Summary
The specificity of a horizontally transmitted microbial symbiosis is often defined by molecular communication between host and microbe during initial engagement, which can occur in discrete stages. In the symbiosis between Steinernema nematodes and Xenorhabdus bacteria, previous investigations focused on bacterial colonization of the intestinal lumen (receptacle) of the nematode infective juvenile (IJ), as this was the only known persistent, intimate, and species-specific contact between the two. Here we show that bacteria colonize the anterior intestinal cells of other nematode developmental stages in a species-specific manner. Also, we describe three processes that only occur in juveniles that are destined to become IJs. First, a few bacterial cells colonize the nematode pharyngeal-intestinal valve (PIV) anterior to the intestinal epithelium. Second, the nematode intestine constricts while bacteria initially remain in the PIV. Third, anterior intestinal constriction relaxes and colonizing bacteria occupy the receptacle. At each stage, colonization requires X. nematophila symbiosis region 1 (SR1) genes and is species-specific: X. szentirmaii, which naturally lacks SR1, does not colonize unless SR1 is ectopically expressed. These findings reveal new aspects of Xenorhabdus bacteria interactions with and transmission by their Steinernema nematode hosts, and demonstrate that bacterial SR1 genes aid in colonizing nematode epithelial surfaces.
Keywords: Symbiosis, Mutualism, Host Range Specificity, Outer membrane protein, NilB, Symbiosis Region 1
Introduction
Most plants and animals are in contact with diverse bacterial species within their native environments (e.g. (Pace, 1997)). From these varied microbial assemblages, host plants and animals engage in intimate associations with specific microbes by concomitantly recruiting certain partners while excluding non-partners. Such associations can be maintained between generations by vertical or horizontal transmission of the specific microbial partner(s) (symbiont) to progeny: Vertically transmitted symbionts are supplied directly from parent to offspring, typically through the germline, while horizontal acquisition occurs from the environment (reviewed in (Bright and Bulgheresi, 2010)). In the latter case, within the environment, host offspring may be exposed to many different types of bacteria, forcing the need for selection of specific partners from diverse non-partner species. The molecular and cellular events that allow recognition and acquisition of specific partners from a background of non-partners are beginning to be revealed through the study of several experimentally tractable, naturally occurring, mutually beneficial associations between plants or animals and bacteria (Parniske, 2008; Popp and Ott, 2011; Ruby, 2008), such as that between entomopathogenic Steinernema nematodes and their Xenorhabdus bacterial symbionts.
Steinernema nematodes horizontally acquire their beneficial Xenorhabdus bacterial partners. The nematode infective juvenile (IJ), a modified J3 or dauer stage of the nematode, carries colonizing bacteria in the receptacle, a structure at the anterior of the nematode intestine ((Bird and Akhurst, 1983; Poinar, 1966; Snyder et al., 2007; Wouts, 1980) see also Fig. 1). P (parental) generation IJs infect insects, penetrating through natural openings and releasing colonizing bacteria into the insect hemocoel (Richards and Goodrich-Blair, 2009). Together the nematode and the bacteria kill the insect and the bacteria divide and provide nutrition, directly or indirectly, to infecting nematodes, which moult from a modified J3 stage into adults that reproduce. F1-generation eggs hatch and moult through juvenile to adult stages. Reproduction continues through two or three generations (Wang and Bedding, 1996) until conditions inside the cadaver, including low nutrient availability and high nematode density (Popiel et al., 1989), cue IJ development. The IJ receptacle is formed as the lumen between two nucleated intestinal cells, and becomes recolonized with bacteria (Martens and Goodrich-Blair, 2005; Snyder et al., 2007). The IJ migrates away from the insect cadaver seeking a new living host insect to infect. This growth cycle can be recapitulated in the laboratory on artificial media (Hirao et al., 2010; Hirao and Ehlers, 2010; Vivas and Goodrich-Blair, 2001; Wang and Bedding, 1998).
Fig. 1.
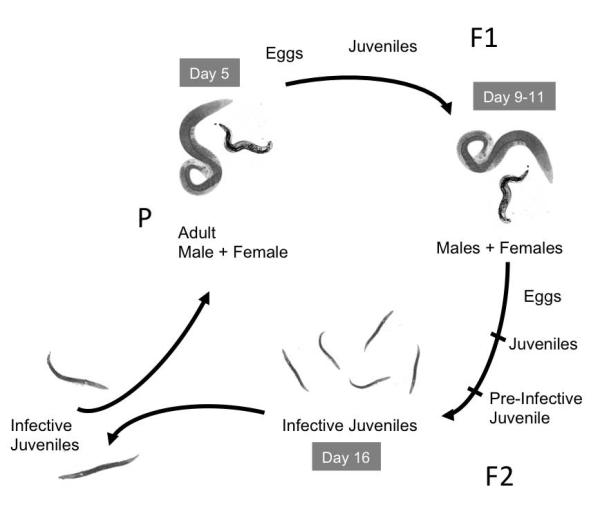
Model for nematode reproduction. In the wild, nematode IJs penetrate an insect host, release their bacteria, and develop into reproductive adults that give rise to F1 progeny that also develop into adults and reproduce. The resulting F2 progeny develop into pre-IJs that reacquire their symbiotic bacteria, develop into IJs, and leave the insect host in search of new prey. In this study, nematodes were grown on lipid agar medium in the laboratory, and the life cycle followed the same scheme. Nematode development was not synchronized within the population, but days indicated give approximate timing of generations during growth on laboratory medium. The figure is adapted from (Wang and Bedding, 1996).
Previous studies revealed early events in and features of X. nematophila colonization of the receptacle in S. carpocapsae IJs. The population of bacteria within a S. carpocapsae IJ receptacle is founded by one or a few colonizing X. nematophila cells: most individual IJs that form in the presence of multiple clonal variants of wild type X. nematophila are colonized by only one clone (Martens et al., 2003). In newly formed (immature) IJs, an oligo-colonized state has been observed, in which fewer than 10 individual bacterial cells can be seen within the receptacle (Martens et al., 2003), often adhering to a nematode-derived anucleate cluster of spherical bodies collectively called the intravesicular structure (IVS) (Kim et al., 2012; Martens and Goodrich-Blair, 2005). A mucus-like substance surrounding the IVS stains positively with wheat-germ agglutinin, suggesting the presence of N-acetyl glucosamine or N-acetyl neuraminic acid (Martens and Goodrich-Blair, 2005). As the IJ matures over the course of approximately one week, the few colonizing bacteria within the receptacle grow to a final population of ~ 50-200 bacterial cells (Martens et al., 2003). X. nematophila mutants defective in the synthesis of certain vitamins or amino acids fail to grow within the receptacle, and ultimately are cleared from this site (Martens et al. 2005), suggesting the host is capable of eliminating non-cooperative, non-functional symbionts.
Entry into the Steinernema nematode receptacle is limited to specific partner Xenorhabdus bacteria. For example, S. feltiae IJs show preference for colonization by X. bovienii strains while S. carpocapsae is highly specific for colonization by X. nematophila (Akhurst, 1983; Chapuis et al., 2009; Cowles and Goodrich-Blair, 2008; Sicard et al., 2004). In the latter association, molecular determinants, nematode intestine localization (nil) factors A, B, and C, have been identified that contribute to specificity. These three genes are encoded on a 3.5-kB genetic locus called symbiosis region 1 (SR1) that is not found in other Xenorhabdus species, and each is independently necessary for nematode receptacle localization (Heungens et al., 2002). Additionally, SR1 is sufficient to confer entry into the nematode receptacle on otherwise non-colonizing Xenorhabdus species (Cowles and Goodrich-Blair, 2008), albeit with lower levels of colonization than X. nematophila. An X. nematophila mutant lacking SR1 is not able to colonize the nematode receptacle, but certain mutations in nilA or nilB lead to a partial colonization defect (Heungens et al., 2002; Cowles and Goodrich-Blair, 2008; Bhasin et al., 2012). Analysis of these mutants indicated that the nil genes function during entry into and growth within the nematode receptacle. Characterization of the Nil factors has demonstrated that NilB is an outer membrane beta barrel protein (Bhasin et al., 2012) and NilC is an outer-membrane-tethered periplasmic lipoprotein (Cowles and Goodrich-Blair, 2004), suggesting that they may interact directly with host surfaces to mediate host colonization.
Experiments that described S. carpocapsae IJ colonization events did not reveal if individual clones of colonizing X. nematophila are selected during initiation or outgrowth. Furthermore, the events leading up to the oligo-colonized state of the immature IJ, including the process by which non-partner bacteria are excluded, have not been elucidated. To lend insights into these questions, we characterized X. nematophila colonization of S. carpocapsae adult and juvenile stages that precede IJ formation. We further interrogated the specificity of these events in S. carpocapsae using a X. nematophila non-colonizing ΔSR1 mutant and a non-colonizing, non-native Xenorhabdus species, X. szentirmaii. Finally, we demonstrate the conservation of the colonization process among members of the Steinernema genus by investigating early host-association events of S. feltiae and its symbiont, X. bovienii.
Results
X. nematophila bacteria colonize the anterior intestinal epithelia of adult and juvenile stage S. carpocapsae nematodes
To observe colonization events between X. nematophila and S. carpocapsae nematodes we engineered X. nematophila to express the green fluorescent protein (GFP), allowing visualization of single bacterial cells inside nematodes (Martens et al., 2003, Martens et al., 2005, Sugar et al., 2012, Murfin et al., 2012) and categorization of nematodes as colonized or un-colonized as they developed on lipid agar plates. Lack of synchrony in nematode development precluded definitive temporal assignment of events, and the exact timing of nematode development varied within and across generations in multiple experiments. Nonetheless, bacterial localization patterns in the animal host over time (days and nematode generations) were reproducible across multiple experiments relative to nematode developmental changes. Unexpectedly, we observed that adult and juvenile nematodes developing on lipid agar plates were colonized by X. nematophila bacteria. Adults in all generations had bacterial cells clustered on the epithelial cell surface within the anterior intestinal lumen (Fig. 2C-D). As in many nematode taxa, this intestinal region formed a caecum surrounding the basal bulb, a posterior portion of the pharynx that pumps and grinds food (Altun and Hall, 2009), and the pharyngeal-intestinal valve (PIV), a compact set of cells that form a channel linking the pharyngeal and intestinal lumina (Fig. 2A) (Baldwin and Perry, 2004; Altun and Hall, 2005). The bacteria colonizing this region in adults were not likely to be transmitted directly to progeny, since eggs laid in the surrounding media were not visibly associated with bacteria (data not shown). In addition, recently hatched juvenile nematodes lacked anterior intestine localization of bacteria (Fig. 2B). This un-colonized state was transient and soon after hatching juvenile nematodes were apparent with bacteria that localized to the anterior intestinal caecum (AIC) (Fig. 2E-F). Thus, bacterial localization at the AIC occurred in both juvenile and adult stages of nematodes. GFP-expressing bacteria were often visible indiscriminately throughout the intestine of developing nematodes at all stages (data not shown), consistent with the fact that S. carpocapsae is bacteriovorous and may be digesting some X. nematophila cells (Kaya and Gaugler, 1993).
Fig. 2.
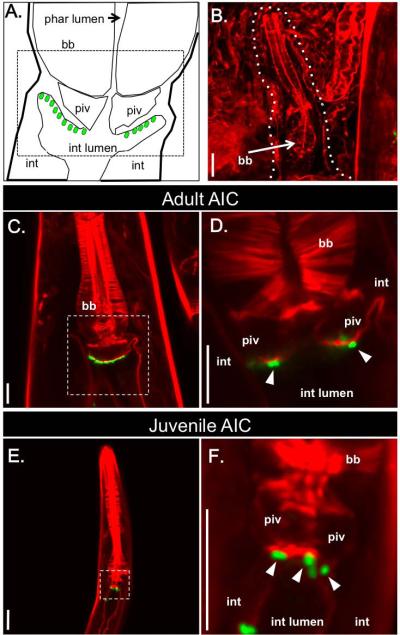
X. nematophila bacteria localize specifically to the anterior intestinal caecum (AIC) in adult and juvenile S. carpocapsae nematodes. S. carpocapsae nematode tissue is stained with rhodamine phalloidin (f-actin; red) and X. nematophila bacteria express green-fluorescent protein (green). A) Schematic diagram representing the pharyngeal region, based on a transmission electron micrograph of a Caenorhabditis elegans hermaphrodite nematode (http://www.wormatlas.org/hermaphrodite/pharynx/Images/phafig11leg.htm). AIC-localized X. nematophila cells are represented by green ovals. B) A recently hatched juvenile nematode lacks AIC-localized X. nematophila. A white dashed line outlines the juvenile, next to an adult nematode (at right). C) An adult S. carpocapsae female with AIC-localized X. nematophila. D) Enlarged image of the boxed region in (C) at a different tissue depth, in which individual bacterial cells expressing GFP are visible (arrowheads); the focal plane of the image in (C) reveals the lumen of the pharyngeal intestinal valve (piv), and the presence of bacteria on the intestinal tissue surface. E) S. carpocapsae juvenile nematode with X. nematophila bacteria localized at the AIC. F) Enlarged image of the nematode shown in E (boxed region) at a different tissue depth with visible GFP-expressing X. nematophila cells (arrowheads) colonizing the AIC. Pharyngeal lumen (phar lumen); basal bulb (bb), pharyngeal-intestinal valve (piv); intestinal cells (int), intestinal lumen (int lumen). Scale bars = 10 μm.
X. nematophila colonize the S. carpocapsae pharyngeal-intestinal valve region of pre-IJs
In the F2 generation, S. carpocapsae juveniles undergo an alternate developmental pathway leading from J2 juveniles to the IJ (Fig. 1). During this generation, prior to IJ development, J2 juveniles (scored based on their size) lacked AIC-localized bacteria and instead had one or a few individual cells associated with the PIV (Fig. 3A,B). At this stage, in nematodes with PIV-localized bacteria, the intestinal space was open (Fig. 3A).
Fig. 3.
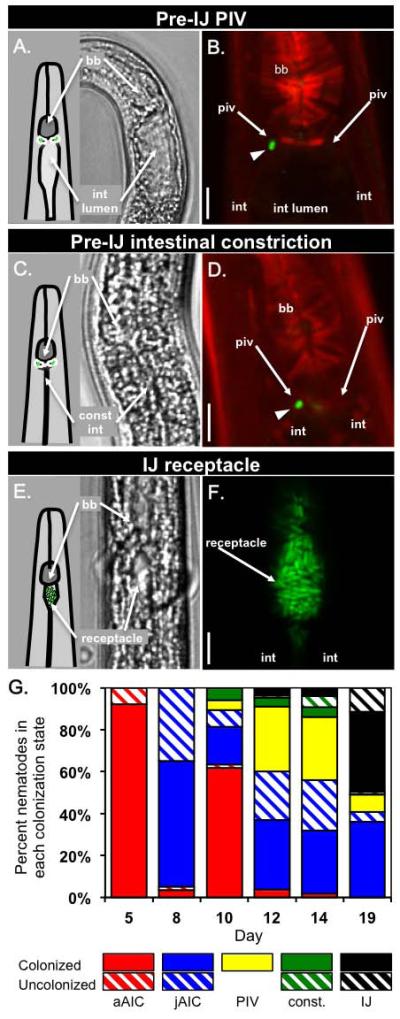
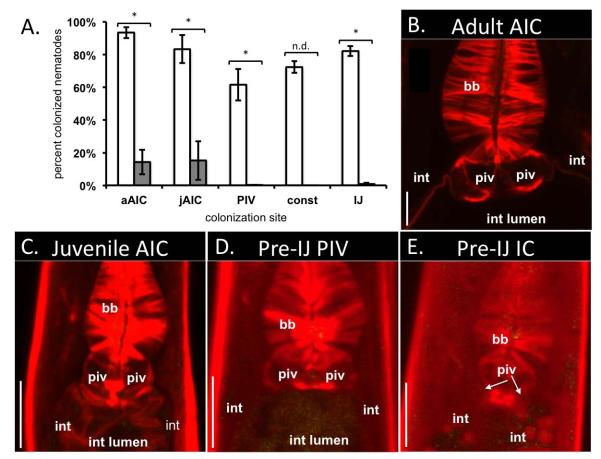
Bacterial colonization and intestinal constriction of nematode pre-IJ stages. DIC images (A, C, E) were taken on a Nikon Eclipse TE300 inverted microscope. Confocal images (B, D, F) of different nematodes at similar stages as in A, C, and E were taken on a Zeiss LSM 510 confocal microscope. A-B) In pre-IJ nematodes with one or a few bacterial cells visible within the PIV region (one bacterial cell is visible in the focal plane in panel B (arrowhead)) the intestinal lumen is open (arrow in A; black space in B). C-D) Pre-IJs in the population exhibit a constricted intestine (const int; compare panels C and A) and bacterial cells are visible in or near the PIV. Note that spheres in panel D are within nematode tissue and not the intestinal lumen. E-F) Subsequently, nematodes display de-constriction of the anterior intestine and X. nematophila bacteria are apparent within the resulting receptacle space. In B and D nematode tissue is stained with rhodamine phalloidin (f-actin; red). In B, D, and F, X. nematophila express green-fluorescent protein (green). G) The average frequency of adults and juveniles with bacteria localized at different tissues over time in a representative experiment (see legend). Abbreviations: basal bulb (bb); pharyngeal-intestinal valve (piv); intestinal lumen (int lumen); intestinal epithelium cells (int); constricted intestine (const int). Legend abbreviations: adult AIC (aAIC), juvenile AIC (jAIC), intestinal constriction (const.). Scale bars = 5 μm.
Using confocal microscopy we sought to determine if PIV-localized bacteria are intracellular within PIV cells or extracellular within a luminal PIV space. A 3-D model of a PIV, constructed from a series of sequential confocal micrographs showed regions devoid of f-actin staining that linked the PIV with the intestinal lumen (Supp. movie 1; the series of images prior to 3-D reconstruction is shown in Supp. movie 2). Bacteria within the PIV had relatively large unstained regions around them, possibly suggesting an extracellular lumenal space or pouch within which the bacteria reside. Further, multiple bacterial cells appeared to be in the process of entering or exiting the PIV without being entirely surrounded by stained actin, suggesting the possibility that the bacteria enter the PIV without being intracellular. Although not conclusive, these data are most consistent with a model that X. nematophila gain entry to the PIV through extracellular channels and colonize an extracellular pouch within the PIV.
Following the appearance of PIV localization, nematodes with constricted intestines were observed (Fig. 3C-D). When nematodes with constricted intestines first appeared, colonizing bacteria remained localized in the PIV: close inspection by rhodamine phalloidin staining of a subset of nematodes with constricted intestines revealed PIV-associated bacteria (Fig. 3D). As nematode development progressed toward the IJ stage the formation of the nematode outer cuticle, which limits penetration by stains and dyes, confounded subsequent observation of stained PIV tissues by confocal microscopy. Therefore, at later time points, we were unable to determine if bacteria within nematodes with constricted intestines occupied the PIV or the constricted intestinal lumen. As the constricted intestine phenotype became less frequent, most nematodes in the population were IJs with colonized receptacles (Fig 3E-F).
The events occurring during IJ development can be categorized into at least three sequential and distinct steps: 1) colonization of the PIV by a few bacterial cells (Fig. 3A-B); 2) constriction of the nematode intestine, with bacteria colonizing the PIV at least through early stages of this process (Fig. 3C-D); 3) expansion of the anterior intestine to form a receptacle with colonizing bacteria (Fig. 3E-F). These events were unique to F2 generation juveniles that would become IJs and were not observed in F1 generation juveniles that moulted from J1 directly through J4 stages into adults. Therefore, they are part of a developmental process specific to IJ development and hereafter we refer to any nematodes that undergo these processes as pre-IJs.
Taken together, these findings demonstrate that X. nematophila bacteria associate with previously unrecognized S. carpocapsae nematodes tissues and life stages. The frequency of each type of colonization changed over time, with AIC localization, PIV localization, PIV colonization during intestinal constriction, and receptacle colonization occurring sequentially (Fig. 3G; a representative experiment). Also, these events occurred in most of the nematodes (Table 1). To assess if these events occur in nematodes that develop inside insects, we co-injected axenic nematodes and GFP-expressing X. nematophila bacteria into Galleria mellonella waxworm larvae and observed the colonization status of nematodes every 2 days from the appearance of F1 nematodes to IJ development. More than 85% of nematodes carried colonizing bacteria at each time point, and each colonization state (adult AIC, juvenile AIC, PIV colonization, intestinal constriction, and IJ receptacle colonization) was observed in at least one time point for each experiment (data not shown). These findings demonstrate that these events occur in nematodes developing within a host and are not an artefact of laboratory culture.
Table 1.
Composite S. carpocapsae colonization dataa
|
X. nematophila
|
X. szentirmaii
|
|||
|---|---|---|---|---|
| SR1+ (wild-type) |
ΔSR1 |
SR1− (wild-type) |
SR1+ |
|
| Adult AIC | 95.4% (350) A | 16.7% (323)B | 14.1% (814) B | 83.4% (825) C |
| Juvenile AIC | 81.4% (1834) A | 10.3% (2374) B | 6.2% (2894) C | 64.8% (2899) D |
| Pre-IJ PIV | 60.4% (546) A | 0.1% (1569) B | 0.1% (2485) B | 15.4% (1069) C |
| Pre-IJ constricted intestine | 66.4% (235) A | 0% (285) B | 0.3% (367) B | 29.1% (492) C |
| IJ receptacle colonization | 79.3% (767) A | 0.5% (765) B | 0.8% (1322) B | 31.7% (1299) C |
Data represent combined total frequency of localization in different nematode stages and tissues from three (X. nematophila) or four (X. szentirmaii) independent experiments. The total number of nematodes counted in each category is indicated in parentheses. Within each nematode life stage, values with the same letter were not significantly different from each other (Fisher’s exact test; P<0.001).
X. nematophila colonization of the AIC and PIV requires SR1 genes
X. nematophila nilA, nilB, and nilC, encoded on SR1, are necessary for X. nematophila and sufficient for other Xenorhabdus species to colonize the IJ receptacle of S. carpocapsae (Heungens et al., 2002; Cowles and Goodrich-Blair, 2008). To assess if SR1 contributes to the colonization events described above, we monitored the localization of a GFP-expressing X. nematophila ΔSR1 deletion mutant in adult, juvenile, and pre-IJ nematodes relative to age-matched nematodes colonized with GFP-expressing X. nematophila. Combined across all experiments and time points, the frequency of S. carpocapsae AIC colonization by the X. nematophila ΔSR1 deletion mutant was significantly lower than the frequency of AIC colonization by wild type X. nematophila (p < 0.05; Fig. 4A-C). When considering individual time points most displayed significant differences between AIC-colonization frequencies of ΔSR1 deletion mutant and wild-type-X. nematophila in age-matched nematodes (p<0.05; data not shown). These data demonstrate that SR1 contributes to colonization of the nematode.
Fig. 4.
X. nematophila requires SR1 for localization to the AIC and PIV. Nematodes were grown on a GFP-expressing X. nematophila ΔSR1 mutant (green; no bacteria visible) or wild type and actin was visualized with rhodamine phalloidin (actin; red). A) Average (+/− S.E.M.) frequency of nematode colonization for adults, juveniles, pre-IJs, and IJs by X. nematophila wild type (Tn7-SR1) (white bars) or a ΔSR1 mutant (gray bars). Significantly fewer nematodes carried colonizing ΔSR1 mutant bacteria than wild type bacteria in each tested colonization state..Statistical significance was determined using a generalized linear mixed effects model (p < 0.05). B, C) Unlike wild type X. nematophila, the ΔSR1 mutant did not localize to the AIC of most adult (B) or juvenile (C) nematodes. D, E) F2 juvenile nematodes that were developmentally similar to pre-IJs did not display PIV colonization (pre-IJ PIV) either prior to (D) or during pre-IJ intestinal constriction (IC) (E). Abbreviations: basal bulb (bb); pharyngeal-intestinal valve (piv); intestinal lumen (int lumen); intestinal epithelium cells (int), adult AIC (aAIC), juvenile AIC (jAIC), intestinal constriction (const), statistics not determined (n.d.). Nematode actin was stained with rhodamine-phalloidin (red) and bacteria expressed green fluorescent protein (green). Size bars: 10 μm.
Juveniles and pre-IJs are not morphologically distinguishable by our methods, so we defined pre-IJs by their occurrence in the F2 generation on wild type X. nematophila lawns concomitant with PIV-colonization and intestinal constriction. We therefore predicted the timing of nematode pre-IJ formation on ΔSR1 mutant lawns based on comparison to wild type controls within each experiment. When pre-IJs were developing on wild-type lawns, nearly all of the juvenile nematodes developing on the ΔSR1 mutant lacked PIV-colonizing bacteria (Fig. 4A,D). This low frequency of PIV colonization demonstrates the ΔSR1 mutant has a severe and significant defect in colonizing the nematode PIV (Fig. 4A). Similarly, no nematodes with visibly constricted intestines had colonizing bacteria (e.g. Fig. 4E; see methods for why statistics were not performed), and only 4 of 765 IJs carried colonizing bacteria. This frequency of IJ receptacle colonization is much higher than those reported for this ΔSR1 mutant within mature IJs (~0.05%) (Bhasin et al., 2012). However, there is precedence for X. nematophila mutants initially colonizing immature IJs then disappearing from the IJ population over time after failing to grow within the receptacle (Martens et al., 2005). At least one of the SR1 genes has been implicated not only in initiation of colonization but also in growth within the receptacle (Cowles and Goodrich-Blair, 2008). Therefore, the relatively high frequency of colonization observed in this study may reflect that the immature IJs we examined have not yet cleared the ΔSR1 mutant from their receptacles. Regardless, the ΔSR1 mutant colonized significantly fewer IJs than wild type X. nematophila (Fig. 4A). These findings indicate that the ΔSR1 mutant is defective in the early stages of nematode colonization during AIC and/or PIV colonization.
We tested if the AIC and PIV colonization defects of the ΔSR1 mutant also occur during nematode development in G. mellonella insect hosts. When we injected G. mellonella with axenic IJs and the GFP-expressing ΔSR1 mutant, we observed similar trends as when nematodes were raised on laboratory media. At the AIC colonization stage, the frequency of nematodes with colonizing ΔSR1 bacteria was higher in insects (up to 50% colonized nematodes) than in vitro conditions, but was still less than the frequency of colonization with wild type (~85%). As pre-IJs and IJs dominated the population in later time points, fewer than 5% of nematodes were colonized (data not shown).
X. nematophila SR1 is sufficient for X. szentirmaii AIC and PIV colonization of S. carpocapsae nematodes
We have shown that SR1 is necessary for normal X. nematophila localization to S. carpocapsae AIC and PIV tissues. Since, other than X. nematophila, SR1 is absent from all Xenorhabdus species tested to date (Heungens et al., 2002; Cowles and Goodrich-Blair, 2008), we reasoned that non-native Xenorhabdus species should not colonize S. carpocapsae AIC and PIV tissues. To test this we cultivated S. carpocapsae on GFP-expressing X. szentirmaii (Fig. 5A). S. carpocapsae developed on X. szentirmaii lawns, in contrast to previous unsuccessful attempts (Cowles and Goodrich-Blair, 2008), possibly due to some unknown aspect of technical variability (e.g. agar thickness, absolute temperature). Over 4 experiments, most adult and juvenile nematodes lacked AIC-localized X. szentirmaii bacteria (Fig. 5A-C), and nearly all late-stage nematodes lacked tissue localized bacteria. Out of 3356 juveniles and pre-IJs observed, 3 nematodes carried PIV-localized bacteria and 1 nematode carried bacteria during intestinal constriction. Of the 1322 progeny IJs observed, 10 carried receptacle-colonizing bacteria (Table 1).
Fig. 5.
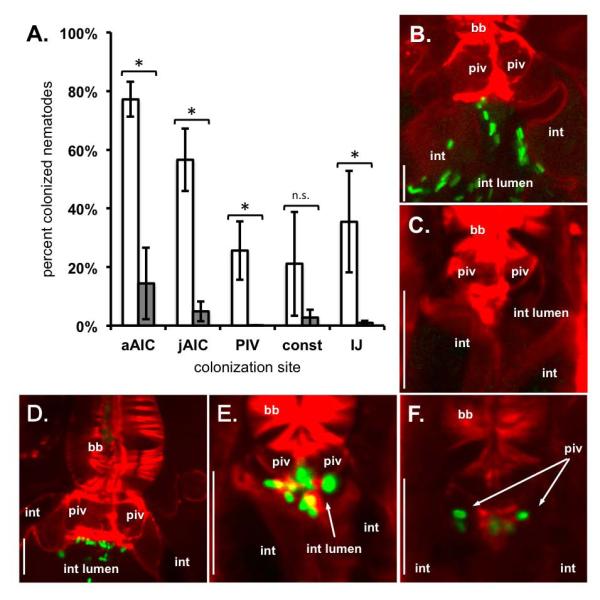
X. nematophila SR1 confers upon X. szentirmaii the ability to colonize the AIC and PIV of S. carpocapsae nematodes. S. carpocapsae nematodes were grown on GFP-expressing X. szentirmaii strains carrying either Tn7/SR1 or empty Tn7. Developing nematodes were visualized by confocal microscopy. S. carpocapsae nematode actin was stained with rhodamine-phalloidin (red) and X. nematophila bacteria expressed green-fluorescent protein (green). A) Average (+/− S.E.M.) frequency of nematode colonization for adults, juveniles, pre-IJs, and IJs by X. szentirmaii Tn7/SR1 (white bars) or X. szentirmaii empty Tn7 (wild type) (grey bars). When X. szentirmaii did not contain SR1, significantly fewer nematodes carried colonizing bacteria in each colonization state except pre-IJs during intestinal constriction (p = 0.14). Statistical significance was determined using a generalized linear mixed effects model (p < 0.05). B, C) X. szentirmaii eTn7 (lacking SR1) does not localize to S. carpocapsae adult (B) or juvenile (C) AIC. D, E) X. szentirmaii Tn7/SR1 (carrying SR1) localizes to the AIC of adult (D) and juvenile (E) nematodes. F) X. nematophila bacteria localize to the pre-IJ PIV. Abbreviations: basal bulb (bb); pharyngeal-intestinal valve (piv); intestinal lumen (int lumen); intestinal epithelium cells (int); adult AIC (aAIC); juvenile AIC (jAIC); intestinal constriction (const); no statistical difference (n.s.). Size bars: 10 μm.
In contrast, when we provided SR1 to GFP-expressing X. szentirmaii, the bacteria localized to specific nematode tissues at significantly higher frequencies during all stages (p < 0.05, Fig. 5A) except intestinal constriction (p = 0.14). Across 4 experiments, 77% of adult nematodes and 57% of juveniles had AIC-colonizing X. szentirmaii carrying SR1. S. carpocapsae AIC colonization by X. szentirmaii-SR1 was not visibly distinguishable from AIC-colonization by X. nematophila bacteria (Fig. 5D-E). Further, as pre-IJs developed, bacteria were localized within the PIV of 26% of juvenile nematodes (Fig. 5F), and colonized 21% of nematodes with constricted intestines and 35% of IJ receptacles. Variation across experiments likely contributed to the fact that differences due to SR1 in X. szentirmaii colonization of nematodes with constricted intestines were not significant. Regardless, our data show that X. szentirmaii carrying SR1 are able to colonize the AIC, PIV, and receptacle of S. carpocapsae nematodes (Table 1).
AIC and PIV colonization are not unique to the S. carpocapsae - X. nematophila mutualism
Since SR1 is necessary for AIC and PIV colonization, is sufficient to confer at least the AIC phenotype, and is specific to X. nematophila, it is possible that the events we have described here are unique to the S. carpocapsae – X. nematophila mutualism. To address this possibility, we examined early colonization events in another Steinernema species, S. feltiae, by its Xenorhabdus symbiont, X. bovienii. X. bovienii strains do not appear to encode SR1 (unpubl. data), its hosts (including S. feltiae) are in different phylogenetic sub-clades than S. carpocapsae (Lee and Stock, 2010), and the receptacle structures of its hosts are distinct from those of S. carpocapsae: X. bovienii host receptacles have a non-cellular envelope (vesicle) that encloses X. bovienii symbionts (Kim et al., 2012). These differences lend strength to the hypothesis that if X. bovienii engages in AIC and PIV colonization during intestinal constriction with its animal host, these events may be conserved among Xenorhabdus/Steinernema associations. Using liver-kidney agar cultivation (a medium that supports higher colonization frequency for S. feltiae nematodes than does lipid agar medium), GFP-expressing X. bovienii bacteria were observed localized at the AIC of both adult and juvenile S. feltiae nematodes (Fig. 6B-C). Further, in the F2 generation when pre-IJs are expected to form, we observed individual PIV-localized X. bovienii (Fig. 6D). PIV colonization was also observed in S. feltiae nematodes with constricted intestines, and in those in which the cuticle and sealed mouth deterred staining (Fig. 6E). In subsequent time points, S. feltiae nematodes were present in which X. bovienii had fully colonized the vesicle within the receptacle (Fig. 6F). Most nematodes carried bacteria localized to these specific tissues (Fig. 6A, Table 2). Overall, our data indicate that different Steinernema nematodes undergo similar events during colonization by their respective Xenorhabdus bacterial symbionts. We note that receptacle-localized X. bovienii were long rods, similar to a previous report of another X. bovienii strain (SS-2004) colonizing Steinernema jolietti nematodes (Sugar et al., 2012), but X. bovienii cells were short rods during AIC and early PIV colonization, suggesting physiological differences of the cells at these stages relative to those in the receptacle.
Fig. 6.
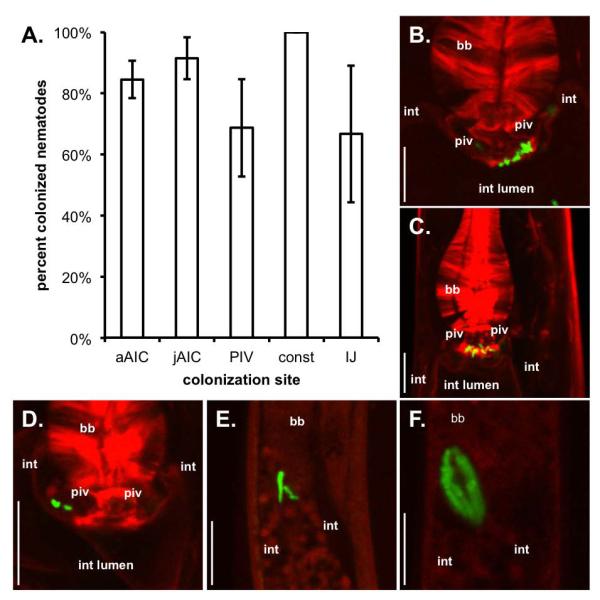
Events in the X. nematophila / S. carpocapsae association are observed in the association between X. bovienii and S. feltiae. Nematodes were grown on a GFP-expressing X. bovienii and bacterial localization to nematode specific tissues was observed over an 11-day period until infective juveniles made up > 50% of the nematode population. A) Average adult, juvenile, pre-IJ, and IJ colonization (+/− S.E.M.). B) Localization of X. bovienii bacteria to the anterior intestinal caecum (AIC) of adult S. feltiae nematodes. C) AIC localization of X. bovienii bacteria in juvenile S. feltiae nematodes. D) Pharyngeal intestinal valve (PIV) colonization by X. bovienii bacteria in pre-infective juvenile (pre-IJ) nematodes. E) X. bovienii localization at the anterior of the constricted pre-IJ intestine (IJ). F) Colonization of the S. feltiae receptacle by X. bovienii bacteria. Abbreviations: basal bulb (bb); pharyngeal-intestinal valve (piv); intestinal lumen (int lumen); intestinal epithelium cells (int). Legend abbreviations: adult AIC (aAIC); juvenile AIC (jAIC); intestinal constriction (const). Nematode actin was stained with rhodamine-phalloidin (red) and bacteria expressed green fluorescent protein (green). Size bars: 10 μm.
Table 2.
Composite frequency of X. bovienii tissue localization in S. feltiaea
| Stage and Tissue |
Frequency of localization |
|---|---|
| Adult AIC | 100.0% (250) |
| Juvenile AIC | 95.7% (3095) |
| Pre-IJ PIV | 71.5% (467) |
| Pre-IJ constricted intestine | 100.0% (222) |
| IJ receptacle colonization | 73.9% (1481) |
Data represent combined totals from two independent experiments, each with three replicates. The total number of counted nematodes is indicated in parentheses.
Discussion
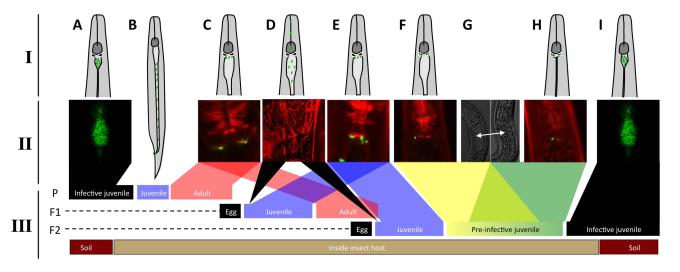
In this study we report heretofore-unrecognized stages of intimate association between Xenorhabdus bacteria and Steinernema nematodes. It has long been held that the Steinernema IJ is the only life stage of the nematode that is specifically colonized by bacteria, and that interactions between other S. carpocapsae life stages are transient and non-specific. Reasons for this supposition include that the IJ carries the bacteria in an easily observed discrete location (the receptacle) while other stages lack obvious specialized structures for harboring bacteria (Bird and Akhurst, 1983; Endo and Nickle, 1995). Also, intimate bacterial colonization of the IJ, in contrast to other life stages, serves a clear and essential function in the symbiosis: transmission of the symbiont between insect hosts. Finally, a practical reason is that IJs, relative to the other stages, can be easily isolated and assessed for internal bacterial content (Martens et al., 2003). In this work we were able to move beyond this technical hurdle by using GFP-expressing bacteria and fluorescence microscopy to monitor bacterial colonization (Murfin et al., 2012; Sugar et al., 2012) throughout the life cycles of two Steinernema species from different phylogenetic sub-clades (Lee and Stock, 2010). Our findings challenge previously held notions by revealing that throughout the reproductive life cycle of both Steinernema species bacteria colonize nematode tissues in the region around the pharyngeal-intestinal junction. Bacterial cells colonized the anterior, but not other areas, of the intestines of adults and juveniles from F1 and F2 generations (Figs. 2 and 6). Then, as IJs formed, three novel events were observed: 1) individual bacterial cell colonization of the PIV of pre-IJs; 2) constriction of the nematode intestine; and 3) de-constriction of only the anterior-most regions of the intestine to form the receptacle, which was colonized by bacteria concomitant with formation of the IJ cuticle.
Taken together with previous findings, our work suggests a temporal model of colonization events occurring during the Steinernema life cycle, summarized in Fig. 7. First, through post-hatching feeding by the nematode (e.g. Fig. 7D), the bacteria must gain access to and survive within the nematode intestine. Next, the bacteria localize to the AIC (Fig. 7C,E). In the F2 generation of juveniles, comprised of pre-IJs that will become IJs, AIC colonization decreases in frequency, while PIV colonization frequency increases, followed by constriction of almost all of the nematode intestine (Fig. 7G, H). At some stage following PIV colonization and intestinal constriction, the anterior of the intestine de-constricts and bacteria are observed attached to the IVS in the receptacle (e.g. (Martens and Goodrich-Blair, 2005)). We were unable to document the transition from PIV to receptacle colonization because of poor penetration of rhodamine phalloidin through the IJ cuticle. However, it is likely that receptacle bacteria are derived from PIV colonizers, since at this stage of the process the nematode is non-feeding and does not have access to external populations of Xenorhabdus bacteria (Bird and Akhurst, 1983; Endo and Nickle, 1995). The transition from PIV to receptacle colonization may occur by bacterial migration, or by a developmental process in which the colonized region of the PIV becomes the nematode receptacle. Following association with the IVS, the bacteria divide and fill the receptacle (Fig. 7I; (Martens et al., 2003)).
Fig. 7.
Spatial and temporal bacterial colonization events during Steinernema development using schematics (I), representative micrographs (II), and a timeline (III) A) A colonized infective juvenile (IJ) (B) releases its bacteria by defecation after insect penetration (see (Snyder et al., 2007)) and moults into adult stages. C) Adults that develop from IJs carry bacteria localized at the anterior intestinal caecum. D) Newly hatched juvenile nematodes lack bacteria that specifically localize to the anterior intestinal caecum. E) After hatching, juvenile (and later, adult) nematodes have bacteria localized to the anterior intestinal caecum. F2 juveniles carry bacteria at the anterior intestinal caecum, and this generation develops into infective juveniles through a pre-IJ stage. The beginning of pre-IJ development is distinguished by F) bacterial colonization of the PIV, and subsequently, intestinal constriction (G and H). I) After relaxation of the anterior intestinal constriction, bacteria are observed localized to the receptacle and over time bacteria grow to completely fill the receptacle (Martens et al., 2003). Through these events IJs acquire a complement of symbionts that they carry as they leave the nutrient depleted insect cadaver in search of a new insect host. Abbreviations: parental generation (P), first generation offspring (F1), second-generation offspring (F2)
The same general events shown in Fig. 7 occur in both S. carpocapsae and S. feltiae, nematodes that are in distinct phylogenetic sub-clades, indicating that the overall processes are likely conserved among Steinernema-Xenorhabdus symbioses. However, since S. carpocapsae and S. feltiae IJ receptacles are colonized preferentially by their native symbionts (Cowles and Goodrich-Blair, 2008; Sicard et al., 2004; Akhurst, 1983; Chapuis et al., 2009) some aspect(s) of the interactions between the nematode and bacteria, likely at the molecular level, must allow for recognition and selection of the native symbiont and exclusion of non-native Xenorhabdus. In S. carpocapsae, specificity occurs during AIC-localization and involves SR1: an X. nematophila ΔSR1 mutant localizes to the AIC in fewer S. carpocapsae nematodes than does wild type (Fig. 4A) and X. szentirmaii colonization of these tissues is allowed by the presence of X. nematophila SR1 (Fig. 5). The ability of the X. nematophilaΔSR1 mutant to initially localize to the AIC of a small number of adult and juvenile nematodes may indicate that SR1-encoded proteins increase the efficiency of binding to these tissues, or that they are necessary for persistence at this location. Characterizing the roles of other bacterial gene products in each of the stages shown in Fig. 7 will help demonstrate what other molecular dialogues are at work as Xenorhabdus symbionts colonize their nematode hosts.
X. nematophila colonization of the PIV is similar to a process that occurs during Photorhabdus luminescens bacterial colonization of the IJ stage of its specific nematode host, Heterorhabditis bacteriophora (Ciche et al., 2008). In both systems, bacteria localize to the PIV, indicating this may be a conserved process in Heterorhabditid and Steinernematid nematodes, despite their relatively distant phylogenetic relationship (Blaxter et al., 1998). However, subsequent events are likely distinct between genera, since the IJ stage of H. bacteriophora lacks a receptacle (instead, bacteria are more widely distributed within a non-constricted intestinal lumen) (Goodrich-Blair and Clarke, 2007). Also, events preceding PIV colonization differ between these two symbioses. P. luminescens bacteria adhere to posterior intestinal cells of adult females prior to invading and replicating within intracellular vacuoles which lyse to inoculate juvenile nematodes inside the maternal body cavity with P. luminescens (Ciche et al., 2008; Stock et al., 2012). In neither S. carpocapsae nor S. feltiae did we observe bacterial binding to or invasion of posterior adult intestinal cells (data not shown).
It was previously demonstrated that Xenorhabdus populations face a bottleneck that leads to the clonality of bacteria within the IJ receptacle (Martens et al., 2003). Clonal symbiont populations have been experimentally demonstrated in multiple mono-specific host-microbe symbioses (Gage, 2002; Martens et al., 2003; Sugar et al., 2012; Wollenberg and Ruby, 2009) and suggested qualitatively in others (Ciche et al., 2008; Kubota et al., 2007; Nussbaumer et al., 2006). This strain bottlenecking may reduce the prevalence of symbionts that reap host rewards without paying the “goods and services” provided to the host (cheaters; (Frank, 1996)). Bottlenecking may result from non-specific (e.g. spatial restriction) and specific (e.g. molecular recognition) processes that restrict symbiont number and type during transmission. Since only few individual cells of X. nematophila and X. bovienii localize to the pre-IJ PIV of their respective nematode hosts, we suggest that PIV colonization is the selective event in nematodes that results in clonal bottlenecking of the symbiont. Based on 3-dimensional reconstructed images of S. carpocapsae colonized PIV we propose a model that bacteria occupy an extracellular pouch within the PIV that is connected to the intestinal lumen by channels (supplemental videos S1 and S2). The channels may restrict entry into the PIV to only one individual bacterial cell on either side (e.g. Fig. 3B) that each then continue dividing within the PIV pouch (e.g. Fig. 6D).
Our findings reveal new insights into the Xenorhabdus-Steinernema association and challenge the long-held assumption that only the IJ stage of the nematode is intimately associated with colonizing bacteria. Previous models also presumed that the key events of colonization initiation occurred within the receptacle. We have shown that X. nematophila occupies at least four distinct S. carpocapsae nematode host niches (intestine, AIC, PIV, receptacle) during inter-generational transmission, each of which may play a role in symbiotic partner selection. Now that these colonization states have been described, further investigation will be necessary to elucidate the temporal and molecular processes and functions of each. For example, is AIC colonization initiated once in the juvenile stage or is it a continuous and dynamic process throughout nematode development? Are the bacteria that colonize the PIV derived from the AIC-localized bacterial population or do they result from a distinct initiation event? Finally, the finding that Xenorhabdus bacteria colonize non-transmission stage juvenile and reproductive adult Steinernema nematodes raises the intriguing possibility that these interactions facilitate bacterial contributions to host nutrition and reproduction. For example, the intimate association between the AIC intestinal epithelia and bacteria may allow nutrient exchange between these cells. The adult and juvenile colonization stages described here also may be essential for reliable transmission of only the proper, cognate symbiont in the nematode IJ. In support of this idea, localization to the AIC and within the PIV of S. carpocapsae nematodes is specific to X. nematophila, suggesting that these are symbiont selective stages at which S. carpocapsae engages in partner choice. Further, since only a few bacterial cells were observed within the PIV, symbiont bottlenecking may occur at this stage. Future studies in this and other model animal-microbe mutualisms will reveal the nature and frequency of selective events encountered by symbionts during transmission.
Experimental Procedures
Strains, media, and growth conditions
Xenorhabdus bacteria used in this study were grown on a roller at 30°C in lysogeny broth (LB) stored in the dark (Xu and Hurlbert, 1990) and supplemented with ampicillin (150 μg/ml) (Table 3). Escherichia coli were grown in LB on a roller at 30°C or 37°C. A GFP-expressing X. nematophila ΔSR1 mutant, HGB1430, was created previously (Bhasin et al., 2012). To create GFP-expressing ΔSR1 Tn7-SR1, a previously created Tn7-SR1 construct (Cowles and Goodrich-Blair, 2008), which inserts into a region (attTn7) that does not impair nematode colonization in X. nematophila, was transferred to HGB1430 by triparental conjugation (Cowles and Goodrich-Blair, 2004). We also transferred the empty Tn7 plasmid (pEVS107) to HGB1430 to create ΔSR1 empty Tn7 (eTn7). To express SR1 in X. szentirmaii, Tn7-SR1 was transferred to X. szentirmaii (HGB836, a gift from A. Fodor) by triparental conjugation to create HGB1323: X. szentirmaii Tn7-SR1. As a negative control, the empty vector was transferred into X. szentirmaii (HGB836) to create HGB1322: X. szentirmaii eTn7. To create GFP-expressing X. szentirmaii strains, pJMC001, which integrates into the X. nematophila chromosome at a site that does not impair nematode colonization (Bhasin et al., 2012; Martens et al., 2003), was conjugated into each of the X. szentirmaii Tn7 strains, and for each a visually green colony was stocked as a GFP-expressing strain. X. bovienii-GFP was created by introduction of GFP in the attTn7 site using mini-Tn7-KSGFP (a gift from T. Ciche; (Teal et al., 2006)). Strains were verified for GFP expression and X. bovienii characteristics (i.e. ampicillin resistance, pigmentation, negative catalase).
Table 3.
Bacterial strains and plasmids used in this study.
| Strain | Plasmid | Species | Comments | Reference/Source |
|---|---|---|---|---|
| HGB283 | pUX-BF13 | E. coli | Triparental mating helper | (Bao et al., 1991) |
| HGB1783 | pJMC001 | E. coli | GFP donor plasmid | (Bhasin et al., 2012) |
| HGB783 | pEVS107/SR1 | E. coli | Tn7/SR1 donor plasmid | (Cowles and Goodrich-Blair, 2004) |
| HGB281 | pEVS107 | E. coli | eTn7 donor plasmid | (Stabb and Ruby, 2002) |
| HGB1262 | pURR25 mini Tn7KS-GFP |
E. coli | Tn7/GFP donor plasmid | D. Lies and D. Newman (Teal et al., 2006) |
| HGB1430 | X. nematophila | GFP expressing, ΔSR1 mutant | (Bhasin et al., 2012) | |
| HGB1508 | X. nematophila | HGB1430 eTn7 | This study | |
| HGB1509 | X. nematophila | HGB1430 Tn7/SR1 | This study | |
| HGB836 | X. szentirmaii | Wild type | A. Fodor | |
| HGB1322 | X. szentirmaii | HGB836 eTn7 | This study | |
| HGB1323 | X. szentirmaii | HGB836 Tn7/SR1 | This study | |
| HGB1786 | X. szentirmaii | HGB1322 GFP expressing, eTn7 |
This study | |
| HGB1787 | X. szentirmaii | HGB1323 GFP expressing, Tn7/SR1 |
This study | |
| HGB1699 | X. bovienii | wild type; symbiont of S. feltiae | This study | |
| HGB1865 | X. bovienii | HGB1699 attTn7::Tn7/GFP | This study |
Nematode cultivations
Nematode cultivations were performed by inoculating surface-sterilized axenic infective juveniles prepared as described previously (Bhasin et al., 2012) to overnight lawns of Xenorhabdus bacteria on lipid agar plates (Vivas and Goodrich-Blair, 2001). For Steinernema feltiae, nematodes were grown on liver-kidney agar (Sicard et al., 2003) instead of lipid agar because nematode colonization frequency is higher on the former. For each bacterial phenotype measured (adult and juvenile AIC-localization, PIV-localization, intestinal localization during intestinal constriction, and receptacle localization), we performed at least two experiments, each with three biological replicates per experiment. We counted a biological replicate as a different lipid agar plate preparation of bacteria and nematodes, and we used the same plate for counts on different days throughout the experiment. We counted approximately 100 nematodes per replicate, and assessed multiple time points per experiment. We display a representative temporal progression for nematodes colonized with each X. nematophila and X. szentirmaii strain in Figure S1. For X. nematophila WT and ΔSR1, we performed three experiments each with three biological replicates per time point except where noted in brackets: Expt. 1 (d 4 [2 replicates for WT] and 7 [2 replicates for WT]), Expt. 2 (d 5, 8, 10, 12 [2 replicates for WT], 14 [2 replicates for ΔSR1], and 19), and Expt. 3 (d 7-12). For X. szentirmaii WT and Tn7-SR1 we performed four experiments, each with three biological replicates per time point except where noted in brackets: Expt. 1 (d 4 [2 replicates for Tn7; 1 replicate for Tn7-SR1] and 7 [2 replicates for Tn7; 1 replicate for Tn7-SR1]), Expt. 2 (d 5, 8, 10, 12, 14, 17, 19, 21, and 24), Expt. 3 (d 10, 12, 14, 15, and 16 [2 replicates for WT]), and Expt. 4 (d 9-15). S. feltiae nematodes grown on X. bovienii were examined in two experiments with three biological replicates per experiment: Expt. 1 (d 8, 10-18) and Expt. 2 (d 8-12). For in vivo co-cultivations, approximately 50 surface-sterilized axenic IJs were co-injected with 200 CFU of log-phase cultures of bacteria (either X. nematophila WT or the ΔSR1 mutant) into Galleria mellonella insects. Two insect cadavers were dissected as biological replicates every other day from 5 to 11 days post injection to collect nematodes for microscopy as described above. Insects were individually dissected in 2mL of PBS, and emergent nematodes were rinsed three times in fresh PBS to remove background bacteria and insect tissues. The entire experiment was performed twice.
Specimen preparation, microscopy, and statistical analysis
To visualize bacterial localization within a nematode host, nematodes were removed from the agar surface, suspended in phosphate buffered saline supplemented with 2 nM levamisole (a paralyzing agent), transferred to a glass slide and viewed by microscopy. Nematodes were viewed on a Nikon Eclipse TE300 inverted microscope (Nikon, Melville, NY, USA) as described previously (Martens et al., 2003) or a Zeiss LSM 510 confocal microscope (Zeiss, Thornwood, NY, USA) as described previously (Sugar et al., 2012). DIC images were collected using MetaMorph software and lightened in Powerpoint 2011 (Microsoft). Confocal images and videos were analyzed using the LSM image browser (Zeiss). Nematodes were individually examined and assessed for relevant phenotypes (e.g. AIC- colonization). PIV-localization was scored positive if a J2 juvenile had one or a few colonizing bacteria bilaterally distributed at the anterior nematode intestine. Bacterial intestinal localization during intestinal constriction was scored as positive if any colonizing bacteria were observed at the anterior nematode intestine when the intestine was tightly constricted during pre-IJ development. While the nematode population was relatively synchronized, our counts represent the percentage of juveniles visible and do not strictly correlate with a single generation (e.g. early F2 juveniles may have been counted along with late F1 juveniles on day 8 or 10).
To observe bacterial colonization with stained nematode tissue, specimens were stained with 6.6 μM rhodamine phalloidin (Sigma) or 0.125 mg/ml Alexa Fluor 633 concanavalin A (Invitrogen; dissolved in 0.1M sodium bicarbonate) (gifts from M. McFall-Ngai). To prevent photobleaching of fluorophores, samples were prepared in the dark. Nematode cultures were resuspended in PBS + 4% final concentration paraformaldehyde and fixed for at least 18 hours. Samples were washed at least three times in PBS, permeabilized in PBS-T (1% final volume Triton X-100) for at least 18 hours, and infiltrated with stains + 1% Triton X-100 for at least 18 hours prior to visualization.
Two different statistical tests were applied to the data. In each case we tested adult AIC, juvenile AIC, pre-IJ PIV, pre-IJ intestinal constriction, and IJ receptacle colonization separately. First, we applied a 2-tailed Fisher’s exact test (http://www.langsrud.com/fisher.htm) to the difference in colonization frequency of wild type- and mutant-colonized nematodes each day that colonization frequencies were assessed (data not shown). We also applied this test to the sum of the frequencies of colonization across all experiments (presented in Table 1). To assess the reproducibility of the statistical differences across experiments, we used a generalized linear mixed effects model (Bates et al., 2012) in R (R, 2012) with a binomial family, with the mutant as a fixed effect, and experiment, day, and replicate as random effects, and day and replicate nested within experiment (presented in Figs. 4A and 5A). We did not perform tests for intestinal constriction in X. nematophila because the ΔSR1 mutant did not colonize any nematodes, and the package does not accurately reflect differences when all of one sample is 0. To calculate the values presented in Figs. 4A, 5A, and 6A (there was no statistical comparison for 6A), the average colonization frequency across all time points was first calculated for each replicate, then replicates were averaged to yield the average colonization frequency for each experiment. Finally, the average colonization frequencies of all experiments were averaged (and the S.E.M. calculated) and presented as percent colonized nematodes in Figs. 4A, 5A, and 6A. Statistical cutoffs were performed at p < 0.05 for all tests.
Because we were unable to distinguish uncolonized (AIC) juvenile nematodes from uncolonized (PIV) pre-IJ nematodes, we assessed the frequency of nematode colonization for each colonization phenotype relative to the same pool of uncolonized juvenile nematodes. For example, at day 10 we observed wild type X. nematophila AIC localization in 23, 7, and 18 S. carpocapsae nematodes from 3 different biological replicates, respectively; we also recorded 6, 5, and 10 nematodes that had no colonizing bacteria, and 7, 0, and 6 nematodes with PIV-colonizing bacteria, respectively. We assessed AIC-localization at 23/29, 7/12, and 18/28 and PIV-localization at 7/13, 0/5, and 6/16. This approach underestimates the frequency of nematode colonization, but was necessary because we were unable to distinguish uncolonized “AIC”-nematodes from uncolonized “PIV”-nematodes. Importantly, despite the underestimation of colonization, we still observed sufficient nematode colonization in each of our tests to identify a significant colonization defect of bacteria that did not carry the SR1 genes (either mutant X. nematophila or wild type X. szentirmaii).
Supplementary Material
Acknowledgements
This work was supported by grants awarded to H.G-B. from the National Science Foundation (IOS-0950873 and IOS-0920631). K.E.M. and J.M.C. were supported by a National Institutes of Health (NIH) National Research Service Award T32 (AI55397 “Microbes in Health and Disease”). J.M.C. was also supported by a National Science Foundation (NSF) Graduate Research Fellowship. E. A. H-H. was supported by grants awarded to Margaret McFall-Ngai (NIH AI50661) and Edward G. Ruby and M. M-N. (NIH RR12294). We are indebted to M. Altura, J. Troll, and M. McFall-Ngai for assistance with confocal microscopy and nematode tissue staining.
Footnotes
The authors have no conflicts of interest to declare.
References
- Akhurst RJ. Neoaplectana-species - specificity of association with bacteria of the genus Xenorhabdus. Exp Parasitol. 1983;55:258–263. doi: 10.1016/0014-4894(83)90020-6. [DOI] [PubMed] [Google Scholar]
- Altun ZF, Hall DH. Handbook of C. elegans anatomy. 2005 [WWW document]. URL http://www.wormatlas.org/ver1/handbook/alimentary/alimentary1.htm.
- Altun ZF, Hall DH. Alimentary system, pharynx. 2009. [WWW document]. URL doi:10.3908/wormatlas.1.3.
- Baldwin JG, Perry RN. Nematode morphology, sensory structure and function. In: Chen ZX, Chen SY, Dickson DW, editors. Nematol Adv Persp. CABI; Cambridge, MA: 2004. pp. 175–257. [Google Scholar]
- Bao Y, Lies DP, Fu H, Roberts GP. An improved Tn7-based system for the single-copy insertion of cloned genes into chromosomes of Gram-negative bacteria. Gene. 1991;109:167–168. doi: 10.1016/0378-1119(91)90604-a. [DOI] [PubMed] [Google Scholar]
- Bhasin A, Chaston JM, Goodrich-Blair H. Mutational analyses reveal overall topology and functional regions of NilB, a bacterial outer membrane protein required for host association in a model of animal-microbe mutualism. J Bacteriol. 2012;194:1763–1776. doi: 10.1128/JB.06711-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird AF, Akhurst RJ. The nature of the intestinal vesicle in nematodes of the family Steinernematidae. Int J Parasitol. 1983;13:599–606. [Google Scholar]
- Bates D, Maechler M, Bolker B. lme4: Linear mixed-effects models using S4 classes. 2012 R packager version 0.999999-0. http://CRAN.R-project.org/package=lme4.
- Blaxter ML, De Ley P, Garey JR, Liu LX, Scheldeman P, Vierstraete A, et al. A molecular evolutionary framework for the phylum Nematoda. Nature. 1998;392:71–75. doi: 10.1038/32160. [DOI] [PubMed] [Google Scholar]
- Bright M, Bulgheresi S. A complex journey: transmission of microbial symbionts. Nat Rev Microbiol. 2010;8:218–230. doi: 10.1038/nrmicro2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapuis E, Emelianoff V, Paulmier V, Le Brun N, Pages S, Sicard M, Ferdy JB. Manifold aspects of specificity in a nematode-bacterium mutualism. J Evol Biol. 2009;22:2104–2117. doi: 10.1111/j.1420-9101.2009.01829.x. [DOI] [PubMed] [Google Scholar]
- Ciche TA, Kim KS, Kaufmann-Daszczuk B, Nguyen KC, Hall DH. Cell invasion and matricide during Photorhabdus luminescens transmission by Heterorhabditis bacteriophora nematodes. Appl Environ Microbiol. 2008;74:2275–2287. doi: 10.1128/AEM.02646-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2012. ISBN 3-900051-07-0, URL http://www.R-project.org/ [Google Scholar]
- Cowles CE, Goodrich-Blair H. Characterization of a lipoprotein, NilC, required by Xenorhabdus nematophila for mutualism with its nematode host. Mol Microbiol. 2004;54:464–477. doi: 10.1111/j.1365-2958.2004.04271.x. [DOI] [PubMed] [Google Scholar]
- Cowles CE, Goodrich-Blair H. The Xenorhabdus nematophila nilABC genes confer the ability of Xenorhabdus spp. to colonize Steinernema carpocapsae nematodes. J Bacteriol. 2008;190:4121–4128. doi: 10.1128/JB.00123-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo BY, Nickle WR. Ultrastructure of anterior and mid-regions of infective juveniles of Steinernema feltiae. Fundam Appl Nematol. 1995;18:271–294. [Google Scholar]
- Frank SA. Host-symbiont conflict over the mixing of symbiotic lineages. Proc R Soc Lond B Biol Sci. 1996;263:339–344. doi: 10.1098/rspb.1996.0052. [DOI] [PubMed] [Google Scholar]
- Gage DJ. Analysis of infection thread development using Gfp- and DsRed-expressing Sinorhizobium meliloti. J Bacteriol. 2002;184:7042–7046. doi: 10.1128/JB.184.24.7042-7046.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich-Blair H, Clarke DJ. Mutualism and pathogenesis in Xenorhabdus and Photorhabdus: two roads to the same destination. Mol Microbiol. 2007;64:260–268. doi: 10.1111/j.1365-2958.2007.05671.x. [DOI] [PubMed] [Google Scholar]
- Heungens K, Cowles CE, Goodrich-Blair H. Identification of Xenorhabdus nematophila genes required for mutualistic colonization of Steinernema carpocapsae nematodes. Mol Microbiol. 2002;45:1337–1353. doi: 10.1046/j.1365-2958.2002.03100.x. [DOI] [PubMed] [Google Scholar]
- Hirao A, Ehlers R-U, Strauch O. Life cycle and population development of the entomopathogenic nematodes Steinernema carpocapsae and S. feltiae (Nematoda, Rhabditida) in monoxenic liquid culture. Nematol. 2010;12:201–210. [Google Scholar]
- Hirao A, Ehlers RU. Influence of inoculum density on population dynamics and dauer juvenile yields in liquid culture of biocontrol nematodes Steinernema carpocapsae and S. feltiae (Nematoda: Rhabditida) Appl Microbiol Biotechnol. 2010;85:507–515. doi: 10.1007/s00253-009-2095-4. [DOI] [PubMed] [Google Scholar]
- Kaya HK, Gaugler R. Entomopathogenic nematodes. Annu Rev Entomol. 1993;38:181–206. [Google Scholar]
- Kim SK, Flores-Lara Y, Patricia Stock S. Morphology and ultrastructure of the bacterial receptacle in Steinernema nematodes (Nematoda: Steinernematidae) J Invertebr Pathol. 2012;110:366–374. doi: 10.1016/j.jip.2012.04.011. [DOI] [PubMed] [Google Scholar]
- Kubota N, Kanemori M, Sasayama Y, Aida M, Fukumori Y. Identification of endosymbionts in Oligobrachia mashikoi (Siboglinidae, Annelida) Microbes Environ. 2007;22:136–144. [Google Scholar]
- Lee MM, Stock SP. A multilocus approach to assessing co-evolutionary relationships between Steinernema spp. (Nematoda: Steinernematidae) and their bacterial symbionts Xenorhabdus spp. (gamma-Proteobacteria: Enterobacteriaceae) Syst Parasitol. 2010;77:1–12. doi: 10.1007/s11230-010-9256-9. [DOI] [PubMed] [Google Scholar]
- Martens EC, Goodrich-Blair H. The Steinernema carpocapsae intestinal vesicle contains a subcellular structure with which Xenorhabdus nematophila associates during colonization initiation. Cell Microbiol. 2005;7:1723–1735. doi: 10.1111/j.1462-5822.2005.00585.x. [DOI] [PubMed] [Google Scholar]
- Martens EC, Heungens K, Goodrich-Blair H. Early colonization events in the mutualistic association between Steinernema carpocapsae nematodes and Xenorhabdus nematophila bacteria. J Bacteriol. 2003;185:3147–3154. doi: 10.1128/JB.185.10.3147-3154.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens EC, Russell FM, Goodrich-Blair H. Analysis of Xenorhabdus nematophila metabolic mutants yields insight into stages of Steinernema carpocapsae nematode intestinal colonization. Mol Microbiol. 2005;51:28–45. doi: 10.1111/j.1365-2958.2005.04742.x. [DOI] [PubMed] [Google Scholar]
- Murfin KE, Chaston JM, Goodrich-Blair H. Visualizing bacteria in nematodes using fluorescent microscopy. JoVE. 2012;68:e4298. doi: 10.3791/4298. 10.3791/4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussbaumer AD, Fisher CR, Bright M. Horizontal endosymbiont transmission in hydrothermal vent tubeworms. Nature. 2006;441:345–348. doi: 10.1038/nature04793. [DOI] [PubMed] [Google Scholar]
- Pace NR. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- Parniske M. Arbuscular mycorrhiza: the mother of plant root endosymbioses. Nat Rev Microbiol. 2008;6:763–775. doi: 10.1038/nrmicro1987. [DOI] [PubMed] [Google Scholar]
- Poinar GO. The presence of Achromobacter nematophilus in the infective stage of a Neoaplectana sp. (Steinernematidae: Nematoda) Nematologica. 1966;12:105–108. [Google Scholar]
- Popiel I, Grove DL, Friedman MJ. Infective juvenile formation in the insect parasite Steinernema feltiae. Parasitol. 1989;99:77–81. [Google Scholar]
- Popp C, Ott T. Regulation of signal transduction and bacterial infection during root nodule symbiosis. Curr Opin Plant Biol. 2011;14:458–467. doi: 10.1016/j.pbi.2011.03.016. [DOI] [PubMed] [Google Scholar]
- Richards GR, Goodrich-Blair H. Masters of conquest and pillage: Xenorhabdus nematophila global regulators control transitions from virulence to nutrient acquisition. Cell Microbiol. 2009;11:1025–1033. doi: 10.1111/j.1462-5822.2009.01322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby EG. Symbiotic conversations are revealed under genetic interrogation. Nat Rev Microbiol. 2008;6:752–762. doi: 10.1038/nrmicro1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicard M, Ferdy JB, Pages S, Le Brun N, Godelle B, Boemare N, Moulia C. When mutualists are pathogens: an experimental study of the symbioses between Steinernema (entomopathogenic nematodes) and Xenorhabdus (bacteria) J Evol Biol. 2004;17:985–993. doi: 10.1111/j.1420-9101.2004.00748.x. [DOI] [PubMed] [Google Scholar]
- Sicard M, Le Brun N, Pages S, Godelle B, Boemare N, Moulia C. Effect of native Xenorhabdus on the fitness of their Steinernema hosts: contrasting types of interaction. Parasitol Res. 2003;91:520–524. doi: 10.1007/s00436-003-0998-z. [DOI] [PubMed] [Google Scholar]
- Snyder H, Stock SP, Kim SK, Flores-Lara Y, Forst S. New insights into the colonization and release processes of Xenorhabdus nematophila and the morphology and ultrastructure of the bacterial receptacle of its nematode host, Steinernema carpocapsae. Appl Environ Microbiol. 2007;73:5338–5346. doi: 10.1128/AEM.02947-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stabb EV, Ruby EG. RP4-based plasmids for conjugation between Escherichia coli and members of the Vibrionaceae. Methods Enzymol. 2002;358:413–426. doi: 10.1016/s0076-6879(02)58106-4. [DOI] [PubMed] [Google Scholar]
- Stock SP, Lee M-M, Flores-Lara Y. The rectal glands of Heterorhabditis bacteriophora (Rhabditida: Heterorhabditidae) hermaphrodites and their role in symbiont transmission. J Inv Pathol. 2012;110:135–138. doi: 10.1016/j.jip.2012.03.018. [DOI] [PubMed] [Google Scholar]
- Sugar DR, Murfin KE, Chaston JM, Andersen AW, Richards GR, deLeon L, et al. Phenotypic variation and host interactions of Xenorhabdus bovienii SS-2004, the entomopathogenic symbiont of Steinernema jollieti nematodes. Environ Microbiol. 2012;14:924–939. doi: 10.1111/j.1462-2920.2011.02663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teal TK, Lies DP, Wold BJ, Newman DK. Spatiometabolic stratification of Shewanella oneidensis biofilms. Appl Environ Microbiol. 2006;72:7324–7330. doi: 10.1128/AEM.01163-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivas EI, Goodrich-Blair H. Xenorhabdus nematophilus as a model for host-bacterium interactions: rpoS is necessary for mutualism with nematodes. J Bacteriol. 2001;183:4687–4693. doi: 10.1128/JB.183.16.4687-4693.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Bedding RA. Population dynamics of Heterorhabditis bacteriophora and Steinernema carpocapsae in in vitro monoxenic solid culture. Fundam Appl Nematol. 1998;21:165–171. [Google Scholar]
- Wang JX, Bedding RA. Population development of Heterorhabditis bacteriophora and Steinernema carpocapsae in the larvae of Galleria mellonella. Fundam Appl Nematol. 1996;19:363–367. [Google Scholar]
- Wollenberg MS, Ruby EG. Population structure of Vibrio fischeri within the light organs of Euprymna scolopes squid from two Oahu (Hawaii) populations. Appl Environ Microbiol. 2009;75:193–202. doi: 10.1128/AEM.01792-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wouts WM. Biology, life cycle, and redescription of Neoaplectana bibionis Bovien, 1937 Nematoda: Steinernematidae. J Nematol. 1980;12:62–72. [PMC free article] [PubMed] [Google Scholar]
- Xu J, Hurlbert RE. Toxicity of irradiated media for Xenorhabdus spp. Appl Environ Microbiol. 1990;56:815–818. doi: 10.1128/aem.56.3.815-818.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.