Abstract
The ability to generate a massive amount of sequencing and genotyping data is transforming the study of human genetic disorders. Driven by such innovation, it is likely that whole exome and whole-genome resequencing will replace regionally focused approaches for gene discovery and clinical testing in the next few years. However, this opportunity brings a significant interpretative challenge to assigning function and phenotypic variance to common and rare alleles. Understanding the effect of individual mutations in the context of the remaining genomic variation represents a major challenge to our interpretation of disease. Here, we discuss the challenges of assigning mutation functionality and, drawing from the examples of ciliopathies as well as cohesinopathies and channelopathies, discuss possibilities for the functional modularization of the human genome. Functional modularization in addition to the development of physiologically-relevant assays to test allele functionality will accelerate our understanding of disease architecture and enable the use of genome-wide sequence data for disease diagnosis and phenotypic prediction in individuals.
Emerging challenges in genomics
The emergence of new genomic technologies is catalyzing the unprecedented production of sequence and genotype information from patients with both rare and common disorders. This, in turn, is expediting the identification of disease-causing genes under traditional mendelian paradigms, such as the recent whole exome sequencing of small numbers of unrelated individuals that identified new variants in two rare mendelian disorders1, 2, and provides promise for a successful transition from haplotypic association to allelic causality in complex traits. Behind these endeavors is the potential to query the extent of variation in normal and disease genomes allowing new insights into the underlying biology of disorders. This approach has been successful for rare traits, where gene and mutation identification have illuminated pathways associated with clinical phenotypes. Many of these studies have been model-free and the resultant functional pathway not obviously linked to the phenotype. The same approach has found some success in complex traits as well. For example, multiple genome-wide studies in large cohorts have linked age-related macular degeneration (AMD) to genes involved in the complement cascade 3 and similar studies in Crohn's disease have revealed an interesting contribution of the autophagy pathway 4, 5.
Despite the perceived differences between rare and complex traits, the fundamental questions of disease architecture and mechanism are identical: How can we categorize the variants and genes to gain a better understanding of the biology? Which alleles drive phenotypes and which alleles modulate the effect of cis and trans genetic lesions? In this review, we will focus on two primary issues that are pertinent to all genetic disorders, irrespective of frequency of disease and alleles, penetrance and expressivity. First, we will explore the idea of mutational load in genetic disease as a potentially accurate predictor of allele pathogenicity and clinical outcomes. We will ask whether ‘modularization’ of the human functional genome might offer advantages and opportunities in solving mechanism of disease, allelic effects and in providing predictive clinical power to genotypic information. Second, we discuss the challenges in establishing the pathogenic potential of alleles with respect to genetic disease when the functional contribution of a particular gene is unknown. To address this question, we offer potential solutions and briefly compare the current tools available for the functional assessment of allele functionality.[SC1]
Modular organization of disease genes
Discussion of modularization of genetic disease is not new. Large informatics-based studies have determined that categorization of phenotypes, such as disorders of the eye or gastrointestinal defects, can be used to create networks of diseases and that these networks are overlapping and interlinked 6. Similarly, genes associated with disease have been classified by protein function to understand how similar proteins might contribute to distinct, but overlapping, phenotypes, as is the case with Stickler syndrome, Marshall syndrome and oto-spondylo-mega-epiphyseal dysplasia (OSMED) syndrome, which are all caused by mutations in collagen genes 7-9. Although useful, such categorization of genes based solely on a narrow phenotypic outcome might have insufficient functional resolution, for instance when significantly different molecular pathways underpin similar phenotypes. For example, non-syndromic retinitis pigmentosa, the progressive loss of photoreceptor or retinal pigment epithelium function, has been linked to mutations in many different genes with functions ranging from photoreceptor specification (CRX) to restoration of visual pigment function (RGR) 10, 11. Likewise, relying exclusively on protein function can be both limiting and hazardous, because most proteins have multiple functions that can contribute to entirely different processes and are often cell or tissue dependent. For example, there is the lack of phenotypic overlap in disorders resulting from fibroblast growth factor receptor 1 (FGFR1) loss-of-function mutations causing Kallmann Syndrome, a disorder characterized by anosmia and hypogonadotrophic hypogonadism 12, and gain-of-function mutations causing Pfeiffer Syndrome, which commonly presents with craniosyntosis and cutaneous syndactyly 13, 14.
A combination of phenotypic and functional modularization offers added value, as demonstrated by overlapping of the human disease network (HDN) with the disease gene network (DGN) where groups of genes contributing to a particular disorder or disorder group are more likely to be involved in similar molecular processes in the cell 15. Under such a paradigm, it is possible to cluster disorders based either on their organellar dysfunction or on their commonalities of signaling defects. Identification of genes by querying candidate pathways has been previously explored. For example, a potential digenic model for inheritance of polycystic ovary syndrome (PCOS) in cortisone reductase deficiency (CRD) patients was established when investigation of genes in the glucocorticoid pathway revealed mutations in hexose-6-phosphate dehydrogenase, a regulator of glucocorticoid availability which acts via a second mutated gene, 11ß-hydroxysteroid dehydrogenase type 1 16. For some disorders, such as those associated with cilia, mitochondria, or peroxisomes, grouping by their organellar site of dysfunction is sufficient to explain the phenotype. In others, such as the cohesinopathies or channelopathies, grouping by cellular functional modalities that are not necessarily organelle-specific might be the best way to capture the breadth and variability of the phenotype.
Organellar grouping of disorders: the ciliopathies
Cilia were first observed in the kidney and the thyroid gland 17, but are now known to be present in nearly all mammalian cells. Extending from the apical surface of the cell and tethered to the basal body, cilia typically consist of nine microtubule doublets around the periphery extending from the basal body, with a subset of cilia containing an additional central microtubule pair that is though to impart motile functions 18. The near ubiquitous presence of cilia in the vertebrate body plan produces a wide range of phenotypes. To date, cell types known to be impacted by ciliary dysfunction include renal and retinal tissues, the neural tube, developing limbs, and the central nervous system 19. It is likely that as our knowledge of these organelles expands, additional tissue defects will also be recognized.
Defects that impact on the function of cilia, directly or indirectly, have been demonstrated in many diseases, including polycystic kidney disease (PKD), nephronophthisis (NPH), Alstrom Syndrome (ALMS), Bardet-Biedl Syndrome (BBS) and Meckel-Gruber syndrome (MKS). The common causality of ciliary dysfunction has led to the grouping of these discrete disorders as the ciliopathies 19. The availability of integrated gene and protein databases for ciliary proteins 20-30 is facilitating both the identification of new genes mutated in these disorders and the recognition of additional clinical entities as ciliopathies. For example, Jeune asphyxiating thoracic dystrophy (JATD), a lethal disorder characterized by a narrow and rigid thoracic cage, was recently characterized as a ciliopathy. The generation of a phenotypic matrix based on the symptoms of known ciliary disorders 19 such as retinal dystrophy, polydactyly, renal cysts, CNS malformations and situs inversus, identified JATD as a candidate ciliopathy. Subsequent cross-referencing of JATD critical regions in the genome with the ciliary proteome led to the identification of causative mutations in IFT80, proving that the ciliopathy module has a robust predictive value and significantly accelerating the identification of the first gene for this disorder 31. Indeed, the observation that JATD is a ciliopathy had a direct impact on other similar phentoypes, in particular short rib polydactyly, which was shown recently to be caused by mutations in a component of the cytoplasmic dynein complex DYNC2H1, which is necessary for proper ciliogenesis 32.
The near-ubiquitous presence of cilia in the mammalian body plan probably underlies the profound pleiotropy of ciliopathy phenotypes, whereas complex genetic interactions between causal and modifying alleles in ciliopathy genes have contributed further to phenotypic variability. This can pose challenges for accurate diagnosis, especially in the absence of reliable genetic or biochemical assays. However, the similarity in disease mechanisms associated with defects in ciliary proteins results in significant overlap in the phenotypes observed across different ciliopathies. For example, defects in retinal and kidney function are observed across a range of ciliopathies owing to defects in photoreceptor and renal cilia 33. In other cell types, dysfunction leads to developmental abnormalities such as polydactyly and mental retardation or pulmonary defects.
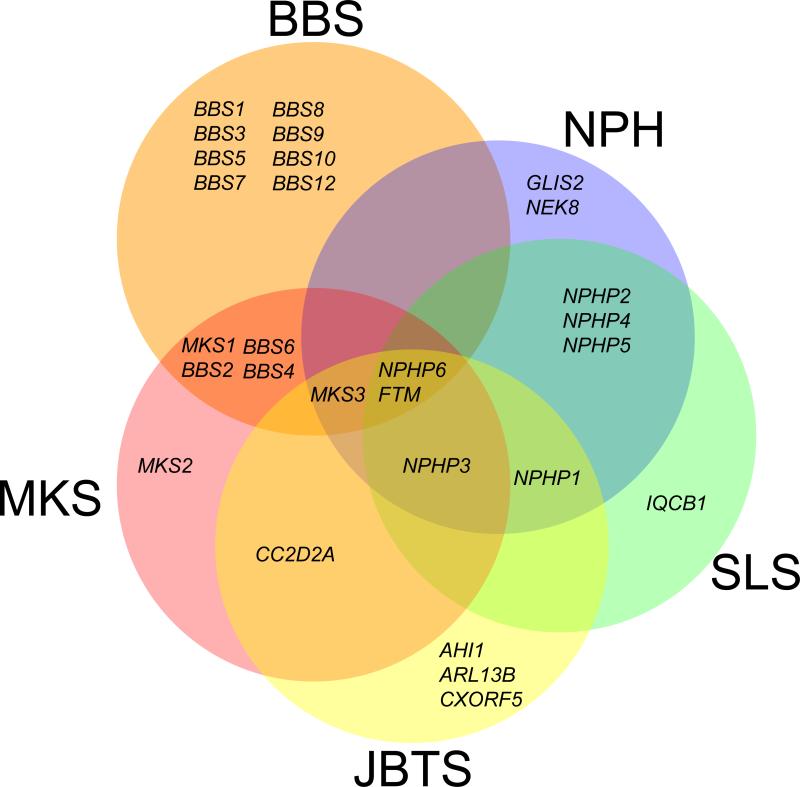
Similar phenotypes are observed in ciliopathies that were originally classified as distinct, unrelated clinical entities. However, not only does the significant phenotypic overlap argue against a compartmentalization of such overlapping disorders, but the underlying genetics raise a compelling grouping argument, because the same genes can contribute alleles to most, if not all, ciliopathies (Figure 1). For example, Bardet-Biedl syndrome (BBS) – a model ciliopathy and a developmental disorder diagnosed on the basis of the presence of obesity, retinal defects, polydactyly, hypogonadism, renal dysfunction and mental retardation 34 – is caused by mutations in 14 genes 23, 35-48, and at least three BBS-interacting loci contribute modifying mutations 49-51. BBS-associated proteins localize primarily at the pericentriolar region and the ciliary axoneme of some cells, and the transition zone and axoneme of sensory neurons in C. elegans 42, 52. Mutations in one or multiple BBS genes can result in clinical phenotypes, pointing to the necessity for protein–protein interactions. Several BBS proteins form a complex that can interact in vitro with the GTPase RAB8 to promote the formation of the ciliary membrane 53, consistent with the idea that multiple genes produce a disease phenotype,
Figure 1. Genetic overlap within the ciliopathy module.
There is significant overlap in the causative genes in disorders associated with ciliary dysfunction. Most genes associated with Nephronophthisis (NPH), Senior-Loken Syndrome (SLS), Joubert Syndrome (JBTS) and Meckel-Gruber Syndrome (MKS) have been associated with at least one other clinically distinct ciliopathy, with the exception of Bardet-Biedl Syndrome (BBS), for which a large proportion of causative genes have not been linked to other ciliopathies. Two genes in particular, NPHP6 and RPGRIP1L (FTM), have been associated across the ciliopathy module.
In addition, defects in BBS proteins lead to defects in the planar-cell polarity (PCP) pathway, an aspect of non-canonical Wnt signaling that regulates the elaboration of structures in three-dimensional space (for a review, see Ref [54]). Bbs4-null mice exhibit defects also seen in mutants for PCP-associated proteins, including exencephaly and rotated cochlear stereociliary bundles. Suppression of bbs4 in the zebrafish PCP mutant trilobite exacerbated the convergent extension defects common to PCP mutants 55. Importantly, many of the phenotypes associated with BBS can be attributed to PCP defects, including defective otoacoustic emissions 55 and renal cystic disease 56-59. This is presumably because the BBS proteins and components of the intraflagellar transport (IFT) machinery such as IFT88 and KIF3A are necessary for regulating Wnt signaling 60 by regulating β-catenin degradation, possibly at the level of proteasome 61.
BBS patients do not exhibit the more severe phenotypes seen in PCP mutants, which include neural tube defects, although the mouse Bbs4 knockout exhibits these features with modest (10-15%) penetrance 62. Neural tube defects, however, feature prominently and are part of the differential diagnosis of a more severe ciliopathy, Meckel-Gruber Syndrome, MKS 63. Importantly, mutations in at least three BBS genes have been reported in antenatal cases diagnosed with Meckel-like syndrome 64 whereas mutations in three genes associated with MKS, MKS1, MKS3 and NPHP6, have been identified in BBS patients 48. As such, BBS and MKS share common phenotypes associated with ciliary dysfunction 65 as well as several phenotypic features and common causal genetic relationships. These observations have cumulatively led to the suggestion that the two disorders represent different positions on a causality continuum caused by ciliary dysfunction and that they should be considered as part of the spectrum of the same clinical entity, a ciliopathy.
Clinical features of BBS overlap with MKS, and Mkks (Bbs6) is associated with both disorders 35, 36. One variant in particular, Y37C, has been reported in the homozygous state in BBS patients, but only in the heterozygous state in MKKS patients, suggesting that MKKS represents a milder, hypomorphic disorder caused by similar pathway defects 36. Although the clinical features common to both disorders include post-axial polydactyly, the Mkks-null mouse exhibits features common to BBS, including obesity and retinal degeneration, and to MKKS, [SC2]most notably hydrometrocolpos, an abnormality of fluid build-up in the female genitalia. This phenotype is also observed in Bbs4-null mice, supporting evidence of overlap between the two disorders 66. These examples highlight the fact that, although the correlation between genotype and phenotype severity has been limited, severity of phenotype can potentially be linked to the nature of mutations in particular module components.
The genetic ’pairing‘ of traditionally discrete clinical disorders is not restricted to these examples; rather, the emerging theme is one in which ciliopathy-causing genes have the capacity to contribute pathogenic alleles to most ciliopathies (Figure 1). For example, mutations in the ciliary gene RPGRIP1L, also known as FTM, have been identified in Joubert syndrome (JS), MKS, BBS, LCA, SLS and NPH patients, and mice ablated for this locus bare the cerebral, renal and hepatic defects associated with both of these disorders [SC3]67. JS also provides examples of this phenomena 51, 68: in addition to mutations in the Jouberin gene (AHI1) that have been reported only in JS patients 69-72, these patients also have mutations in MKS3 (one of the genes associated with Meckel syndrome) 73, which is associated with Nephronopthisis 74. Mutations in another gene, NPHP6, also cause JS, and other ciliopathies including BBS, Meckel syndrome, Nephronophthisis and Leber congential amaurosis (LCA) 48, 75, 76.
An examination of the allelic overlap between ciliopathies illustrates two key points: (i) the historical compartmentalization of the disorders in the ciliopathy continuum is insufficient to explain the genetic observations; and (ii) the genotype at a single locus cannot accurately predict the phenotype. This supports the idea that the ciliopathies share both phenotypes and genetic mechanisms, and give credence to the model where distinct disorders are variations of a spectrum of one disease group caused by genes involved in a limited set of molecular pathways. Application of similar strategies to other organelle-specific groups of disorders, such as the mitochondriopathies, could reveal the contribution of previously unknown genes and/or pathways. Although disorders associated with mitochondrial genes have not been characterized into functional modules, the recent development of a mitochondrial protein interaction database, the MitoInteractome 77, offers the potential to shed light onto novel mechanisms underlying disease and lead to the identification of new genes.
Disorders of other structural modalities: channelopathies
Inherited defects of proteins associated with discrete cellular modalities can also have overlapping clinical phenotypes. Such disorders can also be integrated and unified based on common cellular mechanisms. Perhaps the best-studied examples are the ion channelopathies, disease caused by ion channels. Located on the cell membranes of a wide array of cell types, ion channels are composed of several pore-forming protein subunits that regulate the flow of ions in and out of cells 78. Similar to ciliopathies, mutations in ion channel genes cause disorders of varying severity, often with overlapping clinical presentation. Furthermore, different channelopathies are often caused by mutations in the same genes, suggesting an overlap of biochemical functional defects. For example, the most common channelopathy, cystic fibrosis (CF), is characterized by clinical abnormalities such as bronchitis, asthma, sinusitis, pancreatitis, or gastrointestinal problems 79. However, these symptoms can be presented with a variety of other disorders making definitive diagnosis on the basis of clinical criteria alone somewhat difficult. Mutations in CFTR, which were thought to be exclusively associated with CF, have been identified in another disorder, congenital bilateral absence of the vas deferens (CBAVD), which can also be present in cystic fibrosis patients 80, 81. The overlap in clinical and genetic defects indicates that the development of a biochemical test can produce more accurate diagnoses. Because the biochemical defect underlying cystic fibrosis is known to be abnormal electrolyte transport within cells resulting in excessive salt loss 82, an observation-driven diagnosis can be confirmed by measurement of sodium chloride in sweat 83, 84.
Similarly, mutations in CLCN5, which encodes CIC-5, a renal chloride channel necessary for proper tubular endocytosis of proteins 85, underlie four channelopathies: Dent disease, X-linked hypophosphataemic rickets (XHPR), X-linked recessive nephrolithiasis with renal failure, and low-molecular-weight proteinuria, suggesting similar pathway defects are present in these disorders. Because of similar genetic defects, these disorders are thought to represent a varying spectrum of the same underlying molecular disorder 86, 87. In addition, patients with Dent disease who do not have mutations in CLCN5 have been found to carry mutations in OCLR1, which is associated with another X-linked disorder, Lowe syndrome 88, 89, suggesting the possibility of pathway overlap between the two disorders.
Defects in proteins of specific function: cohesinopathies
Disorders characterized by defects of particular protein complexes can also be grouped together, for example the cohesinopathies 90. Cohesin complexes that bind DNA are thought to serve two functions, mediation of sister-chromatid cohesion and regulation of gene expression (reviewed in 91). Two particular disorders resulting from defects in cohesin and proteins regulating cohesin are Cornelia de Lange (CdLS) Syndrome and Roberts/SC phocomelia (RBS/SC). CdLS, characterized by growth and mental retardation, craniofacial anomalies, and microcephaly, and is caused by mutations in NIPBL, SMC1A and SMC3, genes necessary for loading of cohesin onto DNA 92-95. Exhibiting a similar phenotype to CdLS , Roberts syndrome is also associated with mutations in a gene required for the establishment of cohesion, ESCO2 96, 97. Although these disorders are both characterized by defects in cohesin binding, there is no evidence of defects in cell proliferation, indicating that sister-chromatid cohesion is unaffected. Recent evidence has implicated CTCF, which is required for transcriptional insulation, in regulation of gene transcription by cohesin-regulated promoter insulation 98, suggesting that disorders of cohesin can be attributed to defects in gene expression regulation, specifically in those genes where cohesin binding to DNA is important for regulation of gene expression. Thus, defects in various cohesin proteins could result in expression defects in a defined set of genes. Importantly, some 40% of CdLS patients do not have mutations in NIPBL, SMC1A or SMC3. From a modular perspective, it will be important to understand whether additional cohesin components contribute to the genetic load of the disorder, or whether mutations in downstream transcriptional targets can drive the same phenotype. The cohesinopathy modular idea would predict the former to be true. One also might anticipate that trans-acting mutations in the known cohesinopathy genes might be found in patients with a primary cohesin defect, a hypothesis that is yet to be tested.
Predictive challenges: Modularization and complex disease
The examples above illustrate the potential usefulness of modularization with respect to mendelian or oligogenic traits. This concept may also prove useful for complex disease as well, where the phenotype is the product of interactions between multiple genes and the environment. Though, the evidence for this is currently limited, there are some promising examples. One example is the implication of the complement gene C3 in AMD. Initial genome-wide association studies identified common variants in complement factor H (CFH) and complement factor B (CFB) to be associated with AMD99-107. Based on the potential importance of the complement pathway in giving rise to macular degeneration, Yates et al. investigated the potential contribution of other complement genes and identified a strong association of a non-synonymous polymorphism in complement gene C3108.
A similar notion has been applied to obesity by the evaluation of functional expression networks. A recent study identified a network of genes that are perturbed by susceptibility loci in liver and adipose tissue expression in a mouse population segregating metabolic traits109. Integration of this network with expression networks from other tissue types revealed an enrichment in macrophage genes and led to the identification of three previously unknown candidate obesity genes. These findings highlight that, in addition to modularization based on cellular or molecular functionality, transcriptional networking and other types of interaction matrices will also be useful in the dissemination of the genetic architecture of complex traits. However, one major unanswered question in that regard remains the ability to query such networks in toto. At present, computational limitations and statistical power considerations preclude the systematic assessment of possible epistatic interactions within a complex disease network. It is possible that as resequencing tools become more widely used, there will be a paradigm shift towards resequencing such networks and examining the combinatorial effects of trans alleles.
Functionality of individual variants: effect and context
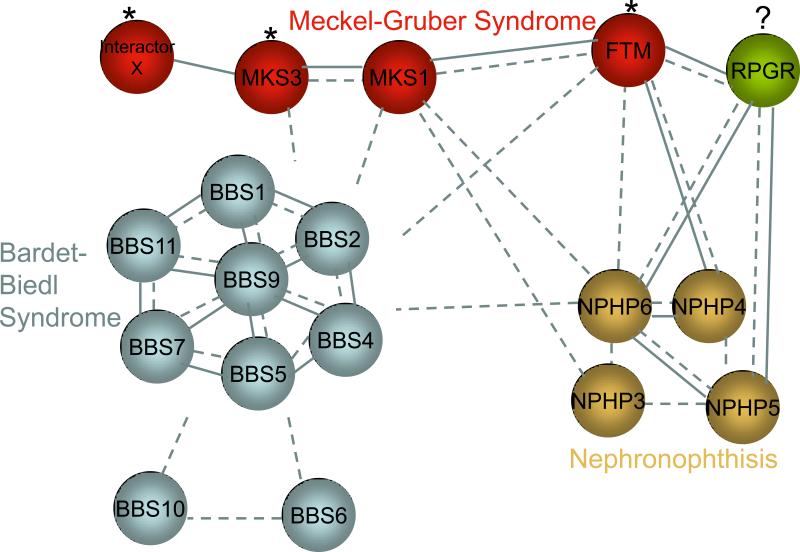
Modularization of genetic disease is likely to be useful in understanding pathways and phenotypic continua. However, understanding the functional consequences of an allele or a group of cis and trans acting alleles within a module is also crucial. Severity of mutation at each locus within a module provides some insight as to which module components might be dispensable and how total mutational load might explain variable penetrance and expressivity. In addition, the relative positions of alleles might also be critical, because proximal functional interactions might be more relevant to disease development and progression than distant ones (Figure 2).
Figure 2. Architecture of interaction across ciliopathy module components.
A simplified depiction of the interaction between components of the ciliopathy module. Genes contributing to the individual disorders have a high likelihood of both functional interactions (dashed lines) and protein-protein interactions (solid lines). The BBSome, for example, comprises physical interactions between BBS proteins. These genes have functional interactions as well because mutations in one BBS gene can modify or be compounded with mutations in other BBS genes. This functional interaction is also true for BBS genes outside of the complex. Likewise, physical and functional interactions are present within Meckel-Gruber or Nephronophthisis genes. In addition, genes contributing to each disorder can also interact with genes contributing to other disorders. For example, FTM (RPGRIP1L) is a modifier (asterisk) of BBS, MKS and NPH, and can physically interact with some proteins underlying those diseases 51. Similarly, MKS3 is a modifier of BBS 48. RPGR is outside of the ciliopathy module but physically interacts with module components and is mutated in some ciliopathy patients 51. This suggests the potential for contribution of RPGR to ciliopathies and incorporation of this gene into the module.
The ciliopathy module represents a useful example, primarily because it is now densely populated by >30 genes and >300 disease-associated alleles. BBS is a less severe phenotype than MKS, although there is overlap between the two in causative genes. Interestingly, characterization of mutation severity revealed that hypomorphic mutations in MKS1 give rise to BBS, suggesting that a homozygous loss of MKS1 results in severe dysfunction, whereas residual protein activity[SC4] from that locus leads to an intermediate phenotype 50. Such a paradigm is abundant in ciliopathies; hypomorphic NPHP3 and MKS3 mutations cause NPH, whereas null alleles cause MKS 74. These observations provide clues about the necessity of individual proteins or nodes within a module and offer initial clues with regard to the buffering ability of individual positions. However, trans interactions and their behaviors are equally important with regard to phenotypic modulation. Mutations in FTM cause a host of ciliopathies with no clear evidence for allelic cis stratification of phenotypes. Likewise, null mutations in NPHP6/CEP290 have been found in patients that cover the entire ciliopathy spectrum, from isolated NPH to MKS. For such examples, either stochastic factors or functionally proximal, trans alleles are likely to determine disease modulation. For instance, although mutations in FTM do not appear to cause the intermediate BBS phenotype, there is a significant enrichment for heterozygous FTM alleles in patients with ciliopathies defined clinically by retinal degeneration. In particular, whereas homozygosity of the A229T allele alone is benign, lesions at other positions in the module can interact genetically to potentiate retinal degeneration 51. [SC5]
This concept has also been explored in the context of cancer modules. Several large sequencing screens in solid tumors from various tissues have used statistical comparison to characterize mutations as ‘driver’ alleles, which confer a clonal growth advantage, or as ‘passenger’ alleles, which do not 110-112; genes carrying driver mutations are categorized as cancer genes. These studies have used either prevalence of mutations 111, 112 or the ratio of non-synonymous to synonymous mutations as the determining factor in this context 110. However, when mutations in FLT3 associated with acute myeloid leukemia (AML) were assayed for differences in effects on kinase activation and downstream signaling, 4 of 9 imparted gain of function and could act as drivers of leukemogenesis, even though these mutations were considered passengers when assessed by statistical methods alone 113. These findings provide evidence that although the importance of genes within a particular module can be garnered by assessing the type of variant, i.e. coding, non-coding, the ultimate determinant of disease mechanism is testing the specific variant function using biological assays.
Concluding remarks
We will shortly have access to genetic variation data both from an increasing catalog of humans of diverse ethnic backgrounds 125 as well as from exomes and eventually genomes of patients with diverse disorders 1, 126. Current practices tend to follow one of two major paths. For rare disorders, the primary focus lies on strict mendelian models, often discarding the variation outside the ’disease gene‘ where two pathogenic alleles might lie (for recessive disorders). At the other end of the spectrum, complex trait genetics remain largely driven by the investigation of alleles of sufficient frequency to empower statistical arguments. We suggest that the first approach, although successful in identifying highly penetrant alleles that drive much of the phenotype, will miss the opportunity to investigate the functional effect of mutations in the context of variation across the genome. As such, it might be important to collate, archive (in the public domain) and analyze all the variation found in patient exome and genome resequencing projects, especially since alleles can appear to be functionally benign in one context but pathogenic in another. Ultimately, one can envision the functional modularization of the entire morbid human genome, wherein it will be possible to conduct functional assays pertinent to a specific module for all the variation detected. We are certainly not in a position to accomplish that goal at present. However, there is no reason to believe that such a goal is unattainable.
It is important to consider the potential limitations of the modular approach. One potentially confounding factor is the spatial and temporal regulation of expression of individual module components by differential expression or tissue-specific splice isoforms. In Usher Syndrome, for example, a functional network including the five causative disease genes has been established based on binding interactions between the proteins127. Different types of mutations in one network component, cadherin 23 (CDH23/USH1D), cause either deafness and blindness associated with Usher syndrome or only nonsyndromic deafness128. Further investigation revealed that the discrepancy in phenotype may result from differential tissue-specific expression CDH23 isoforms in the retina and inner ear 129, 130, suggesting that modularization may not be informative in particular tissue or cell-type contexts. Second, discussions of the modular approach have generally applied to loss of function mutations. For gain of function mutations, however, it might be difficult to assess the contribution of individual components and their impact on the module as a whole. Returning to the FGFR1 example, loss- and gain-of-function mutations produce significantly different phenotypes suggesting that the protein can contribute to different pathways, making functional modularization challenging. Though there is limited functional data to support this notion, informatics based approached have analyzed the differential contribution of disease genes dependent on the nature of mutation. Computational analysis of interaction profiles of mutant proteins in the context of mendelian disorders for example, revealed that perturbations in these networks by complete removal of network components (i.e. loss of function mutations) is predicted to have more deleterious effects on the overall architecture of the network as compared to slight perturbations131. However, mutations conferring a gain of interactions, would also potentially alter the overall network, albeit differently. As the nature of disease gene functionality becomes better understood, the complexity of relationships between them, and the contribution of variation to altering those relationships, will become clearer. It is possible that investigation of variation across functional modules will offer better predictive power with regard to penetrance, expressivity and rate of disease progression and this will help understand the mechanics of the genetic basis of phenotypic variability, which in humans is often complex and ill-defined.
Box 1[JS6]: Tools for prediction of allele function.
The most commonly used method to predict allele functionality has been the use of computational algorithms, which capitalize on some component of protein character to predict the effect that a change will have. Often, these tools are used to predict the character of non-synonymous coding variants using sequence-based or structure-based analyses, but relatively few of them categorize mutations based solely on pathogenic nature 114. Such algorithms include SIFT (Sorting Intolerant From Tolerant) and MAPP (Multi-variant Analysis of Protein Polymorphisms), which are based on conservation of residues in protein families across species 115, 116 or PhD-SNP which utilizes the sequence profile along with conservation information 117. Other programs use a combination of information. For example, the commonly used PolyPhen uses both sequence alignments and structural information 118. Similarly, SNAP (Screening for Non-Acceptable Polymorphisms) considers biochemical properties of particular residues in addition to evolutionary data 119. Such tools have provided rapid, easy methods by which to analyze variants, but the somewhat limited accuracy of prediction (70-80% for most algorithms) makes them insufficient for definitive prediction of function or for clinical diagnostic applications.
Biological assays of variant function remain the gold standard, especially for rare variants for which genetic data are of insufficient resolution and power. Drawing from the ciliopathy group, evaluation of variants already known to underlie disease, such as the BBS1-M390R[SC7] knock-in mouse 120 or in vitro mislocalization of mutant BBS6 121, confirms the association and provides insight into disease mechanism. Evaluation of alleles of ambiguous pathogenic contribution can complement the genetic information to definitively associate variants with disease. For example, a recent study examined 17 mutations in BRCA2 by assessing the ability of wild type and mutant BRCA2 protein to rescue depletion of endogenous protein 122. Using cell viability as a phenotypic readout, Kuznetsov et al. demonstrated the efficiency and accuracy of this application to determine variant neutrality. This concept is potentially true for mutations as well as associated polymorphisms and functional testing of associated polymorphisms have confirmed the functional contribution of such variants. For example, expression of a missense SNP in DDX5, a gene associated with a higher risk of cirrhosis, in hepatic stellate cells revealed enhanced fibrogenic activity via derepression of fibrogenic genes123. Likewise, a study of the effect of a SNP associated with aspirin intolerance in asthma revealed that the high risk variant in CysLTR2[SC8], which promotes inflammation and bronchoconstriction, resulted in higher expression levels and increased mRNA stability in transfected B cells 124. Although examples such as these support the usefulness of biological assays in deciphering allele functionality, it will also be imperative to refine assays to capture multiple functions of proteins to understand the contribution of different alleles to different pathways and phenotypes, Ultimately, rapid and efficient assays will be the necessary tools with which to evaluate novel variants in disease genes found in patients and to refine other tools, including computational predictors, for improved accuracy.
Acknowledgments
We apologize to those colleagues whose work we have been unable to cite owing to space limitations. We thank all the members of the Katsanis laboratory for helpful discussions. This work was supported by grants from the National Institute of Child Health and Development (R01HD04260), the National Institute of Diabetes, Digestive, and Kidney Disorders (R01DK072301 and R01DK075972), and the Macular Vision Research Foundation (to N. Katsanis); and by a Visual NeuroscienceTraining Program fellowship (to N.A. Zaghloul).
Glossary[SC9]
- Functional modularization
the use of modules or collections of biological information about one disease or developmental pathway to aid in the identification of genes for similar or even distinct but linked diseases
- Mutational load
the total of all deleterious mutations across the genome contributing to a genetic trait
- Stickler syndrome
a group of disorders caused by mutations in COL2A1, COL11A1, COL11A2 or COL9A1 and characterized by craniofacial defects including flat mala, hearing loss, myopia or other eye problems
- Marshall syndrome
an autosomal dominant disorder caused by mutations in COL11A1 and presenting with craniofacial defects and myopia. Marshall is distinct from Stickler syndrome in the presence of a flat or retracted midface and the appearance of large eyes
- Oto-spondylo-mega-epiphyseal dysplasia (OSMED) syndrome
an autosomal recessive disorder caused by mutations in COL11A2 or in some reports COL2A1. The disease is characterized by skeletal and craniofacial defects and hearing loss
- Hypogonadotrophic hypogonadism
an absence or reduced functionality of the testes or ovaries
- Cutaneous syndactyly
the appearance fusing together of toes or fingers at the skin, not the bones
- Polycystic kidney disease (PKD)
a disorder that can be autosomal recessive or autosomal dominant (the majority of cases) and characterized by the presence and growth of multiple cysts in the kidneys
- Nephronophthisis (NPH)
an autosomal recessive disorder presenting with polyuria, polydipsia, proteinuria and characterized by multiple renal cysts and fibrosis
- Alstrom Syndrome (ALMS)
an autosomoal recessive disorder caused by mutations in ALMS1 and characterized by obesity, retinal dystrophy, and hearing loss
- Bardet-Biedl Syndrome (BBS)
a pleiotropic disorder caused by mutations in genes localizing to the basal body and cilium and typically characterized by retinal degeneration, obesity, polydactyly, mental retardation, renal dysfunction and hypogonadism
- Meckel-Gruber syndrome (MKS)
a lethal condition resulting in pre- or early post-natal death as a result of neural tube defects and renal cysts and malformations
- Exencephaly
the abnormal development of the brain outside of the skull usually resulting in death of the fetus or newborn
- Otoacoustic emissions
sounds produced in the inner ear as a result of external stimulation and amplification by the cochlea that can be reduced with damage to the inner ear
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ng SB, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ng SB, et al. Exome sequencing identifies the cause of a mendelian disorder. Nat Genet. 42:30–35. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Swaroop A, et al. Unraveling a Multifactorial Late-Onset Disease: From Genetic Susceptibility to Disease Mechanisms for Age-Related Macular Degeneration. Annu Rev Genomics Hum Genet. 2009 doi: 10.1146/annurev.genom.9.081307.164350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parkes M, et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn's disease susceptibility. Nat Genet. 2007;39:830–832. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prescott NJ, et al. A nonsynonymous SNP in ATG16L1 predisposes to ileal Crohn's disease and is independent of CARD15 and IBD5. Gastroenterology. 2007;132:1665–1671. doi: 10.1053/j.gastro.2007.03.034. [DOI] [PubMed] [Google Scholar]
- 6.Hidalgo CA, et al. A dynamic network approach for the study of human phenotypes. PLoS Comput Biol. 2009;5:e1000353. doi: 10.1371/journal.pcbi.1000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ahmad NN, A.-K.L., Knowlton RG, Jimenez SA, Weaver EJ, Maguire JI, Tasman W, Prockop DJ. Stop codon in the procollagen II gene (COL2A1) in a family with the Stickler syndrome (arthro-ophthalmopathy). Proc Natl Acad Sci U S A. 1991;88:6624–6627. doi: 10.1073/pnas.88.15.6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Melkoniemi M, B.H., Manouvrier S, Hennekam R, Superti-Furga A, Kääriäinen H, Pauli RM, van Essen T, Warman ML, Bonaventure J, Miny P, Ala-Kokko L. Autosomal recessive disorder otospondylomegaepiphyseal dysplasia is associated with loss-of-function mutations in the COL11A2 gene. American Journal Human of Genetics. 2000;66:368–377. doi: 10.1086/302750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Steensel MA, B.P., de Waal Malefijt MC, van den Hoogen FH, Brunner HG. Oto- spondylo-megaepiphyseal dysplasia (OSMED): clinical description of three patients homozygous for a missense mutation in the COL11A2 gene. American Journal Human of Genetics. 1997;70:315–323. doi: 10.1002/(sici)1096-8628(19970613)70:3<315::aid-ajmg19>3.3.co;2-y. [DOI] [PubMed] [Google Scholar]
- 10.Freund CL, et al. De novo mutations in the CRX homeobox gene associated with Leber congenital amaurosis. Nat Genet. 1998;18:311–312. doi: 10.1038/ng0498-311. [DOI] [PubMed] [Google Scholar]
- 11.Morimura H, et al. Mutations in RGR, encoding a light-sensitive opsin homologue, in patients with retinitis pigmentosa. Nat Genet. 1999;23:393–394. doi: 10.1038/70496. [DOI] [PubMed] [Google Scholar]
- 12.Dodé C, Levilliers J, Dupont JM, De Paepe A, Le Dû N, Soussi-Yanicostas N, Coimbra RS, Delmaghani S, Compain-Nouaille S, Baverel F, Pêcheux C, Le Tessier D, Cruaud C, Delpech M, Speleman F, Vermeulen S, Amalfitano A, Bachelot Y, Bouchard P, Cabrol S, Carel JC, Delemarre-van de Waal H, Goulet-Salmon B, Kottler ML, Richard O, Sanchez-Franco F, Saura R, Young J, Petit C, Hardelin JP. Loss-of-function mutations in FGFR1 cause autosomal dominant Kallmann syndrome. Nature Genetics. 2003;33:463–465. doi: 10.1038/ng1122. [DOI] [PubMed] [Google Scholar]
- 13.Roscioli T, Flanagan S, Kumar P, Masel J, Gattas M, Hyland VJ, Glass IA. Clinical findings in a patient with FGFR1 P252R mutation and comparison with the literature. American Journal Human of Genetics. 2000;93:22–28. doi: 10.1002/1096-8628(20000703)93:1<22::aid-ajmg5>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 14.Rossi M, Jones RL, Norbury G, Bloch-Zupan A, Winter RM. The appearance of the feet in Pfeiffer syndrome caused by FGFR1 P252R mutation. Clinical Dysmorphology. 2003;12:269–274. doi: 10.1097/00019605-200310000-00012. [DOI] [PubMed] [Google Scholar]
- 15.Goh KI, et al. The human disease network. Proc Natl Acad Sci U S A. 2007;104:8685–8690. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Draper N, et al. Mutations in the genes encoding 11beta-hydroxysteroid dehydrogenase type 1 and hexose-6-phosphate dehydrogenase interact to cause cortisone reductase deficiency. Nat Genet. 2003;34:434–439. doi: 10.1038/ng1214. [DOI] [PubMed] [Google Scholar]
- 17.Zimmermann KW. Beiträge zur Kenntnis einiger Drüsen und Epithelien. Arch. Mikrosk. Anat. 1898;52:552–706. [Google Scholar]
- 18.Satir P. The cilium as a biological nanomachine. FASEB Journal. 1999;13:S235–S237. doi: 10.1096/fasebj.13.9002.s235. [DOI] [PubMed] [Google Scholar]
- 19.Badano JL, Mitsuma N, Beales PL, Katsanis N. The Ciliopathies: An emerging class of Human Genetic Disorders. Annual Review of Genomics and Human Genetics. 2006;7:125–148. doi: 10.1146/annurev.genom.7.080505.115610. [DOI] [PubMed] [Google Scholar]
- 20.Gherman A, Davis EE, Katsanis N. The ciliary proteome database: an integrated community resource for the genetic and functional dissection of cilia. Nature Genetics. 2006;38:961–962. doi: 10.1038/ng0906-961. [DOI] [PubMed] [Google Scholar]
- 21.Pazour GJ, Agrin N, Leszyk J, Witman GB. Proteomic analysis of a eukaryotic cilium. Journal of Cell Biology. 2005;170:103–113. doi: 10.1083/jcb.200504008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stolc V, Samanta MP, Tongprasit W, Marshall WF. Genome-wide transcriptional analysis of flagellar regeneration in Chlamydomonoas reinhardtii indentifies orthologs of ciliary disease genes. Proceedings of the National Academy of Sciences USA. 2005;102:3703–3707. doi: 10.1073/pnas.0408358102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li JB, Gerdes JM, Haycraft CJ, Fan Y, Teslovich TM, May-Simera H, Li H, Blacque OE, Li L, Leitch CC, Lewis RA, Green JS, Parfrey PS, Leroux MR, Davidson WS, Beales PL, Guay-Woodford LM, Yoder BK, Stormo GD, Katsanis N, Dutcher SK. Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell. 2004;117:541–552. doi: 10.1016/s0092-8674(04)00450-7. [DOI] [PubMed] [Google Scholar]
- 24.Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426:570–574. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- 25.Avidor-Reiss T, Maer AM, Koundakjian E, Polyanovsky A, Keil T, Subramaniam S, Zuker CS. Decoding cilia function: defining specialized genes require for compartmentalized cilia biogenesis. Cell. 2004;117:527–539. doi: 10.1016/s0092-8674(04)00412-x. [DOI] [PubMed] [Google Scholar]
- 26.Blacque OE, Perens EA, Boroevich KA, Inglis PN, Li C, Warner A, Khattra J, Holt RA, Ou G, Mah AK, McKay SJ, Huang P, Swoboda P, Jones SJ, Marra MA, Baillie DL, Moerman DG, Shaham S, Leroux MR. Functional genomics of the cilium, a sensory organelle. Cell. 2005;15:935–941. doi: 10.1016/j.cub.2005.04.059. [DOI] [PubMed] [Google Scholar]
- 27.Broadhead R, Dawe HR, Farr H, Griffiths S, Hart SR, Portman N, Shaw MK, Ginger ML, Gaskell SJ, McKean PG, Gull K. Flagellar motility is required for the viability of the bloodstream trypanosome. Nature. 2006;440:224–227. doi: 10.1038/nature04541. [DOI] [PubMed] [Google Scholar]
- 28.Efimenko E, Bubb K, Mak HY, Holzman T, Leroux MR, Ruvkun G, Thomas JH, Swoboda P. Analysis of xbx genes in C. elegans. Development. 2005;132:1923–1924. doi: 10.1242/dev.01775. [DOI] [PubMed] [Google Scholar]
- 29.Keller LC, Romijn EP, Zamora I, Yates JR, 3rd, Marshall WF. Proteomic analysis of isolated chlamydomonas centrioles reveals orthologs of ciliary-disease genes. Current Biology. 2005;15:1090–1098. doi: 10.1016/j.cub.2005.05.024. [DOI] [PubMed] [Google Scholar]
- 30.Ostrowski LE, Blackburn K, Radde KM, Moyer MB, Schlatzer DM, Moseley A, Boucher RC. A proteomic analysis of human cilia: identification of novel components. Molecular and Cellular Proteomics. 2002;1:451–465. doi: 10.1074/mcp.m200037-mcp200. [DOI] [PubMed] [Google Scholar]
- 31.Beales PL, Bland E, Tobin JL, Bacchelli C, Tuysuz B, Hill J, Rix S, Pearson CG, Kai M, Hartley J, Johnson C, Irving M, Elcioglu N, Winey M, Tada M, Scambler PJ. IFT80, which encodes a conserved intraflagellar transport protein, is mutated in Jeune asphyxiating thoracic dystrophy. Nature Genetics. 2007;39:727–729. doi: 10.1038/ng2038. [DOI] [PubMed] [Google Scholar]
- 32.Dagoneau N, et al. DYNC2H1 mutations cause asphyxiating thoracic dystrophy and short rib-polydactyly syndrome, type III. Am J Hum Genet. 2009;84:706–711. doi: 10.1016/j.ajhg.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zariwala MA, Knowles MR, Omran H. Genetic defects in ciliary structure and function. Annual Review of Physiology. 2007;69:423–450. doi: 10.1146/annurev.physiol.69.040705.141301. [DOI] [PubMed] [Google Scholar]
- 34.Beales PL, Elcioglu N, Woolf AS, Parker D, Flinter FA. New criteria for improved diagnosis of Bardet-Biedl syndrome: results of a population survey. Journal of Medical Genetics. 1999;36:437–446. [PMC free article] [PubMed] [Google Scholar]
- 35.Slavotinek AM, Stone EM, Mykytyn K, Heckenlively JR, Green JS, Heon E, Musarella MA, Parfrey PS, Sheffield VC. Mutations in MKKS cause Bardet-Biedl syndrome. Nature Genetics. 2000;26:15–16. doi: 10.1038/79116. [DOI] [PubMed] [Google Scholar]
- 36.Katsanis N, Beales PL, Woods MO, Lewis RA, Green JS, Parfrey PS, Ansley SJ, Davidson WS, Lupski JR. Mutations in MKKS cause obesity, retinal dystrophy and renal malformations associated with Bardet-Biedl syndrome. Nature Genetics. 2000;26:67–70. doi: 10.1038/79201. [DOI] [PubMed] [Google Scholar]
- 37.Mykytyn K, Braun T, Carmi R, Haider NB, Searby CC, Shastri M, Beck G, Wright AF, Iannaccone A, Elbedour K, Riise R, Baldi A, Raas-Rothschild A, Gorman SW, Duhl DM, Jacobson SG, Casavant T, Stone EM, Sheffield VC. Identification of the gene that, when mutated, causes the human obesity syndrome BBS4. Nature Genetics. 2001;28:188–191. doi: 10.1038/88925. [DOI] [PubMed] [Google Scholar]
- 38.Nishimura DY, Searby CC, Carmi R, Elbedour K, Van Maldergem L, Fulton AB, Lam BL, Powell BR, Swiderski RE, Bugge KE, Haider NB, Kwitek-Black AE, Ying L, Duhl DM, Gorman SW, Heon E, Iannaccone A, Bonneau D, Biesecker LG, Jacobson SG, Stone EM, Sheffield VC. Positional cloning of a novel gene on chromosome 16q causing Bardet-Biedl syndrome (BBS2). Human Molecular Genetics. 2001;10:865–874. doi: 10.1093/hmg/10.8.865. [DOI] [PubMed] [Google Scholar]
- 39.Mykytyn K, Nishimura DY, Searby CC, Shastri M, Yen HJ, Beck JS, Braun T, Streb LM, Cornier AS, Cox GF, Fulton AB, Carmi R, Luleci G, Chandrasekharappa SC, Collins FS, Jacobson SG, Heckenlively JR, Weleber RG, Stone EM, Sheffield VC. Identification of the gene (BBS1) most commonly involved in Bardet-Biedl syndrome, a complex human obesity syndrome. Nature Genetics. 2002;31:434–438. doi: 10.1038/ng935. [DOI] [PubMed] [Google Scholar]
- 40.Chiang AP, Nishimura D, Searby C, Elbedour K, Carmi R, Ferguson AL, Secrist J, Braun T, Casavant T, Stone EM, Sheffield VC. Comparative genomic analysis identifies an ADP-ribosylation factor-like gene as the cause of Bardet-Biedl syndrome (BBS3). American Journal Human of Genetics. 2004;75:475–484. doi: 10.1086/423903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fan Y, Esmail MA, Ansley SJ, Blacque OE, Boroevich K, Ross AJ, Moore SJ, Badano JL, May-Simera H, Compton DS, Green JS, Lewis RA, van Haelst MM, Parfrey PS, Baillie DL, Beales PL, Katsanis N, Davidson WS, Leroux MR. Mutations in a member of the Ras superfamily of small GTP-binding proteins causes Bardet-Biedl syndrome. Nature Genetics. 2004;36:989–993. doi: 10.1038/ng1414. [DOI] [PubMed] [Google Scholar]
- 42.Ansley SJ, Badano JL, Blacque OE, Hill J, Hoskins BE, Leitch CC, Kim JC, Ross AJ, Eichers ER, Teslovich TM, Mah AK, Johnsen RC, Cavender JC, Lewis RA, Leroux MR, Beales PL, Katsanis N. Basal body dysfunction is a likely cause of pleiotropic Bardet-Biedl syndrome. Nature. 2003;425:628–633. doi: 10.1038/nature02030. [DOI] [PubMed] [Google Scholar]
- 43.Badano JL, Kim JC, Hoskins BE, Lewis RA, Ansley SJ, Cutler DJ, Castellan C, Beales PL, Leroux MR, Katsanis N. Heterozygous mutations in BBS1, BBS2 and BBS6 have a potential epistatic effect on Bardet-Biedl patients with two mutations at a second BBS locus. Human Molecular Genetics. 2003;12:1651–1659. doi: 10.1093/hmg/ddg188. [DOI] [PubMed] [Google Scholar]
- 44.Nishimura DY, Swiderski RE, Searby CC, Berg EM, Ferguson AL, Hennekam R, Merin S, Weleber RG, Biesecker LG, Stone EM, Sheffield VC. Comparative genomics and gene expression analysis identifies BBS9, a new Bardet-Biedl syndrome gene. American Journal Human of Genetics. 2005;77:1021–1033. doi: 10.1086/498323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stoetzel C, Laurier V, Davis EE, Muller J, Rix S, Badano JL, Leitch CC, Salem N, Chouery E, Corbani S, Jalk N, Vicaire S, Sarda P, Hamel C, Lacombe D, Holder M, Odent S, Holder S, Brooks AS, Elcioglu NH, Silva ED, Rossillion B, Sigaudy S, de Ravel TJ, Lewis RA, Leheup B, Verloes A, Amati-Bonneau P, Megarbane A, Poch O, Bonneau D, Beales PL, Mandel JL, Katsanis N, Dollfus H. BBS10 encodes a vertebrate-specific chaperonin-like protein and is a major BBS locus. Nature Genetics. 2006;38:521–524. doi: 10.1038/ng1771. [DOI] [PubMed] [Google Scholar]
- 46.Chiang AP, Beck JS, Yen HJ, Tayeh MK, Scheetz TE, Swiderski RE, Nishimura DY, Braun TA, Kim KY, Huang J, Elbedour K, Carmi R, Slusarski DC, Casavant TL, Stone EM, Sheffield VC. Homozygosity mapping with SNP arrays identifies TRIM32, an E3 ubiquitin ligase, as a Bardet-Biedl syndrome gene (BBS11). Proceedings of the National Academy of Sciences USA. 2006;103:6287–6292. doi: 10.1073/pnas.0600158103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stoetzel C, Muller J, Laurier V, Davis EE, Zaghloul NA, Vicaire S, Jacquelin C, Plewniak F, Sarda P, Hamel C, de Ravel TJ, Lewis RA, Friederich E, Thibault C, Danse JM, Verloes A, Bonneau D, Katsanis N, Poch O, Mandel JL, Dolfus H. Identification of a novel BBS gene (BBS12) 2007. highlights the major role of a vertebrate specific branch of chaperonin-related proteins in Bardet-Biedl syndrome. American Journal Human of Genetics. 2007;80:1–11. doi: 10.1086/510256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leitch CC, Zaghloul NA, Davis EE, Stoetzel C, Diaz-Font A, Rix S, Al-Fadhel M, Lewis RA, Eyaid W, Banin E, Dollfus H, Beales PL, Badano JL, Katsanis N. Hypomorphic mutations in syndromic encephalocele genes are associated with Bardet-Biedl syndrome. Nature Genetics. 2008;40:443–448. doi: 10.1038/ng.97. [DOI] [PubMed] [Google Scholar]
- 49.Badano JL, Leitch CC, Ansley SJ, May-Simera H, Lawson S, Lewis RA, Beales PL, Dietz HC, Fisher S, Katsanis N. Dissection of epistasis in oligogenic Bardet-Biedl Syndrome. Nature. 2006;439:326–330. doi: 10.1038/nature04370. [DOI] [PubMed] [Google Scholar]
- 50.Leitch CC, et al. Hypomorphic mutations in syndromic encephalocele genes are associated with Bardet-Biedl syndrome. Nat Genet. 2008;40:443–448. doi: 10.1038/ng.97. [DOI] [PubMed] [Google Scholar]
- 51.Khanna H, et al. A common allele in RPGRIP1L is a modifier of retinal degeneration in ciliopathies. Nature Genetics. 2009;41:739. doi: 10.1038/ng.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Blacque OE, Reardon MJ, Li C, McCarthy J, Mahjoub MR, Ansley SJ, Badano JL, Mah AK, Beales PL, Davidson WS, Johnsen RC, Audeh M, Plasterk RH, Baillie DL, Katsanis N, Quarmby LM, Wicks SR, Leroux MR. Loss of C. elegans BBS-7 and BBS-8 protein function results in cilia defects and compromised intraflagellar transport. Genes and Development. 2004;18:1630–1642. doi: 10.1101/gad.1194004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nachury MV, Loktev AV, Zhang Q, Westlake CJ, Peranen J, Merdes A, Slusarski DC, Scheller RH, Bazan JF, Sheffield VC, Jackson PK. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129:1201–1213. doi: 10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- 54.Dale RM, et al. The emerging role of Wnt/PCP signaling in organ formation. Zebrafish. 2009;6:9–14. doi: 10.1089/zeb.2008.0563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ross AJ, May-Simera H, Eichers ER, Kai M, Hill J, Jagger DJ, Leitch CC, Chapple JP, Munro PM, Fisher S, Tan PL, Phillips HM, Leroux MR, Henderson DJ, Murdoch JN, Copp AJ, Eliot MM, Lupski JR, Kemp DT, Dollfus H, Tada M, Katsanis N, Forge A, Beales PL. Disruption of Bardet-Biedl syndrome ciliary proteins perturbs planar cell polarity in vertebrates. Nature Genetics. 2005;37:1135–1140. doi: 10.1038/ng1644. [DOI] [PubMed] [Google Scholar]
- 56.Fischer E, Legue E, Doyen A, Nato F, Nicolas JF, Torres V, Yaniv M, Pontoglio M. Defective planar cell polarity in polycystic kidney disease. Nature Genetics. 2006;38:21–23. doi: 10.1038/ng1701. [DOI] [PubMed] [Google Scholar]
- 57.Simons M.a.W., G. Polycystic kidney disease: cell division without a c(l)ue? Kidney International. 2006;70:854–864. doi: 10.1038/sj.ki.5001534. [DOI] [PubMed] [Google Scholar]
- 58.Patel V, Li L, Cobo-Stark P, Shao X, Somlo S, Lin F, Igarashi P. Acute kidney injury and aberrant planar cell polarity induce cyst formation in mice lacking renal cilia. Human Molecular Genetics. 2008;17:1578–1590. doi: 10.1093/hmg/ddn045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bergmann C, Fliegauf M, Brüchle NO, Frank V, Olbrich H, Kirschner J, Schermer B, Schmedding I, Kispert A, Kränzlin B, Nürnberg G, Becker C, Grimm T, Girschick G, Lynch SA, Kelehan P, Senderek J, Neuhaus TJ, Stallmach T, Zentgraf H, Nürnberg P, Gretz N, Lo C, Lienkamp S, Schäfer T, Walz G, Benzing T, Zerres K, Omran H. Loss of nephrocystin-3 function can cause embryonic lethality, Meckel-Gruber-like syndrome, situs inversus, and renal-hepatic-pancreatic dysplasia. American Journal Human of Genetics. 2008;82:959–970. doi: 10.1016/j.ajhg.2008.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Corbit KC, Shyer AE, Dowdle WE, Gaulden J, Singla V, Chen MH, Chuang PT, Reiter JF. Kif3a constrains beta-catenin-dependent Wnt signalling through dual ciliary and non-ciliary mechanisms. Nature Cell Biology. 2008;10:70–76. doi: 10.1038/ncb1670. [DOI] [PubMed] [Google Scholar]
- 61.Gerdes JM, et al. Disruption of the basal body compromises proteasomal function and perturbs intracellular Wnt response. Nat Genet. 2007;39:1350–1360. doi: 10.1038/ng.2007.12. [DOI] [PubMed] [Google Scholar]
- 62.Eichers ER, Abd-el-Barr MM, Paylor R, Lewis RA, Bi W, Lin X, Meehan TP, Stockton DW, Wu SM, Lindsay E, Justice MJ, Beales PL, Katsanis N, Lupski JR. Phenotypic characterization of Bbs4-null mice reveals age-dependent penetrance and variable expressivity. Human Genetics. 2006;120:211–226. doi: 10.1007/s00439-006-0197-y. [DOI] [PubMed] [Google Scholar]
- 63.Alexiev BA, Lin X, Sun CC, Brenner DS. Meckel-Gruber syndrome: pathologic manifestations, minimal diagnostic criteria, and differential diagnosis. Archives of Pathology and Laboratory Medicine. 2006;130:1236–1238. doi: 10.5858/2006-130-1236-MS. [DOI] [PubMed] [Google Scholar]
- 64.Karmous-Benailly H, Martinovic J, Gubler MC, Sirot Y, Clech L, Ozilou C, Auge J, Brahimi N, Etchevers H, Detrait E, Esculpavit C, Audollent S, Goudefroye G, Gonzales, Tantau J, Loget P, Joubert M, Gaillard D, Jeanne-Pasquier C, Delezoide AL, Peter MO, Plessis G, Simon-Bouy B, Dollfus H, Le Merrer M, Munnich A, Encha-Razavi F, Vekemans M, Attie-Bitach T. Antenatal presentation of Bardet-Biedl syndrome may mimic Meckel syndrome. American Journal Human of Genetics. 2005;75:494–504. doi: 10.1086/428679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dawe HR, et al. The Meckel-Gruber Syndrome proteins MKS1 and meckelin interact and are required for primary cilium formation. Hum Mol Genet. 2007;16:173–186. doi: 10.1093/hmg/ddl459. [DOI] [PubMed] [Google Scholar]
- 66.Fath MA, Mullins RF, Searby C, Nishimura DY, Wei J, Rahmouni K, Davis RE, Tayeh MK, Andrews M, Yang B, Sigmund CD, Stone EM, Sheffield VC. Mkks-null mice have a phenotype resembling Bardet-Biedl syndrome. Human Molecular Genetics. 2005;14:1109–1118. doi: 10.1093/hmg/ddi123. [DOI] [PubMed] [Google Scholar]
- 67.Delous M, Baala L, Salomon R, Laclef C, Vierkotten J, Tory K, Golzio C, Lacoste T, Besse L, Ozilou C, Moutkine I, Hellman NE, Anselme I, Silbermann F, Vesque C, Gerhardt C, Rattenberry E, Wolf MT, Gubler MC, Martinovic J, Encha-Razavi F, Boddaert N, Gonzales M, Macher MA, Nivet H, Champion G, Berthélémé JP, Niaudet P, McDonald F, Hildebrandt F, Johnson CA, Vekemans M, Antignac C, Rüther U, Schneider-Maunoury S, Attié-Bitach T, Saunier S. The ciliary gene RPGRIP1L is mutated in cerebello-oculo-renal syndrome (Joubert syndrome type B) and Meckel syndrome. Nature Genetics. 2007;39:875–881. doi: 10.1038/ng2039. [DOI] [PubMed] [Google Scholar]
- 68.Louie C.M.a.G., J.G. Genetic basis of Joubert syndrome and related disorders of cerebellar development. Human Molecular Genetics. 2005;14:R235–R242. doi: 10.1093/hmg/ddi264. [DOI] [PubMed] [Google Scholar]
- 69.Ferland RJ, Eyaid W, Collura RV, Tully LD, Hill RS, Al-Nouri D, Al-Rumayyan A, Topcu M, Gascon G, Bodell A, Shugart YY, Ruvolo M, Walsh CA. Abnormal cerebellar development and axonal decussation due to mutations in AHI1 in Joubert syndrome. Nature Genetics. 2004;36:1008–1013. doi: 10.1038/ng1419. [DOI] [PubMed] [Google Scholar]
- 70.Dixon-Salazar T, Silhavy JL, Marsh SE, Louie CM, Scott LC, Gururaj A, Al-Gazali L, Al-Tawari AA, Kayserili H, Sztriha L, Gleeson JG. Mutations in the AHI1 gene, encoding jouberin, cause Joubert syndrome with cortical polymicrogyria. American Journal Human of Genetics. 2004;75:979–987. doi: 10.1086/425985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Parisi MA, Doherty D, Eckert ML, Shaw DW, Ozyurek H, Aysun S, Giray O, Al Swaid A, Al Shahwan S, Dohayan N, Bakhsh E, Indridason OS, Dobyns WB, Bennett CL, Chance PF, Glass IA. AHI1 mutations cause both retinal dystrophy and renal cystic disease in Joubert syndrome. J Med Genet. 2006;43:334–339. doi: 10.1136/jmg.2005.036608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Valente EM, Brancati F, Silhavy JL, Castori M, Marsh SE, Barrano G, Bertini E, Boltshauser E, Zaki MS, Abdel-Aleem A, Abdel-Salam GM, Bellacchio E, Battini R, Cruse RP, Dobyns WB, Krishnamoorthy KS, Lagier-Tourenne C, Magee A, Pascual-Castroviejo I, Salpietro CD, Sarco D, Dallapiccola B, Gleeson JG, International JSRD Study Group AHI1 gene mutations cause specific forms of Joubert syndrome-related disorders. Ann Neurol. 2006;59:527–534. doi: 10.1002/ana.20749. [DOI] [PubMed] [Google Scholar]
- 73.Baala L, Romano S, Khaddour R, Saunier S, Smith UM, Audollent S, Ozilou C, Faivre L, Laurent N, Foliguet B, Munnich A, Lyonnet S, Salomon R, Encha-Razavi F, Gubler MC, Boddaert N, de Lonlay P, Johnson CA, Vekemans M, Antignac C, Attie-Bitach T. The Meckel-Gruber syndrome gene, MKS3, is mutated in Joubert syndrome. American Journal Human of Genetics. 2007;80:186–194. doi: 10.1086/510499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Otto EA, et al. Hypomorphic mutations in meckelin (MKS3/TMEM67) cause nephronophthisis with liver fibrosis (NPHP11). J Med Genet. 2009;46:663–670. doi: 10.1136/jmg.2009.066613. [DOI] [PubMed] [Google Scholar]
- 75.den Hollander AI, Koenekoop RK, Yzer S, Lopez I, Arends ML, Voesenek KE, Zonneveld MN, Strom TM, Meitinger T, Brunner HG, Hoyng CB, van den Born LI, Rohrschneider K, Cremers FP. Mutations in the CEP290 (NPHP6) gene are a frequent cause of Leber congenital amaurosis. American Journal Human of Genetics. 2006;79:556–561. doi: 10.1086/507318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sayer JA, Otto EA, O'Toole JF, Nurnberg G, Kennedy MA, Becker C, Hennies HC, Helou J, Attanasio M, Fausett BV, Utsch B, Khanna H, Liu Y, Drummond I, Kawakami I, Kusakabe T, Tsuda M, Ma L, Lee H, Larson RG, Allen SJ, Wilkinson CJ, Nigg EA, Shou C, Lillo C, Williams DS, Hoppe B, Kemper MJ, Neuhaus T, Parisi MA, Glass IA, Petry M, Kispert A, Gloy J, Ganner A, Walz G, Zhu X, Goldman D, Nurnberg P, Swaroop A, Leroux MR, Hildebrandt F. The centrosomal protein nephrocystin-6 is mutated in Joubert syndrome and activates transcription factor ATF4. Nature Genetics. 2006;38:674–681. doi: 10.1038/ng1786. [DOI] [PubMed] [Google Scholar]
- 77.Reja R, et al. MitoInteractome: mitochondrial protein interactome database, and its application in ‘aging network’ analysis. BMC Genomics. 2009;10(Suppl 3):S20. doi: 10.1186/1471-2164-10-S3-S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kass RS. The channelopathies: novel insights into the molecular and genetic mechanisms of human disease. Journal of Clinical Investigation. 2005;115:1986–1989. doi: 10.1172/JCI26011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rosenstein B.J.a.C., G.R. The diagnosis of cystic fibrosis: a consensus statement. Journal of Pediatrics. 1998;132:589–595. doi: 10.1016/s0022-3476(98)70344-0. [DOI] [PubMed] [Google Scholar]
- 80.Surtees R. Inherited ion channel disorders. European Journal of Pediatrics. 2000;159:S199–S203. doi: 10.1007/pl00014403. [DOI] [PubMed] [Google Scholar]
- 81.Dohle GR, Veeze HJ, Overbeek SE, van den Ouweland AMW, Halley DJJ, Weber RFA, Niermeijer MF. The complex relationships between cystic fibrosis and congenital bilateral absence of the vas deferens: clinical, electrophysiological and genetic data. Human Reproduction. 1999;14:371–374. doi: 10.1093/humrep/14.2.371. [DOI] [PubMed] [Google Scholar]
- 82.di Sant'Agnese PA, Darling RC, Perera GA, Shea E. Abnormal electrolyte composition of sweat in cystic fibrosis of the pancreas: clinical significance and relationship to the disease. Pediatrics. 1953;12:549–563. [PubMed] [Google Scholar]
- 83.Gibson L.E.a.C., R.E. A test for concentration of electrolytes in sweat in cystic fibrosis of the pancreas utilizing pilocarpine by iontophoresis. Pediatrics. 1959;23:545–549. [PubMed] [Google Scholar]
- 84.Callen A, Diener-West M, Zeitlin PL, Rubenstein RC. A simplified cyclic adenosine monophosphate-mediated sweat rate test for quantitative measure of cystic fibrosis transmembrane regulator (CFTR) function. Journal of Pediatrics. 2000;137:849–855. doi: 10.1067/mpd.2000.109198. [DOI] [PubMed] [Google Scholar]
- 85.Piwon N, Günther W, Schwake M, Bösl MR, Jentsch TJ. ClC-5 Cl--channel disruption impairs endocytosis in a mouse model for Dent's disease. Nature. 2000;408:369–373. doi: 10.1038/35042597. [DOI] [PubMed] [Google Scholar]
- 86.Scheiman SJ. X-linked hypercalciuric nephrolithiasis: clinical syndromes and chloride channel mutations. Kidney International. 1998;53:3–17. doi: 10.1046/j.1523-1755.1998.00718.x. [DOI] [PubMed] [Google Scholar]
- 87.Scheinman SJ, Guay-Woodward LM, Thakker RV, Warnock DG. Genetic disorders of renal electrolyte transport. New England Journal of Medicine. 1999;340:1177–1187. doi: 10.1056/NEJM199904153401507. [DOI] [PubMed] [Google Scholar]
- 88.Utsch B, Bokenkamp A, Benz MR, Besbas N, Dotsch J, Franke I, Frund S, Gok F, Hoppe B, Karle S, Kuwertz-Broking E, Laube G, Neb M, Nuutinen M, Ozaltin F, Rascher W, Ring T, Tasic V, van Wijk JA, Ludwig M. Novel OCRLI mutations in patients with the phenotype of Dent disease. American Journal of Kidney Disease. 2006;48:942, e941–914. doi: 10.1053/j.ajkd.2006.08.018. [DOI] [PubMed] [Google Scholar]
- 89.Hoopes RRJ, Shrimpton AE, Snohl SJ, Hueber P, Hoppe B, Matyus J, Simckes A, Tasic V, Toenshoff B, Suchy SF, Nussbaum RL, Scheinman SJ. Dent disease with mutations in OCRLI. American Journal Human of Genetics. 2005;76:260–267. doi: 10.1086/427887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu J, Krantz ID. Cohesin and human disease. Annu Rev Genomics Hum Genet. 2008;9:303–320. doi: 10.1146/annurev.genom.9.081307.164211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hagstrom K.A.a.M., B.J. Condensin and cohesin: more than chromosome compactor and glue. Nature Reviews Genetics. 2003;4:520–534. doi: 10.1038/nrg1110. [DOI] [PubMed] [Google Scholar]
- 92.Musio A, Selicorni A, Focarelli ML, Gervasini C, Milani D, Russo S, Vezzoni P, Larizza L. X-linked Cornelia de Lange syndrome owing to SMC1L1 mutations. Nature Genetics. 2006;38:528–530. doi: 10.1038/ng1779. [DOI] [PubMed] [Google Scholar]
- 93.Krantz ID, McCallum J, DeScipio C, Kaur M, Gillis LA, Yaeger D, Jukofsky L, Wasserman N, Bottani A, Morris CA, Nowaczyk MJ, Toriello H, Bamshad MJ, Carey JC, Rappaport E, Kawauchi S, Lander AD, Calof AL, Li HH, Devoto M, Jackson LG. Cornelia de Lange syndrome is caused by mutations in NIPBL, the human homolog of Drosophila melanogaster Nipped-B. Nature Genetics. 2004;36:631–635. doi: 10.1038/ng1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tonkin ET, Wang TJ, Lisgo S, Bamshad MJ, Strachan T. NIPBL, encoding a homolog of fungal Scc2-type sister chromatid cohesion proteins and fly Nipped-B, is mutated in Cornelia de Lange syndrome. Nature Genetics. 2004;36:636–641. doi: 10.1038/ng1363. [DOI] [PubMed] [Google Scholar]
- 95.Deardorff MA, Kaur M, Yaeger D, Rampuria A, Korolev S, Pie J, Gil-Rodríguez C, Arnedo M, Loeys B, Kline AD, Wilson M, Lillquist K, Siu V, Ramos FJ, Musio A, Jackson LS, Dorsett D, Krantz ID. Mutations in cohesin complex members SMC3 and SMC1A cause a mild variant of cornelia de Lange syndrome with predominant mental retardation. American Journal Human of Genetics. 2007;80:485–494. doi: 10.1086/511888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Vega H, Waisfisz Q, Gordillo M, Sakai N, Yanagihara I, Yamada M, van Gosliga D, Kayserili H, Xu C, Ozono K, Jabs EW, Inui K, Joenje H. Roberts syndrome is caused by mutations in ESCO2, a human homolog of yeast ECO1 that is essential for the establishment of sister chromatid cohesion. Nature Genetics. 2005;37:468–470. doi: 10.1038/ng1548. [DOI] [PubMed] [Google Scholar]
- 97.Schüle B, Oviedo A, Johnston K, Pai S, Francke U. Inactivating mutations in ESCO2 cause SC phocomelia and Roberts syndrome: no phenotypegenotype correlation. American Journal Human of Genetics. 2005;77:1117–1128. doi: 10.1086/498695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, Yahata K, Imamoto F, Aburatani H, Nakao M, Imamoto N, Maeshima K, Shirahige K, Peters JM. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 99.Klein RJ, Zeiss C, Chew EY, Tsai J, Sackler RS, Haynes C, Henning AK, SanGiovanni JP, Mane SM, Mayne ST, Bracken MB, Ferris FL, Ott J, Barnstable C, Hoh J. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Haines JL, Hauser MA, Schmidt S, Scott WK, Olson LM, Gallins P, Spencer KL, Kwan SY, Noureddine M, Gilbert JR, Shnetz-Boutaud N, Agarwal A, Postel EA, Pericak-Vance MA. Complement factor H variant increases risk of age-related macular degeneration. Science. 2005;308:419–421. doi: 10.1126/science.1110359. [DOI] [PubMed] [Google Scholar]
- 101.Edwards AO, R.R.I., Abel KJ, Manning A, Panhuysen C, Farrer LA. Complement factor H polymorphism and age-related macular degeneration. Science. 2005;308:421–424. doi: 10.1126/science.1110189. [DOI] [PubMed] [Google Scholar]
- 102.Gold B, et al. Variation in factor B (BF) and complement component 2 (C2) genes is associated with age-related macular degeneration. Nat Genet. 2006;38:458–462. doi: 10.1038/ng1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hageman GS, et al. A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc Natl Acad Sci U S A. 2005;102:7227–7232. doi: 10.1073/pnas.0501536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sepp T, et al. Complement factor H variant Y402H is a major risk determinant for geographic atrophy and choroidal neovascularization in smokers and nonsmokers. Invest Ophthalmol Vis Sci. 2006;47:536–540. doi: 10.1167/iovs.05-1143. [DOI] [PubMed] [Google Scholar]
- 105.Li M, et al. CFH haplotypes without the Y402H coding variant show strong association with susceptibility to age-related macular degeneration. Nat Genet. 2006;38:1049–1054. doi: 10.1038/ng1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hughes AE, et al. A common CFH haplotype, with deletion of CFHR1 and CFHR3, is associated with lower risk of age-related macular degeneration. Nat Genet. 2006;38:1173–1177. doi: 10.1038/ng1890. [DOI] [PubMed] [Google Scholar]
- 107.Maller J, George S, Purcell S, Fagerness J, Altshuler D, Daly MJ, Seddon JM. Common variation in three genes, including a noncoding variant in CFH, strongly influences risk of age-related macular degeneration. Nature Genetics. 2006;38:1055–1059. doi: 10.1038/ng1873. [DOI] [PubMed] [Google Scholar]
- 108.Yates JR, et al. Complement C3 variant and the risk of age-related macular degeneration. N Engl J Med. 2007;357:553–561. doi: 10.1056/NEJMoa072618. [DOI] [PubMed] [Google Scholar]
- 109.Chen Y, et al. Variations in DNA elucidate molecular networks that cause disease. Nature. 2008;452:429–435. doi: 10.1038/nature06757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Greenman C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sjoblom T, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 112.Wood LD, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 113.Frohling S, et al. Identification of driver and passenger mutations of FLT3 by high-throughput DNA sequence analysis and functional assessment of candidate alleles. Cancer Cell. 2007;12:501–513. doi: 10.1016/j.ccr.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 114.Thusberg J, Vihinen M. Pathogenic or not? And if so, then how? Studying the effects of missense mutations using bioinformatics methods. Hum Mutat. 2009;30:703–714. doi: 10.1002/humu.20938. [DOI] [PubMed] [Google Scholar]
- 115.Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Stone E.A.a.S., A. Physicochemical constraint violation by missense substitutions mediates impairment of protein function and disease severity. Genome Research. 2005;15:978–986. doi: 10.1101/gr.3804205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Capriotti E, et al. Predicting the insurgence of human genetic diseases associated to single point protein mutations with support vector machines and evolutionary information. Bioinformatics. 2006;22:2729–2734. doi: 10.1093/bioinformatics/btl423. [DOI] [PubMed] [Google Scholar]
- 118.Ramensky V, B.P., Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Research. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bromberg Y, Rost B. SNAP: predict effect of non-synonymous polymorphisms on function. Nucleic Acids Research. 2007;35:3823–3835. doi: 10.1093/nar/gkm238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Davis RE, S.R., Rahmouni K, Nishimura DY, Mullins RF, Agassandian K, Philp AR, Searby CC, Andrews MP, Thompson S, Berry CJ, Thedens DR, Yang B, Weiss RM, Cassell MD, Stone EM, Sheffield VC. A knockin mouse model of the Bardet-Biedl syndrome 1 M390R mutation has cilia defects, ventriculomegaly, retinopathy, and obesity. Proceedings of the National Academy of Sciences USA. 2007;104:19422–19427. doi: 10.1073/pnas.0708571104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kim JC, O.Y., Badano JL, Esmail MA, Leitch CC, Fiedrich E, Beales PL, Archibald JM, Katsanis N, Rattner JB, Leroux MR. MKKS/BBS6, a divergent chaperonin-like protein linked to the obesity disorder Bardet-Biedl syndrome, is a novel centrosomal component required for cytokinesis. Journal of Cell Science. 2005;118:1007–1020. doi: 10.1242/jcs.01676. [DOI] [PubMed] [Google Scholar]
- 122.Kuznetsov SG, et al. Mouse embryonic stem cell-based functional assay to evaluate mutations in BRCA2. Nat Med. 2008;14:875–881. doi: 10.1038/nm.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Guo J, et al. A DDX5 S480A polymorphism is associated with increased transcription of fibrogenic genes in hepatic stellate cells. J Biol Chem. 2009 doi: 10.1074/jbc.M109.035295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Shin JA, et al. Genetic effect of CysLTR2 polymorphisms on its mRNA synthesis and stabilization. BMC Med Genet. 2009;10:106. doi: 10.1186/1471-2350-10-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hayden EC. International genome project launched. Nature. 2008;451:378–379. doi: 10.1038/451378b. [DOI] [PubMed] [Google Scholar]
- 126.Choi M, et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc Natl Acad Sci U S A. 2009;106:19096–19101. doi: 10.1073/pnas.0910672106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Reiners J, et al. Molecular basis of human Usher syndrome: deciphering the meshes of the Usher protein network provides insights into the pathomechanisms of the Usher disease. Exp Eye Res. 2006;83:97–119. doi: 10.1016/j.exer.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 128.Bork JM, P.L., Riazuddin S, Bernstein SL, Ahmed ZM, Ness SL, Polomeno R, Ramesh A, Schloss M, Srisailpathy CR, Wayne S, Bellman S, Desmukh D, Ahmed Z, Khan SN, Kaloustian VM, Li XC, Lalwani A, Riazuddin S, Bitner-Glindzicz M, Nance WE, Liu XZ, Wistow G, Smith RJ, Griffith AJ, Wilcox ER, Friedman TB, Morell RJ. Usher syndrome 1D and nonsyndromic autosomal recessive deafness DFNB12 are caused by allelic mutations of the novel cadherin-like gene CDH23. American Journal Human of Genetics. 2001;68:26–37. doi: 10.1086/316954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lagziel A, et al. Expression of cadherin 23 isoforms is not conserved: implications for a mouse model of Usher syndrome type 1D. Mol Vis. 2009;15:1843–1857. [PMC free article] [PubMed] [Google Scholar]
- 130.Lagziel A, et al. Spatiotemporal pattern and isoforms of cadherin 23 in wild type and waltzer mice during inner ear hair cell development. Dev Biol. 2005;280:295–306. doi: 10.1016/j.ydbio.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 131.Zhong Q, et al. Edgetic perturbation models of human inherited disorders. Mol Syst Biol. 2009;5:321. doi: 10.1038/msb.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]