Abstract
Kinase suppressor of Ras-1 (KSR-1) is an essential scaffolding protein that coordinates the assembly of the mitogen-activated protein kinase (MAPK) module, consisting of the MAPK kinase kinase Raf, the MAPK kinase MEK (mitogen-activated or extracellular signal–regulated protein kinase kinase), and the MAPK ERK (extracellular signal–regulated kinase) to facilitate activation of MEK and thus ERK. Although KSR-1 is targeted to the cell membrane in part by its atypical C1 domain, which binds to phospholipids, other domains may be involved. We identified another domain in KSR-1 that we termed CC-SAM, which is composed of a coiled coil (CC) and a sterile α motif (SAM). The CC-SAM domain targeted KSR-1 to specific signaling sites at the plasma membrane in growth factor–treated cells, and it bound directly to various micelles and bicelles in vitro, indicating that the CC-SAM functioned as a membrane-binding module. By combining nuclear magnetic resonance spectroscopy and experiments in cultured cells, we found that membrane binding was mediated by helix α3 of the CC motif and that mutating residues in α3 abolished targeting of KSR-1 to the plasma membrane. Thus, in addition to the atypical C1 domain, the CC-SAM domain is required to target KSR-1 to the plasma membrane.
INTRODUCTION
Extracellular signals elicit various biological responses, such as cell proliferation, differentiation, survival, migration, and death (1). Multiprotein complexes assemble in various intracellular compartments to integrate and transmit these signals. Scaffolding proteins play a critical role in the assembly of many multiprotein complexes that drive signal transduction. These scaffolds are often composed of multiple, modular interaction domains, which mediate interactions with specific proteins, lipids, or other ligands and thus function to target signaling components to specific intra-cellular sites (2).
Early in signal transduction mediated by cell surface receptors, many cytosolic proteins are recruited to the plasma membrane, typically to the receptors detecting the incoming stimulus. For example, activated receptor tyrosine kinases (RTKs) recruit various enzymes, adaptor molecules, and docking proteins. Similarly, activated guanosine triphosphatases (GTPases), such as Ras, engage numerous effectors including guanine nucleotide–binding proteins, regulators of GTPase activity, and proteins that contain a Ras-binding domain (RBD) (2, 3). Cytosolic proteins can also directly associate with the membrane through lipid-binding domains, such as PH (pleckstrin homology), FYVE (Fab 1–YOTB–Vac 1–Eea1), or C1 domains, the latter of which binds diacylglycerol (4). Cytosolic proteins often require multiple domains for stable association with the membrane (5, 6). For example, membrane binding by protein kinase C (PKC) requires engagement of both its C1 domain and its C2 domain, which binds acidic phospholipids. Similarly, kinases of the Raf family are recruited to the membrane by two domains: an RBD, which mediates binding to activated Ras, and an atypical C1 domain, which can also bind Ras or bind phospholipid.
Kinase suppressor of Ras (KSR) is a family of scaffold proteins that interacts with the mitogen-activated protein kinase (MAPK) module, which consists of the MAPK kinase kinase Raf, the MAPK kinase MEK (mitogen-activated or extracellular signal–regulated protein kinase kinase), and the MAPK ERK (extracellular signal–regulated kinase). KSR proteins were discovered as signaling molecules through genetic screens performed in Drosophila melanogaster and Caenorhabditis elegans (7–9). Two KSR proteins are present in mammals [KSR-1 and KSR-2 (10)], and all KSR family members contain five distinct domains, known as conserved area 1 (CA1) through CA5 (7). CA1 is a 40–amino acid region unique to KSR proteins, CA2 is a proline-rich domain, CA3 is a cysteine-rich atypical C1 domain (11), CA4 is a serine- and threonine-rich domain that contains at least one FxFP motif for docking to ERK (8), and CA5 constitutes a protein kinase domain that has some catalytic activity in vitro (12), although its precise role in KSR-1 signaling remains to be determined.
In cells that are not exposed to extracellular signals, KSR proteins are found in the cytosol and constitutively interact with MEK (13). CA5 binds MEK, maintaining MEK in an inactive conformation (12). In response to signaling events, KSR bound to MEK rapidly translocates to the plasma membrane, where it interacts with B-Raf, which has been independently recruited to the plasma membrane through interactions with guanosine 5′-triphosphate–bound Ras. Both CA5 and CA1 are required for KSR to bind B-Raf (14). Formation of the KSR–MEK–B-Raf complex facilitates Raf-mediated MEK activation by inducing conformational changes that are thought to expose the MEK activation loop (12). Once the KSR-associated MEK is activated, it can phosphorylate and activate ERK. Subsequently, active ERK binds to the FxFP motif in CA4 of KSR and phosphorylates the KSR–MEK–B-Raf complex, which releases the complex from the plasma membrane and disrupts binding between KSR and B-Raf (14).
The ERK cascade is an essential effector pathway engaged by many signaling events, and the scaffolding activity of KSR plays a critical role in regulating the intensity and duration of ERK cascade signaling that emanates from the plasma membrane. Although the atypical C1 domain of KSR-1 contributes to the membrane binding of KSR-1 (11), it has been unclear how KSR-1 is targeted to the cell surface in response to signaling events. Here, we identified a previously unreported additional conserved region of KSR-1, which we called CA1α, located adjacent to CA1, and showed that amino acids 25 through 170 of KSR-1, which includes both CA1 and CA1α, formed a single, structured module composed of a coiled coil (CC) and a sterile α motif (SAM), which we termed CC-SAM. CC-SAM bound directly to various membrane mimetics, and this module was necessary to target KSR-1 to plasma membrane ruffles in response to growth factor signals. Thus, KSR-1 uses two distinct domains to stably associate with the plasma membrane. This work identified a previously unknown membrane-targeting domain, CC-SAM, which may be present in other membrane-targeted proteins.
RESULTS
CC-SAM is a conserved region in KSR-1
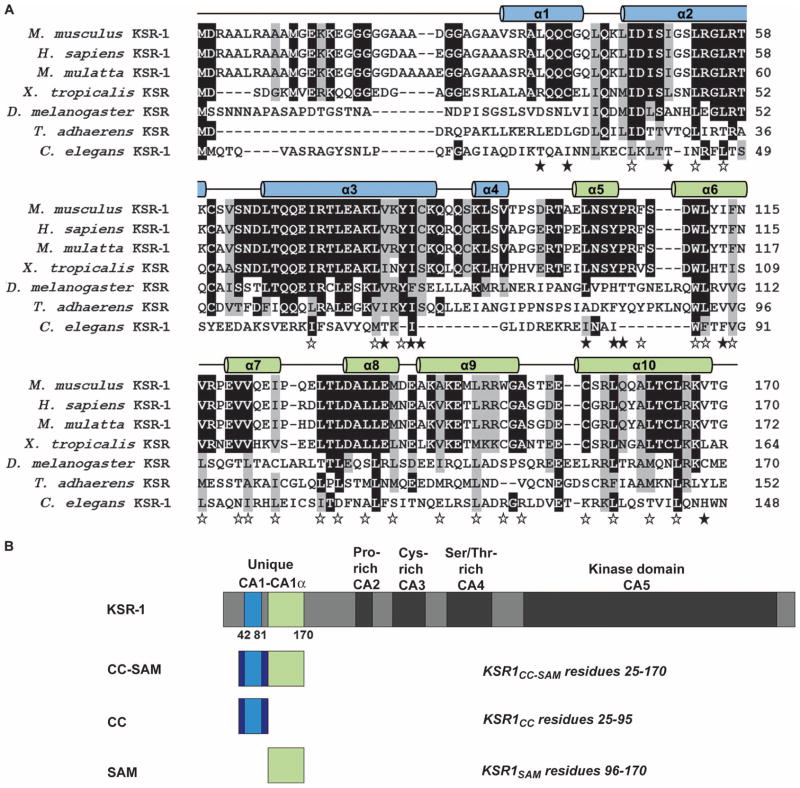
Sequence alignment of KSR proteins from evolutionarily diverse organisms (7) revealed that residues 25 to 95 in mouse KSR-1, which included CA1 (residues 42 to 81), were conserved among vertebrate proteins (Fig. 1A). We found that the residues immediately C-terminal to CA1, residues 82 to 170 in the mouse KSR-1, were also conserved among vertebrate KSR-1 proteins. Because this conserved area is immediately adjacent to CA1, we named this region CA1α (Fig. 1B). Primary sequence comparisons of the CA1α region using BLAST against multiple databases did not reveal any similarities to other proteins of known structure or function, indicating that, as in the case of CA1, the CA1α sequence was unique to the KSR family. Because of the secondary structure and motifs present in the KSR-1 CA1 and CA1α regions (described in detail later in the text), we refer to residues 25 to 170 as the CC-SAM domain.
Fig. 1.
Regions of KSR-1 are conserved across species. (A) Sequence alignment showing conservation of KSR-1 CC (α1 to α4 in blue) and SAM (α5 to α10 in green). Black and gray highlighting indicates sequence identity and similarity, respectively. The 10 α helices are depicted as cylinders above the sequence alignment. Open stars indicate residues that are buried in the individual hydrophobic cores of the CC and SAM domains, whereas filled stars indicate hydrophobic residues that are buried between them. (B) Domain architecture of KSR-1 and constructs used in this study. Conserved areas CA1 to CA5 are indicated. The CC and SAM domains do not exactly correspond to the CA1 and CA1α regions.
The N-terminal portion of KSR-1 targets KSR-1 to plasma membrane ruffles
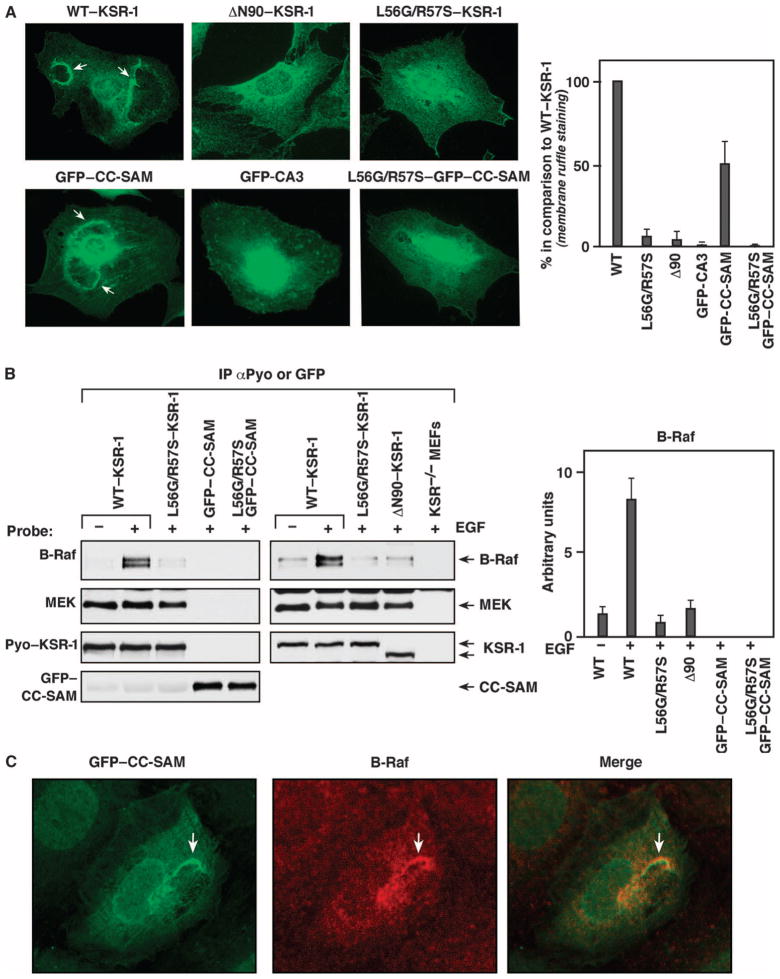
To investigate the contribution of the CC-SAM region to KSR-1 targeting and ERK signaling, we stably expressed wild-type mouse KSR-1, a KSR-1 deletion mutant lacking the first 90 amino acids (ΔN90–KSR-1); green fluorescent protein (GFP) fused to the CC-SAM domain (residues 25 to 170) of KSR-1 (GFP–CC-SAM); or GFP fused to the CA3 domain (GFP-CA3) in ksr-1−/− mouse embryonic fibroblasts (MEFs). A KSR-1 construct (ΔN170–KSR-1) lacking the first 170 amino acids, and thus missing the entire CC-SAM region, did not stably express (fig. S1). We examined the localization of these proteins in response to exposure of the MEFs to epidermal growth factor (EGF). As previously reported (15), we found that wild-type KSR-1 localizes to plasma membrane ruffles in EGF-treated cells (Fig. 2A). In contrast, ΔN90–KSR-1 exhibited diffuse cytoplasmic staining and did not localize to membrane ruffles in cells treated with EGF. Similar to wild-type KSR-1, GFP–CC-SAM accumulated at membrane ruffles in EGF-treated cells (Fig. 2A and fig. S2). Because the atypical C1 domain found in CA3 of KSR-1 plays a role in membrane binding (11, 16), we compared the cellular localization of GFP–CC-SAM to that of GFP-CA3. Although GFP-CA3 was found at the cell surface, it localized in punctate patches throughout the plasma membrane and did not accumulate at membrane ruffles (Fig. 2A).
Fig. 2.
CC-SAM targets KSR-1 to the plasma membrane. (A) Serum-starved ksr-1−/− MEFs stably expressing the indicated KSR-1 proteins were stimulated with EGF, and the localization of the KSR-1 proteins was determined by immunofluorescence staining of at least 200 cells for each construct. Arrows indicate membrane ruffles. The graph on the right shows quantification of three independent localization experiments. Error bars are the SD from the mean. WT, wild type. (B) ksr-1−/− MEFs stably expressing the indicated KSR-1 proteins were serum-starved and then treated with EGF. Pyo–KSR-1 or GFP–CC-SAM proteins were immunoprecipitated and examined for binding of endogenous B-Raf and MEK proteins. The graph on the right shows densitometry performed on three immunoblots. Error bars are the SD from the mean. (C) ksr-1−/− MEFs stably expressing the GFP–CC-SAM protein were stimulated with EGF, and the localization of the GFP–CC-SAM (green) and endogenous B-Raf (red) was determined by immunofluorescence staining (experiments were repeated twice). The merged image shows the colocalization of GFP–CC-SAM and B-Raf in membrane ruffles.
The CC-SAM domain colocalizes with but does not bind B-Raf
Both the CA1 region and the kinase domain (CA5) of KSR-1 are critical for KSR-1 to bind to B-Raf (14, 17). Because B-Raf is recruited to the plasma membrane, we investigated the possibility that GFP–CC-SAM localized to the plasma membrane by binding B-Raf. We examined the ability of wild-type KSR-1 and its truncation mutants to interact with endogenous B-Raf in response to EGF treatment. As a positive control, the KSR-1 proteins were also evaluated for their ability to interact with MEK, which constitutively binds CA5 of KSR-1 (14). B-Raf coimmunoprecipitated with wild-type KSR-1 in lysates from EGF-treated cells (Fig. 2B). Consistent with the inability of ΔN90–KSR-1 to localize to the plasma membrane, only a small amount of B-Raf coimmunoprecipitated with ΔN90–KSR-1 in lysates from EGF-treated cells, equivalent to the amount that coimmunoprecipitated with wild-type KSR-1 in lysates from serum-starved cells.
Mutating conserved leucine and arginine residues in the CC region of KSR-1 (L56G and R57S, hereinafter referred to as LG/RS) disrupts binding between KSR and B-Raf (14). Because the CC-SAM appeared to be critical for targeting KSR-1 to the plasma membrane, which is the location of the interaction of B-Raf with KSR-1 and B-Raf activation, we reasoned that the LG/RS mutations may alter the membrane-targeting ability of the CC-SAM domain. In contrast to wild-type KSR-1 and GFP–CC-SAM, the LG/RS mutants (LG/RS–KSR-1 and LG/RS–CC-SAM) failed to localize to membrane ruffles in EGF-treated MEFs (Fig. 2A). Instead, they were distributed diffusely throughout the cytosol similarly to the ΔN90–KSR-1 mutant. The LG/RS mutants also did not coimmunoprecipitate with B-Raf in lysates from EGF-stimulated cells (Fig. 2B). Despite its membrane localization and the presence of CA1, which is involved in B-Raf binding, GFP–CC-SAM did not coimmunoprecipitate with B-Raf upon growth factor stimulation (Fig. 2B). However, GFP–CC-SAM colocalized with endogenous B-Raf at plasma membrane ruffles of EGF-treated MEFs (Fig. 2C), which suggested that B-Raf and KSR were recruited independently to plasma membrane ruffles. By compromising KSR-1 membrane targeting, LG/RS mutations may impair B-Raf binding. Collectively, these findings were consistent with CA1 and CA5 being required for the stable interaction of KSR-1 and B-Raf and suggested that CC-SAM may function to target KSR-1 to plasma membrane microdomains enriched in B-Raf.
NMR spectroscopy reveals the structure of the KSR-1 CC-SAM
To gain further insight into the role of CC-SAM in KSR-1 signaling, we investigated the three-dimensional (3D) structure of the CC-SAM domain. Nuclear magnetic resonance (NMR) analysis revealed that the first 24 residues were unstructured (fig. S3, A and B); therefore, we used a construct consisting of residues 25 to 170, which includes CA1 and CA1α, for all structural studies (18). We determined the solution structure of this KSR-1 domain using heteronuclear NMR spectroscopy (Table 1, Fig. 3A, and fig. S3C).
Table 1.
Structural statistics of the KSR-1 CC-SAM domain.
| KSR-1 CC-SAM | |
|---|---|
| NMR distance and dihedral constraints | |
| Distance constraints | |
| Total NOE | 3077 |
| Intraresidue | 422 |
| Interresidue | 2655 |
| Sequential (|i − j| = 1) | 807 |
| Medium range (|i − j| < 4) | 779 |
| Long range (|i − j| > 5) | 1069 |
| Structure statistics | |
| Violations (mean ± SD)* | |
| Distance constraints (Å) | 0.0201 ± 0.0008 |
| Maximum distance constraint violation (Å) | 0.28 ± 0.04 |
| Deviations from idealized geometry (mean ± SD)* | |
| Bond lengths (Å) | 0.011± 0.000 |
| Bond angles (°) | 1.35 ± 0.03 |
| Impropers (°) | 1.45 ± 0.08 |
| Average pairwise root mean square deviation (Å)* | |
| Heavy | 1.21 ± 0.10 |
| Backbone | 0.87 ± 0.15 |
| Heavy (residues 33 to 58, 65 to 167) | 1.01 ± 0.08 |
| Backbone (residues 33 to 58, 65 to 167) | 0.61 ± 0.10 |
Mean, SD, and pairwise root mean square deviation were calculated among the 10 lowest energy structural ensembles.
Fig. 3.
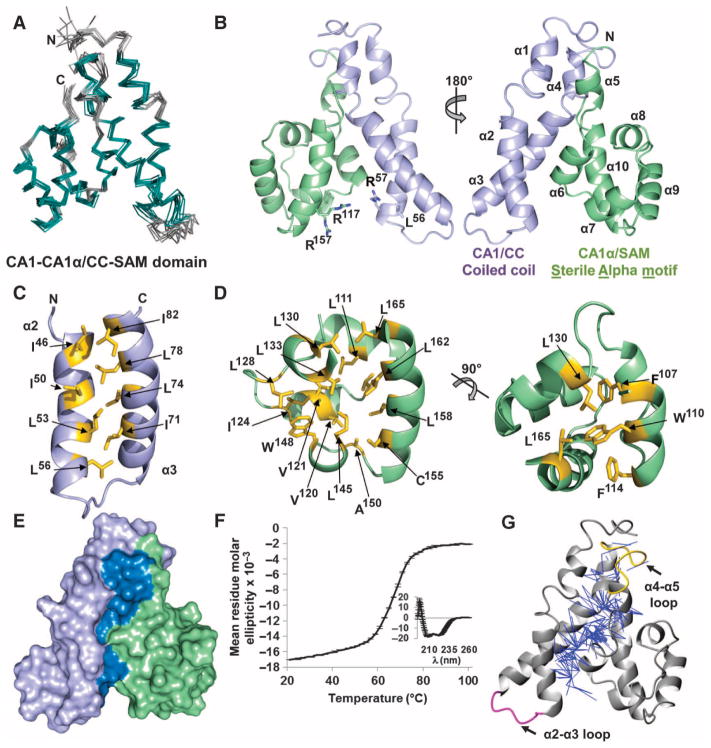
CC interactions with SAM are mediated by hydrophobic residues. (A) Bundle of the 10 lowest energy structures of the KSR-1 CC-SAM domain. The superposition is the best fit of structured regions (residues 32 to 39, 44 to 58, 65 to 83, 87 to 91, 99 to 103, 107 to 114, 118 to 123, 130 to 135, 138 to 148, and 152 to 167 in teal; loops in gray). (B) Lowest energy structure shown as a cartoon (CC, lavender; SAM, green). Left, locations of residues discussed in the text illustrated as sticks. Right, the structure is rotated 180°, and the 10 α helices and the N and C termini are labeled. (C) Hydrophobic residues (yellow sticks) that form the interface between α2 and α3 of the CC. (D) Residues (yellow sticks) that form the hydrophobic core of the SAM region. A 90° rotation of the image appears on the right. (E) Surface representation of the CC-SAM domain, highlighting the interface (dark blue) buried between the CC (lavender) and SAM (green). (F) Thermal denaturation of the CC-SAM domain monitored using CD at 222 nm. The inset depicts the CD spectrum of the CC-SAM domain before thermal denaturation. The error bars in both plots represent the SD from the mean of three independent experiments. (G) Two hundred fifty unambiguous NOE restraints (blue lines) between the CC and SAM.
CC-SAM was an α-helical domain composed of 10 α helices (α1 to α10; Fig. 3B). The CC region spanned residues 25 to 95 and included helices α1 to α4. The defining feature of this region was a short coiled coil formed by helices α2 and α3 (residues 44 to 83), which was stabilized by a hydrophobic interface consisting of residues Ile46, Leu53, Leu56, Ile71, Leu78, and Ile82 (Fig. 3C). Immediately C-terminal to the CC region was a second domain composed of residues 96 to 170, which included helices α5 to α10 (Fig. 3B). Unexpectedly, this region adopted a SAM fold. SAM domains are ubiquitous protein domains that bind kinases, other SAM domains, RNA, or membrane lipids (19, 20). A DALI structural homology search (21) of the SAM domain of KSR-1 revealed that it was most similar to the SAM domains of ephrin A4 (22), SAMSN-1 (SAM domain, SH3 domain, and nuclear localization signals protein 1), and CNK (connector enhancer of KSR) (23) (fig. S4).
Canonical SAM domains have five α helices that pack tightly around a central hydrophobic core (24). Key residues that function to stabilize the SAM fold (25, 26) [Phe107, Trp110, Leu111, Val120, Ile124, and Leu145 in the hydrophobic core of the mouse KSR-1 SAM domain (Fig. 3D)] are conserved in SAM domains in KSRs from multiple species (Fig. 1A). Although the overall sequence conservation of the SAM domain between Drosophila KSR and mammalian KSR-1 is low, these key hydrophobic residues are conserved (Fig. 1A). They are also conserved in mammalian KSR-2 and, although the conservation is weaker, in C. elegans KSR-1. Thus, we predicted that these same regions in fly KSR, nematode KSR-1, and mammalian KSR-2 would also adopt a SAM fold.
The KSR-1 CC and SAM regions form a single modular domain
Whereas most SAM domains exist as a single, independent domain (20), this was not the case for the SAM domain in KSR-1. Instead, the interface between the CC and the SAM domains buried more than 1960 Å2 of solvent-accessible surface area (roughly 20% of the total), and these domains were organized into a single, compact globular fold (Fig. 3E). This interface was stabilized primarily by extensive hydrophobic interactions between Leu36, Cys39, Ile46, Ile50, Val79, Ile82, and Cys83 in the CC domain and Leu101, Pro105, Ile113, and Val168 in the SAM domain, in addition to multiple polar interactions between Arg57, Gln85, Gln86, and Gln87 in the CC domain and Arg97, Leu101, Tyr104, and Val168 in the SAM domain (fig. S5A). The size and chiefly hydrophobic nature of this interface suggested that the CC and the SAM domains did not function as independent domains but instead form a single structural unit.
To further address this possibility, we examined CC-SAM using circular dichroism (CD) polarimetry and NMR spectroscopy. By CD polarimetry, CC-SAM exhibited a two-state thermal denaturation curve with a single transition state at 65°C (Fig. 3F). NMR analysis of instances of the nuclear Overhauser effect (NOE), which allows for the measurement of intra- and intermolecular distances, revealed more than 250 unambiguous NOEs between the CC and the SAM domains (Fig. 3G). These data supported the hypothesis that the CC and the SAM domains form a single structural unit. We also recorded 15N[1H]-NOE data to detect fast time scale picosecond to nanosecond motions in CC-SAM, which would indicate flexibility in regions of this functional unit (fig. S5B). Only the loop connecting the α2 and α3 helices in the CC domain exhibited flexibility. In contrast, the loop between helices α4 and α5, which connects the CC and the SAM regions, was rigid. Additionally, the CC domain, but not the SAM domain, was insoluble when expressed in Escherichia coli. Although the SAM domain could be purified, it behaved as a molten globule, as determined by size exclusion chromatography, CD polarimetry, and NMR spectroscopy (fig. S5C). The instability of the individual CC and SAM regions was consistent with the two-state folding equilibrium of CC-SAM, in which the two domains do not fold independently, but instead, each requires the presence of the other to fold into a single structured domain.
Finally, detailed bioinformatics analysis revealed that the KSR-1 CC-SAM domain defines another family in the SAM clan (http://pfamsvn.sanger.ac.uk/svn/pfam/trunk/Data/Families/PF13543/) and has been given the name KSR-1–SAM. Profile-profile alignment revealed that the CC-SAM domain is present in KSR proteins from Trichoplax adhaerens (XP_002115730) to humans (KSR-1 and KSR-2; Fig. 1A).
The LG/RS mutation destabilizes the CC-SAM fold
The structure of CC-SAM also enabled us to understand the molecular basis for the effects of mutating Leu56 to Gly and Arg57 to Ser on CC-SAM function. Leu56 made weak hydrophobic contacts with Val62, Gln68, and Ile71 in the CC, whereas Arg57 coordinated the interaction between the CC and SAM regions by forming a bipartite hydrogen bond with Glu75 in the CC domain and two hydrogen bonds with the backbone carbonyls of Tyr112 and Ile113 in the SAM domain (fig. S6). According to CD thermal denaturation experiments, the melting point of the KSR-1 CC-SAM with the LG/RS mutations was 44°C, a decrease in 21°C compared to the melting point of the CC-SAM from wild-type KSR-1 (table S1). These data suggested that the LG/RS mutations destabilized the CC-SAM fold and subsequently compromised membrane targeting.
The KSR-1 CC-SAM binds directly to micelles and bicelles
We sought to determine whether CC-SAM of KSR-1 bound RNA, proteins, or membrane lipids. On the basis of the known function of KSR-1 as an ERK scaffold with potential kinase activity, it would not be expected to bind RNA. Consistent with this, the electrostatic surface potential of CC-SAM lacked the positively charged patch that is typically present in RNA binding SAM domains (fig. S7A). We, therefore, performed NMR titration experiments with the CC-SAM– and the KSR-1–interacting proteins ERK and MEK or the SAM domains from CNK (CNKSAM) or Hyphen (HYPSAM) and looked for chemical shift perturbations, which would indicate direct interactions between CC-SAM and the proteins. However, no chemical shift perturbations were observed (fig. S7, B to E). Additionally, GFP–CC-SAM was expressed in 293 cells, immunoprecipitated from cell lysates, and examined for potential protein interactions by mass spectrometry (table S2). Although GFP–CC-SAM was stably expressed in these cells, no specific binding partners could be identified.
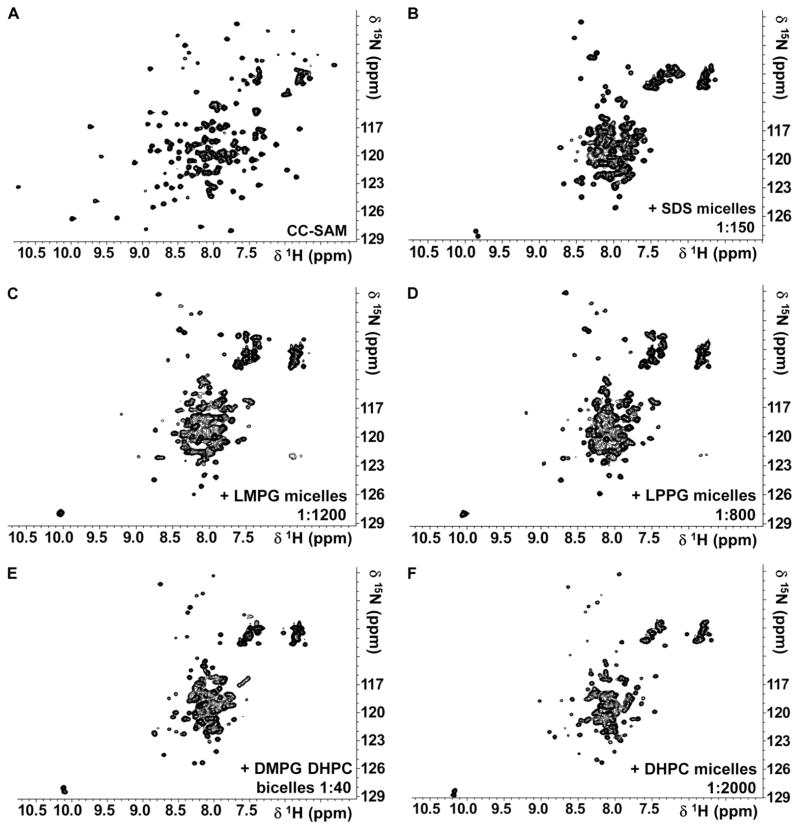
Because SAM domains can bind lipids (19, 27), we examined the interaction of KSR-1 CC-SAM with micelle or bicelle systems (28–32). Micelle and bicelle interactions with CC-SAM were readily followed using NMR spectroscopy because the 2D [1H,15N] heteronuclear single-quantum coherence (HSQC) NMR spectrum of CC-SAM changed in the presence of membrane mimetics. We identified a direct interaction between CC-SAM and lysomyristoylphosphatidylglycerol (LMPG) micelles, lysopalmitoylphosphatidylglycerol (LPPG) micelles, sodium dodecyl sulfate (SDS) micelles, dihexanoylphosphatidylcholine (DHPC) micelles, or bicelles composed of DHPC and dimyristoylphosphatidylglycerol (DMPG) (Fig. 4). The CC-SAM domain had a preferred interaction with anionic membrane surfaces because the interaction of CC-SAM with zwitterionic DHPC micelles was only detected at protein/micelle ratios 20- to 50-fold greater than ratios of protein to anionic micelles or bicelles (Table 2). Indeed, the most prominent change in the 2D [1H,15N] HSQC NMR spectrum of CC-SAM was a twofold reduction of the chemical shift dispersion in the 1HN dimension, which indicated secondary or tertiary structural changes in the CC-SAM upon interaction with membrane mimetics.
Fig. 4.
CC-SAM is a membrane-binding domain. (A to F) 2D [1H,15N] HSQC spectra recorded at 308 K and 500 MHz field strength of (A) 2 mM 15N CC-SAM, (B) 0.55 mM 15N CC-SAM with 82 mM SDS, (C) 0.1 mM 15N CC-SAM with 120 mM LMPG, (D) 0.1 mM 15N CC-SAM with 80 mM LPPG, (E) 0.25 mM 15N CC-SAM with 10 mM DMPG and 30 mM DHPC, and (F) 0.1 mM 15N CC-SAM with 200 mM DHPC.
Table 2.
The KSR-1 CC-SAM is a membrane-interacting domain. All NMR experiments were performed at 308 K and 11.7-T field strength. All bicelles were prepared with q = 0.33.
| Micelle or bicelle | Concentration (mM) | Protein: detergent or lipid ratio | Spectral collapse in δ 1H? | Charge |
|---|---|---|---|---|
| Micelles | ||||
| LMPG | 10, 20, 30, 40, 80, 120, 300 | 1:100*, 1:200, 1:300, 1:400, 1:600, 1:800, 1:1200 | Yes | Anionic |
| LPPG | 10, 20, 30, 40, 80, 120 | 1:100*, 1:200, 1:300, 1:400, 1:800, 1:1200 | Yes | Anionic |
| SDS | 3, 5, 10, 20, 50, 82 | 1:15, 1:25, 1:50*, 1:100, 1:150, 1:250 | Yes | Anionic |
| DHPC | 100, 200 | 1:1000, 1:2000* | Yes | Zwitterionic |
| Isotropic bicelles (1:3) | ||||
| DMPG/DHPC | 10:30 15:45 |
1:20, 1:40*, 1:100, 1:150, 1:200 | Yes | Zwitterionic/anionic |
A strong interaction with the protein was observed starting at this concentration.
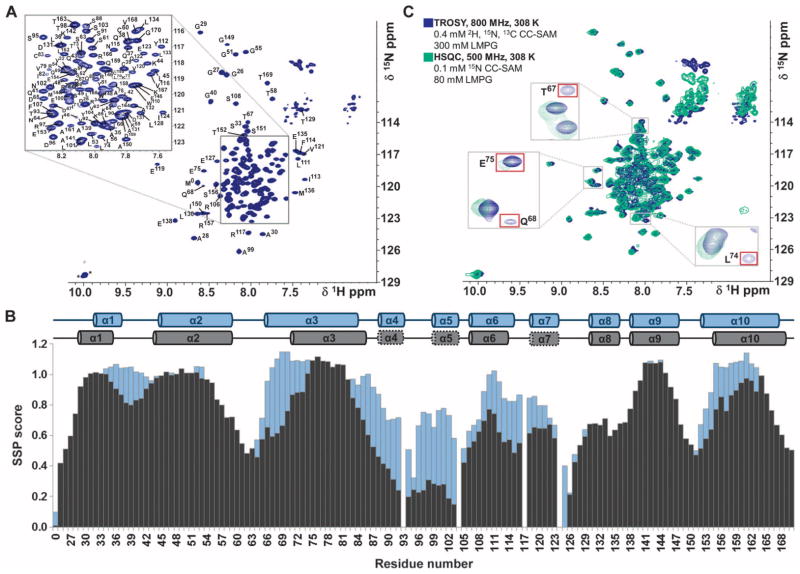
The 2D [1H,15N] HSQC and [1H,15N] transverse relaxation optimized spectroscopy (TROSY) NMR spectra of CC-SAM were similar even in the different membrane mimetic environments (Fig. 4, B to F). Therefore, we analyzed CC-SAM bound to either an LMPG or an SDS micelle, and the results were identical with both types of micelles (Fig. 5 and figs. S8 and S9). These membrane mimetic environments allowed us to use the highest protein concentrations to gather accurate NMR data for the subsequent analysis. N,HN cross peaks in [1H,15N] HSQC spectra of large proteins and protein complexes exhibit line broadening due to rapid transverse relaxation, so we used deuteration and TROSY techniques, which reduce line broadening and thus increase sensitivity, on samples prepared at a 1:750 CC-SAM/LMPG molar ratio (33) to determine the sequence-specific backbone assignment of CC-SAM in LMPG micelles (Fig. 5A). In the micelle- or bicelle-bound state, KSR-1 CC-SAM retained its α-helical structure, as determined by CD polarimetry and NMR chemical shift index calculations based on Cα and Cβ chemical shifts and their translation into secondary structure propensity (SSP) scores (Fig. 5B and figs. S8 and S9). None of the α helices unfolded, and most of the helical regions (all of α1, α8, and α9, and most of α2, α3, α6, and α10) did not exhibit decreased SSP scores when the CC-SAM was bound to LMPG micelles (Fig. 5B). Residues that had the largest decreases in SSP score were located in helices α3, α4, α5, α6, α7, and α10 and included residues that were buried at the interface between the CC and the SAM regions, indicating that this interface was likely disrupted upon micelle binding.
Fig. 5.
CC-SAM domain binds directly to LMPG micelles. (A) Fully annotated 2D [1H,15N] TROSY spectrum of KSR-1 CC-SAM domain in the presence of LMPG micelles. Assigned residues are labeled with the residue name (single-letter code) and number in the sequence. The two asterisks near 10 parts per million (ppm) in the 1H dimension denote Nε1/Hε1 resonances from tryptophan side chains. (B) SSP scores of the free (blue) and micelle-bound (black) CC-SAM domain. Across the top are diagrams of the determined secondary structural elements of free and micelle-bound CC-SAM. In the micelle-bound form, dotted lines around helices α4, α5, and α7 indicate that these helices had the largest loss of helical content upon binding to micelles. (C) Of all the helices in the KSR-1 CC-SAM domain, helix α3 was most tightly associated with lipid micelles. Spectra for 2D [1H,15N] HSQC (green, 500 MHz 1H Lamor frequency) and 2D [1H,15N] TROSY (blue, 800 MHz 1H Lamor frequency) are shown. Fourteen peaks, all from helix α3, were detected only in the TROSY spectrum.
KSR-1 membrane binding is mediated largely by CC helix α3
To determine which residues mediated the direct interaction between CC-SAM and LMPG micelles, we recorded a 2D [1H,15N] HSQC NMR spectrum of 15N-labeled CC-SAM bound to LMPG (Fig. 5C). Although visible in the 2D [1H,15N] TROSY spectrum, 1H,15N resonances for residues 71 to 83 in helix α3 were not observed in the 2D [1H,15N] HSQC spectrum; they underwent severe line broadening, likely due to adopting the overall correlation time of the LMPG micelle, indicating that these residues bound most strongly to LMPG micelles. These data suggested that membrane binding was restricted to a small number of residues in CC-SAM and that the remaining residues in CC-SAM did not interact strongly with the membrane-binding helix α3. Helix α3 contains one arginine and two lysine residues and has a net positive charge [isoelectric point (pI) = 8.2], which would allow for charge compensation of the anionic membrane surface during binding.
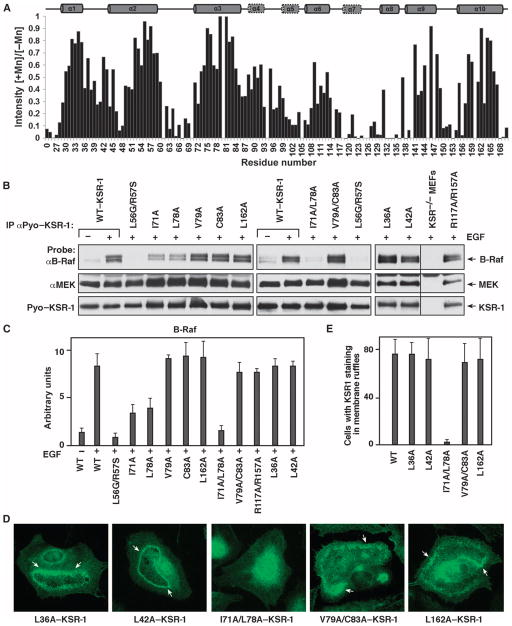
We performed titrations, adding increasing amounts of MnCl2 to micelle-bound CC-SAM, to identify solvent-exposed and micelle-bound parts of CC-SAM. The paramagnetic electrons in Mn2+ induce line broadening (due to paramagnetic relaxation) in resonances corresponding to solvent-exposed residues, whereas micelle-bound residues are protected from interaction with Mn2+. Residues in helices α7 and α8 were most susceptible to the effects of Mn2+, indicating that they were solvent-accessible. Conversely, residues in α1, α2, α3, α9, and α10 were protected (Fig. 6A), and Ile71, Leu78, Val79, and Cys83 in α3 were the most protected.
Fig. 6.
CC-SAM residues Ile71 and Leu78 mediate membrane binding. (A) Residues in α3 were tightly associated with micelles, as monitored by Mn2+ protection. Normalized 2D [1H,15N] HSQC peak intensities in the presence of MnCl2 (I[+Mn]) relative to those obtained in the absence of MnCl2 (I[−Mn]) are plotted for each residue. Peakswith ratios closest to 1 were most protected from the line-broadening effects of Mn2+. (B) ksr-1−/− MEFs stably expressing the indicated KSR-1 proteins were serum-starved and treated with EGF. Pyo–KSR-1 proteins were immunoprecipitated and examined for binding of endogenous B-Raf and MEK proteins. A representative blot from three separate experiments is shown. (C) Quantification of the B-Raf pull-downs is shown in (B). Error bars are the SD of the mean. (D) Serum-starved ksr-1−/− MEFs stably expressing the indicated KSR-1 proteins were stimulated with EGF, and the localization of the KSR-1 proteins was determined by immunofluorescence staining. Staining was performed in three independent experiments, and at least 200 cells were examined per experiment for each KSR-1 construct. Arrows indicate membrane ruffles. (E) Quantification of the localization results is shown in (D). Error bars are the SD of the average.
To determine the importance of the protected residues for membrane binding, we generated a series of KSR-1 proteins with mutations of CC-SAM residues that showed either strong, moderate, or no Mn2+ protection. Using CD polarimetry, we verified that no mutation altered the fold or the stability of CC-SAM (table S1). The point mutants L36A, L42A, V79A, C83A, and L162A and the double-mutants V79A/C83A and R117A/R157A represented KSR-1 with mutations that include residues that exhibited moderate or no Mn2+ protection and thus would likely not affect membrane binding. All of these mutants coimmunoprecipitated with B-Raf in lysates from EGF-treated cells (Fig. 6, B and C, and fig. S10), indicating that binding to B-Raf was unaffected and suggesting that membrane binding was likely also unaffected. However, I71A, L78A, or I71A and L78A (I71A/L78A) KSR-1 mutants, which have mutations in the residues that exhibited the strongest Mn2+ protection, coimmunoprecipitated with reduced amounts of B-Raf in EGF-stimulated cells. The interaction with B-Raf was reduced the most with the I71A/L78A mutant. Consistent with the results of the B-Raf coimmunoprecipitation studies, the L36A, L42A, V79A/C83A, or L162A mutant KSR-1 proteins localized to membrane ruffles in EGF-treated cells, but the I71A/L78A KSR-1 mutant failed to localize to membrane ruffles (Fig. 6, D and E, and fig. S10). Together, the NMR-based data and the analysis of the KSR-1 mutant proteins in cells revealed that helix α3 of CC-SAM was responsible for KSR-1 membrane targeting, and Ile71 and Leu78 were key residues for mediating this interaction (Fig. 7).
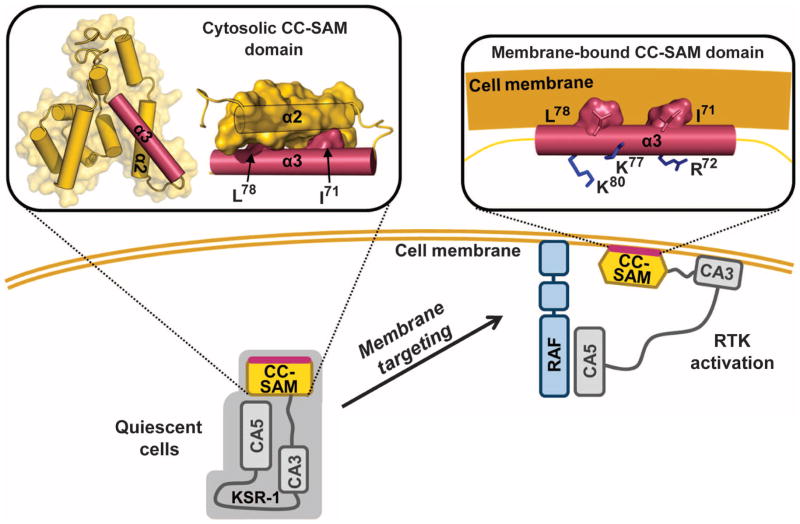
Fig. 7.
Targeting KSR-1 to the plasma membrane ruffles in activated cells requires both the CC-SAM and C1 domains. KSR translocates from the cytosol to the plasma membrane upon RTK activation. CC-SAM, together with the atypical C1 domain (CA3), localizes KSR-1 to B-Raf–rich microdomains in the plasma membrane. CC-SAM membrane binding is mediated by CC helix α3. Residues Ile71 and Leu78 in helix α3 are buried at the interface between helix α2 and helix α3. In the membrane-bound form of the CC-SAM, a structural rearrangement occurs whereby helix α2 releases helix α3, exposing Ile71 and Leu78, so that they can engage in membrane binding. Arg72, Lys77, and Lys80 are positioned such that they compensate for the negatively charged surface of the plasma membrane.
DISCUSSION
The small modular interaction domains found in signaling proteins are essential for linking cell surface receptors to intracellular targets, thus contributing to the regulation of downstream signal transduction pathways. Here, we presented the structure of the CC-SAM domain found in KSR scaffolds, a membrane-binding domain that targeted KSR-1 to plasma membrane ruffles in growth factor–treated cells, a critical step in the regulation of the ERK signaling cascade. The structure revealed that the CC was primarily responsible for membrane binding and that the SAM was likely needed for domain stability.
The high sequence conservation of the KSR CC region, together with the fact that the LG/RS mutant is a dominant suppressor of the rough eye phenotype induced by activated Ras in Drosophila (7), argues that this region has a functional role in KSR signaling. The Drosophila KSR LG/RS mutant exhibits a reduced interaction with D-Raf and cannot facilitate MEK activation (17). In mammalian cells, the LG/RS mutant does not bind B-Raf (14). Our results provided a structural basis for these observations. GFP–CC-SAM colocalized with B-Raf in growth factor–stimulated cells, suggesting a potential interaction with B-Raf. However, coimmunoprecipitation studies (Fig. 2B) and analysis of immunoprecipitated CC-SAM protein complexes by mass spectrometry showed that B-Raf did not associate with CC-SAM. Deletion of only the CC domain was sufficient to nearly abolish the interaction of KSR-1 with B-Raf, consistent with the inability of the ΔN90–KSR-1 mutant to target membranes. Likewise, the LG/RS mutation in helix α2, which disrupted the structure of the CC-SAM domain, also abolished the ability of KSR-1 to interact with B-Raf, consistent with the inability of LG/RS–KSR-1 to target membranes. Although these data suggested that the CC-SAM did not bind B-Raf directly, we cannot exclude the possibility that CC-SAM may still contact B-Raf in the context of the full-length KSR-1 protein.
CC-SAM bound directly to micelles and bicelles, and membrane binding was mediated by helix α3 (Fig. 7). Residues 71 to 83 in helix α3 interacted most strongly with the micelle. In order for this to happen, the CC-SAM domain must change its overall conformation. First, helix α2 must disengage from helix α3 to allow helix α3 to interact with the membrane. Furthermore, helices α1 to α2 and helices α4 to α10 would need to have relatively weak interactions with helix α3. Helix α3 was also important for membrane binding in cells because in stimulated cells, the I71A/L78A double-mutant KSR-1 failed to localize to membrane ruffles. Helix α3 was one of the most highly conserved sequences in KSR proteins, with even the most distal KSR homologs containing only conservative substitutions at residues in this helix (Fig. 1A), highlighting the functional importance of this helix. The CC-SAM domain thus targeted KSR-1 to plasma membrane ruffles in stimulated cells by binding directly to the membrane.
The presence of CC-SAM in KSR-1 may add a new layer of complexity to the regulation of KSR proteins in ERK signaling. The rapid transmission of cell signals requires the dynamic assembly and disassembly of complex, multiprotein signaling hubs (34). The plasma membrane is a key site for the assembly of these signaling hubs, and many proteins, such as PKC and the Raf family kinases, localize to the membrane by simultaneously engaging multiple membrane interaction domains. Here, we showed that CC-SAM bound directly to membranes. Thus, KSR-1 also uses two distinct domains to engage the plasma membrane—the CC-SAM and the atypical C1 domain. SAM domains are highly divergent in sequence and, therefore, are often difficult to identify on the basis of primary sequence alone (35, 36). As a result, many SAM domains have only been identified after their 3D structures were determined. We anticipate that structural studies may also enable divergent CC-SAM domains to be discovered in other proteins.
MATERIALS AND METHODS
Preparation of proteins
KSR-1 was expressed and purified as previously described (18). Briefly, the codon-optimized open reading frames of Mus musculus KSR-1 CC-SAM (residues 1 to 170 and 25 to 170), Homo sapiens CNK2SAM (residues 5 to 84), and D. melanogaster HYPSAM (residues 21 to 98) were synthesized by DNA2.0 and cloned into the pJexpress411 bacterial expression vector. KSR-1 CC-SAM and CNK2SAM were each engineered to have an N-terminal expression, purification, and TEV cleavage tag (Thio-His6-TEV, MGSDKIHHHHHHENLYFQG). HYPSAM was subcloned into a pET28-based bacterial expression vector and engineered with a Thio-His6-MBP-TEV tag at its N terminus to improve solubility (1). Togenerate the CC (residues 25 to 95) and SAM (residues 96 to 170) constructs, inserts were polymerase chain reaction (PCR)–amplified, restriction-digested with Xho I/Nde I, and ligated into the pJexpress411 vector. The CC-SAM L56G/R57S mutant was generated by PCR mutagenesis. ERK2 and MEK1 constructs were produced as previously described (37).
All proteins were expressed and purified with standard methods similar to those previously described (18). Briefly, protein was expressed in BL21 (DE3) Star E. coli cells (Invitrogen). Uniformly 13C/15N- and 15N-labeled protein was expressed in M9 minimal medium supplemented with [13C]-D-glucose (4 g/liter), 15NH4Cl (1 g/liter), or both (Cambridge Isotope Laboratories). Deuterated protein was expressed in 99% D2O and [13C]-D-d7-glucose (4 g/liter) for 2H,15N,13C-labeled protein. Protein was purified with a HisTrap HP column (GE Healthcare), followed by cleavage of the N-terminal tag, Ni2+–nitrilotriacetic acid immobilized metal affinity chromatography, and size exclusion chromatography [Superdex 75 26/60 (GE Healthcare) equilibrated in 20 mM Na phosphate (pH 6.5), 100 mM NaCl, and 0.5 mM tris(2-carboxyl)phosphine (TCEP)]. All proteins were verified to be folded with 1D 1H NMR spectroscopy.
NMR spectroscopy
NMR data used for chemical shift assignments were collected on a Bruker Avance II 500 and 800 MHz spectrometer equipped with a TCI HCN z-gradient cryoprobe at 298 K (CC-SAM, constructs 1 to 170 and 25 to 170) or at 308 K (CC-SAMMB, constructs 25 to 170). NMR measurements of CC-SAM were recorded with either 15N- or 15N,13C-labeled CC-SAM at a final concentration of 2 mM in 20 mM Na phosphate (pH 6.5), 100 mM NaCl, 0.5 mM TCEP, and 90% H2O/10% D2O. Sequence-specific back-bone and side-chain assignments were obtained previously (18). NOE-based distance restraints were assigned from a 3D 15N-resolved [1H,1H]-NOESY (Tm = 70 ms; 800 MHz 1H Lamor frequency), a 3D 13C-resolved [1H,1H]-NOESY (Tm = 70 ms; 800 MHz 1H Lamor frequency), and a 2D [1H,1H]-NOESY (Tm = 70 ms; 100% D2O solution). The ATNOS/CANDID software package (38, 39) was used for automated NOESY peak picking and NOE assignment.
For the sequence-specific backbone assignments of CC-SAM bound to SDS or LMPG micelles, TROSY versions of HNCA, HN(CO)CA, HNCACB, and 3D 15N-resolved [1H,1H]-NOESY (Tm = 150 ms) experiments were collected at 800 MHz field strength and 308 K. CC-SAM in SDS micelles was prepared with 2H,15N,13C-labeled CC-SAM at a final concentration of 0.55 mM in 20 mM Na phosphate (pH 6.5), 82 mM SDS (0.6:1 protein/SDS micelle ratio), 100 mM NaCl, 0.5 mM TCEP, and 90% H2O/10% D2O. Assignments were obtained for 94% of the backbone 15N and 1HN resonances and for 97% of 13Cα and 13Cβ resonances. 1HN and 15N chemical shifts were not obtained for I46, D47, E70, A76, K80, Y81, K84, and two residues in the N-terminal cloning artifact (GH). CC-SAM in LMPG micelles was prepared with 2H,15N,13C-labeled CC-SAM at a final concentration of 0.4 mM in 20 mM Na phosphate (pH 6.5), 300 mM LMPG (0.07:1 protein/LMPG micelle ratio), 100 mM NaCl, 0.5 mM TCEP, and 90% H2O/10% D2O. Assignments were obtained for 94% of the backbone 15N and 1HN resonances and for 97% of 13Cα and 13Cβ resonances. 1HN and 15N chemical shifts were not obtained for Glu70, Lys80, Gln126, Lys140, Leu145, Arg147, Gln160, and two residues in the N-terminal cloning artifact (Gly-His).
For both free CC-SAM constructs, 1 to 170 and 25 to 170, 2D 15N[1H]-NOE experiments were recorded at 500 MHz with a saturation delay of 5 s. The interaction of KSR-1 CC-SAM with ERK2, MEK1, CNK2SAM, and HYPSAM was tested with 15N-labeled KSR-1 CC-SAM and unlabeled ERK2, MEK1, CNK2SAM, and HYPSAM at multiple ratios. All spectra were processed with TopSpin 2.1/3.0/3.1 (Bruker), and data were evaluated with CARA (http://cara.nmr.ch) and the Protein Dynamic Center software (Bruker).
Structure calculation and refinement
Three thousand seventy-seven unambiguous NOESY-derived distance restraints (~20.6 NOE restraints per residue) were used in initial structure calculations with CYANA (38). The 20 structures with the lowest residual CYANA target function were energy-minimized in a water shell with CNS 1.3 using the RECOORD script package (40). One hundred structures were generated for each cycle, and the 10 conformers with the lowest restraint violation energies were selected as the final representative model (fig. S3C). The quality of the final ensemble of lowest energy structures was assessed by WHATCHECK (41), AQUA (42) and NMR-PROCHECK (42) (which are part of the iCing suite; http://nmr.cmbi.ru.nl/cing/iCing.html), and MOLMOL (43). Ramachandran analysis was performed with NMR-PROCHECK on the 10 lowest energy structures and showed that 86.5% of residues were in the most favored regions, 12.4% were in the allowed regions, 0.7% were in the generously allowed regions, and 0.4% were in the disallowed regions.
MnCl2-induced line-broadening experiments in SDS micelles
MnCl2 was titrated into CC-SAM bound to SDS micelles or LMPG micelles at final concentrations of 0.1, 0.2, and 0.4 mM Mn2+. 2D [1H,15N] TROSY spectra were recorded at 500 MHz field strength and 308 K. The final samples consisted of 300 μM 2H,15N–CC-SAM with 45 mM SDS or 75 μM 2H,15N,13C–CC-SAM with 75 mM LMPG in 20 mM Na phosphate (pH 6.5), 100 mM NaCl, and 0.5 mM TCEP. Normalized peak intensities measured in the presence of 0.4 mM MnCl2 ([+Mn]) are plotted relative to those in the absence of MnCl2 ([−Mn]).
Micelle and bicelle preparation
SDS (Sigma), 1-myristoyl-2-hydroxy-sn-glycero-3-phospho-(1′-rac-glycerol) (LMPG; Avanti Polar Lipids Inc.), 1-palmitoyl-2-hydroxy-sn-glycero-3-phospho-(1′-rac-glycerol) (LPPG; Avanti Polar Lipids Inc.), and 1,2-dihexanoyl-sn-glycero-3-phosphocholine (DHPC; Avanti Polar Lipids Inc.) micelles were prepared by solubilizing the solid detergent or lipid in buffer [20 mM Na phosphate (pH 6.5), 100 mM NaCl, and 0.5 mM TCEP]. Isotropic bicelles (q = 0.33) were prepared with DHPC as the short-chain lipid, and either 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC; Avanti Polar Lipids Inc.) or 1,2-dimyristoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (DMPG; Avanti Polar Lipids Inc.) as the long-chain lipid. The long-chain lipid was first hydrated in buffer at 35°C for at least 3 hours and was then vortexed. It was then combined with solubilized DHPC to form bicelles. To improve homogeneity of DMPG/DHPC bicelles, we froze the sample in liquid nitrogen and then warmed it to 35°C twice before the addition of protein.
CD polarimetry
CD spectra for wild-type and mutant CC-SAM proteins were acquired with a Jasco J-815 CD spectrometer equipped with a Peltier temperature control system. CD measurements were carried out at protein concentrations of 10 to 25 μM in 20 mM Na phosphate (pH 6.5), 100 mM NaCl, and 0.5 mM TCEP. Far-ultraviolet wavelength scans from 200 to 260 nm were performed in triplicate at 20°C in a 0.2-cm path length cuvette with a scan rate of 20 nm/min. Thermal denaturation of 25 μM CC-SAM from 20° to 100°C was monitored with the CD signal at 222 nm at a rate of 2°C/min with 2 min of temperature equilibration per data point. A minimum of three replicates were performed per experiment.
Antibodies and cell culture
B-Raf (H-145) polyclonal antibody was obtained from Santa Cruz Biotechnology, MEK1 antibody was purchased from BD Biosciences, and GFP monoclonal antibody was from Roche. N-terminal KSR polyclonal antibody (44) and the antibody recognizing the Pyo epitope (45) have been previously described. EGF and ProLong Gold antifade reagent were obtained from Invitrogen. ksr-1−/− MEF cells (46) were cultured in Dulbecco′s modified Eagle′s medium supplemented with 10% fetal bovine serum, 1% L-glutamine, 1% penicillin/streptomycin, and 1% nonessential amino acids. EGF was used at 100 ng/ml where indicated.
DNA constructs and generation of stable cell lines
pBabe retroviral constructs encoding full-length Pyo-tagged mKSR-1 and mKSR-1–L56G/R57S have been previously described (14, 44). GFP–CC-SAM was generated by isolating the sequence encoding mKSR-1 residues 1 to 180 from pcDNA3–mKSR-1 and inserting it into the pEGFP-N1 vector. The GFP–CC-SAM fusion was then subcloned into the pLPCX retroviral vector. Point mutations were introduced by site-directed muta-genesis and confirmed by DNA sequencing. Stable cell lines were generated by infection of ksr-1−/− MEFs with pBabe-puro or pLPCX-puro retroviruses encoding various KSR-1 proteins and selected with puromycin (2.5 μg/ml).
Cell lysis and coimmunoprecipitation assays
Cells were washed twice in ice-cold phosphate-buffered saline (PBS) and lysed for 15 min in 1% NP-40 buffer [20 mM tris (pH 8.0), 137 mM NaCl, 10% glycerol, 1% NP-40, aprotinin (0.15 U/ml), 1 mM phenylmethylsulfonyl fluoride, 20 μM leupeptin, and 5 mM sodium vanadate]. Lysates were cleared by centrifugation for 15 min (4°C). Equivalent amounts of protein lysate were incubated with the Pyo or GFP antibody and protein G agarose beads for 3 hours at 4°C. Complexes were washed repeatedly with NP-40 lysis buffer, resolved by SDS–polyacrylamide gel electrophoresis, and subjected to immunoblot analysis with the indicated antibodies.
Immunofluorescent staining
ksr-1−/− MEFs stably expressing the indicated proteins were seeded onto 60-mm dishes with glass coverslips at a density of 2 × 105 to 4 × 105 per plate. Cells were grown in complete media at 37°C and 5% CO2 for 28 to 30 hours, at which time cells were serum-starved and incubated until 48 hours after plating. Cells were stimulated for 10 min with EGF, washed twice with ice-cold PBS, fixed with 4% paraformaldehyde, and permeabilized with 0.01% Triton X-100/PBS. Coverslips were blocked in 3% bovine serum albumin and PBS and incubated with either GFP or KSR-1 primary antibodies followed by Alexa Fluor 488 or 568 secondary antibodies. Coverslips were washed extensively in PBS and mounted onto slides with the ProLong Gold antifade reagent. Immunofluorescent staining was visualized with a Leica Microsystems fluorescence microscope.
Supplementary Material
Acknowledgments
We thank A. Godzik (Burnham Institute for Medical Research) for help with the bioinformatics analysis.
Funding: This research was supported by grant RSG-08-067001-LIB from the American Cancer Society to R.P., federal funds from the National Cancer Institute to D.K.M., and NIH grants R01NS056128 to W.P. and R01GM098482 to R.P. NMR (800 MHz) data were recorded at Brandeis University (NIH S10-RR017269). This research is based in part on work conducted using the Rhode Island NSF Experimental Program to Stimulate Competitive Research (EPSCoR) Proteomics Share Resource Facility, supported in part by the NSF/EPSCoR grant 1004057.
Footnotes
www.sciencesignaling.org/cgi/content/full/5/255/ra94/DC1
Fig. S1. Comparison of abundance of wild-type KSR-1 and ΔN170–KSR-1.
Fig. S2. Cellular localization of wild-type or mutant KSR-1 proteins upon stimulation with EGF.
Fig. S3. Characterization of the CC-SAM domain.
Fig. S4. Structural homologs of the KSR-1 SAM domain.
Fig. S5. The CC-SAM domain is a single, structured domain.
Fig. S6. Interactions made by Leu56 and Arg57.
Fig. S7. The CC-SAM domain does not bind RNA- or KSR-1–interacting proteins.
Fig. S8. The KSR-1 CC-SAM domain binds directly to SDS micelles.
Fig. S9. CC-SAM interacts similarly with SDS micelles and LMPG micelles.
Fig. S10. CC-SAM residues Ile71 and Leu78 mediate membrane binding.
Table S1. Residues involved in membrane binding and domain stability.
Table S2. CC-SAM is not a protein interaction domain.
Author contributions: D.K. purified and determined the structure of the KSR-1 CC-SAM domain, performed all NMR titration measurements, and completed the biophysical studies on the wild-type and mutant CC-SAM domains. N.S.-N. and D.R. generated the stable cell lines and performed the coimmunoprecipitation and cellular localization studies. D.K., R.P., D.K.M., and W.P. analyzed and interpreted the data, conceived the experiments, and wrote the manuscript.
Competing interest: The authors declare that they have no competing financial interests.
Data and materials availability: All chemical shifts were deposited in the BioMagResBank (http://www.bmrb.wisc.edu) under accession numbers 17045, 17724, 17725, and 18740. Atomic coordinates have been deposited in the Protein Data Bank (PDB) under the PDB code 2LPE. The plasmid encoding the KSR-1 gene can be obtained from Addgene.
REFERENCES AND NOTES
- 1.Raman M, Chen W, Cobb MH. Differential regulation and properties of MAPKs. Oncogene. 2007;26:3100–3112. doi: 10.1038/sj.onc.1210392. [DOI] [PubMed] [Google Scholar]
- 2.Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300:445–452. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- 3.Katz ME, McCormick F. Signal transduction from multiple Ras effectors. Curr Opin Genet Dev. 1997;7:75–79. doi: 10.1016/s0959-437x(97)80112-8. [DOI] [PubMed] [Google Scholar]
- 4.Stahelin RV. Lipid binding domains: More than simple lipid effectors. J Lipid Res. 2009;50(suppl):S299–S304. doi: 10.1194/jlr.R800078-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williams JG, Drugan JK, Yi GS, Clark GJ, Der CJ, Campbell SL. Elucidation of binding determinants and functional consequences of Ras/Raf-cysteine-rich domain interactions. J Biol Chem. 2000;275:22172–22179. doi: 10.1074/jbc.M000397200. [DOI] [PubMed] [Google Scholar]
- 6.López-Nicolás R, López-Andreo MJ, Marín-Vicente C, Gómez-Fernández JC, Corbalán-García S. Molecular mechanisms of PKCα localization and activation by arachidonic acid. The C2 domain also plays a role. J Mol Biol. 2006;357:1105–1120. doi: 10.1016/j.jmb.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 7.Therrien M, Chang HC, Solomon NM, Karim FD, Wassarman DA, Rubin GM. KSR, a novel protein kinase required for RAS signal transduction. Cell. 1995;83:879–888. doi: 10.1016/0092-8674(95)90204-x. [DOI] [PubMed] [Google Scholar]
- 8.Kornfeld K, Hom DB, Horvitz HR. The ksr-1 gene encodes a novel protein kinase involved in Ras-mediated signaling in C. elegans. Cell. 1995;83:903–913. doi: 10.1016/0092-8674(95)90206-6. [DOI] [PubMed] [Google Scholar]
- 9.Sundaram M, Han M. The C. elegans ksr-1 gene encodes a novel raf-related kinase involved in Ras-mediated signal transduction. Cell. 1995;83:889–901. doi: 10.1016/0092-8674(95)90205-8. [DOI] [PubMed] [Google Scholar]
- 10.Dougherty MK, Ritt DA, Zhou M, Specht SI, Monson DM, Veenstra TD, Morrison DK. KSR2 is a calcineurin substrate that promotes ERK cascade activation in response to calcium signals. Mol Cell. 2009;34:652–662. doi: 10.1016/j.molcel.2009.06.00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou M, Horita DA, Waugh DS, Byrd RA, Morrison DK. Solution structure and functional analysis of the cysteine-rich C1 domain of kinase suppressor of ras (KSR) J Mol Biol. 2002;315:435–446. doi: 10.1006/jmbi.2001.5263. [DOI] [PubMed] [Google Scholar]
- 12.Brennan DF, Dar AC, Hertz NT, Chao WC, Burlingame AL, Shokat KM, Barford D. A Raf-induced allosteric transition of KSR stimulates phosphorylation of MEK. Nature. 2011;472:366–369. doi: 10.1038/nature09860. [DOI] [PubMed] [Google Scholar]
- 13.Denouel-Galy A, Douville EM, Warne PH, Papin C, Laugier D, Calothy G, Downward J, Eychène A. Murine Ksr interacts with MEK and inhibits Ras-induced transformation. Curr Biol. 1998;8:46–55. doi: 10.1016/s0960-9822(98)70019-3. [DOI] [PubMed] [Google Scholar]
- 14.McKay MM, Ritt DA, Morrison DK. Signaling dynamics of the KSR1 scaffold complex. Proc Natl Acad Sci USA. 2009;106:11022–11027. doi: 10.1073/pnas.0901590106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Müller J, Ory S, Copeland T, Piwnica-Worms H, Morrison DK. C-TAK1 regulates Ras signaling by phosphorylating the MAPK scaffold, KSR1. Mol Cell. 2001;8:983–993. doi: 10.1016/s1097-2765(01)00383-5. [DOI] [PubMed] [Google Scholar]
- 16.Michaud NR, Therrien M, Cacace A, Edsall LC, Spiegel S, Rubin GM, Morrison DK. KSR stimulates Raf-1 activity in a kinase-independent manner. Proc Natl Acad Sci USA. 1997;94:12792–12796. doi: 10.1073/pnas.94.24.12792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roy F, Laberge G, Douziech M, Ferland-McCollough D, Therrien M. KSR is a scaffold required for activation of the ERK/MAPK module. Genes Dev. 2002;16:427–438. doi: 10.1101/gad.962902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koveal D, Pinheiro AS, Peti W, Page R. Backbone and side chain 1H, 15N and 13C assignments of the KSR1 CA1 domain. Biomol NMR Assign. 2011;5:39–41. doi: 10.1007/s12104-010-9262-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barrera FN, Poveda JA, González-Ros JM, Neira JL. Binding of the C-terminal sterile α motif (SAM) domain of human p73 to lipid membranes. J Biol Chem. 2003;278:46878–46885. doi: 10.1074/jbc.M307846200. [DOI] [PubMed] [Google Scholar]
- 20.Qiao F, Bowie JU. The many faces of SAM. Sci STKE. 2005;2005:re7. doi: 10.1126/stke.2862005re7. [DOI] [PubMed] [Google Scholar]
- 21.Holm L, Rosenström P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stapleton D, Balan I, Pawson T, Sicheri F. The crystal structure of an Eph receptor SAM domain reveals a mechanism for modular dimerization. Nat Struct Biol. 1999;6:44–49. doi: 10.1038/4917. [DOI] [PubMed] [Google Scholar]
- 23.Rajakulendran T, Sahmi M, Kurinov I, Tyers M, Therrien M, Sicheri F. CNK and HYP form a discrete dimer by their SAM domains to mediate RAF kinase signaling. Proc Natl Acad Sci USA. 2008;105:2836–2841. doi: 10.1073/pnas.0709705105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peterson AJ, Kyba M, Bornemann D, Morgan K, Brock HW, Simon J. A domain shared by the Polycomb group proteins Scm and ph mediates heterotypic and homotypic interactions. Mol Cell Biol. 1997;17:6683–6692. doi: 10.1128/mcb.17.11.6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schultz J, Ponting CP, Hofmann K, Bork P. SAM as a protein interaction domain involved in developmental regulation. Protein Sci. 1997;6:249–253. doi: 10.1002/pro.5560060128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chi SW, Ayed A, Arrowsmith CH. Solution structure of a conserved C-terminal domain of p73 with structural homology to the SAM domain. EMBO J. 1999;18:4438–4445. doi: 10.1093/emboj/18.16.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, Fung KL, Jin DY, Chung SS, Ching YP, Ng IO, Sze KH, Ko BC, Sun H. Solution structures, dynamics, and lipid-binding of the sterile α-motif domain of the deleted in liver cancer 2. Proteins. 2007;67:1154–1166. doi: 10.1002/prot.21361. [DOI] [PubMed] [Google Scholar]
- 28.Welker S, Rudolph B, Frenzel E, Hagn F, Liebisch G, Schmitz G, Scheuring J, Kerth A, Blume A, Weinkauf S, Haslbeck M, Kessler H, Buchner J. Hsp12 is an intrinsically unstructured stress protein that folds upon membrane association and modulates membrane function. Mol Cell. 2010;39:507–520. doi: 10.1016/j.molcel.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 29.Ulmer TS, Bax A. Comparison of structure and dynamics of micelle-bound human α-synuclein and Parkinson disease variants. J Biol Chem. 2005;280:43179–43187. doi: 10.1074/jbc.M507624200. [DOI] [PubMed] [Google Scholar]
- 30.Jaroniec CP, Kaufman JD, Stahl SJ, Viard M, Blumenthal R, Wingfield PT, Bax A. Structure and dynamics of micelle-associated human immunodeficiency virus gp41 fusion domain. Biochemistry. 2005;44:16167–16180. doi: 10.1021/bi051672a. [DOI] [PubMed] [Google Scholar]
- 31.Kim HJ, Howell SC, Van Horn WD, Jeon YH, Sanders CR. Recent advances in the application of solution NMR spectroscopy to multi-span integral membrane proteins. Prog Nucl Magn Reson Spectrosc. 2009;55:335–360. doi: 10.1016/j.pnmrs.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanders CR, Sönnichsen F. Solution NMR of membrane proteins: Practice and challenges. Magn Reson Chem. 2006;44(Spec):S24–S40. doi: 10.1002/mrc.1816. [DOI] [PubMed] [Google Scholar]
- 33.Thévenot C, Grassl B, Bastiat G, Binana W. Aggregation number and critical micellar concentration of surfactant determined by time-dependent static light scattering (TDSLS) and conductivity. Colloids Surf A Physicochem Eng Aspects. 2004;252:105–111. [Google Scholar]
- 34.Scott JD, Pawson T. Cell signaling in space and time: Where proteins come together and when they’re apart. Science. 2009;326:1220–1224. doi: 10.1126/science.1175668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hernandez JA, Phillips AH, Erbil WK, Zhao D, Demuez M, Zeymer C, Pelton JG, Wemmer DE, Rubio LM. A sterile α-motif domain in NafY targets apo-NifDK for iron-molybdenum cofactor delivery via a tethered domain. J Biol Chem. 2011;286:6321–6328. doi: 10.1074/jbc.M110.168732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stathopulos PB, Zheng L, Li GY, Plevin MJ, Ikura M. Structural and mechanistic insights into STIM1-mediated initiation of store-operated calcium entry. Cell. 2008;135:110–122. doi: 10.1016/j.cell.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 37.Francis DM, Różycki B, Tortajada A, Hummer G, Peti W, Page R. Resting and active states of the ERK2:HePTP complex. J Am Chem Soc. 2011;133:17138–17141. doi: 10.1021/ja2075136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Güntert P. Automated NMR structure calculation with CYANA. Methods Mol Biol. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- 39.Herrmann T, Güntert P, Wüthrich K. Protein NMR structure determination with automated NOE-identification in the NOESY spectra using the new software ATNOS. J Biomol NMR. 2002;24:171–189. doi: 10.1023/a:1021614115432. [DOI] [PubMed] [Google Scholar]
- 40.Nederveen AJ, Doreleijers JF, Vranken W, Miller Z, Spronk CA, Nabuurs SB, Güntert P, Livny M, Markley JL, Nilges M, Ulrich EL, Kaptein R, Bonvin AM. RECOORD: A recalculated coordinate database of 500+ proteins from the PDB using restraints from the BioMagResBank. Proteins. 2005;59:662–672. doi: 10.1002/prot.20408. [DOI] [PubMed] [Google Scholar]
- 41.Hooft RW, Vriend G, Sander C, Abola EE. Errors in protein structures. Nature. 1996;381:272. doi: 10.1038/381272a0. [DOI] [PubMed] [Google Scholar]
- 42.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 43.Koradi R, Billeter M, Wüthrich K. MOLMOL: A program for display and analysis of macromolecular structures. J Mol Graph. 1996;14:51–55. 29–32. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 44.Cacace AM, Michaud NR, Therrien M, Mathes K, Copeland T, Rubin GM, Morrison DK. Identification of constitutive and Ras-inducible phosphorylation sites of KSR: Implications for 14-3-3 binding, mitogen-activated protein kinase binding, and KSR overexpression. Mol Cell Biol. 1999;19:229–240. doi: 10.1128/mcb.19.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Therrien M, Michaud NR, Rubin GM, Morrison DK. KSR modulates signal propagation within the MAPK cascade. Genes Dev. 1996;10:2684–2695. doi: 10.1101/gad.10.21.2684. [DOI] [PubMed] [Google Scholar]
- 46.Kortum RL, Lewis RE. The molecular scaffold KSR1 regulates the proliferative and oncogenic potential of cells. Mol Cell Biol. 2004;24:4407–4416. doi: 10.1128/MCB.24.10.4407-4416.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.