Abstract
Transcription initiation is a key event in the regulation of gene expression. RNA polymerase (RNAP), the central enzyme of transcription, is able to efficiently locate promoters in the genome, carry out promoter opening, and initiate RNA synthesis. All the sub-steps of transcription initiation are subject to complex cellular regulation. Understanding the molecular details of each step in the promoter-opening pathway is essential for a complete mechanistic and quantitative picture of gene expression. In this mini-review, primarily using bacterial RNAP as an example, I briefly summarize some of the key recent advances in our understanding of the mechanisms of promoter search and promoter opening.
Keywords: RNA polymerase, transcription, promoter, protein–DNA recognition
Introduction
Transcription is the basis for decoding genetic information stored in DNA. Core RNAP (subunit composition α2ββ’ω) is responsible for all cellular transcription in bacteria.1 Specific transcription initiation at promoter sites requires an additional σ-subunit. Association of core and σ yields the holoenzyme capable of locating promoter sequences, opening DNA to form a transcription bubble, and initiating RNA synthesis.2–4 The primary σ-factors (σ70 in Escherichia coli) feature four structural domains and direct core RNAP to the majority of promoters active during log-phase growth, while alternative σ-factors control specialized promoters activated in response to environmental and intracellular signals.5
RNAP holoenzyme binds DNA along an extensive, positively charged interface formed by various regions of all RNAP subunits (except ω) (Fig. 1A). Some parts of the interface are engaged in non-specific DNA binding, primarily with the sugar–phosphate DNA backbone, while others ensure specific readout of the DNA sequence by interacting with the bases. The overall shape of RNAP resembles a crab claw: the two pincers form the active site cleft with the catalytic Mg2+ located deep in the cleft.6 The backside of the holoenzyme where the σ-subunit is bound features patches of positively charged surface carrying out promoter recruitment through non-specific interactions as well as read-out of dsDNA (double stranded DNA) shape and sequence. After the initial positioning of RNAP on promoter DNA is achieved, a sharp kink (possible after nucleation of melting) brings the downstream DNA in contact with the active site cleft. The cleft is too narrow to accommodate dsDNA, so in order to reach the active site, DNA must unwind. The melting is achieved through interactions of DNA strands with the positively charged surface of the cleft, through both non-specific contacts to the DNA backbone and specific interactions with individual bases in the ssDNA (single stranded DNA).
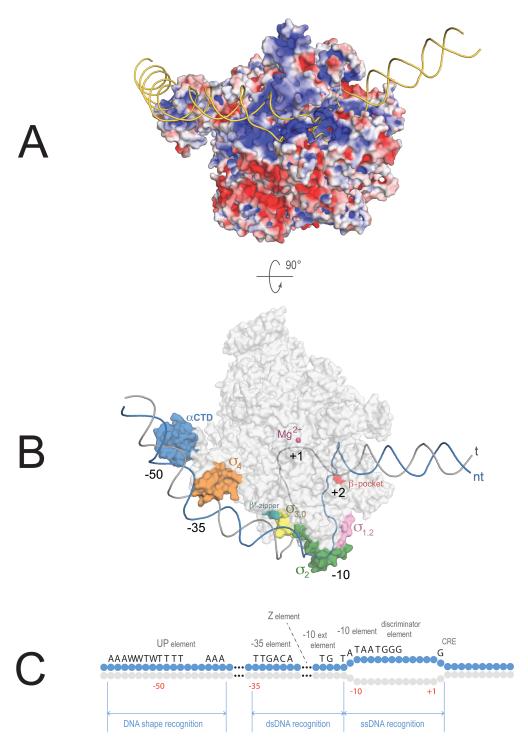
Figure 1.
Model of the open promoter complex. (A) View showing electrostatic surface potential of RNAP in the open promoter complex (red = negative, blue = positive charge). DNA (golden) is bound across a positively charged path on RNAP. RNAP engages the upstream region of promoter DNA (left) in sequence-specific recognition of dsDNA promoter elements. The melted part of the promoter bubble (right) is recognized through sequence-specific contacts with ssDNA. (B) View showing sequence-specific promoter elements and parts of RNAP recognizing them. RNAP shown as a gray transparent surface, except patches involved in sequence specific recognition of promoter DNA. DNA backbone is outlined (non-template strand = blue, template strand = gray). Model was created by combining coordinates from PDB 4G7H, 3UGO, 1LB2, and 1L9Z. (C) Promoter motifs recognized by RNAP holoenzyme with primary σ–factors. Blue circles represent nucleotides of the non-template DNA strand, light gray = template strand. W = A or T. The position with respect to the transcription start-site (+1) is denoted below.
Analysis of promoter sequences revealed several conserved elements important for RNAP binding. The most common and most highly conserved are two hexamers centered 35 and 10 bp upstream of the transcription start site (+1): the −10 element (TATAAT) and the −35 element (TTGACA)7––recognized by domain 2 and 4 of σ, respectively.8,9 Additional promoter elements include the extended −10 element (TG)10 and the discriminator (GGGA),11,12 also recognized by σ-subunit (domain 3 and 2, respectively).13,14 Core subunits also provide DNA-binding specificity and are involved in recognition of the UP-element (upstream promoter element), Z-element and core recognition element (CRE)14–16(Fig. 1C).
With the exception of the indispensable −10 element, other promoter elements may or may not be present. Each element plays its role at a certain step(s) of the promoter-opening pathway as it is recognized by RNAP: some in dsDNA form as they recruit RNAP to the promoter region, while others in ssDNA form concurently with melting. In fact, sequence-specific ssDNA recognition in the region undergoing melting initiates and drives promoter opening. The DNA-binding surfaces of RNAP, therefore, can be viewed as a collection of DNA-binding sites with different roles and DNA-specificities that come into play in a concerted fashion. In the following, I will review recent advances in our understanding of how RNAP finds promoters in the vastness of the genome and how promoter melting occurs, leading to initiation of RNA synthesis.
Promoter search
DNA-binding proteins are able to locate their target sites in an overwhelming excess of non-target DNA. The impressive speed and efficiency of this search, given the size of the macromolecules involved, cannot be explained by simple diffusion in the cytoplasm.17 This needle-in-the-haystack problem has puzzled molecular biologists for decades, and the mechanisms utilized by proteins in search of their target sites on genomic DNA are still being elucidated. To explain this paradox, Berg, Winter, and von Hippel suggested that DNA-binding proteins first use their non-specific DNA-binding affinity to arrive at any binding site on the DNA and in the next step search for their target site by means of thermal diffusion along DNA (reduced available volume for this search explains fast target location).18,19 Along a short DNA stretch, a protein can slide freely or translocate through a series of microscopic dissociation–reassociation events, whereas for larger DNA lengths the search could be manifested through intersegment transfer.
In the case of RNAP, the location of its binding site (promoter search) is perhaps the most enigmatic step in transcription initiation, due to the short-lived nature of the search intermediates. For the bacterial RNA-polymerase (RNAP), the task of locating promoters within an average-sized genome translates into finding a fraction of sequences comprising just a few percent of the available DNA in the cell.20 Evidence for 1D sliding of RNAP along DNA has been obtained both in bulk biochemical assays21,22and in single-molecule experiments.23 More specifically, RNAP has been shown to track a DNA groove as it moves along in search of a promoter site.24 Time-resolved scanning force microscopy revealed that both 1D sliding and 3D hopping accompany RNAP movement along DNA,25 while recent in vivo fluorescent microscopy studies suggest that both mechanisms may operate in living cells.26 Despite the ample evidence that RNAP can slide along the DNA while searching for the promoter site, the observed promoter-association kinetic parameters are perfectly explained by a 3D-diffusion mechanism alone, and a recent study argued that 3D collisions may indeed be the only mechanism operating in the cell, given the high in vivo concentrations of RNAP holoenzyme.27 It is still possible that low-copy transcription factors or RNAPs with alternative σ-factors (with just a few tens of molecules per cell) resort to facilitated diffusion in order to reach their target sites.
The structure of the bacterial nucleoid and the distribution of RNAP are dynamic and can be influenced by environmental conditions.28 Similar to transcription factories in eukaryotes, bacteria can organize promoters in specific cellular locations, which in turn can modulate promoter strength.29 Active promoters often reside within regions of the bacterial genome containing multiple overlapping promoter-like sequences, which could play roles in channeling RNAP into the promoter region.30 For eukaryotic transcription factors, a mechanism was proposed that would rapidly engage a “treadmilling” transcription factor (i.e., in a state of continual binding and dissociation from its target site) and convert it to a more stable binding state, allowing for a clutch-like genomic response to developmental or environmental cues.31 The latter work also introduced an important methodology to study the binding dynamics of a transcription factor as a true predictor of its strength. Studies of how the presence of additional promoter-like sequences affect RNAP binding turnover dynamics at true promoter sites could shed light on their role and explain the abundance of such sequences in bacterial genomes.
Base excision repair (BER) proteins, a classic model for the studies of target site search process, are able to locate isolated damaged bases in the genome with an efficiency that cannot be explained even by facilitated diffusion in a restricted volume. It was proposed that redox-active [4Fe–4S] clusters present in some DNA repair proteins are able to sense charge-conducting properties of DNA (electron propagation along an intact base stack that would be disrupted in the case of a DNA lesion) and dramatically improve the efficiency of damage detection via this mechanism.32 In addition to DNA repair factors, Fe–S clusters were found to be essential components of various nucleic acid processing enzymes such as DNA polymerases, helicases, glycosylases, primases, nucleases, and transcription factors.33 RNAPs from some archea, plants, and protozoa contain a Fe–S cluster that is required for RNAP assembly and has been proposed to play a role in sensing redox state of the cell.34 The location of the cluster near DNA in the modeled RNAP–promoter complex and its high conservation among several evolutionary distant RNAPs suggest the attractive but, at this point, speculative possibility that it may be involved in DNA-mediated redox signaling either directly or via a transcription factor.
The initial phase of promoter search by the bacterial RNAP may involve indirect readout of DNA (shape recognition). Variations in DNA sequence create unique conformational signatures with distinct geometrical helix parameters and deformability depending on local patterns of interactions between stacked bases. Whole-genome analyses argue that the topographical landscape of DNA molecular shape formed as a result of these interactions is conserved and can be subject to evolutionary selection35 much like protein shape. This can provide an efficient means for fast shape readout.36 For example, RNAP binding to UP-element involves recognition of a narrow minor groove,15,37 while −10 hexamer DNA was proposed to have altered structure even in the absence of DNA-binding proteins.38,39
While the quest for the characterization of the elusive promoter search intermediates is ongoing, one should keep in mind the highly mobile nature of the DNA helix, in which base flipping and non-canonical base pairing occur frequently and can be recognized by proteins. The detection of transient Hoogsteen base pairs forming within duplex DNA40 points to the possibility for multiple layers of a protein–DNA readout code in addition to simple linear sequence.
In many cases, the promoter search by RNAP is modulated by the actions of activators and repressors (reviewed in Ref. 41). These protein factors can modify the RNAP DNA-binding surface by either supplying it with additional DNA-recognition determinants or by blocking existing surfaces. Alternatively, the modulating factors can change the conformation of the promoter DNA by either making it more attractive to RNAP or by obstructing existing promoter sites. The majority of activators and repressors act early in the initiation pathway (usually at the promoter recruitment phase), although recent studies of the bacteriophage T7 Gp2 inhibitor and the Mycobacterium tuberculosis transcriptional modulator CarD argue that these factors affect the promoter-opening step.42,43
Promoter opening
Following the recruitment phase of the promoter search, involving recognition of DNA shape and dsDNA sequence, RNAP unwinds about 1.3 turns of the DNA (from −11 to +3), forming the open promoter complex. At this stage, RNAP specifically recognizes individual bases in the non-template strand of the promoter DNA––this activity underlies the melting capabilities of RNAP since the contacts with ssDNA bases can only be established during or after DNA unwinding.
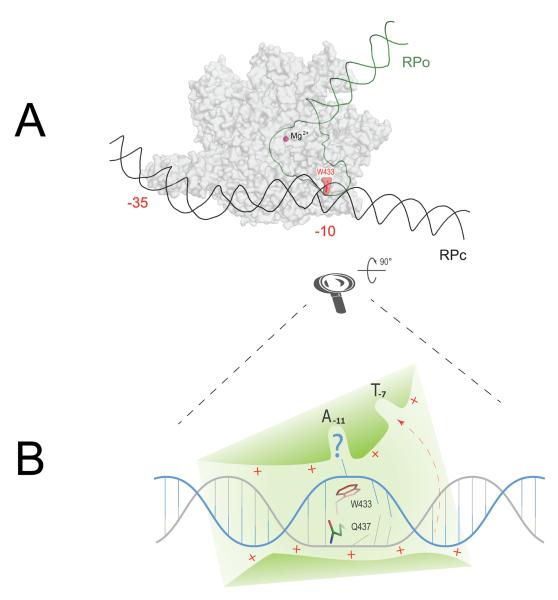
Promoter melting is triggered by the recognition of the −10 element, which occurs as the two most conserved bases of the element (A–11 and T–7) are flipped out of the double-stranded DNA and into complementary protein pockets of the σ2 domain.9,14,44 Structural modeling and biochemical data suggest a hypothetical timeline of this key event in promoter opening (Fig. 2): directed by upstream sequence-specific promoter–RNAP contacts and electrostatic interactions with the sugar–phosphate backbone, the −10 element DNA is loaded in a shallow positively charged trough formed by the surface of σ2, σ3 and parts of the β subunit. A prominent Trp residue (σ W433 in E. coli) located at the bottom of the trough precludes binding of an intact B-form DNA helix, acting as a wedge disrupting the −11 base pair and initiating the flipping of −11A into its pocket.9,45 This absolutely conserved Trp may be acting, therefore, as a functional analog of the “interrogating residue” of DNA-binding proteins that recognize flipped-out bases.46,47
Figure 2.
Structural modeling of −10 element recognition as the first step in promoter opening. (A) Schematic comparison of closed (RPc) and open (RPo) promoter complexes and suggested wedge role for W433 as an initiator of promoter melting. RNAP shown as a gray semi-transparent surface. DNA in RPc is shown black, downstream portion of promoter DNA after the melting (RPo) is green. The wedge W433 is highlighted in red. Catalytic Mg2+ = purple sphere. −35 and −10 promoter elements are labeled. (B) Schematic close-up of the first step in −10 element recognition. DNA directed in the shallow trough on RNAP surface (green) via a steric and electrostatic fit. Non-template strand is shown in blue, template strand is gray. The helix invasion by W433 disrupts one of the base pairs and flips out the non-template base into A-11 pocket, thereby making the upstream neighboring template-strand base accessible for H-bonding with Q437 (this amino acid residue was implicated in −12 base pair recognition in genetic screens.58,59 Following the recognition of the T–12A–11 step, DNA untwisting will proceed downstream, accompanied by T–7 flipping out into the respective σ-pocket (shown by red dashed arrow).
Remarkably, even though dsDNA recognition elements recruit RNAP to the promoter and align the −10 element with the recognition surface of σ2, specific recognition of the −10 element and initiation of melting (at least in vitro) has been shown to occur in their absence. Promoter fragments having only the −10 and discriminator elements (both recognized in ssDNA form) support transcription in vitro,11 and DNA fragments with only the −10 element are recognized by RNAP through specific interaction with the nt-strand.9,48 These examples suggest that RNAP distorts the DNA helix as it searches for the −10 element within dsDNA. The prominent position of Trp433 at the cusp of the protein–DNA interface at the origin of the melting suggests that as the dsDNA is threaded through the trough during the search process, the Trp wedge can flip bases out to be sampled in the A–11 pocket (Fig. 2).
Using structural modeling along with biochemical evidence, Feklistov and Darst9 challenged the previous concept of sequence-specific recognition of the double-stranded −10 element in the closed promoter complex, suggesting that binding of RNAP to the −10 element DNA may involve an intermediate with locally distorted duplex where the DNA helix is pre-opened to facilitate readout of the flipped bases. The early intermediates of promoter opening observed on various promoters have been termed closed on the basis of their non-reactivity toward MnO4– treatment (an assay used to reveal unstacked, solvent-exposed thymine bases). This technique fails to detect opening in cases where the thymines in the melted region remain stacked and/or protected by contacts with the protein. Therefore, new methods for assessing the state of the DNA helix in early melting intermediates need to be developed.
After initiation of melting at the A-11 position, the promoter bubble grows in the downstream direction. Sequence specific recognition of additional ssDNA elements (discriminator, −6 to −4; and core recognition element, −4 to +2) at some promoters may facilitate promoter opening at this stage (Fig. 1C). Therefore, the checkpoints in the process of promoter search do not stop at the recruitment phase but continue all the way to the formation of the fully open promoter bubble. Even when the bubble is fully formed, sequence-dependent conformational fluctuations of the open complex can fine-tune the choice of the transcription start site.49
The exact sequence of events leading to the formation of the open promoter bubble is currently a subject of debate. Data obtained at the bacteriophage λ PR promoter at sub-physiological temperatures (used to slow down the opening reaction) argue that after establishing upstream promoter contacts, RNAP readily bends promoter DNA (at about 90°) at the −10 element and inserts the downstream duplex in the active site cleft.50 In the next slow step, RNAP opens the DNA and readjusts contacts with emerging stretches of ssDNA as well as with the downstream DNA duplex.51–53 Real time X-ray–generated hydroxyl-radical footprinting data at the bacteriophage T7A1 promoter under physiological temperatures suggests that DNA opening originates outside of the cleft while DNA bends and enters the cleft later in the pathway.54,55 It is not impossible that even for a particular promoter both pathways may coexist. It is also possible that, depending on promoter sequence, one or the other pathway may prevail, which could present an opportunity for differential regulation in vivo. Notably, even for the same promoter sequence, DNA opening pathway can go through different structural intermediates depending on the experimental temperature.55
Overall, promoter melting is driven by RNAP affinity toward the final state (i.e., the conformation of promoter DNA existing in RPo). At the opening step, RNAP can be envisioned as an isomerization machine utilizing binding free energy to bend promoter DNA around its surface and unwind about 13 base pairs of the dsDNA, placing the t-strand near the active site ready for coding of the transcript sequence.50 Opening of the promoter DNA is accompanied by closing of the RNAP clamp,56 allowing RNAP to acquire a tight grip on the downstream DNA to assure processivity of transcription. This process may involve refolding of several structural domains of RNAP at the downstream parts of the cleft.50,53 The driving force of this dramatic rearrangement is supplied by electrostatic and steric complementarity between the positively charged surface of RNAP and the negatively charged DNA backbone, while the accuracy ensuring precise register of transcription comes from sequence-specific contacts to the dsDNA and ssDNA promoter elements (Fig. 1B).
Conclusions
Between the moment of association of RNAP core enzyme with one of the cellular σ-factors and the moment of synthesis of the first phosphodiester bond, RNAP is engaged in a complex multi-step process of promoter search. Each sub-step of this process can be modulated by regulatory protein factors or small molecules.46,57 The transcriptional output of a promoter is determined not just by how efficiently it is located and melted by RNAP, but also by the ease of promoter escape. Mechanistic understanding of the role of individual promoter elements in determining promoter output is a prerequisite for building a detailed quantitative model of bacterial gene expression. Recent structural and biochemical studies have advanced our understanding of the open promoter complex organization, although many important promoter search/opening intermediates still await characterization.
Acknowledgements
I would like to thank Ruth Saecker and Anastasia Sevostyanova for helpful comments on this manuscript. I am especially grateful to Seth A. Darst for inspiring mentorship, engaging discussions and support. My postdoctoral research was supported, in part, by a Merck Postdoctoral Fellowship at The Rockefeller University and NIH R01 GM053759 Grant to Seth A. Darst.
References
- 1.Darst SA. Bacterial RNA polymerase. Curr Opin Struct Biol. 2001;11:155–162. doi: 10.1016/s0959-440x(00)00185-8. [DOI] [PubMed] [Google Scholar]
- 2.Vassylyev DG, Sekine S-I, Laptenko O, Lee J, Vassylyeva MN, Borukhov S, Yokoyama S. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 A resolution. Nature. 2002;417:712–719. doi: 10.1038/nature752. [DOI] [PubMed] [Google Scholar]
- 3.Murakami KS, Masuda S, Darst SA. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 A resolution. Science. 2002a;296:1280–1284. doi: 10.1126/science.1069594. [DOI] [PubMed] [Google Scholar]
- 4.Murakami KS. The X-ray Crystal Structure of Escherichia Coli RNA Polymerase Sigma70 Holoenzyme. J Biol Chem. 2013;288:9126–9134. doi: 10.1074/jbc.M112.430900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gruber TM, Gross CA. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol. 2003;57:441–466. doi: 10.1146/annurev.micro.57.030502.090913. [DOI] [PubMed] [Google Scholar]
- 6.Murakami KS, Darst SA. Bacterial RNA polymerases: the wholo story. Curr Opin Struct Biol. 2003;13:31–39. doi: 10.1016/s0959-440x(02)00005-2. [DOI] [PubMed] [Google Scholar]
- 7.Shultzaberger RK, Chen Z, Lewis KA, Schneider TD. Anatomy of Escherichia coli sigma70 promoters. Nucleic Acids Res. 2007;35:771–788. doi: 10.1093/nar/gkl956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell EA, Muzzin O, Chlenov M, Sun JL, Olson CA, Weinman O, Trester-Zedlitz ML, Darst SA. Structure of the bacterial RNA polymerase promoter specificity sigma subunit. Mol Cell. 2002;9:527–539. doi: 10.1016/s1097-2765(02)00470-7. [DOI] [PubMed] [Google Scholar]
- 9.Feklistov A, Darst SA. Structural Basis for Promoter −10 Element Recognition by the Bacterial RNA Polymerase σ Subunit. Cell. 2011;147:1257–1269. doi: 10.1016/j.cell.2011.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barne KA, Bown JA, Busby SJW, Minchin SD. Region 2.5 of the Escherichia coli RNA polymerase sigma70 subunit is responsible for the recognition of the “extended-10” motif at promoters. EMBO J. 1997;16:4034–4040. doi: 10.1093/emboj/16.13.4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feklistov A, Barinova N, Sevostyanova A, Heyduk E, Bass I, Vvedenskaya I, Kuznedelov K, Merkienẹ E, Stavrovskaya E, Klimasauskas S, et al. A Basal Promoter Element Recognized by Free RNA Polymerase σ Subunit Determines Promoter Recognition by RNA Polymerase Holoenzyme. Mol Cell. 2006;23:97–107. doi: 10.1016/j.molcel.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 12.Haugen SP, Berkmen MB, Ross W, Gaal T, Ward C, Gourse RL. rRNA promoter regulation by nonoptimal binding of sigma region 1.2: an additional recognition element for RNA polymerase. Cell. 2006;125:1069–1082. doi: 10.1016/j.cell.2006.04.034. [DOI] [PubMed] [Google Scholar]
- 13.Murakami KS, Masuda S, Campbell EA, Muzzin O, Darst SA. Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex. Science. 2002b;296:1285–1290. doi: 10.1126/science.1069595. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y, Feng Y, Chatterjee S, Tuske S, Ho MX, Arnold E, Ebright RH. Structural basis of transcription initiation. Science. 2012;338:1076–1080. doi: 10.1126/science.1227786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benoff B, Yang H, Lawson CL, Parkinson G, Liu J, Blatter E, Ebright YW, Berman HM, Ebright RH. Structural basis of transcription activation: the CAP-alpha CTD-DNA complex. Science. 2002;297:1562–1566. doi: 10.1126/science.1076376. [DOI] [PubMed] [Google Scholar]
- 16.Yuzenkova Y, Tadigotla VR, Severinov K, Zenkin N. A new basal promoter element recognized by RNA polymerase core enzyme. EMBO J. 2011 doi: 10.1038/emboj.2011.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Riggs AD, Bourgeois S, Cohn M. The lac repressor-operator interaction. 3. Kinetic studies. J Mol Biol. 1970;53:401–417. doi: 10.1016/0022-2836(70)90074-4. [DOI] [PubMed] [Google Scholar]
- 18.Berg OG, Winter RB, Hippel, von PH. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry. 1981;20:6929–6948. doi: 10.1021/bi00527a028. [DOI] [PubMed] [Google Scholar]
- 19.Hippel, von PH, Berg OG. Facilitated target location in biological systems. J Biol Chem. 1989;264:675–678. [PubMed] [Google Scholar]
- 20.Wang F, Greene EC. Single-Molecule Studies of Transcription: From One RNA Polymerase at a Time to the Gene Expression Profile of a Cell. J Mol Biol. 2011;412:814–831. doi: 10.1016/j.jmb.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park CS, Wu FY, Wu CW. Molecular mechanism of promoter selection in gene transcription. II. Kinetic evidence for promoter search by a one-dimensional diffusion of RNA polymerase molecule along the DNA template. J Biol Chem. 1982;257:6950–6956. [PubMed] [Google Scholar]
- 22.Ricchetti M, Metzger W, Heumann H. One-dimensional diffusion of Escherichia coli DNA-dependent RNA polymerase: a mechanism to facilitate promoter location. Proc Natl Acad Sci USA. 1988;85:4610–4614. doi: 10.1073/pnas.85.13.4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kabata H, Kurosawa O, Arai I, Washizu M, Margarson SA, Glass RE, Shimamoto N. Visualization of single molecules of RNA polymerase sliding along DNA. Science. 1993;262:1561–1563. doi: 10.1126/science.8248804. [DOI] [PubMed] [Google Scholar]
- 24.Sakata-Sogawa K, Shimamoto N. RNA polymerase can track a DNA groove during promoter search. Proc Natl Acad Sci USA. 2004;101:14731–14735. doi: 10.1073/pnas.0406441101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bustamante C, Guthold M, Zhu X, Yang G. Facilitated target location on DNA by individual Escherichia coli RNA polymerase molecules observed with the scanning force microscope operating in liquid. J Biol Chem. 1999;274:16665–16668. doi: 10.1074/jbc.274.24.16665. [DOI] [PubMed] [Google Scholar]
- 26.Bratton BP, Mooney RA, Weisshaar JC. Spatial distribution and diffusive motion of RNA polymerase in live Escherichia coli. J Bacteriol. 2011;193:5138–5146. doi: 10.1128/JB.00198-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang F, Redding S, Finkelstein IJ, Gorman J, Reichman DR, Greene EC. The promoter-search mechanism of Escherichia coli RNA polymerase is dominated by three-dimensional diffusion. Nat Struct Mol Biol. 2012;20:174–181. doi: 10.1038/nsmb.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cabrera JE, Jin DJ. The distribution of RNA polymerase in Escherichia coli is dynamic and sensitive to environmental cues. Mol Microbiol. 2003;50:1493–1505. doi: 10.1046/j.1365-2958.2003.03805.x. [DOI] [PubMed] [Google Scholar]
- 29.Sánchez-Romero M-A, Lee DJ, Sánchez-Morán E, Busby SJW. Location and dynamics of an active promoter in Escherichia coli K-12. Biochem. J. 2012;441:481–485. doi: 10.1042/BJ20111258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huerta AM, Collado-Vides J. Sigma70 promoters in Escherichia coli: specific transcription in dense regions of overlapping promoter-like signals. J Mol Biol. 2003;333:261–278. doi: 10.1016/j.jmb.2003.07.017. [DOI] [PubMed] [Google Scholar]
- 31.Lickwar CR, Mueller F, Hanlon SE, McNally JG, Lieb JD. Genome-wide protein-DNA binding dynamics suggest a molecular clutch for transcription factor function. Nature. 2012;484:251–255. doi: 10.1038/nature10985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sontz PA, Muren NB, Barton JK. DNA Charge Transport for Sensing and Signaling. Acc. Chem. Res. 2012;45:1792–1800. doi: 10.1021/ar3001298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.White MF, Dillingham MS. Iron-sulphur clusters in nucleic acid processing enzymes. Curr Opin Struct Biol. 2012;22:94–100. doi: 10.1016/j.sbi.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 34.Hirata A, Murakami KS. Archaeal RNA polymerase. Curr Opin Struct Biol. 2009;19:724–731. doi: 10.1016/j.sbi.2009.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parker SCJ, Hansen L, Abaan HO, Tullius TD, Margulies EH. Local DNA topography correlates with functional noncoding regions of the human genome. Science. 2009;324:389–392. doi: 10.1126/science.1169050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rohs R, Jin X, West SM, Joshi R, Honig B, Mann RS. Origins of Specificity in Protein-DNA Recognition. Annu Rev Biochem. 2010;79:233–269. doi: 10.1146/annurev-biochem-060408-091030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gourse RL, Ross W, Gaal T. UPs and downs in bacterial transcription initiation: the role of the alpha subunit of RNA polymerase in promoter recognition. Mol Microbiol. 2000;37:687–695. doi: 10.1046/j.1365-2958.2000.01972.x. [DOI] [PubMed] [Google Scholar]
- 38.Drew HR, Weeks JR, Travers AA. Negative supercoiling induces spontaneous unwinding of a bacterial promoter. EMBO J. 1985;4:1025–1032. doi: 10.1002/j.1460-2075.1985.tb03734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spassky A, Rimsky S, Buc H, Busby SJW. Correlation between the conformation of Escherichia coli −10 hexamer sequences and promoter strength: use of orthophenanthroline cuprous complex as a structural index. EMBO J. 1988;7:1871–1879. doi: 10.1002/j.1460-2075.1988.tb03020.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nikolova EN, Kim E, Wise AA, O’Brien PJ, Andricioaei I, Al-Hashimi HM. Transient Hoogsteen base pairs in canonical duplex DNA. Nature. 2011;470:498–502. doi: 10.1038/nature09775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee DJ, Minchin SD, Busby SJW. Activating transcription in bacteria. Annu. Rev. Microbiol. 2012;66:125–152. doi: 10.1146/annurev-micro-092611-150012. [DOI] [PubMed] [Google Scholar]
- 42.James E, Liu M, Sheppard C, Mekler V, Cámara B, Liu B, Simpson P, Cota E, Severinov K, Matthews S, et al. Structural and mechanistic basis for the inhibition of Escherichia coli RNA polymerase by T7 Gp2. Mol Cell. 2012;47:755–766. doi: 10.1016/j.molcel.2012.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bae B, Campbell E, Darst S, et al. manuscripts in preparation.
- 44.Liu X, Bushnell DA, Kornberg RD. Lock and key to transcription: σ-DNA interaction. Cell. 2011;147:1218–1219. doi: 10.1016/j.cell.2011.11.033. [DOI] [PubMed] [Google Scholar]
- 45.Tomsic M, Tsujikawa L, Panaghie G, Wang Y, Azok J, deHaseth PL. Different roles for basic and aromatic amino acids in conserved region 2 of Escherichia coli sigma(70) in the nucleation and maintenance of the single-stranded DNA bubble in open RNA polymerase-promoter complexes. J Biol Chem. 2001;276:31891–31896. doi: 10.1074/jbc.M105027200. [DOI] [PubMed] [Google Scholar]
- 46.Lee S, Bowman BR, Ueno Y, Wang S, Verdine GL. Synthesis and structure of duplex DNA containing the genotoxic nucleobase lesion N7-methylguanine. J Am Chem Soc. 2008;130:11570–11571. doi: 10.1021/ja8025328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yi C, Chen B, Qi B, Zhang W, Jia G, Zhang L, Li CJ, Dinner AR, Yang C-G, He C. Duplex interrogation by a direct DNA repair protein in search of base damage. Nat Struct Mol Biol. 2012;19:671–676. doi: 10.1038/nsmb.2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Niedziela-Majka A, Heyduk T. Escherichia coli RNA polymerase contacts outside the - 10 promoter element are not essential for promoter melting. J Biol Chem. 2005;280:38219–38227. doi: 10.1074/jbc.M507984200. [DOI] [PubMed] [Google Scholar]
- 49.Robb NC, Cordes T, Hwang LC, Gryte K, Duchi D, Craggs TD, Santoso Y, Weiss S, Ebright RH, Kapanidis AN. The Transcription Bubble of the RNA Polymerase-Promoter Open Complex Exhibits Conformational Heterogeneity and Millisecond-Scale Dynamics: Implications for Transcription Start-Site Selection. J Mol Biol. 2012 doi: 10.1016/j.jmb.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saecker RM, Record MT, deHaseth PL. Mechanism of bacterial transcription initiation: RNA polymerase - promoter binding, isomerization to initiation-competent open complexes, and initiation of RNA synthesis. J Mol Biol. 2011;412:754–771. doi: 10.1016/j.jmb.2011.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Davis CA, Bingman CA, Landick R, Record MT, Saecker RM. Real-time footprinting of DNA in the first kinetically significant intermediate in open complex formation by Escherichia coli RNA polymerase. Proc Natl Acad Sci USA. 2007;104:7833–7838. doi: 10.1073/pnas.0609888104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gries TJ, Kontur WS, Capp MW, Saecker RM, Record MT. One-step DNA melting in the RNA polymerase cleft opens the initiation bubble to form an unstable open complex. Proceedings of the National Academy of Sciences. 2010;107:10418–10423. doi: 10.1073/pnas.1000967107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Drennan A, Kraemer M, Capp M, Gries T, Ruff E, Sheppard C, Wigneshweraraj S, Artsimovitch I, Record MT. Key Roles of the Downstream Mobile Jaw of Escherichia coli RNA Polymerase in Transcription Initiation. Biochemistry. 2012;51:9447–9459. doi: 10.1021/bi301260u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sclavi B, Zaychikov E, Rogozina A, Walther F, Buckle M, Heumann H. Real-time characterization of intermediates in the pathway to open complex formation by Escherichia coli RNA polymerase at the T7A1 promoter. Proc Natl Acad Sci USA. 2005;102:4706–4711. doi: 10.1073/pnas.0408218102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rogozina A, Zaychikov E, Buckle M, Heumann H, Sclavi B. DNA melting by RNA polymerase at the T7A1 promoter precedes the rate-limiting step at 37 degrees C and results in the accumulation of an off-pathway intermediate. Nucleic Acids Res. 2009;37:5390–5404. doi: 10.1093/nar/gkp560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chakraborty A, Wang D, Ebright YW, Korlann Y, Kortkhonjia E, Kim T, Chowdhury S, Wigneshweraraj S, Irschik H, Jansen R, et al. Opening and closing of the bacterial RNA polymerase clamp. Science. 2012;337:591–595. doi: 10.1126/science.1218716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Haugen SP, Ross W, Gourse RL. Advances in bacterial promoter recognition and its control by factors that do not bind DNA. Nat. Rev. Microbiol. 2008;6:507–519. doi: 10.1038/nrmicro1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kenney TJ, York K, Youngman P, Moran CPJ., Jr. Genetic evidence that RNA polymerase associated with sA factor uses a sporulation-specific promoter in Bacillus subtilis. Proc Natl Acad Sci USA. 1989;86:9109–9113. doi: 10.1073/pnas.86.23.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Waldburger C, Gardella T, Wong R, Susskind MM. Changes in conserved region 2 of Escherichia coli sigma 70 affecting promoter recognition. J Mol Biol. 1990;215:267–276. doi: 10.1016/s0022-2836(05)80345-6. [DOI] [PubMed] [Google Scholar]