Abstract
In diagnostic studies without a gold standard, the assumption on the dependence structure of the multiple tests or raters plays an important role in model performance. In case of binary disease status, both conditional independence and crossed random effects structure have been proposed and their performance investigated. Less attention has been paid to the situation where the true disease status is ordinal, with the exception of Wang et al [1] who assumed conditionally independent multiple tests when studying traditional Chinese medicine and Wang and Zhou [2] who assumed a normal subject random effect but a fixed rater effect. In this paper, we propose crossed subject- and rater-specific random effects to account for the dependence structure and assess the robustness of the proposed model to misspecification in the random effects distributions. The models are applied to data from the Physician Reliability Study which focuses on assessing the diagnostic accuracy in a population of raters for the staging of endometriosis, a gynecological disorder in women. Using this new methodology, we estimate the probability of a correct classification and show that regional experts can more easily classify the intermediate stage than resident physicians.
Keywords: Random effects models, Endometriosis, MCEM algorithm
1. Introduction
In public health and medical research, it is of importance to study the diagnostic accuracy of new tests or raters (e.g., [3, 4]). This is usually done by comparing the diagnostic results from the tests or raters with the true disease status (i.e., the gold standard). In many cases, however, a gold standard may not be measured due to cost constraints, concerns of the invasive nature of the diagnostic procedure, or a lack of biotechnology to obtain a definitive result. There has been extensive literature on estimating diagnostic accuracy without a gold standard, with the latent class approach (LCA) widely being used (e.g., [4, 5, 6]).
It is usually assumed in LCA that the multiple tests or raters are independent conditional on the true disease status. However, the conditional independence assumption may not be valid in practice and models with such an assumption may not fit the data well. Alternatively, Qu et al [7] developed a LCA by including normally distributed subject-specific random effects to model conditional dependence among binary tests. Albert and Dodd [8] demonstrated that, when the unknown disease status is binary, the model is weakly identified in the random effects distribution in the sense that different random effects distributions may fit the data equally well.
LCA can also be utilized when the true outcome of interest is ordinal rather than binary. Wang et al [1] extended the work of Zhou et al [6] on binary outcome to ordered multiple symptom categories and applied it to data from traditional Chinese medicine. In a further extension, Wang and Zhou [2] incorporated normal subject-specific random effects while assuming fixed effects for the raters. In this paper, we are interested in relaxing the conditional independence assumptionin Wang and Zhou [1] by proposing crossed subject- and rater-specific random effects to account for the dependence structure in the data. We are interested in assessing the robustness of the proposed models to misspecifications in the Gaussian random effects by considering a mixture of normals for both subject- and rater-specific random effects.
This article is motivated by the Physician Reliability Study (PRS) [9] that investigated the reliability of endometriosis between different physicians and settings. In the PRS, 12 physicians in obstetric and gynecology (OB/GYN) separately reviewed participant clinical information (digital intra-uterus image taken during laparoscopy, surgeon notes, MRI and histopathology reports) and assessed the endometriosis staging. Each physician conducted the review in a sequence of four settings, with each successive setting having an additional piece of clinical information to the reviewing physicians. In this article, we evaluate the diagnostic accuracy of 8 physicians (4 regional experts and 4 residents) who are practicing at the same medical center (Utah) when each of them reviewed the digital images (setting 1). Our interest here is evaluating the diagnostic accuracy in the population of these physicians; hence we treat physicians as a random rather than a fixed effect.
Endometriosis is a gynecological disorder in women that occurs when cells from the lining of the uterus grow in other area of the uterus. The cause of endometriosis is unknown and the accurate staging of the disease is subject to substantial errors. In this article, we focus on the 5 stagings of endometriosis: no endometriosis, stage I (minimal), stage II (mild), stage III (moderate) and stage IV (severe). In PRS, 79 subjects have complete staging results from the 8 physicians of interest and constitute the study sample. Among the 632(= 79 × 8) reviews, 155(25%) are no endometriosis while 250(40%), 136(21%), 63(10%) and 28(4%) are stages I to IV, respectively. Table 1 presents the averaged conditional sample proportions of endometriosis staging by one physician given the staging by another that are based on 10000 bootstrapped samples (drawn with replacement from the original data set) of the diagnostic results of two arbitrary physicians. As an indication of agreement, the kappa statistics is estimated to be 0.379.
Table 1.
Average conditional sample proportions of the endometriosis staging rated by one physician given the staging by another physician in the PRS data. Based on 10000 bootstrapped samples of the diagnostic results of arbitrary two physicians on all 79 subjects.
| Physician 2 | ||||||
|---|---|---|---|---|---|---|
|
| ||||||
| No endo | Stage I | Stage II | Stage III | Stage IV | ||
| Physician 1 | No ENDO | 0.686 | 0.180 | 0.076 | 0.080 | 0.061 |
| Stage I | 0.225 | 0.570 | 0.400 | 0.144 | 0.062 | |
| Stage II | 0.049 | 0.202 | 0.371 | 0.237 | 0.102 | |
| Stage III | 0.028 | 0.039 | 0.126 | 0.371 | 0.326 | |
| Stage IV | 0.011 | 0.009 | 0.027 | 0.168 | 0.449 | |
More specific substantive questions include (1) do the physicians have worse diagnostic accuracy at higher stages (moderate and severe) than at lower stages (no disease and minimal)? (2) are the extreme stages (no disease, minimal and severe) easier to diagnose than the middle stages (mild and moderate)? (3) how accurate are the physicians at correctly staging endometriosis? Off by only 1 stage? Off by 2 stages? (4) do the two groups of physicians (regional experts and residents) have different misclassification matrices in diagnosing endometriosis? From a statistical methodological perspective, we are interested in evaluating the robustness of our proposed crossed random effects model with respect to misspecification to the random effects distributions.
The remainder of this article is organized as follow. In Section 2, we propose a latent class model with crossed random effects for estimating the diagnostic accuracy of ordinal tests for ordinal true disease status. In Section 3, we present a Monte-Carlo EM algorithm for parameter estimation. In Section 4, we perform simulation studies to assess the convergence of the maximum likelihood estimators (MLE) obtained from a Monte-Carlo EM algorithm and assess the operating characteristics of the procedure. In Section 5, we apply the proposed model to data from the PRS in the staging of endometriosis. Conclusions and discussions are provided in Section 6.
2. Methods
Let Yij denote the ordinal diagnostic test result of the stages of endometriosis (Yij = 0, …, K) for the ith subject by the jth rater (i.e. physician), i = 1, …, I and j = 1, …, J. In the PRS, there are four stages of endometriosis and we have 79 subjects who had complete diagnoses from four regional experts and four residents; thus K = 4, I = 79 and J = 8. We denote Di as the true disease status of the ith subject, where for each subject Di = 0 denotes absence of endometriosis and Di = 1, 2, 3 and 4 denote the four stages of endometriosis respectively. Investigators are interested in estimating the average accuracy across physician group rather than physician-specific accuracy. We parameterize the cumulative probability by the following model with two crossed random effects ({bi}, {cj}) and p covariates:
| (1) |
where are subject-specific random effects with scale parameters are rater-specific random effects with scale parameters are monotonically nondecreasing cut-points for disease status di, is the ith row of the I × p matrix Xj, which is the design matrix of the jth rater, and βdi is the di-th column of βp × D, the matrix of coefficients for the covariates. Let
be a matrix of cut-off points with rows corresponding to the rating and columns corresponding to the true unknown category.
The model specifications above differ from that in Wang and Zhou [2] in that the rater-specific effect is assumed to be random in addition to the subject-specific effect. This is important since in PRS the physicians were chosen from a population of OBGYNs and it is of interest to obtain the average estimate across the population. The random effects and can each be assumed to follow a standard normal distribution. In addition, to allow for a more flexible random effects structure, we can also consider mixture of normals (MixNs). Define Gb and Gc to be random variables from a Bernoulli distribution with success probabilities λb and λc respectively, then
| (2) |
where φ(•, μ, ν2) is the density of normal distribution with mean μ and variance ν2. For model identification, we impose the restrictions on gb(•):
with similar restrictions for gc(•). Let πdi = P (Di = di) and note that πd (d = 0, 1, …, D) is the prevalence of stage d of the disease. The probability that rater j rates subject i as k given the true disease stage and random effects bi and cj can be expressed as
| (3) |
As in the PRS, we assume D = K = 4, which means that the number of rating categories (i.e. stages) is the same as the number of true categories. Similar to Wang et al [1], extensions to the case when D ≠ K can be considered. Given random effects and , the ratings are each conditionally independent and have a multinomial distribution. Thus, the conditional distribution of each measurement is
Let θ = (γ, σ, τ, π, β)’ be a vector of the unknown parameters. The likelihood is given by
| (4) |
Once we obtain which is the MLE of θ from (4), for covariate X = x, we obtain the MLE of by (1), where we use and to denote the generic test and generic disease status respectively rather than any specific rater or subject.
We consider the misclassification matrix as a measure of diagnostic accuracy, where the (K + 1) × (D + 1) misclassification matrix has elements given by
| (5) |
in the (k + 1)th row and (d + 1)th column of the matrix. The misclassification matrix is square (D = K) and it can be estimated by (5) once we obtain . By the invariance property of MLE, we can estimate the probability of a correct classification as , where is a function of .
3. Maximum Likelihood Estimation: MCEM Algorithm
In this section, we introduce the method for obtaining the MLEs of the model parameters. Due to the high dimensional integration and summation, it can be challenging to use the classical EM algorithm to estimate (4). Instead, we follow Wei and Tanner [10] in using the Monte-Carlo EM (MCEM) algorithm. The MCEM incorporates a Monte Carlo implementation into the E-step of EM algorithm so that the required expectation can be approximated by the average of the Monte-Carlo samples from the target distribution. By using the Metropolis-Hasting algorithm to draw samples from target distributions, McCulloch [11] developed maximum likelihood algorithms for generalized linear mixed models. Robert and Casella [12] gave examples of R implementations using the MCEM algorithm. Here, we treat the true disease status , subject-specific random effects and rater-specific as missing data. For notational brevity, let be the observed data, be the missing data and W* = (Y*, Z*)’ be the complete data. Accordingly, the complete-data likelihood is
| (6) |
For our purpose, we can obtain the kernel of the target distribution by Bayes Rule:
where for d = 0, 1, …, D. We implement the MCEM algorithm as follows:
Start by initial values θ(0) = (γ(0), σ(0), τ(0), π(0), β(0))’ and set r = 0.
- Generate M groups of values z*(1), z*(2), …, z*(M) from the target distribution f(z*|y*, θ(r)) and choose θ(r+1) to maximize
which is the Monte-Carlo estimate of
where θ(r) is the parameter value from the rth iteration and set r = r + 1. The maximization is subject to the restrictions that elements in each column of matrix γ are nondecreasing from the top to the bottom, all the elements in σ, τ and π are nonnegative and that all the elements in π sum up to one. R function nloptr can be employed for such constrained maximization. We generate sample z*(m) (m = 1, …, M) from the target distribution by Metropolis-Hasting algorithm:- Let denote the candidate distribution with being the empirical distribution of di and and being t distribution centered at the means of bi and cj sampled from the rth iteration of the EM algorithm respectively. Choose z0 from the support of the target distribution.
- For n = 1, 2, …: generate Un ~ uniform(0, 1) and . Set
As n → ∞, zn converges to z* ~ f(z*|y*, θ) in distribution. In order to generate random effects bi’s and cj’s from the candidate distribution, we choose 30 as the degrees of freedom of the t distributions mentioned in part (a) so that they do not have heavy tails.
In the simulation studies and the analysis of the PRS data presented in Sections 4 and 5, we run the algorithms 200 steps. We take M = 50 for iterations r = 1, …, 50, M = 200 for iterations r = 51, …, 80 and M = 1000 for iterations r = 81, …, 200. Final estimates are computed by averaging over the last ten iterations.
We wish to compare different random effects distributions for {bi}, {cj} in (2) using penalized likelihood method such as the Akaike information criterion (AIC). However, the likelihood is difficult to evaluate given the high dimension of the random effects. We use the simulated likelihood approach by Geyer and Thompson [13] to overcome this difficulty. Specifically, let be the MLE obtained from the MCEM algorithm and in (4) consider
where the expectation is taken over all random effects {bi}, {cj} and {di}. The maximized likelihood can be approximated by
where , and are the t-th simulated realizations from the density functions gb for the i-th subject, gc for the j-th rater and mass functions multinomial for the i-th subject respectively, T is the total number of such simulated realizations and
by (3). The Monte-Carlo estimate of the Akaike Information Criteria can be used to compare different models for the random effects, where npar is the number of parameters in the model. In the simulation studies and the analysis of the PRS data, we take T = 5000
Proving global identifiability for complex latent class models such as the one we are proposing is a difficult and challenging problem. In fact, at the very least, these models lack identifiability due to a label-switching problem. Practitioners need to be cautious when applying these methods to be sure that results are scientifically sensible. Various authors have worked on the issue of local identifiability (e.g., [14, 15]). This latter form of identifiability can be proved by showing that the Hessian matrix is non-singular. Unfortunately this is difficult for our model since the Hessian matrix can only be estimated by using Monte-Carlo method and its estimate can be unstable when the number of parameters is large.
4. Simulation Study
In this section, we perform simulation studies to assess the convergence and operating characteristics of the MCEM algorithm. For simplicity, we focus on the case of a homogeneous group of physicians (i.e., no covariates in the model). For notational brevity, we therefore write in place of . Data were simulated according to model (1) with random effects following distribution (2). All simulations were performed with I = 100 subjects and J = 10 raters. The true prevalences are 0.3, 0.25, 0.2, 0.15, 0.1 for no endometriosis, stage I to IV respectively, and the true misclassification matrix is as in Table 2. These values are chosen to be close to the parameter estimates in the PRS shown in Section 5. Based on the parameters in Table 2, the true probability of a correct classification is .
Table 2.
True misclassification matrix in the simulation studies.
| d = 0 | d = 1 | d = 2 | d = 3 | d = 4 | ||
|---|---|---|---|---|---|---|
| P(Y = k|D = d) | k = 0 | 0.7 | 0.2 | 0.25 | 0.05 | 0.15 |
| k = 1 | 0.2 | 0.55 | 0.46 | 0.28 | 0.1 | |
| k = 2 | 0.05 | 0.2 | 0.25 | 0.35 | 0.15 | |
| k = 3 | 0.04 | 0.03 | 0.03 | 0.21 | 0.35 | |
| k = 4 | 0.01 | 0.02 | 0.01 | 0.11 | 0.25 |
4.1. Convergence
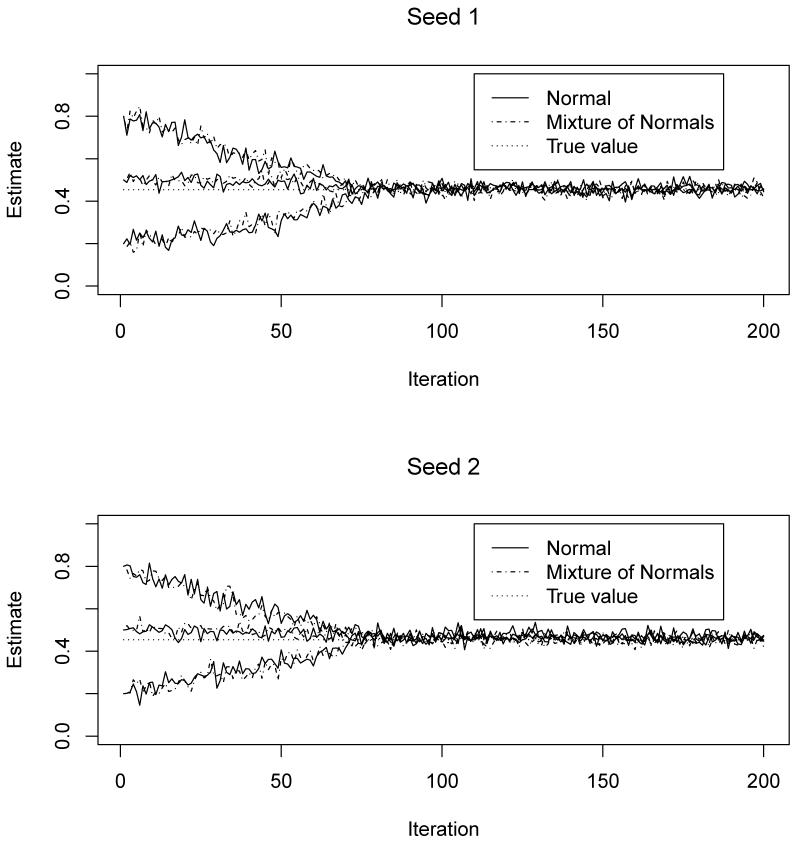
To assess the convergence of the MCEM algorithm and the Monte-Carlo variations, we simulated data sets based on random effects with normal and MixNs respectively. We obtained the MLEs of the parameters by assuming that the random effects follow the true corresponding distributions. The algorithm was performed based on two different seeds and three different starting values of the probability of a correct classification: 0.8, 0.2 and 0.5, for larger than, smaller than and close to the true value respectively. We show in Figure 1 that the convergence of the estimated probability of a correct classification is insensitive to the choice of either the starting values or the seeds of the MCEM algorithm. This empirically suggests that the model is locally identifiable. The convergence of each element in θ is similar to that of and is not presented here to save space. Specifically, the algorithm converges at around the 80th iteration and appears stable thereafter with tolerable Monte-Carlo variations. This is probably because we increased the size of Monte-Carlo samples from the target distribution in the E-step from 200 to 1000 at the 80th iteration. Similar results are observed regardless of whether the distributions of random effects are normal or MixNs. We performed similar simulations using different parameter values and observed the same results (data not shown). For the algorithm to reach the approximate convergence (within Monte-Carlo error), it took approximately 16 hours on an NIH linux cluster with a 2.8 GHz Intel X5660 processor.
Figure 1.
Convergence of the MCEM algorithm by seeds and starting values under true random effect distributions.
4.2. Operating characteristics
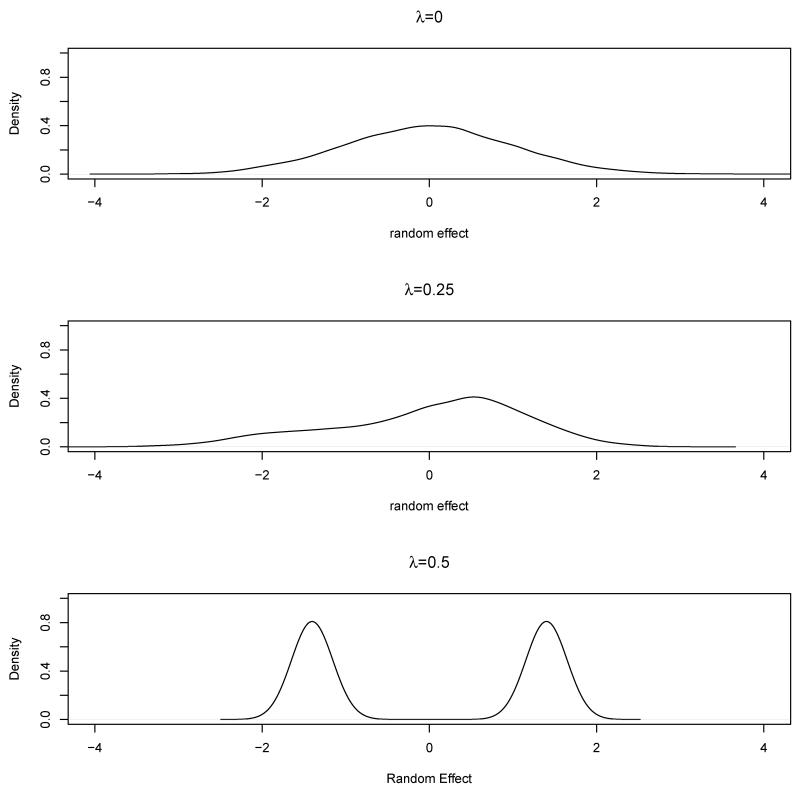
To assess the operating characteristics of the MCEM algorithm, we simulated different data sets based on different random effects distributions. More specifically, in (2), we choose λ = λb = λc = 0, 0.25 or 0.5 with the corresponding distributions shown in Figure 2. For each λ, we generated 200 data sets and obtained the MLEs of model parameters using each individual data set.
Figure 2.
The distribution of random effects under different λ values.
In Table 3, we summarize the estimated prevalence, with the standard error in parentheses. When λ = 0, the data are simulated according to normally distributed subject-specific and rater-specific random effects. When a normal distribution is assumed as the working model, the estimates are mostly unbiased. This is also true when MixNs are assumed since normal random effects are special cases of MixN random effects. In contrast, when λ ≠ 0, the data are simulated according to a model with the subject and rater-specific random effects following MixNs. When MixNs are assumed as the working model, the estimates are nearly unbiased. However, when normal distributions are assumed, most estimates are biased. Moreover, the biases are more profound when λ = 0.5 where the misspecification of the random effects distribution is more severe than when λ = 0.25 (see Figure 2). The estimated misclassification matrix in Table 4 shows a similar pattern. For example, when λ = 0, the estimates of P (Y = 4|D = 4) are nearly unbiased under both random effects distributions, 0.262 and 0.271 for normal and MixNs respectively compared to the true value of 0.25; when λ ≠ 0 and MixNs are assumed as the working model, the estimates are mostly unbiased: 0.237 and 0.231 for λ = 0.25 and 0.5 respectively. When normal distributions are assumed, the estimates are biased: 0.326 and 0.381 for λ = 0.25 and 0.5 respectively. Clearly the bias is larger (0.381) when λ = 0.5 than that (0.326) when λ = 0.25. When normal distributions are assumed for the random effects in the true model and either normal or MixNs in the working model, the estimated probabilities of a correct classification (standard errors) are 0.466(0.087) and 0.472(0.127) respectively, and both are close to the true probability of a correct classification, 0.454. However, when λ = 0.25 or 0.5, namely the true distributions of the random effects are MixNs, fitting a working model with normal random effects distribution resulted in a biased probability of a correct classification: 0.548(0.095) and 0.602(0.105), respectively (compared with a true value of 0.454). In comparison, the corresponding estimates under a correctly specified working models are 0.466(0.139) and 0.464(0.146) respectively.
Table 3.
Average estimated prevalence πd (standard deviation) in the simulation study based on different assumptions on the distributions of random effects. Based on 200 simulated data sets. The true prevalences are 0.3, 0.25, 0.2, 0.15 and 0.1 for no endometriosis, Stage I to IV, respectively.
| λ | Working random effects | No ENDO | Stage I | Stage II | Stage III | Stage IV |
|---|---|---|---|---|---|---|
| 0 | Normal | 0.314(0.10) | 0.239(0.08) | 0.223(0.12) | 0.138(0.03) | 0.086(0.03) |
| 0 | MixN | 0.320(0.12) | 0.231(0.14) | 0.228(0.14) | 0.129(0.07) | 0.092(0.09) |
| 0.25 | Normal | 0.360(0.08) | 0.205(0.14) | 0.271(0.08) | 0.112(0.11) | 0.052(0.05) |
| 0.25 | MixN | 0.311(0.10) | 0.236(0.16) | 0.221(0.16) | 0.141(0.14) | 0.091(0.09) |
| 0.5 | Normal | 0.401(0.08) | 0.179(0.12) | 0.272(0.08) | 0.109(0.11) | 0.039(0.04) |
| 0.5 | MixN | 0.318(0.18) | 0.231(0.23) | 0.216(0.07) | 0.139(0.13) | 0.096(0.10) |
Table 4.
Estimated misclassification matrices in simulation studies based on different distributions of random effects (subject, rater). In the body of the table, we list averageP̂(Y = k|D = d), the elements of misclassification matrix with their standard deviations based on 200 simulated data sets in parentheses. The true parameters are in Table 2, rows 2-6.
| λ = 0 | Normal | d = 0 | d = 1 | d = 2 | d = 3 | d = 4 |
| k = 0 | 0.718(0.11) | 0.178(0.05) | 0.236(0.10) | 0.043(0.04) | 0.143(0.12) | |
| k = 1 | 0.181(0.08) | 0.532(0.09) | 0.443(0.06) | 0.298(0.09) | 0.093(0.09) | |
| k = 2 | 0.068(0.07) | 0.212(0.10) | 0.276(0.05) | 0.334(0.15) | 0.167(0.09) | |
| k = 3 | 0.027(0.03) | 0.036(0.04) | 0.036(0.04) | 0.209(0.12) | 0.335(0.07) | |
| k = 4 | 0.006(0.01) | 0.042(0.04) | 0.009(0.01) | 0.116(0.12) | 0.262(0.17) | |
|
| ||||||
| MixN | d = 0 | d = 1 | d = 2 | d = 3 | d = 4 | |
| k = 0 | 0.726(0.19) | 0.172(0.12) | 0.227(0.13) | 0.038(0.04) | 0.139(0.14) | |
| k = 1 | 0.175(0.16) | 0.523(0.12) | 0.435(0.14) | 0.306(0.09) | 0.088(0.09) | |
| k = 2 | 0.072(0.07) | 0.226(0.13) | 0.286(0.07) | 0.329(0.17) | 0.175(0.15) | |
| k = 3 | 0.020(0.02) | 0.042(0.04) | 0.042(0.04) | 0.219(0.16) | 0.327(0.08) | |
| k = 4 | 0.007(0.01) | 0.037(0.04) | 0.010(0.01) | 0.108(0.11) | 0.271(0.18) | |
|
| ||||||
| λ = 0.25 | Normal | d = 0 | d = 1 | d = 2 | d = 3 | d = 4 |
| k = 0 | 0.781(0.12) | 0.151(0.08) | 0.212(0.16) | 0.032(0.03) | 0.099(0.10) | |
| k = 1 | 0.132(0.08) | 0.633(0.11) | 0.428(0.09) | 0.321(0.11) | 0.072(0.07) | |
| k = 2 | 0.073(0.07) | 0.150(0.14) | 0.322(0.13) | 0.305(0.11) | 0.191(0.14) | |
| k = 3 | 0.011(0.01) | 0.050(0.05) | 0.036(0.04) | 0.295(0.14) | 0.312(0.08) | |
|
| ||||||
| k = 4 | 0.003(0.00) | 0.016(0.02) | 0.002(0.00) | 0.047(0.05) | 0.326(0.17) | |
| MixN | d = 0 | d = 1 | d = 2 | d = 3 | d = 4 | |
| k = 0 | 0.721(0.13) | 0.173(0.16) | 0.231(0.17) | 0.041(0.04) | 0.167(0.17) | |
| k = 1 | 0.171(0.17) | 0.539(0.13) | 0.441(0.13) | 0.296(0.13) | 0.091(0.09) | |
| k = 2 | 0.063(0.06) | 0.218(0.14) | 0.272(0.17) | 0.332(0.18) | 0.174(0.14) | |
| k = 3 | 0.022(0.02) | 0.040(0.04) | 0.033(0.03) | 0.230(0.18) | 0.331(0.13) | |
| k = 4 | 0.023(0.02) | 0.030(0.03) | 0.023(0.02) | 0.101(0.10) | 0.237(0.21) | |
|
| ||||||
| λ = 0.5 | Normal | d = 0 | d = 1 | d = 2 | d = 3 | d = 4 |
| k = 0 | 0.802(0.10) | 0.122(0.10) | 0.156(0.16) | 0.028(0.03) | 0.046(0.05) | |
| k = 1 | 0.096(0.06) | 0.681(0.13) | 0.403(0.07) | 0.322(0.11) | 0.054(0.05) | |
| k = 2 | 0.079(0.08) | 0.129(0.13) | 0.394(0.11) | 0.271(0.09) | 0.213(0.14) | |
| k = 3 | 0.011(0.01) | 0.053(0.05) | 0.036(0.04) | 0.336(0.14) | 0.306(0.06) | |
| k = 4 | 0.012(0.01) | 0.015(0.01) | 0.011(0.01) | 0.043(0.04) | 0.381(0.15) | |
|
| ||||||
| MixN | d = 0 | d = 1 | d = 2 | d = 3 | d = 4 | |
| k = 0 | 0.719(0.19) | 0.175(0.11) | 0.236(0.24) | 0.032(0.03) | 0.171(0.17) | |
| k = 1 | 0.168(0.17) | 0.531(0.15) | 0.438(0.13) | 0.335(0.15) | 0.103(0.10) | |
| k = 2 | 0.067(0.07) | 0.183(0.16) | 0.268(0.20) | 0.278(0.18) | 0.169(0.17) | |
| k = 3 | 0.024(0.02) | 0.037(0.04) | 0.039(0.04) | 0.236(0.17) | 0.326(0.08) | |
| k = 4 | 0.022(0.02) | 0.074(0.07) | 0.019(0.02) | 0.119(0.12) | 0.231(0.23) | |
Since it is important to specify the distribution of random effects correctly, we are interested in the empirical rates that the penalized likelihood criteria select the true model. By simulation, Zhang et al [16] showed that when the true disease status and outcomes from raters are both binary, penalized likelihood criteria empirically select the true model at the rate of 55%, which is slightly higher than 50%. Here, when the true disease status and outcomes are both ordinal, the empirical rate that penalized likelihood criteria select the true model is 74%, 73% and 76% for λ = 0, 0.25 and 0.5 respectively. Thus, compared to the method of dichotomizing ordinal data, the proposed method is empirically more likely to select the true model and estimate the parameters accurately. This is likely due to the increased information contained in the ordinal data.
5. Application
We now apply the proposed modeling framework to data from the Physician Reliability Study [9]. Eight physicians (4 residents and 4 regional experts) reviewed digital images on 79 subjects. Scientific substantive questions include: (1) how did these physicians stage endometriosis? Were they more likely on target or within one stage? (2) are the physicians better at diagnosing some particular stages as compared to others? (3) are the findings in questions (1) and (2) different for the 4 residents as compared with the 4 regional experts?
With respect to the 8 physicians, we first ignore their difference, treating them as a homogeneous group. Later we will account for their difference (residents vs regional experts) in the modeling framework.
Among all the four possible combinations of subject and rater-specific random effects the AIC is the smallest (3219.47) for the model with normal subject-specific random effects and MixN rater-specific random effects. Under the best model, the prevalences of the five stages are estimated at 0.321(0.11), 0.238(0.08), 0.211(0.03), 0.151(0.06) and 0.079(0.06) respectively, and the estimated misclassification matrix (with the bootstrap standard errors based on 1000 bootstrapped samples in parentheses) is presented in Table 5. The estimated probability of a correct classification is . In Table 5, the diagonal elements represent the average diagnostic accuracy across the population of the physicians for each true endometriosis stage. For example, when the true stage of endometriosis is mild (d = 2), the average probability that the physicians would correctly stage the disease is 0.289. In each row, the elements adjacent to the diagonal elements are important since they represent the situation when the physicians categorize the diagnostic result only one stage off from the truth. In the PRS, the physicians are more likely to underestimate the severity by one stage than overestimate by one stage. Moreover, it appears that these physicians are better at diagnosing lower stages of endometriosis (no endometriosis and mild stage) than higher ones, with the correction classification of 75%, 63%, 29%, 26% and 29% for the five stages (from low to high), respectively.
Table 5.
Estimated misclassification matrix in the PRS based on normal subject-specific random effects and MixN rater-specific random effects when pooling the 8 physicians. In the body of the table, we list each element with the bootstrap standard errors based on 1000 bootstrapped samples in parentheses.
| d = 0 | d = 1 | d = 2 | d = 3 | d = 4 | ||
|---|---|---|---|---|---|---|
| P̂(Y = k|D = d) | k = 0 | 0.754(0.11) | 0.155(0.08) | 0.256(0.12) | 0.087(0.06) | 0.114(0.11) |
| k = 1 | 0.163(0.08) | 0.628(0.07) | 0.421(0.08) | 0.237(0.09) | 0.122(0.08) | |
| k = 2 | 0.045(0.04) | 0.157(0.13) | 0.289(0.09) | 0.315(0.12) | 0.133(0.09) | |
| k = 3 | 0.034(0.03) | 0.046(0.05) | 0.028(0.03) | 0.262(0.08) | 0.342(0.12) | |
| k = 4 | 0.004(0.00) | 0.014(0.01) | 0.006(0.01) | 0.099(0.09) | 0.289(0.10) |
Since there are two physician groups (residents and regional experts) serving as raters, it is of interest to see whether the above conclusions hold when we account for the difference between the two groups of physicians with different levels of experience. In doing so, we utilize a modified version of model (1):
where I(•) is an indicator function.
Among all the four combinations of random effects distributions, the model assuming normal subject-specific random effects and normal rater-specific random effects had the smallest AIC (3211.36), with the probability of a correct classification for resident and regional expert being 0.463 and 0.489 respectively. The estimated prevalences and coefficients for the group indicator as well as the misclassification matrices for both groups (with the bootstrap standard errors based on 1000 bootstrapped samples in parentheses) are listed in Tables 6 and 7 respectively. By including the group indicator as covariate, we have smaller AIC than when the two groups are treated as homogeneous, suggesting that the resident and regional expert group might be different at staging endometriosis. More specifically, we can see from Table 7 that when the true disease stage is mild (d = 2), the probability that regional experts make the right judgement is 0.342, which is significantly higher than 0.151, the probability that residents make the right judgement. This difference in the physician type may also explain why a MixNs distribution is needed for the rater-specific random effects when the 8 physicians are treated homogeneously. It appears that failure to include an important binary rater-specific covariate will induce a bimodal structure in the random effects distribution.
Table 6.
Estimated prevalence and coefficients for the group indicator (1 = resident, 0 = regional expert) based on normal subject- and rater-specific random effects in the PRS. In the body of the table, we report bootstrap standard errors based on 1000 bootstrapped samples in parentheses. The coefficients that are more than two estimated standard errors from 0 are in bold.
| d = 0 | d = 1 | d = 2 | d = 3 | d = 4 | |
|---|---|---|---|---|---|
| 0.290(0.04) | 0.275(0.04) | 0.159(0.03) | 0.183(0.03) | 0.093(0.02) | |
| −0.035(0.02) | 0.054(0.03) | − 0.146(0.04) | −0.022(0.05) | − 0.133(0.06) |
Table 7.
Estimated misclassification matrices in PRS based on normal subject- and rater-specific random effects. In the body of the table, we report P̂(Y = k|D = d) with their bootstrap standard errors based on 1000 bootstrapped samples in parentheses for each value of the covariate (resident and regional expert group) respectively.
| d = 0 | d = 1 | d = 2 | d = 3 | d = 4 | |
|---|---|---|---|---|---|
| Residents | |||||
| k = 0 | 0.693(0.05) | 0.196(0.04) | 0.213(0.07) | 0.060(0.02) | 0.118(0.07) |
| k = 1 | 0.203(0.04) | 0.622(0.05) | 0.611(0.07) | 0.307(0.07) | 0.090(0.06) |
| k = 2 | 0.052(0.03) | 0.166(0.03) | 0.151(0.05) | 0.348(0.07) | 0.185(0.06) |
| k = 3 | 0.051(0.04) | 0.015(0.02) | 0.013(0.01) | 0.224(0.05) | 0.327(0.07) |
| k = 4 | 0.001(0.00) | 0.001(0.00) | 0.013(0.01) | 0.061(0.04) | 0.280(0.09) |
| Regional Experts | |||||
| k = 0 | 0.748(0.05) | 0.171(0.04) | 0.256(0.07) | 0.062(0.02) | 0.141(0.07) |
| k = 1 | 0.170(0.04) | 0.562(0.04) | 0.344(0.06) | 0.244(0.06) | 0.087(0.05) |
| k = 2 | 0.041(0.02) | 0.189(0.04) | 0.342(0.07) | 0.298(0.06) | 0.089(0.05) |
| k = 3 | 0.034(0.02) | 0.049(0.02) | 0.047(0.02) | 0.218(0.05) | 0.430(0.11) |
| k = 4 | 0.006(0.01) | 0.028(0.02) | 0.012(0.01) | 0.177(0.06) | 0.253(0.10) |
6. Discussion
In this article, we have described a method for evaluating diagnostic accuracy without gold standard by latent class approach with crossed random effects for ordinal tests. This work generalizes Wang and Zhou [2] to allow for the rater effect to be treated as random rather than fixed and for the incorporation of flexible random effect distributions. We show that estimate of diagnostic accuracy may be sensitive to assumptions on the random effects distributions and that unlike for the binary case (see Zhang et al [16]), penalized likelihood comparisons can be used for assessing the adequacy of these distributions.
The proposed method was motivated by the Physician Reliability Study in staging endometriosis. To that end, we have found that it is important to incorporate covariates in the modeling framework to capture heterogeneity in raters. Substantively, we found that regional experts make better diagnoses than the residents when the true disease is mild, that physicians in PRS are more likely under-staging than over-staging, and that they are better at diagnosing lower stages of endometriosis than higher ones. These interesting findings can be useful in helping training physicians in the field.
In the analysis of the PRS, the covariate is binary, making it easy to present the misclassification matrix for both realizations of the covariate. For more complicated case of covariates, we recommend plotting the elements of the misclassification matrix as a function of changes in these covariates. In particular, since we can write the misclassification matrix in a close-form as presented in the Appendix, we can plot each element of it against a continuous covariate.
One feature in both our simulation study and the application is that the number of raters is small. With such small numbers of random effects, estimate of the corresponding variance component (τd) can be biased. We indeed observed this bias in a separate simulation study with J = 10 and near unbiasedness with J = 100. For example, with the true τ4 = 0.1, we obtained when J = 10, reflecting substantial bias, and when J = 100, reflecting little bias. Estimates of the subject-specific variance component in both cases are unbiased. Interestingly, regardless of J = 10 or 100, the estimated misclassification matrices, the primary focus of our work, are unbiased.
Acknowledgement
We thank the associate editor and reviewers for providing for the constructive comments. This research was supported by the Intramural Research Program of the National Institutes of health, Eunice Kennedy Shriver National Institute of Child Health and Human Development. We thank the Center for Information Technology, the National Institutes of Health, for providing access to the high performance computational capabilities of the Biowulf cluster.
APPENDIX: CLOSED FORM EXPRESSION OF EQUATION (5)
The closed form expression of the element in the misclassification matrix can be written as
| (7) |
For the special case of normal random effects with λb = λc = 1, μ1b = μ1c = 0 and νb = νc = 1, we have the following closed form of the misclassification matrix for an arbitrary realization of covariate x.
Equation (7) can be derived by considering the following.
Note that since , we have given Gb = 1 and similarly given Gc = 1. It follows that
where the last equation simply extends the argument by Qu, Tan and Kutner [7]. Similarly, we can obtain , and . By using , we obtain .
References
- 1.Wang Z, Zhou XH, Wang M. Evaluation of Diagnostic Accuracy in Detecting Ordered Symptom Statuses without a Gold Standard. Biostatistics. 2011;12(3):567–581. doi: 10.1093/biostatistics/kxq075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Z, Zhou XH. Random effects models for assessing diagnostic accuracy of traditional Chinese doctors in absence of a gold standard. Statistics in Medicine. 2012;31:661–671. doi: 10.1002/sim.4275. [DOI] [PubMed] [Google Scholar]
- 3.Pepe MS. The Statistical Evaluation of Medical Tests for Classification and Prediction. 1st edn Oxford University Press; New York: 2003. [Google Scholar]
- 4.Hui SL, Walter SD. Estimating the error rates of diagnostic tests. Biometrics. 1980;36:167–171. [PubMed] [Google Scholar]
- 5.Hui SL, Zhou XH. Evaluation of diagnostic tests without a gold standard. Statistical Methods in Medical Research. 1998;7:354–370. doi: 10.1177/096228029800700404. [DOI] [PubMed] [Google Scholar]
- 6.Zhou XH, Castelluccio P, Zhou C. Nonparametric estimation of ROC curves in the absence of a gold standard. Biometrics. 2005;61:600–609. doi: 10.1111/j.1541-0420.2005.00324.x. [DOI] [PubMed] [Google Scholar]
- 7.Qu Y, Tan M, Kutner MH. Random Effects Models in Latent Class Analysis for Evaluating Accuracy of Diagnostic Tests. Biometrics. 1996;52:797–810. [PubMed] [Google Scholar]
- 8.Albert PS, Dodd LE. A cautionary note on the robustness of latent class models for estimating diagnostic error without a gold standard. Biometrics. 2004;60:427–435. doi: 10.1111/j.0006-341X.2004.00187.x. [DOI] [PubMed] [Google Scholar]
- 9.Schliep K, Stanford JB, Zhang B, Chen Z, Dorais JK, Johnstone EB, Hammoud AO, Varner MW, Buck Louis GM, Peterson CM. on behalf of the ENDO Study Working Group. Inter- and intra-rater reliability in the diagnosis and staging of endometriosis: the ENDO Study. 2012. Submitted manuscript. [Google Scholar]
- 10.Wei GCG, Tanner MA. A Monte-Carlo implementation of the E-M algorithm and the poor man’s data augmentation algorithms. Journal of the American Statistical Association. 1990;85:699–704. [Google Scholar]
- 11.McCulloch CE. Maximum Likelihood Algorithms for Generalized Linear Mixed Models. Journal of the American Statistical Association. 1997;92:162–170. [Google Scholar]
- 12.Robert CP, Casella G. Introducing Monte Carlo Methods with R. 1st edn Springer; New York: 2010. [Google Scholar]
- 13.Geyer CJ, Thompson EA. Constrained Monte Carlo Maximum Likelihood for Dependent Data (with discussion) Journal of the Royal Statistical Society. Series B. 1992;54(3):657–699. [Google Scholar]
- 14.Bandeen-Roche K, Miglioretti DL, Zeger SL, Rathouz PJ. Latent Variable Regression for Multiple Discrete Outcomes. Journal of the American Statistical Association. 1997;92:1375–1386. [Google Scholar]
- 15.Jones G, Johnson W, Hanson T, Christensen R. Identifiability of Models for Multiple Diagnostic Testing in the Absence of a Gold Standard. Biometrics. 2010;66:855–863. doi: 10.1111/j.1541-0420.2009.01330.x. [DOI] [PubMed] [Google Scholar]
- 16.Zhang B, Chen Z, Albert PS. Estimating Diagnostic Accuracy of Raters without a Gold Standard by Exploiting a Group of Experts. Biometrics. 2012 doi: 10.1111/j.1541-0420.2012.01789.x. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]