Abstract
The intracellular parasite Toxoplasma gondii is a leading cause of congenital neurological defects. To cause disease, it must reiterate its lytic cycle through host cell invasion, replication,and parasite egress. This requires the parasite to sense changes in its environment and switch between the non-motile (for replication) and motile (for invasion and egress) states appropriately. Recently, we discovered a previously unknown mechanism of motility regulation in T. gondii, mediated by a lysine methyltransferase, AKMT (for Apical complex lysine (K) methyltransferase). When AKMT is absent, activation of motility is inhibited, which compromises parasite invasion and egress, and thus severely impairs the lytic cycle. Although the methyltransferase activity of AKMT has been established, the phylogenetic relationship of AKMT with other better studied lysine methyltransferases (KMTs) was not known. Also unknown was the functional relationships between different domains of AKMT. In this work we carried out phylogenetic analyses, which show that AKMT orthologs form a new subfamily of KMTs. We systematically generated truncation mutants of AKMT, and discovered that the predicted enzymatic domain alone is a very poor enzyme and cannot complement the function of AKMT in vivo. Interestingly, the N- and C-terminal domains of the AKMT have drastically different impacts on its enzyme activity, localization as well as in vivo function. Our results thus reveal that AKMT is an unusual, parasite-specific enzyme and identified regions and interactions within this novel lysine methyltransferase that can be used as drug targets.
Keywords: lysine methyltransferase, AKMT, motility, Toxoplasma gondii, Plasmodium, KMT phylogeny
1. INTRODUCTION
The phylum Apicomplexa contains thousands of species, many of which are serious health threats to humans [1]. Each year, more than 1 million lives are claimed by Plasmodium falciparum infection. Another apicomplexan parasite, Toxoplasma gondii is one of the most successful human parasites. Nearly 20% of the global population is permanently infected by T. gondii In otherwise healthy individuals, extracellular parasites are quickly eliminated by the immune system, thus preventing the expansion of the population, but live parasites persist within cysts in brain, skeletal and cardiac muscle. These latent parasites can reactivate and regenerate the rapidly replicating form of the parasite if the immune system is compromised, as in AIDS patients [2–8]. The consequences are devastating, including the development of toxoplasmic encephalitis, a lethal disease if left untreated [9–15]. Another important clinical setting for toxoplasmosis is the infection of unprotected fetus [16–25]. Congenital toxoplasmosis results in severe physical and mental defects and is a leading cause of congenital neurological defects in the U.S.
To cause disease, T. gondii (as well as other apicomplexan parasites) must reiterate its lytic cycle through host cell invasion, parasite replication, and parasite egress, which requires that the parasite switch between non-motile and motile states according to changes in its environment [26–39]. Despite its importance in parasite physiology and the pathogenesis of toxoplasmosis, little is known about how this switch is regulated. Recently we discovered a key regulator of parasite motility, a lysine methyltransferase named AKMT (for Apical complex lysine (K) methyltransferase) (GenBank: XP_002370918; ToxoDB: TGME49_016080) [40]. When AKMT is absent, the parasite remains largely immotile. Both invasion and egress, and thus the complete lytic cycle, are inhibited. Importantly, the lysine methyltransferase activity of AKMT is required for its function. Interestingly, we found that the localization of AKMT in T gondii is sensitive to a potent motility-stimulating signal - parasite cytoplasmic [Ca2+] increase,with AKMT departing from the apical complex just before the onset of parasite motility and egress, suggesting that parasite motility might be regulated through the precise temporal control of AKMT localization [40].
All members of the lysine (K) methyltransferase (KMT) family, except for the DOT1 family, contain a SET domain, which, together with clusters of zinc binding cysteines in the pre and/or post-SET region, binds to the co-factor S-adenosyl-L-methionine (SAM) and catalyzes the methyl transfer reaction to specific lysine(s) on the substrate [41, 42]. In this study, we carried out phylogenetic analyses and found that AKMT orthologs form a clade distinct from those of other known KMT families. To further understand the properties of this unusual enzyme, we examined factors that might affect AKMT function in vitro and in vivo. We found that although AKMT localization in the parasite is sensitive to intra-parasite [Ca2+], the enzymatic activity of AKMT is not. Truncation analyses showed that the predicted enzymatic domain [the SET domain plus the cysteine cluster (SET-cys)] by itself is only weakly active as a lysine methyltransferase, mislocalizes and cannot complement AKMT function in the parasite.The truncation remains a poor enzyme when both the SET-cys and the N-terminal domain of AKMT are included, although the N-terminal domain does seem to have a moderate contribution to AKMT function and localization in vivo. In contrast, when the C-terminal domain AKMT is included together with SET-cys, it yields a very active enzyme, which can also activate motility during egress, thus revealing the importance of this module to AKMT function.
2. MATERIALS AND METHODS
2.1 Phylogenetic analysis
The phylogenetic tree was built using distance-based neighbor-joining method in PHYLIP, a part of Unipro-UGENE (V 1.11) package with the Dayhoff PAM substitution model [43]. An alignment generated by T-Coffee [44] using the SET and post-SET domain of KMTs comprising of apicomplexan KMTs as well as those from 10 other species including Homo sapiens, Mus musculus, Gallus gallus, Xenopus laevis, Drosophila melanogaster, Danio rerio,Caenorhabditis elegans, Dictyostelium discoideum, Saccharomyces cerevisiae, and Neurospora crassa. The trees were evaluated with 1,000 bootstrap replicates. Use of alternative tree building methods did not significantly affect the results (not shown).
The accession numbers as well as the sequences used for building the phylogenetic tree are included in the Supplementary Table I.
2.2 Construction of expression plasmids
2.2.1 T. gondii expression plasmids
pmin-eGFP-AKMT-full-length was created as described earlier [40]. AKMT truncation constructs pmin-eGFP-Nt, pmin-eGFP-Nt-SET-cys, pmin-eGFP-SET-cys, pmin-eGFP-SET-cys- Ct, and pmin-eGFP-cys-Ct were created by PCR amplification of truncated AKMT sequence by primer combinations shown in Supplemental Table II, followed by digestion with BglII and AflII and subsequent cloning into pmin-eGFP-AKMT-full-length replacing the full-length AKMT/BglII-AflII fragment. To create ptub-mTagRFP-T-TgTUBA1, a DNA fragment containing mTagRFP-T with a C-terminal SGLR linker was amplified by PCR with corresponding primers (Supplemental Table II) with pmin-mTagRFP-T-TgCentrin2_v2 (a kind gift from Dr. John Murray, University of Pennsylvania) as the template, and digested with restriction enzymes NheI and BglII. The digested fragment was then subcloned into the NheI- BglII site of ptub-mCherryFP-TgTUBA1 (A kind gift from Dr. John Murray, University of Pennsylvania) to replace mCherryFP fragment with mTagRFP-T_SGLR linker.
2.2.2 Bacterial expression plasmids
pET22b–FLAG-AKMT-full-length was created as described earlier [40]. AKMT truncation constructs pET22b-FLAG-Nt-SET-cys and pET22b-FLAG-SET-cys were created by amplifying AKMT coding sequence from pET22b-FLAG-AKMT-full-length as template and using primer combinations shown in Supplemental Table II. The PCR product was digested with NheI and EcoRI and was subcloned into the NheI and EcoRI sites of pET22b-FLAG-AKMT-full-length. To create pET22B-FLAG-SET-cys-Ct, a DNA fragment (flanked with NheI-EcoRI) encoding 301–709 aa of AKMT was first synthesized and cloned into the PUC57 simple plasmid to generate pUC-Simple-SET-cys-Ct (constructed by Genescript, Inc, New Jersey, U.S.A). The fragment was then isolated via NheI-EcoRI digestion and then subcloned into the NheI-EcoRI sites of pET22B-FLAG-AKMT-full-length.
2.3 Parasite culture and transfection
T. gondii Δakmt tachyzoites generated in a previous study [40] were used in all experiments. The parasites were maintained by continuous passage in human foreskin fibroblasts (HFFs) as previously described [40]. For each T. gondii transfection, 30–50 µg of DNA was electroporated into extracellular parasites harvested from 1 T12.5 flask (~2–3 ×107 parasites) and suspended in apotassium-based "intracellular buffer" (120 mM KCl; 0.15 mM CaCl2; 10 mM KH2PO4/K2HPO4 pH 7.6; 25 mM HEPES-KOH pH 7.6; 2 mM K2EGTA pH 7.6; 5 mM MgCl2, pH adjusted with KOH) [45] containing 2 mM K2ATP and 5 mM glutathione. Electroporation was performed using a Harvard Apparatus BTX-BCM630 electroporator (Holliston, MA) with the electroporation voltage set at 1400 V, capacitance set at 50 µF, and resistance set at 25Ω.
2.4 Protein expression and purification
FLAG-tagged full-length and truncation constructs of AKMT were purified from BL21 E. coli expressing corresponding plasmids as described previously [40]. Briefly, bacteria were induced with 1 mM IPTG for ~20 hours at 16°C and lysed in lysis buffer (8 mM Tris-Acetate pH 7.5, 7 mM Tris base pH unadjusted, 100 mM KAcetate, 1 mM MgAcetate) containing 0.5% Triton-100 (TX-100), protease inhibitors (0.25 mM phenylmethanesulfonyl fluoride, 10 µg/ml tosylphenylalanine chloromethyl ketone, 10 µg/ml N-tosyl- L-lysine chloromethyl ketone, 10 µg/ml p- toluenesulfonyl-L-arginine methyl ester) and 4mM DTT. The FLAG tagged protein was then purified using anti-FLAG M2 agarose (CAT# A2220: Sigma) and eluted with 100 µg/ml FLAG-peptide (CAT# 3290: Sigma) in lysis buffer containing 4 mM DTT. The eluted fractions were pooled and the pooled fractions were then concentrated 18–25 fold using Amicon centrifugal filters (10000 – 50000 MWCO, Millipore). Glycerol was added to concentrated proteins to a final concentration of 36% and stored at −20°C.
2.5 Protein lysine(K) methyltransferase (PKMT) assay
PKMT assays were performed as described [40, 46, 47] with some modifications. For activity assays with recombinant full-length and truncated AKMT proteins (FLAG-Nt-SET-cys, FLAG-SET-cys-Ct, and FLAG-SET-cys), 30 µl reactions were set up with 0.2 – 1 µg of full-length or truncated AKMT proteins, 2 µg of Xenopus histone H3.3 (CAT# 14–411: Millipore), 0.5mM DTT, 2.5 – 5 µM 3H-S-adenosyl-L-methionine (3H-SAM) (CAT# NET155V250UC: Perkin Elmer) in "intracellular buffer" [45]. Reactions were incubated at 30°C for 60 minutes, and then stopped by adding LDS sample buffer and reducing agent and 10 min 70°C heating. Proteins in the reactions were resolved on 4–12% gradient bis-tris NuPAGE gels (CAT# NP0332: Invitrogen). For detecting the methylated histone by autoradiography, proteins resolved on NuPAGE gels were transferred to PVDF. To visualize total proteins, the PVDF membranes were first washed twice in DPBS for 5 minutes each, stained with amido black staining solution (0.1% amido black 10B, 45% methanol, 10% acetic acid) for 5 – 10 min and rinsed briefly 4 – 6 times in destaining solution (90% methanol and 2% acetic acid). The membranes were then air-dried,sprayed with EN3HANCE Spray (CAT# 6NE970C: Perkin Elmer) per manufacturer’s instructions and exposed to X-ray film for 2 –18 hr as necessary to detect the 3H signal incorporated into the histone H3.3 protein. For determining the effect of free [Ca2+] on AKMT activity (Figure 2), free [Ca2+] was buffered by EGTA and calculated using Theo Schoenmakers Chelator program [48].
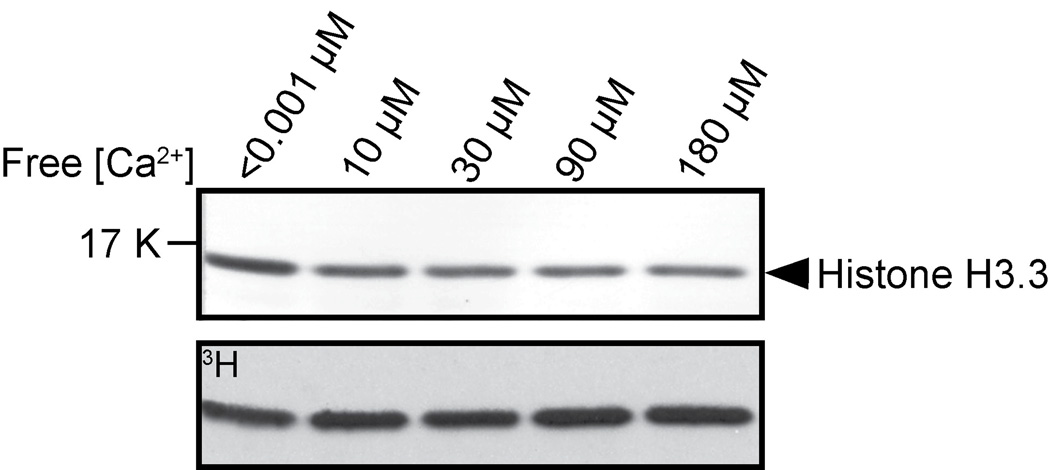
Figure 2. The effect of free calcium concentration on AKMT lysine methyltransferase activity.
In vitro methyltransferase assays were performed with 2 µg Xenopus histone H3.3, 2.5 µM 3H-SAM and 0.2 µg of recombinant FLAG-AKMT-full-length in the presence of < 0.001 µM , 10 µM, 30 µM, 90 µM, and 180 µM free Ca2+. Top: Blot stained with amido black showing total histone H3.3. Bottom: Autoradiograph showing 3H signals.
2.6 Immunofluorescence
For immunolabeling, parasites grown in HFF monolayer were fixed in 3.7% formaldehyde in DPBS for 15 minutes and permeabilized with 0.25% TX-100 in DPBS for 15 minutes. Cells were blocked with 1% BSA in DPBS for 30 minutes and then incubated in primary and subsequently secondary antibody solutions for 1 hr each. All incubations were performed at room temperature. Antibody dilutions were as follows: mouse anti-IMC1, 1:1000 (a kind gift from Dr. Gary Ward, University of Vermont); goat anti-mouse Alexa 568 (CAT# A11031: Molecular Probes-Invitrogen), 1:1000.
2.7 Fluorescence Imaging and line-scan analyses
2.7.1 Wide-field deconvolution
3-D image stacks were collected at 35–37°C at z-increments of 0.2 µm using an Applied Precision Delta Vision imaging station constructed on an Olympus inverted microscope base. A 100X oil immersion lens (1.4 NA), and immersion oil of refractive index 1.524 was used for the imaging. Deconvolved images were computed using the point-spread functions and software supplied by the manufacturer. The brightness and contrast of images used in the final figures were optimized for color prints. The line-scan analysis tool in Metamorph software (Molecular Devices) was used for obtaining fluorescence intensity (expressed as relative intensity) profiles.The area of analysis was defined by a 3 pixel line width and a line length drawn to cover the apical region of the parasite.
2.7.2 Structured-illumination (SIM) based super-resolution imaging
3D image stacks of live parasites maintained in CO2 independent media (Sku# RR060058:Invitrogen) containing 1% heat-inactivated bovine calf serum (CO2 independent imaging media) were collected at room temperature at z-increments of 0.125 µm using an the OMX imaging station (Applied precision-GE). A 100X oil immersion lens (1.4 NA), and immersion oil of refractive index 1.516 was used for the imaging. Deconvolved images were computed using the point-spread functions and software supplied by the manufacturer.
2.8 Live cell imaging and analyses of egress
Induced egress assay as described [40] was performed on Δakmt parasites transfected with eGFP-tagged full-length and truncated AKMT constructs for assessing functional complementation. Briefly, transfected parasites were added to MatTek dishes containing a confluent HFF monolayer and grown for ~35–40 hours. Cells were washed once and incubated in 1.5 ml of CO2 independent imaging media. 0.5 ml of 20 µM A23187 in CO2 independent imaging media was then added to the MatTek dishes for a final concentration of 5 µM to induce egress. Images were collected at 35–37°C for 10–40 minutes on an Applied Precision Delta Vision imaging station equipped with an environmental chamber. Vacuoles that did not show any significant changes in the refractivity or morphology of the host nor the parasites (likely due to inadequate exposure to the A23187) were excluded from the analysis.
3. RESULTS
3.1 AKMT orthologs form a clade distinct from other KMTs
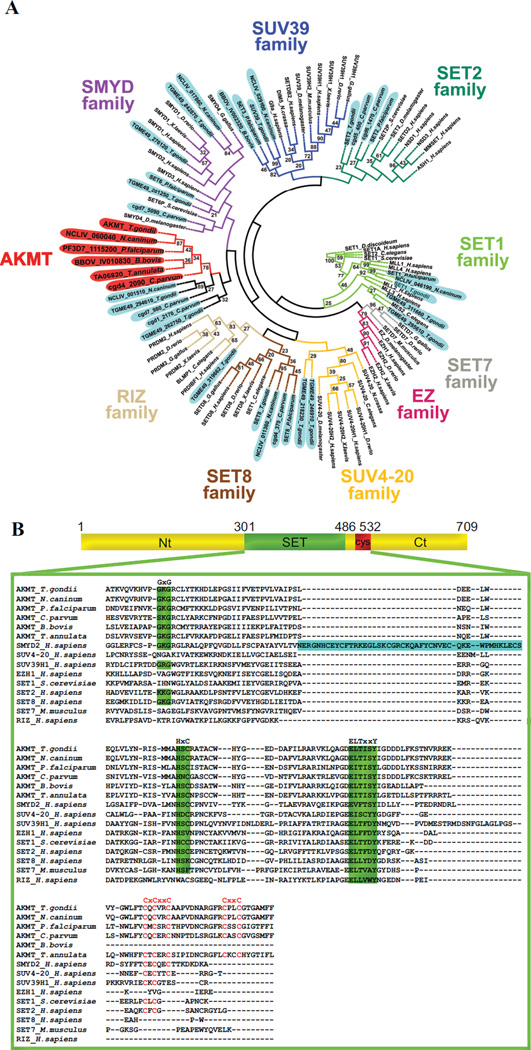
The KMTs have been classified into multiple subfamilies based on distinct features in the amino acid sequences of various SET domains. Determining the phylogenetic relationship of AKMT orthologs with respect to other KMTs will help us understand its function (if it belongs to a certain subfamily) or reveal its novelty (if the AKMT orthologs form a distinct cluster).There were two previous phylogenetic analyses of apicomplexan KMTs [49, 50].However, those studies either focused on KMTs in only Plasmodium sps [49] or did not include AKMT orthologs [50].We therefore carried out phylogenetic analysis of SET and post-SET domains in AKMT orthologs as well as those in members of known KMT families using a distance based neighbor-joining method [43, 51] (Figure 1A, Supplementary Table I).Our analysis shows that the TgAKMT is contained within a cluster that includes its apicomplexan orthologs as well as several other putative apicomplexan KMTs that share significant homology with the AKMTs in the C-terminal region of SET domain and in the post-SET cysteine cluster. This AKMT-containing clade is adjacent to that of SMYDs (Figure 1A), a family of KMTs involved in transcriptional regulation for oncogenesis as well as developmental processes such as muscular differentiation [52–55]. This is consistent with the results from a previous phylogenetic analysis of Plasmodium KMTs [49].However, although clustered closely to SMYDs, multiple sequence alignment (Figure 1B) shows that AKMT orthologs form a distinct subfamily. For instance, they lack a MYND zinc finger domain within the SET domain, a unique and fundamental feature of the SMYDs. Further, the highly-conserved post-SET cysteine cluster in AKMT orthologs (CXCX2CX11CX2C) contains two extra cysteines and is considerably longer than that (CXCX2C) in SMYDs. Besides the unusual enzymatic core, the regions in AKMT N- or C-terminal to this domain are also novel and no significant homology is found outside Apicomplexa.
Figure 1. Phylogenetic analyses of KMTs.
A) Phylogenetic analysis of SET and post-SET domains from AKMT orthologs and KMTs representing several other subfamilies of SET-domain containing methyltransferases. The tree shows that AKMT orthologs form a distinct clade close to the SMYD subfamily. Gene identifiers used are either accession numbers or, when available, annotated gene names in publicly accessible databases (e.g. GenBank, EuPathDB or ToxoDB). The names for apicomplexan KMTs are shaded (AKMTs in red and others in light blue). The name and branches of each subfamily are color-coded. Bootstrap percentages higher than 20 are displayed.
B) Protein sequence alignment of AKMT SET and post-SET domains with representative members of the KMT superfamily. Green highlighting indicates highly conserved motifs found in SET domain-containing proteins. Letters highlighted in red indicate cysteine cluster. Note that although AKMTs and SMYDs clusters are closely placed in the phylogenetic analysis, they each have some distinct features. SMYDs have a MYND zinc finger domain (highlighted in blue) within the SET domain, a unique and fundamental feature of this subfamily. The post-SET cysteine cluster in AKMTs (CXCX2CX11CX2C) contains 2 more cysteine residues and is significantly longer than that (CXCX2C) in SMYDs. Accession numbers of the proteins used in the alignment are: AKMT_T.gondii: TGME49_016080, AKMT_N.caninum: NCLIV_060040, AKMT_P.falciparum: PF3D7_1115200, AKMT_C.parvum: cgd4_2090, AKMT_B.bovis: BBOV_IV010830, AKMT_T.annulata: TA06820, SMYD2_H.sapiens: NP_064582, SUV4-20H1_H.sapiens: NP_060105, SUV39H1_H.sapiens: NP_003164, EZH1_H.sapiens: NP_001982, SET1_S.cerevisiae: EDN62358, SETD2_H.sapiens: NP_054878, SETD8_H.sapiens: Q9NQR1, SETD7_Mus musculus: NP_542983, RIZ_H.sapiens: Q13029.
3.2 The enzymatic activity of AKMT in vitro is not sensitive to free [Ca2+]
Previous studies have shown that the motility activation leading to parasite egress can be induced by calcium flux into the parasite [56, 57] and we have shown that the lysine methyltransferase activity of AKMT is required for the parasite motility and that the localization of AKMT is sensitive to intra-parasite [Ca2+] [40]. It is therefore conceivable that calcium might regulate the activity of AKMT. To test this hypothesis, we examined the effect of varying concentrations of free calcium on the methyltransferase activity of AKMT, using an artificial substrate (histone H3.3) and 3H labeled cofactor SAM in an in vitro methylation assay (Figure 2). We found that the methylation of the substrate by AKMT is not affected by varying concentrations of free calcium, suggesting that calcium flux does not have a direct effect on regulating enzyme activity of AKMT. However, it is still possible that its activity is indirectly regulated by calcium in vivo, via for example, the activity of calcium binding proteins, such as calmodulins, which are known to regulate the activity of many signaling proteins, including the calmodulin dependent mediated kinases [58–62] .
3.3 The predicted enzymatic domain (SET and post-SET cysteine cluster) only exhibits weak methyltransferase activity
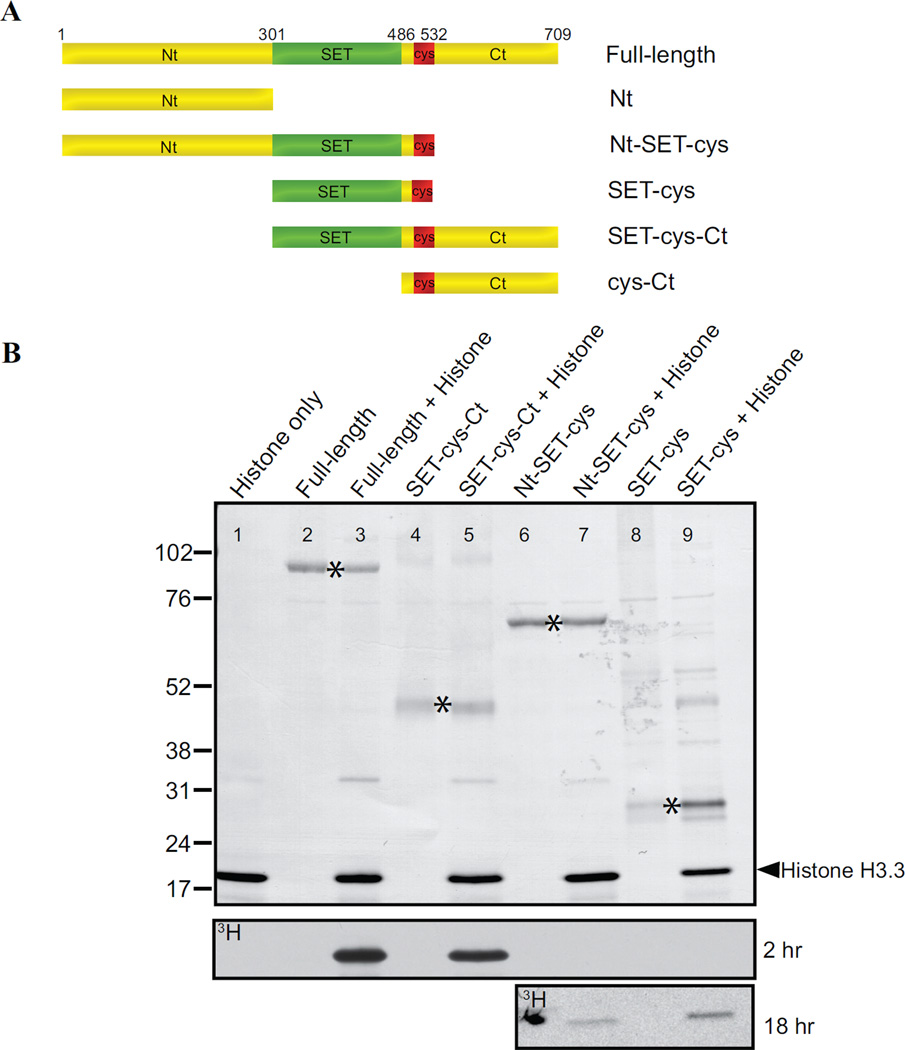
The SET domain (SET; 301–486 aa) and post-SET cysteine cluster (cys; 487–532 aa) are the only recognizable features in AKMT (Figure 1B). Although we have previously shown that a single point mutation within the SET domain (H447V) abolishes the enzyme activity in vitro as well as the motility regulation function in vivo [40], it was not known whether this predicted enzymatic core is sufficient for these functions. Also not known is how other domains of AKMT contribute to its function. To address these questions, we generated recombinant proteins with N-terminal FLAG-tag for the truncations that include the SET domain and the cysteine cluster (Figure 3A). We found that SET-cys (301–532 aa) exhibits only weak methyltransferase activity (Figure 3B), producing signal only detectable upon prolonged exposures. The inclusion of the N-terminal domain (Nt; 2–300 aa) together with SET-cys in the truncation (Nt-SET-cys; 2–532 aa) does not rescue the enzyme activity. In contrast, SET-cys and the C-terminal domain (Ct;533–709 aa) together (SET-cys-Ct; 301–709aa) constitute a robust enzyme, with an activity level similar to that of full-length AKMT under the condition of this assay (Figure 3B). Therefore SET-cys is necessary [40], but not sufficient for AKMT enzyme activity. Further, the N- and C- terminal domains of AKMT have drastically different contributions to the enzyme activity in vitro, with the N-terminal domain being largely dispensable and the C-terminal domain being very crucial.
Figure 3. In vitro analyses of the lysine methyltransferase activity of AKMT truncations.
A) Schematic representation of the full-length and truncated AKMT proteins used in the study. Full-length: 2–709 aa; Nt: 2–300 aa; Nt-SET-cys: 2–532 aa; SET-cys: 301–532 aa; SET-cys-Ct: 301–709 aa; cys-Ct: 487–709 aa.
B) In vitro methyltransferase assays were performed on histone H3.3 with FLAG tagged recombinant full-length and truncated AKMT constructs that contain the putative enzymatic core, the SET-cys domain. Top: Blot stained with amido black showing total protein in the reactions: histone H3.3 alone (lane 1), full-length alone (lane 2), full-length + histone H3.3 (lane 3), SET-cys-Ct alone (lane 4), SET-cys-Ct + histone H3.3 (lane 5), Nt-SET-cys alone (lane 6), Nt-SET-cys + histone H3.3 (lane 7), SET-cys alone (lane 8), and SET-cys + histone H3.3 (lane 9). The protein truncations are indicated by an asterisk and the position of histone H3.3 is indicated by the arrowhead. Bottom: Autoradiographs showing 3H signals of the same blot after 2 hour (for all lanes) and 18 hour (for lanes 6–9) exposures. The irregularly-shaped dark spot observed (lane 6) in the 18 hr exposure panel is an artifact.
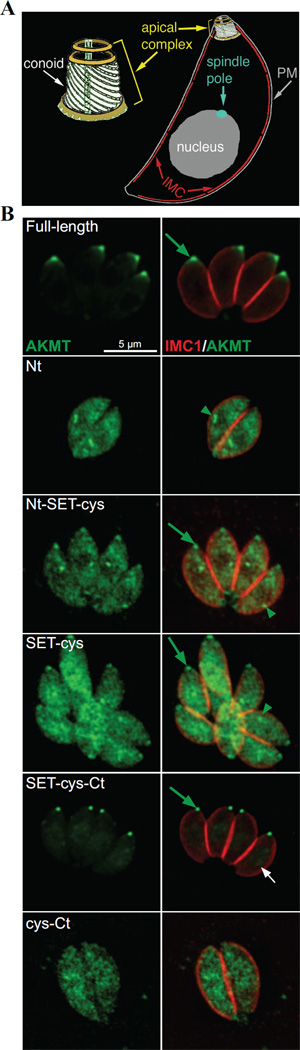
3.4 The C-terminal domain of AKMT is required for the proper localization of AKMT
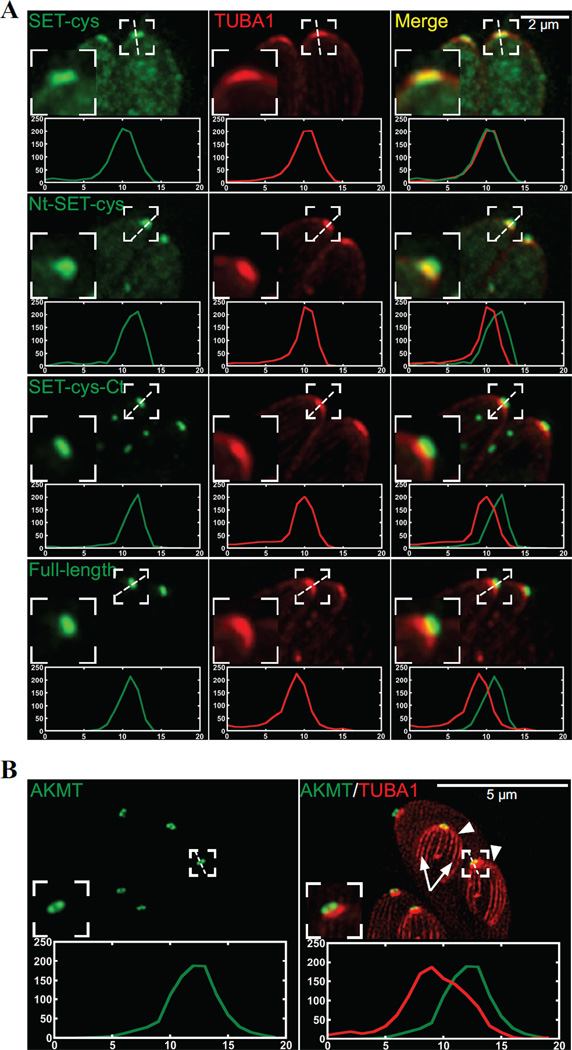
How do these modules each contribute to AKMT behavior in the parasite? To address this question, we first examined the localization of various AKMT truncations by transiently expressing fusions with N-terminal eGFP-tag. To eliminate the interference from endogenous AKMT, we expressed these fusion proteins in the AKMT knockout (Δakmt) mutant (Figure 4). In intracellular parasites, full-length AKMT is concentrated in the apical complex (Figure 4). For both Nt-SET-cys and SET-cys, which displayed weak enzyme activity in vitro, a significant amount of the fusion proteins were found in the parasite body. They were also localized to a perinuclear foci, likely to be the spindle pole of the parasite. Nt-SET-cys and SET-cys are incorporated into the apical complex with different consistency. Nt-SET-cys was incorporated into apical complex in all transfected cells examined, but the apical localization of SET-cys was only detectable in some (Figure 4) but not others (data not shown). In contrast to Nt-SET-cys and SET-cys, SET-cys-Ct, an active enzyme in vitro, is incorporated very efficiently into the apical complex, largely recapitulating the localization of the full-length AKMT (Figure 4).Although unlike full-length AKMT, some eGFP-SET-cys-Ct enters the nucleus (Figure 4B, white arrow), perhaps due to its smaller size. Interestingly, the C-terminal 223 aa (cys-Ct) exhibited a completely cytosolic distribution, indicating that an efficient targeting of AKMT to the apical complex requires the coordination of at least two domains (SET-cys and Ct), and either part on its own fails to properly localize. Upon closer examination, we noticed that when SET-cys was incorporated into the apical complex, the shape of its apical labeling appeared to be different from those of other SET-cys containing constructs. To confirm this, we co-localized eGFP-tagged full-length or truncated AKMT proteins (Nt-SET-cys, SET-cys, and SET-cys-Ct) with a fluorescently tagged T.gondii α1-Tubulin (TubA1), which marks the conoid- a tubulin-containing substructure within the apical complex [63] (Figure 5, cf. Figure 4A). We found that while the apical labeling of SET-cys is restricted to the conoid in most parasites analyzed (78%, n=18), the full-length AKMT, SET-cys-Ct, or Nt-SET-cys is found in a region anterior to the conoid (Figure 5A). Using a structured illumination (SIM) based super-resolution imaging, we further confirmed that AKMT indeed resides in a ring-like structure anterior to the conoid (Figure 5B). Thus, compared with SET-cys alone, there is a change in apical labeling pattern when the N- and/or C-terminal domain is combined with SET-cys. As the N- and C- terminal domains of AKMT themselves do not target to the apical complex (cf. Figure 4), this suggests that new interfaces form when SET-cys and the N- or C-terminal domains interact. This allows for the association with a different set of proteins in the apical complex and results in a shift in apical localization. Further, consistent with our previous hypothesis [40], AKMT localization to the apical complex is greatly dependent on AKMT enzyme activity, for which the interaction between SET-cys and the C-terminal domain is critical.
Figure 4. Cellular localization of the AKMT truncations.
A) Cartoon drawing showing several membrane and cytoskeletal structures referred to in the text. For clarity, cortical microtubules of the parasite are not shown. PM: Plasma Membrane. IMC: Inner Membrane Complex
B) Immunofluorescence of intracellular parasites expressing eGFP-tagged full-length, Nt, Nt-SET-cys, SET-cys, SET-cys-Ct, or cys-Ct in the Δakmt background. Similar to full-length AKMT, SET-cys-Ct is incorporated into the apical complex (green arrow) very efficiently. SET-cys-Ct also shows faint nuclear localization (white arrow). Nt, Nt-SET-cys and SET-cys are predominantly distributed in the cytoplasm and found in a perinuclear foci (green arrowheads), likely to be the spindle pole. Nt-SET-cys is incorporated into the apical complex in all transfected cells examined, but the apical localization of SET-cys was only detectable in some (as shown), but not others (data not shown). cys-Ct is cytoplasmic. Each image is the sum projection of a deconvolved 3D stack. Red: anti-IMC1, outlining the parasite periphery; Green: eGFP fluorescence. The host cells occupy the dark space. They are not visible in these images, because they are not fluorescently labeled.
Figure 5. Colocalization of the AKMT truncations and T. gondii α1- tubulin.
A) Intracellular Δakmt parasites co-expressing mTAG-RFP-TgTUBA1 with eGFP tagged SET-cys, Nt-SET-cys, SET-cys-Ct, or full-length. To facilitate the line-scan analyses, maximum intensity projections of deconvolved 3-D stacks are used. Red: mTAG-RFP-TgTUBA1; Green: eGFP fluorescence. Insets: 2X.
B) Structured illumination-based super-resolution imaging of dividing intracellular parasites co-expressing mCherryFP-TgTUBA1 and eGFP-AKMT-full length in the Δakmt background. Arrowheads: daughters. Arrows: daughter cortical microtubules. Contrast of the mCherryFP-TgTUBA1 image is adjusted to display the daughter cortical microtubules. Insets: 2X. Line-scans represent fluorescence intensity variation along the region indicated by the white dotted lines. Green:AKMT. Red:TUBA1. X-axis: distance in pixels. Y-axis: relative intensity in arbitrary units.
3.5 The C-terminal domain of AKMT is crucial to the motility regulating function of AKMT
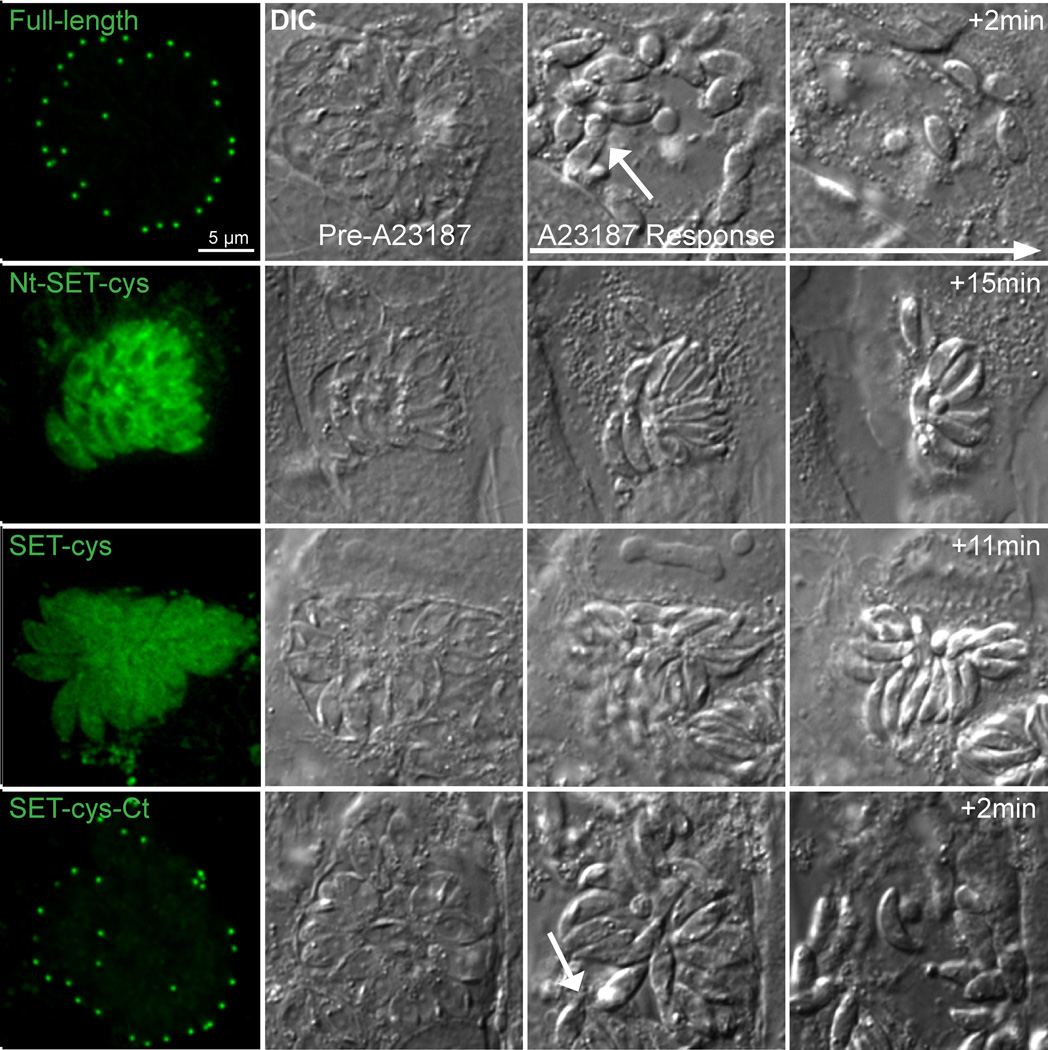
How do the localization and enzymatic activity of these truncation products correlate with their function in vivo? Our previous study showed that Δakmt parasites display a motility activation defect [40]. As a result, these parasites fail to exit the vacuole actively during egress, although they are able to secret pore-forming proteins [e.g. Toxoplasma perforin-like protein 1 [39] like the wild-type parasites to permeablize the host cell membrane [40]. To test the functionality of the AKMT truncations, we transfected eGFP fusion of those that contain the enzymatic core (SET-cys, Nt-SET-cys, and SET-cys-Ct), into Δakmt parasites and examined their ability to rescue the phenotype of the Δakmt parasites during calcium ionophore (A23187) induced egress (Figure 6). We found that upon A23187 exposure, in a majority of the vacuoles containing parasites expressing SET-cys (97%, n=31 vacuoles) or Nt-SET-cys (78%, n=38 vacuoles), no active parasite dispersion occurred despite efficient permeabilization of the surrounding membrane (as indicated by the increase in refractivity of the parasites and contraction of the host cell) (Figure 6). In 22% of vacuoles containing parasites expressing Nt- SET-cys, active egress was observed. However, in these cases, the egress was often incomplete, i.e. only a minority of the parasites actively exit with most parasites in the vacuole remaining immobile. It also usually occurred with a significant delay after membrane permeabilization. The slightly higher level of complementation in parasites expressing eGFP-Nt-SET-cys compared with the those expressing eGFP-SET-cys suggests that despite having no apparent effect on in vitro enzyme activity, the N-terminal domain of AKMT contributes, albeit very modestly, to motility activation in vivo. In contrast to the low level of complementation by Nt-SET-cys, complete and active egress occurred in a majority of vacuoles (~75%, n=39) containing the eGFP-SET-cys-Ct expressing parasites. Further, in these vacuoles, egress and membrane permeabilization occurred nearly simultaneously (Figure 6), similar to parasites expressing full-length AKMT (98% active egress, n=37). eGFP-SET-cys-Ct-expressing parasites that failed to actively egress (25%) all had a very high level expression of the truncation (data not shown),where the truncation is no longer predominantly localized to the apical complex, but also found at a high level in the cytoplasm and nucleus (likely due to the saturation of the binding sites in the apical complex). We suspect that the lack of egress in these vacuoles reflects an indirect effect of over-expression induced toxicity. The C-terminal domain of AKMT therefore is crucial to not only the enzymatic activity, but also the motility regulating function of AKMT in the parasite.
Figure 6. Functional complementation by AKMT truncations in Δakmt parasites.
Images show the representative results from induced egress experiments of Δakmt parasites expressing eGFP tagged full-length, Nt-SET-cys, SET-cys, SET-cys-Ct. Active dispersion occurred in most vacuoles containing parasites expressing eGFP-tagged full-length or SET-cys-Ct upon 5 µM A23187 exposure. In these vacuoles, egress and membrane permeabilization occurred nearly simultaneously (White arrows in the full-length and SET-cys-Ct “A23187 response” panels indicate moving parasites). However, most parasites expressing eGFP tagged SET-cys and Nt-SET-cys failed to actively exit the vacuole despite efficient vacuole permeabilization (as evidenced by the increase in refractivity of the culture).
4. DISCUSSION
Lysine methylation was first extensively studied in histones [64–70] and later in other nuclear proteins, such as p53, for its role in transcription control [53, 71, 72]. In T. gondii, the first characterized histone lysine methyltransferase (KMT) was TgSET8, which is capable of mono-, di- and trimethylating histone H4 and was found to associate with hetereochromatic domains in chromatin immunoprecipitation experiments [50]. Another interesting nuclear KMT identified in T. gondii is TgKMTox, which forms a complex with peroxiredoxin-1 and might be involved in oxidative stress responses by regulating the expression of relevant genes [70].Recently, a histone H3 lysine 4 methyltransferase, PfSET10, has been shown to specifically associate with the var gene locus in Plasmodium falciparum. The disruption of its function appears to affect the expression level of certain var genes [73]. Although AKMT can methylate histones in vitro, several observations in our previous work make it highly unlikely that AKMT regulates motility by controlling gene expression [40]. First, canonical histone lysine methyltransferases are chromatin-bound proteins concentrated in the nucleus, whereas AKMT is undetectable in the nucleus. Furthermore, inhibition of transcription and translation has no effect on AKMT regulatory function. In recent years, it becomes increasingly clear that the lysine methylation of non-nuclear proteins is common. These proteins span a variety of functions,including a chloroplast protein (Rubisco, for ribulose- 1,5-bisphosphate carboxylase/oxygenase) [74, 75], a mitochondrial protein (cytochrome C) [76, 77], a growth factor receptor (vascular endothelial growth factor receptor 1) [78], calmodulin [79], an ATP-Synthase [80] and several ribosomal proteins [81]. However, the purpose for these modifications remains poorly understood. AKMT provides a clear example that lysine methylation is directly involved in an important cellular function (motility regulation) other than transcriptional control [40]. Our previous work demonstrated that AKMT and its enzyme activity are crucial in regulating the motility state switch of T. gondii, an essential step in the lytic cycle of this parasite. Here we further characterized this protein and found that the AKMT orthologs form a new subfamily. We also discovered that besides the SET domain containing enzymatic core, the C-terminal 177 aa of AKMT are also required for AKMT functions. These two modules together constitute a protein that is necessary and sufficient to function as a lysine methyltransferase, to efficiently localize the protein to the apical complex, as well as to regulate motility regulation during induced egress.
In our phylogenetic analysis, the AKMT and SMYD families branch off from a common node. However, they have clearly diverged from each other during evolution before the speciation of the apicomplexan parasites. Further, orthologs of AKMT are found in the genomes of other apicomplexan parasites such as Plasmodium sps, but not in animals. AKMT thus represents a new subclass of lysine methyltransferases distinct from those in the host, therefore could be a target for treatment and preventive measures.
SET and post-SET domains from certain KMTs, such as SET8 or ASH1, form an active methyltransferase [82, 83]. SET-cys in AKMT, which contains the predicted SET domain and the post-SET cysteine cluster, displays only very poor methyltransferase activity in vitro. The addition of the N-terminal domain of AKMT to SET-cys appears to have little effect on the enzymatic activity of AKMT in vitro when histone H3.3 is used as the artificial substrate.However, compared with SET-cys, Nt-SET-cys displays a slightly higher level of in vivo complementation and its localization pattern within the apical complex is also more similar to that of wild-type AKMT allele, suggesting that the N-terminal domain might serve minor modulating role for AKMT function via regulating its in vivo activity and/or localization. In contrast to the very modest contribution of the N-terminal domain to AKMT function, the C-terminal 177 aa of AKMT is clearly important. The truncation (SET-cys-Ct) that includes both SET-cys and Ct to a large extent mimics the in vitro activity, in vivo complementation and localization of the wild-type allele. This reveals that the C-terminal domain contains novel features that coordinate with the enzymatic core for processing substrates, which in turn is critical for protein localization and function in vivo. Importantly, this domain is well conserved within Apicomplexa. The C-terminal domain of T. gondii AKMT shares ~44% identity with the P. falciparum ortholog in the corresponding region, but no significant homology is found with any animal proteins. Thus the novel SET and the C-terminal domain, as well as the interaction between these two modules provide promising targets for designing drug for treating or preventing infection by this large group of important parasites.
In sum, AKMT is a novel KMT with interesting features both in terms of its domain structure as well as its function in vivo. Future work including comprehensive identification of substrates, interaction partners, and small-molecule inhibitors will further elucidate how this new class of KMTs functions as a key enzyme and a motility regulator, which will facilitate a deeper understanding of KMT biology in general.
Supplementary Material
Highlights.
AKMT orthologs form a clade distinct from other KMTs.
The predicted core enzymatic domain of AKMT only has weak activity in vitro.
The C-terminal domain of AKMT is crucial for its enzyme activity in vitro.
The C-terminal domain of AKMT is crucial for its motility regulating function.
ACKNOWLEDGEMENTS
We thank Dr. Gary Ward (University of Vermont) for the mouse anti-IMC1 antibody, Dr. John Murray (University of Pennsylvania) for the ptub-mCherryFP-TubA1 and the pmin-mTagRFP-T-TgCentrin2-v2 plasmids, and Dr. Jim Power and the LMIC at Indiana University for imaging support. This study was supported by March of Dimes (6-FY12-258) and NIH-NIAID (R01-AI098686).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Levine ND. Progress in taxonomy of the Apicomplexan protozoa. J Protozool. 1988;35:518–520. doi: 10.1111/j.1550-7408.1988.tb04141.x. [DOI] [PubMed] [Google Scholar]
- 2.Holliman RE. Toxoplasmosis and the acquired immune deficiency syndrome. J Infect. 1988;16:121–128. doi: 10.1016/s0163-4453(88)93847-9. [DOI] [PubMed] [Google Scholar]
- 3.Fleming AF. Opportunistic infections in AIDS in developed and developing countries. Trans R Soc Trop Med Hyg. 1990;84(Suppl 1):1–6. doi: 10.1016/0035-9203(90)90446-l. [DOI] [PubMed] [Google Scholar]
- 4.Belanger F, Derouin F, Grangeot-Keros L, Meyer L. Incidence and risk factors of toxoplasmosis in a cohort of human immunodeficiency virus-infected patients: 1988–1995. HEMOCO and SEROCO Study Groups. Clin Infect Dis. 1999;28:575–581. doi: 10.1086/515147. [DOI] [PubMed] [Google Scholar]
- 5.Vastagh I, Jelencsik I, Barsi P, Balint K, Szlavik J, Szirmai I. Single cerebral Toxoplasma abscess: first manifestation of HIV infection. Eur J Neurol. 1999;6:725–726. doi: 10.1046/j.1468-1331.1999.660725.x. [DOI] [PubMed] [Google Scholar]
- 6.Pereira-Chioccola VL, Vidal JE, Su C. Toxoplasma gondii infection and cerebral toxoplasmosis in HIV-infected patients. Future Microbiol. 2009;4:1363–1379. doi: 10.2217/fmb.09.89. [DOI] [PubMed] [Google Scholar]
- 7.Emina MO, Odjimogho SE. Ocular problems in HIV and AIDS patients in Nigeria. Optom Vis Sci. 2010;87:979–984. doi: 10.1097/OPX.0b013e3181fef198. [DOI] [PubMed] [Google Scholar]
- 8.Dubey JP, Tiao N, Gebreyes WA, Jones JL. A review of toxoplasmosis in humans and animals in Ethiopia. Epidemiol Infect. 2012;140:1935–1938. doi: 10.1017/S0950268812001392. [DOI] [PubMed] [Google Scholar]
- 9.Chaddha DS, Kalra SP, Singh AP, Gupta RM, Sanchetee PC. Toxoplasmic encephalitis in acquired immunodeficiency syndrome. J Assoc Physicians India. 1999;47:680–684. [PubMed] [Google Scholar]
- 10.Martin-Blondel G, Alvarez M, Delobel P, Uro-Coste E, Cuzin L, Cuvinciuc V, et al. Toxoplasmic encephalitis IRIS in HIV-infected patients: a case series and review of the literature. J Neurol Neurosurg Psychiatry. 2011;82:691–693. doi: 10.1136/jnnp.2009.199919. [DOI] [PubMed] [Google Scholar]
- 11.Subauste CS, Ajzenberg D, Kijlstra A. Review of the series "Disease of the year 2011: toxoplasmosis" pathophysiology of toxoplasmosis. Ocul Immunol Inflamm. 2011;19:297–306. doi: 10.3109/09273948.2010.605198. [DOI] [PubMed] [Google Scholar]
- 12.Basavaprabhu A, Soundarya M, Deepak M, Satish R. CNS toxoplasmosis presenting with obstructive hydrocephalus in patients of retroviral disease--a case series. Med J Malaysia. 2012;67:214–216. [PubMed] [Google Scholar]
- 13.Tan IL, McArthur JC. HIV-associated neurological disorders: a guide to pharmacotherapy. CNS Drugs. 2012;26:123–134. doi: 10.2165/11597770-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 14.Ganiem AR, Dian S, Indriati A, Chaidir L, Wisaksana R, Sturm P, et al. Cerebral Toxoplasmosis Mimicking Subacute Meningitis in HIV-Infected Patients; a Cohort Study from Indonesia. PLoS Negl Trop Dis. 2013;7:e1994. doi: 10.1371/journal.pntd.0001994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramirez-Crescencio MA, Velasquez-Perez L. Epidemiology and trend of neurological diseases associated to HIV/AIDS. Experience of Mexican patients 1995–2009. Clin Neurol Neurosurg. 2013 doi: 10.1016/j.clineuro.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 16.Boyer K, Hill D, Mui E, Wroblewski K, Karrison T, Dubey JP, et al. Unrecognized ingestion of Toxoplasma gondii oocysts leads to congenital toxoplasmosis and causes epidemics in North America. Clin Infect Dis. 2011;53:1081–1089. doi: 10.1093/cid/cir667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delair E, Latkany P, Noble AG, Rabiah P, McLeod R, Brezin A. Clinical manifestations of ocular toxoplasmosis. Ocul Immunol Inflamm. 2011;19:91–102. doi: 10.3109/09273948.2011.564068. [DOI] [PubMed] [Google Scholar]
- 18.McLeod R, Boyer K, Roizen N, Stein L, Swisher C, Holfels E, et al. The child with congenital toxoplasmosis. Curr Clin Top Infect Dis. 2000;20:189–208. [PubMed] [Google Scholar]
- 19.McLeod R, Kieffer F, Sautter M, Hosten T, Pelloux H. Why prevent, diagnose and treat congenital toxoplasmosis? Mem Inst Oswaldo Cruz. 2009;104:320–344. doi: 10.1590/s0074-02762009000200029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mets MB, Holfels E, Boyer KM, Swisher CN, Roizen N, Stein L, et al. Eye manifestations of congenital toxoplasmosis. Am J Ophthalmol. 1996;122:309–324. doi: 10.1016/s0002-9394(14)72057-4. [DOI] [PubMed] [Google Scholar]
- 21.Mets MB, Holfels E, Boyer KM, Swisher CN, Roizen N, Stein L, et al. Eye manifestations of congenital toxoplasmosis. Am J Ophthalmol. 1997;123:1–16. doi: 10.1016/s0002-9394(14)70986-9. [DOI] [PubMed] [Google Scholar]
- 22.Noble AG, Latkany P, Kusmierczyk J, Mets M, Rabiah P, Boyer K, et al. Chorioretinal lesions in mothers of children with congenital toxoplasmosis in the National Collaborative Chicago-based, Congenital Toxoplasmosis Study. Sci Med (Porto Alegre) 2010;20:20–26. [PMC free article] [PubMed] [Google Scholar]
- 23.Olariu TR, Remington JS, McLeod R, Alam A, Montoya JG. Severe congenital toxoplasmosis in the United States: clinical and serologic findings in untreated infants. Pediatr Infect Dis J. 2011;30:1056–1061. doi: 10.1097/INF.0b013e3182343096. [DOI] [PubMed] [Google Scholar]
- 24.Roizen N, Kasza K, Karrison T, Mets M, Noble AG, Boyer K, et al. Impact of visual impairment on measures of cognitive function for children with congenital toxoplasmosis: implications for compensatory intervention strategies. Pediatrics. 2006;118:e379–e390. doi: 10.1542/peds.2005-1530. [DOI] [PubMed] [Google Scholar]
- 25.Swisher CN, Boyer K, McLeod R. Congenital toxoplasmosis. The Toxoplasmosis Study Group. Semin Pediatr Neurol. 1994;1:4–25. [PubMed] [Google Scholar]
- 26.Dobrowolski JM, Carruthers VB, Sibley LD. Participation of myosin in gliding motility and host cell invasion by Toxoplasma gondii. Mol Microbiol. 1997;26:163–173. doi: 10.1046/j.1365-2958.1997.5671913.x. [DOI] [PubMed] [Google Scholar]
- 27.Lingelbach K, Joiner KA. The parasitophorous vacuole membrane surrounding Plasmodium and Toxoplasma: an unusual compartment in infected cells. J Cell Sci. 1998;111(Pt 11):1467–1475. doi: 10.1242/jcs.111.11.1467. [DOI] [PubMed] [Google Scholar]
- 28.Sheffield HG, Melton ML. The fine structure and reproduction of Toxoplasma gondii. J Parasitol. 1968;54:209–226. [PubMed] [Google Scholar]
- 29.Moudy R, Manning TJ, Beckers CJ. The loss of cytoplasmic potassium upon host cell breakdown triggers egress of Toxoplasma gondii. J Biol Chem. 2001;276:41492–41501. doi: 10.1074/jbc.M106154200. [DOI] [PubMed] [Google Scholar]
- 30.Morrissette NS, Sibley LD. Cytoskeleton of apicomplexan parasites. Microbiol Mol Biol Rev. 2002;66:21–38. doi: 10.1128/MMBR.66.1.21-38.2002. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin AM, Liu T, Lynn BC, Sinai AP. The Toxoplasma gondii parasitophorous vacuole membrane: transactions across the border. J Eukaryot Microbiol. 2007;54:25–28. doi: 10.1111/j.1550-7408.2006.00230.x. [DOI] [PubMed] [Google Scholar]
- 32.Mondragon R, Frixione E. Ca(2+)-dependence of conoid extrusion in Toxoplasma gondii tachyzoites. J Eukaryot Microbiol. 1996;43:120–127. doi: 10.1111/j.1550-7408.1996.tb04491.x. [DOI] [PubMed] [Google Scholar]
- 33.Lovett JL, Marchesini N, Moreno SN, Sibley LD. Toxoplasma gondii microneme secretion involves intracellular Ca(2+) release from inositol 1,4,5-triphosphate (IP(3))/ryanodine-sensitive stores. J Biol Chem. 2002;277:25870–25876. doi: 10.1074/jbc.M202553200. [DOI] [PubMed] [Google Scholar]
- 34.Carruthers VB, Sibley LD. Sequential protein secretion from three distinct organelles of Toxoplasma gondii accompanies invasion of human fibroblasts. Eur J Cell Biol. 1997;73:114–123. [PubMed] [Google Scholar]
- 35.Stommel EW, Ely KH, Schwartzman JD, Kasper LH. Toxoplasma gondii: dithiol-induced Ca2+ flux causes egress of parasites from the parasitophorous vacuole. Exp Parasitol. 1997;87:88–97. doi: 10.1006/expr.1997.4187. [DOI] [PubMed] [Google Scholar]
- 36.Hoff EF. Carruthers VB. Is Toxoplasma egress the first step in invasion? Trends Parasitol. 2002;18:251–255. doi: 10.1016/s1471-4922(02)02240-7. [DOI] [PubMed] [Google Scholar]
- 37.Carruthers VB, Sherman GD, Sibley LD. The Toxoplasma adhesive protein MIC2 is proteolytically processed at multiple sites by two parasite-derived proteases. J Biol Chem. 2000;275:14346–14353. doi: 10.1074/jbc.275.19.14346. [DOI] [PubMed] [Google Scholar]
- 38.Carruthers VB, Sibley LD. Mobilization of intracellular calcium stimulates microneme discharge in Toxoplasma gondii. Mol Microbiol. 1999;31:421–428. doi: 10.1046/j.1365-2958.1999.01174.x. [DOI] [PubMed] [Google Scholar]
- 39.Kafsack BF, Pena JD, Coppens I, Ravindran S, Boothroyd JC, Carruthers VB. Rapid membrane disruption by a perforin-like protein facilitates parasite exit from host cells. Science. 2009;323:530–533. doi: 10.1126/science.1165740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heaslip AT, Nishi M, Stein B, Hu K. The motility of a human parasite, Toxoplasma gondii, is regulated by a novel lysine methyltransferase. PLoS Pathog. 2011;7:e1002201. doi: 10.1371/journal.ppat.1002201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dillon SC, Zhang X, Trievel RC. Cheng X. The SET-domain protein superfamily: protein lysine methyltransferases. Genome Biol. 2005;6:227. doi: 10.1186/gb-2005-6-8-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheng X, Collins RE, Zhang X. Structural and sequence motifs of protein (histone) methylation enzymes. Annu Rev Biophys Biomol Struct. 2005;34:267–294. doi: 10.1146/annurev.biophys.34.040204.144452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Felsenstein J. PHYLIP - Phylogeny Inference Package (Version 3.2) Cladistics. 1989;Vol. 5:164–166. [Google Scholar]
- 44.Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 45.van den Hoff MJ, Moorman AF. Lamers WH. Electroporation in 'intracellular' buffer increases cell survival. Nucleic Acids Res. 1992;20:2902. doi: 10.1093/nar/20.11.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fingerman I, Du H, Briggs S. In Vitro Histone Methyltransferase Assay. Cold Spring Harb Protoc. 2008 doi: 10.1101/pdb.prot4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tan X, Rotllant J, Li H, De Deyne P, Du SJ. SmyD1, a histone methyltransferase, is required for myofibril organization and muscle contraction in zebrafish embryos. Proc Natl Acad Sci U S A. 2006;103:2713–2718. doi: 10.1073/pnas.0509503103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schoenmakers TJ, Visser GJ, Flik G, Theuvenet AP. CHELATOR: an improved method for computing metal ion concentrations in physiological solutions. Biotechniques. 1992;12:870–874. 6–9. [PubMed] [Google Scholar]
- 49.Cui L, Fan Q, Cui L, Miao J. Histone lysine methyltransferases and demethylases in Plasmodium falciparum. International journal for parasitology. 2008;38:1083–1097. doi: 10.1016/j.ijpara.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sautel CF, Cannella D, Bastien O, Kieffer S, Aldebert D, Garin J, et al. SET8-mediated methylations of histone H4 lysine 20 mark silent heterochromatic domains in apicomplexan genomes. Molecular and cellular biology. 2007;27:5711–5724. doi: 10.1128/MCB.00482-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 52.Gottlieb PD, Pierce SA, Sims RJ, Yamagishi H, Weihe EK, Harriss JV, et al. Bop encodes a muscle-restricted protein containing MYND and SET domains and is essential for cardiac differentiation and morphogenesis. Nat Genet. 2002;31:25–32. doi: 10.1038/ng866. [DOI] [PubMed] [Google Scholar]
- 53.Huang J, Perez-Burgos L, Placek BJ, Sengupta R, Richter M, Dorsey JA, et al. Repression of p53 activity by Smyd2-mediated methylation. Nature. 2006;444:629–632. doi: 10.1038/nature05287. [DOI] [PubMed] [Google Scholar]
- 54.Saddic LA, West LE, Aslanian A, Yates JR, 3rd, Rubin SM, Gozani O, et al. Methylation of the retinoblastoma tumor suppressor by SMYD2. J Biol Chem. 2010;285:37733–37740. doi: 10.1074/jbc.M110.137612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hamamoto R, Furukawa Y, Morita M, Iimura Y, Silva FP, Li M, et al. SMYD3 encodes a histone methyltransferase involved in the proliferation of cancer cells. Nat Cell Biol. 2004;6:731–740. doi: 10.1038/ncb1151. [DOI] [PubMed] [Google Scholar]
- 56.Lovett JL, Sibley LD. Intracellular calcium stores in Toxoplasma gondii govern invasion of host cells. J Cell Sci. 2003;116:3009–3016. doi: 10.1242/jcs.00596. [DOI] [PubMed] [Google Scholar]
- 57.Wetzel DM, Chen LA, Ruiz FA, Moreno SN, Sibley LD. Calcium-mediated protein secretion potentiates motility in Toxoplasma gondii. J Cell Sci. 2004;117:5739–5748. doi: 10.1242/jcs.01495. [DOI] [PubMed] [Google Scholar]
- 58.Pezzella N, Bouchot A, Bonhomme A, Pingret L, Klein C, Burlet H, et al. Involvement of calcium and calmodulin in Toxoplasma gondii tachyzoite invasion. Eur J Cell Biol. 1997;74:92–101. [PubMed] [Google Scholar]
- 59.Pezzella-D'Alessandro N, Le Moal H, Bonhomme A, Valere A, Klein C, Gomez-Marin J, et al. Calmodulin distribution and the actomyosin cytoskeleton in Toxoplasma gondii. J Histochem Cytochem. 2001;49:445–454. doi: 10.1177/002215540104900404. [DOI] [PubMed] [Google Scholar]
- 60.Sugi T, Kato K, Kobayashi K, Pandey K, Takemae H, Kurokawa H, et al. Molecular analyses of Toxoplasma gondii calmodulin-like domain protein kinase isoform 3. Parasitol Int. 2009;58:416–423. doi: 10.1016/j.parint.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 61.Kieschnick H, Wakefield T, Narducci CA, Beckers C. Toxoplasma gondii attachment to host cells is regulated by a calmodulin-like domain protein kinase. J Biol Chem. 2001;276:12369–12377. doi: 10.1074/jbc.M011045200. [DOI] [PubMed] [Google Scholar]
- 62.Lourido S, Shuman J, Zhang C, Shokat KM, Hui R, Sibley LD. Calcium-dependent protein kinase 1 is an essential regulator of exocytosis in Toxoplasma. Nature. 2010;465:359–362. doi: 10.1038/nature09022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hu K, Roos DS, Murray JM. A novel polymer of tubulin forms the conoid of Toxoplasma gondii. J Cell Biol. 2002;156:1039–1050. doi: 10.1083/jcb.200112086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, et al. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 65.Krogan NJ, Kim M, Tong A, Golshani A, Cagney G, Canadien V, et al. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Molecular and cellular biology. 2003;23:4207–4218. doi: 10.1128/MCB.23.12.4207-4218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schaft D, Roguev A, Kotovic KM, Shevchenko A, Sarov M, Shevchenko A, et al. The histone 3 lysine 36 methyltransferase, SET2, is involved in transcriptional elongation. Nucleic Acids Res. 2003;31:2475–2482. doi: 10.1093/nar/gkg372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 68.Gissot M, Kelly KA, Ajioka JW, Greally JM, Kim K. Epigenomic modifications predict active promoters and gene structure in Toxoplasma gondii. PLoS Pathog. 2007;3:e77. doi: 10.1371/journal.ppat.0030077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sautel CF, Ortet P, Saksouk N, Kieffer S, Garin J, Bastien O, et al. The histone methylase KMTox interacts with the redox-sensor peroxiredoxin-1 and targets genes involved in Toxoplasma gondii antioxidant defences. Mol Microbiol. 2009;71:212–226. doi: 10.1111/j.1365-2958.2008.06519.x. [DOI] [PubMed] [Google Scholar]
- 71.Chuikov S, Kurash JK, Wilson JR, Xiao B, Justin N, Ivanov GS, et al. Regulation of p53 activity through lysine methylation. Nature. 2004;432:353–360. doi: 10.1038/nature03117. [DOI] [PubMed] [Google Scholar]
- 72.Shi X, Kachirskaia I, Yamaguchi H, West LE, Wen H, Wang EW, et al. Modulation of p53 function by SET8-mediated methylation at lysine 382. Mol Cell. 2007;27:636–646. doi: 10.1016/j.molcel.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Volz JC, Bartfai R, Petter M, Langer C, Josling GA, Tsuboi T, et al. PfSET10, a Plasmodium falciparum methyltransferase, maintains the active var gene in a poised state during parasite division. Cell host & microbe. 2012;11:7–18. doi: 10.1016/j.chom.2011.11.011. [DOI] [PubMed] [Google Scholar]
- 74.Houtz RL, Poneleit L, Jones SB, Royer M, Stults JT. Posttranslational modifications in the amino- terminal region of the large subunit of ribulose- 1,5-bisphosphate carboxylase/oxygenase from several plant species. Plant Physiol. 1992;98:1170–1174. doi: 10.1104/pp.98.3.1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zheng Q, Simel EJ, Klein PE, Royer MT, Houtz RL. Expression, purification, and characterization of recombinant ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit nepsilon-methyltransferase. Protein Expr Purif. 1998;14:104–112. doi: 10.1006/prep.1998.0936. [DOI] [PubMed] [Google Scholar]
- 76.DeLange RJ, Glazer AN, Smith EL. Identification and location of episilon-N-trimethyllysine in yeast cytochromes c. J Biol Chem. 1970;245:3325–3327. [PubMed] [Google Scholar]
- 77.Pollock WB, Rosell FI, Twitchett MB, Dumont ME, Mauk AG. Bacterial expression of a mitochondrial cytochrome c. Trimethylation of lys72 in yeast iso-1-cytochrome c and the alkaline conformational transition. Biochemistry. 1998;37:6124–6131. doi: 10.1021/bi972188d. [DOI] [PubMed] [Google Scholar]
- 78.Kunizaki M, Hamamoto R, Silva FP, Yamaguchi K, Nagayasu T, Shibuya M, et al. The lysine 831 of vascular endothelial growth factor receptor 1 is a novel target of methylation by SMYD3. Cancer Res. 2007;67:10759–10765. doi: 10.1158/0008-5472.CAN-07-1132. [DOI] [PubMed] [Google Scholar]
- 79.Takemori N, Komori N, Thompson JN, Jr, Yamamoto MT, Matsumoto H. Novel eye-specific calmodulin methylation characterized by protein mapping in Drosophila melanogaster. Proteomics. 2007;7:2651–2658. doi: 10.1002/pmic.200700343. [DOI] [PubMed] [Google Scholar]
- 80.Katz ML, Christianson JS, Norbury NE, Gao CL, Siakotos AN, Koppang N. Lysine methylation of mitochondrial ATP synthase subunit c stored in tissues of dogs with hereditary ceroid lipofuscinosis. J Biol Chem. 1994;269:9906–9911. [PubMed] [Google Scholar]
- 81.Porras-Yakushi TR, Whitelegge JP, Clarke S. Yeast ribosomal/cytochrome c SET domain methyltransferase subfamily: identification of Rpl23ab methylation sites and recognition motifs. J Biol Chem. 2007;282:12368–12376. doi: 10.1074/jbc.M611896200. [DOI] [PubMed] [Google Scholar]
- 82.An S, Yeo KJ, Jeon YH, Song JJ. Crystal structure of the human histone methyltransferase ASH1L catalytic domain and its implications for the regulatory mechanism. J Biol Chem. 2011;286:8369–8374. doi: 10.1074/jbc.M110.203380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yin Y, Liu C, Tsai SN, Zhou B, Ngai SM, Zhu G. SET8 recognizes the sequence RHRK20VLRDN within the N terminus of histone H4 and mono-methylates lysine 20. J Biol Chem. 2005;280:30025–30031. doi: 10.1074/jbc.M501691200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.