Abstract
Insect oocytes grow in close association with the ovarian follicular epithelium (OFE), which escorts the oocyte during oogenesis and is responsible for synthesis and secretion of the eggshell. We describe a transcriptome of OFE of the triatomine bug Rhodnius prolixus, a vector of Chagas disease, to increase our knowledge of the role of FE in egg development. Random clones were sequenced from a cDNA library of different stages of follicle development. The transcriptome showed high commitment to transcription, protein synthesis, and secretion. The most abundant cDNA was a secreted (S) small, proline-rich protein with maximal expression in the vitellogenic follicle, suggesting a role in oocyte maturation. We also found Rp45, a chorion protein already described, and a putative chitin-associated cuticle protein that was an eggshell component candidate. Six transcripts coding for proteins related to the unfolded protein response (UPR) by were chosen and their expression analyzed. Surprisingly, transcripts related to UPR showed higher expression during early stages of development and downregulation during late stages, when transcripts coding for S proteins participating in chorion formation were highly expressed. Several transcripts with potential roles in oogenesis and embryo development are also discussed. We propose that intense protein synthesis at the FE results in reticulum stress (RS) and that lowering expression of a set of genes related to cell survival should lead to degeneration of follicular cells at oocyte maturation. This paradoxical suppression of UPR suggests that ovarian follicles may represent an interesting model for studying control of RS and cell survival in professional S cell types.
Keywords: Reticulum stress, Rhodnius prolixus, Transcriptome, Chorion, Oocyte, Apoptosis
1. Introduction
Making viable eggs that will survive in the environment independently of the maternal body is a key step in the reproduction process of all oviparous animals, including insects. In their way from oocyte precursor to the mature egg—a process frequently completed in just a few hours, these cells accumulate large amounts of reserves and increase about a thousand-fold in size, followed by isolation from the external world by formation of the egg shell. Egg development in insect ovaries is divided into three main phases: pre-vitellogenic growth, vitellogenic growth (also called vitellogenesis), and choriogenesis. Pre-vitellogenic growth occurs in the anterior end of the ovariole, in the case of the hemipteran telotrophic ovary, in a lanceolate structure called tropharium (Atella et al., 2005). Despite their astonishing growth rates, oocytes themselves show very modest synthetic activity (Wallace et al., 1972), and their development is supported mainly by other cells either from the ovary or from extra-ovarian tissues (i.e., fat body).
Early growth of oocytes takes place in close association with other cell types such as the surrounding follicular epithelium (FE) and nurse cells. These are linked to oocytes by means of cytoplasmic projections/bridges called trophic cords that are used to provide both proteins and RNA. Trophic cords are retained until the early phases of vitellogenic growth (Telfer et al., 1982). During vitellogenesis, oocytes rapidly accumulate large amounts of lipids, glycogen, RNA, and yolk proteins. Vitellin, the most common yolk protein, derives from a hemolymphatic lipoprotein called vitellogenin (Vtg), which in most cases is synthesized in the fat body and is taken up by oocytes by receptor-mediated endocytosis. Synthesis of Vtg by ovarian FE (OFE) cells has been reported in cycloraphan dipteran ovaries (Postlethwait et al., 1980) as well as in the triatomine Rhodnius prolixus (Melo et al., 2000). Also, non-vitellin yolk proteins such as the 30-kDa protein from lepidopteran eggs are synthesized by FE. Upon completion of vitellogenesis, fully grown eggs are separated from the external world by the chorion, a protective proteinaceous layer synthesized and secreted by follicle cells at the very end of oocyte development inside the ovary. After choriogenesis, FE cells degenerate and die, exhibiting several features characteristic of programmed cell death, in a process that is believed to be important to prevent blockage of the ovarioles and to support proper egg development (Nezis et al., 2006; Nezis et al., 2002).
The endoplasmic reticulum (ER) is the gate to the exocytotic pathway, where newly synthesized secretory (S) proteins must fold and acquire their correct functional tri-dimensional structure. In recent years, it has been recognized that frequently a large portion of nascent polypeptide chains fail to fold appropriately, eventually leading to accumulation of misfolded proteins in the ER—what is called reticulum stress (RS) (Banhegyi et al., 2007). Eukaryotic cells respond to ER stress by increasing transcription of genes for ER resident chaperones and proteins that help either to accelerate flux in the secretory pathway or to promote destruction of misfolded proteins (Kohno et al., 1993; Mori et al., 1992; Romisch, 2005). Collectively, this process has been termed the unfolded protein response (UPR). To eliminate organelles or unhealthy cells, autophagy or apoptotic cell death take place whenever the UPR does not occur properly or is not sufficient to deal with the ER stress challenge (Lai et al., 2007).
To provide insights into the molecular framework involved in development of the insect ovary, we describe here a transcriptome of the most abundant transcripts found in the follicle of the triatomine Rhodnius prolixus, a vector of Trypanosoma cruzi, the causative agent of Chagas disease. Several messages related to oocyte and embryo development were identified, and expression of selected transcripts during follicle development was studied. We propose that failure in triggering UPR during choriogenesis leads to death of FE cells as part of the developmental program of the insect egg.
2. Materials and methods
2.1. Insects and tissues
R. prolixus were from a colony maintained at 28°C and 70% relative humidity. Insects used here were adult mated females having their second blood meal after their imaginal molt. Ovarioles were dissected free of tracheae and ovarian sheath. Ovarian follicles at vitellogenic stages (0.5–2.0 mm) or initiating choriogenesis (about 2 mm) were opened with tweezers and the oocyte content discarded. Due to their reduced size, pre-vitellogenic follicles were separated from tropharium and used both for library and quantitative real-time PCR (qPCR). Tropharium samples were used only for qPCR and were not included in the cDNA library.
2.2. cDNA library and sequencing
mRNA was isolated from using the Micro FastTrack mRNA isolation kit (Invitrogen, Carlsbad, CA), and the PCR-based cDNA library was made following the instructions for the SMART cDNA library construction kit (Clontech, Palo Alto, CA) according to the manufacturer’s instructions. cDNA from follicle cells isolated as described above were fractionated in three size intervals using a gel filtration column, ligated separately to the λ TriplEx 2 vector, packaged with packing extract Gigapack III Gold (Stratagene, La Jolla, CA), and then joined in a single pool. The obtained phage library was mixed with log phase XL-1 Blue cells (Stratagene) and plated in agar medium containing IPTG and X-gal for blue/white screening. A sample of individual white lysis plates was chosen randomly and the cloned insert amplified using PT2F1 (5′-AAG TAC TCT AGC AAT TGT GAG C-3′, upstream) and PT2R1 (5′ CTC TTC GCT ATT ACG CCA GCT G-3′, downstream) primers and sequenced in a DNA sequencing instrument ABI 377 (Applied Biosystems, Foster City, CA) using the BigDye Terminator 3.0 kit (Applied Biosystems) and the primer PT2F3 (5′-TCT CGG GAA GCG CGC CAT TGT-3′, downstream a PT2F1) as described elsewhere (Valenzuela et al., 2002).
2.3. Bioinformatics
The sequences obtained were cleaned from vector and primer sequences and assembled into contigs using CAP3 software (Huang and Madan, 1999) mastered by an in-house bioinformatics program (Ribeiro et al., 2007). The protein coding potential of the contigs and singletons obtained was examined using the blastx algorithm (Altschul and Gish, 1996) to search the non-redundant protein (NR) and Gene Ontology (GO) (Ashburner et al., 2000) databases. As the translation mRNA of proteins that are coded by mitochondrial DNA involves the use of an alternate genetic code, we have also compared the transcripts with a database created from mitochondrial DNA sequences. Essentially, these analyses were performed using a program pipeline already described in the literature (Valenzuela et al., 2003). To get hints on the possible biologic role of the proteins obtained from the translated contigs, rpsblast was used to search for conserved protein domains in the Pfam (Bateman et al., 2000), SMART (Schultz et al., 2000), COG (Tatusov et al., 2003), and conserved domains (CDD) (Marchler-Bauer et al., 2002) databases. This information is particularly important, because we kept in our analysis open-reading frames that lack the initial methionine (5′-truncated), stop codon (3′-truncated), or both (fragment) and not only full-length coding sequences. Although the BLAST result against GO gives insight to a putative function of the transcripts, in the process of manual annotation the transcripts were classified into one of the following fifteen classes: protein synthesis, protein export machinery, transcription machinery, transcription factor, proteasome machinery, extracellular matrix (ECM)/S protein/ovarian function, protein modification machinery, energy metabolism, signal transduction, nuclear regulation, transporter, cytoskeletal/cell adhesion, intermediate metabolism, unknown (U), and unknown conserved.
To confirm that the expressed sequence tags (ESTs) derived from the Rhodnius genome, these were also BLASTed against the R. prolixus genome. Supercontigs from the R. prolixus assembled genome (release 1.0.1, from January 2009) were downloaded from http://genome.wustl.edu/genomes/view/rhodnius_prolixus/. The FASTA files of these supercontigs were broken into 50-kb fragments with 10 kb from previous sequence, and a database was created. This procedure was taken because BLAST does not work well if subjects in the database are very long sequences (http://blast.ncbi.nlm.nih.gov/Blast.cgi?CMD=Web&PAGE_TYPE=BlastDocs).
This formatted database was used to blastn the ESTs to the genome of the insect. The few ESTs not found in any genome trace file were disregarded unless they code for a sequence very similar to a known protein.
After assembly and comparison to databases, sequences coding for proteins larger than 50 amino acids (aa) and/or that showed best matches to NR with e-values lower than 10−5 were further analyzed. As polypeptides produced by the FE cells that are components of the chorion must be secreted, we tried to identify translation products that were potentially secreted. Segments of the six-frame translations of the EST having a methionine in the first 100 predicted amino acids were submitted to the SignalP server (Nielsen et al., 1997). Multiple alignments of proteins of interest were performed using ClustalW (Thompson et al., 2002).
2.4. qPCR
Total RNA was isolated using TRIzol reagent (Invitrogen) from isolated tropharium or ovarian follicles that were either pre-vitellogenic, vitellogenic, or that had already entered choriogenesis. RNA extracted from oocyte content of vitellogenic or chorionated oocytes was also extracted and analyzed together with previous preparations. Tissues were obtained as described above. Total RNA was quantified in a spectrophotometer, and 1 μg was treated with 1 Unit of DNase (RNAse free; Invitrogen) for 30 min at 37°C. Reactions were stopped by adding 1 μl of 20 mM EDTA and heating for 10 min at 65°C. cDNA synthesis was performed from the DNase-treated RNA using the high-capacity cDNA reverse transcription kit from Applied Biosystems.
All primers (Supplemental Table S3) were made using Primer 3 (http://frodo.wi.mit.edu/primer3/) and Oligoanalyzer software (http://www.idtdna.com/analyzer/applications/oligoanalyzer/).
Before qPCR reactions, all primers pairs were tested by conventional PCR using a 40 cycle reaction (94°C, 30 s denaturation; 60°C, 30 s annealing; 72°C, 30 s – Taq extension) in an Applied Biosystems 2400 thermocycler and analyzed in 2% agarose gel.
Real-time qPCR was performed in an AB 7500 (Applied Biosystems) using power SYBR-green PCR master mix. The comparative Ct method (Livak and Schmittgen, 2001; Pfaffl, 2001) was used to compare changes in gene expression levels. Actin gene expression was used as endogenous control, using primers based on an actin cDNA sequence described elsewhere (Ribeiro et al., 2004): RpAct F 50-AGTAGCTGCATGGGTTGTAG-3(forward) and RpAct R 50-CAACATACATTGCTGGACTG-30 (reverse) (Alves-Bezerra et al., 2010).
2.5. Apoptosis TUNEL assay
Vitellogenic follicles never showed evidence of cell death, a feature characteristic of the end of choriogenesis. Late follicles were first screened by Trypan blue exclusion assay to check for the presence of dead cells. The FEs of selected oocytes were carefully dissected, fixed for 1 h in 4% paraformaldehyde, and treated for the terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) assay using the ApopTagPlus kit (Chemicon, Temecula CA) following manufacturer’s instructions. Briefly, the cells were treated with proteinase K (20 μg/ml) to pretreat the tissue, followed by incubations in 3% H2O2 to quench endogenous peroxidase. Samples were then incubated for 2 h at 37°C in a solution of terminal deoxynucleotidyl transferase. After washing, samples were incubated at room temperature for 1 h with anti-digoxigenin antibody conjugated with peroxidase, washed with PBS, and developed with diaminobenzidine. Slides were observed in an Observer.Z1 fluorescence microscope equipped with a CCD camera model AxioCAM MRM (Carl Zeiss, Inc., Thornwood, NJ).
3. Results and Discussion
3.1. FC cDNA from R. prolixus was isolated, sequenced, and used to build a sequence database
Clones (total of 494) were sequenced and used to assemble a database (see Supplemental Table S1) that, after clusterization, yielded 341 unique sequences (320 singletons and 21 contigs). Approximately 71 % of all sequences showed matches with an e-value < 10−5 to the NR protein database. All pertinent information was included in an Excel spreadsheet (Supplemental Table S1) and used for manual annotation of sequences (included in column O). The annotated sequences were grouped into 15 major functional classes (Fig. 1; Supplemental Tables S1, column P and TableS2, column AM). The overall picture of the transcriptome showed a tissue highly committed to transcription, protein synthesis, and secretion—functions that accounted for more than two-thirds of all transcripts. Only contigs that gave a positive match with the R. prolixus genome were included in this analysis (Assembly 1.0, available at http://genome.wustl.edu/tools/blast/Rhodnius_prolixus-1.0.1). The results of the BLAST of the contigs against the genome can be found in column Q of Supplemental Table S1.
Figure 1.
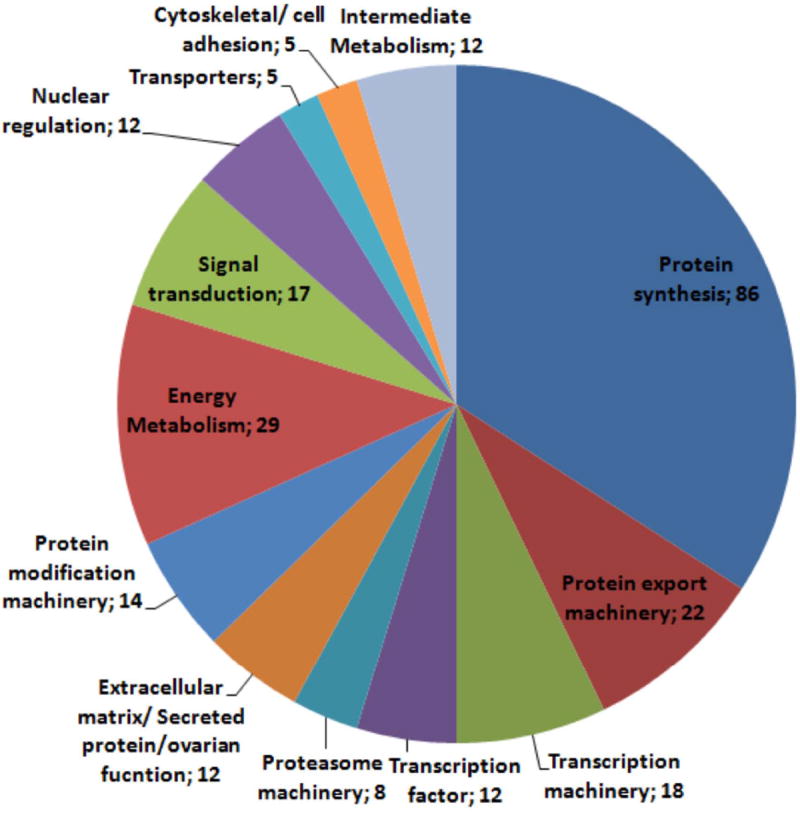
Functional classification of transcripts from ovary follicular epithelium library
The 252 transcripts that matched a known protein database with an evalue < 10-5 were grouped in 13 major functional classes: Protein synthesis, Protein export machinery, Transcription machinery, Transcription factor, Proteasome machinery, Extracellular matrix/Secreted protein/ovarian function, Protein modification machinery, Energy Metabolism, Signal transduction, Nuclear regulation, Transporters, Cytoskeletal/ cell adhesion, Intermediate Metabolism.
3.2. Secreted (S) proteins and genes possibly related to follicle and oocyte development
S proteins represent a contribution of follicle cells to oocyte content or to eggshell. Searching the open reading frames for candidate S proteins showing a signal peptide—and as a help to guide annotation—the contigs from Supplemental Table S1 were six-frame translated and the resulting peptides submitted to http://www.cbs.dtu.dk/services/SignalP/. The prediction results (Supplemental Table S1 column K in Spreadsheet ALL) were manually examined, and those in which the initial methionine was present were transferred to Supplemental Table S1, spreadsheet SignalP. We identified eight contigs coding for S proteins (Supplemental Fig. 3) and another seven coding for membrane proteins. Three of those contigs are composed of multiple reads but have no reliable match in any available protein database.
The most highly expressed message (contig 1, herein named rpSPRP, for small proline-rich protein) accounted for about 8% of all transcripts. This cDNA codes for a protein of 137 aa, which is expected to be converted into a mature protein of 118 aa a residues and an estimate mass of 13.2 kDa. It has a low-complexity sequence in which the most conspicuous feature is the high proline content (21 residues; about 18%). Although similar proteins have not been described in insects, in mammalian tissues, SPRPs were identified as important components of epithelial tissue (Nemes and Steinert, 1999). SPRPs have been shown to take part in formation of the so-called cornified cell envelope, which is a hard layer made of protein/lipid polymer that stays on the exterior of dead cornified cells of the skin. Several proteins (including SPRPs) are covalently linked by protein crosslinking to form this layer, which is thought to be important in conferring biomechanical properties to the cornified cell envelope that allow it to function as a protective barrier, which is the essence of the squamous epithelial tissue, a physiological role that is also performed by insect egg chorion. Analysis of gene expression shows that rpSPRP expression is basically limited to the epithelium of vitellogenic follicles (Fig. 2A). This could result from either rpSPRP performing a role in yolk formation or being a component of follicle ECM or, alternatively, could be explained by accumulation of rpSPRP during vitellogenic growth followed by incorporation into eggshell only later, during choriogenesis.
Figure 2.
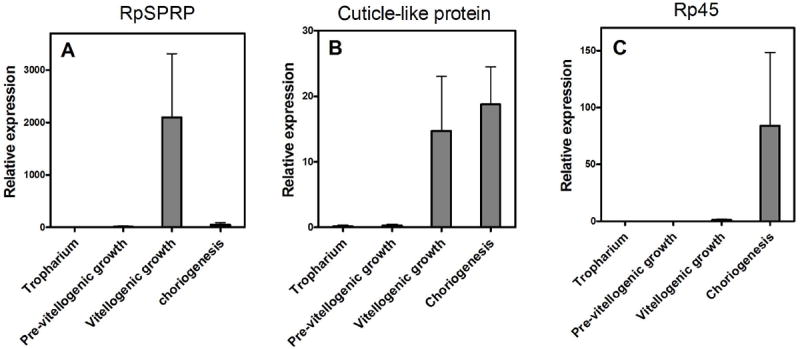
Expression of selected secreted proteins during developmental stages of ovarian follicle
Tropharium or follicular epithelium from follicles at pre-vitellogenesis, vitellogenesis or from follicles already engaged in chorion formation were dissected and mRNA levels of three transcripts showing signal peptides were evaluated by Q-PCR. Data show are mean ± SEM (n=4).
Another putative S protein (contig 162) showed significant homology to chitin-binding protein components of the insect cuticle that has a non-cysteine-based chitin-binding domain, thereby being a typical chorion protein candidate (Supplemental Fig. 2B). It has been shown that chitin is an important component of the insect egg shell (Moreira et al., 2007); therefore, chitin-binding proteins made by FE may also participate in formation of chorion. This mRNA coded for a 125- aa precursor and a small 105-residue mature protein, which was about equally expressed by epithelia of both vitellogenic and choriogenic follicles (Fig. 2B), reinforcing the point that some chorion proteins may be expressed before actual formation of the egg envelope. Contigs 232 and 296 (Supplemental Tables S1 and S2) codify for proteins with a strong candidate signal peptide (Supplemental Fig. S3) that shows a low complexity amino acid profile with relatively high abundance of glycine (26 and 20% of the amino acid residues of the mature protein, respectively). Glycine-rich proteins are frequently found among components of ECM, being identified also as components of the cornified envelope of mammalian skin such as loricrin (Nemes and Steinert, 1999). Glycine-rich domains are also found in spider fibroin, where repeats of GA or GS resembling those found in contigs 232 and 296 are thought to be involved in determination of mechanical properties of silk web (Gosline et al., 2002; Gosline et al., 1999).
A partial sequence for one of the secreted proteins (Rp45, contig 97; Supplemental Table S1 and Supplemental Fig. 1) had already been reported, being described as a component of Rhodnius eggshell (Bouts et al., 2007). As expected for a major chorion component, evaluation of expression of this message by qPCR showed that this protein is exclusively expressed during choriogenesis (Fig. 2C), confirming the developmental profile described before (Bouts et al., 2007).
One mechanism that has been shown to produce hardening of the egg chorion is the oxidative crosslinking of proteins promoted by the action of peroxidases, such as the ovoperoxidases (Edens et al., 2001; Heinecke and Shapiro, 1990; Margaritis, 1985), which need a source of hydrogen peroxide to catalyze the formation of dityrosine. This task could well be fulfilled by the extracellular SOD coded by contig 417 (Supplemental Tables S1 and S2).
Contig 531 codes for a putative S protein (Table S1, Supplemental Fig. S3) that would fit in the class of odorant-binding proteins, a group that already has several members that do not bind odor molecules but other types of hydrophobic ligands, such as heme as the Rhodnius heme-binding protein (Dansa-Petretski et al., 1995; Paiva-Silva et al., 2002) or biogenic amines such as the D7 family of proteins found in the saliva of Diptera (Calvo et al., 2006). One interesting candidate ligand for this protein would be juvenile hormone, as the Drosophila take-out protein—also a member of the odorant-binding protein family—has been shown to bind juvenile hormone and modulate development of the male reproductive tract (Dauwalder et al., 2002; Noriega et al., 2006).
Contig 306 codes from a homolog of Jagunal (Supplemental Tables S1 and S2), an evolutionarily conserved transmembrane ER protein that is found in both oocytes and follicular cells of the Drosophila ovary, where it has been related to reorganization of the ER during follicle development and ER-derived vesicle trafficking (Lee and Cooley, 2007). Jagunal comes from a Korean word that means “small egg,” as mutations for this gene are lethal and were shown to lead to abnormal follicle morphology.
During oogenesis, the FE is exposed to mechanical stretching due both to the large rate of growth of the vitellogenic oocyte and to contraction of the ovarian sheath muscle. Ezrin/radixin/moesin (ERM) proteins (Supplemental Tables S1 and S2, contig 124) are located at the cell cortex and interact with actin filaments, bridging them with the plasma membrane, and play a major role in this process. The dramatic changes in cell volume and shape that occur during oocyte development and the opening of large spaces between epithelial cells are needed to expose the oocyte surface to hemolymph, thus allowing receptor-mediated uptake of vitellogenin, the yolk protein precursor that is 80% of the protein content of mature Rhodnius eggs (Oliveira et al., 1986). This opening of intercellular spaces, termed patency, is known to be under the control of juvenile hormone (Davey, 2007) and requires extensive rearrangement of cytoskeleton and cell volume. ERM proteins (Supplemental Tables S1 and S2, contig 124) are also involved in formation of microvilli, cell-cell adhesion, cell shape, and membrane trafficking and seem to be part of this process. In addition to rearrangement of the cytoskeleton, these proteins integrate diverse signaling pathways, are phosphorylated by protein kinases, are capable of binding phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2), and interact with small GTP-binding proteins (Niggli and Rossy, 2008).
A VASA-like transcript (Supplemental Tables S1 and S2, contig 322) was found in this transcriptome and might play a role in oocyte development. VASA proteins—DEAD-box RNA-binding proteins with an RNA dependent helicase activity—are required for pole determination and completion of oogenesis in Drosophila (Abdelhaleem, 2005; Styhler et al., 1998; Tinker et al., 1998). In Drosophila, VASA expression takes place in the oocyte, germ cells, and nurse cells (Raz, 2000); however, in the lizard Podarcis sicula, VASA protein is localized also in a group of follicle cells of somatic origin that perform functions that in Drosophila are characteristic of nurse cells, transferring molecules and eventually their entire cytoplasm to the oocytes by means of intercellular bridges (Maurizii et al., 2009). Because cytoplasmic connections between nurse cells and oocyte are broken during mid vitellogenesis (Bjornsson and Huebner, 2004), it is interesting to speculate that VASA may enable FE cells from the telotrophic ovary of hemipterans to perform functions that in Drosophila are restricted to nurse cells.
3.3. Protein synthesis, secretion, and control of cell death
Global analysis of functional classification of transcripts (Fig.1, Supplemental Tables S1 and S2) showed that close to two-thirds of all proteins expressed that had a functional category (excluding proteins with unknown function) were somehow related to transcription, translation, protein turnover, and vesicular traffic, reflecting the immense metabolic effort accomplished by follicular cells in providing their contribution to egg maturation (Fig. 1). This picture fits well with the abundant ER that occupies almost all the cytosol of binucleated FE cells (Atella et al., 2005). In the last few years, the concept of RS has been well established, defining a link between intense protein synthesis/secretion and control of programmed cell death. Proteins targeted for secretion enter the ER as unfolded polypeptides with reduced cysteines. Maturation of these proteins in the secretory pathway involves oxidative formation of the disulphide bond and reshuffling; hence, proper protein folding is highly dependent on fine tuning of redox equilibrium in the ER. The so-called unfolded-protein response (UPR) frequently observed during cellular stress conditions—including increased protein secretion—is intrinsically related to the mechanisms that control cell death and survival and has been the subject of an extensive literature recently reviewed (Tabas and Ron, 2011). Several transcripts identified here are potentially involved in the control of cell death (Supplemental Table S1, Supplemental Fig. S2) and could play an important role in the preservation of FE cells facing a condition of RS. Initially, we hypothesized that this would mean that control of RS would be a major priority of follicular cells that were actively engaged in secretion of oocyte yolk and chorion proteins. This led us to evaluate expression of a selected set of genes by qPCR. Measurement of the expression of six transcripts probably involved in the control of UPR and prevention of programmed cell death showed expression to be increased during early stages of oogenesis and repressed at late stages, especially in the choriogenic follicle (Fig. 3; defender against cell death [DAD]-like, HSP90, DNAJ/HSP40, Sec-24, Sec-61, and 14-3-3ε). The Dad gene (contig 259, Fig. 3A, Supplemental Fig. S2, panel D) (Kelleher and Gilmore, 1997) is a subunit of an oligosaccharyltransferase, and mutants with truncated forms of DAD were shown to present increased apoptosis (Nakashima et al., 1993), while higher expression led to survival (Sugimoto et al., 1995), reflecting the importance of N-glycosylation for protein traffic in the ER-Golgi pathway and the link between ER stress and cell death. In a similar way, Sec24 (contig 73, Fig. 3F and Supplemental Fig. S2, panel B) is an essential component of the coat protein complex (COPII) that works in secretory vesicle formation and is involved in selection of newly synthesized proteins for export (Pagano et al., 1999; Peng et al., 2000). Sec61 (contig 46, Fig. 3E, Supplemental Fig. S2, panel A) also participates in assembly of the secretory highway in the Sec61 complex that constitutes a channel for signal peptide-dependent protein import and retrograde transport of misfolded proteins out of the ER (Robson and Collinson, 2006). Chaperones of the Hsp90 family (contig 488, Fig. 2E) and the DNAJ/Hsp40 (a co-chaperone of HSP70) (contig 516, Supplemental Fig. S2, panel F) are also well known for their participation in protein folding and secretion (Qiu et al., 2006). The Hsp70/DNAJ chaperone has been shown to act in UPR, preventing apoptosis induced by CHOP (Gotoh et al., 2004). 14-3-3ε proteins (contig 446, Supplemental Fig. S2, panel C) homologs are downregulated at late oogenesis (Fig. 3 D). Proteins of this class have been implicated in several signal transducing pathways (Fu et al., 2000) including cell-cycle control in Drosophila (Su et al., 2001) and definition of oocyte and epithelial cell polarity (Benton and St Johnston, 2003), although their precise role in cell death control and survival is less well defined concerning molecular mechanisms. Evidence relating 14-3-3ε proteins to apoptosis show that these proteins are capable of associating with BAD and that cleavage by caspase 3 releases BAD, which in turn associates with Bcl-2 (Qi and Martinez, 2003).
Figure 3.
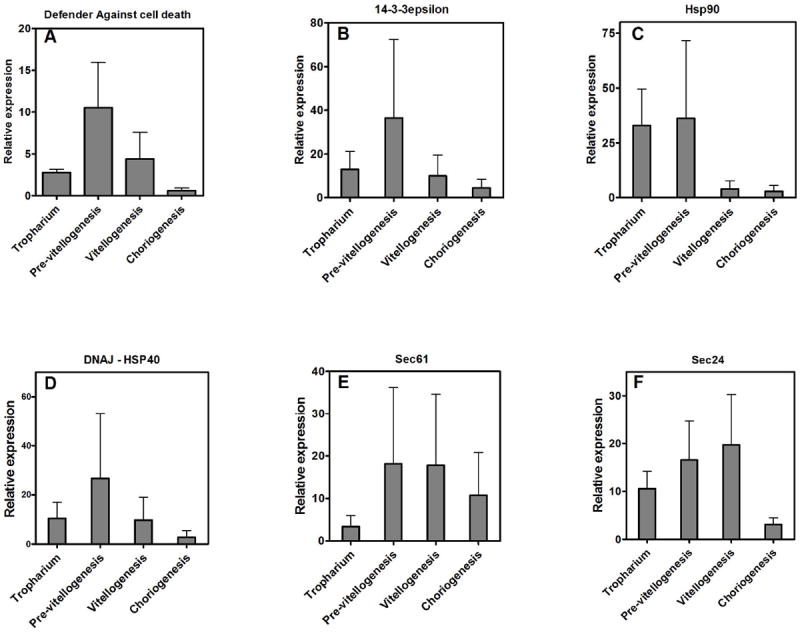
Expression of selected genes related to reticulum stress during development of ovarian follicles
In most eukaryotic cells, the expected response of being exposed to conditions that induce UPR is increased expression of survival transcripts, as observed in the fungi Aspergillus nidans (Guillemette et al., 2007). Lee et al. (2003) have described a similar set of genes, the transcription of which is induced by XBP-1 and that are thought to be the chaperone effectors of the UPR in the ER; however, we show here that FE at late developmental stages not only does not increase expression of this group of genes but rather shows a pronounced trend to lower its transcription (Fig. 3).
In contrast to the more common pattern of cellular response of triggering the UPR gene set to preserve cell integrity, these results show downregulation of UPR-related genes when there is intense ER traffic. After follicle cells complete chorion secretion, they degenerate and die, forming a residual body, referred to by some authors as corpus luteum (Baum et al., 2005; Wigglesworth, 1953). Therefore, programmed death of follicle cells should be taken as part of the oogenesis developmental program. In accordance with this (Nezis et al., 2006; Nezis et al., 2002) reported the occurrence of programmed cell death in degeneration of ovarian follicle cells of higher Diptera. Therefore, here we propose as a working hypothesis that death of cells in the ovarian follicle is fueled by the RS owing to its effort to synthesize chorion proteins. In line with this hypothesis, we found evidence that clusters of cells from the FE die in the end of oogenesis, as showed by Trypan blue staining exclusion assay in choriogenic oocytes (Fig. 4, A and B, arrows). Additionally, it was possible to detect TUNEL-positive cells randomly dispersed in the epithelium of choriogenic oocytes (Fig. 4 E–H, arrows) (1.5 ± 0.9 TUNEL-positive cells/ 400 μm2) and not in the epithelium of vitellogenic oocytes (Fig. 4, C and D). The process of cell death seems to be very rapid and is apparently triggered by local interactions between cells, as Trypan blue-stained cells were always found as clusters and TUNEL-positive cells were only found in the epithelium of oocytes at late choriogenesis. It was described in Drosophila that follicle cells are interconnected with intercellular bridges (gap-junction type) that are confined to arrays of no more than eight cells, and that several of these independent clusters are found in the epithelium (Woodruff and Tilney, 1998) (Haglund et al., 2010). The authors discuss that cell-to-cell movement throughout the epithelium of global cytosolic regulatory molecules cannot occur via these intercellular bridges, but weak signals affecting only one or a few cells in each cluster would be amplified by spread through the intercellular bridges. Thus, we cannot rule out the possibility that the signaling events that trigger cell death at late oogenesis in R. prolixus may be associated with these types of intimate connections between the follicle cells, resulting in the cluster patterning that we observed by Trypanblue staining.
Figure 4.
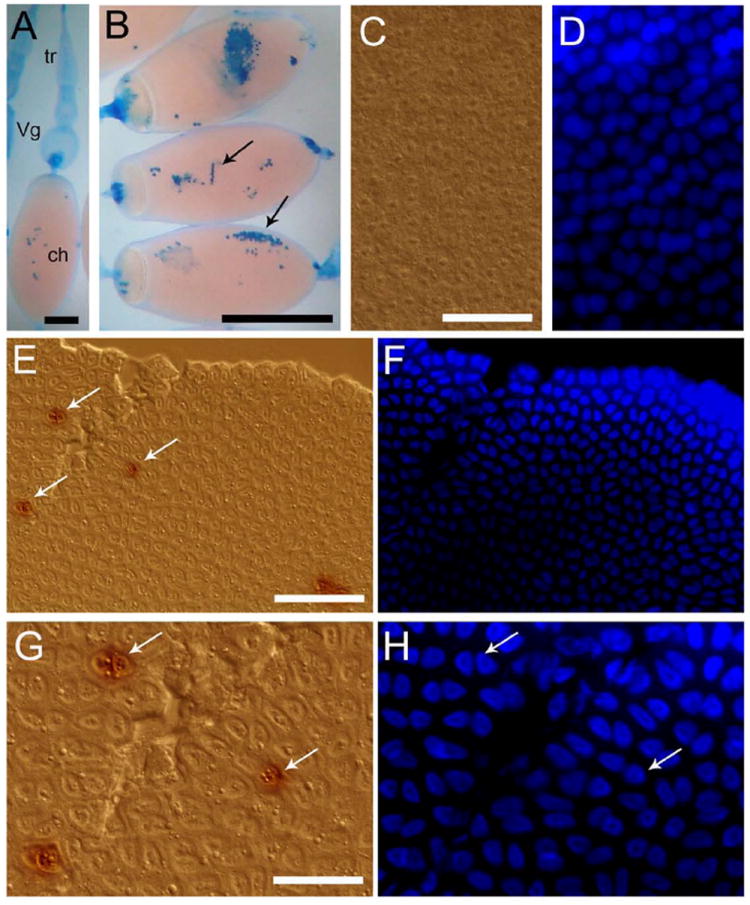
Apoptotic cell death occurs at the end of chorion formation
A, B, Trypan blue exclusion assay. Arrows indicate regions with dead cells. Tr (tropharium), Vg (vitellogenic follicle) and ch (choriogenic oocyte). Bars: 1 mm, 2 mm, respectively. C, D, Follicular epithelium of vitelogenic oocytes after TUNEL assay. Bar: 120 μm. E, F, Follicular epithelium of choriogenic oocytes showing labeled nuclei (arrows) after TUNEL assay. Bar: 120 μm. G, H, Detail of the TUNEL label in different nuclei. Bar: 50 μm. Note that the epithelial cells are binucleated.
4. Conclusions
The field of developmental biology is heavily biased toward embryogenesis. Egg components that are synthesized and properly assembled during oogenesis constitute the starting point for growth of a viable embryo. Here we present evidence that in R. prolixus at late stages of egg formation, programmed death of follicle cells occurs and we propose that this simultaneity is not merely a fortuitous coincidence. Instead, we propose that the death of follicle cells is a crucial part of the oogenesis developmental program and occurs to help in building of the chorion, a protective coat that will increase the biologic success of insect progeny. Our data show that downregulation of expression of cell survival-inducing transcripts occurs simultaneously with the increased expression of secreted proteins. Although we do not have evidence to prove a cause-consequence relationship between these two events, their simultaneity leads us to hypothesize that development of endoplasmic RS is the driving force for FE cell death, which marks the termination of oogenesis. In this case, FE cell survival would not serve to increase the biologic success of this organism. On the other hand, death and degeneration of these cells fit the survival program of this species’ offspring. This unconventional regulation of UPR in ovarian cells is an interesting model for research on the relation between RS and cell death control.
Supplementary Material
Hyperlinked Microsoft Excel file with assembled EST’s and various database comparisons. This file can also be found at http://rhodnius.iq.ufrj.br/English/index.php?option=com_content&view=article&id=4&Itemid=5. This spreadsheet has three worksheets. The first one is named ALL and has all the information on the analysis of the sequences studied. The one named SinalP has concise information on sequences bearing a signaling sequence directing their corresponding protein to secretion or to membrane. The third worksheet has a detailed information on contend of the table.
Hyperlinked Microsoft Excel file with assembled EST’s and various database comparisons. This file can also be found at http://rhodnius.iq.ufrj.br/ as supplemental data.
Primers used for Q-PCR
- RMPLFEMC-97 was aligned with R. prolixus RP45 (RHOPRO_122720928-truncated at 3 prime) showing extremely high identity.
- RMPLFEMC-162 -a cuticular protein with chitin-binding domain (non-cysCBD) -was aligned with arthropod orthologous proteins, including:
- DROPSE_125980123 (Drosophila pseudoobscurapseudoobscura),
- DROWIL_195440352 (Drosophila willistoni),
- AEDAEG_157135722 (Aedes aegypti),
- CULQUI_170058444 (Culexquinquefasciatus),
- PEDHUM_242005146 (Pediculushumanuscorporis)
- TRICAS_270008195(Triboliumcastaneum)
The number attached to scientific name abbreviation is the NCBI gi number.
- RMPLFEMC-46 -SEC61, gamma subunit -fragment [Rhodnius prolixus]
- RMPLFEMC-73 -Vesicle coat complex COPII, subunit SEC24/subunit SFB2/subunit SFB3 -truncated at 5 prime [Rhodnius prolixus]
- RMPLFEMC-446 -Multifunctional chaperone (14-3-3 family) -truncated at 3 prime [Rhodnius prolixus]
- RMPLFEMC-259 -Defender against cell death protein/oligosaccharyltransferase, epsilon subunit [Rhodnius prolixus]
- RMPLFEMC-488 -heat shock protein 90 -truncated at 3 prime [Rhodnius prolixus]
- RMPLFEMC-516 -Molecular chaperone (DnaJ superfamily) -truncated at 3 prime [Rhodnius prolixus]
- AEDAEG, Aedes aegypti;
- ANOGAM, Anopheles gambiae;
- ACYPIS, Acyrthosiphonpisum;
- APIMEL, Apis mellifera;
- CULQUI, Culex quinquefasciatus;
- HARAXY, Harmonia axyridis;
- IXOSCA, Ixodes scapularis;
- NASVIT, Nasonia vitripennis;
- PEDHUM, Pediculus humanus corporis;
- RHOPRO, Rhodnius prolixus;
- TRICAS, Tribolium castaneum;
- TRIVAP, Trialeuro desvaporariorum.
The numbers following species name abbreviation is the gi number of the corresponding protein.
Signal peptide sequences from Rhodnius follicular secreted proteins.
- RMPLFEMC-1-rpSPRP,
- RMPLFEMC-97-RP45,
- RMPLFEMC-98-Putativethioesteraseprotein,
- RMPLFEMC-232-glycine-richprotein1,
- RMPLFEMC-277-Hypotheticalsecretedprotein,
- RMPLFEMC-296-glycine-richprotein2,
- RMPLFEMC-417-Superoxide dismutase (Cu/Zn)SOD1,
- RMPLFEMC-531-Putative OBP/JHbindingprotein.
Acknowledgments
We thank S. R. Cássia for technical assistance and NIAID intramural editor B. R. Marshall. This work was supported by grants from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Fundação de Amparo a Pesquisa de Estado do Rio de Janeiro (FAPERJ), Fundação Universitária José Bonifácio (FUJB), INCT-Entomologia Molecular, and Howard Hughes Medical Institute (HHMI, to PLO). JMCR was supported by the Intramural Research Program of the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Because JMCR is a government employees and this is a government work, the work is in the public domain in the United States. Notwithstanding any other agreements, the NIH reserves the right to provide the work to PubMedCentral for display and use by the public, and PubMedCentral may tag or modify the work consistent with its customary practices. You can establish rights outside of the U.S. subject to a government use license.
References
- Abdelhaleem M. RNA, helicases: Regulators of differentiation. Clinical Biochemistry. 2005;38:499–503. doi: 10.1016/j.clinbiochem.2005.01.010. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W. Local alignment statistics. Methods Enzymol. 1996;266:460–480. doi: 10.1016/s0076-6879(96)66029-7. [DOI] [PubMed] [Google Scholar]
- Alves-Bezerra M, Majerowicz D, Grillo LA, Tremonte H, Almeida CB, Braz GR, Sola-Penna M, Paiva-Silva GO, Gondim KC. Serotonin regulates an acyl-CoA-binding protein (ACBP) gene expression in the midgut of Rhodnius prolixus. Insect Biochem Mol Biol. 2010;40:119–125. doi: 10.1016/j.ibmb.2010.01.002. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atella GC, Gondim KC, Machado EA, Medeiros MN, Silva-Neto MA, Masuda H. Oogenesis and egg development in triatomines: a biochemical approach. An Acad Bras Cienc. 2005;77:405–430. doi: 10.1590/s0001-37652005000300005. [DOI] [PubMed] [Google Scholar]
- Banhegyi G, Baumeister P, Benedetti A, Dong D, Fu Y, Lee AS, Li J, Mao C, Margittai E, Ni M, Paschen W, Piccirella S, Senesi S, Sitia R, Wang M, Yang W. Endoplasmic reticulum stress. Ann N Y Acad Sci. 2007;1113:58–71. doi: 10.1196/annals.1391.007. [DOI] [PubMed] [Google Scholar]
- Bateman A, Birney E, Durbin R, Eddy SR, Howe KL, Sonnhammer EL. The Pfam protein families database. Nucleic Acids Res. 2000;28:263–266. doi: 10.1093/nar/28.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum JS, St George JP, McCall K. Programmed cell death in the germline. Semin Cell Dev Biol. 2005;16:245–259. doi: 10.1016/j.semcdb.2004.12.008. [DOI] [PubMed] [Google Scholar]
- Benton R, St Johnston D. Drosophila PAR-1 and 14-3-3 inhibit Bazooka/PAR-3 to establish complementary cortical domains in polarized cells. Cell. 2003;115:691–704. doi: 10.1016/s0092-8674(03)00938-3. [DOI] [PubMed] [Google Scholar]
- Bjornsson CS, Huebner E. Extracellular H+ dynamics during oogenesis in Rhodnius prolixus ovarioles. Journal of Experimental Biology. 2004;207:2835–2844. doi: 10.1242/jeb.01089. [DOI] [PubMed] [Google Scholar]
- Bouts DM, Melo AC, Andrade AL, Silva-Neto MA, Paiva-Silva Gde O, Sorgine MH, da Cunha Gomes LS, Coelho HS, Furtado AP, Aguiar EC, de Medeiros LN, Kurtenbach E, Rozental S, Cunha ESNL, de Souza W, Masuda H. Biochemical properties of the major proteins from Rhodnius prolixus eggshell. Insect Biochem Mol Biol. 2007;37:1207–1221. doi: 10.1016/j.ibmb.2007.07.010. [DOI] [PubMed] [Google Scholar]
- Calvo E, Mans BJ, Andersen JF, Ribeiro JM. Function and evolution of a mosquito salivary protein family. J Biol Chem. 2006;281:1935–1942. doi: 10.1074/jbc.M510359200. [DOI] [PubMed] [Google Scholar]
- Dansa-Petretski M, Ribeiro JM, Atella GC, Masuda H, Oliveira PL. Antioxidant role of Rhodnius prolixus heme-binding protein. Protection against heme-induced lipid peroxidation. J Biol Chem. 1995;270:10893–10896. doi: 10.1074/jbc.270.18.10893. [DOI] [PubMed] [Google Scholar]
- Dauwalder B, Tsujimoto S, Moss J, Mattox W. The Drosophila takeout gene is regulated by the somatic sex-determination pathway and affects male courtship behavior. Genes Dev. 2002;16:2879–2892. doi: 10.1101/gad.1010302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey K. The interaction of feeding and mating in the hormonal control of egg production in Rhodnius prolixus. Journal of Insect Physiology. 2007;53:208–215. doi: 10.1016/j.jinsphys.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Edens WA, Sharling L, Cheng G, Shapira R, Kinkade JM, Lee T, Edens HA, Tang X, Sullards C, Flaherty DB, Benian GM, Lambeth JD. Tyrosine cross-linking of extracellular matrix is catalyzed by Duox, a multidomain oxidase/peroxidase with homology to the phagocyte oxidase subunit gp91phox. Journal of Cell Biology. 2001;154:879–891. doi: 10.1083/jcb.200103132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Subramanian RR, Masters SC. 14-3-3 proteins: structure, function, and regulation. Annu Rev Pharmacol Toxicol. 2000;40:617–647. doi: 10.1146/annurev.pharmtox.40.1.617. [DOI] [PubMed] [Google Scholar]
- Gosline J, Lillie M, Carrington E, Guerette P, Ortlepp C, Savage K. Elastic proteins: biological roles and mechanical properties. Philos Trans R Soc Lond B Biol Sci. 2002;357:121–132. doi: 10.1098/rstb.2001.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosline JM, Guerette PA, Ortlepp CS, Savage KN. The mechanical design of spider silks: from fibroin sequence to mechanical function. J Exp Biol. 1999;202:3295–3303. doi: 10.1242/jeb.202.23.3295. [DOI] [PubMed] [Google Scholar]
- Gotoh T, Terada K, Oyadomari S, Mori M. hsp70-DnaJ chaperone pair prevents nitric oxide- and CHOP-induced apoptosis by inhibiting translocation of Bax to mitochondria. Cell Death Differ. 2004;11:390–402. doi: 10.1038/sj.cdd.4401369. [DOI] [PubMed] [Google Scholar]
- Guillemette T, van Peij NN, Goosen T, Lanthaler K, Robson GD, van den Hondel CA, Stam H, Archer DB. Genomic analysis of the secretion stress response in the enzyme-producing cell factory Aspergillus niger. BMC Genomics. 2007;8:158. doi: 10.1186/1471-2164-8-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haglund K, Nezis IP, Lemus D, Grabbe C, Wesche J, Liestol K, Dikic I, Palmer R, Stenmark H. Cindr interacts with anillin to control cytokinesis in Drosophila melanogaster. Curr Biol. 2010;20:944–950. doi: 10.1016/j.cub.2010.03.068. [DOI] [PubMed] [Google Scholar]
- Heinecke JW, Shapiro BM. Superoxide peroxidase activity of ovoperoxidase, the cross-linking enzyme of fertilization. J Biol Chem. 1990;265:9241–9246. [PubMed] [Google Scholar]
- Huang X, Madan A. CAP3: A DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher DJ, Gilmore R. DAD1, the defender against apoptotic cell death, is a subunit of the mammalian oligosaccharyltransferase. Proc Natl Acad Sci U S A. 1997;94:4994–4999. doi: 10.1073/pnas.94.10.4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno K, Normington K, Sambrook J, Gething MJ, Mori K. The promoter region of the yeast KAR2 (BiP) gene contains a regulatory domain that responds to the presence of unfolded proteins in the endoplasmic reticulum. Mol Cell Biol. 1993;13:877–890. doi: 10.1128/mcb.13.2.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai E, Teodoro T, Volchuk A. Endoplasmic reticulum stress: signaling the unfolded protein response. Physiology (Bethesda) 2007;22:193–201. doi: 10.1152/physiol.00050.2006. [DOI] [PubMed] [Google Scholar]
- Lee AH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol. 2003;23:7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Cooley L. Jagunal is required for reorganizing the endoplasmic reticulum during Drosophila oogenesis. Journal of Cell Biology. 2007;176:941–952. doi: 10.1083/jcb.200701048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Panchenko AR, Shoemaker BA, Thiessen PA, Geer LY, Bryant SH. CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res. 2002;30:281–283. doi: 10.1093/nar/30.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margaritis LH. The egg-shell of Drosophila melanogaster III. Covalent crosslinking of the chorion proteins involves endogenous hydrogen peroxide. Tissue Cell. 1985;17:553–559. doi: 10.1016/0040-8166(85)90031-x. [DOI] [PubMed] [Google Scholar]
- Maurizii MG, Cavaliere V, Gamberi C, Lasko P, Gargiulo G, Taddei C. Vasa protein is localized in the germ cells and in the oocyte-associated pyriform follicle cells during early oogenesis in the lizard Podarcis sicula. Development Genes and Evolution. 2009;219:361–367. doi: 10.1007/s00427-009-0295-7. [DOI] [PubMed] [Google Scholar]
- Melo AC, Valle D, Machado EA, Salerno AP, Paiva-Silva GO, Cunha ESNL, de Souza W, Masuda H. Synthesis of vitellogenin by the follicle cells of Rhodnius prolixus. Insect Biochem Mol Biol. 2000;30:549–557. doi: 10.1016/s0965-1748(00)00023-0. [DOI] [PubMed] [Google Scholar]
- Moreira MF, Dos Santos AS, Marotta HR, Mansur JF, Ramos IB, Machado EA, Souza GH, Eberlin MN, Kaiser CR, Kramer KJ, Muthukrishnan S, Vasconcellos AM. A chitin-like component in Aedes aegypti eggshells, eggs and ovaries. Insect Biochem Mol Biol. 2007;37:1249–1261. doi: 10.1016/j.ibmb.2007.07.017. [DOI] [PubMed] [Google Scholar]
- Mori K, Sant A, Kohno K, Normington K, Gething MJ, Sambrook JF. A 22 bp cis-acting element is necessary and sufficient for the induction of the yeast KAR2 (BiP) gene by unfolded proteins. EMBO J. 1992;11:2583–2593. doi: 10.1002/j.1460-2075.1992.tb05323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima T, Sekiguchi T, Kuraoka A, Fukushima K, Shibata Y, Komiyama S, Nishimoto T. Molecular cloning of a human cDNA encoding a novel protein, DAD1, whose defect causes apoptotic cell death in hamster BHK21 cells. Mol Cell Biol. 1993;13:6367–6374. doi: 10.1128/mcb.13.10.6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemes Z, Steinert PM. Bricks and mortar of the epidermal barrier. Exp Mol Med. 1999;31:5–19. doi: 10.1038/emm.1999.2. [DOI] [PubMed] [Google Scholar]
- Nezis IP, Stravopodis DJ, Margaritis LH, Papassideri IS. Programmed cell death of follicular epithelium during the late developmental stages of oogenesis in the fruit flies Bactrocera oleae and Ceratitis capitata (Diptera, Tephritidae) is mediated by autophagy. Dev Growth Differ. 2006;48:189–198. doi: 10.1111/j.1440-169X.2006.00856.x. [DOI] [PubMed] [Google Scholar]
- Nezis IP, Stravopodis DJ, Papassideri I, Robert-Nicoud M, Margaritis LH. Dynamics of apoptosis in the ovarian follicle cells during the late stages of Drosophila oogenesis. Cell Tissue Res. 2002;307:401–409. doi: 10.1007/s00441-001-0498-3. [DOI] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- Niggli V, Rossy J. Ezrin/radixin/moesin: versatile controllers of signaling molecules and of the cortical cytoskeleton. Int J Biochem Cell Biol. 2008;40:344–349. doi: 10.1016/j.biocel.2007.02.012. [DOI] [PubMed] [Google Scholar]
- Noriega FG, Ribeiro JM, Koener JF, Valenzuela JG, Hernandez-Martinez S, Pham VM, Feyereisen R. Comparative genomics of insect juvenile hormone biosynthesis. Insect Biochem Mol Biol. 2006;36:366–374. doi: 10.1016/j.ibmb.2006.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira PL, Gondim KC, Guedes DM, Masuda H. Uptake of Yolk Proteins in Rhodnius-Prolixus. Journal of Insect Physiology. 1986;32:859–866. [Google Scholar]
- Pagano A, Letourneur F, Garcia-Estefania D, Carpentier JL, Orci L, Paccaud JP. Sec24 proteins and sorting at the endoplasmic reticulum. J Biol Chem. 1999;274:7833–7840. doi: 10.1074/jbc.274.12.7833. [DOI] [PubMed] [Google Scholar]
- Paiva-Silva GO, Sorgine MHF, Benedetti CE, Meneghini R, Almeida IC, Machado EA, Dansa-Petretski M, Yepiz-Plascencia G, Law JH, Oliveira PL, Masuda H. On the biosynthesis of Rhodnius prolixus heme-binding protein. Insect Biochemistry and Molecular Biology. 2002;32:1533–1541. doi: 10.1016/s0965-1748(02)00074-7. [DOI] [PubMed] [Google Scholar]
- Peng R, De Antoni A, Gallwitz D. Evidence for overlapping and distinct functions in protein transport of coat protein Sec24p family members. J Biol Chem. 2000;275:11521–11528. doi: 10.1074/jbc.275.15.11521. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postlethwait JH, Bownes M, Jowett T. Sexual phenotype and vitellogenin synthesis in Drosophila melanogaster. Developmental Biology. 1980;79:379–387. doi: 10.1016/0012-1606(80)90123-2. [DOI] [PubMed] [Google Scholar]
- Qi W, Martinez JD. Reduction of 14-3-3 proteins correlates with increased sensitivity to killing of human lung cancer cells by ionizing radiation. Radiat Res. 2003;160:217–223. doi: 10.1667/rr3038. [DOI] [PubMed] [Google Scholar]
- Qiu XB, Shao YM, Miao S, Wang L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell Mol Life Sci. 2006;63:2560–2570. doi: 10.1007/s00018-006-6192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz E. The function and regulation of vasa-like genes in germ-cell development. Genome Biol. 2000;1:REVIEWS1017. doi: 10.1186/gb-2000-1-3-reviews1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro JM, Andersen J, Silva-Neto MA, Pham VM, Garfield MK, Valenzuela JG. Exploring the sialome of the blood-sucking bug Rhodnius prolixus. Insect Biochem Mol Biol. 2004;34:61–79. doi: 10.1016/j.ibmb.2003.09.004. [DOI] [PubMed] [Google Scholar]
- Ribeiro JM, Arca B, Lombardo F, Calvo E, Phan VM, Chandra PK, Wikel SK. An annotated catalogue of salivary gland transcripts in the adult female mosquito, Aedes aegypti. BMC Genomics. 2007;8:6. doi: 10.1186/1471-2164-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson A, Collinson I. The structure of the Sec complex and the problem of protein translocation. EMBO Rep. 2006;7:1099–1103. doi: 10.1038/sj.embor.7400832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romisch K. Endoplasmic reticulum-associated degradation. Annu Rev Cell Dev Biol. 2005;21:435–456. doi: 10.1146/annurev.cellbio.21.012704.133250. [DOI] [PubMed] [Google Scholar]
- Schultz J, Copley RR, Doerks T, Ponting CP, Bork P. SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 2000;28:231–234. doi: 10.1093/nar/28.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styhler S, Nakamura A, Swan A, Suter B, Lasko P. vasa is required for GURKEN accumulation in the oocyte, and is involved in oocyte differentiation and germline cyst development. Development. 1998;125:1569–1578. doi: 10.1242/dev.125.9.1569. [DOI] [PubMed] [Google Scholar]
- Su TT, Parry DH, Donahoe B, Chien CT, O’Farrell PH, Purdy A. Cell cycle roles for two 14-3-3 proteins during Drosophila development. J Cell Sci. 2001;114:3445–3454. doi: 10.1242/jcs.114.19.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto A, Hozak RR, Nakashima T, Nishimoto T, Rothman JH. dad-1, an endogenous programmed cell death suppressor in Caenorhabditis elegans and vertebrates. EMBO J. 1995;14:4434–4441. doi: 10.1002/j.1460-2075.1995.tb00122.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabas I, Ron D. Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat Cell Biol. 2011;13:184–190. doi: 10.1038/ncb0311-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, Rao BS, Smirnov S, Sverdlov AV, Vasudevan S, Wolf YI, Yin JJ, Natale DA. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telfer WH, Huebner E, Smith DS. The cell biology of vitellogenic follicles in Hyalophora and Rhodnius. In: King RC, Akai H, editors. Insect ultrastructure. Plenum Press; New York: 1982. [Google Scholar]
- Thompson JD, Gibson TJ, Higgins DG. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics. 2002;Chapter 2(Unit 2 3) doi: 10.1002/0471250953.bi0203s00. [DOI] [PubMed] [Google Scholar]
- Tinker R, Silver D, Montell DJ. Requirement for the vasa RNA helicase in gurken mRNA localization. Developmental Biology. 1998;199:1–10. doi: 10.1006/dbio.1998.8941. [DOI] [PubMed] [Google Scholar]
- Valenzuela JG, Francischetti IM, Pham VM, Garfield MK, Ribeiro JM. Exploring the salivary gland transcriptome and proteome of the Anopheles stephensi mosquito. Insect Biochem Mol Biol. 2003;33:717–732. doi: 10.1016/s0965-1748(03)00067-5. [DOI] [PubMed] [Google Scholar]
- Valenzuela JG, Pham VM, Garfield MK, Francischetti IM, Ribeiro JM. Toward a description of the sialome of the adult female mosquito Aedes aegypti. Insect Biochem Mol Biol. 2002;32:1101–1122. doi: 10.1016/s0965-1748(02)00047-4. [DOI] [PubMed] [Google Scholar]
- Wallace RA, Nickol JM, Ho T, Jared DW. Studies on amphibian yolk. X. The relative roles of autosynthetic and heterosynthetic processes during yolk protein assembly by isolated oocytes. Developmental Biology. 1972;29:255–272. doi: 10.1016/0012-1606(72)90066-8. [DOI] [PubMed] [Google Scholar]
- Wigglesworth VB. The principles of insect physiology. 5. Methuen; London: 1953. [Google Scholar]
- Woodruff RI, Tilney LG. Intercellular bridges between epithelial cells in the Drosophila ovarian follicle: a possible aid to localized signaling. Dev Biol. 1998;200:82–91. doi: 10.1006/dbio.1998.8948. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Hyperlinked Microsoft Excel file with assembled EST’s and various database comparisons. This file can also be found at http://rhodnius.iq.ufrj.br/English/index.php?option=com_content&view=article&id=4&Itemid=5. This spreadsheet has three worksheets. The first one is named ALL and has all the information on the analysis of the sequences studied. The one named SinalP has concise information on sequences bearing a signaling sequence directing their corresponding protein to secretion or to membrane. The third worksheet has a detailed information on contend of the table.
Hyperlinked Microsoft Excel file with assembled EST’s and various database comparisons. This file can also be found at http://rhodnius.iq.ufrj.br/ as supplemental data.
Primers used for Q-PCR
- RMPLFEMC-97 was aligned with R. prolixus RP45 (RHOPRO_122720928-truncated at 3 prime) showing extremely high identity.
- RMPLFEMC-162 -a cuticular protein with chitin-binding domain (non-cysCBD) -was aligned with arthropod orthologous proteins, including:
- DROPSE_125980123 (Drosophila pseudoobscurapseudoobscura),
- DROWIL_195440352 (Drosophila willistoni),
- AEDAEG_157135722 (Aedes aegypti),
- CULQUI_170058444 (Culexquinquefasciatus),
- PEDHUM_242005146 (Pediculushumanuscorporis)
- TRICAS_270008195(Triboliumcastaneum)
The number attached to scientific name abbreviation is the NCBI gi number.
- RMPLFEMC-46 -SEC61, gamma subunit -fragment [Rhodnius prolixus]
- RMPLFEMC-73 -Vesicle coat complex COPII, subunit SEC24/subunit SFB2/subunit SFB3 -truncated at 5 prime [Rhodnius prolixus]
- RMPLFEMC-446 -Multifunctional chaperone (14-3-3 family) -truncated at 3 prime [Rhodnius prolixus]
- RMPLFEMC-259 -Defender against cell death protein/oligosaccharyltransferase, epsilon subunit [Rhodnius prolixus]
- RMPLFEMC-488 -heat shock protein 90 -truncated at 3 prime [Rhodnius prolixus]
- RMPLFEMC-516 -Molecular chaperone (DnaJ superfamily) -truncated at 3 prime [Rhodnius prolixus]
- AEDAEG, Aedes aegypti;
- ANOGAM, Anopheles gambiae;
- ACYPIS, Acyrthosiphonpisum;
- APIMEL, Apis mellifera;
- CULQUI, Culex quinquefasciatus;
- HARAXY, Harmonia axyridis;
- IXOSCA, Ixodes scapularis;
- NASVIT, Nasonia vitripennis;
- PEDHUM, Pediculus humanus corporis;
- RHOPRO, Rhodnius prolixus;
- TRICAS, Tribolium castaneum;
- TRIVAP, Trialeuro desvaporariorum.
The numbers following species name abbreviation is the gi number of the corresponding protein.
Signal peptide sequences from Rhodnius follicular secreted proteins.
- RMPLFEMC-1-rpSPRP,
- RMPLFEMC-97-RP45,
- RMPLFEMC-98-Putativethioesteraseprotein,
- RMPLFEMC-232-glycine-richprotein1,
- RMPLFEMC-277-Hypotheticalsecretedprotein,
- RMPLFEMC-296-glycine-richprotein2,
- RMPLFEMC-417-Superoxide dismutase (Cu/Zn)SOD1,
- RMPLFEMC-531-Putative OBP/JHbindingprotein.


