SUMMARY
Chromosomal instability in early cancer stages is caused by stress on DNA replication. The molecular basis for replication perturbation in this context is currently unknown. We studied the replication dynamics in cells in which a regulator of S phase entry and cell proliferation, the Rb-E2F pathway, is aberrantly activated. Aberrant activation of this pathway by HPV-16 E6/E7 or cyclin E oncogenes significantly decreased the cellular nucleotide levels in the newly transformed cells. Exogenously supplied nucleosides rescued the replication stress and DNA damage and dramatically decreased oncogene-induced transformation. Increased transcription of nucleotide biosynthesis genes, mediated by expressing the transcription factor c-myc, increased the nucleotide pool and also rescued the replication-induced DNA damage. Our results suggest a model for early oncogenesis in which uncoordinated activation of factors regulating cell proliferation leads to insufficient nucleotides that fail to support normal replication and genome stability.
INTRODUCTION
Oncogenesis drives cell proliferation by interfering with various cellular pathways. One such pathway is the retinoblastoma (Rb) E2F pathway (Rb-E2F), an important regulator of cell proliferation. Rb is a key player in cell-cycle regulation, restricting cell proliferation by direct inhibition of the E2F family of transcription factors (van den Heuvel and Dyson, 2008). One oncogene deregulating the Rb-E2F pathway is the human papillomavirus (HPV) E7, which binds and degrades Rb. This degradation leads to aberrant activation of the E2F family of proteins, which promotes cell proliferation and DNA synthesis. HPV infection is the major cause of cervical cancer in women.
Another oncogene that aberrantly activates the Rb-E2F pathway is cyclin E, which is overexpressed in many cancers, including carcinomas, lymphomas, leukemias, and sarcomas (Akli and Keyomarsi, 2003). Cyclin E regulates S phase entry by Rb phosphorylation and inactivation, facilitating E2F release. Overexpression of cyclin E greatly accelerates S phase entry in cultured cells (Ohtsubo and Roberts, 1993).
Aberrant activation of the Rb-E2F pathway leads to the formation of DNA double-strand breaks (DSBs) and chromosomal instability (Frame et al., 2006; Pickering and Kowalik, 2006). This leads to senescence or apoptosis, providing a barrier to cancer progression. In order to overcome this barrier, the high-risk HPV expresses the E6 oncoprotein, which induces proteasomal degradation of p53, thus avoiding cell-cycle arrest or apoptosis. Hence, together, E6 and E7 enable proliferative activity of HPV-infected cells (Duensing and Münger, 2004). In cancers involving cyclin E overexpression, additional mutations abrogating the DNA damage response are required to overcome the apoptosis/senescence barrier (Gorgoulis et al., 2005).
Chromosomal instability is a hallmark feature of nearly all solid tumors and adult onset leukemias. This instability develops at early stages of cancer neoplasia and can be detected even in premalignant lesions (Duensing and Münger, 2003; Klausner, 2002). Different mechanisms have been suggested to explain this genome instability, including shortening of telomeres (Plug-DeMaggio et al., 2004), defective DNA damage repair (Duensing and Münger, 2002), oxidative stress (Bartkova et al., 2010), and chromosomal segregation errors (Duensing et al., 2000). These mechanisms contribute mainly to chromosomal changes, which appear in more advanced stages of cancer development (Gorgoulis et al., 2005). In early stages, the instability was found preferentially at fragile sites, genomic regions sensitive to replication perturbation (Gorgoulis et al., 2005). More recently, genomic instability in early cancer stages was shown to be caused by stress on the DNA replication, conferred by oncogene expression (Bartkova et al., 2006; Di Micco et al., 2006). However, the molecular events leading to the formation of replication stress, DSBs, and chromosomal instability shortly after oncogene expression are still unclear.
Here, we show that viral (HPV-16 E6/E7) or cellular (cyclin E) oncogenes, leading to abnormal activation of the Rb-E2F pathway, enforce cell proliferation with an insufficient pool of nucleotides to support normal DNA replication. This leads to replication perturbation, DNA damage, and genome instability. Importantly, an exogenous supply of nucleosides rescues the replication stress, decreases the replication-induced DNA damage, and reduces transformation of the cells expressing the oncogenes. We suggest that the low-nucleotide pool is a result of an unbalanced activation of nucleotide biosynthesis genes. We further show that activation of the cellular nucleotide biosynthesis pathways increases the nucleotide pool and rescues the replication-induced DNA damage. Altogether, our study reveals that nucleotide insufficiency plays an important role in the replication stress and genomic instability caused by aberrant activation of the Rb-E2F pathway.
RESULTS
As a first step in investigating the early events leading to DNA damage and genomic instability in cancer development, we studied the effect of aberrant activation of the Rb-E2F pathway by HPV-16 E6/E7 oncoproteins. For this, we used primary keratinocytes derived from adult skin biopsies, which have a very poor proliferation capacity ex vivo. The cells were infected with a LXSN-based retroviral vector, which did not affect cell proliferation or DNA replication (Figures S1A and S1B available online). Keratinocytes from the same donor were infected with the LXSN vector containing the HPV-16 E6 and E7 viral genes. We verified the expression of E6/E7 by RT-qPCR using primers for each of these viral genes. Using quantitative real-time PCR (RT-qPCR), we found that the expression level of E6 and E7 in these keratinocyte cells was ~2 times lower, compared to the level in a cell line originating from HPV16-induced cervical cancer (CaSki) (Figure S1C), showing that the E6/E7 expression in keratinocytes is at physiological levels. As expected, E6 and E7 transcripts were detected only in keratinocytes infected with the E6/E7 vector and not in the primary keratinocytes of the same donor (data not shown). Importantly, the E6/E7-expressing cells continued to proliferate at least 100 days, indicating their successful immortalization. These results show that E6/E7 genes were expressed and enforced continuous proliferation of the infected keratinocytes. In order to investigate early events induced by E6/E7 expression, the experiments were performed in newly transformed cells 2–6 weeks following E6/E7 infection and before anaphase bridges and micronuclei are visible.
HPV-16 E6/E7 Expression Generates Stress on the Cellular DNA Replication
In order to investigate the effect of HPV on the cellular DNA replication, we took advantage of the DNA combing approach, which enables replication analysis on single DNA molecules. The newly synthesized DNA labeled with IdU and CldU was detected by fluorescent antibodies (green and red, respectively) (Figure 1A). The replication dynamics were studied in normal primary keratinocytes obtained from adult skin samples of two individuals and in keratinocytes from the same donors expressing E6/E7 proteins for 2–6 weeks.
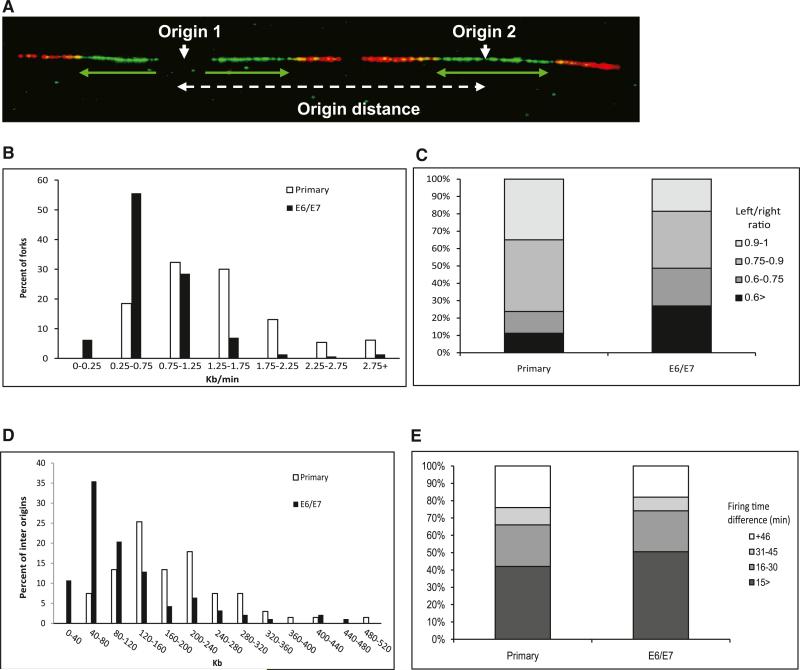
Figure 1. Replication Dynamics in Primary and HPV-16 E6/E7-Expressing Keratinocytes.
(A) Example of a single combed DNA molecule labeled with IdU (green) and CldU (red), showing replication from two adjacent origins. Green arrows, orientation and length of fork progression.
(B) Fork rate (Kb/min) distribution. White bars, primary keratinocytes (n = 130); black bars, keratinocytes expressing E6/E7 (n = 144).
(C) Progression ratio between left and right forks emerging from the same origin. Primary keratinocytes (n = 156); keratinocyte-expressing E6/E7 (n = 194).
(D) Origin density distribution. White bars, primary keratinocytes (n = 67); black bars, keratinocytes expressing E6/E7 (n = 93).
(E) Differences in firing time between adjacent origins. The groups represent different relative firing time in minutes. Primary keratinocytes (n = 50); keratinocytes expressing E6/E7 (n = 89).
See also Figure S1, Figure S2, Table S1, and Table S2.
First, we analyzed the effect of E6/E7 expression on the cellular DNA replication fork rate (Figure 1B). The results show a dramatic decrease in the mean replication rate, from 1.3 ± 0.06 Kb/min (n = 130) in human primary keratinocytes to 0.75 ± 0.05 Kb/min (n = 144) in cells expressing E6/E7 (p < 1.5 × 10–14). Importantly, a dramatic increase in the percentage of very slow forks was observed following E6/E7 expression (Figure 1B). Similar results were obtained in the analysis of keratinocytes from another donor (Figure S2A). These results indicate that, shortly following expression of the viral E6/E7 proteins, there is a significant decrease in DNA fork progression rate.
The two replication forks emerging from the same origin tend to exhibit the same replication rate (Conti et al., 2007). However, under replication stress conditions, perturbed fork progression might lead to asymmetric progression of the outgoing forks (Di Micco et al., 2006). Hence, we compared the progression of the left and right forks emerging from the same origin in primary and E6/E7-expressing keratinocytes from the same donor. As previously suggested (Conti et al., 2007), a symmetric progression of the two forks was considered when the ratio of left and right forks was > 0.75. The analysis revealed a significantly decreased symmetry between the right and left fork progression in the E6/E7-expressing cells compared to the primary cells (p < 3 × 10–6). In the primary replicating cells, the mean ratio was 0.81 ± 0.01 (n = 156), and only 23% of the replication bubbles showed asymmetric dynamics. In contrast, in the E6/E7-expressing cells, a poor symmetry was found with a mean ratio of 0.71 ± 0.01 (n = 194), and 47% of the origins showed asymmetric replication progression (Figure 1C and Figure S2C). This indicates that, a short time following E6/E7 expression, the processivity of the DNA replication machinery is perturbed.
Previous studies indicate that slow fork progression correlates with increased number of active origins (Anglana et al., 2003; Courbet et al., 2008; Ge et al., 2007). Hence, we studied the effect of E6/E7 proteins on the origin density by measuring the distance between two adjacent origins. The results showed that the mean origin distance in the primary keratinocytes was 187 ± 26 Kb (n = 67), whereas in the E6/E7-expressing keratinocytes, it was significantly shorter, only 121 ± 24 Kb (n = 93) (p < 5 × 10–5). Furthermore, a dramatic increase in the fraction of molecules with very short origin distance was observed following E6/E7 expression (Figure 1D). Similar results were obtained from other donors (Figure S2B and data not shown). These results indicate that E6/E7 expression leads to activation of an increased number of origins. Altogether, the replication dynamic results show that, in newly transformed E6/E7-expressing cells, the host cell DNA replication is dramatically perturbed.
Chronic HPV-16 E6/E7 Expression Causes a Change in the Programmed Activation of Dormant Origins
Dormant origins activation could occur in S phase to enable completion of DNA synthesis in perturbed replicons (Ge et al., 2007). In this case, a difference in the firing time of adjacent origins is expected. Alternatively, dormant origins could already be licensed in G1 as a cellular response to chronic replication stress (Courbet et al., 2008). In this case, a synchronic replication time between adjacent origins is expected.
In order to study the dynamics of origin firing as a result of the viral E6/E7 expression, we estimated the relative firing time of every two adjacent origins in the analyzed fibers (Figure S2D). In primary keratinocytes, the mean difference in firing time between two adjacent origins was 26 ± 3.1 min (n = 50). No significant difference (p > 0.95) in firing time was observed in primary keratinocytes expressing E6/E7 (mean of 26 ± 3.8 min [n = 89]) (Figure 1E). These results support the possibility of programmed activation of dormant origins in G1 prior to the replication initiation in S phase.
HPV-16 E6/E7 Expression Leads to Loss of Heterozygosity at Common Fragile Sites
Chromosomal instability can lead to loss of heterozygosity (LOH). Previous studies have demonstrated that, in early stages of cancer development, the instability occurs preferentially in genomic regions that are sensitive to replication stress, defined as fragile sites (Bartkova et al., 2005; Di Micco et al., 2006; Gorgoulis et al., 2005; Tsantoulis et al., 2008). Later in cancer development, the instability is found along the entire genome (Gorgoulis et al., 2005). LOH analysis was performed using single-nucleotide polymorphism (SNP) array. The heterozygosity in primary keratinocytes was compared to that in keratinocytes expressing E6/E7 for 100 or 250 days.
The results showed that, 100 days following expression of E6/E7, 1.4% of the informative SNPs in the entire genome revealed LOH, indicating that E6/E7 expression leads to genome instability. The instability is dramatically increased with time, and after 250 days, 17% of the informative SNPs showed LOH. In the fragile sites, the most replication-sensitive sites (FRA3B, FRA16D, and FRA7G) revealed 4.1% LOH after 100 days, which is significantly higher than found in the entire genome (p < 0.0024). After 250 days, even less-sensitive fragile sites showed significantly higher LOH frequency (p < 9 × 10–4) (Table S1), indicating a preferential instability along common fragile sites. In several fragile sites, a dramatic LOH of 31% (FRA11E) and 50% (FRA9G) was found. These results indicate that, in early stages of cancer development, the replication stress caused by E6/E7 expression leads to increased genomic instability at the most sensitive fragile sites, followed by instability at other fragile regions. As previously shown, in later stages of cancer development, a massive genomic instability is found in fragile and nonfragile regions in the genome (Gorgoulis et al., 2005).
Copy number variation (CNV) analysis of the same experiment revealed after 100 days a loss of chromosome 8p and a gain of the entire chromosome 20 in the E6/E7-expressing cells. Interestingly, chromosome 20q amplification is recurrently observed in cervical carcinoma cells (Tsantoulis et al., 2008). Analysis of cells expressing E6/E7 for 250 days revealed additional amplifications, which frequently appear in cervical cancer (Table S2), mainly in chromosomes 8q, 7p, and 11q (Harris et al., 2003). Overall, the LOH and CNV results indicate that similar instability events are found in ex vivo and in vivo cell proliferation and transformation processes.
A Low-Nucleotide Pool in Cells Expressing HPV-16 E6/E7 Leads to Replication Stress, Genome Instability, and Tumorigenicity
The replication dynamics is largely determined by the size and balance of the nucleotide pool (Anglana et al., 2003; Courbet et al., 2008). Hence, we studied the effect of E6/E7 on the nucleotide pool using the high-performance liquid chromatography (HPLC) method. Our results show a 2- to 5-fold decrease in the level of the four dNTPs following E6/E7 expression for 2–4 weeks (Figure 2A and Table S3A). Interestingly, a 2-fold decrease was also found in the level of all ribonucleotides (rNTPs) (Figure S3A and Table S3B), indicating that the nucleotide biosynthesis is disturbed upstream in the pathway. These results suggest that the replication perturbation and genomic instability found in the E6/E7-expressing cells result from an insufficient nucleotide pool required to support extensive proliferation.
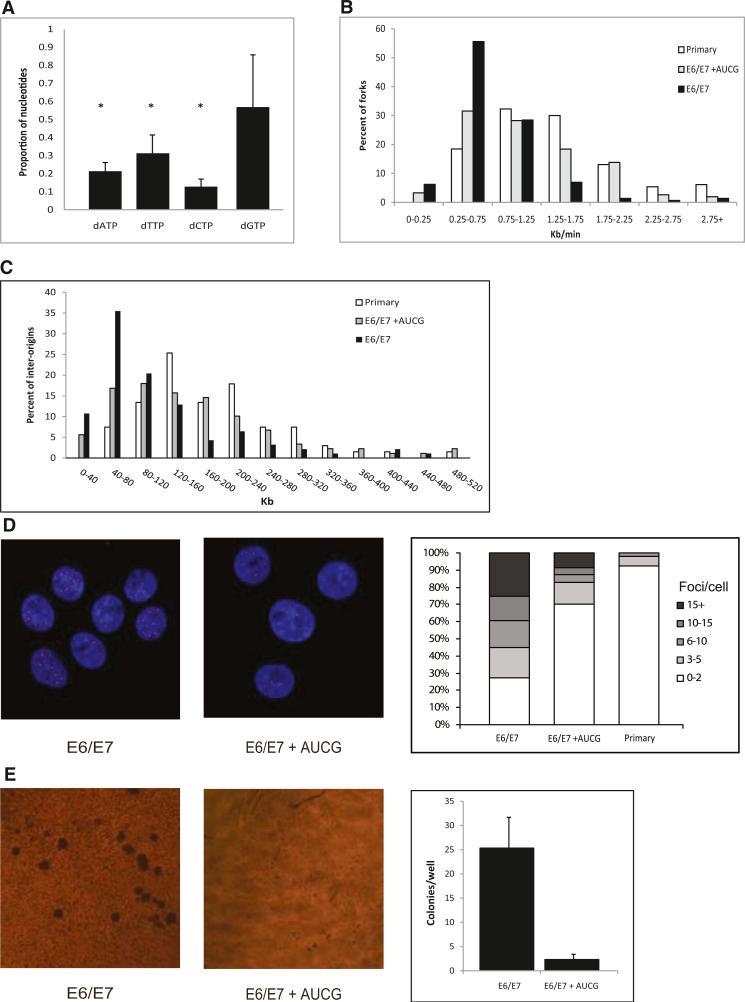
Figure 2. The Effect of an Exogenous Supply of Nucleosides on the Replication Dynamics, DNA Damage, and Transformation in HPV-16 E6/E7-Expressing Cells.
(A) The nucleotide levels in keratinocyte cells expressing E6/E7 normalized to the level in primary cells of the same donor. *p < 0.05. The levels are expressed as mean fold change ± SEM (n = 3).
(B) Fork rate (Kb/min) distribution. White bars, primary keratinocytes (n = 130); black bars, keratinocytes expressing E6/E7 (n = 150); gray bars, keratinocytes expressing E6/E7 grown for 48 hr with exogenous supply of nucleosides (n = 155).
(C) Origin density (Kb) distribution. White bars, primary keratinocytes (n = 67); black bars, keratinocytes expressing E6/E7 (n = 93); gray bars, keratinocytes expressing E6/E7 grown for 48 hr with exogenous supply of nucleosides (n = 89).
(D) (Left) Examples of nuclei with γH2AX foci following expression of E6/E7 grown in a normal medium (E6/E7) and in a medium supplied with exogenous nucleosides (E6/E7+AUCG). The nuclei were stained with DAPI. (Right) Percent of nuclei with the indicated number of γH2AX foci. Primary keratinocyte cells (n = 51); keratinocytes expressing E6/E7 (n = 52); keratinocytes expressing E6/E7 supplied with exogenous nucleosides (n = 70).
(E) (Left) Examples of anchorage-independent growth in soft agar. (Right) Mean number of colonies per a soft agar plate. Cells expressing E6/E7; cells expressing E6/E7 supplied with exogenous nucleosides. The number of colonies per plate is expressed as mean ± SEM (n = 3).
See also Figure S3 and Figure S4, and Table S3.
In order to test this hypothesis, we analyzed whether exogenous supply of nucleosides can modulate the replication dynamics in E6/E7-expressing cells. For this, primary keratinocytes and keratinocytes expressing E6/E7 for 2–4 weeks were grown for 48 hr in a medium supplemented with the four nucleosides (A, U, C, and G). Evaluating the replication dynamics using DNA combing revealed that exogenous supply of nucleosides to keratinocytes expressing E6/E7 led to a dramatic increase in the replication rate to an average of 1.1 ± 0.06 Kb/min (n = 155), as compared to 0.75 ± 0.05 Kb/min (n = 150) (p = 8 × 10–7) in the same cells grown in a normal medium (Figure 2B). Moreover, the average replication rate after supply of exogenous nucleosides reached 90% of the mean fork rate in primary keratinocytes. Similar results were obtained using cells from another donor (Figure S3B). It is important to note that exogenous supply of nucleosides had no effect on the replication rate and interorigin distances of normal primary keratinocyte cells (1.2 ± 0.04 Kb/min [n = 83]; 129 ± 11 Kb [n = 42]) (Figures S3D and S3E).
Subsequently, we studied the effect of exogenous nucleosides supply on the interorigin distances. In accordance with the replication rate, E6/E7-expressing cells supplied with nucleosides showed a dramatic increase in the average interorigin distance compared to that of E6/E7 cells grown in a normal medium, 165 ± 12 Kb (n = 89) and 120 ± 10 Kb (n = 93) (p = 8 × 10–7), respectively. Whereas E6/E7-expressing keratinocytes exhibited in 45% of the cases an origin distance < 80 Kb, after supply of exogenous nucleosides, the fraction of these distances was only 22% (Figure 2C). Similar results were obtained using cells from another donor (Figure S3C).
Our LOH results revealed that replication-induced DSBs are generated following E6/E7 expression. Hence, we further investigated whether exogenous supply of nucleosides can decrease these replication-induced DSBs. For this, we analyzed the formation of γH2AX foci, known to localize at DSBs (Rogakou et al., 1998). Immunofluorescence analysis using antibodies directed against γH2AX was performed in cells expressing E6/E7 for 2–6 weeks, grown in a normal medium and in a medium supplemented with exogenous nucleosides. The analysis showed a significant decrease in the number of γH2AX foci following exogenous nucleoside supply compared to cells grown in a normal growth medium, 4 ± 1 (n = 70) and 9 ± 1.1 (n = 52) foci per nucleus, respectively (Figure 2D) (p < 7 × 10–4). Importantly, a high level of γH2AX foci (>10 foci per nuclei) was observed in 40% of the nuclei from cells expressing E6/E7 grown in a normal keratinocyte medium, whereas such a high focus level was found in only 13% of the cells supplemented with nucleosides for 48 hr. Furthermore, 70% of supplemented nuclei revealed a very low focus level (0–2), similar to that found in > 90% of primary keratinocytes.
Keratinocyte cells expressing the full-length HPV-16 genome (F1 cells) for 6 weeks revealed replication stress and DNA damage similar to that observed in cells expressing only HPV-16 E6/E7 (Figures S4A–S4C). Importantly, both the replication stress and DNA damage in these cells could be rescued by exogenous supply of nucleosides (Figures S4A–S4C), indicating that the replication-induced DNA damage indeed results from the expression of the HPV-16 E6/E7 oncogenes and is not affected by expression of other viral proteins.
We then studied whether the nucleotide pool can modulate oncogene-induced cell transformation. We tested this using the common in vitro transformation assay that measures anchorage-independent growth in soft agar. The cells grown in the normal medium showed a significant (p = 0.02) anchorage-independent growth in soft agar, whereas cells grown in the nucleoside-enriched medium formed rare and small colonies: 25.3 ± 6 and 2.3 ± 1 colonies per plate, respectively (n = 3) (Figure 2E). It is important to note that exogenous supply of nucleosides to E6/E7-expressing cells did not affect cell proliferation (Figure S4D), indicating that the decrease in soft agar colony formation, following exogenous nucleoside supply, is not due to a decrease in the rate of cell proliferation.
Altogether, our results show that, following E6/E7 expression, a low-nucleotide pool causes replication perturbation, leading to genomic instability and increased tumorigenic potential.
A Low-Nucleotide Pool in Cells Expressing Cyclin E Leads to Replication Stress and Genome Instability
We further investigated whether aberrant activation of the Rb-E2F pathway by cellular oncogenes also leads to an insufficient nucleotide pool, required for normal DNA replication. For this, we expressed the human cyclin E, which is frequently overex-pressed in human precancerous and cancerous lesions. Cyclin E has a major role in the control of G1/S transition as it mediates Rb phosphorylation, which inactivates Rb and facilitates E2F release. Like the viral E7 oncogene, cyclin E activity leads to G1 shortening and forces cells to enter S phase (Hwang and Clurman, 2005).
Using retroviral infection, we expressed the human cyclin E in human primary fibroblasts from healthy donors or immortalized foreskin fibroblasts (BJ cells). The expression of cyclin E was verified by RT-qPCR and western blot (Figure S5A). First, we measured the nucleotide levels in BJ cells expressing cyclin E or an empty control vector for 2–4 weeks, before senescence prevents cell divisions in the tissue cultured cells. The results revealed a 50% decrease (p = 0.03) in the rNTP pool following cyclin E expression (Figure 3A and Table S3D). Importantly, the dNTP levels decreased dramatically to a level that is below detection (Table S3C). A similar decrease was found in primary fibroblast cells (Table S3E and data not shown).
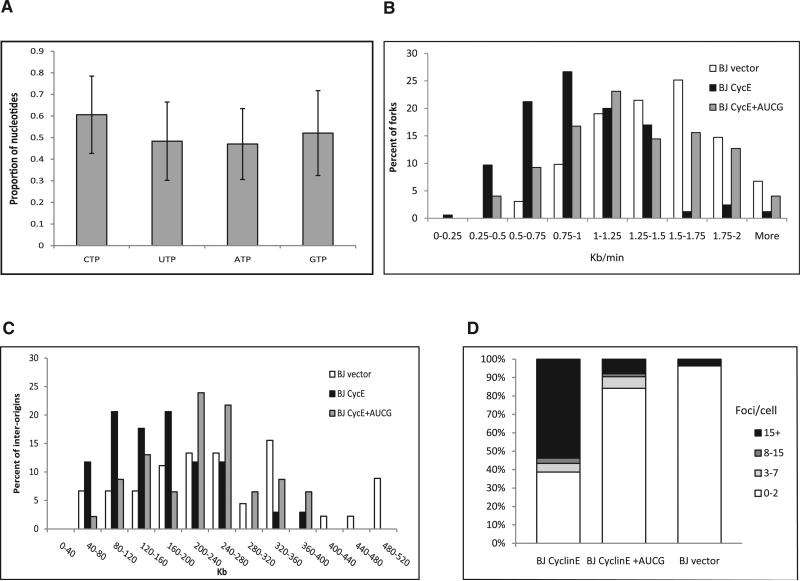
Figure 3. The Effect of an Exogenous Supply of Nucleosides on the Replication Dynamics and DNA Damage of BJ Cells Expressing Cyclin E.
(A) rNTP pool following cyclin E expression in BJ cells. The levels are expressed as mean fold change ± SEM (n = 4).
(B) Fork rate distribution (Kb/min). White bars, BJ cells (n = 163); black bars, BJ expressing cyclin E (n = 165); gray bars, BJ expressing cyclin E grown for 48 hr with exogenous supply of nucleosides (n = 173).
(C) Origin density distribution (Kb). White bars, BJ cells (n = 45); black bars, BJ expressing cyclin E (n = 34); gray bars, BJ expressing cyclin E grown for 48 hr with exogenous supply of nucleosides (n = 46).
(D) Percent of nuclei with the indicated number of γH2AX foci. BJ cells (n = 57); BJ expressing cyclin E (n = 110); BJ expressing cyclin E grown for 48 hr with exogenous supply of nucleosides (n = 64).
Next, we analyzed the effect of exogenously supplied nucleosides on the replication dynamics of cells expressing cyclin E. The analysis revealed a significant perturbation in the replication dynamics following cyclin E expression. The mean fork rate in BJ cells expressing the empty vector was 1.5 ± 0.03 Kb/min (n = 163), whereas following cyclin E expression, the rate was significantly slower (1.0 ± 0.03 Kb/min, n = 165) (p < 3 × 10–28). The fraction of very slow forks (<0.75 Kb/min) was 10 times higher (Figure 3B). Exogenous supply of the four nucleosides for 48 hr increased fork rate to 1.3 ± 0.03 Kb/min (n = 173), which constitutes an 87% recovery (p = 6 × 10–6). Similar results were found in primary fibroblasts from an adult donor expressing the cyclin E oncogene. Fork progression rate decreased from 1.2 ± 0.06 Kb/min (n = 125) in the primary cells to 0.8 ± 0.06 Kb/min (n = 62) (p < 2 × 10–7). Exogenous nucleoside supply increased the fork rate to 1.1 ± 0.05 Kb/min (n = 80), a 92% recovery (p = 0.0002) (Figure S5B).
Subsequently, we analyzed the interorigin distance. The analysis in BJ cells revealed a significant decrease from 289 ± 25 Kb (n = 45) to 168 ± 14 Kb (n = 34) following cyclin E expression (p < 2.5 × 10–5). Importantly, exogenous nucleoside supply increased the distance to 237 ± 14 Kb (n = 46) (p < 4 × 10–4) (Figure 3C). Similar results were found in primary fibroblasts. The interorigin distance decreased from 172 ± 12 Kb (n = 47) to 127 ± 15 Kb (n = 27) (p = 0.01) following cyclin E expression, and exogenous nucleoside supply increased the interorigin distance to 159 ± 11 (n = 42) (Figure S5C). These results indicate that cyclin E overexpression leads to replication stress that can be rescued by supply of exogenous nucleosides.
Next, we studied the effect of exogenous nucleoside supply on the DSBs induced by cyclin E overexpression. The analysis revealed a significant increase in the level of DSBs following cyclin E expression, as measured by γH2AX foci: 7.7 ± 1.5 foci/cell (n = 35) to 18.5 ± 1.9 foci/cell (n = 39) in fibroblasts expressing cyclin E (p = 5 × 10–5) (Figure S5D) and 1.2 ± 1.5 foci/cell (n = 57) to 27 ± 2.5 (n = 110) foci/cell in BJ cells expressing cyclin E (p = 1 × 10–15) (Figure 3D). As found in E6/E7-expressing cells, exogenous nucleoside supply decreased the extent of DNA damage in the cyclin E expressing cells to 3.5 ± 2.1 foci/cell in BJ cells (n = 64, p = 8 × 10–13) and 3.3 ± 0.8 foci/cell in primary fibroblasts (n = 44) (p = 5 × 10–9) (Figure 3D and Figure S5D). Altogether, these results indicate that activation of the Rb-E2F pathway by cellular or viral oncogenes results in an insufficient nucleotide pool, leading to replication stress and DNA damage. Importantly, this replication-induced DNA damage can be rescued by exogenous supply of nucleosides.
Activation of the Nucleotide Biosynthesis Pathways Rescues the Replication Stress and Genome Instability
We aimed to further understand the molecular basis for the low-nucleotide pool in cells enforced to proliferate by oncogene expression. Cell proliferation depends on coordinated activation of the different nucleotide metabolic genes (Liu et al., 2008; Mannava et al., 2008), which are tightly regulated by the transcription factors and master regulators of cell proliferation. Hence, we hypothesized that the low-nucleotide pool in oncogene-expressing cells results from insufficient activation of the nucleotide biosynthesis pathways. In order to test this hypothesis, we performed unbiased whole-transcriptome analysis of BJ cells in comparison to BJ cells expressing cyclin E. The analysis revealed that the most significantly upregulated pathways pertain to cell cycle (p = 1.52 × 10–15) and DNA replication (p = 3.2 × 10–12) (Table 1). Importantly, cyclin E expression failed to upregulate the nucleotide biosynthesis pathways. The expression levels of eight important genes in the purine and pyrimidine biosynthesis pathways—PPAT, DHODH, ADSL, CTPS, NME1, IMPDH2, TS, and RRM2—are shown in Figure S6A. We further verified these results using RT-qPCR in both BJ and keratinocyte cells compared to cells expressing cyclin E or E6/E7, respectively. The results revealed that, following oncogene expression, the level of most genes was not altered (six and seven genes, respectively) (Figure 4A, black bars, and Figure S6B). These results suggest that Rb-E2F aberrant activation enforces cell proliferation but fails to activate the nucleotide biosynthesis pathway, leading to insufficient pool of nucleotides required for normal replication.
Table 1.
The Significant Upregulated Pathways following the Expression of Cyclin E, c-Myc, or Coexpression of Both Genes in BJ Cells
| Term | P Value | Fold Enrichment | FDR | |
|---|---|---|---|---|
| Cyclin E | Cell cycle | 1.42 × 10–18 | 8.693907512 | 1.52 × 10–15 |
| DNA replication | 2.98 × 10–15 | 16.8358209 | 3.20 × 10–12 | |
| Oocyte meiosis | 1.32 × 10–8 | 5.962686567 | 1.4 × 10–5 | |
| Systemic lupus erythematosus | 5.45 × 10–6 | 5.183660644 | 0.00580449 | |
| Progesterone-mediated oocyte maturation | 1.08 × 10–5 | 5.34784899 | 0.011501479 | |
| c-Myc | DNA replication | 7.02 × 10–15 | 10.63095238 | 8.34 × 10–12 |
| Cell cycle | 4.50 × 10–13 | 4.953161593 | 5.36 × 10–10 | |
| Pyrimidine metabolism | 6.07 × 10–10 | 4.876691729 | 7.24 × 10–7 | |
| Purine metabolism | 4.18 × 10–9 | 3.68627451 | 4.98 × 10–6 | |
| Oocyte meiosis | 4.30 × 10–6 | 3.543650794 | 0.005126052 | |
| Progesterone-mediated oocyte maturation | 1.36 × 10–5 | 3.791596639 | 0.016195283 | |
| Cyclin E c-Myc | DNA replication | 2.12 × 10–17 | 12.87671233 | 2.45 × 10–14 |
| Cell cycle | 2.89 × 10–11 | 4.939591287 | 3.34 × 10–8 | |
| Pyrimidine metabolism | 3.78 × 10–11 | 5.611535689 | 4.37 × 10–8 | |
| Purine metabolism | 8.97 × 10–10 | 4.090249799 | 1.04 × 10–6 | |
| Oocyte meiosis | 1.17 × 10–5 | 3.648401826 | 0.013464979 | |
| Nucleotide excision repair | 1.19 × 10–5 | 5.794520548 | 0.01377895 | |
| Mismatch repair | 3.33 × 10–5 | 8.061941632 | 0.038459564 |
Pathways discussed in the text are set in italics.
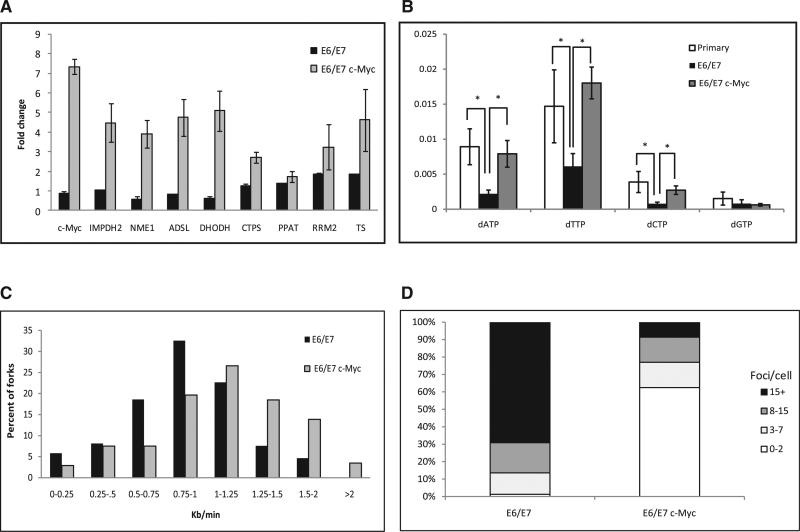
Figure 4. The Effect of c-myc Expression on the Expression of the Nucleotide Biosynthesis Genes, Nucleotide Pools, Replication Dynamics, and DNA Damage.
(A) Fold change in the expression level of c-Myc targets from the nucleotide biosynthesis pathways. Black bars, expression of E6/E7 normalized to the level in primary keratinocytes; gray bars, expression of c-myc in E6/E7-expressing keratinocytes normalized to the level in cells expressing only E6/E7. The levels are expressed as mean fold change ± SEM (n = 3).
(B) Nucleotide levels in primary keratinocytes, keratinocytes expressing E6/E7, and keratinocytes coexpressing E6/E7 and c-myc. *p < 0.05. The levels are expressed as mean fold change ± SEM (n > 3).
(C) Fork rate distribution (Kb/min). Black bars, keratinocytes expressing E6/E7 (n = 173); gray bars, keratinocytes expressing E6/E7 with enhanced nucleotide biosynthesis by c-Myc (n = 175).
(D) Percent of nuclei with the indicated number of γH2AX foci in cells expressing E6/E7 (n = 80) and in keratinocytes expressing E6/E7 with enhanced nucleotide biosynthesis by c-Myc (n = 103).
See also Figure S6.
Nucleotide biosynthesis was recently found to be regulated by c-Myc, a master regulator of many genes that are involved in DNA replication and cell proliferation (Liu et al., 2008; Mannava et al., 2008). However, the expression of c-myc is not altered following E6/E7 or cyclin E expression (Figure 4A, black bars, and Figure S6B) (Gross-Mesilaty et al., 1998). Whole-transcriptome analysis of BJ cells expressing retroviral vector containing the human c-myc cDNA for 3 weeks revealed, as expected, that the most significantly upregulated pathways are DNA replication (p = 8.34 × 10–12), cell cycle (p = 5.36 × 10–10), pyrimidine (p = 7.24 × 10–7), and purine (p = 4.98 × 10–6) metabolism (Table 1 and Figure S6A). It is worth noting that primary keratinocytes expressing c-myc are unable to proliferate; hence, the whole-transcriptome analysis could not be performed in these cells.
It is important to note that Rb-E2F aberrant activation fails to activate many important genes from the nucleotide biosynthesis pathways, whereas c-myc significantly upregulates the purine and pyrimidine metabolism pathways. Hence, it was interesting to study the effect of upregulation of nucleotide biosynthesis pathways by increased c-myc expression in aberrantly activated Rb-E2F cells. For this, we expressed c-myc in BJ and keratinocyte cells expressing cyclin E or E6/E7 for 2–4 weeks. Whole-transcriptome analysis in the BJ cells coexpressing cyclin E and c-myc revealed that the most significantly activated pathways are DNA replication (p = 2.45 × 10–14), cell cycle (p = 3.34 × 10–8), pyrimidine (p = 4.37 × 10–8), and purine metabolism (p = 1.04 × 10–6) (Table 1 and Figure S6A). These results indicate that c-myc expression upregulates the nucleotide biosynthesis in cells forced to proliferate by aberrant activation of the Rb-E2F pathway. We verified these results using RT-qPCR analysis in keratinocyte cells coexpressing E6/E7 and c-myc. The analysis revealed a 2- to 5-fold increase in the transcript levels of important nucleotide biosynthesis c-Myc targets (IMPDH2, NME1, ADSL, DHODH, CTPS, PPAT, RRM2, and TS), compared to levels in cells from the same donor expressing only E6/E7 for the same time period (Figure 4A, gray bars). We further tested whether this activation of biosynthesis genes can modulate the nucleotide levels in the E6/E7-expressing keratinocytes. Indeed, HPLC analysis revealed ~3.5-fold increase in the levels of dATP, dTTP, and dCTP in cells expressing E6/E7 upon activation of the nucleotide biosynthesis pathways (Figure 4B and Table S3).
We then analyzed the replication dynamics in these cells. The results showed a significant increase in the replication rate, from 0.86 ± 0.06 Kb/min (n = 173) in the E6/E7-expressing cells to 1.14 ± 0.05 Kb/min (n = 175) in the E6/E7- and c-myc-expressing cells (p < 4 × 10–8) (Figure 4C). Activation of the nucleotide biosynthesis in BJ cells overexpressing cyclin E also led to a higher fork progression rate (from 0.84 ± 0.04 Kb/min [n = 72] to 0.97 ± 0.03 Kb/min [n = 104] [p = 0.005]) (Figure S6C).
We further hypothesized that the increased levels of the nucleotides and the rescue of the replication stress would lead to a decrease in the DSBs in these cells. To test this, we measured γH2AX foci formation. The results show a highly significant (p < 5 × 10–24) decrease in γH2AX foci formation in keratinocytes expressing E6/E7, in which nucleotide biosynthesis was upregulated by c-Myc (4.4 ± 0.6 foci per cell, n = 103) compared to cells expressing only the E6 and E7 genes (19.6 ± 1 foci per cell, n = 80) (p < 5 × 10–24) (Figure 4D). BJ cells expressing cyclin E revealed similar results. Upregulation of the nucleotide biosynthesis led to a decrease in γH2AX foci (1.3 ± 0.3 foci per cell, n = 48) as compared to the control cells expressing cyclin E (3.0 ± 0.5 foci per cell, n = 64) (p < 0.009) (Figure S6D). It is important to note that c-Myc activates a large number of genes; hence, additional effects of c-Myc cannot be excluded.
Another mechanism that may lead to a low-nucleotide pool in cells expressing E6/E7 oncoproteins is E6-mediated p53 inactivation, which inhibits the expression of the ribonucleotide reductase M2 B (RRM2B) gene. RRM2B is required for deoxynucleotide biosynthesis during DNA repair (Kimura et al., 2003; Tanaka et al., 2000). In order to test this possibility, we infected keratinocyte cells expressing E6/E7 for 3 weeks with a retroviral vector containing the human RRM2B cDNA. First, we verified the RRM2B expression using RT-qPCR (Figure S6E). We then analyzed the effect of RRM2B expression on the replication dynamics. Our results show no effect of RRM2B expression on the replication stress caused by the E6/E7 expression. In primary keratinocytes, the mean replication rate was 1.2 ± 0.04 Kb/min (n = 169), and the mean interorigin distance was 132 ± 8 Kb (n = 67). Following ~3 weeks of E6/E7 expression in cells from the same donor, the replication rate was 0.54 ± 0.2 Kb/min (n = 161), and the interorigin distance was 69 ± 4 Kb (n = 94). In cells expressing E6/E7 and RRM2B, the mean rate was 0.59 ± 0.04 Kb/min (n = 60), and the mean interorigin distance was 79 ± 7Kb (n = 32). These results indicate that the RRM2B expression did not rescue the replication stress conferred by the expression of the E6/E7 oncogenes (Figures S6F and S6G). Aberrant activation of the Rb-E2F pathway by cyclin E does not lead to p53 degradation; hence, it seems unlikely that RRM2B plays an important role in the mechanism leading to a low-nucleotide pool in these cells.
In summary, our results show that activation of the Rb-E2F pathway fails to activate many of the nucleotide biosynthesis genes. Upregulation of the nucleotide biosynthesis pathways increases the nucleotide pool, rescues the replication stress, and decreases the DNA damage.
DISCUSSION
Here, we show that the basis for genome instability induced by oncogenes activating the Rb-E2F pathway is uncoordinated S phase entry, leading to insufficient factors required for normal DNA replication. We revealed that cells are forced to proliferate with an insufficient pool of nucleotides to support normal DNA replication. Under these conditions, the replication machinery fails to achieve regular rate and processivity, resulting in DNA damage and genome instability.
A great effort has been made in the last decade to understand the mechanisms leading to genomic instability in cancer development. Different models have suggested that this instability is caused by shortening of telomeres (Plug-DeMaggio et al., 2004), hypoxia (Coquelle et al., 1998), defective DNA damage repair (Duensing and Münger, 2002), chromosomal segregation errors (Duensing et al., 2000), abrogated mitotic checkpoints (Thompson et al., 1997), and reactive oxygen species (Klaunig and Kamendulis, 2004). More recently, oncogene-induced DNA replication stress was suggested as a model (Halazonetis et al., 2008). According to this model, the mechanism by which oncogenes induce genomic instability in early stages of cancer development is by causing replication stress leading to DNA double-strand breaks. However, the molecular basis for this oncogene-enforced replication stress remained largely unknown.
In our study, we investigated the mechanisms leading to DNA damage in cells expressing the HPV-16 viral oncogenes E6/E7 or the cellular oncogene cyclin E, both activating the Rb-E2F pathway, a master regulator of cell proliferation and DNA replication. Our results show that abnormal activation of the Rb-E2F pathway by viral or cellular oncogenes leads to perturbed progression of the replication forks and a poor symmetry between forks emerging from the same origin (Figures 1B and 1C, Figure 3B, Figure S2A, and Figure S5B). This indicates a poor processivity of the replication machinery, which leads to replication fork collapse and DSBs (Figure 2D, Figure 3D, and Figure S5D). The LOH analysis provides further evidence for perturbed replication by showing preferential instability at common fragile sites, known to be sensitive to replication stress conditions (Tsantoulis et al., 2008).
Previous studies showed the importance of the nucleotide pool for normal DNA replication (Anglana et al., 2003; Courbet et al., 2008). Hydroxyurea, an inhibitor of ribonucleotide reductase, lowers the dNTP pool and leads to slow DNA replication rate (Anglana et al., 2003), short interorigin distance (Ge et al., 2007), formation of DSBs (Saintigny et al., 2001), and expression of fragile sites (Yan et al., 1987). Our results show that the nucleotide pool is reduced dramatically following expression of the Rb-E2F-activating oncogenes (Figure 2A, Figure 3A, and Table S3). Furthermore, an increase of the nucleotide pool rescued the perturbed replication and the replication-induced DNA damage in the oncogene-expressing cells (Figure 2, Figure 3, Figures S3B and S3C, Figures S4A–S4C, and Figures S5B–S5D). Importantly, exogenous nucleoside supply decreased the anchorage-independent growth of the cells in soft agar (Figure 2E), suggesting reduced tumorigenic potential. Altogether, our study indicates that induction of nucleotide synthesis by oncogene-induced Rb-E2F expression is insufficient to support the extensive proliferation of the cells, leading to replication-induced DNA damage and increased tumorigenicity.
As previously suggested, the nucleotide pool size may modulate the replication in two distinct ways. It can activate dormant origins during S phase (Ge et al., 2007), leading to asynchronous origin firing. Alternatively, a poor nucleotide pool can determine origin choice in G1, when origins are committed to become active during S phase. Recently, Courbet et al. (2008) studied the replication dynamics of cells exposed to aphidicolin, an inhibitor of DNA polymerase α and δ that affects the replication rate in S phase. Using DNA combing, they revealed an asynchronous firing of adjacent origins, as expected by the first model (Ge et al., 2007). Here, we demonstrate a modulation of origin firing in pre-S phase. We suggest that, under a chronic exposure to a low-nucleotide pool, the cells compensate for the DNA replication stress by determining an increased origin density during G1, rather than by activating the dormant origins during S phase (Figure 1E).
Oncogenic activation of the Rb-E2F pathway by cyclin E or HPV-16 E6/E7 causes upregulation of many genes involved in cell proliferation and DNA replication, among which are many E2F targets (Garner-Hamrick et al., 2004; Hwang and Clurman, 2005). Our results (Figure 4A, Figures S6A and S6B, and Table 1) suggest that the low-nucleotide pool in the cells results from uncoordinated activation of the nucleotide biosynthesis pathways and cell cycle. Recently, it was shown that c-Myc is an important regulator of nucleotide biosynthesis and that its expression increases the nucleotide levels and cell proliferation (Liu et al., 2008; Mannava et al., 2008). Indeed, coexpression of cyclin E and c-myc revealed that the most significantly upregulated pathways are cell cycle and DNA replication, as well as purine and pyrimidine metabolism (Table 1, Figure 4A, and Figures S6A and S6B). This leads to an increased nucleotide pool, resulting in a rescue of the replication stress and DNA damage in these cells (Figure 4B and Table S3).
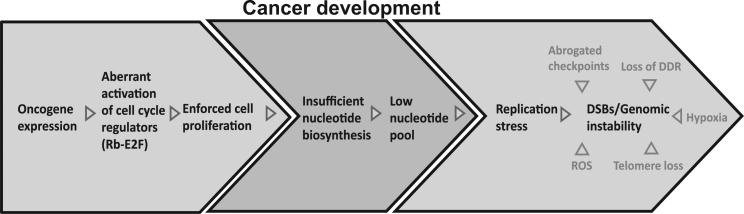
Altogether, we suggest that activation of the Rb-E2F pathway by cellular or viral oncogenes leads, in early stages of cancer development, to a change in the transcription of many genes involved in cell-cycle control, forcing cell proliferation and DNA replication. This leads to an uncoordinated entry to S phase, with aberrant activation of the nucleotide biosynthesis pathways. As a result, DNA replication starts with a low-nucleotide pool, leading to replication stress and DNA damage. Our results indicate that elevating the cellular nucleotide pool can rescue this oncogene-induced replication stress, reduce DNA damage, and decrease tumorigenicity of cells overexpressing these oncogenes. In more advanced stages of cancer development, additional mechanisms likely lead to chromosomal instability, such as telomere shortening, defective DNA damage repair, chromosomal segregation errors, and oxidative stress (Figure 5).
Figure 5. A Model for the Events Leading to Genomic Instability in Early Stages of Cancer Development.
Oncogene expression forces cell proliferation by aberrant activation of cell-cycle regulators (Rb-E2F). Insufficient activation of the nucleotide biosynthesis pathways results in a low-nucleotide pool that fails to support normal DNA replication. This leads to replication stress and promotes genomic instability during early stages of cancer development. Additional factors contribute to genomic instability in different stages of tumorigenesis, such as reactive oxygen species (ROS), telomere loss, hypoxia, abrogated mitotic checkpoints, and loss of the DNA damage response (DDR).
Disease progression in cancer is complex and multifactorial and depends on acquiring adaptive mutations. In many cancers, c-myc is highly expressed at a later stage in development (Grandori et al., 2000). This leads to enhanced proliferation and reduced DNA damage (Gordan et al., 2008). Apparently, high expression of c-myc in advanced stages of cancer development enables cells to proliferate more efficiently and increases the tumorigenicity of the cells (Narisawa-Saito et al., 2008). Whereas in many types of cancer, genomic instability may initiate with uncoordinated cellular pathways leading to insufficient nucleotides, in other cancers such as those with high expression of c-myc at early stages, chromosomal instability may be caused by other mechanisms.
The nucleotide pool is also affected by environmental factors such as folate, which is an essential vitamin involved in the metabolism of many cellular components, including the synthesis of dTTP. Hence, proper levels of folate in the diet are required for normal DNA replication (Rampersaud et al., 2002). In recent years, many studies showed a correlation between folate deficiency and the development of several types of cancer, including HPV-induced cervical carcinoma (García-Closas et al., 2005; Rampersaud et al., 2002). Our replication dynamics analysis showed that the expression of the HPV oncogenes and cyclin E leads to insufficient nucleotide pool. Together with a low folate intake, the replication is expected to be highly perturbed. This may lead to an increased risk for carcinogenesis due to a high rate of DSB formation and genomic instability.
In summary, replication stress and the resulting genomic instability are prominent driving forces of early stages of cancer development. Our results shed new light on the molecular basis of oncogene-induced replication stress, showing that oncogene-induced cell proliferation may fail to achieve a sufficient nucleotide pool to support normal DNA replication. Our results suggest a new model of uncoordinated regulation of essential pathways required for the production of a balanced level of cell proliferation factors. This raises the possibility for the development of new approaches for protection against precancerous development.
EXPERIMENTAL PROCEDURES
Molecular Combing
Unsynchronized cells were pulse labeled for 30 min by medium containing 100 μM of the thymidine analog iododeoxyuridine (IdU). At the end of the first labeling period, the cells were washed twice with a warm medium and pulse labeled once more for 30 min with medium containing 100 μM of another thymidine analog chlorodeoxyuridine (CldU). Cells were then harvested, and genomic DNA was extracted, combed, and analyzed as previously described (Herrick and Bensimon, 1999; Lebofsky et al., 2006). The primary antibody for fluorescence detection of IdU was mouse anti-BrdU (Becton Dickinson), and the secondary antibody was goat anti-mouse Alexa Fluor 488 (Invitrogen). The primary antibody for fluorescence detection of CldU was rat anti-CldU (Novus Biologicals). The secondary antibody was goat anti-rat Alexa Fluor 594 (Invitrogen). The length of the replication signals and the distances between origins were measured in micrometers and converted to kilo bases according to a constant and sequence-independent stretching factor (1 μm = 2 Kb), as previously reported (Herrick and Bensimon, 1999).
Nucleotide Pool Analysis
Keratinocytes were harvested, and cellular nucleotides were extracted with 0.4 N perchloric acid and neutralized with potassium chloride. Deoxynucleotides were separated from ribonucleotides using a boronate affinity column (Shewach, 1992). Deoxynucleotides were analyzed by HPLC using UV absorbance at 254 and 281 nm for identification and quantitation as previously described (Flanagan et al., 2007).
Soft Agar Assay
Following E6/E7 expression in primary keratinocytes, cells were grown in two different growth conditions: (1) normal keratinocyte medium and (2) nucleoside-enriched medium (50 nM of A, U, C, and G). After 3–4 weeks, the cells were trypsinized, and 250K cells were plated in 0.3% low gelling temperature agarose (Agarose, Type VII, Sigma) to a final volume of 1 ml per well; on the top of the 3% agar layer, 0.5 ml medium was added (normal medium or enriched with nucleosides). Colonies were counted after 4–6 weeks of growth.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Prof. Levana Sherman for the LXSN vectors and for helpful advice, Dr. Tzipora Shlomai for the F1 cells, and Prof. Jiri Bartek for the pbabepuro-cyclin E vector. This work was partially supported by a grant from the Nachmias Foundation of the Hebrew University, France-Israel program of the Israeli Ministry of Science, and the Israel Ministry of Health.
Footnotes
ACCESSION NUMBERS
The data for whole-transcriptome analysis has been deposited in the GEO DataSets under ID code GSE28266.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures, six figures, and three tables and can be found with this article online at doi:10.1016/j.cell.2011.03.044.
REFERENCES
- Akli S, Keyomarsi K. Cyclin E and its low molecular weight forms in human cancer and as targets for cancer therapy. Cancer Biol. Ther. 2003;2(4, Suppl 1):S38–S47. [PubMed] [Google Scholar]
- Anglana M, Apiou F, Bensimon A, Debatisse M. Dynamics of DNA replication in mammalian somatic cells: nucleotide pool modulates origin choice and interorigin spacing. Cell. 2003;114:385–394. doi: 10.1016/s0092-8674(03)00569-5. [DOI] [PubMed] [Google Scholar]
- Bartkova J, Horejsí Z, Koed K, Krämer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, et al. DNA damage response as a candidate anti-cancerbarrierinearly humantumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, Vassiliou LV, Kolettas E, Niforou K, Zoumpourlis VC, et al. Onco-gene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- Bartkova J, Hamerlik P, Stockhausen MT, Ehrmann J, Hlobilkova A, Laursen H, Kalita O, Kolar Z, Poulsen HS, Broholm H, et al. Replication stress and oxidative damage contribute to aberrant constitutive activation of DNA damage signalling in human gliomas. Oncogene. 2010;29:5095–5102. doi: 10.1038/onc.2010.249. [DOI] [PubMed] [Google Scholar]
- Conti C, Saccà B, Herrick J, Lalou C, Pommier Y, Bensimon A. Replication fork velocities at adjacent replication origins are coordinately modified during DNA replication in human cells. Mol. Biol. Cell. 2007;18:3059–3067. doi: 10.1091/mbc.E06-08-0689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coquelle A, Toledo F, Stern S, Bieth A, Debatisse M. A new role for hypoxia in tumor progression: induction of fragile site triggering genomic rearrangements and formation of complex DMs and HSRs. Mol. Cell. 1998;2:259–265. doi: 10.1016/s1097-2765(00)80137-9. [DOI] [PubMed] [Google Scholar]
- Courbet S, Gay S, Arnoult N, Wronka G, Anglana M, Brison O, Debatisse M. Replication fork movement sets chromatin loop size and origin choice in mammalian cells. Nature. 2008;455:557–560. doi: 10.1038/nature07233. [DOI] [PubMed] [Google Scholar]
- Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, Schurra C, Garre’ M, Nuciforo PG, Bensimon A, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–642. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- Duensing S, Münger K. The human papillomavirus type 16 E6 and E7 oncoproteins independently induce numerical and structural chromo-some instability. Cancer Res. 2002;62:7075–7082. [PubMed] [Google Scholar]
- Duensing S, Münger K. Centrosomes, genomic instability, and cervical carcinogenesis. Crit. Rev. Eukaryot. Gene Expr. 2003;13:9–23. doi: 10.1615/critreveukaryotgeneexpr.v13.i1.20. [DOI] [PubMed] [Google Scholar]
- Duensing S, Münger K. Mechanisms of genomic instability in human cancer: insights from studies with human papillomavirus oncoproteins. Int. J. Cancer. 2004;109:157–162. doi: 10.1002/ijc.11691. [DOI] [PubMed] [Google Scholar]
- Duensing S, Lee LY, Duensing A, Basile J, Piboonniyom S, Gonzalez S, Crum CP, Munger K. The human papillomavirus type 16 E6 and E7 oncoproteins cooperate to induce mitotic defects and genomic instability by uncoupling centrosome duplication from the cell division cycle. Proc. Natl. Acad. Sci. USA. 2000;97:10002–10007. doi: 10.1073/pnas.170093297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan SA, Robinson BW, Krokosky CM, Shewach DS. Mismatched nucleotides as the lesions responsible for radiosensitization with gemcitabine: a new paradigm for antimetabolite radiosensitizers. Mol. Cancer Ther. 2007;6:1858–1868. doi: 10.1158/1535-7163.MCT-07-0068. [DOI] [PubMed] [Google Scholar]
- Frame FM, Rogoff HA, Pickering MT, Cress WD, Kowalik TF. E2F1 induces MRN foci formation and a cell cycle checkpoint response in human fibroblasts. Oncogene. 2006;25:3258–3266. doi: 10.1038/sj.onc.1209352. [DOI] [PubMed] [Google Scholar]
- García-Closas R, Castellsagué X, Bosch X, González CA. The role of diet and nutrition in cervical carcinogenesis: a review of recent evidence. Int. J. Cancer. 2005;117:629–637. doi: 10.1002/ijc.21193. [DOI] [PubMed] [Google Scholar]
- Garner-Hamrick PA, Fostel JM, Chien WM, Banerjee NS, Chow LT, Broker TR, Fisher C. Global effects of human papillomavirus type 18 E6/E7 in an organotypic keratinocyte culture system. J. Virol. 2004;78:9041–9050. doi: 10.1128/JVI.78.17.9041-9050.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge XQ, Jackson DA, Blow JJ. Dormant origins licensed by excess Mcm2-7 are required for human cells to survive replicative stress. Genes Dev. 2007;21:3331–3341. doi: 10.1101/gad.457807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordan JD, Lal P, Dondeti VR, Letrero R, Parekh KN, Oquendo CE, Greenberg RA, Flaherty KT, Rathmell WK, Keith B, et al. HIF-alpha effects on c-Myc distinguish two subtypes of sporadic VHL-deficient clear cell renal carcinoma. Cancer Cell. 2008;14:435–446. doi: 10.1016/j.ccr.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, Venere M, Ditullio RA, Jr., Kastrinakis NG, Levy B, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- Grandori C, Cowley SM, James LP, Eisenman RN. The Myc/ Max/Mad network and the transcriptional control of cell behavior. Annu. Rev. Cell Dev. Biol. 2000;16:653–699. doi: 10.1146/annurev.cellbio.16.1.653. [DOI] [PubMed] [Google Scholar]
- Gross-Mesilaty S, Reinstein E, Bercovich B, Tobias KE, Schwartz AL, Kahana C, Ciechanover A. Basal and human papillomavirus E6 oncoprotein-induced degradation of Myc proteins by the ubiquitin pathway. Proc. Natl. Acad. Sci. USA. 1998;95:8058–8063. doi: 10.1073/pnas.95.14.8058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- Harris CP, Lu XY, Narayan G, Singh B, Murty VV, Rao PH. Comprehensive molecular cytogenetic characterization of cervical cancer cell lines. Genes Chromosomes Cancer. 2003;36:233–241. doi: 10.1002/gcc.10158. [DOI] [PubMed] [Google Scholar]
- Herrick J, Bensimon A. Single molecule analysis of DNA replication. Biochimie. 1999;81:859–871. doi: 10.1016/s0300-9084(99)00210-2. [DOI] [PubMed] [Google Scholar]
- Hwang HC, Clurman BE. Cyclin E in normal and neoplastic cell cycles. Oncogene. 2005;24:2776–2786. doi: 10.1038/sj.onc.1208613. [DOI] [PubMed] [Google Scholar]
- Kimura T, Takeda S, Sagiya Y, Gotoh M, Nakamura Y, Arakawa H. Impaired function of p53R2 in Rrm2b-null mice causes severe renal failure through attenuation of dNTP pools. Nat. Genet. 2003;34:440–445. doi: 10.1038/ng1212. [DOI] [PubMed] [Google Scholar]
- Klaunig JE, Kamendulis LM. The role of oxidative stress in carcinogenesis. Annu. Rev. Pharmacol. Toxicol. 2004;44:239–267. doi: 10.1146/annurev.pharmtox.44.101802.121851. [DOI] [PubMed] [Google Scholar]
- Klausner RD. The fabric of cancer cell biology-Weaving together the strands. Cancer Cell. 2002;1:3–10. doi: 10.1016/s1535-6108(02)00020-x. [DOI] [PubMed] [Google Scholar]
- Lebofsky R, Heilig R, Sonnleitner M, Weissenbach J, Bensimon A. DNA replication origin interference increases the spacing between initiation events in human cells. Mol. Biol. Cell. 2006;17:5337–5345. doi: 10.1091/mbc.E06-04-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YC, Li F, Handler J, Huang CR, Xiang Y, Neretti N, Sedivy JM, Zeller KI, Dang CV. Global regulation of nucleotide biosynthetic genes by c-Myc. PLoS ONE. 2008;3:e2722. doi: 10.1371/journal.pone.0002722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannava S, Grachtchouk V, Wheeler LJ, Im M, Zhuang D, Slavina EG, Mathews CK, Shewach DS, Nikiforov MA. Direct role of nucleotide metabolism in C-MYC-dependent proliferation of melanoma cells. Cell Cycle. 2008;7:2392–2400. doi: 10.4161/cc.6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narisawa-Saito M, Yoshimatsu Y, Ohno S, Yugawa T, Egawa N, Fujita M, Hirohashi S, Kiyono T. An in vitro multistep carcinogenesis model for human cervical cancer. Cancer Res. 2008;68:5699–5705. doi: 10.1158/0008-5472.CAN-07-6862. [DOI] [PubMed] [Google Scholar]
- Ohtsubo M, Roberts JM. Cyclin-dependent regulation of G1 in mammalian fibroblasts. Science. 1993;259:1908–1912. doi: 10.1126/science.8384376. [DOI] [PubMed] [Google Scholar]
- Pickering MT, Kowalik TF. Rb inactivation leads to E2F1-mediated DNA double-strand break accumulation. Oncogene. 2006;25:746–755. doi: 10.1038/sj.onc.1209103. [DOI] [PubMed] [Google Scholar]
- Plug-DeMaggio AW, Sundsvold T, Wurscher MA, Koop JI, Klingelhutz AJ, McDougall JK. Telomere erosion and chromosomal instability in cells expressing the HPV oncogene 16E6. Oncogene. 2004;23:3561–3571. doi: 10.1038/sj.onc.1207388. [DOI] [PubMed] [Google Scholar]
- Rampersaud GC, Bailey LB, Kauwell GP. Relationship of folate to colorectal and cervical cancer: review and recommendations for practitioners. J. Am. Diet. Assoc. 2002;102:1273–1282. doi: 10.1016/s0002-8223(02)90281-6. [DOI] [PubMed] [Google Scholar]
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- Saintigny Y, Delacôte F, Varès G, Petitot F, Lambert S, Averbeck D, Lopez BS. Characterization of homologous recombination induced by replication inhibition in mammalian cells. EMBO J. 2001;20:3861–3870. doi: 10.1093/emboj/20.14.3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewach DS. Quantitation of deoxyribonucleoside 5′-triphosphates by a sequential boronate and anion-exchange high-pressure liquid chromatographic procedure. Anal. Biochem. 1992;206:178–182. doi: 10.1016/s0003-2697(05)80030-2. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Arakawa H, Yamaguchi T, Shiraishi K, Fukuda S, Matsui K, Takei Y, Nakamura Y. A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature. 2000;404:42–49. doi: 10.1038/35003506. [DOI] [PubMed] [Google Scholar]
- Thompson DA, Belinsky G, Chang TH, Jones DL, Schlegel R, Münger K. The human papillomavirus-16 E6 oncoprotein decreases the vigilance of mitotic checkpoints. Oncogene. 1997;15:3025–3035. doi: 10.1038/sj.onc.1201495. [DOI] [PubMed] [Google Scholar]
- Tsantoulis PK, Kotsinas A, Sfikakis PP, Evangelou K, Sideridou M, Levy B, Mo L, Kittas C, Wu XR, Papavassiliou AG, Gorgoulis VG. Oncogene-induced replication stress preferentially targets common fragile sites in preneoplastic lesions. A genome-wide study. Onco-gene. 2008;27:3256–3264. doi: 10.1038/sj.onc.1210989. [DOI] [PubMed] [Google Scholar]
- van den Heuvel S, Dyson NJ. Conserved functions of the pRB and E2F families. Nat. Rev. Mol. Cell Biol. 2008;9:713–724. doi: 10.1038/nrm2469. [DOI] [PubMed] [Google Scholar]
- Yan ZA, Li XZ, Zhou XT. The effect of hydroxyurea on the expression of the common fragile site at 3p14. J. Med. Genet. 1987;24:593–596. doi: 10.1136/jmg.24.10.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.