Abstract
Biotechnological production of high value chemical products increasingly involves engineering in vivo multi-enzyme pathways and host metabolism. Recent approaches to these engineering objectives have made use of molecular tools to advance de novo pathway identification, tunable enzyme expression, and rapid pathway construction. Molecular tools also enable optimization of single enzymes and entire genomes through diversity generation and screening, whole cell analytics, and synthetic metabolic control networks. In this review, we focus on advanced molecular tools and their applications to engineered pathways in host organisms, highlighting the degree to which each tool is generalizable.
Introduction
Chemical biotechnology is the use of biocatalysts in engineered systems to produce bulk and fine chemicals [1]. Three waves of biocatalysis have been described: first, realization that biological components could be used for chemical transformations; second, development of genetic engineering techniques needed for industrial production of proteins; and third, development of directed evolution-enabled enzyme engineering [2]. The coming wave relies not only on further improvements in protein engineering and DNA synthesis technologies, but also critically on our ability to engineer controlled, multi-enzyme pathway systems. Although isolated enzymes are widely used industrially today, whole cells are a more feasible system for multi-enzyme pathways. The introduction of heterologous pathways into a host organism and metabolic flux optimization toward the product of interest is a synergistic application of concepts from metabolic engineering and synthetic biology [3]. In this review, we describe selected contributions of metabolic engineering, synthetic biology, systems biology, and protein engineering to chemical biotechnology to improve the productivity of multi-enzyme pathways. These fields have provided advanced molecular tools for de novo pathway identification, tuned pathway construction, diversity generation and screening, genome-scale identification of optimization targets, and dynamic pathway control. Here, we focus on such molecular tools developed, improved, and applied in new contexts over the past few years.
Enhanced tools for precise biosynthetic pathway construction
Engineered biosynthetic pathways require composition of genetically encoded expression devices that support precise and tunable levels of pathway enzymes. Both the number of characterized control elements—such as ribosome binding sites (RBSs), promoters, and terminators—and the degree to which those control elements can be made to behave in a predictable manner under a range of contexts have expanded. Additionally, improved and new methods have been developed to assemble these control elements with enzymes to construct biosynthetic pathways.
One challenge to the rational design of genetically encoded elements is that they often behave in a context-dependent manner, exhibiting properties that depend on the combination of other elements used in the device or exhibiting off-target perturbations of the biological host. This challenge has been addressed by designing for context and then iteratively optimizing to improve behavior. Contextual features to consider range from the specific (e.g., DNA sequence surrounding the element) to the holistic (e.g., environmental growth conditions). For example, the impact of oxygen and glucose conditions on constitutive yeast promoter activities was characterized to permit design for these culture conditions [4]. Known variation in tRNA availabilities among hosts has been used to reduce the host-dependence of protein expression via codon optimization [5, 6]. Alternatively, insulated elements have been developed that behave robustly in varying contexts. For example, researchers have developed an insulated constitutive bacterial promoter library with relative protein production rates that span two orders of magnitude and are independent of the coding sequence of the expressed protein [7]. New insulating elements that use RNA processing to reduce the context dependence of genes in multi-gene operons have also been introduced [8].
Quantitative modeling and characterization of control elements have enabled researchers to create larger libraries of elements that exhibit predictable behaviors when integrated into gene expression devices. For example, a thermodynamic model of bacterial translation initiation was developed and used to forward design synthetic RBSs with a 47% chance of exhibiting protein expression levels within 2.3-fold of the desired level [9]. In addition, libraries of novel gene control elements have been developed using evolutionary and screening strategies. For example, a set of Rnt1p-cleavable hairpins provides post-transcriptional tuning of protein expression levels ranging from 8-84% of a control construct without a hairpin [10]. In vivo scaffolds are another set of synthetic control elements and act post-translationally to improve pathway flux by spatially co-localizing enzymes to RNA [11], DNA [12], protein [13, 14], cell surface [15], or a specific organelle [16].
Along with the diversification and insulation of genetic control elements have come faster, more reliable methods to construct biosynthetic pathways. Notably, an eight-gene biosynthetic pathway was assembled into a shuttle vector or yeast artificial chromosome in a single transformation with over 50% efficiency (Figure 1a) [17]. Pathways can also be integrated iteratively, which may increase the accessible library size for testing variants in a multi-gene pathway [18]. The construction and transplantation of a chemically synthesized bacterial genome showcased the cumulative advances of in vitro enzyme-mediated assembly and in vivo transformation-associated-recombination in yeast [19]. Similar techniques have been used to replace chromosome arms in yeast with circular or linear synthetic versions [20]. In E. coli, researchers have developed “recombineering” methods that use phage proteins to facilitate recombination-based genetic engineering. Rec-mediated recombineering was developed for efficient recombination between linear PCR products and linearized plasmids, which complements efficient lambda phage Red-mediated recombination between linear PCR products and circular plasmids [21].
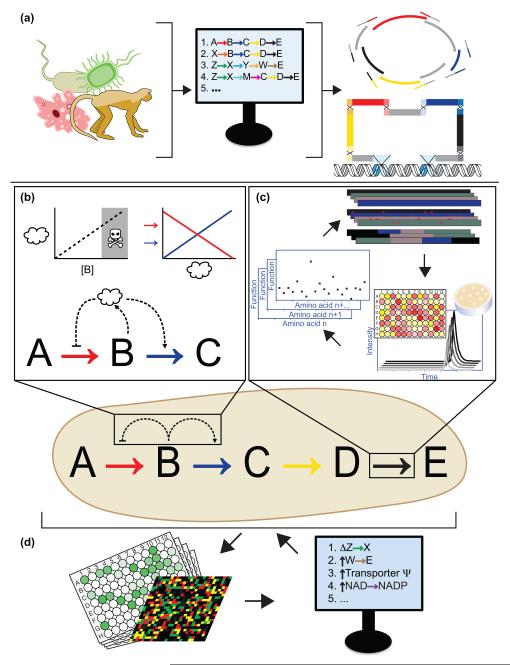
Figure 1.
Molecular tools to advance engineering of multi-enzyme biochemical pathways for chemical biotechnology. (a) Genetic and genomic databases are used with predictive algorithms to design pathways, which are then genetically constructed in vitro and/or in vivo. (b) Dynamic control elements allow enzyme expression levels to vary in response to small molecule concentrations. (c) Rounds of diversity generation and screening, informed by machine learning and design of experiment algorithms, generate optimized enzyme variants. (d) Whole cell read-outs provide data for systems level analysis and prediction of specific changes to enable global phenotypic improvements.
Improved tools for de novo pathway identification
Engineered biosynthetic pathways were once painstakingly pieced together from a single organism’s cDNA to mimic natural biosynthesis strategies, and optimization of host platforms involved serial knockouts or overexpression of targeted genes. However, modern bioinformatic tools allow rapid mining of huge sequence repositories for functions of interest and even prediction of multiple possible pathways to produce a desired small molecule (Figure 1a). Databases such as BioCyc [22], Kyoto Encyclopedia of Genes and Genomes (KEGG) [23], Rhea [24], and Braunschweig Enzyme Database (BRENDA) [25] facilitate search and display of metabolic pathways. These databases and others are populated by proteins whose discovery is accelerated by functional prediction algorithms, e.g., global biochemical reconstruction using sampling (GLOBUS), that take a systems biology approach to adjusting functional predictions generated by homology and secondary structure [26]. Genome-scale metabolic models of several industrially useful host organisms are available and reviewed elsewhere [27]. These stoichiometric models are used in computational predictions of routes from central metabolites to a product of interest, implementing a variety of rules for reaction qualifications (e.g., known enzymes, metabolites, or reaction chemistries) and scoring schemes (e.g., route length, number of enzymes known, complexity of transformations, flux balance analysis) to rank possible pathways [28]. For example, researchers used in-house pathway prediction and ranking software with a genome-scale E. coli model to identify two possible pathways to the non-natural product 1,4-butanediol [29].
In a creative use of genomic information, an alkane-producing operon was discovered via subtractive genomic analysis of 11 cyanobacteria strains and used in E. coli to produce a mixture of alkanes and alkenes [30]. Enzymes variants can be rationally selected when enzymes with similar functions from many organisms are available. In the reverse engineering of the β- oxidation cycle for production of n-butanol, enzymes were chosen based on kinetic parameters and co-factor specificity [31, 32]. The combination of genomic information and library-based cloning strategies is a rapid way to obtain rare enzymatic activities. In one example, a 30 kb Vibrio splendidus genome fragment, suspected by homology to encode transporters needed for alginate utilization, was cloned into E. coli by screening a genomic fosmid library [33].
Tools for molecular diversity generation and screening
Directed evolution is a powerful method for optimizing biosynthetic pathways and requires molecular tools for generating and screening diversity (Figure 1c). Tools for generating diversity at the single-gene level, such as error-prone PCR, DNA shuffling, saturation mutagenesis, and site-directed mutagenesis, have been used to generate enzyme libraries of varying size for some time. Screening for small molecule production depends on physical characteristics of the substrates, cofactors, products, or strains. Screens employ selection, growth, colorimetric, fluorescent, or UV readouts in a high-throughput format; or direct quantification by a separation method coupled to a detection method, like LC-MS, in lower-throughput formats. Recent work to engineer E. coli for increased levopimaradiene production provides examples of both small and large library generation and screening [34]. One enzyme in the pathway, levopmaradiene synthase, was iteratively subjected to site-directed and single-site saturation mutagenesis at amino acid positions selected from a structural homology model, and resulting changes in productivity were analyzed by GC-MS. A second enzyme, geranylgeranyl diphosphate synthase, was subjected to error-prone PCR, expressed in the context of a lycopene-producing strain, and then screened visually for production of the red pigment characteristic of lycopene. Together, these two optimized enzymes, along with higher expression of four upstream endogenous enzymes, increased levopimaradiene production from 0.15 mg/L to 700 mg/L [34].
Advances in DNA synthesis support the design of synthetic oligonucleotides and genes; thus smaller libraries with defined variation and a higher fraction of functional proteins can be built, permitting lower-throughput, higher-sensitivity screens (Figure 1c). Several commercial DNA synthesis companies offer tailored synthetic libraries, including specified amino acid frequencies at each site, truncations of varied length, mixed-and-matched domains from similar proteins, and specific frequencies of random mutations in all or a subset of amino acid sites. Structure or structural homology, phylogeny, and known amino acid sequence-function relationships often inform the design of these tailored libraries. For example, a computational structure-guided recombination method was used to divide cellulases into domains, and a subset of all possible chimeras was synthesized to sample the effect of each domain on thermostability and activity [35]. Linear regression analysis and machine learning were then applied to the results from the first chimera library and used to design a second small synthetic library, which yielded three variants with similar activities to the parent and greatly enhanced thermostability.
Several tools for generating directed diversity at the genome level have been developed. Some genome alteration methods build on recombineering E. coli. For example, a process termed “multiplex automated genome engineering” (MAGE) introduces changes mediated by lambda phage Red and directed by synthetic oligonucleotides to generate up to 15 billion genetically distinct E. coli cell populations in three days of automated transformation cycles [36]. Since MAGE is most efficient with a pool of ~10 oligonucleotides, a hierarchical conjugation assembly genome engineering (CAGE) method was developed to combine altered portions of the genome into a single strain using E. coli donor-receptor strains engineered for site-specific recombination [37]. A trackable multiplex recombineering (TRMR) method was developed to introduce thousands of targeted and barcoded changes into the bacterial genome such that fitness phenotypes can be mapped back to the specific change in the genome [38]. This approach was recently used with MAGE to combinatorially alter the ribosome binding sequences of thousands of genes and map resulting growth phenotypes to their accompanying genomic changes [39]. In yeast, homologous recombination has been used for generating diversity via transformation with libraries of synthetic oligonucleotides flanked by homology regions [40]. Additionally, homologous recombination has been combined with an inducible double-stranded break system and sexual reproduction to generate rounds of diversity in vivo by mating rather than by transformation [41].
Optimal enzymes or control elements for a biosynthetic pathway can only be identified through appropriate screens; thus, generalizable, high-throughput screens are highly desirable. For example, enzyme activity can be assayed indirectly by monitoring consumption or production of a UV-active cofactor like NAD(P)H or using either a general colorimetric assay for pH or an organic functional group. However, these assays can be difficult to conduct in vivo and may not have the necessary sensitivity and signal-to-noise ratio to detect small changes in enzyme function. Recently, a screening strategy was described that combines yeast display of an enzyme library, chemoenzymatic conjugation of one substrate to the cell surface, incubation with a second substrate with an affinity handle, fluorescent staining of or conjugation to the affinity handle, and fluorescence-activated cell sorting (FACS) to select cells expressing enzymes capable of forming bonds between the two substrates [42]. Although this method is not dependent on physical properties of the substrates or product, it does require that both substrates be amenable to chemical conjugation and that the enzyme being selected tolerate surface display and have a sufficiently flexible active site. In an alternative approach, an in vivo RNA-based biosensor couples intracellular small-molecule product concentrations to levels of a genetically encoded reporter like GFP, such that large enzyme or pathway libraries can be rapidly screened by FACS [43]. This method depends on selection of an appropriate RNA aptamer to the small molecule target of interest and integration of the aptamer into the RNA biosensor platform, rather than on any innate property of the substrate, product, or enzyme.
Small molecule biosensors that can be linked to gene expression are particularly powerful in the context of the recently developed in vivo phage-assisted continuous evolution (PACE) [44]. In PACE, host E. coli cells express two plasmids, one for continuous mutagenesis and one on which the expression of a protein necessary for phage survival is linked to a desired protein activity. The host E. coli is infected by selection phage that encode the protein library, and the culture is maintained in a turbidostat such that phage that encode protein library members that activate the expression of the phage-survival protein are enriched over time.
Systems biology analysis for host-level optimization
As tools and methods for identifying, building, and optimizing heterologous metabolic pathways have grown in number, the need to understand host metabolism at a systems level to support further optimization has emerged. To meet this need, systems biology has developed techniques to support “-ome-level” interrogation of cellular behavior and quantitative models for analysis of whole-cell metabolic networks (Figure 1d) [45-47]. The use of systems-level measurements has expanded researchers’ abilities to identify gene targets to improve engineered strains. For example, transcriptome profiling has been used to identify sources of stress in yeast strains harboring evolved P450 monooxygenases. By analyzing the global transcriptional response across a series of evolved enzyme variants, researchers identified heme depletion as the major limiting factor for optimized monooxygenase activity at high expression. Subsequent overexpression of cellular machinery for producing heme increased the productivity of the highest activity evolved variant by 2.3-fold [48]. In another study, global transcript, metabolite, and genotype measurements were used to identify traits associated with higher yeast growth rates on galactose, an industrially relevant sugar disfavored by native yeast metabolism. Researchers compared two previously engineered strains and three newly evolved strains against a parent strain and identified specific favorable mutations that arose in gene targets unpredictably related to carbohydrate sensing and catabolism [49].
Improvements in stoichiometric genome-scale metabolic networks with constraint-based models like flux balance analysis (FBA) or minimization of metabolic adjustment (MOMA) have enabled several recent model-guided strain optimization efforts, in which predicted modifications of host metabolism improved product yields (Figure 1d) [50]. For example, OptGene, a genetic search algorithm for non-linear optimization [51], and MOMA were used to identify knockout targets within a stoichiometric model of S. cerevisiae that would lead to increased sesquiterpene production [52, 53]. Deletion of the predicted target glutamate dehydrogenase GDH1 and overexpression of the NADH-dependent glutamate dehydrogenase gene GDH2 to repair the consequent growth defect, resulted in a strain with nearly triple the total sesquiterpene titer [53]. Using similar computational methods, researchers simulated the effect of gene knockouts on the relationship between growth rate and polylactic acid production rate, identifying three knockout targets that were combined with two rationally selected overexpression targets to increase overall polymer accumulation by 3.7-fold [54]. An engineered strain of E. coli for the production of 1,4-butanediol (BDO) was improved by the introduction of targeted changes to the host genome guided by a whole-cell metabolic model [29, 55, 56]. The changes made to host metabolism included knockouts of key enzymes, deletion of a global regulator, and a point mutation in an enzyme to destroy the allosteric inhibition of the native citrate synthase, leading to over 95% of carbon flux being directed to the BDO pathway as measured by 13C labeling [29].
Molecular control elements for dynamic pathway regulation
Taking a cue from natural regulatory networks, researchers have begun designing dynamic regulation for engineered biosynthetic pathways (Figure 1b) [57, 58]. For example, a theoretical analysis detailed parameter constraints necessary to build a biological proportional-integral control network capable of perfect adaptation and modeled the effects of these parameters in a two-promoter gene network [59]. An in silico model of dynamic control in a biofuel production pathway concluded that efflux pumps under control of a biofuel-responsive promoter would limit toxicity [60]. However, implementation of these in silico designs is currently limited by availability and tunability of the molecular components required to build dynamic controllers.
Genetically encoded sensors developed for selection and screening can potentially be coupled to activators to create synthetic molecular dynamic controllers [61]. While conceptually simple, building such a device can be practically challenging and demands parameter tuning to obtain desired behavior [62]. For example, nine distinct parameters were involved in the design of static ribozyme-regulated expression devices [63]. Dynamic controllers sense and process a molecular input and generate a gene-regulatory output. Open-loop controllers sense an external molecular input, such as the IPTG supplied to an IPTG-inducible promoter, while closed-loop controllers sense a molecular input associated with the pathway of interest, such as an intermediate in an engineered biosynthetic pathway [61]. In a recent implementation of open-loop control, researchers used the glucose-responsive promoters HXT1 and HXT2 to control the expression level of squalene synthase, ERG9, resulting in a 2-fold increase in α-santalene production in yeast, which was improved by further host metabolic engineering and bioprocess optimization strategies [64]. In addition to concentrations of small molecules, open-loop controllers can respond to autoinduction by cellular quorum sensing. By engineering the native quorum sensing regulon to initiate the expression of T7 polymerase, researchers demonstrated an autoinduction “switch” that serves as a late stage E. coli heterologous protein expression system [65].
Because closed-loop control requires response to a signal associated with the biosynthetic pathway, it can be more complex to engineer. The first example of a closed-loop metabolic controller was a synthetic regulon built using parts of the acetyl phosphate-responsive promoter glnAP2 applied to the control of enzyme expression levels in the lycopene biosynthesis pathway. As metabolic flux to acetyl phosphate competes with that to lycopene, this controller served to divert flux to the lycopene pathway, increasing productivity 3-fold [66]. In a recent example, researchers built a dynamic closed-loop controller from the natural fatty-acid sensing transcription factor FadR, which binds to and represses a recombinant promoter [67]. High cellular concentrations of fatty acids lead to increased pools of acyl-CoA that bind FadR, which then releases its DNA binding region and allows transcription from the engineered promoter. This promoter was used to control the expression of pdc and adhB for ethanol production, fadD for fatty acyl-CoA production, and atfA for fatty acid ethyl ester (FAEE) production. The dynamic control system effectively prevents the build-up of ethanol, a toxic pathway intermediate, and over-production of acyl-CoA, which consumes fatty acids needed for other processes. FAEE yield increased by 3-fold to 28% of the theoretical maximum under the dynamic control scheme. These systems demonstrate the promise of dynamic control strategies for engineered biosynthetic pathways, and show that repurposing of native systems can be a useful starting point for building dynamic controllers [67].
Conclusion
Enhancing our ability to engineer controlled, multi-enzyme biosynthetic pathways in whole-cell hosts is essential to expanding the spectrum of fine and bulk products that can be made with chemical biotechnology. Molecular tools for pathway identification, rapid and precise pathway construction, directed evolution for component optimization, systems-level models and measurements for host optimization, and dynamic pathway control advance these efforts. While a convergent set of tools and methodologies that allow researchers to build pathways to synthesize molecules of interest with ease is desired, developing generally applicable tools is an overarching challenge in chemical biotechnology. As shown in Table 1, solutions to improve pathway productivity are often specific to the host organism, the small molecule product, or even the particular strain. Much work remains to gain the predictive understanding necessary to build a biosynthetic pathway “off-the-shelf” and will involve amassing particular solutions to find generalizable patterns for strategic construction and optimization as well as developing generalizable, robust biological components and system composition methods.
Table 1.
Generalizability of selected molecular tools for chemical biotechnology.
| Role | Molecular tool | Host cell(s) | Generalizability | References |
|---|---|---|---|---|
| Expression tuning |
Constitutive promoters |
S. cerevisiae | Applications in yeast | [4] |
| Expression tuning and robustness |
Codon optimization |
Any | Any | [5, 6] |
| Expression robustness |
Insulated bacterial promoters |
E. coli | Any bacterial system where a 160 bp promoter is acceptable and elements in the 5’- UTR are not needed |
[7] |
| Expression robustness |
Clustered regularly interspaced short palindromic repeat (CRISPR) RNA processing |
E. coli, B.
subtilis, S. cerevisiae |
Any organism in which the Csy4-based processing platform can be functionally expressed |
[8] |
| Expression tuning |
Synthetic ribosome binding sites |
E. coli | Applications in E. coli | [9] |
| Expression tuning |
RNA control modules based on Rnt1p hairpins |
S. cerevisiae | Applications in yeast | [10] |
| Enzyme scaffolding |
RNA-enzyme assemblies |
E. coli | Applications in prokaryotes in which enzymes are amenable to tagging with aptamer binding domains |
[11] |
| Enzyme scaffolding |
DNA-enzyme assemblies |
E. coli | Applications in prokaryotes in which enzymes are amenable to tagging with zinc-finger proteins |
[12] |
| Enzyme scaffolding |
Protein scaffold- enzyme assemblies |
E. coli, S.
cerevisiae |
Applications in which enzymes are amenable to tagging with protein scaffold binding domains |
[13, 15, 68, 69] |
| Enzyme scaffolding |
Protein microcompart- ments |
E. coli | Potential applications in prokaryotes for enzymes that are amenable to tagging with binding domains or localization tags that direct them to protein microcompartments |
[70-73] |
| Enzyme scaffolding |
Host-heterologous protein fusions for localization |
S. cerevisiae (in this example) |
Any application in which a suitable host localization protein can be identified and a functional host protein-heterologous enzyme chimera can be generated |
[14] |
| Enzyme scaffolding |
Organelle- targeting tags |
S. cerevisiae | Applications in eukaryotes in which an appropriate organelle tag is available and the enzyme(s) is amenable to fusion |
[16] |
| Genetic assembly |
One step homologous recombinatio- nbased plasmid assembly or integration |
S. cerevisiae | Applications requiring assembly of 8-10 DNA segments in yeast |
[17] |
| Genetic assembly |
Iterative homologous recombination- based integration |
S. cerevisiae | Applications in yeast in which one-at-a-time integration and selection marker rescue is desired, such as pathway library construction |
[18] |
| Genetic assembly |
Combined in vitro and in vivo assembly for genome-scale constructs |
S. cerevisiae | Applications requiring assembly of multi kB- MBconstructs in yeast |
[19, 74] |
| Genetic assembly |
Construction of synthetic chromosome arms and replacement of endogenous arms |
S. cerevisiae | Applications in yeast in which the incorporation of large/many synthetic fragments and/or deletion of many native elements for stability is desired |
[20] |
| Genetic assembly |
Recombination of linear fragments |
E. coli | Applications in bacteria | [21] |
| Genome scale diversity generation |
Multiplex automated genome engineering |
E. coli | Applications in bacteria where genome-scale diversity generation is useful for optimization and a suitable screening method is available to evaluate the resulting large library |
[36] |
| Genome scale diversity generation |
Conjugation assembly genome engineering |
E. coli | Applications in bacteria where the combination of defined genomic fragments from multiple lineages into a single genome is desired |
[37] |
| Genome scale diversity generation |
Continuous recombination |
E. coli | Applications in bacteria where continuous recombination is useful for optimization (e.g. by combining multiple lineages) and a suitable screening method is available to evaluate the large library generated |
[75] |
| Genome scale diversity generation |
Homologous recombination of synthetic oligonucleotide libraries |
S. cerevisiae | Applications in yeast | [40] |
| Genome scale diversity generation |
Inducible doublestranded break and sexual reproduction |
S. cerevisiae | Applications in yeast where desired changes can be obtained in a manageable number of manual rounds |
[41] |
| Diversity screening |
Yeast 3-hybrid chemical complementation to screen for bond- forming enzymes |
S. cerevisiae | Applications in yeast where enzyme tolerates surface display and two substrates between which a covalent bond is formed are amenable to chemical conjugation |
[42] |
| Diversity screening |
Small moleculeresponsive synthetic RNA switch |
S. cerevisiae | Applications where RNA that binds the molecule is available or can be selected and incorporated into a self-cleaving genetic device |
[43] |
| Genome scale diversity generation and screening |
Phage-assisted continuous evolution |
E. coli | Applications in bacteria where the activity of the enzyme to be evolved can be linked to gene expression |
[44] |
| Whole cell gene expression measurement |
DNA microarray measurement of global transcriptional response to heterologous protein expression |
S. cerevisiae | Applications in yeast where host metabolism or stress response is limiting to strain productivity |
[48] |
| Model-guided host optimization |
Target gene knockouts and promoter substitutions based on model prediction |
S. cerevisiae | Applications in yeast where host metabolism is limiting to strain productivity |
[53] |
| Model-guided host optimization |
Target gene knockouts and gene overexpression based on model prediction |
E. coli | Applications in E. coli where host metabolism is limiting to strain productivity |
[54] |
| Dynamic control of gene expression |
Glucose responsive promoter |
S. cerevisiae | Applications in yeast where linking enzyme expression to glucose levels improves productivity |
[64] |
| Dynamic control of gene expression |
Rewired quorum sensing regulon |
E. coli | Applications in E. coli where cell-density dependent expression of proteins is desired |
[65] |
| Dynamic control of gene expression |
Fatty acid responsive transcriptional regulator |
E. coli | Applications in bacteria where gene expression tied to cellular free fatty acid pools is desired and cross-talk between the transcriptional regulator and native transcriptional control is not limiting |
[67] |
Highlights.
□ Novel molecular tools advance the productivity of engineered multi-enzyme biochemical pathways in live cells.
□ These tools enable precise, rapid pathway construction and static and dynamic control of enzyme expression levels.
□ Molecular tools facilitate enzyme or pathway optimization via directed evolution and systems biology approaches.
□The generalizability of each tool depends on its function, context dependence, and host requirements.
Acknowledgements
C.D.S. is supported by funds from the National Institutes of Health, National Science Foundation, Defense Advanced Research Projects Agency, and the Bill and Melinda Gates Foundation. S.G. is supported by a National Science Foundation Graduate Research Fellowship and a Stanford Graduate Research Fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference Annotations [7]*The authors developed a bacterial promoter library with relative protein production rates that are independent of the coding sequence of the expressed protein. This is one of the first examples of a context-independent biological control element.
[11]**Researchers engineered an RNA-based scaffolding system capable of forming 1, 2, or 3-D assemblies in vivo and binding tagged enzymes with aptamers.
[16]*By fusing plant sesquiterpene synthases to mitochondrial targeting sequences, the authors co-localized these enzymes with host enzymes that produce sesquiterpene precursors and thereby improved sesquiterpene biosynthesis in engineered yeast.
[20]**In this work, researchers replaced entire chromosome arms in yeast with designed synthetic versions. This is the first example of 10-100,000 bp sections of synthetic DNA being constructed to replace native sequence in a eukaryote.
[29]*The authors identified a biocatalytic route to 1,4-butanediol from central metabolism using pathway prediction software, built the suggested pathways, and improved overall titers by using a whole-cell metabolic model of E. coli to predict engineering targets. This is one of the first successful applications of de novo pathway prediction software.
[31]*Researchers improved titers of n-butanol by carefully identifying and characterizing enzymes from genetic databases with optimal kinetic parameters. This work is an excellent example of how genetic information can be rationally implemented for pathway design.
[33]**Genetic databases were used to identify a 36 kb Vibrio splendidus genome fragment suspected to encode transporters needed for alginate utilization, and this large fragment was cloned into E. coli by screening a genomic fosmid library. The researchers also engineered an alginate lyase secretion system and heterologous ethanol pathway to produce ethanol from macroalgae feedstocks.
[37]*The authors used conjugative genome assembly in E. coli to combine 32 defined sections of different strain genomes into a single recoded E. coli genome.
[43]**An in vivo RNA-based biosensor was used to couple intracellular small-molecule product concentrations to GFP levels, allowing a large enzyme library to be rapidly screened by plate-based fluorescence readings and FACS. This is one of the first screening methods that does not depend on any innate property of the substrate, product, or enzyme.
[44]**In the system developed here, the expression of a protein necessary for phage survival is linked to a desired biomolecule activity. The host E. coli is infected by selection phage that encode the biomolecule library, and the culture is maintained in a turbidostat such that phage that encode biomolecule library members that activate the expression of the phage-survival protein are enriched over time.
[48]**By measuring changes to global transcriptional patterns by DNA microarrays, heme depletion was discovered to be the principal source of stress for a yeast strain expressing evolved P450 monooxygenase enzymes. Subsequently alleviating this stress by overexpression of the genes responsible for heme biosynthesis showed recovered activity for the evolved P450 at high copy expression.
[53]**Model-guided analysis identified gene knockouts for increase in cellular availability of NADPH for sesquiterpene synthesis. The analysis also suggested a gene target to overexpress – this targeted change offset the growth impairment in the engineered strain versus wild type.
[54]**Targeted changes to the native E. coli metabolism were identified by a whole cell E. coli metabolic model. These modifications increased flux toward lactyl-CoA production, increasing lactate fraction of polymer as well as total polymer content of the cells by dry weight %.
[64]**Expression of a key biosynthetic enzyme in the isoprenoid biosynthesis pathway was tied to media glucose concentration by use of a glucose responsive promoter.
[65]*Heterologous protein expression can be tied to cell concentration by use of modified quorum sensing components.
[67]**The native E. coli transcriptional regulator FadR was used to control ethanol overproduction and FAEE synthase modules relative to pools of free fatty acids in the E. coli host strain. This approach achieved higher titers than tuning relative transcriptional strengths of the different modules with various constitutive promoters.
References
- 1.Petersen M. Chemical biotechnology industrial applications and recent advances - Invited overview. Curr Opin Biotechnol. 1999;10:593–594. [Google Scholar]
- 2.Bornscheuer UT, Huisman GW, Kazlauskas RJ, Lutz S, Moore JC, Robins K. Engineering the third wave of biocatalysis. Nature. 2012;485:185–194. doi: 10.1038/nature11117. [DOI] [PubMed] [Google Scholar]
- 3.Nielsen J, Keasling JD. Synergies between synthetic biology and metabolic engineering. Nat Biotechnol. 2011;29:693–695. doi: 10.1038/nbt.1937. [DOI] [PubMed] [Google Scholar]
- 4.Sun J, Shao Z, Zhao H, Nair N, Wen F, Xu JH. Cloning and characterization of a panel of constitutive promoters for applications in pathway engineering in Saccharomyces cerevisiae. Biotechnol Bioeng. 2012;109:2082–2092. doi: 10.1002/bit.24481. [DOI] [PubMed] [Google Scholar]
- 5.Kudla G, Murray AW, Tollervey D, Plotkin JB. Coding-sequence determinants of gene expression in Escherichia coli. Science. 2009;324:255–258. doi: 10.1126/science.1170160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Welch M, Villalobos A, Gustafsson C, Minshull J. Designing genes for successful protein expression. Methods Enzymol. 2011;498:43–66. doi: 10.1016/B978-0-12-385120-8.00003-6. [DOI] [PubMed] [Google Scholar]
- 7.Davis JH, Rubin AJ, Sauer RT. Design, construction and characterization of a set of insulated bacterial promoters. Nucleic Acids Res. 2011;39:1131–1141. doi: 10.1093/nar/gkq810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qi L, Haurwitz RE, Shao W, Doudna JA, Arkin AP. RNA processing enables predictable programming of gene expression. Nat Biotechnol. 2012;30:1002–1006. doi: 10.1038/nbt.2355. [DOI] [PubMed] [Google Scholar]
- 9.Salis HM, Mirsky EA, Voigt CA. Automated design of synthetic ribosome binding sites to control protein expression. Nat Biotechnol. 2009;27:946–950. doi: 10.1038/nbt.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Babiskin AH, Smolke CD. A synthetic library of RNA control modules for predictable tuning of gene expression in yeast. Mol Syst Biol. 2011;7:471. doi: 10.1038/msb.2011.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delebecque CJ, Lindner AB, Silver PA, Aldaye FA. Organization of intracellular reactions with rationally designed RNA assemblies. Science. 2011;333:470–474. doi: 10.1126/science.1206938. [DOI] [PubMed] [Google Scholar]
- 12.Conrado RJ, Wu GC, Boock JT, Xu H, Chen SY, Lebar T, Turnsek J, Tomsic N, Avbelj M, Gaber R, et al. DNA-guided assembly of biosynthetic pathways promotes improved catalytic efficiency. Nucleic Acids Res. 2012;40:1879–1889. doi: 10.1093/nar/gkr888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dueber JE, Wu GC, Malmirchegini GR, Moon TS, Petzold CJ, Ullal AV, Prather KL, Keasling JD. Synthetic protein scaffolds provide modular control over metabolic flux. Nat Biotechnol. 2009;27:753–759. doi: 10.1038/nbt.1557. [DOI] [PubMed] [Google Scholar]
- 14.Albertsen L, Chen Y, Bach LS, Rattleff S, Maury J, Brix S, Nielsen J, Mortensen UH. Diversion of flux toward sesquiterpene production in Saccharomyces cerevisiae by fusion of host and heterologous enzymes. Appl Environ Microbiol. 2011;77:1033–1040. doi: 10.1128/AEM.01361-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun J, Wen F, Si T, Xu JH, Zhao H. Direct conversion of xylan to ethanol by recombinant Saccharomyces cerevisiae strains displaying an engineered minihemicellulosome. Appl Environ Microbiol. 2012;78:3837–3845. doi: 10.1128/AEM.07679-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farhi M, Marhevka E, Masci T, Marcos E, Eyal Y, Ovadis M, Abeliovich H, Vainstein A. Harnessing yeast subcellular compartments for the production of plant terpenoids. Metab Eng. 2011;13:474–481. doi: 10.1016/j.ymben.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 17.Shao Z, Zhao H. DNA assembler, an in vivo genetic method for rapid construction of biochemical pathways. Nucleic Acids Res. 2009;37:e16. doi: 10.1093/nar/gkn991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wingler LM, Cornish VW. Reiterative Recombination for the in vivo assembly of libraries of multigene pathways. Proc Natl Acad Sci U S A. 2011;108:15135–15140. doi: 10.1073/pnas.1100507108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gibson DG, Glass JI, Lartigue C, Noskov VN, Chuang RY, Algire MA, Benders GA, Montague MG, Ma L, Moodie MM, et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science. 2010;329:52–56. doi: 10.1126/science.1190719. [DOI] [PubMed] [Google Scholar]
- 20.Dymond JS, Richardson SM, Coombes CE, Babatz T, Muller H, Annaluru N, Blake WJ, Schwerzmann JW, Dai J, Lindstrom DL, et al. Synthetic chromosome arms function in yeast and generate phenotypic diversity by design. Nature. 2011;477:471–476. doi: 10.1038/nature10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fu J, Bian X, Hu S, Wang H, Huang F, Seibert PM, Plaza A, Xia L, Muller R, Stewart AF, et al. Full-length RecE enhances linear-linear homologous recombination and facilitates direct cloning for bioprospecting. Nat Biotechnol. 2012;30:440–446. doi: 10.1038/nbt.2183. [DOI] [PubMed] [Google Scholar]
- 22.Caspi R, Altman T, Dale JM, Dreher K, Fulcher CA, Gilham F, Kaipa P, Karthikeyan AS, Kothari A, Krummenacker M, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2010;38:D473–479. doi: 10.1093/nar/gkp875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kotera M, Hirakawa M, Tokimatsu T, Goto S, Kanehisa M. The KEGG databases and tools facilitating omics analysis: latest developments involving human diseases and pharmaceuticals. Methods Mol Biol. 2012;802:19–39. doi: 10.1007/978-1-61779-400-1_2. [DOI] [PubMed] [Google Scholar]
- 24.Alcantara R, Axelsen KB, Morgat A, Belda E, Coudert E, Bridge A, Cao H, de Matos P, Ennis M, Turner S, et al. Rhea--a manually curated resource of biochemical reactions. Nucleic Acids Res. 2012;40:D754–760. doi: 10.1093/nar/gkr1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scheer M, Grote A, Chang A, Schomburg I, Munaretto C, Rother M, Sohngen C, Stelzer M, Thiele J, Schomburg D. BRENDA, the enzyme information system in 2011. Nucleic Acids Res. 2011;39:D670–676. doi: 10.1093/nar/gkq1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Plata G, Fuhrer T, Hsiao TL, Sauer U, Vitkup D. Global probabilistic annotation of metabolic networks enables enzyme discovery. Nat Chem Biol. 2012;8:848–854. doi: 10.1038/nchembio.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feist AM, Herrgard MJ, Thiele I, Reed JL, Palsson BO. Reconstruction of biochemical networks in microorganisms. Nat Rev Microbiol. 2009;7:129–143. doi: 10.1038/nrmicro1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Medema MH, van Raaphorst R, Takano E, Breitling R. Computational tools for the synthetic design of biochemical pathways. Nat Rev Microbiol. 2012;10:191–202. doi: 10.1038/nrmicro2717. [DOI] [PubMed] [Google Scholar]
- 29.Yim H, Haselbeck R, Niu W, Pujol-Baxley C, Burgard A, Boldt J, Khandurina J, Trawick JD, Osterhout RE, Stephen R, et al. Metabolic engineering of Escherichia coli for direct production of 1,4-butanediol. Nat Chem Biol. 2011;7:445–452. doi: 10.1038/nchembio.580. [DOI] [PubMed] [Google Scholar]
- 30.Schirmer A, Rude MA, Li X, Popova E, del Cardayre SB. Microbial biosynthesis of alkanes. Science. 2010;329:559–562. doi: 10.1126/science.1187936. [DOI] [PubMed] [Google Scholar]
- 31.Bond-Watts BB, Bellerose RJ, Chang MC. Enzyme mechanism as a kinetic control element for designing synthetic biofuel pathways. Nat Chem Biol. 2011;7:222–227. doi: 10.1038/nchembio.537. [DOI] [PubMed] [Google Scholar]
- 32.Dellomonaco C, Clomburg JM, Miller EN, Gonzalez R. Engineered reversal of the beta-oxidation cycle for the synthesis of fuels and chemicals. Nature. 2011;476:355–359. doi: 10.1038/nature10333. [DOI] [PubMed] [Google Scholar]
- 33.Wargacki AJ, Leonard E, Win MN, Regitsky DD, Santos CN, Kim PB, Cooper SR, Raisner RM, Herman A, Sivitz AB, et al. An engineered microbial platform for direct biofuel production from brown macroalgae. Science. 2012;335:308–313. doi: 10.1126/science.1214547. [DOI] [PubMed] [Google Scholar]
- 34.Leonard E, Ajikumar PK, Thayer K, Xiao WH, Mo JD, Tidor B, Stephanopoulos G, Prather KL. Combining metabolic and protein engineering of a terpenoid biosynthetic pathway for overproduction and selectivity control. Proc Natl Acad Sci U S A. 2010;107:13654–13659. doi: 10.1073/pnas.1006138107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heinzelman P, Snow CD, Smith MA, Yu X, Kannan A, Boulware K, Villalobos A, Govindarajan S, Minshull J, Arnold FH. SCHEMA recombination of a fungal cellulase uncovers a single mutation that contributes markedly to stability. J Biol Chem. 2009;284:26229–26233. doi: 10.1074/jbc.C109.034058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang HH, Isaacs FJ, Carr PA, Sun ZZ, Xu G, Forest CR, Church GM. Programming cells by multiplex genome engineering and accelerated evolution. Nature. 2009;460:894–898. doi: 10.1038/nature08187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Isaacs FJ, Carr PA, Wang HH, Lajoie MJ, Sterling B, Kraal L, Tolonen AC, Gianoulis TA, Goodman DB, Reppas NB, et al. Precise manipulation of chromosomes in vivo enables genome-wide codon replacement. Science. 2011;333:348–353. doi: 10.1126/science.1205822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Warner JR, Reeder PJ, Karimpour-Fard A, Woodruff LB, Gill RT. Rapid profiling of a microbial genome using mixtures of barcoded oligonucleotides. Nat Biotechnol. 2010;28:856–862. doi: 10.1038/nbt.1653. [DOI] [PubMed] [Google Scholar]
- 39.Sandoval NR, Kim JY, Glebes TY, Reeder PJ, Aucoin HR, Warner JR, Gill RT. Strategy for directing combinatorial genome engineering in Escherichia coli. Proc Natl Acad Sci U S A. 2012;109:10540–10545. doi: 10.1073/pnas.1206299109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pirakitikulr N, Ostrov N, Peralta-Yahya P, Cornish VW. PCRless library mutagenesis via oligonucleotide recombination in yeast. Protein Sci. 2010;19:2336–2346. doi: 10.1002/pro.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Romanini DW, Peralta-Yahya P, Mondol V, Cornish VW. A heritable recombination system for synthetic Darwinian evolution in yeast. ACS Synth Biol. 2012;1:602–609. doi: 10.1021/sb3000904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen I, Dorr BM, Liu DR. A general strategy for the evolution of bond-forming enzymes using yeast display. Proc Natl Acad Sci U S A. 2011;108:11399–11404. doi: 10.1073/pnas.1101046108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Michener JK, Smolke CD. High-throughput enzyme evolution in Saccharomyces cerevisiae using a synthetic RNA switch. Metab Eng. 2012;14:306–316. doi: 10.1016/j.ymben.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 44.Esvelt KM, Carlson JC, Liu DR. A system for the continuous directed evolution of biomolecules. Nature. 2011;472:499–503. doi: 10.1038/nature09929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Covert MW, Schilling CH, Famili I, Edwards JS, Goryanin II, Selkov E, Palsson BO. Metabolic modeling of microbial strains in silico. Trends Biochem Sci. 2001;26:179–186. doi: 10.1016/s0968-0004(00)01754-0. [DOI] [PubMed] [Google Scholar]
- 46.Lee JW, Kim TY, Jang YS, Choi S, Lee SY. Systems metabolic engineering for chemicals and materials. Trends Biotechnol. 2011;29:370–378. doi: 10.1016/j.tibtech.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 47.Mahadevan R, Burgard AP, Famili I, Van Dien S, Schilling CH. Applications of metabolic modeling to drive bioprocess development for the production of value-added chemicals. Biotechnol Bioproc E. 2005;10:408–417. [Google Scholar]
- 48.Michener JK, Nielsen J, Smolke CD. Identification and treatment of heme depletion attributed to overexpression of a lineage of evolved P450 monooxygenases. Proc Natl Acad Sci U S A. 2012;109:19504–19509. doi: 10.1073/pnas.1212287109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hong KK, Vongsangnak W, Vemuri GN, Nielsen J. Unravelling evolutionary strategies of yeast for improving galactose utilization through integrated systems level analysis. Proc Natl Acad Sci U S A. 2011;108:12179–12184. doi: 10.1073/pnas.1103219108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Segre D, Vitkup D, Church GM. Analysis of optimality in natural and perturbed metabolic networks. Proc Natl Acad Sci U S A. 2002;99:15112–15117. doi: 10.1073/pnas.232349399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Patil KR, Rocha I, Forster J, Nielsen J. Evolutionary programming as a platform for in silico metabolic engineering. BMC Bioinformatics. 2005;6:308. doi: 10.1186/1471-2105-6-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Forster J, Famili I, Fu P, Palsson BO, Nielsen J. Genome-scale reconstruction of the Saccharomyces cerevisiae metabolic network. Genome Res. 2003;13:244–253. doi: 10.1101/gr.234503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Asadollahi MA, Maury J, Patil KR, Schalk M, Clark A, Nielsen J. Enhancing sesquiterpene production in Saccharomyces cerevisiae through in silico driven metabolic engineering. Metab Eng. 2009;11:328–334. doi: 10.1016/j.ymben.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 54.Jung YK, Kim TY, Park SJ, Lee SY. Metabolic engineering of Escherichia coli for the production of polylactic acid and its copolymers. Biotechnol Bioeng. 2010;105:161–171. doi: 10.1002/bit.22548. [DOI] [PubMed] [Google Scholar]
- 55.Reed JL, Vo TD, Schilling CH, Palsson BO. An expanded genome-scale model of Escherichia coli K-12 (iJR904 GSM/GPR) Genome Biol. 2003;4:R54. doi: 10.1186/gb-2003-4-9-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Burgard AP, Pharkya P, Maranas CD. Optknock: a bilevel programming framework for identifying gene knockout strategies for microbial strain optimization. Biotechnol Bioeng. 2003;84:647–657. doi: 10.1002/bit.10803. [DOI] [PubMed] [Google Scholar]
- 57.Bennett MR, Pang WL, Ostroff NA, Baumgartner BL, Nayak S, Tsimring LS, Hasty J. Metabolic gene regulation in a dynamically changing environment. Nature. 2008;454:1119–1122. doi: 10.1038/nature07211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zaslaver A, Mayo AE, Rosenberg R, Bashkin P, Sberro H, Tsalyuk M, Surette MG, Alon U. Just-in-time transcription program in metabolic pathways. Nat Genet. 2004;36:486–491. doi: 10.1038/ng1348. [DOI] [PubMed] [Google Scholar]
- 59.Ang J, Bagh S, Ingalls BP, McMillen DR. Considerations for using integral feedback control to construct a perfectly adapting synthetic gene network. J Theor Biol. 2010;266:723–738. doi: 10.1016/j.jtbi.2010.07.034. [DOI] [PubMed] [Google Scholar]
- 60.Dunlop MJ, Keasling JD, Mukhopadhyay A. A model for improving microbial biofuel production using a synthetic feedback loop. Syst Synth Biol. 2010;4:95–104. doi: 10.1007/s11693-010-9052-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Michener JK, Thodey K, Liang JC, Smolke CD. Applications of genetically-encoded biosensors for the construction and control of biosynthetic pathways. Metab Eng. 2012;14:212–222. doi: 10.1016/j.ymben.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang F, Keasling J. Biosensors and their applications in microbial metabolic engineering. Trends Microbiol. 2011;19:323–329. doi: 10.1016/j.tim.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 63.Carothers JM, Goler JA, Juminaga D, Keasling JD. Model-driven engineering of RNA devices to quantitatively program gene expression. Science. 2011;334:1716–1719. doi: 10.1126/science.1212209. [DOI] [PubMed] [Google Scholar]
- 64.Scalcinati G, Knuf C, Partow S, Chen Y, Maury J, Schalk M, Daviet L, Nielsen J, Siewers V. Dynamic control of gene expression in Saccharomyces cerevisiae engineered for the production of plant sesquitepene alpha-santalene in a fed-batch mode. Metab Eng. 2012;14:91–103. doi: 10.1016/j.ymben.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 65.Tsao CY, Hooshangi S, Wu HC, Valdes JJ, Bentley WE. Autonomous induction of recombinant proteins by minimally rewiring native quorum sensing regulon of E. coli. Metab Eng. 2010;12:291–297. doi: 10.1016/j.ymben.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 66.Farmer WR, Liao JC. Improving lycopene production in Escherichia coli by engineering metabolic control. Nat Biotechnol. 2000;18:533–537. doi: 10.1038/75398. [DOI] [PubMed] [Google Scholar]
- 67.Zhang F, Carothers JM, Keasling JD. Design of a dynamic sensor-regulator system for production of chemicals and fuels derived from fatty acids. Nat Biotechnol. 2012;30:354–359. doi: 10.1038/nbt.2149. [DOI] [PubMed] [Google Scholar]
- 68.Moon TS, Dueber JE, Shiue E, Prather KL. Use of modular, synthetic scaffolds for improved production of glucaric acid in engineered E. coli. Metab Eng. 2010;12:298–305. doi: 10.1016/j.ymben.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 69.Wang YC, Yu O. Synthetic scaffolds increased resveratrol biosynthesis in engineered yeast cells. J Biotechnol. 2012;157:258–260. doi: 10.1016/j.jbiotec.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 70.Bonacci W, Teng PK, Afonso B, Niederholtmeyer H, Grob P, Silver PA, Savage DF. Modularity of a carbon-fixing protein organelle. Proc Natl Acad Sci U S A. 2012;109:478–483. doi: 10.1073/pnas.1108557109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Choudhary S, Quin MB, Sanders MA, Johnson ET, Schmidt-Dannert C. Engineered protein nano-compartments for targeted enzyme localization. PLoS One. 2012;7:e33342. doi: 10.1371/journal.pone.0033342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fan CG, Cheng SQ, Liu Y, Escobar CM, Crowley CS, Jefferson RE, Yeates TO, Bobik TA. Short N-terminal sequences package proteins into bacterial microcompartments. Proc Natl Acad Sci U S A. 2010;107:7509–7514. doi: 10.1073/pnas.0913199107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fan CG, Cheng SQ, Sinha S, Bobik TA. Interactions between the termini of lumen enzymes and shell proteins mediate enzyme encapsulation into bacterial microcompartments. Proc Natl Acad Sci U S A. 2012;109:14995–15000. doi: 10.1073/pnas.1207516109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gibson DG. Synthesis of DNA fragments in yeast by one-step assembly of overlapping oligonucleotides. Nucleic Acids Res. 2009;37:6984–6990. doi: 10.1093/nar/gkp687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Winkler J, Kao KC. Harnessing recombination to speed adaptive evolution in Escherichia coli. Metab Eng. 2012;14:487–495. doi: 10.1016/j.ymben.2012.07.004. [DOI] [PubMed] [Google Scholar]