Abstract
Age-associated loss of muscle mass, or sarcopenia, contributes directly to frailty and an increased risk of falls and fractures among the elderly. Aged mice and elderly adults both show decreased muscle mass as well as relatively low levels of the fat-derived hormone leptin. Here we demonstrate that loss of muscle mass and myofiber size with aging in mice is associated with significant changes in the expression of specific miRNAs. 57 miRNAs were altered with aging in the mice and many of these miRNAs are, for the first time, reported to be associated specifically with age-related muscle atrophy. These include miR-221, previously identified in studies of myogenesis and muscle development as playing a role in the proliferation and terminal differentiation of myogenic precursors. We also treated aged mice with recombinant leptin, to determine whether leptin therapy could improve muscle mass and alter the miRNA expression profile of aging skeletal muscle. Leptin treatment significantly increased hindlimb muscle mass and extensor digitorum longus fiber size in aged mice. Furthermore, the expression of 37 miRNAs was altered in muscles of leptin treated mice. In particular, leptin treatment increased the expression of miR-31 and miR-223, miRNAs known to be elevated during muscle regeneration and repair. These findings suggest that aging in skeletal muscle is associated with marked changes in the expression of specific miRNAs, and that nutrient-related hormones such as leptin may be able to reverse muscle atrophy and alter the expression of atrophy-related miRNAs in aging skeletal muscle.
Keywords: sarcopenia, aging, microRNAs, miR-31, miR-221, miR-223
1. Introduction
It is estimated that approximately 5-8% of muscle mass is lost per decade of life after about age 30, and this rate of decline accelerates after age 65 [1]. Age-related muscle atrophy, or sarcopenia, significantly increases morbidity and mortality among the elderly because muscle weakness and postural instability are major contributors to falls and fractures [2, 3]. The cytokine-like hormone leptin is an important factor linking food intake with energy expenditure and body composition [4, 5]. Leptin is secreted from fat cells (adipocytes), but muscle is also a primary source of leptin [6], and serum leptin levels increase with increased muscle mass [7]. Older (~85 yrs) populations of frail, continuing care patients are observed to show low serum leptin, low bone mass, and muscle atrophy, suggesting that altered leptin signaling is indeed likely to play a major role in sarcopenia and the anorexia of aging [8]. Leptin deficiency itself is associated with decreased muscle mass [9], and the functional characteristics of skeletal muscle in leptin-deficient ob/ob mice resemble those of aged rodents [10]. Leptin receptors are abundantly expressed in peripheral tissues such as skeletal muscle, liver, and bone [11]. Leptin receptors have been identified in human skeletal muscle [12], their expression is elevated with disuse atrophy [13], and leptin-deficiency increases expression of the muscle-wasting protein myostatin [14].
Recent studies suggest that alterations in the expression of muscle-specific microRNAs (miRNAs) may play a role in several muscle disorders. miRNAs are short (~22 nucleotides) molecules that bind to complemetary sequences of specific target mRNAs and inhibit translation. Accumulating evidence indicates that miRNAs regulate essential biological functions, such as cellular differentiation, proliferation, and apoptosis, and have become one of the most important gene regulators in eukaryotic organisms [15-17]. A number of miRNAs have been identified that are tissue-specific, and may therefore be involved in tissue development, disease, and regeneration. For example, the expression of the muscle-specific miRNA miR-206 has been found to decrease with aging and increase with mechanical stimulation [18]. Local injection of miR-206 can accelerate muscle regeneration [19], whereas miR-133 promotes myoblast proliferation and miR-1 can suppress myoblast proliferation [20]. A broad molecular profiling approach examining the expression patterns of miRNAs in primary muscular disorders identified 18 miRNAs that were associated with specific diseases such as Duchenne and Becker muscular dystrophies [21]. These findings suggest that miRNAs may represent potential therapeutic targets for muscle-related diseases [22].
We have identified an animal model, the aged C57BL/6 mouse, that shares a number of key features in common with the aging human musculoskeletal system: an age-related decline in serum leptin, decreased muscle mass, and loss of bone density [23]. Here we test the hypothesis that loss of skeletal muscle mass with age in this model is associated with marked changes in the expression of muscle-specific miRNAs. We also determine whether recombinant leptin therapy can increase muscle mass and myofiber hypertrophy in this animal model, and whether leptin treatment can alter the miRNA expression profile that accompanies age-associated muscle atrophy.
2. Materials and methods
2.1. Animals and treatments
C57BL/6 mice 12 and 24 months of age were obtained from the National Institute on Aging colony maintained by Taconic Farms. 24 mice per age group were divided into control mice and those given subcutaneous injections of leptin (10 μg/day) for 10 days. Mice were euthanized after the 10 day treatment period and body weight and quadriceps mass were recorded, and the extensor digitorum longus (EDL; predominantly type II, or fast-twitch fibers) and soleus (primarily type I, or slow-twitch fibers) dissected free, embedded in OCT medium, and snap frozen. Cryostat sections of the EDL and soleus were stained alternately with H&E or anti-laminin antibody (rabbit anti mouse, Sigma L9393) and muscle fiber cross-sectional areas measured using SigmaScan.
2.2. Real-time miRNA profiling
Quadriceps muscles were dissected free and snap frozen in liquid nitrogen. Total RNA of quadriceps muscles was extracted using a mirRNA isolation kit (Qiagen) and mouse miRNAs were analyzed by the TaqMan miRNA arrays using 7900HT Real-Time PCR System (Applied Biosystems, Foster City, CA). For each reaction, 300ng-500ng of total RNA was added to RT enzyme mixture with 10X Megaplex RT primers to produce cDNA. The specific primers and TaqMan assay probes for the miRNAs were distributed on the 384-well micro-fluidic card (rodent V1 cards) that allows for 384 simultaneous PCR reactions, including up to 379 miRNA genes as well as endogenous controls. Normalization was performed with the small nuclear RNAs (snRNAs) U87, which are stably expressed in most tissues, as well as small RNA RNU202. For data analysis, we used RQ Manager SDS 2.3 software from ABI (version 1.2). The cycle number at which the reaction crossed an arbitrarily placed threshold (CT) was determined for each miRNA and expression of each miRNA was calculated with the relative amount of each miRNA to endogenous controls U87 or RNU202 using the equation 2−ΔCt where CT =(CTmiRNA-CTU87) or (CTmiRNA-CTrun202). A heat map was generated by Cluster 3.0 and Tree view software based on each miRNA’s relative expression amount. We used CT = 35 as a cutoff. The assay was run in duplicate for each case to allow for assessment of technical variability. Comparisons were run for muscles of aged (24 mo) versus young (12 mo) mice, and for aged mice treated with leptin versus aged mice receiving saline, by calculating 2−ΔΔCT. Predicted gene targets of miRNAs were identified using TargetScan and PicTar software.
3. Results
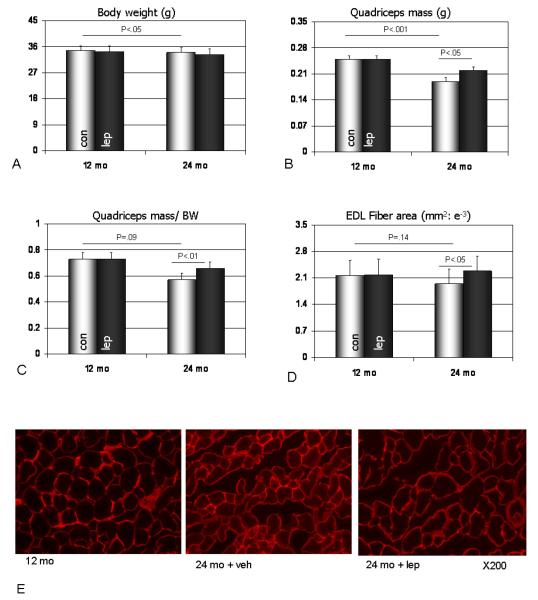
Body weight data demonstrate that the aged mice were significantly smaller than the younger mice, and that leptin treatment did not significantly alter body weight in mice of either age group (Fig. 1a). Quadriceps muscle weights were significantly lower in the aged mice, and were also slightly (but not significantly) lower when normalized to body weight (Fig. 1b, c). Leptin treatment did, however, significantly increase quadriceps muscle mass both absolutely (Fig. 1b) and relative to body mass (Fig. 1c) in the aged mice but not in the younger mice. Muscle fiber cross-sectional areas of the extensor digitorum longus muscle (EDL) were slightly lower in aged mice, and leptin treatment significantly increased EDL fiber area in the aged mice but not the young mice (Fig. 1d, Fig. 1e). Muscle fiber cross-sectional areas of the soleus muscle were similar between young and aged mice, and leptin treatment produced a slight but non-significant increase in soleus fiber area in mice from each age group.
Fig. 1.
Data on body weight (a), quadriceps mass (b), quadriceps mass normalized by body weight (c), and cross-sectional area of extensor digitorum longus muscle fibers (d) in adult (12 mo) and aged mice (24 mo) receiving saline (con) or recombinant leptin (lep; 10 μg/day). (e) Cryostat sections of the extensor digitorum longus (EDL) stained with a Cy3-conjugated anti-laminin antibody from adult mice receiving saline (12 mo), aged mice receiving saline (24 mo + veh), and aged mice treated with leptin (24 mo + lep). Note the relatively larger size of the EDL fibers in aged mice receiving leptin compared to the fibers of aged mice receiving saline.
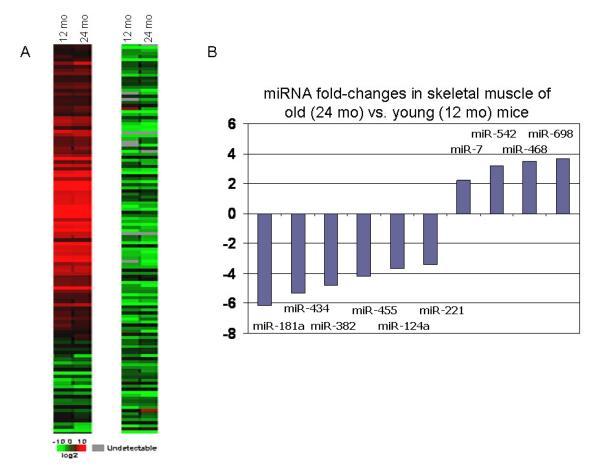
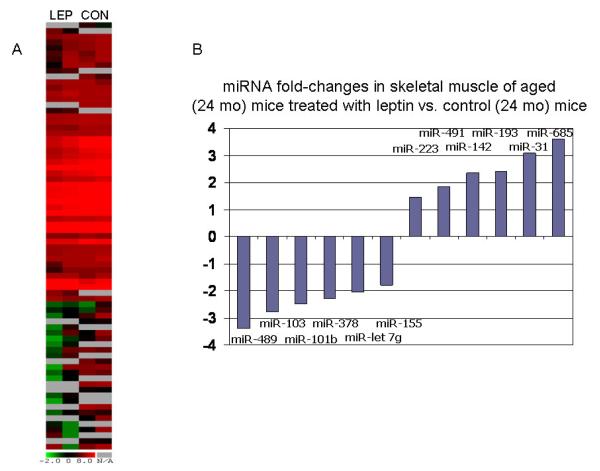
To test if miRNAs are involved in aging muscle development, we first used TaqMan RT-PCR miRNA array to compare the miRNA expression profiles between mice 12- and 24-months of age. miRNA profilings revealed that relative expression of 57 miRNAs was significantly altered quadriceps muscle samples from aged mice compared to those from younger adult mice. Approximately 36 miRNAs were decreased whereas only 21 miRNAs were increased (Fig. 2). The muscle-specific miRNA miR-206 was upregulated about 2-fold in aged mice compared to young. To test if the miRNA expression profile of aged skeletal muscle might be altered with leptin treatment, we further analyzed miRNA gene expression profiles in the muscles from vehicle- or leptin-treated mice. As shown in Fig. 3, 37 genes were changed in leptin-treated aged mice compared to control aged mice, including 7 upregulated and 30 downregulated miRNAs. Interestingly, leptin treatment reversed the expression of several miRNAs in muscles from aged mice. miR-685 and miR-142-3p were all downregulated in quadriceps muscles of aged mice compared to younger 12 month-old mice, whereas leptin treatment significantly increased the expression of these miRNAs relative to control aged mice (Fig. 3B). Likewise, leptin treatment treatment also decreased expression of several miRNAs whose expression was increased in muscles of aged mice compared to muscles from younger mice. miR-155 was increased in muscle of aged mice, but leptin treatment significantly decreased miR-155 expression compared to vehicle-treated controls (Fig. 3B).
Fig. 2.
Heat map (a) and miRNA expression changes > 1-fold (b) in quadriceps muscles from aged mice (24 mo) compared to adult mice (12 mo).
Fig. 3.
Heat map (a) and miRNA expression changes > 1-fold (b) in quadriceps muscles from leptin-treated aged mice compared to vehicle (saline)-treated aged mice.
4. Discussion
To date, the tissue specific expression of three miRNAs--miR-1, miR-133a, and miR-206--has been consistently associated with skeletal muscle. These miRNAs have also been shown to induce significant effects on muscle development and myogenesis by targeting myogenic factors such as mef2, SRF, and myostatin [22]. Interestingly, our profiling approach reveals that miRNA miR-206 was significantly up-regulated with age in the mice (Fig. 2). miR-206 can induce muscle hypertrophy [19], and its increased expression with muscle atrophy in aging may indicate an adaptive, compensatory response to antagonize other catabolic signals. The data presented here also identify additional miRNAs that may be involved in age-associated muscle atrophy. Specifically, miR-698 and -468 were highly upregulated with age and miR-434, -455, and -382, -181a, and -221 were downregulated. The strongest predicted target for miR-698 is cardiotrophin 1, a molecule that maintains the undifferentiated state of skeletal myoblasts by inhibiting myogenic regulatory factors, myocyte enhance factors, and myogenin [24]. Likewise, miR-221 was recently found to be downregulated upon myogenic differentiation, and overexpression of miR-221 delays withdrawal of differentiating myoblasts from the cell cycle and suppresses the expression of myogenin [25]. These findings raise the possibility that with aging, and its concomitant increase in miR-698 expression and decline in miR-221 expression, molecular cues favoring the terminal differentiation of myoblasts are increased in skeletal muscle. Furthermore, the strongest predicted PicTar target for miR-181a, which is downregulated more than six-fold in aging muscle, is the type IIA activin receptor (ActRIIA or Acvr2a). This receptor is a primary receptor for activin A [26], which has been shown to inhibit the proliferation of muscle-derived stem cells by phosphorylating Smad 2/3 [27]. Thus, aging in mouse skeletal muscle is associated with changes in several miRNAs that together favor a decreased proliferative potential of myogenic precursors and a tendency toward terminal myogenic differentiation. This age-associated change in skeletal muscle is similar to that previously postulated for aging bone, where a general decrease in stem cell number and proliferative potential has been implicated in the development of osteoporosis [28, 29].
Leptin treatment increased the relative mass of quadriceps muscles in aged mice as well as the fiber size of skeletal muscle fibers in the extensor digitorum longus (Fig. 1). These results are consistent with previous work in leptin-deficient ob/ob mice, where recombinant leptin therapy increased muscle mass in part by suppressing myostatin and Foxo3a expression [14, 30]. As noted in the introduction, populations of aging adults show low serum leptin levels [8], and we have also found that aging in mice is accompanied by a decline in serum leptin [23]. Previous work has indicated that leptin resistance, via downregulation of central (hypothalamic) leptin receptors, increases with age [31]. Our data suggest that, at least in the case of skeletal muscle, leptin resistance may not necessarily increase with age. That is, exogenous leptin is capable of inducing a significant anabolic response in skeletal muscle, as well as producing changes in the expression of specific miRNAs. MicroRNAs increased in aged muscle with leptin treatment include miRNAs previously identified as playing a role in muscle regeneration. Greco et al [32] demonstrated that specific miRNAs were upregulated in the inflammatory (miR-miR-222, -223), degenerative (miR-1, -29c, -135a), and regenerative (miR-206, -34c, -31, -335, 449, and -494) phases of muscle damage and regeneration in Duchenne muscular dystrophy. miR-223 and miR-31 were both upregulated in skeletal muscles from leptin-treated aged mice (Fig. 3), suggesting that leptin can activate molecular pathways involved in muscle repair and regeneration. miRNAs that were downregulated in skeletal muscles from leptin-treated animals include several that have been previously described as playing a role in mesenchymal stem cell (MSC) differentiation. These include miR-489, known to be downregulated during the osteogenic differentiation of MSCs [33], and miR-103, which is specifically localized to bone marrow populations of MSCs [34] and can induce adipogenic differentiation when expressed ectopically [35]. The strong downregulation of these genes associated with MSCs suggests that exogenous leptin may significantly alter the expression profile of muscle-derived stem cells.
In conclusion, our data reveal that as C57BL6 mice age they lose skeletal muscle mass, and are therefore a useful animal model for studying the development of age-associated pathologies of the musculoskeletal system such as osteoporosis and sarcopenia [23]. Loss of muscle mass with age in these mice resembles human age-associated muscle loss in that fast-twitch fibers, which are abundant in the extensor digitorum longus, are reduced in size more so than slow-twitch fibers, which are more numerous in the soleus muscle. Age-associated loss of muscle mass in this mouse model was accompanied by specific changes in the expression pattern of miRNAs. To date, only one other study [18] has investigated the miRNA changes associated with sarcopenia. Our miRNA expression data indicate that several miRNAs are altered with aging and may contribute to a decreased proliferative potential of myogenic precursors and a tendency toward terminal myogenic differentiation. While leptin therapy increased muscle mass in the aged mice, it can regulate 37 miRNA gene expression, but only reverse three dysregulated miRNAs in aged mice, miR-685, miR-142-3p and miR-155, suggesting that other therapeutic approaches need to be investigated in order to target certain miRNAs identified in aged muscle. Future studies should be directed at developing nutritional interventions that might alter the expression of specific miRNAs associated with the onset and progression of muscle wasting and frailty in aged rodents and elderly human populations. At the very least, new data emerging from the study of miRNAs in musculoskeletal diseases suggest that many promising therapeutic targets remain to be identified.
Aging is associated with muscle atrophy and loss of muscle mass, known as the sarcopenia of aging.
We demonstrate that age-related muscle atrophy is associated with marked changes in miRNA expression in muscle.
Treating aged mice with the adipokine leptin significantly increased muscle mass and the expression of miRNAs involved in muscle repair.
Recombinant leptin therapy may therefore be a novel approach for treating age-related muscle atrophy.
Acknowledgments
Funding for this research was provided by the Medical College of Georgia School of Medicine, the Henry Ford Health System (J8005), and the National Institutes of Health (NIAMS AR 049717).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no conflict of interest and no financial disclosures.
References
- [1].Greenlund L, Nair K. Sarcopenia--consequences, mechanisms, and potential Therapies. Mech. Ageing. Dev. 2003;124:287–99. doi: 10.1016/s0047-6374(02)00196-3. [DOI] [PubMed] [Google Scholar]
- [2].Nguyen N, Pongchaiyakul C, Center J, Eisman J, Nguyen T. Identification of high-risk individuals for hip fracture: a 14-year prospective study. J. Bone Miner. Res. 2005;20:1921–1928. doi: 10.1359/JBMR.050520. [DOI] [PubMed] [Google Scholar]
- [3].Jarvinen T, Sievanen H, Khan K, Heinonen A, Kannus P. Shifting the focus in fracture prevention from osteoporosis to falls. Br. Med. J. 2008;336:124–126. doi: 10.1136/bmj.39428.470752.AD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Flier J. What’s in a name? In search of leptin’s physiologic role. J. Clin. Endocrinol. Metabol. 1988;83:1407–1413. doi: 10.1210/jcem.83.5.4779. [DOI] [PubMed] [Google Scholar]
- [5].Hamrick MW. Leptin, bone mass, and the thrifty phenotype. J. Bone Miner. Res. 2004;19:1607–1611. doi: 10.1359/JBMR.040712. [DOI] [PubMed] [Google Scholar]
- [6].Wang J, Liu R, Hawkins M, Barzilai N, Rossetti L. A nutrient-sensing pathway regulations leptin gene expression in muscle and fat. Nature. 1998;393:684–688. doi: 10.1038/31474. [DOI] [PubMed] [Google Scholar]
- [7].Fernandez-Real H, Vayreda M, Casamitjana R, Gonzalez-Huix F, Ricart W. The fat-free mass compartment influences serum leptin in men. Eur. J. Endocrinol. 2000;142:25–29. doi: 10.1530/eje.0.1420025. [DOI] [PubMed] [Google Scholar]
- [8].Hubbard R, O’Mahony M, Calver B, Woodhouse K. Nutrition, inflammation, and leptin levels in aging and frailty. JAGS. 2008;56:279–284. doi: 10.1111/j.1532-5415.2007.01548.x. [DOI] [PubMed] [Google Scholar]
- [9].Hamrick MW, Pennington C, Newton D, Xie D, Isales CM. Leptin deficiency produces contrasting phenotypes in bones of the limb and spine. Bone. 2004;34:376–383. doi: 10.1016/j.bone.2003.11.020. [DOI] [PubMed] [Google Scholar]
- [10].Warmington S, Rolan R, McBennett S. Functional and histological characteristics of skeletal muscle and the effects of leptin in the genetically obese (ob/ob) mouse. Int. J. Obes. Rel. Metab. Disord. 2000;24:1040–1050. doi: 10.1038/sj.ijo.0801357. [DOI] [PubMed] [Google Scholar]
- [11].Margetic S, Gazzola C, Pegg G, Hill R. Leptin: a review of its peripheral actions and interactions. Int. J. Obes. Rel. Metab. Disord. 2002;26:1407–33. doi: 10.1038/sj.ijo.0802142. [DOI] [PubMed] [Google Scholar]
- [12].Guerra B, Santana A, Fuentes T, Delgado-Guerra S, Cabrera-Socorro A, Dorado C, Calbet J. Leptin receptors in human skeletal muscle. J. Appl. Physiol. 2007;102:1786–1792. doi: 10.1152/japplphysiol.01313.2006. [DOI] [PubMed] [Google Scholar]
- [13].Chen Y, Gregory C, Scarborough M, Shi R, Walter G, Vandenborne K. Transcriptional pathways associated with skeletal muscle disuse atrophy in humans. Physiol. Genomics. 2007;31:510–520. doi: 10.1152/physiolgenomics.00115.2006. [DOI] [PubMed] [Google Scholar]
- [14].Allen D, Clearly A, Speaker K, Lindsay S, Uyenishi J, Reed J, Madden M, Mehan E. Myostatin, activin receptor IIB, and follistatin-like-3 gene are altered in adipose tissue and skeletal muscle of obese mice. Am. J. Phys. Endocrinol. Metab. 2008;294:E918–27. doi: 10.1152/ajpendo.00798.2007. [DOI] [PubMed] [Google Scholar]
- [15].Hatfield S, Shcherbata H, Fischer K, Nakahara K, Carthew R, Ruohola-Baker H. Stem cell division is regulated by the microRNA pathway. Nature. 2005;435:974–978. doi: 10.1038/nature03816. [DOI] [PubMed] [Google Scholar]
- [16].Plasterk R. MicroRNAs in animal development. Cell. 2006;124:877–881. doi: 10.1016/j.cell.2006.02.030. [DOI] [PubMed] [Google Scholar]
- [17].He X, Eberhart J, Postlethwait J. MicroRNAs and micromanaging the skeleton in disease, development, and evolution. J Cell Mol Med. 2009;13:606–18. doi: 10.1111/j.1582-4934.2009.00696.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Drummond M, McCarthy J, Fry C, Esser K, Rasmussen B. Aging differentially affects human skeletal muscle microRNA expression at rest and after an anabolic stimulus of resistance exercise and essential amino acids. Am. J. Physiol. Endrocrinol. Metab. 2008;295:E1333–40. doi: 10.1152/ajpendo.90562.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Nakasa T, Ishikawa M, Shi M, Shibuya H, Adachi N, Ochi M. Acceleration of muscle regeneration by local injection of muscle-specific microRNAs in rat skeletal muscle injury model. J Cell Mol Med. 2009 Sep 14; doi: 10.1111/j.1582-4934.2009.00898.x. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Van Rooij E, Liu N, Olson E. microRNAs flex their muscles. Trends Genetics. 2008;24:159–66. doi: 10.1016/j.tig.2008.01.007. [DOI] [PubMed] [Google Scholar]
- [21].Eisenberg I, Eran A, Nishino I, Moggio M, Lamperti C, Amato A, Lidov H, Kang P, North K, Mitrani-Rosenbaum S, Flanigan K, Neely L, Whitney D, Beggs A, Kohane I, Kunkel L. Distinctive patterns of microRNA expression in primary muscular disorders. Proc Natl Acad Sci (USA) 2007;104:17016–17021. doi: 10.1073/pnas.0708115104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Chen J, Callis T, Wang D-Z. microRNAs and muscle disorders. J. Cell Science. 2009;122:13–20. doi: 10.1242/jcs.041723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Hamrick MW, Ding K, Pennington C, Chao Y, Wu Y, Howard B B, Immel D, Borlongan C, McNeil P, Bollag W, Curl W, Yu J, Isales CM. Age-related loss of muscle mass and bone strength in mice is associated with a decline in physical activity and serum leptin. Bone. 2006;39:845–853. doi: 10.1016/j.bone.2006.04.011. [DOI] [PubMed] [Google Scholar]
- [24].Miyake T, Alli N, Aziz A, Knudson J, Fernando P, Megeney L, McDermott J. Cardiotrophin-1 maintains the undifferentiated state in skeletal myoblasts. J. Biol. Chem. 2009;294:19679–93. doi: 10.1074/jbc.M109.017319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Cardinali B, Castellani L, Fasanaro P, Basso A, Alema S, Martelli F, Falcone G. Microrna-221 and microrna-222 modulate differentiation and maturation of skeletal muscle cells. PLoS One. 2009;4:e7607. doi: 10.1371/journal.pone.0007607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Tsuchida K, Nakatani M, Uezumi A, Murakami T, Cui X. Signal transduction pathway through activin receptors as a therapeutic target of musculoskeletal diseases and cancer. Endocr. J. 2008;55:11–21. doi: 10.1507/endocrj.kr-110. [DOI] [PubMed] [Google Scholar]
- [27].Nomura T, Ueyama T, Ashihara E, Tateishi K, Asada S, Nakajima N, Isodono K, Takahashi T, Matsubara H, Oh H. Skeletal muscle-derived progenitors capable of differentiating into cardiomyocytes proliferate through myostatin-independent TGF-ß family signaling. Biochem. Biophys. Res. Comm. 2008;365:863–869. doi: 10.1016/j.bbrc.2007.11.087. [DOI] [PubMed] [Google Scholar]
- [28].Stolzing A, Scutt A. Age-related impairment of mesenchymal progenitor cell function. Aging Cell. 2006;5:213–224. doi: 10.1111/j.1474-9726.2006.00213.x. [DOI] [PubMed] [Google Scholar]
- [29].Zhang W, Ou G, Hamrick M, Hill W, Borke J, Wenger K, Chutkan N, Yu J, Mi Q, Isales CM, Shi X. Age-related changes in the osteogenic differentiation potential of mouse bone marrow stromal cells. J. Bone Mine.r Res. 2008;23:1118–28. doi: 10.1359/JBMR.080304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Sainz N, Rodriguez A, Catalan V, Becerril S, Ramirez B, Gomez-Ambrosi J, Hruhbeck H. Leptin administration favors muscle mass accretion by decreasing Fox03a and increasing PGC-1alpha in ob/ob mice. PLoS One. 2009;4:e6808. doi: 10.1371/journal.pone.0006808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Fernández-Galaz C, Fernández-Agulló T, Pérez C, Peralta S, Arribas C, Andrés A, Carrascosa J, Ros M. Long-term food restriction prevents ageing-associated central leptin resistance in Wistar rats. Diabetologia. 2002;45:997–1003. doi: 10.1007/s00125-002-0851-4. [DOI] [PubMed] [Google Scholar]
- [32].Greco S, DeSimone M, Colussi C, Zaccagnini G, Fasanaro P, Pescatori M, Cardani R, Perbellini R, Isaia E, Sale P, Meola G, Capogrossi M, Gaetano C, Martelli F. Common micro-RNA signature in skeletal muscle damage and regeneration induced by Duchenne muscular dystrophy and acute ischemia. FASEB J. 2009;23:3335–46. doi: 10.1096/fj.08-128579. [DOI] [PubMed] [Google Scholar]
- [33].Schoolmeesters A, Eklund T, Leake D, Vermeulen A, Smith Q, Force-Aldred S, Fedorov Y. Functional profiling reveals critical role for miRNA in differentiation of human mesenchymal stem cells. PLoS One. 2009;4:e5606. doi: 10.1371/journal.pone.0005605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Liu S, Fu R, Yu H, Li K, Tsai C, Shyu W, Lin S. MicroRNAs regulation modulated self-renewal and lineage differentiation of stem cells. Cell Transplant. 2009;18:1039–45. doi: 10.3727/096368909X471224. [DOI] [PubMed] [Google Scholar]
- [35].Xie H, Lim B, Lodish H. MicroRNAs induced during adipogenesis that accelerate fat cell development are downregulated in obesity. Diabetes. 2009;58:1050–57. doi: 10.2337/db08-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]