Abstract
Background
Oral cancer in the form of squamous cell carcinoma (OSCC) is typically detected in advanced stages when treatment is complex and may not be curative. The need for surgical biopsy may contribute to delays in diagnosis and impede early detection. Multiple studies of RNA from surgically obtained tumor samples have revealed many genes differentially expressed with this disease. We sought to determine if the identified mRNAs could be used as markers by a noninvasive detection system for OSCC using RNA from brush cytology.
Methods
Levels of mRNAs from 21 genes known to be differentially expressed in head and neck squamous cell carcinoma surgical samples, compared to controls, were shown to be quantifiable in oral brush cytology samples. These mRNAs were quantified in a training set of 14 tumor and 20 nonmalignant brush cytology samples from tobacco/betel nut users. The measurement of two additional mRNAs and analysis using support vector machines produced an algorithm for class prediction of these cancers.
Results
This OSCC classifier based on the levels of 5 mRNAs in RNA from brush cytology initially showed 0.93 sensitivity and 0.91 specificity in differentiating OSCC from benign oral mucosal lesions based on leave-one-out cross-validation. When used on a test set of 19 samples from 6 OSCCs and 13 nonmalignant oral lesions we found misclassification of only one OSCC and one benign lesion.
Conclusions
This shows the promise of using RNA from brush cytology for early OSCC detection and the potential for clinical usage of this noninvasive classifier.
Keywords: oral squamous cell carcinoma, brush cytology, RNA, gene expression, tobacco
Introduction
Oral squamous cell carcinoma (OSCC), by far the most common oral cancer, is typically detected in advanced stages when treatment is complex and may not be curative (1). In contrast, early stage OSCC responds well to treatment, making improvements in early detection important. Testing for OSCC carcinoma is appropriate with suspicious oral lesions without a known cause that endure for more than 2 weeks, especially in the patient with high risk behavior such as tobacco usage. The patient is asked to make an appointment with a surgeon, at which time a surgical biopsy is taken. This allows for histopathologic examination of the tissue to determine abnormalities in epithelial cell and tissue morphology and architecture that correspond to OSCC. These include changes in nuclear cytoplasmic ratios, cellular and nuclear pleomorphism, and mucosal architecture changes, the location of mitotic cells and disruption of the basement membrane of the epithelium. Based on the histopathology, a diagnosis of malignancy (OSCC), premalignancy (carcinoma in situ, dysplasia), or nonmalignancy is made.
The last 15 years have seen an increase in methodologies for detection of possible oral malignancies (2, 3). These include fluoroscopy, toluidine blue staining, and laser tomography. In addition, screening methods using brush cytology have sought to obtain cells from oral lesions. These can then be examined after staining for changes in cell size and nuclear cytoplasmic ratio that can occur with malignancy, thus eliminating surgical biopsy as a screening method. Despite these advances, the vast majority of OSCCs in the U.S. and worldwide are still detected by the patient or during a visual oral examination by a dentist or a health care provider, usually during the advanced stages of the disease process, and then verified after biopsy and tissue histopathologic examination.
Global gene expression analysis of tissue obtained by surgical biopsy of OSCC, and more generally head and neck squamous cell carcinoma, has defined a large group of genes that show changes in expression with these diseases (4-11). Despite efforts to dissect out non-tumor tissue in these studies, there are typically large amounts of mesenchymal cells from stroma, in addition to epithelium. Studies that focused on oral cancers have produced gene expression-based classifiers for OSCC with preliminary external validation (5, 10, 11). This points to the promise that gene expression analysis of OSCC tissue may provide accurate diagnosis. Limiting the usage of this methodology for oral cancer screening has been the lack of a demonstrated ability to differentiate malignant versus benign pathology, validation by external groups, and the difficulty in standardization of tissue collection. The requirement for surgical acquisition of tissue with biopsy is also a limiting factor. For these and other reasons, the many published DNA microarray gene expression studies of OSCC tissue have unfortunately been an “endpoint of research endeavor “ … and not “screening tools for identifying potential genes for validation and further investigation “ as observed by Choi and Chen (12).
Brush cytology offers a noninvasive method of cell acquisition from oral lesions that allows RNA isolation from mucosal cells. Work in the field has shown that this RNA is amenable to RT-PCR analysis (13, 14) that provides reproducible measurements of gene expression (15-17). This noninvasive approach to obtaining RNA from oral tissue has recently been used to provide a window to changes in gene expression that occur with tobacco usage in benign tissue (15, 18, 19). We and others have described changes in gene expression that can be detected in RNA from OSCC lesions obtained by brush cytology (16, 17, 20, 21). Perhaps because of the challenges of performing global gene expression analysis with RNA from brush cytology, an externally validated gene expression-based classifier for this disease using this noninvasive approach has never been published (15, 18). Our focus here is OSCC associated with long-term exposure to tobacco and to a lesser degree betel nut, two powerful mutagenic compositions. These cancers may be separate from human papilloma virus (HPV)-associated head and neck tumors, which are typically not linked to long-term tobacco usage and occur with higher frequency in the oropharynx than the oral cavity (22). We tested if genes shown to be differentially expressed in surgical samples of head and neck squamous cell carcinoma and normal tissue were also differentially expressed in RNA from brush cytology of OSCC and benign lesions. The goal is to find a classifier that uses RNA from brush cytology to noninvasively allow gene expression-based identification of these tumors.
Material and methods
Clinical sampling
Brush cytology samples were collected from 34 former and current tobacco and betel nut users, who presented with oral lesions, typically just prior to taking a diagnostic biopsy. Patients were seen in the Oral and Maxillofacial Surgery Clinic, the Multidisciplinary Head and Neck Cancer Clinic, and the Otolaryngology Clinic in the University of Illinois Medical Center (16). The person who took the brush cytology sample was the same person who performed the biopsy in all except one case. Samples from an earlier study, along with those from additional patients, make up the 14 OSCC samples (Table 1) and 20 benign controls of the classifier training set. Benign samples include mucosal lesions, such as lichen planus and leukoplakia, all without dysplasia and are described in Kolokythas et al. (16). Nineteen additional patients from the same clinics provided RNA from brush cytology of 6 OSCC and 13 benign lesions to make up the validation set (Tables 2 and 3). We note that while keratocystic odontogenic tumors and ameloblastomas are typically not confused with OSCC, in both cases, in this study mucosal perforation led to differential diagnoses that included malignancy. Similarly, the salivary tumor presented with mucosal ulceration suggestive of malignancy. All diagnoses were verified by histopathologic examination of surgically obtained tumor tissue for OSCCs and scalpel biopsy material for nonmalignancies, with the exception of one leukoplakic lesion that resolved on cessation of smoking over a one-year follow-up. All subjects provided consent to participate in accordance with guidelines of the Office for the Protection of Research Subjects of the University of Illinois at Chicago.
Table 1.
Training Set: Patient data and OSCC details tumor samples
| Sample* | Site† | Sex | Age | Tobacco/Betel‡ | TNM | Grade |
|---|---|---|---|---|---|---|
| Classification | ||||||
| OSCC 1 | UG | M | 45 | Tob and Bet | T1NOMO | Gr 1 |
| OSCC 2 | LG | M | 46 | F-Tob | T4aN0M0 | Gr 2 |
| OSCC 3 | UG | M | 65 | F-Tob | Ta1N1M0 | Gr 1 |
| OSCC 4 | FOM | M | 55 | Tob | T2N0M0 | Gr 2 |
| OSCC 5 | Bu | M | 64 | Tob | T4aN0M0 | Gr 2 |
| OSCC 6 | LG | M | 38 | Tob | T4aN2bM0 | Gr 2 |
| OSCC 7 | FOM | M | 64 | Tob | T1N0M0 | Gr 2 |
| OSCC 8 | T | M | 81 | Tob | T2N0M0 | Gr 1 |
| OSCC 9 | Bu | M | 53 | Tob | T2N0M0 | Gr 2 |
| OSCC 10 | FOM-TM | M | 60 | Tob | T4aN0M0 | Gr 1 |
| OSCC 11 | T | M | 64 | F-Tob | T1N0M0 | Gr 3 |
| OSCC 12 | P | M | 61 | Tob | T3N0M0 | Gr 2 |
| OSCC 56 | UG | M | 84 | F-Tob | T4aNoMo | Gr 2 |
| OSCC 63 | LG | M | 85 | Tob | T4bN3M1 | Gr 1 |
OSCC1 – OSCC12 previously described (Kolokythas et al, 2011).
T, tongue; P, palate; LG/UG, lower/upper gingival; FOM, floor of mouth; Bu, buccal mucosa.
Tob, Tobacco user; Bet, betel nut user, if former user than F-Tob or F-Bet.
Table 2.
RT-PCR analysis of gene expression using RNA from brush cytology of OSCC versus
| Parametric p-value* |
FDR† | Fold-change in mRNA level |
Gene | Number of samples tested‡ |
|---|---|---|---|---|
| 0.0003012 | 0.00663 | 0.21 | LAMC2 | 13T 24N |
| 0.0014582 | 0.0107 | 2.49 | KRT17 | 14T 24N |
| 0.0054184 | 0.0298 | 9.43 | IL8 | 14T 23N |
| 0.0098977 | 0.0435 | 4.46 | ANXA1 | 14T 24N |
| 0.017282 | 0.0585 | 3.33 | ECM1 | 14T 24N |
| 0.0908235 | 0.225 | 0.43 | IL6 | 10T 15N |
| 0.0922038 | 0.225 | 3.76 | MAL | 13T 21N |
| 0.1030077 | 0.227 | 1.83 | MMP12 | 11T 22N |
| 0.1568689 | 0.314 | 3.18 | CXCL1 | 14T 24N |
| 0.1817851 | 0.333 | 2.32 | TGM3 | 13T 22N |
| 0.2025812 | 0.343 | 2.24 | EMP | 11T 17N |
| 0.2489573 | 0.378 | 2.42 | MMP1 | 10T 21N |
| 0.2765658 | 0.378 | 2.14 | SPINK5 | 12T 18N |
| 0.3184763 | 0.378 | 1.57 | SCEL | 12T 18N |
| 0.3228818 | 0.378 | 1.39 | CEACAM1 | 12T 17N |
| 0.3414761 | 0.378 | 2.32 | KRT4 | 10T 18N |
| 0.3435379 | 0.378 | 2.03 | KRT13 | 12T 22N |
| 0.2574326 | 0.412 | 0.51 | PPL | 11T 15N |
| 0.2991023 | 0.422 | 0.44 | CSTB1 | 11T 15N |
| 0.5283724 | 0.554 | 1.62 | ISG15 | 13T 22N |
| 0.8653252 | 0.865 | 0.96 | ALDH9 | 11T 17N |
Sorted by p-value of the univariate test
FDR is the false discovery rate, a measure of the probability that the data point is truly differentially expressed.
Number of samples tested refers to number of tumor samples (T) and nonmalignant samples (N) that provided expression data for these mRNAs.
Table 3.
Validation set: Patient data and OSCC details for tumor samples
| Sample | Site* | Sex | Age | Tobacco/Betel† | TNM | Grade |
|---|---|---|---|---|---|---|
| Classification | ||||||
| OSCC 81 | RMF | M | 61 | Tob | T4aN0M0 | Gr 2 |
| OSCC 102 | LG | M | 46 | Tob | T4bN3M0 | Gr 1 |
| OSCC 127 | FOM | M | 52 | Tob | T1N0M0 | Gr 1 |
| OSCC 204 | T | F | 76 | Tob | T1N0M0 | Gr 1 |
| OSCC 213 | T | F | 55 | Tob | T1N0M0 | Gr 3 |
| OSCC 216 | FOM | M | 67 | Tob | T2N0M0 | Gr 3 |
T, tongue; SP, soft palate; HP, Hard palate; LG/UG, lower/upper gingival; FOM, floor of mouth; Bu, buccal mucosa, LM, lip mucosa.
Tob, Tobacco user; Bet, betel nut user, if former user than F-Tob or F-Bet.
Brush cytology
Brush cytology was performed on patients as they presented in the clinic just prior to biopsy. Care was taken to sample only epithelium in the case of ulcerative lesions. Internal controls were not used and only samples from lesions were taken. Samples were immediately placed in Trizol (Invitrogen, Carlsbad, CA, USA), mixed, and frozen. We used a cervical cytology brush with RNA purification as described in Schwartz et al. (17).
Gene target selection
Nearly 30 studies have compared gene expression in surgically obtained tumor tissue versus control mucosa to define genes differentially expressed with OSCC. A survey by Choi and Chen reported 65 genes consistently differentially expressed in OSCC in multiple studies (12). Similar characterization of gene expression in surgically obtained tumor tissue in studies that focused on OSCC revealed 5 additional genes differentially expressed within this subset of cancers (4, 8, 23-25). These 70 genes are strong candidates to contribute to an OSCC classifier.
Due to the limited amount of RNA from each brush sample, in some cases less than 150 ng, the focus was on mRNAs with greater than average expression levels in the epithelium, as tested by global gene expression analysis of 3 tumor brush cytology samples using hybridization to Affymetrix Human Genome U133 Plus 2.0 microarrays. Thirty-four of the 70 head and neck squamous cell carcinoma genes were poorly expressed or undetectable, which was verified by RT-PCR analysis of a subset of 6 mRNAs (data not shown). Three mRNAs were not testable using RT-PCR analysis, due to sequence and 12 more were tested with one or two different sets of primers but did not consistently form a product or made multiple products during RT-PCR analysis and SYBR green detection and were not examined further. This left 21 genes that were tested for expression in brush samples from OSCC and benign control patients.
RT-PCR
RNA from brush cytology was converted to cDNA and quantitative RT-PCR was carried out using the iCycler iQ (Bio-Rad, Hercules, CA) and SYBR Green fluorescence as described (16). Values were normalized to the geometric mean of the controls, GAPD, RPLPO, and RPL4. Primers for these mRNAs, and those to detect the target mRNAs, were designed using Primer Express to give products of approximately 100 bases; sequences were previously published and/or included in the supplemental data section (supplemental table)(16).
Statistical analysis, class comparison, and class prediction using RNA from brush cytology
The class comparison function of BRB-Array Tools 3.9 (two-sample t-test with random variance model) allowed the determination of mRNA of genes that discriminate between RNA from cytology samples obtained from OSCC and nonmalignant lesions with a maximum allowed proportion of false positive genes of 0.1 (R. Simon and A. Pang Lam (http://linus.nci.nih.gov/BRB-ArrayTools.html).
A gene expression-based OSCC classifier was generated using the class predictor function of BRB-Array Tools as described (16, 26). Briefly, normalized mRNA levels for the 21 plus 2 additional targets were log2 transformed and entered for each sample of the training set. Due to the small number of genes tested, genes with measured levels significantly different between the classes at 0.01 significance level were used for class prediction. The program used 7 separate algorithms to generate optimized predictors while simultaneously performing leave-one-out cross-validation of the generated classifiers.
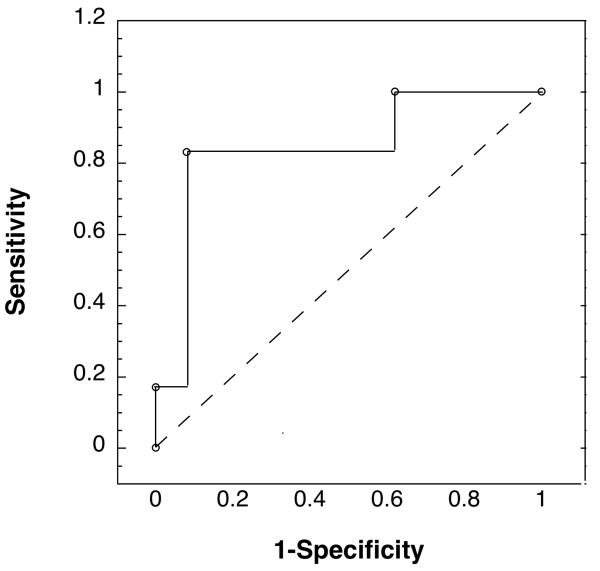
A receiver operating characteristic (ROC) curve was obtained on the 19-subject validation set by varying the decision threshold of the 5-gene OSCC classifier algorithm and plotting the effect on sensitivity and significance (Fig. 2).
Figure 2.
The ROC curve shows the accuracy of the 5-gene classifier for tobacco/betel associated OSCC when tested on RNA from brush cytology of the validation set of 6 OSCCs and 13 benign lesions at different thresholds for OSCC detection. The 5 genes were B2M, KRT17, ANXA2, LAMC2, and IL8.
Results
Brush oral cytology samples were obtained from 34 patients, 14 with OSCC and 20 with nonmalignant oral mucosal lesions (see Kolokythas et al. 2011 and Table 1), all tobacco or betel nut users (16). Diagnoses were verified with histopathology of biopsy and surgical tissue, with one exception. Based on the number of genes interrogated (<35), an observed within class standard deviation of 0.7, and cutoff for differential expression of 2-fold between classes, we estimated, a minimum of 11 samples from each group was sufficient to define a classifier with a tolerance of 0.10 (27).
Of the 21 mRNAs shown in the literature to be at different levels in surgically obtained head and neck squamous cell carcinoma or OSCC versus healthy tissue, we found that 5 of the genes showed differential expression in brush cytology RNA samples from OSCC versus benign lesions (Fig. 2). This was defined as a fold change of over 2× and an FDR of below 0.1. Interestingly, one of these mRNAs, ANXA1, showed an increase in levels in OSCC brush cytology samples, while earlier studies that examined RNA isolated from surgically obtained tissue, had indicated a decrease (12).
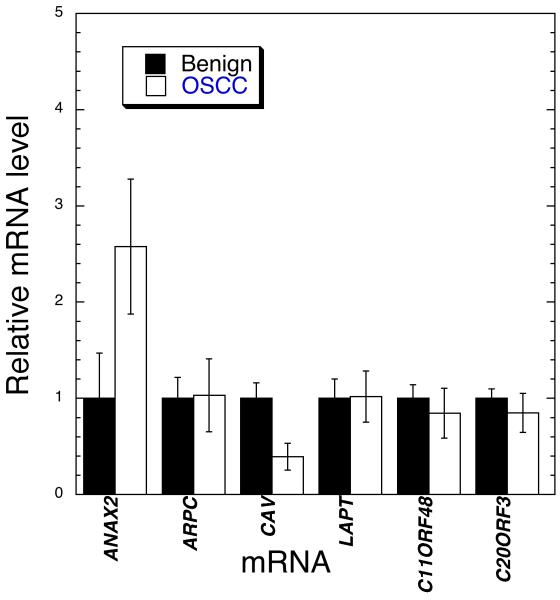
In an effort to increase the number of marker mRNAs for OSCC using RNA from brush cytology we also tested for changes in expression of 6 more genes, LAPT, C20orf3, ARPC1, C11orf48, ANXA2, and CAV1. These were arbitrarily chosen among the many genes reported to be differentially expressed on the RNA level in tissue from head and neck squamous cell carcinoma and nonmalignant or normal sites in a single study (8, 23, 25). A preliminary analysis of RNA from oral cytology samples from 7 OSCC lesion and 8 nonmalignant oral mucosal lesions was done (Fig. 1). Of the mRNAs analyzed those from the ANXA2 and CAV1 genes showed statistically significant or near significant changes in levels based on the Student’s t-test (supplemental data). Results of RT-PCR for quantitation of ANXA2, when tested against the entire training set, showed a 4.08 change in levels in OSCC samples with an FDR of 0.101. We did not pursue CAV1 mRNA analysis due to the low levels of signal.
Figure 1.
Expression of the above genes was measured in RNA from cytology of a subset of 7 OSCC samples and 6 benign samples using qRT-PCR. Three housekeeping genes were used as controls for total RNA level. Shown are means plus and minus standard error of the means. ANXA2 and CAV showed differences that attained or approached statistical significance based on the Student’s t-test, P < 0.08 and P < 0.017, respectively.
To perform class prediction for OSCC using the RNA from cytology samples, BRB-Array Tools was used for leave-one-out cross-validation, simultaneously testing 7 classifiers for their ability to differentiate OSCC from nonmalignant samples based on the expression levels of the 21 genes in table 1. Two additional genes were tested, ANXA2, based on its earlier identification as a marker for OSCC (23) and our findings here, and B2M, a gene we have shown to be enriched in RNA from brush cytology of OSCC in earlier studies (16, 17). We found that 3 out of 7 methods, compound covariate predictor, diagonal linear discriminant analysis, and 1-nearest neighbor, showed approximately 89% accuracy in identifying these samples. Support vector machines generated a classifier that showed the highest level of success in correct classification of OSCC and nonmalignant lesions using RNA from cytology samples in the training set with 92% accuracy. The permutation p value for the support vector machine generated classifier was <0.0001 indicating that zero out of 10000 classifiers generated from samples with randomly permuted class labels provided a classifier this accurate. This OSCC classifier was based on the levels of 5 mRNAs in RNA from brush cytology, B2M, KRT17, ANXA2, LAMC2, and IL8.
The support vector machines-generated classifier based on the levels of these 5 mRNAs was then tested on an independent validation set of 19 samples of RNA from brush cytology of 6 OSCC lesion and 13 benign lesions (Tables 3 and 4). In this preliminary external validation, the classifier made two errors, incorrectly classifying one tumor and one nonmalignant lesion. This classifier had 89% accuracy, sensitivity of 0.83, and specificity of 0.92. The ROC curve in figure 2 illustrates the accuracy of the classifier at various cut-offs and indicates an area under the curve (AUC) of 0.83, indicating while it worked well there is room for improvement.
Table 4.
Validation set: Patient data and pathologic appearance of benign lesions
| Sample | Site* | Sex | Age | Tobacco/Betel† | Diagnosis |
|---|---|---|---|---|---|
| BL66 | SP | M | 93 | F-Tob | Pleomorphic adenoma |
| BL93 | UG | F | 63 | Tob | Keratocystic Odontogenic Tumor |
| BL115 | Bu | M | 22 | Bet/Tob | Submucous Fibrosis |
| BL120 | LG | M | 55 | Tob | Periodontal cyst |
| BL130 | LG | M | 32 | F-Tob | Ameloblastoma |
| BL142 | Bu | M | 53 | Tob | Lichen planus |
| BL151 | HP | M | 88 | Tob | Nicotinic stomatitis |
| BL158 | T | M | 65 | Tob | Lichen planus, candidiasis |
| BL29 | Bu | F | 66 | Tob | Hyperplastic foliat papilloma |
| BL225 | Bu | F | 56 | Tob | Lichen planus |
| BL2337 | LM | M | 52 | Tob | Salivary gland tumor |
| BL242 | SP | F | 39 | Tob | Papilloma |
| BL278 | G | M | 57 | Tob | Verruciform xanthoma, mild |
| dysplasia |
T, tongue; SP, soft palate; HP, Hard palate; LG/UG, lower/upper gingival; FOM, floor of mouth; Bu, buccal mucosa, LM, lip mucosa.
Tob, Tobacco user; Bet, betel nut user, if former user than F-Tob or F-Bet.
Discussion
We took advantage of the many studies of surgical samples from head and neck cancer that have analyzed gene expression changes occurring with this disease to narrow the set of mRNAs tested (4-11). We studied in detail the expression of 21 genes identified in multiple studies and found 5 were differentially expressed in RNA from brush cytology samples of tobacco and betel nut associated OSCCs (Table 2). Most of the genes did not show differential expression and this may be due to several possible reasons: (1) many of the earlier studies did not differentiate oral, oropharyngeal, and laryngeal tumors, which have different etiologies and response to treatment (12, 28, 29); (2) in almost all studies no effort was made to focus on oral tumors associated with a consistent risk factor such as tobacco usage; and (3) many of these earlier studies compared gene expression in surgically obtained tumor versus histopathologically normal tissue from the same subject or normal subjects (5, 11). In the present study RNA from brush cytology was taken from malignancies, with controls from pathologic, but benign, oral mucosal lesions of other subjects. A final explanation for differences in gene expression might be the make-up of the cells in brush cytology samples versus those in surgically obtained biopsy tissue. As we, and others, have shown, brush oral cytology samples are almost exclusively epithelial cells (13, 17). Brush samples from friable lesions may contain blood cells but brushing is done to minimize their contribution. In contrast, surgical biopsies routinely have a range of stromal cells in addition to epithelium unless laser microdissection is done. These stromal cells include fibroblasts, immune cells, endothelial cells, and blood cells. Homogeneous cells obtained by brush cytology allow sensitive detection of changes in gene expression of the epithelium, but only detect changes in that tissue. For these reasons, we did not expect all 21 genes tested to be differentially expressed in RNA from brush cytology sampled OSCC versus benign mucosa.
The focus in this study is on genes already linked to head and neck squamous cell carcinoma or OSCC as shown in surgically obtained tumor tissues. This limited the number of features analyzed and greatly lowered the chance for overfitting the data, which can occur with true global gene expression analysis (27, 30). We tested approximately 30 mRNAs versus the greater than 20,000 typically tested in global gene expression analysis. This reduced the number of samples required to produce a statistically valid gene expression-based class predictor. The focus was tumors of a similar etiology, tobacco or betel nut-associated OSCC, with samples from a single cell type, epithelial cells, which further decreased sample needs (31, 32). While an examination of global gene expression may indicate more marker RNAs for OSCC in brush cytology samples it would require many more samples. A possible disadvantage of the present approach is that most studies that identify genes associated with OSCC report on genes that are differentially expressed and not on those genes that are ideally useful for class prediction. An ideal class predictor gene may show smaller differential expression between the two groups but be more consistently differentially expressed (31). mRNAs that show changes that complement other mRNAs in the goal of class prediction would be optimal. One of our future aims is to expand the field of tested genes to create a superior OSCC classifier that uses RNA from brush cytology.
Half of the patients in the training set had early stage disease (T1N0 or T2N0), suggesting the classifier would be accurate with both early and advanced stage OSCC. The ROC curve with an AUC of 0.83 for the validation set demonstrated the value of the 5-gene OSCC classifier but also indicated its errors (Fig. 2). The one OSCC misclassified, OSCC213, was from an early stage tumor (T1N0), though somewhat unusual in that it was grade 3. If one relaxes the classifier cut-off value for OSCC identification in order to identify OSCC213 correctly and provide a sensitivity of 1.0, the specificity would drop to an unacceptable value of 0.38. Variability in site selection or the types of cells acquired during the brush cytology is a possible explanation; though a second backup brush cytology sample gave the same result (data not shown). As described earlier, efforts were made to only sample intact epithelium in the case of ulcerative lesions so to avoid contamination of samples with stromal cells. Also, this OSCC may have been distinct. While all OSCCs were associated with tobacco or betel usage, this oral tumor may have been caused not by either of these mutagens but have a distinct etiology or be different in other ways that result in a different global gene expression pattern. This phenomenon has been well described in other cancers though not yet in OSCC (33, 34). Control sample BL225, a benign sample from a patient with lichen planus, was misidentified by the classifier as an OSCC. While lichen planus is reported to progress to OSCC at elevated rates, analysis of the biopsy did not show dysplastic or neoplastic changes (35). Given that a second brush cytology sample from the lesion gave the same anomalous expression pattern, possible explanations include difficulty in sampling some hyperkeratotic lesions, or a gene expression pattern in this benign inflammatory lesion that shares some features with malignancy. This patient is under close observation.
This external validation of an OSCC classifier that relies on RNA from brush cytology has not been reported before even as a preliminary finding. The sensitivity and specificity of this prototype OSCC classifier was 0.91 and 0.93 with the training set using leave-one-out cross-validation, and 0.83 and 0.92 with the small validation set of subjects. In research studies done over the years, an alternative screen for OSCC using brush cytology samples and then microscopic examination of the cells, revealed sensitivity for dysplasia and early tumors of 0.70 - 1 and specificity of 0.90 - 1 (1, 36-41). In the future we will need to expand the number of subjects in both sets to further test and optimize the gene expression-based classifier for brush oral cytology cells and to determine if it is superior in early oral malignancy detection compared to microscopic examination of these cells. We note that unlike the microscopic examination of cells, gene expression analysis of RNA from brush cytology does not require morphologic examination of the cells, but relies on gene expression measurement that has been proposed to potentially provide information on tumor staging and prognosis (42). The immediate goals are to provide validation with a larger data set focusing on early stage tumors and potential malignancies where early detection allows superior prognosis.
Supplementary Material
Acknowledgements
The authors thank Dr. Zarema Arbieva of the Core Genomics Facility at the University of Illinois at Chicago for performing the DNA microarray experiments on these non-ideal samples. We also thank Alexander J. Adami for meticulously compiling and organizing the list of genes shown to be differentially expressed in head and neck and OSCC studies in the literature. This work was supported by NIH Grant (NCI) CA139137 to Joel Schwartz.
References
- 1.Lingen MW, Kalmar JR, Karrison T, Speight PM. Critical evaluation of diagnostic aids for the detection of oral cancer. Oral Oncol. 2008;44:10–22. doi: 10.1016/j.oraloncology.2007.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mehrotra R, Gupta DK. Exciting new advances in oral cancer diagnosis: avenues to early detection. Head Neck Oncol. 2011;3:33. doi: 10.1186/1758-3284-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steele TO, Meyers A. Early detection of premalignant lesions and oral cancer. Otolaryngol Clin North Am. 2011;44:221–9. vii. doi: 10.1016/j.otc.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 4.Chen C, Mendez E, Houck J, et al. Gene expression profiling identifies genes predictive of oral squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 2008;17:2152–62. doi: 10.1158/1055-9965.EPI-07-2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi P, Jordan CD, Mendez E, et al. Examination of oral cancer biomarkers by tissue microarray analysis. Arch Otolaryngol Head Neck Surg. 2008;134:539–46. doi: 10.1001/archotol.134.5.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kondoh N, Ohkura S, Arai M, et al. Gene expression signatures that can discriminate oral leukoplakia subtypes and squamous cell carcinoma. Oral Oncol. 2007;43:455–62. doi: 10.1016/j.oraloncology.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 7.Kuriakose MA, Chen WT, He ZM, et al. Selection and validation of differentially expressed genes in head and neck cancer. Cell Mol Life Sci. 2004;61:1372–83. doi: 10.1007/s00018-004-4069-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tomioka H, Morita K, Hasegawa S, Omura K. Gene expression analysis by cDNA microarray in oral squamous cell carcinoma. J Oral Pathol Med. 2006;35:206–11. doi: 10.1111/j.1600-0714.2006.00410.x. [DOI] [PubMed] [Google Scholar]
- 9.Villaret DB, Wang T, Dillon D, et al. Identification of genes overexpressed in head and neck squamous cell carcinoma using a combination of complementary DNA subtraction and microarray analysis. Laryngoscope. 2000;110:374–81. doi: 10.1097/00005537-200003000-00008. [DOI] [PubMed] [Google Scholar]
- 10.Whipple ME, Mendez E, Farwell DG, Agoff SN, Chen C. A genomic predictor of oral squamous cell carcinoma. Laryngoscope. 2004;114:1346–54. doi: 10.1097/00005537-200408000-00006. [DOI] [PubMed] [Google Scholar]
- 11.Ziober AF, Patel KR, Alawi F, et al. Identification of a gene signature for rapid screening of oral squamous cell carcinoma. Clin Cancer Res. 2006;12:5960–71. doi: 10.1158/1078-0432.CCR-06-0535. [DOI] [PubMed] [Google Scholar]
- 12.Choi P, Chen C. Genetic expression profiles and biologic pathway alterations in head and neck squamous cell carcinoma. Cancer. 2005;104:1113–28. doi: 10.1002/cncr.21293. [DOI] [PubMed] [Google Scholar]
- 13.Spira A, Beane J, Schembri F, et al. Noninvasive method for obtaining RNA from buccal mucosa epithelial cells for gene expression profiling. Biotechniques. 2004;36:484–7. doi: 10.2144/04363RN03. [DOI] [PubMed] [Google Scholar]
- 14.Spivack SD, Hurteau GJ, Jain R, et al. Gene-environment interaction signatures by quantitative mRNA profiling in exfoliated buccal mucosal cells. Cancer Res. 2004;64:6805–13. doi: 10.1158/0008-5472.CAN-04-1771. [DOI] [PubMed] [Google Scholar]
- 15.Sridhar S, Schembri F, Zeskind J, et al. Smoking-induced gene expression changes in the bronchial airway are reflected in nasal and buccal epithelium. BMC Genomics. 2008;9:259. doi: 10.1186/1471-2164-9-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kolokythas A, Schwartz JL, Pytynia KB, et al. Analysis of RNA from brush cytology detects changes in B2M, CYP1B1 and KRT17 levels with OSCC in tobacco users. Oral Oncol. 2011;47:532–6. doi: 10.1016/j.oraloncology.2011.03.029. [DOI] [PubMed] [Google Scholar]
- 17.Schwartz JL, Panda S, Beam C, Bach LE, Adami GR. RNA from brush oral cytology to measure squamous cell carcinoma gene expression. J Oral Pathol Med. 2008;37:70–7. doi: 10.1111/j.1600-0714.2007.00596.x. [DOI] [PubMed] [Google Scholar]
- 18.Kupfer DM, White VL, Jenkins MC, Burian D. Examining smoking-induced differential gene expression changes in buccal mucosa. BMC Med Genomics. 2010;3:24. doi: 10.1186/1755-8794-3-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spira A. Upper airway gene expression in smokers: the mouth as a “window to the soul” of lung carcinogenesis? Cancer Prev Res (Phila) 2010;3:255–8. doi: 10.1158/1940-6207.CAPR-10-0013. [DOI] [PubMed] [Google Scholar]
- 20.Ries J, Mollaoglu N, Toyoshima T, et al. A novel multiple-marker method for the early diagnosis of oral squamous cell carcinoma. Dis Markers. 2009;27:75–84. doi: 10.3233/DMA-2009-0652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Toyoshima T, Koch F, Kaemmerer P, Vairaktaris E, Al-Nawas B, Wagner W. Expression of cytokeratin 17 mRNA in oral squamous cell carcinoma cells obtained by brush biopsy: preliminary results. J Oral Pathol Med. 2009;38:530–4. doi: 10.1111/j.1600-0714.2009.00748.x. [DOI] [PubMed] [Google Scholar]
- 22.Hennessey PT, Westra WH, Califano JA. Human papillomavirus and head and neck squamous cell carcinoma: recent evidence and clinical implications. J Dent Res. 2009;88:300–6. doi: 10.1177/0022034509333371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Estilo CL, P OC, Talbot S, et al. Oral tongue cancer gene expression profiling: Identification of novel potential prognosticators by oligonucleotide microarray analysis. BMC Cancer. 2009;9:11. doi: 10.1186/1471-2407-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.West M, Ginsburg GS, Huang AT, Nevins JR. Embracing the complexity of genomic data for personalized medicine. Genome Res. 2006;16:559–66. doi: 10.1101/gr.3851306. [DOI] [PubMed] [Google Scholar]
- 25.Ye H, Yu T, Temam S, et al. Transcriptomic dissection of tongue squamous cell carcinoma. BMC Genomics. 2008;9:69. doi: 10.1186/1471-2164-9-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simon R, Lam A, Li M-C, Ngan M, Menenzes S, Zhao Y. Analysis of Gene Expression Data Using BRB-Array Tools. Cancer Informatics. 2007;2:11–7. [PMC free article] [PubMed] [Google Scholar]
- 27.Dobbin KK, Zhao Y, Simon RM. How large a training set is needed to develop a classifier for microarray data? Clin Cancer Res. 2008;14:108–14. doi: 10.1158/1078-0432.CCR-07-0443. [DOI] [PubMed] [Google Scholar]
- 28.Yu YH, Kuo HK, Chang KW. The evolving transcriptome of head and neck squamous cell carcinoma: a systematic review. PLoS One. 2008;3:e3215. doi: 10.1371/journal.pone.0003215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Belbin TJ, Schlecht NF, Smith RV, et al. Site-specific molecular signatures predict aggressive disease in HNSCC. Head Neck Pathol. 2008;2:243–56. doi: 10.1007/s12105-008-0071-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.West M, Blanchette C, Dressman H, et al. Predicting the clinical status of human breast cancer by using gene expression profiles. Proc Natl Acad Sci U S A. 2001;98:11462–7. doi: 10.1073/pnas.201162998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simon RM. Interpretation of genomic data: Questions and answers. Seminars in Hematology. 2008;45:196–204. doi: 10.1053/j.seminhematol.2008.04.008. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Venet D, Pecasse F, Maenhaut C, Bersini H. Separation of samples into their constituents using gene expression data. Bioinformatics. 2001;17(Suppl 1):S279–87. doi: 10.1093/bioinformatics/17.suppl_1.s279. [DOI] [PubMed] [Google Scholar]
- 33.Lohavanichbutr P, Houck J, Fan W, et al. Genomewide gene expression profiles of HPV-positive and HPV-negative oropharyngeal cancer: potential implications for treatment choices. Arch Otolaryngol Head Neck Surg. 2009;135:180–8. doi: 10.1001/archoto.2008.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 35.Mithani SK, Mydlarz WK, Grumbine FL, Smith IM, Califano JA. Molecular genetics of premalignant oral lesions. Oral Dis. 2007;13:126–33. doi: 10.1111/j.1601-0825.2006.01349.x. [DOI] [PubMed] [Google Scholar]
- 36.Patton LL, Epstein JB, Kerr AR. Adjunctive techniques for oral cancer examination and lesion diagnosis: a systematic review of the literature. J Am Dent Assoc. 2008;139:896–905. doi: 10.14219/jada.archive.2008.0276. quiz 93-4. [DOI] [PubMed] [Google Scholar]
- 37.Perez-Sayans M, Reboiras-Lopez MD, Gayoso-Diz P, et al. Non-computer-assisted liquid-based cytology for diagnosis of oral squamous cell carcinoma. Biotech Histochem. 2012;87:59–65. doi: 10.3109/10520295.2011.575797. [DOI] [PubMed] [Google Scholar]
- 38.Poate TW, Buchanan JA, Hodgson TA, et al. An audit of the efficacy of the oral brush biopsy technique in a specialist Oral Medicine unit. Oral Oncol. 2004;40:829–34. doi: 10.1016/j.oraloncology.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 39.Sciubba JJ. Improving detection of precancerous and cancerous oral lesions. Computer-assisted analysis of the oral brush biopsy. U.S. Collaborative OralCDx Study Group. J Am Dent Assoc. 1999;130:1445–57. doi: 10.14219/jada.archive.1999.0055. [DOI] [PubMed] [Google Scholar]
- 40.Seijas-Naya F, Garcia-Carnicero T, Gandara-Vila P, et al. Applications of OralCDx(R) methodology in the diagnosis of oral leukoplakia. Med Oral Patol Oral Cir Bucal. 2012;17:e5–9. doi: 10.4317/medoral.17275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scheifele C, Schmidt-Westhausen AM, Dietrich T, Reichart PA. The sensitivity and specificity of the OralCDx technique: evaluation of 103 cases. Oral Oncol. 2004;40:824–8. doi: 10.1016/j.oraloncology.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 42.Lallemant B, Evrard A, Chambon G, et al. Gene expression profiling in head and neck squamous cell carcinoma: Clinical perspectives. Head Neck. 2010;32:1712–9. doi: 10.1002/hed.21491. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.