Abstract
Autoimmune thyroid diseases (AITD) have become increasingly recognized as a complication of interferon-alpha (IFNα) therapy in patients with chronic Hepatitis C virus (HCV) infection. Interferon-induced thyroiditis (IIT) can manifest as clinical thyroiditis in approximately 15% of HCV patients receiving IFNα and subclinical thyroiditis in up to 40% of patients, possibly resulting in either dose reduction or discontinuation of IFNα treatment. However, the exact mechanisms that lead to the development of IIT are unknown and may include IFNα-mediated immune-recruitment as well as direct toxic effects on thyroid follicular cells. We hypothesized that IIT develops in genetically predisposed individuals whose threshold for developing thyroiditis is lowered by IFNα. Therefore, our aim was to identify the susceptibility genes for IIT. We used a genomic convergence approach combining genetic association data with transcriptome analysis of genes upregulated by IFNα. Integrating results of genetic association, transcriptome data, pathway, and haplotype analyses enabled the identification of 3 putative loci, SP100/110/140 (2q37.1), HLA (6p21.3), and TAP1 (6p21.3) that may be involved in the pathogenesis of IIT. Immune-regulation and apoptosis emerged as the predominant mechanisms underlying the etiology of IIT.
Keywords: Interferon, thyroiditis, autoimmunity, Thyroid
1. INTRODUCTION
Interferon alpha (IFNα) is a critical cytokine in the host immune response to viral infections and tumors, and plays an important role in the cross talk between the innate and adaptive immune responses [1]. Moreover, IFNα is a powerful therapeutic agent for a variety of infectious and malignant diseases. Intriguingly, therapy with IFNα has been associated with several autoimmune complications, most notably autoimmune thyroiditis [2]. The association of IFNα therapy with the development autoimmune thyroiditis (i.e. interferon induced thyroiditis [IIT]) has been well documented, most commonly when IFNα is used as a therapy for chronic hepatitis C virus (HCV) infection [3]. Numerous epidemiological studies have shown that IIT is quite common with subclinical thyroid disease developing in approximately 40% of HCV patients receiving a course of IFNα, while clinical disease (i.e. Hashimoto’s thyroiditis [HT] or Graves’ disease [GD]) develops in up to 20% of patients [4]. The complications of hypo- and hyper-thyroidism can be serious (e.g. cardiac arrhythmias) and could lead to dose reduction or cessation of therapy [5]. Therefore, current treatment algorithms recommend that HCV patients receiving IFNα therapy undergo periodic thyroid function testing throughout their treatment course [3].
The mechanisms by which IFNα induces thyroiditis are still unknown. We have recently shown that IFNα induces thyroiditis by both direct toxic effects on the thyroid and by immune recruitment mechanisms [6]. The fact that only 20–40% of HCV patients receiving IFNα therapy develop IIT suggests that HCV and IFNα act synergistically to induce thyroid disease in genetically predisposed individuals. Therefore, we hypothesized that genetic susceptibility plays a major role in the etiology of interferon induced thyroiditis. Indeed, the presence of thyroid autoantibodies, a biomarker of genetic susceptibility to autoimmune thyroid diseases (AITD), is the strongest risk factor for development of thyroiditis in HCV patients receiving IFNα [7]. However, so far, only a few small studies have analyzed genes that may be associated with IIT.
The goal of the current study was to identify the susceptibility genes for interferon induced thyroiditis in HCV patients. We performed a comprehensive genetic analysis in a cohort of well-characterized IIT patients comparing them to HCV patients that received IFNα therapy but did not develop thyroiditis (designated “HCV controls”) as well as to healthy individuals that were not infected by HCV (designated “healthy controls”). Our data point toward a key role for immune-regulatory and apoptosis related genes and pathways in the genetic susceptibility and pathogenesis of IIT.
2. PATIENTS AND METHODS
2.1 Study Subjects
The project was approved by the Mount Sinai School of Medicine Institutional Review Board. 326 patients with a diagnosis of chronic Hepatitis C virus (HCV) infection, who were treatment-eligible, were retrospectively screened for the development of thyroid disease. Demographics, co-infection with HIV, HCV genotype, and viral response to IFNα treatment were recorded based on the documented clinical information that was available at the time of chart review. None of the patients had documented evidence of other forms of liver disease including hepatitis B virus infection, non-alcoholic fatty liver disease, or alcoholic liver disease. Of the 326 patients who were screened, 245 individuals fulfilled our inclusion criteria: 1) confirmed infection with chronic HCV; 2) received at least one 12 week course of IFNα therapy; 3) had documented clinical, biochemical, and antibody information to enable the appropriate diagnosis and classification of thyroiditis. We excluded 81 patients based on the following criteria: 1) prior history of thyroid disease (i.e., Graves’ disease, Hashimoto’s thyroiditis, or primary hypothyroidism); 2) expired at the time of chart review; 3) received less than 12 weeks of IFNα therapy; 4) did not ultimately receive IFNα treatment. Following at least one 12 week course of IFNα therapy, 53 individuals (21.6%) were confirmed to have developed interferon-induced thyroiditis (IIT cases), while 192 did not develop thyroiditis (HCV controls). This frequency of IIT is consistent with the literature [3]. The patient characteristics are shown in table 1. Plasma and DNA samples for the included cases and controls were obtained from Mount Sinai Biobank (Charles R. Bronfman Institute for Personalized Medicine, Mount Sinai School of Medicine, New York, NY). 851 healthy Caucasians individuals, who were previously genotyped using the immunochip (see below) with no known personal history of autoimmune disease, liver disease, or thyroid disease, served as control subjects (provided by Dr. Peter K. Gregersen, Feinstein Institute for Medical Research, North Shore-Long Island Jewish Health System, Manhasset, NY).
Table 1.
Clinical characteristics of patients with chronic Hepatitis C virus who developed thyroiditis (IIT cases) compared to patients with chronic Hepatitis C virus without evidence of thyroiditis (HCV controls)
| IIT Cases | HCV Controls | |
|---|---|---|
| Total (n = 245) | 53 | 192 |
| Gender (p < 0.006) | ||
| Male | 25 (47%) | 130 (68%) |
| Female | 28 (53%) | 62 (32%) |
| Ethnicity1 (p = NS) | ||
| Caucasian | 44 (83%) | 134 (70%) |
| Non-Caucasian/unknown | 9 (17%) | 58 (30%) |
| Average age at enrollment (yrs) | 52.4 | 53.7 |
| IIT diagnosis | N/A | |
| Hashimoto's thyroiditis | 18 (34%) | |
| Graves' disease | 4 (8%) | |
| Other inflammatory thyroiditis2 | 31 (58%) | |
| HCV/HIV co-infection | 6 (11%) | 55 (29%) |
| HCV genotype3 (p = NS) | ||
| Genotype 1 | 74% | 76% |
| Genotype 2 | 17.3% | 11.3% |
| Genotype 3 | 0 | 7.8% |
| Genotype 4 | 8.7% | 4.9% |
| No. of IFNα treatment responders3 | 50% | 39% |
Note that since a significant number of self-reported ethnicities were unknown, we used the PCA to confirm and match ethnicities between the IIT patients and the HCV controls (see text)
These include destructive thyroiditis and primary myxedema
Percentage based on available clinical information (HCV genotype: n = 23 (IIT cases), n = 141 (HCV controls); No. of IFNα treatment responders: n = 24 (IIT cases), n = 145 (HCV controls))
2.2 Diagnosis and classification of IIT
The clinical spectrum of IIT includes autoimmune (Graves’ disease, Hashimoto’s thyroiditis, or presence of thyroid antibodies without clinical dysfunction) and non-autoimmune (destructive thyroiditis or primary myxedema) thyroiditis [3]. Patients with GD or HT were diagnosed based on previously established clinical and biochemical criteria [8]. Those patients that developed thyroid antibodies (TAb, anti-TPO and/or anti-Tg) without evidence of clinical disease as a result of IFNα therapy were classified as subclinical, autoimmune thyroiditis. Patients who developed clinical thyroid dysfunction, defined by initial hyperthyroid phase followed by gradual resolution (sometimes with hypothyroid phase) without the presence of thyroid Abs, were diagnosed as non-autoimmune inflammatory (destructive) thyroiditis. Individuals with clinical symptoms and biochemical evidence of hypothyroidism, without the presence of thyroid Abs, were classified as non-autoimmune hypothyroidism, or primary myxedema.
2.3 Laboratory assessment of HCV infection and thyroid function testing
The diagnosis of hepatitis C virus was confirmed and quantitatively assessed by the presence of HCV-RNA, with an assay detectable lower limit of 600 IU/ml (developed by Roche Diagnostics Corporation; adapted and optimized by the Molecular Diagnostics Laboratory at Mount Sinai Medical Center, New York, NY). HCV viral genotype was determined by RT-PCR and reverse hybridization (line probe) assay (Quest Diagnostics Incorporated, USA). Plasma levels of antibodies to TPO and Tg were measured using a radioimmunoassay kit (KRONUS, Star, ID, USA) according to manufacturer’s protocol. Briefly, patients’ plasma samples were incubated with I125 labeled TPO or Tg. Samples were centrifuged and the supernatant was removed by decanting or aspirating the liquid component. The amount of antibody present is directly proportional to the amount of radioactivity in the remaining pellet, which was read by a gamma counter. Calibrators were tested in the same assay with unknown patient plasma samples and antibody concentration was calculated using a standard concentration curve.
2.4 Genotyping
Genotyping of the cases and controls was performed at the Feinstein Institute for Medical Research, North Shore-Long Island Jewish Health System, Manhasset, NY. DNA samples were genotyped using the illumina infinium immunochip, a customized chip enriched for genes known to be associated with most of the major autoimmune and inflammatory disorders. Initiated by the Wellcome Trust Case-Control Consortium, the immunochip contains 9,021 genes covering 196,524 polymorphisms (718 insertion/deletions, 195,806 SNPs) with approximately 3,000 SNPs for each autoimmune disease [9]. PLINK (http://pngu.mgh.harvard.edu/purcell/plink/) was used to perform quality control (QC) and association analysis [10]. Firstly, SNPs with less than 95% SNP call rate were removed as part of quality control. Of 196,524 SNPs, 189,672 passed the initial QC step. From 189,672 markers, 182,832 SNPs, including 175,368 non-HLA SNPs, were successfully genotyped and used in association analysis comparing all IIT cases (n = 53) to HCV controls (n = 190) (2 HCV control samples were removed for ‘missingness’). 185,425 SNPs were used in the analysis comparing only Caucasian IIT cases (n = 44) to healthy, Caucasian controls (n = 851) (see below).
2.5 Principal Component Analysis (PCA)
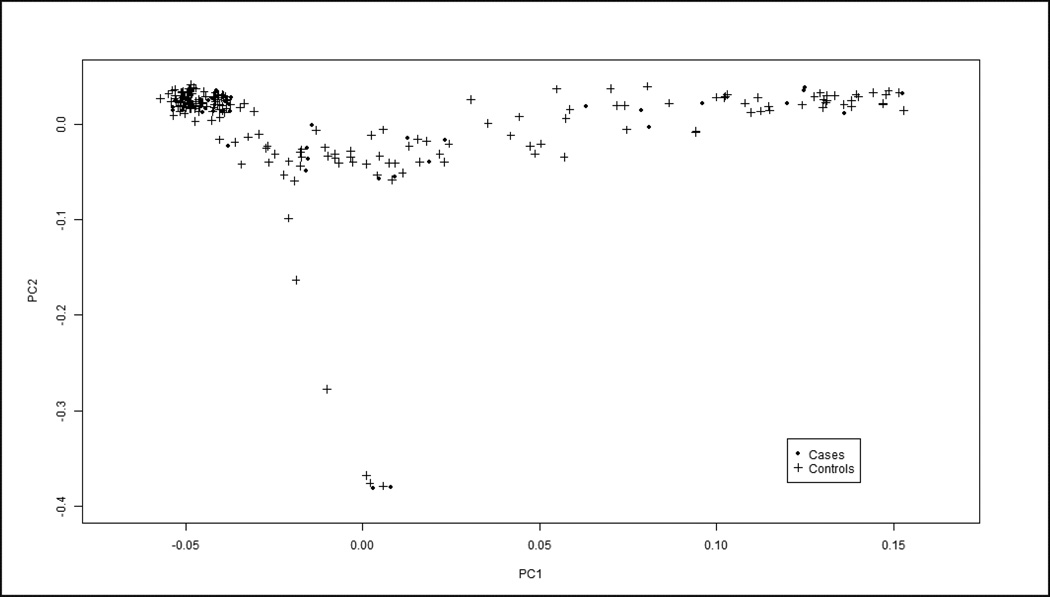
EIGENSOFT [11] was used to perform principal component analysis (PCA) on the data to investigate the sample distribution based on ethnicity and experimental batch and to ensure that our cases and controls were appropriately matched to avoid effects of population stratification. PCA is a method that analyzes genotypes of ancestry informative markers and aligns them with genotypes known to be prevalent among different ethnic groups [12]. Although our IIT cases (n = 53) and HCV controls (n = 190) were ethnically heterogeneous, PCA demonstrated our datasets to be appropriately matched with similar graphical distributions (figure 1) enabling us to compare the groups in the association studies. The PCA plot revealed that the majority of our IIT cases were Caucasian, while the remainder was comprised of other ethnicities, mostly Hispanic-Latino (table 1). Those samples falling in close proximity or inside the Caucasian cluster were selected for further analysis in which we compared only the Caucasian IIT cases (n = 44) to healthy, Caucasian controls (n = 851), representing a more ethnically homogeneous dataset. Using healthy, Caucasian controls also enabled us to use a much larger Caucasian control group than the subset of Caucasian HCV controls, thus increasing the power of our analysis. PCA confirmed the matching of our Caucasian IIT patients to healthy, Caucasian controls.
Figure 1.
Principal Component Analysis (PCA) was performed to extrapolate ethnicity based on genotype information of our study population. Although our two HCV datasets were ethnically heterogeneous, both IIT cases (dots) and HCV controls (crosses) showed similar distributions and while they did not cluster at any one corner of the graph (as would happen in an ethnically homogenous sample), their compatibility enabled us to compare them in our association analyses. Moreover, the PCA showed that the lambda score, or genomic inflation factor, was 1 which implies that the data is not confounded by effects of population stratification.
2.6 Association analysis
Association analysis in PLINK was performed by logistic regression using the first 2 principal components as covariates to adjust for the effect of population stratification. The Cochran-Armitage trend test was applied because it has the advantage that it is not influenced by the presence or absence of Hardy-Weinberg equilibrium for any tested SNP. For the two groups analyzed (the complete dataset and the Caucasian subset), the 100 SNPs (complete dataset) and 512 SNPs (Caucasian subset) that reached the highest statistical significance (see below) were selected and annotated with gene and autoimmune pathway information only if the SNP was within an interval that went from 40Kb upstream to 10Kb downstream of the pathway-associated gene. Ideally, when choosing the appropriate cutoff value for statistical significance when evaluating approximately 10,000 genes and 200,000 SNPs, a statistical threshold of a p-value less than 1 × 10−5, based on Bonferroni correction, would be the appropriate cut-off value for genome wide significance. However, few SNPs were identified above this genome wide threshold, most likely attributable to the relatively small number of individuals included in our study. Therefore, we used a significance threshold of a p value less than 1 × 10−3. This adjustment would increase the likelihood of identifying false positive associations. Since we analyzed approximately 10,000 genes we would expect to detect 10 signals by chance alone (false positives) when the threshold is set to a p value less than 1 × 10−3. To further narrow down the markers to only those that represent a true association with IIT, we filtered our associated genes to include only genes that contained multiple associated SNPs and applied the approach of genomic convergence (see below).
2.7 Genomic convergence analysis
To identify genes that are more likely to play an important role in susceptibility to IIT, we used the method of genomic convergence, an approach that has been successfully used in previous studies [13, 14]. The principle of the genomic convergence approach is to combine and align data obtained using several genetic techniques. Genes that are identified using more than one approach are more likely to influence the disease process of interest. Association analysis alone, even when performed on very large datasets, can yield false positive results. However, combining the association data with gene expression data is more likely to pinpoint genes that contribute to the etiology of disease. Therefore, to identify genes that are important to the etiology of IIT we merged the case-control association data with the transcriptome data looking for genes that were both associated with IIT and upregulated by IFNα. This allowed us to prioritize those genes that demonstrated significance both in the genetic analysis and in the expression analysis, thereby increasing the likelihood that these genes are involved in the pathogenesis of IIT.
2.8 Peripheral blood mononuclear cell isolation and transcriptome analysis
To determine the effects of IFNα on gene expression, peripheral blood mononuclear cells (PBMCs) were isolated from venous blood of a healthy donor by Ficoll-Paque density gradient centrifugation (GE Healthcare, Pittsburgh, PA) and incubated for 48 hours. After 48 hours incubation, one set of cells was exposed to IFNα (5,000U/ml) while the other set remained untreated, and then allowed to incubate for a further 24 hours. All experiments were performed in triplicates. RNA was extracted from PBMCs, using TRIzol (Invitrogen, Carlsbad, CA), and quantified. RNAseq was used to analyze the PBMC transcriptome from PBMCs incubated with and without IFNα treatment. Transcriptome analysis and Ingenuity Pathway Analysis were performed as previously described [6]. Results from gene expression profiling of PBMCs were then integrated with results of genetic association analysis (see above description of the genomic convergence analysis).
2.9 Haplotype analysis
Haplotype analysis was performed on genes that displayed the strongest association with IIT (by genomic convergence analysis) and contained multiple associated SNPs. Haploview program (http://broad.mit.edu/mpg/haploview/) was used to determine haplotype frequency and identify LD patterns based on the concept that alleles of markers within the same gene often demonstrate statistical dependence [15]. D’ values were used as a measure to quantify the degree of LD between 2 or more polymorphisms. The haplotype analysis enabled us to identify specific SNP haplotypes that may be associated with IIT.
2.10 Pathway analysis
We performed pathway analysis in order to define functional networks that may underlie IIT etiology and identify interactions between genes associated with IIT and genes upregulated by IFNα in PBMCs. Using Ingenuity Pathway Analysis (IPA) system version 8.6 (http://www.ingenuity.com/), analysis was performed on results of the genetic association analysis and on differentially expressed transcripts from the transcriptome analysis. The fold change of these genes were expressed as log2-ratio and subsequently used in IPA. The probability that a pathway was significantly enriched, when compared to the genome database, was based on the calculated p-value using Fisher’s exact test. The pathways that were up- or downregulated were then ranked accordingly. To identify the pathways that are likely to be most relevant in the pathogenesis of IIT, we merged IPA results from genetic association and PBMC transcriptome analyses.
3. RESULTS
3.1 Immunochip genetic association analysis
To identify susceptibility genes for IIT, 2 separate comparisons were performed:
All HCV patients who developed thyroiditis (IIT cases, n = 53) were compared to all patients with HCV who did not develop thyroiditis (HCV controls, n = 190), as they were shown to be closely matched by PCA (figure 1).
Caucasian HCV patients who developed thyroiditis (IITc cases, n = 44, based on PCA analysis, see 2. Patients and Methods) were compared to healthy, Caucasian controls (n = 851).
3.1.1 Association Analysis of all IIT cases compared to HCV controls
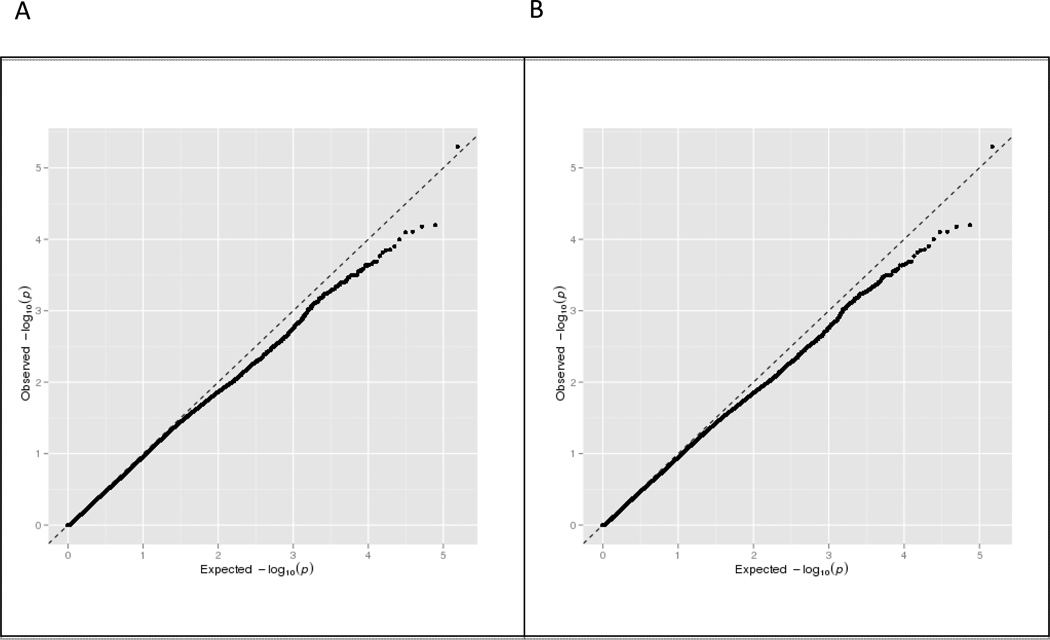
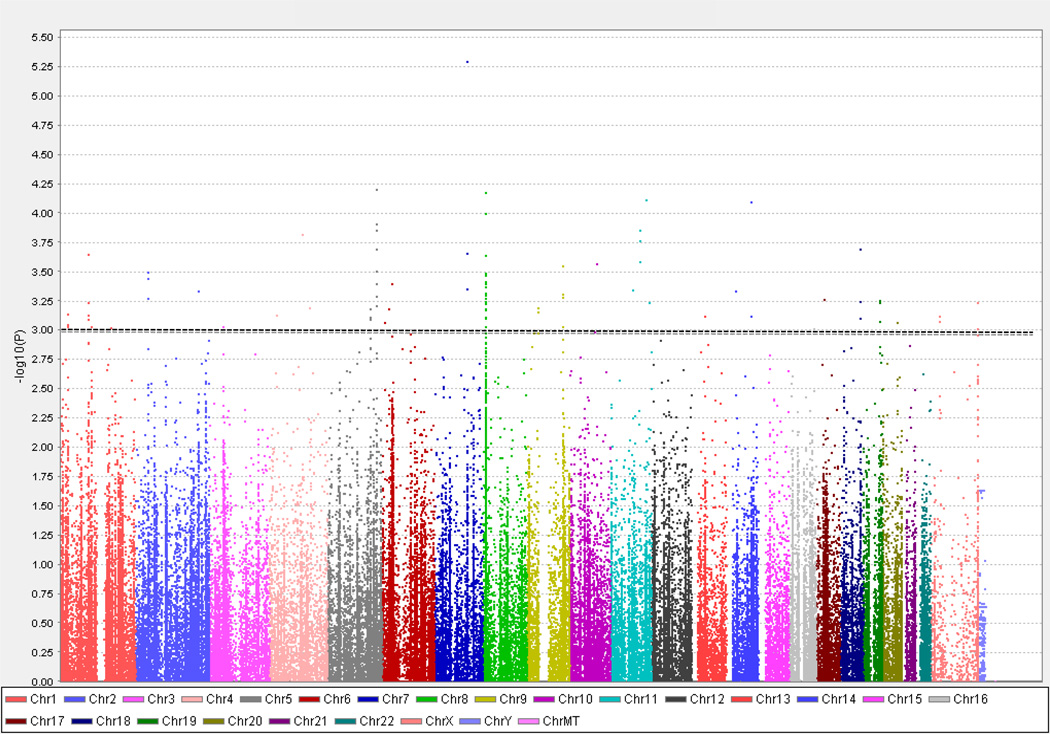
We identified 100 SNPs within 69 genes that had a p value less than 1 × 10−3. All genes within this significance threshold were comprised of non-HLA genes. The quantile-quantile (QQ) plot, which illustrates results of the association analyses for all the tested SNPs, suggested that there were multiple SNPs that were associated with IIT (figure 2). After removal of all HLA locus SNPs that were analyzed, no significant change was observed in the QQ plot distribution implying that non-HLA genes are likely to be associated with IIT. As described above (see 2. Patients and Methods), setting a higher significance threshold of p value less than 1 × 10−3 enabled the identification of several SNPs that may be associated with IIT (figure 3), although using this p-value threshold would be expected to yield on the average 10 false positive genes. However, by focusing on genes that contained multiple associated SNPs and using the genomic convergence method (see below), we identified genes that have a much greater likelihood of being associated with IIT.
Figure 2.
Quantile-quantile (QQ) plots for genetic association analysis comparing all IIT cases (n = 53) to HCV controls (n = 190) that passed quality controls. The expected values (x-axis), from a theoretical x2 distribution, are plotted against the observed p-values (y-axis) for each SNP. If no SNP showed association all points would remain on or very near to the baseline (dotted line) which represents the null distribution. Panel A shows results of association analysis for all SNPs that were tested on the immunochip. The QQ plot demonstrates deviation from the null towards the upper extreme end of the line implying that multiple SNPs show association with IIT when compared to controls. Panel B shows QQ plot after removal of all SNPs in the HLA region. After removing the HLA SNPs the QQ plot still demonstrates deviation from the null towards the upper extreme end of the line suggesting that non-HLA genes are also likely to be associated with IIT.
Figure 3.
Manhattan plot of immunochip association analysis data comparing all IIT cases (n = 53) to HCV controls (n = 190). 182,832 SNPs that passed the quality control testing are sorted by chromosomal location [x- axis] and are plotted against the −log10(p value) [y-axis], with the height of each point corresponding to the strength of association with disease. The dotted line indicates a p-value < 1 × 10−3 (the threshold we chose as our cutoff; see text) and the SNPs that show association with IIT with p < 1 × 10−3 are displayed above this line.
3.1.2 Association Analysis comparing Caucasian IIT cases compared to Caucasian controls
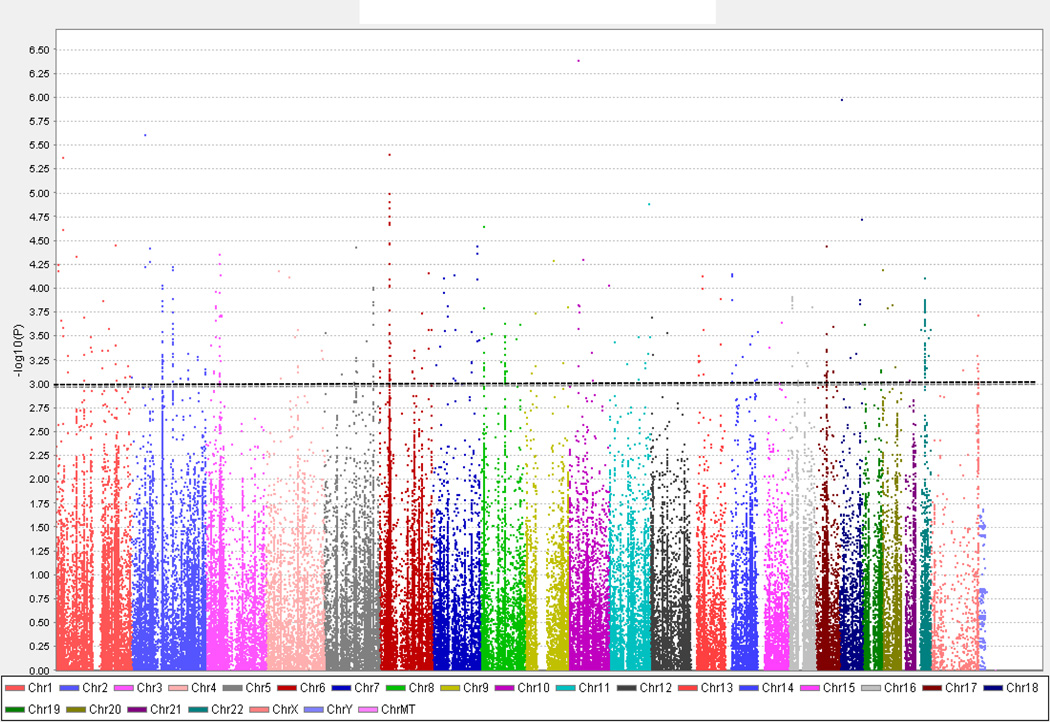
Based on a cutoff p-value less than 1 × 10−3, we identified 512 SNPs in 475 genes that showed association with IIT in Caucasians. 11 of the 475 genes that were associated belong to the HLA gene family (4 MHC Class I and 7 MHC Class II genes), while the remaining 464 were comprised of non-HLA genes. In this Caucasian-only dataset analysis, strong association with HLA (MHC Class I) was identified (HLA-A, p = 2.9 × 10−4; HLA-B, p = 3.0 × 10−4; HLA-C, p = 9.3 × 10−5, HLA-G, p = 3.3 × 10−5) and each HLA type contained multiple associated SNPs (figure 4). The Q-Q plot of this association analysis (data not shown) showed similar distribution to the analysis described above, comparing IIT cases to HCV controls, and suggested that genes outside the HLA complex locus also likely play an important role in the etiology of IIT.
Figure 4.
Manhattan plot of immunochip association analysis data comparing only Caucasian IIT cases (n = 44) to healthy, Caucasian controls not infected with HCV (n = 851). 185,425 SNPs that passed the quality control testing are sorted by chromosomal location [x- axis] and are plotted against the −log10(p value) [y-axis], with the height of each point corresponding to the strength of association with disease. The dotted line indicates a p-value < 1 × 10−3 (the threshold we chose as our cutoff; see text), and the SNPs that show association with IIT with p < 1 × 10−3 are displayed above this line.
3.2 Transcriptome analysis
We hypothesized that genes predisposing to IIT are regulated by IFNα. Therefore, we performed a transcriptome analysis to determine the genes specifically up- or down-regulated by IFNα and then used genomic convergence (see below) to integrate the transcriptome data with the genetic association data. For the transcriptome analysis total RNA was extracted from PBMCs incubated with or without IFNα for 24 hours, and RNAseq was used to analyze the transcriptome changes induced by IFNα (see 2. Patients and Methods). We identified 1,212 genes that were differentially expressed by IFNα (539 up-regulated and 673 down-regulated), with p-value < 5 × 10−2 and fold change > 1.5. Using Gene Ontology (GO), the biological processes determined to be most over-represented included immune response (GO:0006955), response to wounding (GO:0009611), and defense response (GO:0006952) (p < 6.6 × 10−13 and enrichment > 11.3%). Most notably, the genes involved in these pathways, with considerable overlap among the various GO subcategories, included IFN-inducible genes (IFIT family, SP100/110/140 gene cluster), chemokine and chemokine receptors (CXCL10, CCR1), interleukins (IL8, IL1RN), toll-like receptors (TLR3), and complement component and subcomponents (C1S, C1R, SERPING1).
3.3 Genomic convergence analysis
To narrow down the putative genes that may be contributing to the etiology of IIT, we used the genomic convergence method (see 2. Patients and Methods) integrating results of genetic association with PBMC transcriptome analyses. To reduce the likelihood of excluding associated genes that may be potentially important in IIT etiology, for the convergence analysis we used a less stringent significance threshold of p-value less than 5 × 10−3. Our genomic convergence analysis demonstrated 54 genes that were found to be both genetically associated (p < 0.005), in either of the 2 datasets, and differentially upregulated in PBMCs exposed to IFNα (p < 0.01, q < 0.001, > 1.5 fold change), while 44 genes were downregulated (data not shown). Of the 54 genes that were both associated with IIT and upregulated by IFNα in PBMCs, we narrowed down the list further by including only genes that contained at least 2 associated SNPs (see supplementary information); this resulted in 21 genes that were both associated with IIT and upregulated by IFNα. Of the 21 genes, only 5 were associated with both datasets comparing all IIT cases to HCV controls and comparing Caucasian IIT cases to healthy Caucasian controls. These included SP100/110/140, HLA (MHC Class I), TAP1, LILRA3, and INS genes. Interestingly, 4 of these 5 genes (SP100/110/140 complex, HLA, TAP1, LILRA3) are immune response genes. While its role in the development of IIT remains unclear, insulin (INS) has been shown to be expressed in various extrapancreatic tissues [16, 17], including immune organs such as the spleen.
Our analysis filtered genes that had multiple associated SNPs regardless of whether the SNPs were in linkage disequilibrium (LD) or not. However, SNPs within a gene that are associated with IIT and are not in LD represent independent associations with the disease, thereby increasing the likelihood that this gene influences the etiology of the disease. Therefore, we examined the associated SNPs within the 5 genes that were identified by genomic convergence for LD patterns using Haploview (http://broad.mit.edu/mpg/haploview/). The LD analysis demonstrated that in 3 of the associated genes at least 2 SNPs were not in LD - SP100/110/140, HLA (MHC Class I), TAP1, making these three genes very likely to play a major role in the etiology of IIT.
3.4 Haplotype analysis of putative IIT genes
Five IIT susceptibility genes (SP100/110/140 complex, HLA, TAP1, LILRA3, and INS) were identified based on genomic convergence and on having more than one associated SNP (see above). Therefore, we next performed a haplotype analysis on them to examine whether specific SNP haplotypes within these 5 genes were associated with IIT. Haplotype analysis was performed using Haploview. As before, we compared both datasets, i.e., all IIT cases to HCV controls and Caucasian IIT cases to healthy, Caucasian controls. All of the putative IIT susceptibility genes had at least one unique haplotype (table 2) that showed statistically significant association with IIT (p < 0.005).
Table 2.
Haplotype analysis of putative IIT susceptibility genes: genes with multiple-associated SNPs that were found to be both associated with interferon-induced thyroiditis and upregulated by IFNα in PBMCs (see text).
| Gene | Chr | IIT Cases vs HCV Controls | IIT Cases vs Healthy Controls Gene (Caucasians) | ||
|---|---|---|---|---|---|
| Haplotypes | p-value | Haplotypes | p-value | ||
| SP100/110/140 | 2q37.1 | rs55796826-rs4972946-rs17330341-rs1972707 (1kb) | 6.4 × 10−5 | rs59559342-rs2033157-rs13007094-rs73106397 (1kb) | 2 × 10−4 |
| rs57841972-rs4972946-rs17330341-rs6743287-rs1972707 (1kb) | 1 × 10−4 | rs13007094-rs73106397-rs116469870 (2kb) | 7 × 10−4 | ||
| rs115474882-rs78723887-rs56317957-rs76029471 (1kb) | 3 × 10−4 | rs2114590-rs34855578-rs13395371-rs9989735-rs9989792 (1kb) | 1.6 × 10−3 | ||
| rs933957-rs6731421-rs12615773, rs56017039-rs6713654-rs2396744 (3kb) | 1.5 × 10−3 | rs3813464-rs4973304-rs10498245-rs10200874 (1kb) | 3.5 × 10−3 | ||
| HLA-A/B/C | 6p21.3 | HLA-C: rs9264921-rs364415 (<1kb) | 1.3 × 10−3 | HLA-A rs1611493-rs386014-rs9259852 (10kb) | 9 × 10−4 |
| HLA-B rs9501587-rs9266636-rs9266638, rs2244020-rs2507980 (<1kb) | 6.9 × 10−5 | ||||
| HLA-C rs12199223-rs3134745-rs3132486-rs3130696-rs9264731 (1kb) | 2.3 × 10−6 | ||||
| rs2596503-2596501 (<1kb); rs2523554-rs2596574-rs9266381-rs6457401 (3kb) | 4.4 × 10−4 | ||||
| TAP1 | 6p21.3 | rs2239701-rs2071465-rs4148870-rs2071552 (1kb) | 3.2 × 10−3 | rs6934645-rs12206377-rs260547 (3kb) | 5.9 × 10−5 |
| LILRA3 | 19q13.4 | rs75085835-rs117299550-rs7257187 (2kb) | 4.1 × 10−3 | rs36012517-rs73061042-rs11084333-rs73061043 (1kb) | 7 × 10−4 |
| INS | 11p15.5 | rs7114836-rs7126857-rs7127162(rs71472148)-rs10743180 (<1kb) | 1.3 × 10−3 | rs7126857-rs7127162 (rs71472148)-rs10743180-rs10840588 (<1kb) | 4.5 × 10−3 |
3.5 Pathway analysis of genetically associated SNPs
To provide insight into the mechanisms by which IIT may develop and to better define the networks through which these genes may interact, pathway analysis was performed on the 2 datasets used in the genetic association analyses (see 2. Patients and Methods). Using Ingenuity Pathway Analysis system (IPA), several pathways were found to be significantly associated with IIT (p < 0.05, > 1.3 fold change). As expected, the interferon signaling pathway was associated in both analyses (IIT cases compared to both HCV controls and to healthy, Caucasian controls). IPA also revealed strong association with the following pathways: antigen presentation pathway, NFκb signaling, IL-10 signaling, and apoptosis-related pathways. Several other pathways were specifically associated in the Caucasian analysis (Caucasian IIT cases compared to healthy Caucasian controls) including IL-6 signaling, pattern recognition receptor signaling, and complement system related pathways. Interestingly, our previous work has shown that the interferon signaling, antigen presentation, pattern recognition receptor signaling, and complement pathways were also upregulated in both the thyroids of IFNα transgenic mice and in human thyroid follicular cells exposed to IFNα for 24 hours; IL-6 signaling was increased only in the transgenic mice [6]. Taken together these data support them as major pathways contributing to the pathogenesis of IIT.
3.6 Pathway analysis of differentially expressed genes
Ingenuity Pathway Analysis (IPA) was performed to identify the pathways significantly upregulated in PBMCs exposed to IFNα (p < 0.05, > 1.3 fold change). As expected, interferon signaling pathway was found to be most highly upregulated (−log (p-value) = 7.49). Other upregulated pathways included pattern recognition receptors, retinoic acid mediated apoptosis signaling, and complement system. To a lesser degree, there was also upregulation of NFκb signaling, toll-like receptor signaling, IL-10 signaling, and IL-6 signaling. Furthermore, in our previous study, an apoptosis-related pathway (i.e., retinoic acid mediated apoptosis signaling) was also demonstrated to be increased specifically in human thyroid follicular cells following exposure to IFNα, a finding not observed in the thyroids of transgenic mice [6].
3.7 Convergence analysis of pathway data
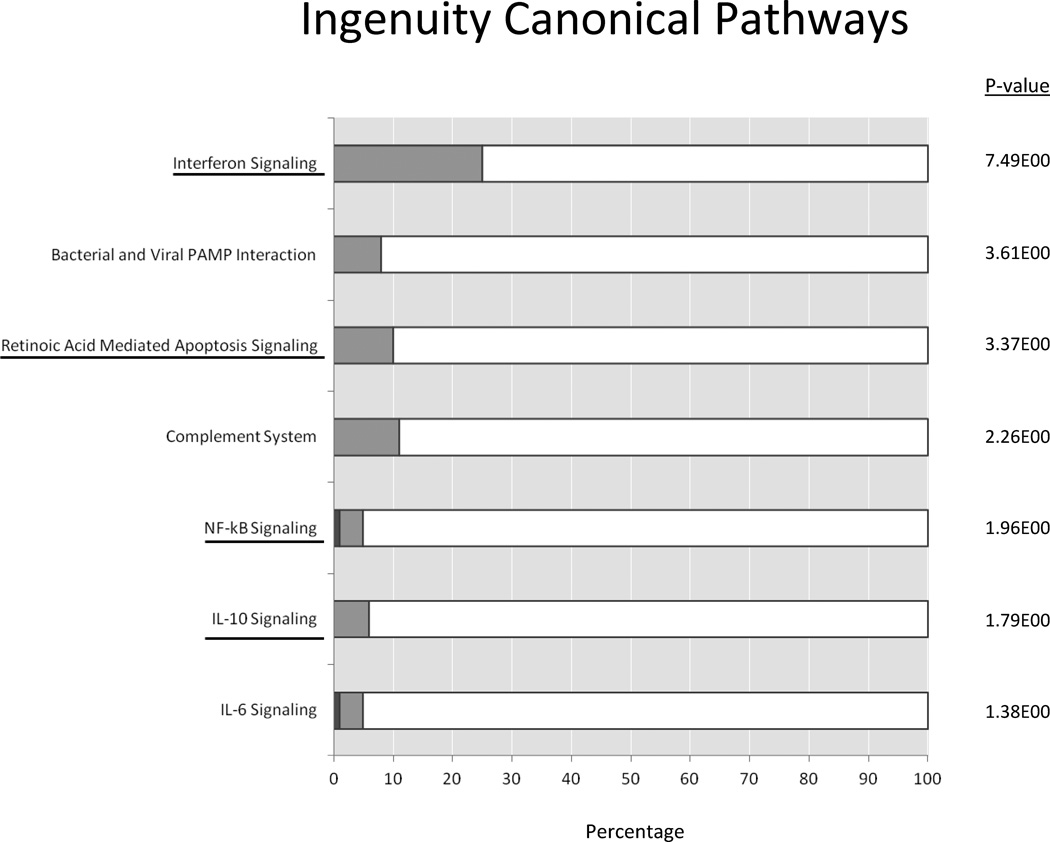
To identify the most important pathways involved in the etiology of IIT we performed genomic convergence of the pathway analyses of associated genes and differentially regulated genes. Integration of the two pathway analyses revealed that 7 groups of pathways were both genetically associated with IIT and significantly upregulated in PBMCs exposed to IFNα. These 7 pathways included interferon signaling, pattern recognition receptor related, apoptosis, complement, NFκB signaling, IL-10 signaling, and IL-6 signaling pathways. Figure 5 depicts one representative pathway in each group identified by genomic convergence that was upregulated by IFNα in PBMCs. Of these 7 pathways, four (figure 5, underlined) were associated with IIT in both datasets compared in the genetic association analyses and were also found to be upregulated in PBMCs exposed to IFNα. These included interferon signaling, apoptosis, NFκb signaling, and IL-10 signaling. Thus, these are likely the most important pathways involved in the predisposition to and pathogenesis of IIT.
Figure 5.
Ingenuity pathway analysis (IPA) of PBMC RNAseq expression data. Shown are 7 pathways that belonged to groups of pathways identified by genomic convergence to be both genetically associated with IIT and significantly upregulated by IFNα in PBMCs. The percentage of genes that were upregulated are shown in light gray and genes that were downregulated are shown in dark gray, with the −log(p-value) shown at the right side of each bar. Interferon signaling pathway was found to be the most significantly upregulated. Other groups of pathways found to be associated with IIT and upregulated by IFNα in PBMCs included pattern recognition receptor related, apoptosis, complement, NFκB signaling, IL-10 signaling, and IL-6 signaling pathways. Four of these groups of pathways were found to be strongly associated with IIT in both datasets (all IIT cases vs. HCV controls and Caucasian IIT cases vs. healthy, Caucasian controls). These include interferon signaling, apoptosis related pathways, NFκB signaling, and IL-10 signaling (underlined).
4. DISCUSSION
Autoimmunity is thought to develop in certain, predisposed individuals through a complex interaction between environmental and genetic factors, likely involving epigenetic modulation. While recognized to be an effective therapeutic agent, IFNα is one causal factor that likely offsets the fine balance of the normal immune state to trigger autoimmune responses through upregulation of certain genes and signaling pathways. IFNα has been associated with the initiation and/or exacerbation of a range of autoimmune diseases including Systemic Lupus Erythematosus and type 1 diabetes [18]. This unintended complication of IFNα therapy therefore offers a human model for studying the role of IFNα in the etiology of autoimmunity [19]. In this manuscript we used IIT as a model to study the causal relationship between IFNα and autoimmunity. It is unclear why only about 20% of individuals treated with IFNα for HCV develop clinical IIT while 80% of patients do not, but genetic susceptibility seems a plausible explanation. Indeed, it has been suggested that immune regulation and responsiveness are quantitative genetic traits [20]. Thus, we hypothesized that in individuals harboring susceptibility genes for autoimmune thyroiditis, exposure to HCV infection and IFNα therapy may trigger thyroiditis (reviewed in [3, 21]).
In this manuscript we dissected the genetic predisposition to IIT using a genomic convergence approach, and identified several immune-regulatory genes and pathways that were associated with IIT. Specifically, loci with at least 2 associated SNPs not in LD that showed both genetic association and upregulation by IFNα were more likely to be involved in IIT etiology. These genes include unique, non-MHC genes, SP100/110/140 gene cluster (2q37.1), and MHC genes previously shown to be associated with AITD, HLA (6p21.3) and TAP1 (6p21.3). The HLA region contains numerous immune response genes that are associated with various autoimmune diseases including AITD [22]. In addition, several other non-MHC loci have been associated with AITD including 1) immune-regulatory genes (CD25, CD40, CTLA4, FOXP3, and PTPN22) and 2) thyroid-specific genes (Thyroglobulin (Tg) and TSHR). Results of our genetic analysis however showed weak association of IIT with the known AITD susceptibility genes except HLA (data not shown) and when we applied the genomic convergence approach these genes were not supported as putative IIT susceptibility genes. These data, therefore, raise the question of whether IIT represents a unique disease entity or a form of AITD that is triggered by IFNα.
The results of our analysis provide new insight into the genetic etiology of IIT. Interestingly, the non-MHC genes that demonstrated strong association with IIT have not been previously shown to be associated with AITD, but have been linked to other autoimmune conditions. This suggests that, with the exception of HLA, the genetic susceptibility to IIT is distinct from that of AITD. Among the non-MHC genes that were highly prioritized by our convergence approach is the SP100/110/140 gene cluster. SP100 is a promyelocytic leukemia (PML) nuclear body (NB) related protein that has been strongly implicated as a target antigen in patients with primary biliary cirrhosis (PBC); antibodies to SP100 occur in up to 40% of affected individuals (reviewed in [23]). Recently, SP140 was also identified as a highly specific autoantigen in patients with PBC [23]. Although, the exact function is not yet clearly defined, studies suggest that PML NB related proteins (SP100/110/140, PML) may be involved in the regulation of gene transcription and in cell growth, differentiation, apoptosis, and anti-viral defense [24]. Indeed, SP100/110/140 genes are known to be inducible by interferons [24, 25].
Not unexpectedly the MHC gene complex showed the strongest association with IIT. As previously reported, MHC Class I genes were also upregulated by IFNα suggesting a major role for MHC Class I genes in the etiology of IIT (reviewed in [3]). An earlier study performed in Japan examined the relationship between HLA antigens and subsequent development of thyroid disease in HCV patients treated with IFNα [26]. They showed an increased frequency of HLA-A2 in individuals who developed AITD during or after IFNα therapy. The TAP1 gene, located within the MHC gene complex, also showed association with IIT. TAP1, or transporter associated with antigen processing, is a multi-membrane spanning protein that allows for efficient cytosolic peptide loading onto MHC Class I molecules [27]. Indeed, polymorphisms of the TAP1 gene have shown association with other autoimmune diseases including type 1 diabetes [28] and psoriasis [29]. The association of both MHC class I and TAP1 genes with IIT suggests that peptide presentation by the MHC class I pathway plays a major role in the etiology of IIT and possible other autoimmune diseases. Interestingly, TAP1 has also been associated with rheumatoid arthritis [30] and Graves’ disease [27, 31], which are primarily MHC Class II-mediated diseases. Although the exact mechanisms are unknown, it is possible that TAP1 may also be responsible for presentation of cytosolic peptides to MHC Class II-restricted T cells [32] or may be related to defective MHC Class I expression [30]. In contrast to the evidence supporting MHC Class I-related genes in the development of IIT, MHC Class II genes specifically DRβ1-Arg74 are the primary genes predisposing to GD and HT [33, 34]. Sequencing of the HLA-DRβ1 locus enabled us to identify these amino acid variants (i.e., Arg74/Glu74) located in pocket 4 of the HLA-DR peptide binding cleft and presented a mechanism by which pathogenic peptides (i.e., Tg-derived) can bind, thereby conferring susceptibility to AITD. Previously, we have shown that an interaction between DRβ1-Arg74 and specific Tg variants resulted in an increased risk of developing GD, with a higher combined OR than would be produced by either gene variant alone [34].
Interferon induced thyroiditis (IIT) provides an ideal model for studying the interactions between genetic and environmental factors to trigger autoimmunity. Previous work by our group has shown direct effects of both HCV and IFNα on thyroid cells [6, 21, 35]. Here we show, for the first time, that these effects most likely occur in genetically predisposed individuals. In addition to immune genes (i.e., SP100/110/140, MHC Class I, TAP1), we identified 4 pathways that were both upregulated by IFNα and genetically associated with IIT: Interferon signaling, apoptosis, NFκb, and IL-10 signaling pathways (figure 5). These genes and pathways may also play a role in the pathogenesis of other autoimmune diseases and future studies will be needed to confirm our results. Furthermore, we previously identified several pathways that were found to be upregulated in both the thyroids of IFNα transgenic mice and human thyrocytes exposed to IFNα [6], which we have now similarly shown to be genetically associated with IIT in individuals with chronic HCV infection. More specifically, pathway analyses performed on both associated SNPs data and IFNα inducible transcriptome data demonstrated upregulation of apoptotic pathways that were also uniquely increased in human thyrocytes following exposure to IFNα [6]. Indeed, there is a plethora of data describing the role of apoptosis in the pathogenesis of thyroid autoimmunity [36] and autoimmunity in general [37]. These results suggest a predominant mechanism by which thyrocyte destruction occurs in IIT, possibly mediated by SP100/110/140 (see above), favoring the development of HT that represents the most common autoimmune, clinical manifestation of IIT, as also confirmed in our study (table 1). Thus, it is possible that in genetically susceptible individuals HCV activates this gene to trigger specific cellular responses, such as apoptosis, which are further exacerbated by IFNα. Although the exact nature through which these genes, and respective pathways, interact with environmental factors to induce autoimmunity is still uncertain, a possible mechanism may be through epigenetics. Epigenetic effects generally include any non-DNA sequence-encoded effects on gene expression that are long lasting, or mitotically stable. This may occur through alterations in DNA methylation, histone modifications, and microRNA activity [38]. Indeed, IFNα was shown to induce an increase in transcription of Tg through histone modifications [39]. In addition to increased expression of autoantigens, such as Tg, IFNα can lead to T-cell activation that, in the setting of cytokine response, can trigger autoimmunity. There is now abundant evidence that epigenetics play a major role in the development of various complex diseases, including type 1 diabetes and rheumatoid arthritis (reviewed in [38]). Epigenetic effects, therefore, may explain how IFNα triggers thyroid autoimmunity in genetically predisposed individuals and/or in the setting of viral infections where there is an increased production of IFNα.
The main weakness of our study is the small size of our cohort of IIT patients (n = 53). This small sample size was, in part, due to the rigid inclusion criteria of our study. In view of our small sample size it is possible that some of the gene-loci we found to be associated with IIT may be false positives. While no statistical test can differentiate false positive associations from true associations even when sample size is large, one approach to increase the likelihood that an association is a true positive is to use genomic convergence. Our analysis strongly suggested that immune genes and pathways, in particular HLA and apoptosis related genes, play a major role in the genetic susceptibility to IIT. How these susceptibility genes interact with HCV and IFNα to trigger thyroiditis is still not known, and will require further investigation.
Supplementary Material
Research Highlights.
Thyroiditis is a major complication of interferon-alpha therapy
Our aim was to map genes predisposing to thyroiditis in IFNα treated HCV patients
Using a genomic convergence approach we identified 3 putative gene-loci
Our data suggest a key role for immune-regulatory and apoptosis related mechanisms
Acknowledgements
This work was supported in part by a VA Merit award 1I01BX002031 from the Department of Veterans Affairs, and by grants DK61659, and DK073681 from NIDDK (to YT). We thank Dr. David A. Greenberg (Battelle Center for Mathematical Medicine and Department of Neurology, Nationwide Children’s Hospital, Columbus, Ohio) for his suggestions and expert review of this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Hall JC, Rosen A. Type 1 interferons: crucial participants in disease amplification in autoimmunity. Nat Rev Rheumatol. 2010;6:40–49. doi: 10.1038/nrrheum.2009.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tomer Y, Blackard JT, Akeno N. Interferon alpha treatment and thyroid dysfunction. Endocrinol Metab Clin North Am. 2007;36(4):1051–1066. doi: 10.1016/j.ecl.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mandac JC, Chaudhry S, Sherman KE, Tomer Y. The clinical and physiological spectrum of interferon-alpha induced thyroiditis: toward a new classification. Hepatology. 2006;43(4):661–672. doi: 10.1002/hep.21146. [DOI] [PubMed] [Google Scholar]
- 4.Mammen JS, Ghazarian SR, Pulkstenis E, Subramanian GM, Rosen A, Ladenson PW. Phenotypes of interferon-a-induced thyroid dysfunction among patients treated for hepatitis C are associated with pretreatment serum TSH and female sex. J Clin Endocrinol Metab. 2012;97(9):3270–3276. doi: 10.1210/jc.2012-1026. [DOI] [PubMed] [Google Scholar]
- 5.Russo MW, Fried MW. Side effects of therapy for chronic hepatitis C. Gastroenterology. 2003;124(6):1711–1719. doi: 10.1016/s0016-5085(03)00394-9. [DOI] [PubMed] [Google Scholar]
- 6.Akeno N, Smith EP, Stefan M, Huber AK, Zhang W, Keddache M, et al. IFN-a mediates the development of autoimmunity both by direct tissue toxicity and through immune cell recruitment mechanisms. J Immunol. 2011;186(8):4693–4706. doi: 10.4049/jimmunol.1002631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roti E, Minelli R, Giuberti T, Marchelli S, Schianchi C, Gardini E, et al. Multiple changes in thyroid function in patients with chronic active HCV hepatitis treated with recombinant interferon-alpha. Am J Med. 1996;101(5):482–487. doi: 10.1016/s0002-9343(96)00259-8. [DOI] [PubMed] [Google Scholar]
- 8.Tomer Y, Hasham A, Davies TF, Stefan M, Concepcion E, Keddache M, et al. Fine mapping of loci linked to autoimmune thyroid disease identifies novel susceptibility genes. J Clin Endocrinol Metab. 2013;98(1):E144–E152. doi: 10.1210/jc.2012-2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cortes A, Brown MA. Promise and pitfalls of the immunochip. Arth Res Ther. 2011;13(1):101. doi: 10.1186/ar3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2(12):e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 13.Li YJ, Oliveira SA, Xu P, Martin ER, Stenger JE, Scherzer CR, et al. Glutathione S-transferase omega-1 modifies age-at-onset of Alzheimer disease and Parkinson disease. Hum Mol Genet. 2003;12(24):3259–3267. doi: 10.1093/hmg/ddg357. [DOI] [PubMed] [Google Scholar]
- 14.Krug T, Gabriel JP, Taipa R, Fonseca BV, Domingues-Montanari S, Fernandez-Cadenas I, et al. TTC7B emerges as a novel risk factor for ischemic stroke through the convergence of several genome-wide approaches. J Cereb Blood Flow Metab. 2012;32(6):1061–1072. doi: 10.1038/jcbfm.2012.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao H, Pfeiffer R, Gail MH. Haplotype analysis in population genetics and association studies. Pharmacogenomics. 2003;4(2):171–178. doi: 10.1517/phgs.4.2.171.22636. [DOI] [PubMed] [Google Scholar]
- 16.Rosenzweig JL, Havrankova J, Lesniak MA, Brownstein M, Roth J. Insulin is ubiquitous in extrapancreatic tissues of rats and humans. Proc Natl Acad Sci USA. 1980;77(1):572–576. doi: 10.1073/pnas.77.1.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kojima H, Fujimiya M, Matsumura K, Nakahara T, Hara M, Chan L. Extrapancreatic insulin-producing cells in multiple organs in diabetes. Proc Natl Acad Sci USA. 2004;101(8):2458–2463. doi: 10.1073/pnas.0308690100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burdick LM, Somani N, Somani AK. Type 1 IFNs and their role in the development of autoimmune diseases. Expert Opin Drug Saf. 2009;8(4):459–472. doi: 10.1517/14740330903066726. [DOI] [PubMed] [Google Scholar]
- 19.Oppenheim Y, Ban Y, Tomer Y. Interferon induced autoimmune thyroid disease (AITD): a model for human autoimmunity. Autoimmun Rev. 2004;3(5):388–393. doi: 10.1016/j.autrev.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 20.Cho JH, Gregersen PK. Genomics and the multifactorial nature of human autoimmune disease. N Engl J Med. 2011;365(17):1612–1623. doi: 10.1056/NEJMra1100030. [DOI] [PubMed] [Google Scholar]
- 21.Akeno N, Blackard JT, Tomer Y. HCV E2 protein binds directly to thyroid cells and induces IL-8 production : a new mechanism for HCV induced thyroid autoimmunity. J Autoimmun. 2008;31(4):339–344. doi: 10.1016/j.jaut.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Invernizzi P, Gershwin ME. The genetics of human autoimmune disease. J Autoimmun. 2009;33(3–4):290–299. doi: 10.1016/j.jaut.2009.07.008. [DOI] [PubMed] [Google Scholar]
- 23.Granito A, Yang WH, Muratori L, Lim MJ, Nakajima A, Ferri S, et al. PML nuclear body component Sp140 is a novel autoantigen in primary biliary cirrhosis. Am J Gastrotenterol. 2010;105(1):125–131. doi: 10.1038/ajg.2009.596. [DOI] [PubMed] [Google Scholar]
- 24.Regad T, Chelbi-Alix MK. Role and fate of PML nuclear bodies in response to interferon and viral infections. Oncogene. 2001;20(49):7274–7286. doi: 10.1038/sj.onc.1204854. [DOI] [PubMed] [Google Scholar]
- 25.Grotzinger T, Sternsdorf T, Jensen K, Will H. Interferon-modulated expression of genes encoding the nuclear-dot-associated proteins SP100 and promyelocytic leukemia protein (PML) Eur J Biochem. 1996;238(2):554–560. doi: 10.1111/j.1432-1033.1996.0554z.x. [DOI] [PubMed] [Google Scholar]
- 26.Kakizaki S, Takagi H, Murakami M, Takayama H, Mori M. HLA antigens in patients with interferon-α-induced autoimmune thyroid disorders in chronic hepatitis C. J Hepatol. 1999;30(5):794–800. doi: 10.1016/s0168-8278(99)80131-7. [DOI] [PubMed] [Google Scholar]
- 27.Chen RH, Chen WC, Chen CC, Tsai CH, Tsai FJ. Association between the TAP1 gene codon 637 polymorphism and Graves’ disease. Endocrine. 2004;25(2):137–140. doi: 10.1385/ENDO:25:2:137. [DOI] [PubMed] [Google Scholar]
- 28.Yan G, Fu Y, Faustman DL. Reduced expression of Tap1 and Lmp2 antigen-processing genes in the nonobese diabetic (NOD) mouse due to a mutation in their shared bidirectional promoter. J Immunol. 1997;159(6):3068–3080. [PubMed] [Google Scholar]
- 29.Hohler T, Weinmann A, Schneider PM, Rittner C, Schopf RE, Knop J, et al. TAP-polymorphisms in juvenile onset psoriasis and psoriatic arthritis. Hum Immunol. 1996;51(1):49–54. doi: 10.1016/s0198-8859(96)00156-5. [DOI] [PubMed] [Google Scholar]
- 30.Zhang SL, Chabod J, Penfornis A, Reviron D, Tiberghien P, Wendling D, et al. TAP1 and TAP2 gene polymorphism in rheumatoid arthritis in a population in eastern France. Eur J Immunogenet. 2002;29(3):241–249. doi: 10.1046/j.1365-2370.2002.00307.x. [DOI] [PubMed] [Google Scholar]
- 31.Rau H, Nicolay A, Usadel KH, Finke R, Donner H, Walfish PG, et al. Polymorphisms of TAP1 and TAP2 genes in Graves’ disease. Tissue Antigens. 1997;49(1):16–22. doi: 10.1111/j.1399-0039.1997.tb02704.x. [DOI] [PubMed] [Google Scholar]
- 32.Malnati MS, Marti M, LaVaute T, Jaraquemada D, Biddison W, DeMars R, et al. Processing pathways for presentation of cytosolic antigen to MHC Class II-restricted T cells. Nature. 1992;357(6380):702–704. doi: 10.1038/357702a0. [DOI] [PubMed] [Google Scholar]
- 33.Jacobson EM, Huber A, Tomer Y. The HLA gene complex in thyroid autoimmunity: from epidemiology to etiology. J Autoimmun. 2008;30(1–2):58–62. doi: 10.1016/j.jaut.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tomer Y, Huber A. The etiology of autoimmune thyroid disease: a story of genes and environment. J Autoimmun. 2009;32(3–4):231–239. doi: 10.1016/j.jaut.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blackard J, Kong L, Huber A, Tomer Y. Hepatitis C virus infection of a thyroid cell line: Implications for pathogenesis of HCV and thyroiditis. Thyroid. 2012 Dec 23; doi: 10.1089/thy.2012.0507. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang SH, Baker JR. The role of apoptosis in thyroid autoimmunity. Thyroid. 2007;17(10):975–979. doi: 10.1089/thy.2007.0208. [DOI] [PubMed] [Google Scholar]
- 37.Lleo A, Selmi C, Invernizzi P, Podda M, Gershwin ME. The consequences of apoptosis in autoimmunity. J Autoimmun. 2008;31(3):257–262. doi: 10.1016/j.jaut.2008.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu Q. The critical importance of epigenetics in autoimmunity. J Autoimmun. 2013 Jan 31; doi: 10.1016/j.jaut.2013.01.010. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 39.Stefan M, Jacobson EM, Huber AK, Greenberg DA, Li CW, Skrabanek L, et al. Novel variant of thyroglobulin promoter triggers thyroid autoimmunity through an epigenetic interferon alpha-modulated mechanism. J Biol Chem. 2011;286(36):31168–31179. doi: 10.1074/jbc.M111.247510. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.