liu AY, True LD. Characterization of prostate cell types by CD cell surface molecules. Am J Pathol. 2002;160:37–43. doi: 10.1016/S0002-9440(10)64346-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 78.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Glinsky GV, Glinskii AB, Stephenson AJ, Hoffman RM, Gerald WL. Gene expression profiling predicts clinical outcome of prostate cancer. J Clin Invest. 2004;113:913–923. doi: 10.1172/JCI20032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Goldstein AS, Zong Y, Witte ON. A two-step toward personalized therapies for prostate cancer. Sci Transl Med. 2011;3:72–77. doi: 10.1126/scitranslmed.3002169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Valdez JM, Zhang L, Su Q, Dakhova O, Zhang Y, Shahi P, et al. Notch and TGFβ form a reciprocal positive regulatory loop that suppresses murine prostate basal stem/progenitor cell activity. Cell Stem Cell. 2012;11:676–688. doi: 10.1016/j.stem.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]