Abstract
NKX3.1 is a homeobox gene that codes for a haploinsufficient prostate cancer tumor suppressor. NKX3.1 protein levels are down regulated in the majority of primary prostate cancer tissues. NKX3.1 expression in PC-3 cells increased IGFBP-3 mRNA expression 10-fold as determined by expression microarray analysis. In both stably and transiently transfected PC-3 cells and in LNCaP cells NKX3.1 expression increased IGFBP-3 mRNA and protein expression. In prostates of Nkx3.1 gene-targeted mice Igfbp-3 mRNA levels correlated with Nkx3.1 copy number. NKX3.1 expression in PC-3 cells attenuated the ability of IGF-I to induce phosphorylation of IGF-IR, IRS-1, PI3-kinase, and AKT. The effect of NKX3.1 on IGF-I signaling was not seen when cells were exposed to long-R3-IGF-I, an IGF-1 variant peptide that does not bind to IGFBP-3. Additionally, siRNA-induced knock down of IGFBP-3 expression partially reversed the attenuation of IGF-1R signaling by NKX3.1 and abrogated NKX3.1 suppression of PC-3 cell proliferation. Thus there is a close relationship in vitro and in vivo between NKX3.1 and IGFBP-3. The growth suppressive effects of NKX3.1 in prostate cells are mediated, in part, by activation of IGFBP-3 expression.
Adenocarcinoma of the prostate, like many epithelial malignancies, initiates in luminal epithelial cells in prostatic ducts that acquire the precursor or gatekeeper mutations required for development of the malignant phenotype. Early in prostate cancer a region of 8p21.2 is lost in the majority of cancers (1). At least one target for 8p21.2 loss is the homeobox gene NKX3.1 that is expressed specifically in prostate luminal epithelial cells. NKX3.1 undergoes progressive loss of protein expression during prostate cancer progression from hormone-dependence to hormone-independence and metastatic disease (2, 3).
The NKX3.1 gene is not subject to somatic mutation in prostate cancer (4, 5). Gene targeting studies in mice showed that Nkx3.1 haploinsufficiency alone can predispose to prostate epithelial dysplasia and can cooperate with other oncogenic mutations to augment prostate carcinogenesis (6, 7). In gene-targeted mice decreased Nkx3.1 expression is accompanied by decreased expression of genes under the regulation of the Nkx3.l homeoprotein (8). We have recently shown that diminished levels of NKX3.1 expression in primary human prostate cancer and intraepithelial neoplasia correlated with the degree of gene inactivation by deletion, methylation, or both. Not only is NKX3.1 down regulated in preinvasive prostate cancer, but NKX3.1 expression is reduced in regions of inflammatory atrophy that are precursors for malignant transformation (9). Inflammatory cytokines in these lesions can induce ubiquitination of NKX3.1 and protein loss (10). Therefore NKX3.1 may play a role in premalignant events in the prostate gland by modulating gene expression to increase the susceptibility of prostate epithelial cells to malignant transformation. We have sought to characterize the gene expression program activated by NKX3.1 in human cells. Here we show in vitro and in vivo NKX3.1 activates expression of insulin-like growth factor binding protein-3 (IGFBP-3), a known growth suppressor protein and down regulator of insulin-like growth factor-I (IGF-I) activity.
IGFBP-3 is one of six IGFBPs that bind to and modulate the activity of IGFs. IGFBP-3 is a highly abundant serum protein and therefore affects the physiologic bioavailability of circulating IGF-I (11). In the pericellular environment IGFBP-3 is thought to be proapoptotic and to counteract the proliferative effects of IGF-I (12). Pericellular proteases cleave IGFBP-3, thus releasing IGF-I to bind to the type I IGF receptor (IGF-1R). For example, prostate specific antigen (PSA) is a metalloproteinase that cleaves IGFBP-3 to yield at least seven proteolytic fragments, some of which retain the ability to bind IGF-I, albeit with lower affinity than the intact protein (13–16). The interaction of IGFBP-3 with cells is more complex than suggested by its interactions with IGF-I. IGFBP-3 stimulates cells directly as demonstrated by the biological effects of IGFBP-3 mutant proteins that lack IGF-I binding (17). Interestingly, although IGFBP-3 expression was not identified in a high throughput expression analysis of Nkx3.1 gene targeted mice (8), IGFBP-3 was identified as a major target of down regulation in prostate cancer compared to nonmalignant prostate tissue (18). We now present data demonstrating a role for IGFBP-3 in growth suppression by NKX3.1. We propose that IGFBP-3 expression represents an important mechanistic link between the tumor suppression effects of NKX3.1 and the pro-survival and proliferative effects of IGF-I, a peptide growth factor that has been implicated in prostate carcinogenesis.
Materials and Methods
Expression Array Analysis
Total RNA from stable PC-3(NKX3.1) and PC-3(pcDNA3.1) cells was harvested using the RNeasy Miniprep kit (Qiagen, Valencia, CA). First strand cDNA synthesis from total RNA was carried out using the GeneChip® T7-Oligo(dT) primer kit (Affymetrix, Santa Clara, CA). Second strand cDNA synthesis was performed using the SuperScript™ Choice System (Invitrogen, Carlsbad, CA). The cDNA was then processed using the GeneChip® Sample Cleanup Module (Affymetrix). Amplification and biotin labeling of antisense cRNA was carried out using the BioArray High Efficiency RNA Transcript Labeling system (Affymetrix). Finally, the GeneChip® Sample Cleanup Module (Affymetrix) was utilized to cleanup the biotinylated cRNA before it was sent out for analysis on an Affymetrix U-133 array system.
Cell Culture and Reagents
The prostate cancer cell lines PC-3 and LNCaP, and the A172 human glioblastoma cell line were obtained from the American Type Culture Collection, Rockville, MD. PC-3 and A172 cell lines are grown in Modified IMEM (Invitrogen, Carlsbad, CA) containing 10% FBS. LNCaP cells are grown in Modified IMEM with phenol red (Invitrogen) containing 10% fetal bovine serum (FBS). The PC-3 cells stably expressing the NKX3.1 expression vector are continuously grown in Modified IMEM (Invitrogen) containing 10% FBS and 1.2mg/ml G418 (Invitrogen). LNCaP cells were serum starved overnight in IMEM supplemented with 5% charcoal charcoal-stripped calf serum (CCS) and treated with 10nM R1881 for 48hr before harvesting.
Plasmids and Transfection
Full length NKX3.1 was cloned into the mammalian expression vector, pcDNA3.1 (Invitrogen) as previously described (19). The PTEN expression vector, cloned into pcDNA3.1 was a kind gift from Charles Sawyers, Memorial Sloan-Kettering Cancer Center, NY, NY (20). Transient and stable transfections were carried out in 75 cm2 cell culture flasks (Corning Inc., Corning, NY). Briefly, PC-3 and LNCaP prostate cancer cells were grown to 40–60% confluence and 4μg of plasmid DNA was transfected into the cell lines using Lipofectamine and Plus reagents (Invitrogen) in Opti-MEM (Invitrogen). After a 4hr incubation, the media was replaced with IMEM containing 10% FBS for an additional 24 hours. The PC-3 clones that stably express NKX3.1 were derived by transfection. After 4hr incubation in transfection reagent, PC-3 cells were trypsinized and seeded at a 1:30 density in Falcon Integrid 20mm grid tissue culture dishes (Becton Dickinson, Franklin Lakes, NJ) in modified IMEM containing 10% FBS and 1.2mg/ml G418 (Invitrogen). The media was replaced every 4 days until colonies derived from a single cell could be seen with a light microscope. Single clone colonies were isolated with sterile cloning disks (Scienceware, Pequannock, NJ) soaked in 0.25% Trypsin-EDTA (Invitrogen) and grown to confluence in 6-well tissue culture dishes (Corning) for further study.
Western Blot Analysis
Cells were grown to 60–80% confluence and media was aspirated from the tissues culture dish. Immediately following media aspiration, lysis buffer was pipetted directly onto the cell monolayer and cells were scraped from the tissue culture flask. Cells were lysed with radio-immunoprecipitation assay (RIPA) buffer containing complete mini protease inhibitors (Roche, Nutley, NJ) and/or phosphatase inhibitors (Cell Signaling, Danvers, MA) followed by brief sonication to complete lysis. Sixty to ninety μg of total cell lysate was boiled in Novex® 2X Tris-glycine SDS sample buffer (Invitrogen) containing β-mercaptoethanol for six minutes and resolved on a 10–20% Tris-glycine SDS-PAGE gel (Invitrogen) Protein was then transferred onto a nitrocellulose membrane (Bio-Rad, Hercules, CA) and probed with primary antibody concentrations, as follows; β-Actin (Sigma, St. Louis, MO) 1:10,000; NKX3.1 (2) 1:2000; IGFBP-3 (sc-9028, Santa Cruz Biotechnology, Santa Cruz, CA) 1:8,000, AKT (#9272, Cell Signaling) 1:7500, phospho-AKT Thr308 (#9275, Cell Signaling) 1:7500, at 4°C overnight, followed by three washes in PBST. HRP conjugated Goat-anti-Rabbit and Goat-anti-Mouse (ImmunoPure® antibodies, Pierce Biotechnology, Rockford, IL) secondary antibodies in 1% milk or 1% BSA were applied for 1 hour at room temperature. Signal detection was performed with Super-Signal West Pico Chemiluminescent Substrate (Pierce Biotechnology).
Reverse Transcriptase PCR analysis
Total RNA was extracted using the RNeasy Mini Kit (Qiagen) and cells were homogenized using the Qiashredder (Qiagen) method. 125–250ng of RNA was added to the RT-PCR master mix from One-step RT-PCR kit (Qiagen) (includes 5x buffer, DNTPs, and Taq polymerase). The following primers were used in the RT-PCR reactions: β-Actin (Fwd 5′-GGC CAC GGC TGC TTC-3′ and Rev 5′-GTT GGC GTA CAG GTC TTT GC-3′); NKX3.1 (Fwd 5′-GCC GCA CGA GCA GCC AGA GAC A-3′ and Rev 5′-TTC AGG GCC GGC AAA GAG GAG TG-3′); IGFBP-3 (Fwd 5′-CGC CAG CTC CAG GAA ATG-3′ and Rev 5′-GCA TGC CCT TTC TTG ATG ATG-3′); IGFBP-4 (Fwd 5′-TTA GCC CAA GAG GTC TGA GC-3′ and Rev 5′-CTG TGC TTC AAG TCT TCC TTT G-3′); Lamin A/C (Fwd 5′-AAC TTC AGG ATG AGA TGC TGC G-3′ and Rev 5′-GTC CAG AAG CTC CTG GTA CTC GT-3′). RT-PCR was performed in a Techne Techgene PCR machine; 30 min. at 50°; 15 min. at 94°; 22–30 cycles of 30s–1 min. at 94°, 30s–1 min. at melting temperatures of 55°–65°, and 30s–1 min. at 72°; followed by 15 min. at 72°. Samples were stored on ice until and mixed with 10x Blue Juice gel loading buffer (Invitrogen) and run on a 1.5% agarose gel containing 0.1μg/ml ethidium bromide in TAE buffer. Gels were imaged on a luminometer and recorded using a Kodak 1D digital camera.
Real-time RT-PCR analysis of murine prostate RNA
Frozen anterior prostates from three individual mice of each of the following genotypes: Nkx3.1+/+, Nkx3.1+/−, and Nkx3.1−/− of animals both 4 and 12 months of age, were generously provided by Cory Abate-Shen, Columbia University, New York, NY (6). mRNA was extracted using Qiagen RNeasy mini kit (Qiagen Inc.). The Real-Time Quantitative PCR TaqMan Assays were performed on the ABI PRISM 7700 Sequence Detection System (SDS) equipment (Applied Biosystems, Foster City, California). The primers and probe were selected for igfbp-3 using Primer Express software (Applied Biosystems). The primer sequences were: forward 5′-GCAGGCAGCCTAAGCACC-3′, reverse 5′-CCTCCTCGGACTCACTGAT-3′. The probe sequence (TCCCCTCCCAACCTGCTCCAGG) was labeled at the 5′ end with the reporter molecule 6-carboxyfluorescein (FAM) and at the 3′ with the quencher BHQ-1. Amplification of commercially available endogenous VIC labeled control, Rodent-GAPDH (Applied Biosystems), was used to standardize the amount of sample DNA added. Dilutions of DNA from the cell line (LNCaP) were used to construct standard curves for the target gene and endogenous control. TaqMan Universal PCR Master Mix was combined with 100ng of sample DNA, 900nM final concentration of primers and 100nM final concentration of the probe. All samples were analyzed as replicates of 4 wells. Relative quantitation of the data from 7700 SDS was performed using SDS 2.1 Software (Applied Biosystems).
IGF-IR Activation and Signaling
Cells were plated in 100mm culture dishes and washed twice with 1x phosphate buffered saline (PBS) before being serum starved for 14–16 hours in Modified IMEM containing 1.2mg/ml G418 (Invitrogen). Cells were then washed once with PBS and treated for 3 minutes with 100pM IGF-I (a gift from Dr. J. Toretsky, Georgetown University, Washington, DC) or Long-R3-IGF-I (GroPep) in IMEM at 37°C. The media was immediately aspirated and cells were scraped from the flask and suspended in 2X Cell Lysis Buffer (Cell Signaling) containing phosphatase inhibitors and protease inhibitors by using Complete Mini tablets (Roche). Western blot analysis was completed as described above using anti-IGF-I Receptor β (#3027, Cell Signaling), anti-Phospho-IGF-I Receptor (Tyr1131) (#3021, Cell Signaling), anti-IRS-1 (06-248, Upstate), anti-IRS-1[pY612] (44-816G, Biosource), anti-PI3K p85 (#4257, Cell Signaling), and anti-phospho-PI3K [pY458] (#4228, Cell Signaling) primary antibodies. Bands were quantified by Scion Imager software and p-values were assessed from triplicate experiments by t-test analysis using Prism Graphpad software. [* indicates p-value of <0.05, ** indicates p-value of <0.005, *** indicates p-value of <0.001].
Cell Proliferation Assay
PC-3, PC-3(pcDNA3.1), and PC-3(NKX3.1) cells were seeded in triplicate in 96-well plates at a concentration of 4000 cells per well in IMEM containing 10% FBS (PC-3) or 10% FBS plus 1.2mg/ml G418 [PC-3(pcDNA3.1) and PC-3(NKX3.1)] and incubated for 24 hr at 37°C. At 24, 48, 72, and 96 hr after seeding, wells were trypsinized, suspended in IMEM, and immediately counted in a Beckman Coulter Z1 cell counter. Doubling times were calculated using Microsoft Excel and p-values were calculated by ANOVA.
Tumor Xenografts
Animal studies were carried out under the approved protocol AAAA-7422 as per Columbia University’s Institutional Animal Care and Use Committee guidelines and approval. Cell lines were grown to 80% confluence in IMEM + 10% FBS + 1.2mg/ml G418 in a Hyperflask (Corning) and trypsinized with 0.25% Trypsin-EDTA (Invitrogen). Cells were resuspended in IMEM containing 10% FBS to deactivate trypsin and washed twice with PBS. Cells were then counted and resuspended in PBS at a concentration of 3×107cells/ml. Cell suspensions (150μl) were injected into 5-week old female NCr-Nude mice (Taconic Farms, Hudson, NY) on their ventral surface and tumors were measured in two dimensions once a week. We performed 20 inoculations per cell line. All measurements were performed by one observer (E.M.). Once the tumors reached 500mm3 or if illness was observed, mice were sacrificed and tumors were dissected and stored in 10% buffered formalin for paraffin embedding or in RNAlater (Qiagen) at −80°C for western blot analysis.
Immunohistochemistry
Cells grown under tissue culture conditions were embedded in 1% agarose before sectioning and staining. Paraffin-embedding and sectioning of the tumor xenografts and agarose cell plugs was performed by the Molecular Pathology Shared Resource of the Herbert Irving Comprehensive Cancer Center at Columbia University. Slides were microwaved for 5 minutes and immersed in two washes of xylene, followed by successive washes in 100%, 90%, and 70% ethanol and followed by a 5 minute wash in PBS. Slides were immersed in 10mM citrate buffer at pH 6.0 and steamed in a Black and Decker vegetable steamer for 40min. Slides were fully cooled to room temperature and washed once with PBS before the blocking step, horse serum in PBS (1:66, Pierce) for 30 min. at room temperature. NKX3.1 primary antibody (1:500, Zymed) was applied for 1 hour at room temperature followed by biotin-anti-mouse secondary antibody for 30 min. (1:200, Vector Laboratories, Burlingame, CA). This was followed by application of Vectastain Elite ABC kit (Vector Laboratories) and Vector VIP substrate kit (Vector Laboratories). Methyl green (Vector Laboratories) was used as a nuclear counter stain.
siRNA knockdown of IGFBP-3
All siRNA oligonucleotides were purchased from Dharmacon, derived from sequences published by Stewart et al (21). An siRNA duplex directed against nucleotides 603-623 of IGFBP-3 mRNA (reference sequence NM_000598) (5′-AAU CAU CAU CAA GAA AGG GCA -3′), a mismatch siRNA sequence that differs from the IGFBP-3 siRNA oligonucleotide by one base pair (5′-AAU CAU CUA CAA GAA AGG GCA -3′), and the lamin A/C positive control sequence from Dharmacon (5′-GGU GGU GAC GAU GUG GGC U -3′). This single siRNA sequence was the only one of eleven siRNA sequences tested. The ten IGFBP-3 siRNA sequences that had no effect are shown in Supplementary Table 1. 2×105 cells were plated, in triplicate, in a 6-well plate in IMEM + 10%FBS + 1.2mg/ml G418 the night before the exposure to the oligonucleotides. Cells were transfected with 20uM of siRNA oligonucleotide using Lipofectamine (Invitrogen) in Opti-MEM (Invitrogen). The media was changed back to IMEM + 10%FBS + 1.2mg/ml G418 3.5 hr post transfection and knockdown of IGFBP-3 mRNA was assayed at 24 and 96 hr by RT-PCR. In the cell proliferation assay, knockdown was allowed to proceed for 24 hr before the initial cell count. Assessment of IGF-IR activation and downstream signaling 1×106 cells were plated in IMEM + 10%FBS + 1.2mg/ml G418 the night before exposure to the oligonucleotides. Cells pretreated with oligonucleotide for up to 16 hr were washed twice with PBS and serum starved for 14–16 hours. Cells were further washed with PBS and treated with 100pM IGF-I for 3 minutes at 37°C.
Results
Effect of NKX3.1 on gene expression in PC-3 prostate cancer cells
To identify genes whose expression is affected by NKX3.1 we initially generated independently transfected clones of PC-3 prostate cancer cells chosen because PC-3 cells express essentially no NKX3.1 protein and express NKX3.1 mRNA at about 1/250 the level seen in LNCaP cells (our unpublished data). Two derivative PC-3 cell lines transfected with the pcDNA3.1 empty expression vector and two derivative NKX3.1 expressing clones were analyzed using the Affymetrix U-133 expression arrays. Approximately 99% of the signals obtained were regulated concordantly between the two control clones and the two NKX3.1-expressing clones. The cDNAs that were discordant between the two control clones or between the two NKX3.1-expressing clones were eliminated from the analysis and the concordant clone expression levels were averaged across the PC-3 control cells and the PC-3(NKX3.1) cells. Then the two mean expression levels were compared. Using a cut-off of 1.4-fold up- or down- regulation, 984 transcripts were identified (Supplementary Table 2). Two separate IGFBP-3 probes were activated 9.22- and 10.23-fold in PC-3 cells expressing NKX3.1 compared to PC-3 control transfectants. The IGFBP-3 message difference between control and NKX3.1-expressing cells were the 6th and 9th highest increase of the 508 up regulated transcripts.
Effect of NKX3.1 on IGFBP-3 expression in vitro
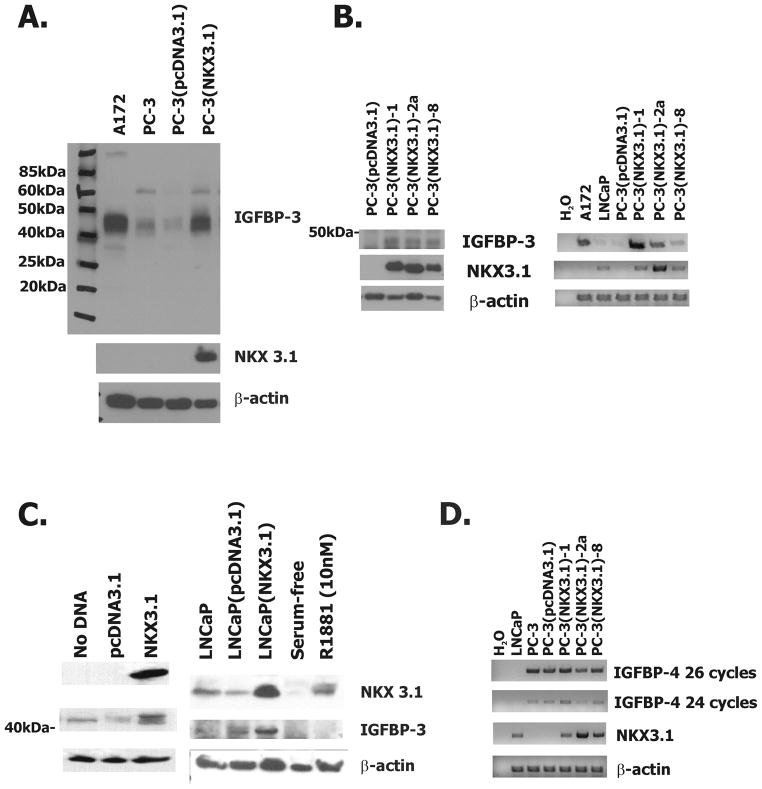
To validate the effect of NKX3.1 on IGFBP-3 expression and determine whether protein expression was also affected we performed western blotting on a PC-3 clone expressing NKX3.1. NKX3.1 expression was clearly seen in transfected cells and IGFBP-3 expression was ~10-fold activated compared to control transfected cells (Figure 1A). In a separate transfection experiment additional PC-3 clones expressing NKX3.1 were isolated. A marked increase was observed for clone 1 in Fig 1a; similar but smaller increases were observed in protein and mRNA in two other clones (Figures 1B).
Figure 1.
NKX3.1 up regulates the expression of IGFBP-3 in prostate cancer cell lines.
A, Western blot analysis of cell extracts from stably expressing PC-3(pcDNA3.1) and PC-3(NKX3.1)-1 cell clones that have been analyzed by expression array. B, On the left is western blot analysis of extracts from additional PC-3 clones expressing NKX3.1. One the right is RT-PCR analysis of PC-3(pcDNA3.1) and PC-3(NKX3.1) clones for NKX3.1 and IGFBP-3 expression. A172 cell extract is a positive control for IGFBP-3 expression and LNCaP cell extract is used as a positive control for NKX3.1 expression. C, On the left is western blot analysis of extracts from PC-3 cells transiently transfected with an NKX3.1 expression vector. On the right is western blot analysis of extracts from LNCaP cells transiently transfected with an NKX3.1 expression vector or serum starved in 5% CCS supplemented media overnight and treated with 10nM R1881. D, RT-PCR analysis of IGFBP-4 mRNA expression in PC-3(pcDNA3.1) and PC-3(NKX3.1) clones.
To demonstrate that the apparent induction of IGFBP-3 expression by NKX3.1 was not an adaptation of the cells during clonal selection we performed western blotting on PC-3 cells transiently transfected with NKX3.1 and again observed increase expression of IGFBP-3 (Figure 1C). The relationship between expression of NKX3.1 and expression of IGFBP-3 was not exclusive to PC-3 cells as increasing NKX3.1 expression in LNCaP cells by transfection of an NKX3.1 expression vector also increased expression of IGFBP-3 (Figure 1C). Note that exposure to the synthetic androgen R1881 activated NKX3.1 expression, but not IGFBP-3. This may be due to proliferative signals of R1881 that interfere with the activation of IGFBP-3 by NKX3.1. Alternatively, LNCaP cells may have down regulated expression of IGFBP-3 as an adaptation to growth with continuous expression of NKX3.1. However, it should be noted that differences in the media in which PC-3 and LNCaP cells were cultured may have also contribute to the differences in IGFBP-3 expression in these two cell lines. We also noted a single probe for IGFBP-4 was activated 4-fold by NKX3.1 in the expression array analysis. However, increased expression of IGFBP-4 mRNA was not seen in multiple other PC-3 clones engineered to express NKX3.1 (Figure 1D). LNCaP cells have been shown not to express IGFBP-4 and are used as a negative control in this western blot while PC-3 cells have been shown to express IGFBP-4 positive (22). No other IGF binding protein mRNAs were found to be activated in the expression array.
Expression of IGFBP-3 in prostate tissues correlates with expression of NKX3.1
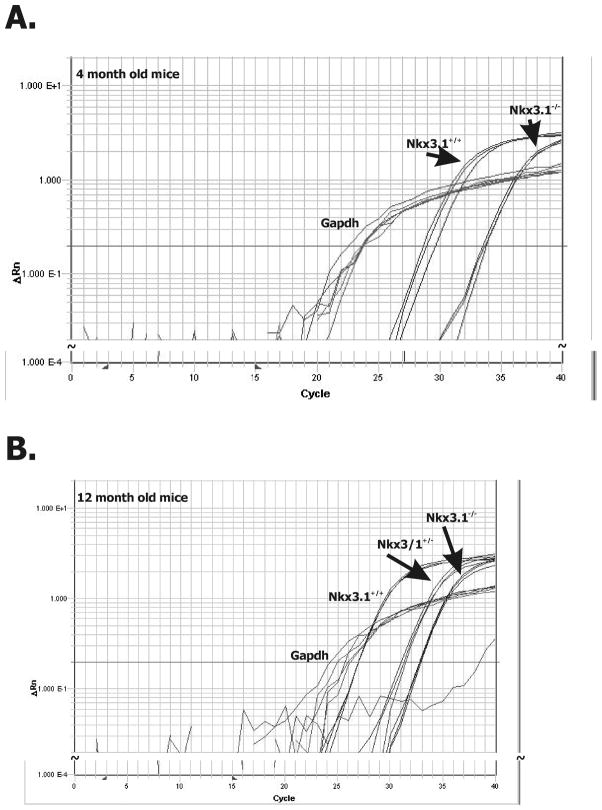
To determine whether the correlation of NKX3.1 and IGFBP-3 expression could also be observed in vivo, we analyzed prostate mRNA in Nkx3.1 gene targeted mice. We have published quantitation of Nkx3.1 protein in intact, Nkx3.1+/−, and Nkx3.1−/− mice, showing that levels of Nkx3.1 protein correlated with gene copy number in Nkx3.1-targeted mice (3). We performed real-time RT-PCR of RNA extracted from these mice. Data in Figure 2 show that Igfbp-3 mRNA expression was related to Nkx3.1 gene copy number in murine prostates. In each reaction Gapdh was used as a control and was invariant between the different strains. We found that in murine prostates Igfbp-3 expression levels correlated with Nkx3.1 copy number and thus with Nkx3.1 expression (Figure 2).
Figure 2.
NKX3.1 and IGFBP-3 expression in mouse prostate.
Real-time RT-PCR analysis of mRNA extracts of prostatic tissue of Nkx3.1 gene targeted mice at (A) 4 and (B) 12 months of age. Anterior prostates were analyzed from three separate animals of each genotype at each time point. Gapdh expression was used as a loading control. Each one of the multiple assay lines in each group represent the results of a single tissue sample. The results correlated with Nkx3.1 genotype.
Even though we had demonstrated an increase in IGFBP-3 mRNA as a result of NKX3.1 expression in cultured human cells, we were unable to show an effect of NKX3.1 on luciferase reporter constructs that contained regions from the IGFBP-3 gene promoter (data not shown) (23). This is not entirely surprising since NKX3.1 by itself does not contribute to the formation of a transcriptional complex and suppresses transcription from reporter constructs engineered with the NKX3.1 cognate DNA binding domain (19).
The effect of NKX3.1 on IGF-I signaling
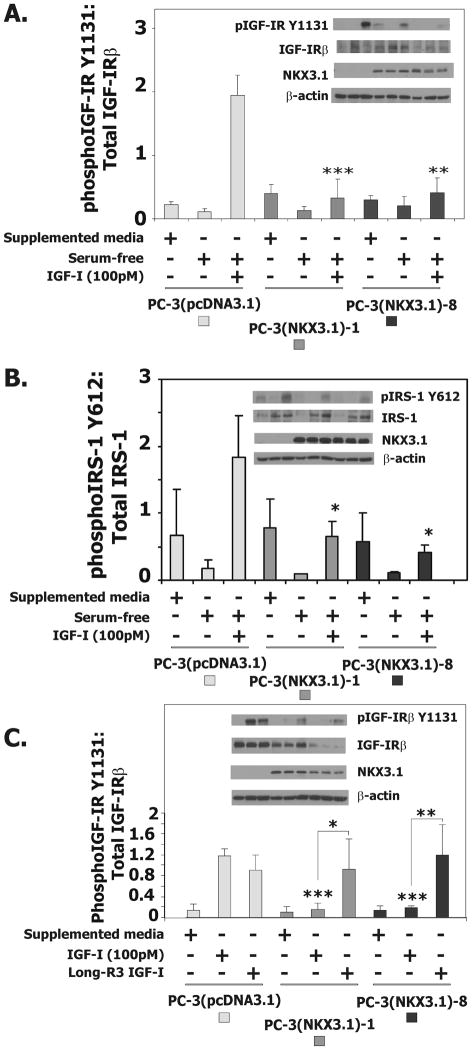
To determine whether the induction of IGFBP-3 expression by NKX3.1 affected IGF-I signaling we examined the response of the IGF-IR to IGF-I in derivative PC-3 cells. In the presence of serum-free medium IGF-I induced IGF-IR phosphorylation at tyrosine 1131 within three minutes of exposure to the ligand (24). Phosphorylation of IGF-IR was diminished in cells expressing NKX3.1 (Figure 3A). The effect of NKX3.1 on IGF-IR activation also could be seen on downstream signaling targets. IGF-I-induced phosphorylation of IRS-1, a target of both the insulin receptor and IGF-IR, was diminished in cells expressing NKX3.1 (Figure 3B). PC-3 derivative cells were also treated with the long-R3-IGF-I that has minimal binding to IGFBP-3 (25, 26). Long-R3-IGF-I activated phosphorylation of IGF-IR equally well in derivative PC-3 cells regardless of NKX3.1 expression (Figure 3C). This result is consistent with the notion that NKX3.1 mediates inhibition of IGF-I signaling via increased expression of IGFBP-3.
Figure 3.
NKX3.1 expression inhibits the IGF-1-mediated phosphorylation of the IGF-1R and IRS-1 in PC-3 cells. A, Western blot analysis of extracts from PC-3(pcDNA3.1), PC-3(NKX3.1)-1 and PC-3(NKX3.1)-8 stable cell clones serum starved for 16 hours and treated with 100pM IGF-I. The histogram of IGF-IR activation is based upon three separate experiments. B, Western blot analysis of extracts from PC-3(pcDNA3.1), PC-3(NKX3.1)-1 and PC-3(NKX3.1)-8 clones serum starved for 16 hours and treated with 100pM IGF-I. The histogram of IRS-1 activation is based upon three separate experiments. C, Western blot analysis of extracts from PC-3(pcDNA3.1), PC-3(NKX3.1)-1, and PC-3(NKX3.1)-8 clones serum starved for 16 hours and treated with 100pM IGF-1 or Long-R3-IGF-I. The histogram of IGF-IR activation is based upon three separate experiments. Statistical comparisons are indicated by asterisks as explained in Materials and Methods. Comparisons are versus PC-3(pcDNA3.1) treated with IGF-I unless otherwise indicate by brackets.
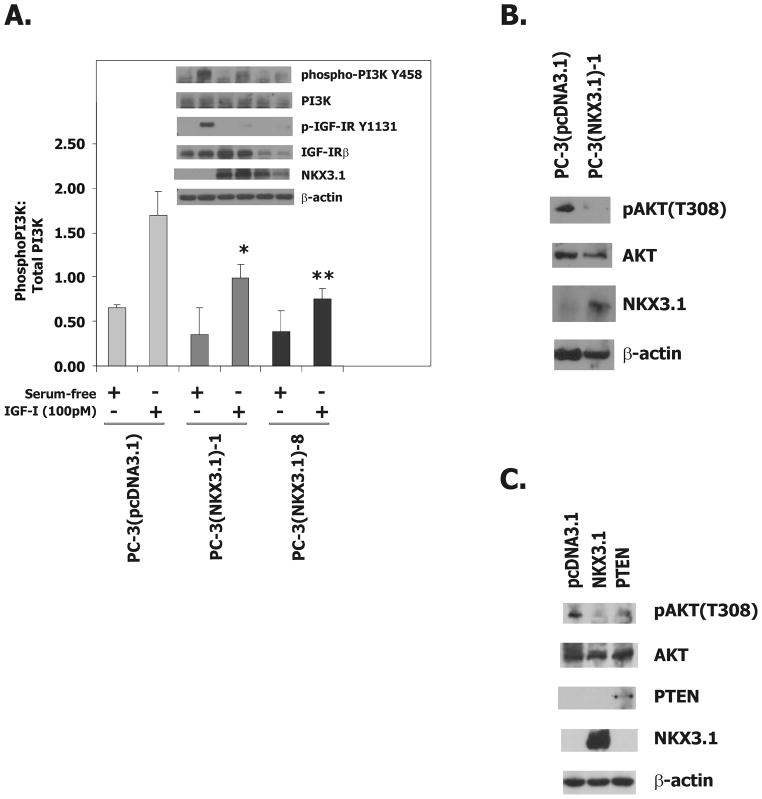
We examined the effect of NKX3.1 expression on the phosphorylation of PI3 kinase and its downstream target AKT. PI3 kinase phosphorylation was diminished to some degree by expression of NKX3.1 in two PC-3(NKX3.1) clones (Figure 4A). AKT phosphorylation was also decreased in a PC-3(NKX3.1) clone grown in serum-supplemented media which contains IGF-1 from the fetal bovine serum (Figure 4B). We compared the effect of NKX3.1 expression to phosphatase and tensin homolog (PTEN) expression in transient transfection of PC-3 cells and saw comparable degrees of reduction in p-AKT (Figure 4C).
Figure 4.
NKX3.1 attenuates IGF-1R downstream signaling.
A, Western blot analysis of extracts from PC-3(pcDNA3.1), PC-3(NKX3.1)-1, and PC-3(NKX3.1)-8 clones serum starved for 16 hours and treated with 100pM IGF-I for 3 minutes. The histogram of PI-3K activation is based upon three separate experiments. Statistical comparisons are indicated by asterisks. Comparisons are versus PC-3(pcDNA3.1) treated with IGF-I. B, Western blot analysis of cell extracts from PC-3(pcDNA3.1) and PC-3(NKX3.1)-1 cells grown in media containing 10% FBS. C, Western blot analysis of cell extracts of PC-3 cells transiently transfected with either a NKX3.1 or a PTEN expression vector.
Growth suppression by NKX3.1 is mediated by IGFBP-3
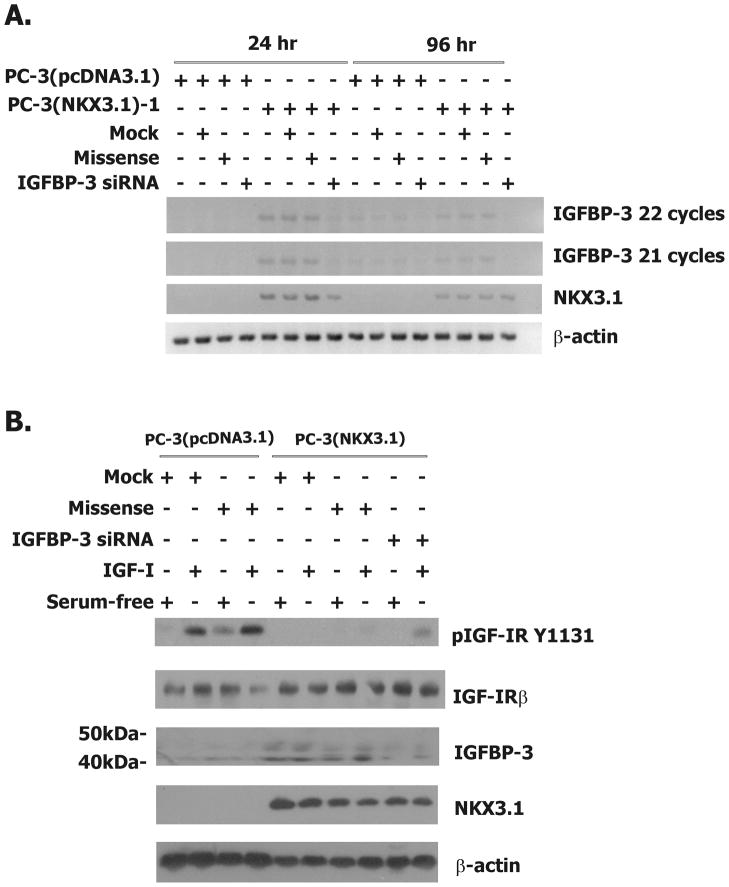
In vitro growth of the parental and derivative PC-3 cells was assessed by cell counting over 96 hours. As shown in the upper section of Table 1, NKX3.1 expression decreased cell proliferation in three independent clones. The doubling times for PC-3 cells expressing NKX3.1 ranged from 25–60% above the doubling times for control cells. To determine whether this effect of NKX3.1 expression on cell proliferation was due to IGFBP-3 expression we performed growth experiments in the presence of an siRNA oligonucleotide and control oligonucleotides for IGFBP-3 knockdown (21). The IGFBP-3 siRNA treatment decreased IGFBP-3 expression at both 24 and 96 hours after cells were exposed to the oligonucleotide (Figure 5A). Growth curves of control and NKX3.1-expressing PC-3 cells were done in the presence of transfection reagent, missense oligonucleotide, and IGFBP-3 siRNA. Only the IGFBP-3 siRNA restored PC-3 proliferative rate to the level of the controls (Table 1, lower section). Thus IGFBP-3 mediates, at least in part, in vitro growth suppression by NKX3.1. Consistent with this finding, the IGFBP-3 siRNA reversed the suppression of IGF-IR phosphorylation induced by NKX3.1 (Figure 5B). IGFBP-3 knockdown was accomplished with a single IGFBP-3 siRNA oligonucleotide as described in Materials and Methods. Of the eleven sequences tested only one induced substantial IGFBP-3 knockdown. This one effective siRNA sequence has no identifiable homology with sequences in other genes, as confirmed by a BLAST search against the entire human genome sequence.
Table 1.
Effect of IGFBP-3 Knock Down on Cell Proliferation
| Cell Line | Treatment | Doubling Time (hr) | p-value |
|---|---|---|---|
| PC-3 | None | 25.34 ± 1.91 | NS |
| PC-3(pcDNA3.1) | None | 24.26 ± 2.05 | reference |
| PC-3(NKX3.1)-1 | None | 32.24 ± 5.91 | 0.0037 |
| PC-3(NKX3.1)-2a | None | 38.88 ± 7.82 | 0.0002 |
| PC-3(NKX3.1)-8 | None | 30.24 ± 2.55 | 0.002 |
| PC-3(pcDNA3.1) | None | 23.6 ± 2.52 | reference |
| PC-3(pcDNA3.1) | Mock | 25.5 ± 1.23 | NS |
| PC-3(pcDNA3.1) | Missense oligo | 24.4 ± 1.86 | NS |
| PC-3(pcDNA3.1) | IGFBP-3 siRNA oligo | 26.4 ± 1.40 | NS |
| PC-3(NKX3.1)-1 | None | 32.9 ± 1.33 | 0.0092 |
| PC-3(NKX3.1)-1 | Mock | 31.7 ± 2.49 | 0.0013 |
| PC-3(NKX3.1)-1 | Missense oligo | 32.0 ± 3.12 | 0.0022 |
| PC-3(NKX3.1)-1 | IGFBP-3 siRNA oligo | 24.9 ± 2.56 | NS |
The cell doubling time, in hours, of parental PC-3 cells, PC-3(pcDNA3.1) cells, and PC-3(NKX3.1) clones was assayed by cell counting. Cells were counted at 24, 48, 72, and 96 hours post-seeding and doubling times were calculated. In the lower section of the table PC-3(pcDNA3.1) and PC-3(NKX3.1)-1 cells were treated with transfection reagent alone, the missense siRNA oligonucleotide, and the IGFBP-3 siRNA oligonucleotide for 24 hours before the first cell count was taken. p-values were calculated in comparison to the PC-3(pcDNA3.1) cell doubling time, using ANOVA. NS = not significant.
Figure 5.
IGFBP-3 knock down in PC-3(NKX3.1) cells.
A, RT-PCR analysis of mRNA from PC-3(pcDNA3.1) and PC-3(NKX3.1)-1 cells that were treated with transfection reagent alone, missense oligo, or the IGFBP-3 siRNA oligo at 24 and 96 hours post-transfection. B. Western blot analysis of extracts from PC-3(pcDNA3.1) and PC-3(NKX3.1)-1 cells treated with transfection reagent alone, missense oligo, or the IGFBP-3 siRNA oligo for 96 hours, after which they were serum starved for 16 hours and treated with 100pM IGF-I.
We also performed xenograft experiments where PC-3(NKX3.1) clones -1, -2, and -8 and PC-3(cDNA3.1) cells were inoculated into female NCr/nu mice. In every instance we observed tumor growth of derivative PC-3 cells (Supplementary Figure 1). Each tumor type had lost expression in vivo of the NKX3.1 transgene as shown by immunohistochemical analysis. Thus there was a selection in xenografts for loss of NKX3.1 expression preventing us from observing any growth suppressive effects of NKX3.1 on PC-3 cells in vivo.
Discussion
NKX3.1 is important for prostate epithelial cell development, growth control, and differentiation (6, 27). Murine Nkx3.1 is haploinsufficient and loss of a single allele manifests a phenotype similar to homozygous deletion, but with longer latency (6). In early human prostate cancer we have found that NKX3.1 expression is down regulated over a broad range, suggesting a complex effect on the development of human prostate cancer (3). It is important to define the pathways of tumorigenesis that are affected by NKX3.1. We argue here expression of IGFBP-3 is downstream of NKX3.1 and speculate that IGFBP-3 regulates IGF-I action in prostate epithelial cells.
IGFs are peptide growth factors that bind to the IGF receptor-I (IGFR-I) to regulate cell growth, differentiation, and apoptosis (12). IGFs are present in abundance in the circulation and may exert systemic effects and effects locally on cells. Circulating IGF-I is bound mainly to IGFBP-3, one of the most abundant serum proteins (28). Although IGFBP-3 can inhibit the interaction of IGF-I with its receptor at the cellular level, serum IGFBP-3 serves to stabilize circulating IGF-I (29). Serum levels of both proteins vary with age, nutrition, and hormonal status (30). The interaction of serum IGF-I and IGFBP-3 and prostate cancer risk has been studied by a number of investigators (29). The majority of studies have found an association between higher IGF-I levels and prostate cancer (31–39). Some investigators have not been able to confirm these findings (40–42).
NKX3.1 haploinsufficiency affects cell transformation, at least in part, by downstream effects on transcriptional targets. NKX3.1 binds to DNA and suppresses expression of genes downstream from cognate DNA binding sites (19). We have yet to identify a promoter that is transcriptionally activated by direct binding of NXK3.1 to its cognate DNA –TAAGTA- sequence. In fact, our experiments with reporter constructs containing NKX3.1 binding DNA suggested that NKX3.1 alone cannot initiate assembly of a transcriptional complex (19). NKX3.1 is known to interact with other transcription factors such as serum response factor (SRF) and serves as a synergistic coactivator of promoters with serum response elements such as smooth muscle γ-actin (43). Therefore the effect of NKX3.1 on gene expression is complex and is likely mediated by a number of cofactors. Because other NK homeodomain protein family members like Nkx2.5 interact with SRF, the physical interaction of NKX3.1 and SRF has been studied as a model for transcription factor interactions of NKX3.1. SRF is a widely expressed transcription factor involved in orchestrating disparate programs of gene expression linked to muscle differentiation and cellular growth (44). It is likely that different targets of transcriptional activation are affected by the interaction of NKX3.1 with several different transcription factors.
We have shown an indirect link between NKX3.1 and transcription of IGFBP-3 mRNA. In cultured cells with NKX3.1 over expression we have shown a mechanistic link between NKX3.1, IGFBP-3 expression, IGF-1R activation, and cell proliferation. IGF1-R signaling is complex and affected by many factors that regulate IGF-1 availability and intracellular signaling downstream from the IGF-1R. Therefore, the interaction between NKX3.1 and IGFBP-3 expression in vivo is likely to be part a more complex system regulating the effect of IGF-1 on prostate epithelial cells. The IGFBPs have functions that can compensate for one another making it difficult to determine specific functions of a single IGF binding protein by studies of gene-targeted mice (45, 46). Whether the same compensatory activation of IGF binding proteins occurs in prostate epithelial cell that have reduced NKX3.1 expression was not determined.
Down regulation of NKX3.1 protein per se is sufficient to predispose cells to malignant transformation. In addition we have described a family in which hereditary prostate cancer cosegregated with a T164A missense mutation in the NKX3.1 homeodomain that reduced DNA binding by 95% (47). Haploinsufficiency is a reflection of the dominant nature of regulation by NK family members. Both missense and truncation mutations in NKX2.5 are autosomal dominant determinants of congenital cardiac abnormalities (48, 49). Similarly, mutations in NKX2.1/TTF cause pulmonary and thyroid developmental abnormalities (50). Paradoxically, NKX2.1 is amplified in a subset of lung cancers and NKX2.1 over expression contributes to cell transformation and oncogenesis (51). We argue that tumor suppression by NKX3.1 is exerted in a relative manner by modulation of down stream targets to different degrees. Our finding that IGFBP-3 expression in human prostate cancer cells correlates quantitatively with NKX3.1 expression levels is reminiscent of findings that Nkx3.1 gene dosage determines the degree of transcriptional effects in gene targeted mice (8). Further studies will identify additional NKX3.1 targets and elucidate their role in prostate cancer suppression and, perhaps, prevention.
Supplementary Material
Acknowledgments
Supported by NIEHS grant ES09888 to EPG and by DOD grants W81XWH-07-1-0263 to EPG and PC-05-0590 to EM
Contributor Information
Erin Muhlbradt, Email: eem2126@columbia.edu.
Ekaterina Asatiani, Email: easatia@yahoo.com.
Elizabeth Ortner, Email: liz.ortner@yahoo.com.
Antai Wang, Email: aw94@georgetown.edu.
Reference List
- 1.Swalwell JI, Vocke CD, Yang Y, et al. Determination of a minimal deletion interval on chromosome band 8p21 in sporadic prostate cancer. Genes Chromosomes Cancer. 2002;33:201–5. doi: 10.1002/gcc.10015. [DOI] [PubMed] [Google Scholar]
- 2.Bowen C, Bubendorf L, Voeller HJ, et al. Loss of NKX3. 1 expression in human prostate cancers correlates with tumor progression. Cancer Res. 2000;60:6111–5. [PubMed] [Google Scholar]
- 3.Asatiani E, Huang WX, Wang A, et al. Deletion, methylation, and expression of the NKX3. 1 suppressor gene in primary human prostate cancer. Cancer Res. 2005;65:1164–73. doi: 10.1158/0008-5472.CAN-04-2688. [DOI] [PubMed] [Google Scholar]
- 4.Voeller HJ, Augustus M, Madlike V, et al. Coding region of NKX3. 1, prostate-specific homeobox gene on 8p21, is not mutated in human prostate cancers. Cancer Res. 1997;57:4455–9. [PubMed] [Google Scholar]
- 5.Ornstein DK, Cinquanta M, Weiler S, et al. Expression studies and mutational analysis of the androgen regulated homeobox gene nkx3. 1 in benign and malignant prostate epithelium. J Urol. 2001;165:1329–34. [PubMed] [Google Scholar]
- 6.Bhatia-Gaur R, Donjacour AA, Sciavolino PJ, et al. Roles for Nkx3. 1 in prostate development and cancer. Genes and Development. 1999;13:966–77. doi: 10.1101/gad.13.8.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim MJ, Cardiff RD, Desai N, et al. Cooperativity of Nkx3. 1 and Pten loss of function in a mouse model of prostate carcinogenesis. Proc Natl Acad Sci U S A. 2002;99:2884–9. doi: 10.1073/pnas.042688999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Magee JA, Abdulkadir SA, Milbrandt J. Haploinsufficiency at the Nkx3.1 locus. A paradigm for stochastic, dosage-sensitive gene regulation during tumor initiation. Cancer Cell. 2003;3:273–83. doi: 10.1016/s1535-6108(03)00047-3. [DOI] [PubMed] [Google Scholar]
- 9.Bethel CR, Faith D, Li X, et al. Decreased NKX3. 1 protein expression in focal prostatic atrophy, prostatic intraepithelial neoplasia, and adenocarcinoma: association with gleason score and chromosome 8p deletion. Cancer Res. 2006;66:10683–90. doi: 10.1158/0008-5472.CAN-06-0963. [DOI] [PubMed] [Google Scholar]
- 10.Markowski MC, Bowen C, Gelmann EP. Inflammatory cytokines induce phosphorylation and ubiquitination of prostate suppressor protein NKX3. 1. Cancer Res. 2008;68:6896–901. doi: 10.1158/0008-5472.CAN-08-0578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Firth SM, Baxter RC. Cellular actions of the insulin-like growth factor binding proteins. Endocr Rev. 2002;23:824–54. doi: 10.1210/er.2001-0033. [DOI] [PubMed] [Google Scholar]
- 12.Khandwala HM, McCutcheon IE, Flyvbjerg A, Friend KE. The effects of insulin-like growth factors on tumorigenesis and neoplastic growth. Endocr Rev. 2000;21:215–44. doi: 10.1210/edrv.21.3.0399. [DOI] [PubMed] [Google Scholar]
- 13.Cohen P, Graves HC, Peehl DM, et al. Prostate-specific antigen (PSA) is an insulin-like growth factor binding protein-3 protease found in seminal plasma. J Clin Endocrinol Metab. 1992;75:1046–53. doi: 10.1210/jcem.75.4.1383255. [DOI] [PubMed] [Google Scholar]
- 14.Koistinen H, Paju A, Koistinen R, et al. Prostate-specific antigen and other prostate-derived proteases cleave IGFBP-3, but prostate cancer is not associated with proteolytically cleaved circulating IGFBP-3. Prostate. 2002;50:112–8. doi: 10.1002/pros.10039. [DOI] [PubMed] [Google Scholar]
- 15.Fielder PJ, Rosenfeld RG, Graves HC, et al. Biochemical analysis of prostate specific antigen-proteolyzed insulin-like growth factor binding protein-3. Growth Regul. 1994;4:164–72. [PubMed] [Google Scholar]
- 16.Cohen P, Peehl DM, Graves HC, Rosenfeld RG. Biological effects of prostate specific antigen as an insulin-like growth factor binding protein-3 protease. J Endocrinol. 1994;142:407–15. doi: 10.1677/joe.0.1420407. [DOI] [PubMed] [Google Scholar]
- 17.Hong J, Zhang G, Dong F, Rechler MM. Insulin-like growth factor (IGF)-binding protein-3 mutants that do not bind IGF-I or IGF-II stimulate apoptosis in human prostate cancer cells. J Biol Chem. 2002;277:10489–97. doi: 10.1074/jbc.M109604200. [DOI] [PubMed] [Google Scholar]
- 18.Rhodes DR, Barrette TR, Rubin MA, Ghosh D, Chinnaiyan AM. Meta-analysis of microarrays: interstudy validation of gene expression profiles reveals pathway dysregulation in prostate cancer. Cancer Res. 2002;62:4427–33. [PubMed] [Google Scholar]
- 19.Steadman DJ, Giuffrida D, Gelmann EP. DNA-binding sequence of the human prostate-specific homeodomain protein NKX3. 1. Nucleic Acids Res. 2000;28:2389–95. doi: 10.1093/nar/28.12.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu X, Senechal K, Neshat MS, Whang YE, Sawyers CL. The PTEN/MMAC1 tumor suppressor phosphatase functions as a negative regulator of the phosphoinositide 3-kinase/Akt pathway. Proc Natl Acad Sci U S A. 1998;95:15587–91. doi: 10.1073/pnas.95.26.15587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stewart LV, Weigel NL. Role of insulin-like growth factor binding proteins in 1alpha,25-dihydroxyvitamin D(3)-induced growth inhibition of human prostate cancer cells. Prostate. 2005;64:9–19. doi: 10.1002/pros.20212. [DOI] [PubMed] [Google Scholar]
- 22.Kimura G, Kasuya J, Giannini S, et al. Insulin-like growth factor (IGF) system components in human prostatic cancer cell-lines: LNCaP, DU145, and PC-3 cells. Int J Urol. 1996;3:39–46. doi: 10.1111/j.1442-2042.1996.tb00628.x. [DOI] [PubMed] [Google Scholar]
- 23.Walker GE, Wilson EM, Powell D, Oh Y. Butyrate, a histone deacetylase inhibitor, activates the human IGF binding protein-3 promoter in breast cancer cells: molecular mechanism involves an Sp1/Sp3 multiprotein complex. Endocrinology. 2001;142:3817–27. doi: 10.1210/endo.142.9.8380. [DOI] [PubMed] [Google Scholar]
- 24.Hernandez-Sanchez C, Blakesley V, Kalebic T, Helman L, LeRoith D. The role of the tyrosine kinase domain of the insulin-like growth factor-I receptor in intracellular signaling, cellular proliferation, and tumorigenesis. J Biol Chem. 1995;270:29176–81. doi: 10.1074/jbc.270.49.29176. [DOI] [PubMed] [Google Scholar]
- 25.Francis GL, Ross M, Ballard FJ, et al. Novel recombinant fusion protein analogues of insulin-like growth factor (IGF)-I indicate the relative importance of IGF-binding protein and receptor binding for enhanced biological potency. J Mol Endocrinol. 1992;8:213–23. doi: 10.1677/jme.0.0080213. [DOI] [PubMed] [Google Scholar]
- 26.King R, Wells JR, Krieg P, et al. Production and characterization of recombinant insulin-like growth factor-I (IGF-I) and potent analogues of IGF-I, with Gly or Arg substituted for Glu3, following their expression in Escherichia coli as fusion proteins. J Mol Endocrinol. 1992;8:29–41. doi: 10.1677/jme.0.0080029. [DOI] [PubMed] [Google Scholar]
- 27.Abdulkadir SA, Magee JA, Peters TJ, et al. Conditional loss of Nkx3. 1 in adult mice induces prostatic intraepithelial neoplasia. Mol Cell Biol. 2002;22:1495–503. doi: 10.1128/mcb.22.5.1495-1503.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones JI, Clemmons DR. Insulin-like growth factors and their binding proteins: biological actions. Endocr Rev. 1995;16:3–34. doi: 10.1210/edrv-16-1-3. [DOI] [PubMed] [Google Scholar]
- 29.Pollak MN, Schernhammer ES, Hankinson SE. Insulin-like growth factors and neoplasia. Nat Rev Cancer. 2004;4:505–18. doi: 10.1038/nrc1387. [DOI] [PubMed] [Google Scholar]
- 30.Rajaram S, Baylink DJ, Mohan S. Insulin-like growth factor-binding proteins in serum and other biological fluids: regulation and functions. Endocr Rev. 1997;18:801–31. doi: 10.1210/edrv.18.6.0321. [DOI] [PubMed] [Google Scholar]
- 31.Harman SM, Metter EJ, Blackman MR, Landis PK, Carter HB. Serum levels of insulin-like growth factor I (IGF-I), IGF-II, IGF-binding protein-3, and prostate-specific antigen as predictors of clinical prostate cancer. J Clin Endocrinol Metab. 2000;85:4258–65. doi: 10.1210/jcem.85.11.6990. [DOI] [PubMed] [Google Scholar]
- 32.Chan JM, Stampfer MJ, Ma J, et al. Insulin-like growth factor-I (IGF-I) and IGF binding protein-3 as predictors of advanced-stage prostate cancer. J Natl Cancer Inst. 2002;94:1099–106. doi: 10.1093/jnci/94.14.1099. [DOI] [PubMed] [Google Scholar]
- 33.Chokkalingam AP, Pollak M, Fillmore CM, et al. Insulin-like growth factors and prostate cancer: a population-based case-control study in China. Cancer Epidemiol Biomarkers Prev. 2001;10:421–7. [PubMed] [Google Scholar]
- 34.Oliver SE, Gunnell D, Donovan J, et al. Screen-detected prostate cancer and the insulin-like growth factor axis: results of a population-based case-control study. Int J Cancer. 2004;108:887–92. doi: 10.1002/ijc.11631. [DOI] [PubMed] [Google Scholar]
- 35.Platz EA, Pollak MN, Leitzmann MF, et al. Plasma insulin-like growth factor-1 and binding protein-3 and subsequent risk of prostate cancer in the PSA era. Cancer Causes Control. 2005;16:255–62. doi: 10.1007/s10552-004-3484-8. [DOI] [PubMed] [Google Scholar]
- 36.Stattin P, Bylund A, Rinaldi S, et al. Plasma insulin-like growth factor-I, insulin-like growth factor-binding proteins, and prostate cancer risk: a prospective study. J Natl Cancer Inst. 2000;92:1910–7. doi: 10.1093/jnci/92.23.1910. [DOI] [PubMed] [Google Scholar]
- 37.Stattin P, Rinaldi S, Biessy C, et al. High levels of circulating insulin-like growth factor-I increase prostate cancer risk: a prospective study in a population-based nonscreened cohort. J Clin Oncol. 2004;22:3104–12. doi: 10.1200/JCO.2004.10.105. [DOI] [PubMed] [Google Scholar]
- 38.Li L, Yu H, Schumacher F, Casey G, Witte JS. Relation of serum insulin-like growth factor-I (IGF-I) and IGF binding protein-3 to risk of prostate cancer (United States) Cancer Causes Control. 2003;14:721–6. doi: 10.1023/a:1026383824791. [DOI] [PubMed] [Google Scholar]
- 39.Wolk A, Mantzoros CS, Andersson SO, et al. Insulin-like growth factor 1 and prostate cancer risk: a population-based, case-control study. J Natl Cancer Inst. 1998;90:911–5. doi: 10.1093/jnci/90.12.911. [DOI] [PubMed] [Google Scholar]
- 40.Janssen JA, Wildhagen MF, Ito K, et al. Circulating free insulin-like growth factor (IGF)-I, total IGF-I, and IGF binding protein-3 levels do not predict the future risk to develop prostate cancer: results of a case-control study involving 201 patients within a population-based screening with a 4-year interval. J Clin Endocrinol Metab. 2004;89:4391–6. doi: 10.1210/jc.2004-0232. [DOI] [PubMed] [Google Scholar]
- 41.Woodson K, Tangrea JA, Pollak M, et al. Serum insulin-like growth factor I: tumor marker or etiologic factor? A prospective study of prostate cancer among Finnish men. Cancer Res. 2003;63:3991–4. [PubMed] [Google Scholar]
- 42.Chen C, Lewis SK, Voigt L, et al. Prostate carcinoma incidence in relation to prediagnostic circulating levels of insulin-like growth factor I, insulin-like growth factor binding protein 3, and insulin. Cancer. 2005;103:76–84. doi: 10.1002/cncr.20727. [DOI] [PubMed] [Google Scholar]
- 43.Carson JA, Fillmore RA, Schwartz RJ, Zimmer WE. The Smooth Muscle gamma -Actin Gene Promoter Is a Molecular Target for the Mouse bagpipe Homologue, mNkx3-1, and Serum Response Factor. J Biol Chem. 2000;275:39061–72. doi: 10.1074/jbc.M006532200. [DOI] [PubMed] [Google Scholar]
- 44.Miano JM. Serum response factor: toggling between disparate programs of gene expression. J Mol Cell Cardiol. 2003;35:577–93. doi: 10.1016/s0022-2828(03)00110-x. [DOI] [PubMed] [Google Scholar]
- 45.Ning Y, Schuller AG, Bradshaw S, et al. Diminished growth and enhanced glucose metabolism in triple knockout mice containing mutations of insulin-like growth factor binding protein-3, -4, and -5. Mol Endocrinol. 2006;20:2173–86. doi: 10.1210/me.2005-0196. [DOI] [PubMed] [Google Scholar]
- 46.Wood TL, Rogler LE, Czick ME, Schuller AGP, Pintar JE. Selective Alterations in Organ Sizes in Mice with a Targeted Disruption of the Insulin-Like Growth Factor Binding Protein-2 Gene. Mol Endocrinol. 2000;14:1472–82. doi: 10.1210/mend.14.9.0517. [DOI] [PubMed] [Google Scholar]
- 47.Zheng SL, Ju JH, Chang BL, et al. Germ-Line Mutation of NKX3. 1 Cosegregates with Hereditary Prostate Cancer and Alters the Homeodomain Structure and Function. Cancer Res. 2006;66:69–77. doi: 10.1158/0008-5472.CAN-05-1550. [DOI] [PubMed] [Google Scholar]
- 48.Benson DW, Silberbach GM, Kavanaugh-McHugh A, et al. Mutations in the cardiac transcription factor NKX2. 5 affect diverse cardiac developmental pathways. J Clin Invest. 1999;104:1567–73. doi: 10.1172/JCI8154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kasahara H, Lee B, Schott JJ, et al. Loss of function and inhibitory effects of human CSX/NKX2. 5 homeoprotein mutations associated with congenital heart disease. J Clin Invest. 2000;106:299–308. doi: 10.1172/JCI9860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iwatani N, Mabe H, Devriendt K, Kodama M, Miike T. Deletion of NKX2. 1 gene encoding thyroid transcription factor-1 in two siblings with hypothyroidism and respiratory failure. J Pediatr. 2000;137:272–6. doi: 10.1067/mpd.2000.107111. [DOI] [PubMed] [Google Scholar]
- 51.Weir BA, Woo MS, Getz G, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–8. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.