Abstract
Arsenic (As) is a ubiquitous element that is widespread in the environment and causes numerous health problems. Biomethylation of As has implications for its mobility and toxicity. Photosynthetic organisms may play a significant role in As geochemical cycling by methylating it to different As species, but little is known about the mechanisms of methylation. Methylated As species have been found in many photosynthetic organisms, and several arsenite S-adenosylmethionine (SAM) methyltransferases have been characterized in cyanobacteria and algae. However, higher plants may not have the ability to methylate As. Instead, methylated arsenicals in plants probably originate from microorganisms in soils and the rhizosphere. Here, we propose possible approaches for developing ‘smart’ photosynthetic organisms with an enhanced and sensitive biomethylation capacity for bioremediation and safer food.
As contamination
As is a ubiquitous contaminant in the environment because of the use of As-laden groundwater and mining and smelting of base metals. As-contaminated water affects the health of millions of people, particularly in Southeast Asia [1,2]. Food supply is another source of human As exposure. In China, rice (Oryza sativum) has been estimated to be the largest contributor (approximately 60%) of inorganic As (iAs) ingestion through food consumption. Incremental lifetime cancer risk from food iAs intake is 106 per 100 000 for adult individuals and the median population cancer risk is 177 per 100 000 varying between regions [3]. Similar cancer risks have also been reported in some Bangladesh areas where As in drinking water is not a significant problem [4]. It is estimated that 35 million–77 million people have been chronically exposed to As through drinking water in Bangladesh alone [5]. Human exposure to As can lead to an array of diseases [6,7], such as bladder, skin and lung cancers [8,9], diabetes [10], cardiovascular disease [11], developmental disorders [12,13], neurological disorders [14,15], metabolic disorders [16] and dermal disease [17]. A recent large-scale epidemiological study in Bangladesh showed that exposure to As from drinking water contributed 21% and 24% to all-cause and chronic-disease mortalities, respectively [5]; although As ingestion from food was not included in this study.
As speciation plays a significant role in its behavior and fate in the environment, and different As species differ greatly in their toxicity to cells. As exists primarily as inorganic arsenate [As(V)] and arsenite [As(III)] in the environment, but can also be methylated into mono-, di-or trimethylarsines [18]. As(III) is ten times more soluble, mobile and toxic than is As(V) [19]. By contrast, trimethylarsine [TMA(III)] is almost nontoxic [20] at moderate concentrations and can be volatilized. It is also relatively stable and can travel considerable distances in the environment [21]. Thus, As volatilization is a process that should be further explored to aid bioremediation of As-contaminated environments. Previous studies of As biomethylation focused on bacteria, archaea and mammals. During the past few years, more attention has been paid to cyanobacteria, algae and plants. In this review, we highlight the recent progress that has been made in understanding As biomethylation by photosynthetic organisms. This knowledge will be important for putting future As bioremediation into practice. Biotransformation of As to volatile and/or less toxic species may alleviate the toxicity of As-contaminated environments and also has the potential to mitigate As toxicity in food.
As biomethylation
Biomethylation of As is widespread in nature; it has been observed in bacteria, archaea, fungi, algae, plants, animals and humans. The biotransformation of As(III) to garlic-odored TMA(III) by fungi was first documented during the 1890s [18]. In fact, sequencing of multiple fungal genomes identified genes encoding homologs of As(III) methyltransferase (ArsM) in many fungi. These genes are often in gene clusters adjacent to other genes encoding As-resistance proteins. For example, the As resistance gene cluster in Aspergillus fumigatus contains genes encoding As resistance protein H (ArsH), ArsM, arsenical compound resistance 3 (ACR3; annotated ArsB) and ArsC, suggesting that ArsM has a role in As resistance. As biomethylation has been found in bacteria, such as Pseudomonas spp. [22], anaerobic archaea, such as methanogens [23], and halo-philes, such as Halobacterium sp. NRC-1 [24]. Methylated As species have also been detected in urine samples from humans who have consumed inorganic As-containing water [25–27]. As biomethylation and biovolatilization have also been observed in a model protozoan Tetrahymena thermophila [28].
Molecular mechanisms of As biomethylation
Although As biomethylation has been known for a long time, the detailed molecular mechanisms have only been studied intensively over the past few years. Despite some progress, many mechanistic details of the pathway remain elusive. Frederick Challenger proposed a scheme in which alternating reduction of arsenicals containing pentavalent As and oxidative methylation of arsenicals containing trivalent As lead to a series of methylated arsenical species [18,29,30]. At the time that the Challenger mechanism was proposed, neither the methyl donor nor the reducing equivalents were known. Later, it was generally accepted that SAM is the methyl donor [18]. Many findings relating to the pathway of methylation of As species have been derived from studying the mammalian ArsM, AS3MT [30,31]. An alternative conceptual model to the Challenger mechanism was proposed in which As–glutathione complexes are substrates for sequential methyl group transfer to the arsenicals [32]. However, this model could not explain how other reductants, such as Tris(2-carboxyethyl) phosphine (TCEP), would still be able to support methylation of As as they do. Nevertheless, current models cannot explicitly describe the As biomethylation process, and the underlying mechanism has yet to be further investigated.
In bacteria, the ArsM is homologous to mammalian AS3MT. It is probable that ArsM functions similarly to AS3MT. The ArsM from Rhodopseudomonas palustris was shown to confer As(III) resistance when expressed in an As-sensitive strain of Escherichia coli [33]. Further chemical speciation analysis showed that the purified enzyme can convert As(III) to dimethylarsenate [DMA(V)], TMA(III) and TMA(III) oxide (TMAO) in the presence of SAM and glutathione (GSH). Neither monomethylarsenite [MAs(III)] nor monomethylarsenate [MAs(V)] were observed in the reaction. Why this ArsM produced dimethyl and trimethyl species, but no monomethyl species needs to be studied further. Methylation of As by ArsM resulted in the loss of As from both the medium and the cells through volatilization.
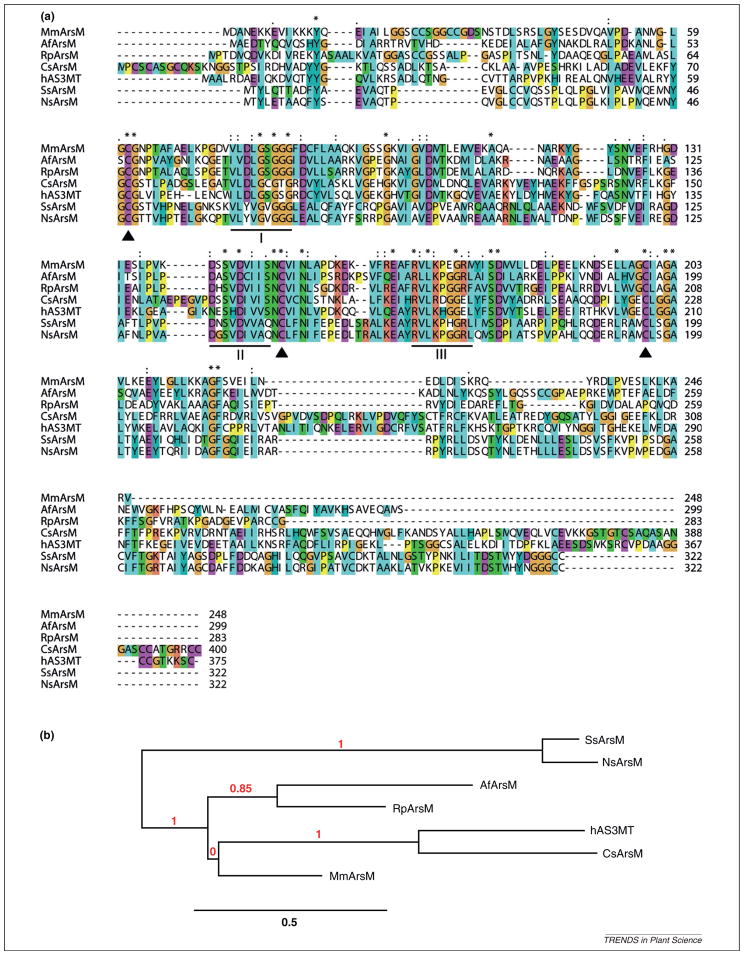
The multiple-sequence alignment of bacterial, archaeal, algal, fungal, cyanobacterial and mammalian As methyltransferase illustrates conserved motifs and cysteine residues as well as regions of considerable variability (Figure 1a). Although the lengths of these proteins range from 248 to 400 amino acids, the conserved region is limited to a core of approximately 150 amino acids. Three sequence motifs common to many SAM-dependent methyltransferases that methylate proteins or small molecules can be identified [30,34]. Some residues of these motifs make direct contact with SAM [35]. These three motifs are conserved in As methyltransferases across all species. There are three fully conserved cysteines, corresponding to residues 48, 143 and 195 in Synechocystis sp. PCC6803, which are probably involved in As binding, because of the affinity of the thiolate to the metalloid As(III). It is noteworthy that Cys 143 is located within the consensus sequence of the SAM-dependent methyltransferase domain, whereas Cys 48 and 195 are approximately 15 residues upstream and downstream of this domain, respectively. We hypothesize that the core region with three conserved cysteines and the SAM-dependent methyltransferase domain are required for As methyltransferases. Outside the core region, there are almost no similarities. The core region is critical for methyl group transfer to As, whereas the rest of the protein is species specific, and its function needs to be investigated further.
Figure 1.
Relationships of the arsenite methyltransferases (ArsM) from different species. (a) Multi-sequence alignment of ArsMs. Conserved cysteines are indicated by solid triangle. Motifs I, II and III are underlined and are involved in interactions with S-adenosylmethionine (SAM). The sequences of seven species are (species names are given in parentheses): AfArsM (Aspergillus fumigatus A1163); Czars (Cyanidioschyzon sp. 5508); hAS3MT (Homo sapiens); MmArsM (Methanosarcina mazei Go1); NsArsM (Nostoc sp. PCC 7120); RpArsM (Rhodopseudomonas palustris CGA009); and SsArsM (Synechocystis sp. PCC 6803). (b) Phylogenetic tree (produced by Phylogeny.fr [68]) comparing ArsMs.
The phylogenetic analysis (Figure 1b) shows that bacterial ArsM is more closely related to fungal ArsM, whereas mammalian AS3MT is grouped with eukaryotic algal ArsM. The distant relationship between cyanobacterial ArsMs and other groups was unexpected. The evolution of ArsM therefore remains an open question.
Whether As biomethylation is a pathway of detoxification is still being debated. Some of the intermediate products, MAs(III) and DMA(III), are more toxic than inorganic As [36]. This leads to some doubt over whether As biomethylation is a detoxification mechanism [30]. However, the pentavalent intermediate products, MAs(V) and DMA(V), are much less toxic than inorganic As [36]. Moreover, the end product, TMA(III), is almost nontoxic [20] and the fact that it can be volatilized can decrease the As concentration in cells. In addition, arsM is often regulated by arsenical resistance operon repressor (ArsR) and adjacent to or part of an ars operon in bacteria and archaea. In fungi, arsM is mostly encoded as part of a gene cluster with other genes encoding As resistance proteins. Therefore, it is reasonable to believe that As biomethylation is indeed a detoxification mechanism.
Methylation of As in cyanobacteria and algae
The presence of methylated As species has been reported for numerous photosynthetic organisms, including cyano-bacteria, algae and plants. When the marine green alga Chlorella salina was exposed to 1μM As(V) or As(III), the amount of MAs(V) and DMA(V) was 9–12% of total intra-cellular As [37]. A more recent study on the short-term metabolic processes of As(V) in a freshwater green alga, Chlamydomonas reinhardtii, showed that As(III), As(V) and MAs(V) concentrations in the cells increased from 30 min up to 1 h, and then leveled off [38]. DMA(V) and oxo-arsenosugar-glycerol levels also increased continuously for 24 h. These results indicated that C. reinhardtii can rapidly biotransform As(V) into oxo-arsenosugar-glycerol and then oxo-arsenosugar-phosphate. The reduction of As(V) to As(III) and methylation of As(III) to DMA(V) is coupled in this process, and MAs(V) was observed to be a possible intermediate. This is different from Rhodopseudomonas palustris, in which no monomethylated species were observed.
When the marine brown alga (seaweed) Fucus serratus was treated with different concentrations of As(V) for up to 19 weeks, As(V), As(III), MAs(V), DMA(V) and four arseno-sugars were detected [39]. The proportion of As species found in F. serratus depended on the As(V) concentration that it was exposed to. When the As(V) concentration was at 100 μg As(V)/L, the major product was As(III), with smaller quantities of methylated species and traces of arsenosugars. At 20 μg As(V)/L, DMA(V) and arsenosugars were major As metabolites, with small quantities of As(III) and MAs(V). If one assumes that methylation of As(III) is the rate-limiting step and not As(V) reduction, one would predict As(III) to accumulate at higher As(V) concentrations. These results suggest that, at higher As(V) concentration, the As(III) methylation process becomes saturated, leading to the accumulation of As(III) and subsequent toxicity [39]. However, at this point, it is not known whether F. serratus actually has a gene encoding an ArsM. Fucus serratus is often intimately associated with specific fungi and it is possible that these fungi carry out the methylation reaction [40].
Thermophilic algae have also been shown to methylate and volatilize As. Cyanidioschyzon sp. isolate 5508, isolated from thermal wells in Yellowstone National Park, is capable of oxidizing As(III) to As(V), reducing As(V) to As(III) and then methylating As(III) to form TMAO and DMAs(V) [41]. Two As methyltransferase genes from Cyanidioschyzon sp. isolate 5508, CmarsM7 and CmarsM8, were expressed in an As(III)-hypersensitive strain of E. coli and conferred resistance to As(III). Purified enzymes of the two recombinant CmArsMs were shown to transform As(III) into MAs(III), DMA(V), TMAO and TMA gas, with an optimum temperature of 60–70°C. These thermophilic capabilities of Cyanidioschyzon sp. isolate 5508 could be further explored for the development of ‘green’ bioreactors using solar energy to remove As from contaminated water.
Cyanobacteria, prokaryotic photosynthetic organisms, are ubiquitous in environments ranging from soil and water to deserts. They are responsible for 20–30% of photosynthesis on Earth and are major contributors to the global oxygen cycle [42]. Because of their rapid growth rates and their ability to adapt rapidly to environmental changes, cyanobacteria are often key players in toxic algal blooming in various aquatic environments. Methylated As species have been found in a wide range of cyanobacteria and algae. A list of cyanobacteria and algae in which methylated arsenicals were detected is shown in Table 1. As biotransformation and volatilization has also been observed in three cyanobacteria under laboratory conditions [43]. These three cyanobacteria, Microcystis sp. PCC7806, Nostoc sp. PCC7120 and Synechocystis sp. PCC6803, accumulated large amounts of As when treated with 10μM sodium As(III). Methylated species of As(III) were detected following exposure to higher As(III) concentrations for 14 days. Volatile arsenicals were detected when the cyanobacteria were treated with As(V) for 6 weeks. Two ArsM homologs were cloned and purified. The enzymes have been shown to methylate As(III) in vitro with TMA(III) as the end product [43]. Because cyanobacteria are ubiquitous and often abundant in the environment, their potential in regulating As biogeochemistry should not be underestimated; however, their role in biomethylating As has yet to be characterized under field conditions. Systems biology approaches to manipulating the expression of related genes in cyano-bacteria may result in operationally viable bioremediation solutions.
Table 1.
Cyanobacteria and algae in which methylated As species have been detected
| Species | Refs |
|---|---|
| Alsidium corallinum | [60] |
| Ascophyllum nodosum | [61] |
| Caulerpa racemosa | [62] |
| Chlamydomonas reinhardtii | [38] |
| Chlorella salina | [37] |
| Chlorella vulgaris | [63,64] |
| Cladophora prolifera | [60] |
| Closterium aciculare | [65] |
| Codium vermilara | [60] |
| Cyanidioschyzon sp. isolate 5508 | [41] |
| Cystoseira mediterranea | [60] |
| Enteromorpha compressa | [60] |
| Fucus serratus | [39] |
| Fucus sp. | [66] |
| Fucus vesiculosus | [61] |
| Fucus virsoides | [66] |
| Gelidium abbottiorum | [62] |
| Gelidium sp. 2 | [66] |
| Halopteris scoparia | [60] |
| Laminaria digitata | [61] |
| Melanosiphen intestinalis | [67] |
| Microcystis sp. PCC7806 | [43] |
| Nostoc sp. PCC7120 | [43] |
| Plocamium corallorhiza | [62] |
| Sargassum fulvellum | [60] |
| Sargassum piluliferum | [67] |
| Synechocystis sp. PCC6803 | [43] |
| Ulva lactuca | [62] |
| Ulva rigida | [60] |
As biomethylation in higher plants
Whether higher plants can methylate As is not known. Although analyses of numerous plant species have shown that methylated arsenicals do exist in plant tissues, a corresponding enzyme has not been found. Methylated As species, such as MAs(V), DMA(V) and TMAO, have been detected in many plant samples grown in the field [44]. Among these plant samples, rice has been intensively studied for As speciation. MAs(V) and DMA(V) have often been detected in vegetative tissues and grains of rice plants [45]. As(III) and DMA(V) have been detected in USA grain [46]. In these earlier publications, authors suggested that methylation occurs within rice. Although inorganic As is the dominant form, DMA can account for up to 60% of total As in some rice species [47]. By contrast, no methylated As species have been detected in extracts of wheat (Triticum aestivum) grain [48].
Recently, more studies seem to support the hypothesis of a microbial (including algal) origin of methylated As species in the environment. In a non-sterile solution culture experiment with rice, DMA(V) was detected in the solution amended by MAs(V) or As(III) [49]. However, DMA(V) concentration in the solution decreased dramatically when the antibacterial agent chloramphenicol was included, suggesting that rice rhizosphere-associated bacteria are involved in the formation of DMA(V) in rice. However, chloroplasts can also be sensitive to chloramphenicol and more data are needed. Not surprisingly, in hydroponic cultures where only inorganic As was added, only small amounts of methylated As species, usually less than 1% of total As content, were found in plant tissues [50], suggesting that de novo As methylation activity, if it exists, is limited. Results from our own laboratory also demonstrated that rice fed with inorganic As in hydroponic culture contains methylated As forming less than 3% of the total As, possibly because of contamination of bacteria or algae in the system (M.Z. Zheng et al., unpublished). Indeed, in the most recent study using a sterile hydroponic system, no methylated As was detected in plants fed with inorganic As [51].
A methyltransferase responsible for As methylation in plants has yet to be found. However, As methylation activities have been determined from leaf and root extracts of Acrostic tenuous [52]. In the in vitro assay using 3H-labeled SAM as the methyl donor, leaf extract showed methylation activity with either As(III) or As(V) as the substrate. However, root extract only showed methylation when exposed to As(III). The early product was MAs(V), but over a long assay time, DMA(V) accumulated to levels exceeding MAs(V). This may be the strongest evidence to date suggesting that plants methylate As. However, unless more rigorously controlled experiments are repeated by more laboratories, it should be taken with a pinch of salt.
In a microarray study in which rice was exposed to As(V) in hydroponic solution, a gene annotated as a methyltransferase (Os02g51030) was unregulated [53]. Although this gene contains a UbiE/Coq5 family protein motif, which is also present in the arsM genes of bacteria and archaea [33], it does not have the three conserved cysteines present in all As methyltransferases. Therefore, it is probably not an As methyltransferase, and whether the rice genome contains an As methyltransferase has yet to be determined. Overall, it is difficult to come to a general conclusion regarding the ability of higher plants to methylate As(III); given the limited plant species tested so far, it is clear that plants are unlikely to harbor an As(III) SAM methyltransferase gene in great numbers. Conversely, no gene encoding an As(III)-methyltransferase with significant homology to ArsM/AS3MT has been detected in rice. Our preliminary analyses have not found any protein containing the core region sequence that is present in ArsM/AS3MT in higher plants.
If methylated As species originated from microbial activities, how did they enter plants? Recent studies have shown that plants take up undissociated methylated As species through the nodulin 26-like intrinsic (NIP) aquaporin channels [54]. The NIP aquaporin Lsi1 mediates the uptake of MAs(V) and DMA(V) into rice roots [55]. The lsi1 mutant has lost 80% and 50% of the uptake capacity for MAs(V) and DMA(V), respectively, compared with wild-type rice.
Because rice contributes to substantial inorganic As ingestion by humans for whom rice is a stable part of their diet, it is of vital importance to either reduce As accumulation in rice grain or transform proportionally more As into volatile, methylated species. Given that rice itself does not seem to have an arsM-like gene, in a recent study, a bacterial arsM gene was, for the first time, successfully expressed in rice [56]. MAs(III) and DMA(V) were detected in the roots and shoots of the transgenic rice, and the transgenic rice gave off tenfold more volatile arsenicals. At this stage, this is merely a proof of concept, because the amount of volatile arsenicals produced during the 12-day period was so low that it only represented 0.06% of the total As in the rice plants. However, it suggests that it will be possible to engineer plants genetically to volatilize As, thus aiding bioremediation of As-contaminated soils. Furthermore, producing rice with lower content of As (because of overexpression of ArsM) on As-contaminated fields should also be possible.
Regulation of arsM gene expression
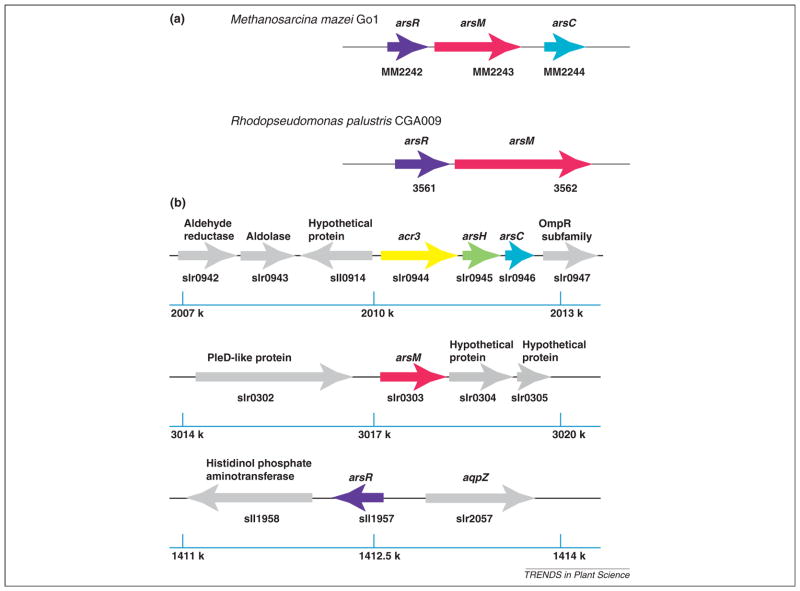
In bacteria and archaea, ars operons (including arsM) are usually under the tight control of ArsR (Figure 2a). When As(III) is not present, ArsR binds to the operator/promoter region of the operon, repressing the expression of downstream genes. When ArsR senses As(III), it binds to it and goes through conformational changes, resulting in dissociation from the operator/promoter region. The downstream ars genes are then expressed. Given that there are many methyltransferase genes in a typical free-living organism, one way to find arsM genes is to determine whether they are downstream of an arsR gene [33]. However, preliminary genomic analysis did not reveal arsM in cyanobacteria being under control of an adjacent ArsR repressor. In Synechocystis sp. PCC 6803 (Figure 2b), although arsR is located far away from arsC, arsC is still regulated by ArsR [57]. DNase I footprinting experiments indicated that ArsR binds to two 17-bp direct repeats, with each in the form ATCAA(N)6TT-GAT, in the region from nucleotides −34 to +17 of the promoter-operator. The arsM gene is adjacent to neither arsC nor arsR; furthermore, the upstream sequence of arsM does not have the 17-bp repeats typical of ArsR-regulated genes. Therefore, it is unlikely that arsM is regulated by ArsR and so how ArsM expression is regulated has yet to be investigated. One hypothesis is that ArsM is constitutively expressed.
Figure 2.
Regulation of genes encoding arsenite methyltransferase (arsM). Genes are shown as arrows, with right or left transcriptional directions indicated. The ars genes are shown in colors; non-ars genes are shown in gray. The rulers show the positions in the genome. (a) The arsM genes appear to be regulated by arsenical resistance operon repressor (ArsR)-type repressors in Methanosarcina mazei Go1 and Rhodopseudomonas palustris CGA009. (b) The arsM gene of Synechocystis sp. PCC 6803 may not be regulated by ArsR.
Concluding remarks
The biomethylation of metal(loid)s, such as As, selenium and mercury, has a fundamental impact on the global biogeochemistry of these trace elements. Research on the biomethylation of As by bacteria and mammals has provided insight into the molecular mechanisms responsible. However, less is known about these processes in cyanobacteria and algae, and almost nothing is known in higher plants, or if they even occur in these organisms. No enzyme responsible for As methylation has been found in plants. Understanding the mechanism is the first step to harnessing the biomethylation process for bioremediation and the production of safe food.
How different methyltransferases methylate As to different extents is also poorly understood. Deciphering the enzyme structure will be helpful in understanding the process. ArsM from the thermophilic alga Cyanidioschyzon sp. 5508 has been crystallized [58] and its structure is expected to be published soon. Linking methyltransferase structures from bacteria, cyanobacteria and mammals to their functions would further understanding of the mechanisms underlining As methylation, thus providing insights for manipulating this process for bioremediation and the production of safe food.
Current evidence supporting the existence of As methyltransferases in plants is very weak. If there is no such methyltransferase, one way to manipulate As biomethylation for phytoremediation and safe food is to use transgenic plants expressing bacterial homologs of the enzyme, such as has been demonstrated for rice [56]. However, a more suitable enzyme is needed for this to work more efficiently. An alternative way is to explore the rhizosphere-associated bacteria by selecting bacteria with strong As methylation activity, or engineering bacteria to give off more volatile arsenicals.
Why higher plants appear not to have As methyltransferases remains an open question. Perhaps higher plants do not benefit much from having an As methyltransferase; thus, such a methyltransferase has not been selected during evolution. The transgenic rice with a bacterial arsM gene expressed is the same as the nontransgenic control in appearance and growth [56]. After exposure to As(III) or As(V), no significant differences were observed in biomass or total As accumulation between the transgenic rice and the nontransgenic control.
It would be interesting to see whether other factors besides gene regulation may affect methylation of As(III). For example, in rice, shoot phosphorus putatively has a negative relationship with grain As [59]. There are many factors, both biotic and abiotic, that influence the fate and speciation of As. Understanding and manipulating the flux and fate of As in the environment therefore requires more holistic approaches, such as systems biology. It is anticipated that the integration of various components influencing biomethylation may be essential in developing operationally viable systems for environmental bioremediation and food safety. Each component involved in the As biomethylation process and their interactions should be studied to provide a full picture at the system level; then, existing or new control elements can be built into genomes to achieve the desired outputs of bioremediation and food safety.
Acknowledgments
Research at Zhu’s labs is financially supported by the Natural Science Foundation of China (41090282 and 41090284) and the Ministry of Science and Technology (for international cooperation, nos. 2009DFB90120 and s2012zr0012).
References
- 1.Nordstrom DK. Public health. Worldwide occurrences of arsenic in ground water. Science. 2002;296:2143–2145. doi: 10.1126/science.1072375. [DOI] [PubMed] [Google Scholar]
- 2.Christen K. The arsenic threat worsens. Environ Sci Technol. 2001;35:286A–291A. doi: 10.1021/es012394f. [DOI] [PubMed] [Google Scholar]
- 3.Li G, et al. Inorganic arsenic in Chinese food and its cancer risk. Environ Int. 2011;37:1219–1225. doi: 10.1016/j.envint.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 4.Sohel N, et al. Arsenic in drinking water and adult mortality: a population-based cohort study in rural Bangladesh. Epidemiology. 2009;20:824–830. doi: 10.1097/EDE.0b013e3181bb56ec. [DOI] [PubMed] [Google Scholar]
- 5.Argos M, et al. Arsenic exposure from drinking water, and all-cause and chronic-disease mortalities in Bangladesh (HEALS): a prospective cohort study. Lancet. 2010;376:252–258. doi: 10.1016/S0140-6736(10)60481-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jomova K, et al. Arsenic: toxicity, oxidative stress and human disease. J Appl Toxicol. 2011;31:95–107. doi: 10.1002/jat.1649. [DOI] [PubMed] [Google Scholar]
- 7.Agusa T, et al. Exposure, metabolism, and health effects of arsenic in residents from arsenic-contaminated groundwater areas of Vietnam and Cambodia: a review. Rev Environ Health. 2010;25:193–220. doi: 10.1515/reveh.2010.25.3.193. [DOI] [PubMed] [Google Scholar]
- 8.Miller WH, Jr, et al. Mechanisms of action of arsenic trioxide. Cancer Res. 2002;62:3893–3903. [PubMed] [Google Scholar]
- 9.Rossman TG. Mechanism of arsenic carcinogenesis: an integrated approach. Mutat Res. 2003;533:37–65. doi: 10.1016/j.mrfmmm.2003.07.009. [DOI] [PubMed] [Google Scholar]
- 10.Diaz-Villasenor A, et al. Arsenic-induced alteration in the expression of genes related to type 2 diabetes mellitus. Toxicol Appl Pharmacol. 2007;225:123–133. doi: 10.1016/j.taap.2007.08.019. [DOI] [PubMed] [Google Scholar]
- 11.Navas-Acien A, et al. Arsenic exposure and cardiovascular disease: a systematic review of the epidemiologic evidence. Am J Epidemiol. 2005;162:1037–1049. doi: 10.1093/aje/kwi330. [DOI] [PubMed] [Google Scholar]
- 12.Liu Y, et al. The relationship between mental retardation and developmental delays in children and the levels of arsenic, mercury and lead in soil samples taken near their mother’s residence during pregnancy. Int J Hyg Environ Health. 2010;213:116–123. doi: 10.1016/j.ijheh.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rossman TG, Klein CB. Genetic and epigenetic effects of environmental arsenicals. Metallomics. 2011;3:1135–1141. doi: 10.1039/c1mt00074h. [DOI] [PubMed] [Google Scholar]
- 14.Calderon J, et al. Exposure to arsenic and lead and neuropsychological development in Mexican children. Environ Res. 2001;85:69–76. doi: 10.1006/enrs.2000.4106. [DOI] [PubMed] [Google Scholar]
- 15.Uede K, Furukawa F. Skin manifestations in acute arsenic poisoning from the Wakayama curry-poisoning incident. Br J Dermatol. 2003;149:757–762. doi: 10.1046/j.1365-2133.2003.05511.x. [DOI] [PubMed] [Google Scholar]
- 16.Xue P, et al. Prolonged inorganic arsenite exposure suppresses insulin-stimulated AKT S473 phosphorylation and glucose uptake in 3T3-L1 adipocytes: involvement of the adaptive antioxidant response. Biochem Biophys Res Commun. 2011;407:360–365. doi: 10.1016/j.bbrc.2011.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCarty KM, et al. Arsenic methylation, GSTT1, GSTM1, GSTP1 polymorphisms, and skin lesions. Environ Health Perspect. 2007;115:341–345. doi: 10.1289/ehp.9152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bentley R, Chasteen TG. Microbial methylation of metalloids: arsenic, antimony, and bismuth. Microbiol Mol Biol Rev. 2002;66:250–271. doi: 10.1128/MMBR.66.2.250-271.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Herreweghe S, et al. Solid phase speciation of arsenic by sequential extraction in standard reference materials and industrially contaminated soil samples. Environ Pollut. 2003;122:323–342. doi: 10.1016/s0269-7491(02)00332-9. [DOI] [PubMed] [Google Scholar]
- 20.Cullen WR. The toxicity of trimethylarsine: an urban myth. J Environ Monit. 2005;7:11–15. doi: 10.1039/b413752n. [DOI] [PubMed] [Google Scholar]
- 21.Mestrot A, et al. Atmospheric stability of arsine and methylarsines. Environ Sci Technol. 2011;45:4010–4015. doi: 10.1021/es2004649. [DOI] [PubMed] [Google Scholar]
- 22.Shariatpanahi M, et al. Biotransformation of the pesticide sodium arsenate. J Environ Sci Health B. 1981;16:35–47. doi: 10.1080/03601238109372237. [DOI] [PubMed] [Google Scholar]
- 23.Michalke K, et al. Production of volatile derivatives of metal(loid)s by microflora involved in anaerobic digestion of sewage sludge. Appl Environ Microbiol. 2000;66:2791–2796. doi: 10.1128/aem.66.7.2791-2796.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang G, et al. Arsenic resistance in Halobacterium sp. strain NRC-1 examined by using an improved gene knockout system. J Bacteriol. 2004;186:3187–3194. doi: 10.1128/JB.186.10.3187-3194.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aposhian HV, et al. Occurrence of monomethylarsonous acid in urine of humans exposed to inorganic arsenic. Chem Res Toxicol. 2000;13:693–697. doi: 10.1021/tx000114o. [DOI] [PubMed] [Google Scholar]
- 26.Le XC, et al. Speciation of key arsenic metabolic intermediates in human urine. Anal Chem. 2000;72:5172–5177. doi: 10.1021/ac000527u. [DOI] [PubMed] [Google Scholar]
- 27.Mandal BK, et al. Identification of dimethylarsinous and monomethylarsonous acids in human urine of the arsenic-affected areas in West Bengal, India. Chem Res Toxicol. 2001;14:371–378. doi: 10.1021/tx000246h. [DOI] [PubMed] [Google Scholar]
- 28.Yin XX, et al. Rapid biotransformation of arsenic by a model protozoan Tetrahymena thermophila. Environ Pollut. 2011;159:837–840. doi: 10.1016/j.envpol.2010.12.033. [DOI] [PubMed] [Google Scholar]
- 29.Challenger F. Biological methylation. Adv Enzymol Relat Subj Biochem. 1951;12:429–491. doi: 10.1002/9780470122570.ch8. [DOI] [PubMed] [Google Scholar]
- 30.Thomas DJ, et al. Arsenic (+3 oxidation state) methyltransferase and the methylation of arsenicals. Exp Biol Med. 2007;232:3–13. [PMC free article] [PubMed] [Google Scholar]
- 31.Lin S, et al. A novel S-adenosyl-L-methionine:arsenic(III) methyltransferase from rat liver cytosol. J Biol Chem. 2002;277:10795–10803. doi: 10.1074/jbc.M110246200. [DOI] [PubMed] [Google Scholar]
- 32.Hayakawa T, et al. A new metabolic pathway of arsenite: arsenic–glutathione complexes are substrates for human arsenic methyltransferase Cyt19. Arch Toxicol. 2005;79:183–191. doi: 10.1007/s00204-004-0620-x. [DOI] [PubMed] [Google Scholar]
- 33.Qin J, et al. Arsenic detoxification and evolution of trimethylarsine gas by a microbial arsenite S-adenosylmethionine methyltransferase. Proc Natl Acad Sci USA. 2006;103:2075–2080. doi: 10.1073/pnas.0506836103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kagan RM, Clarke S. Widespread occurrence of three sequence motifs in diverse S-adenosylmethionine-dependent methyltransferases suggests a common structure for these enzymes. Arch Biochem Biophys. 1994;310:417–427. doi: 10.1006/abbi.1994.1187. [DOI] [PubMed] [Google Scholar]
- 35.Kozbial PZ, Mushegian AR. Natural history of S-adenosylmethionine-binding proteins. BMC Struct Biol. 2005;5:19. doi: 10.1186/1472-6807-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Styblo M, et al. Comparative toxicity of trivalent and pentavalent inorganic and methylated arsenicals in rat and human cells. Arch Toxicol. 2000;74:289–299. doi: 10.1007/s002040000134. [DOI] [PubMed] [Google Scholar]
- 37.Karadjova IB, et al. The biouptake and toxicity of arsenic species on the green microalga Chlorella salina in seawater. Aquat Toxicol. 2008;87:264–271. doi: 10.1016/j.aquatox.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 38.Miyashita S, et al. Rapid biotransformation of arsenate into oxo-arsenosugars by a freshwater unicellular green alga, Chlamydomonas reinhardtii. Biosci Biotechnol Biochem. 2011;75:522–530. doi: 10.1271/bbb.100751. [DOI] [PubMed] [Google Scholar]
- 39.Geiszinger A, et al. Arsenic biotransformation by the brown macroalga, Fucus serratus. Environ Toxicol Chem. 2001;20:2255–2262. doi: 10.1897/1551-5028(2001)020<2255:abbtbm>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 40.Zuccaro A, et al. Detection and identification of fungi intimately associated with the brown seaweed Fucus serratus. Appl Environ Microbiol. 2008;74:931–941. doi: 10.1128/AEM.01158-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qin J, et al. Biotransformation of arsenic by a Yellowstone thermoacidophilic eukaryotic alga. Proc Natl Acad Sci USA. 2009;106:5213–5217. doi: 10.1073/pnas.0900238106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pisciotta JM, et al. Light-dependent electrogenic activity of cyanobacteria. PLoS ONE. 2010;5:e10821. doi: 10.1371/journal.pone.0010821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yin XX, et al. Biotransformation and volatilization of arsenic by three photosynthetic cyanobacteria. Plant Physiol. 2011;156:1631–1638. doi: 10.1104/pp.111.178947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meharg AA, Hartley-Whitaker J. Arsenic uptake and metabolism in arsenic resistant and nonresistant plant species. New Phytol. 2002;154:29–43. [Google Scholar]
- 45.Heitkemper DT, et al. Determination of total and speciated arsenic in rice by ion chromatography and inductively coupled plasma mass spectrometry. J Anal At Spectrom. 2001;16:299–306. [Google Scholar]
- 46.Zavala YJ, et al. Arsenic in rice: II. Arsenic speciation in USA grain and implications for human health. Environ Sci Technol. 2008;42:3861–3866. doi: 10.1021/es702748q. [DOI] [PubMed] [Google Scholar]
- 47.Wu C, et al. Arsenic accumulation and speciation in rice are affected by root aeration and variation of genotypes. J Exp Bot. 2011;62:2889–2898. doi: 10.1093/jxb/erq462. [DOI] [PubMed] [Google Scholar]
- 48.Zhao FJ, et al. Accumulation, distribution, and speciation of arsenic in wheat grain. Environ Sci Technol. 2010;44:5464–5468. doi: 10.1021/es100765g. [DOI] [PubMed] [Google Scholar]
- 49.Arao T, et al. Effects of arsenic compound amendment on arsenic speciation in rice grain. Environ Sci Technol. 2011;45:1291–1297. doi: 10.1021/es1033316. [DOI] [PubMed] [Google Scholar]
- 50.Quaghebeur M, Rengel Z. The distribution of arsenate and arsenite in shoots and roots of Holcus lanatus is influenced by arsenic tolerance and arsenate and phosphate supply. Plant Physiol. 2003;132:1600–1609. doi: 10.1104/pp.103.021741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lomax C, et al. Methylated arsenic species in plants originate from soil microorganisms. New Phytol. 2012;193:665–672. doi: 10.1111/j.1469-8137.2011.03956.x. [DOI] [PubMed] [Google Scholar]
- 52.Wu JH, et al. Methylation of arsenic in vitro by cell extracts from bentgrass (Acrostic tenuous): effect of acute exposure of plants to arsenate. Funct Plant Biol. 2002;29:73–80. doi: 10.1071/PP01022. [DOI] [PubMed] [Google Scholar]
- 53.Norton GJ, et al. Rice–arsenate interactions in hydroponics: whole genome transcriptional analysis. J Exp Bot. 2008;59:2267–2276. doi: 10.1093/jxb/ern097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao FJ, et al. Arsenic as a food chain contaminant: mechanisms of plant uptake and metabolism and mitigation strategies. Annu Rev Plant Biol. 2010;61:535–559. doi: 10.1146/annurev-arplant-042809-112152. [DOI] [PubMed] [Google Scholar]
- 55.Li RY, et al. The rice aquaporin Lsi1 mediates uptake of methylated arsenic species. Plant Physiol. 2009;150:2071–2080. doi: 10.1104/pp.109.140350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meng XY, et al. Arsenic biotransformation and volatilization in transgenic rice. New Phytol. 2011;191:49–56. doi: 10.1111/j.1469-8137.2011.03743.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lopez-Maury L, et al. Arsenic sensing and resistance system in the cyanobacterium Synechocystis sp. strain PCC 6803. J Bacteriol. 2003;185:5363–5371. doi: 10.1128/JB.185.18.5363-5371.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marapakala K, et al. Crystallization and preliminary X-ray crystallographic analysis of the ArsM arsenic(III) S-adenosylmethionine methyltransferase. Acta Crystallogr Sect F: Struct Biol Cryst Commun. 2010;66:1050–1052. doi: 10.1107/S1744309110027661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lu Y, et al. Arsenic accumulation and phosphorus status in two rice (Oryza sativa L.) cultivars surveyed from fields in South China. Environ Pollut. 2010;158:1536–1541. doi: 10.1016/j.envpol.2009.12.022. [DOI] [PubMed] [Google Scholar]
- 60.Llorente-Mirandes T, et al. Measurement of arsenic compounds in littoral zone algae from the Western Mediterranean Sea. Occurrence of arsenobetaine. Chemosphere. 2010;81:867–875. doi: 10.1016/j.chemosphere.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 61.Nischwitz V, Pergantis SA. Improved arsenic speciation analysis for extracts of commercially available edible marine algae using HPLC-ES-MS/MS. J Agric Food Chem. 2006;54:6507–6519. doi: 10.1021/jf060971j. [DOI] [PubMed] [Google Scholar]
- 62.Misheer N, et al. Seaweeds along KwaZulu-Natal Coast of South Africa–4: elemental uptake by edible seaweed Caulerpa racemosa (sea grapes) and the arsenic speciation. J Environ Sci Health A: Tox Hazard Subst Environ Eng. 2006;41:1217–1233. doi: 10.1080/10934520600656489. [DOI] [PubMed] [Google Scholar]
- 63.Salgado SG, et al. Optimisation of sample treatment for arsenic speciation in alga samples by focussed sonication and ultrafiltration. Talanta. 2006;68:1522–1527. doi: 10.1016/j.talanta.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 64.Garcia Salgado S, et al. Determination of soluble toxic arsenic species in alga samples by microwave-assisted extraction and high performance liquid chromatography-hydride generation-inductively coupled plasma-atomic emission spectrometry. J Chromatogr A. 2006;1129:54–60. doi: 10.1016/j.chroma.2006.06.083. [DOI] [PubMed] [Google Scholar]
- 65.Hasegawa H, et al. Biosynthesis and release of methylarsenic compounds during the growth of freshwater algae. Chemosphere. 2001;43:265–272. doi: 10.1016/s0045-6535(00)00137-5. [DOI] [PubMed] [Google Scholar]
- 66.Slejkovec Z, et al. Arsenosugars and other arsenic compounds in littoral zone algae from the Adriatic Sea. Chemosphere. 2006;63:1098–1105. doi: 10.1016/j.chemosphere.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 67.Hirata S, Toshimitsu H. Determination of arsenic species and arsenosugars in marine samples by HPLC-ICP-MS. Anal Bioanal Chem. 2005;383:454–460. doi: 10.1007/s00216-005-3413-z. [DOI] [PubMed] [Google Scholar]
- 68.Dereeper A, et al. Phylogeny. fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]