Abstract
The Anopheles gambiae salivary gland protein 6 (gSG6) is a small protein specifically found in the salivary glands of adult female mosquitoes. We report here the expression of a recombinant form of the protein and we show that in vivo gSG6 is expressed in distal-lateral lobes and is secreted with the saliva while the female mosquito probes for feeding. Injection of gSG6 dsRNA into adult An. gambiae females results in decreased gSG6 protein levels, increased probing time and reduced blood feeding ability. gSG6 orthologs have been found so far only in the salivary glands of Anopheles stephensi and Anopheles funestus, both members of the Cellia subgenus. We report here the gSG6 sequence from five additional anophelines, four species of the An. gambiae complex and Anopheles freeborni, a member of the subgenus Anopheles. We conclude that gSG6 plays some essential blood feeding role and was recruited in the anopheline subfamily most probably after the separation of the lineage which gave origin to Cellia and Anopheles subgenera.
Keywords: Anopheles gambiae, gSG6, salivary glands, saliva, blood feeding
INTRODUCTION
The saliva of hematophagous arthropods carries a large number of pharmacologically active compounds whose main role is to facilitate blood feeding by helping to overcome host physiological responses such as hemostasis, inflammation and immunity (Ribeiro 1995). Besides its physiological role in blood feeding, vector saliva affects pathogen transmission, as is the case for Leishmania, where vector saliva may considerably enhance transmission (Titus and Ribeiro 1988). Furthermore, immunization with vector saliva or salivary proteins can give rise to protective immune responses against parasite development in murine models of leshmaniasis (Kamhawi et al. 2000) and malaria (Donovan et al. 2007; Fonseca et al. 2007). These observations point out the possible use of vector salivary proteins as vaccine components for vector-borne diseases (Gomes et al. 2008; Titus et al. 2006; Valenzuela et al. 2001).
In the last ten years the increasing power of large scale genomic, transcriptomic and proteomic analyses allowed the accumulation of a considerable amount of information on the salivary secretions of several blood sucking arthropods (Ribeiro and Francischetti 2003). As far as mosquitoes are concerned, the analysis of salivary transcriptomes of a number of Anopheles (Arcà et al. 2005; Calvo et al. 2004; Calvo et al. 2007a; Valenzuela et al. 2003), Aedes (Arcà et al. 2007; Ribeiro et al. 2007) and Culex (Ribeiro et al. 2004) species led to the discovery of several novel protein families, providing some clues on the evolution of blood feeding and revealing the complexity of mosquito salivary secretions. Interestingly, a large group of anopheline- and culicine-specific salivary proteins was identified, which testifies their unusually fast evolutionary rate (see discussion) {Arcà, 2007 #395; Valenzuela, 2003 #361}. In addition, the large number of proteins for which we cannot even postulate a physiological role emphasizes how much we still have to learn concerning the role of salivary proteins in blood feeding, pathogen transmission and manipulation of host responses.
So far the salivary transcriptome of the Afrotropical malaria vector Anopheles gambiae is the best characterized among anopheline mosquitoes: it consists of almost 3000 ESTs classified in over 850 contigs. Its analysis allowed to build up a catalogue comprising at least 72 bona fide secreted salivary proteins, and expression analysis allowed to identify 47 proteins whose expression is specific or enriched in the mosquito salivary glands (Arcà et al. 2005). Intriguingly, the function of more than half of these proteins is presently unknown, suggesting that functional analysis of the An. gambiae salivary protein repertoire might lead to the discovery of several novel pharmacological activities.
The gSG6 protein was originally identified in An. gambiae in the form of a transcript specifically expressed in adult female salivary glands and predicted to encode a small secretory protein (Lanfrancotti et al. 2002). The corresponding 10 kDa protein was indeed found highly expressed in salivary glands of adult females (Francischetti et al. 2002). Both tissue-specific expression pattern and its secretory nature were indicative of a likely role of gSG6 in blood feeding. Orthologs have been found in the female salivary glands of two additional anophelines, Anopheles stephensi and Anopheles funestus (Calvo et al. 2007a; Valenzuela et al. 2003), but were not retrieved in the transcriptomes of the Culicinae subfamily members analyzed so far, i.e. Culex pipiens quinquefasciatus, Aedes aegypti and Aedes albopictus (Arcà et al. 2007; Ribeiro et al. 2007; Ribeiro et al. 2004). These observations suggest that most probably gSG6 is a salivary protein specific to members of the Anophelinae subfamily. Sequence comparison to proteins in the databases failed to show any obvious similarity. A weak but potentially relevant match with AcAP6, a small protein with anti-coagulant function from the hematophagous nematode Ancylostoma caninum (Uniprot:Q16939_ANCCA, 24% identity, 65% similarity), has been recorded (Lanfrancotti et al. 2002). An additional match of potential biological significance was found with members of the Tumor Necrosis Factor-alfa receptor family (Uniprot:Q6PI12_MOUSE, 24% identity, 65% similarity). In the framework of an effort to understand evolution and function of mosquito salivary proteins, we expressed gSG6 in recombinant form and used RNA interference to try to get insights into the physiological role of this protein in anopheline mosquitoes.
RESULTS
Production of a recombinant gSG6 in Pichia pastoris
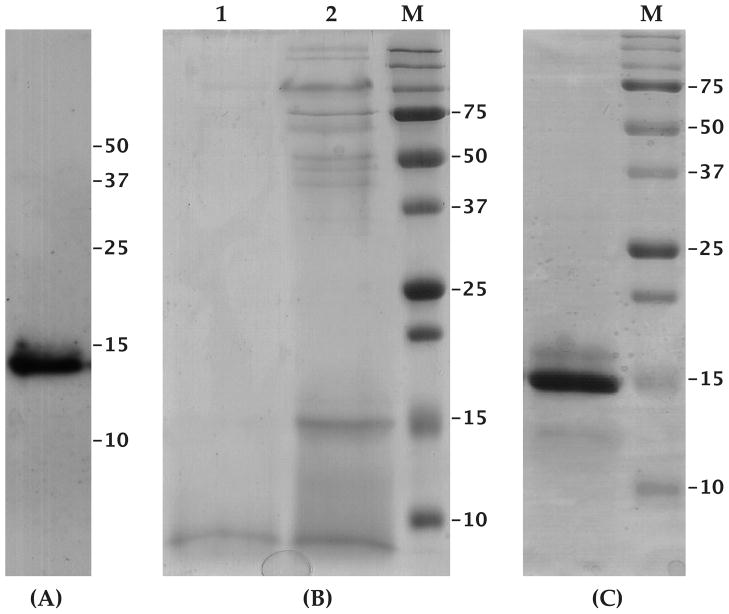
The gSG6 protein was expressed in secreted form in the yeast Pichia pastoris. Typically, maximum expression was achieved approximately twenty-four hours after methanol induction and the recombinant gSG6 was detectable in the supernatant by western analysis (Figure 1A) or by Coomassie staining after concentration by ultra-filtration (Figure 1B). Highly enriched recombinant gSG6 was already obtained after a single affinity purification step and residual contaminants were removed by ion-exchange chromatography, yielding a protein purified to homogeneity (Figure 1C), as also confirmed by mass spectrometry analysis (supplementary Figure S1). Overall, in different preparations about 2.5 mg of purified protein per liter of culture were obtained. Careful examination of the Coomassie stained gel in Figure 1C shows two additional bands. The weak band of apparently higher molecular mass is most likely the result of incomplete reduction of cysteine residues, whereas the hardly detectable band of faster electrophoretic mobility corresponds to a shorter version of the recombinant protein, lacking the last 23 C-terminal residues. These interpretations are supported by mass spectrometry analysis and by immuno-staining with both anti-c-myc and anti-gSG6 antibodies (not shown).
Figure 1. Expression of the recombinant gSG6 protein in Pichia pastoris.
(A) Recombinant gSG6 detected by the anti-c-myc mAb in Pichia culture supernatant (10 μl) after fractionation by SDS-PAGE and transfer to nitrocellulose filter. (B) Pichia culture supernatant fractionated by SDS-PAGE and stained by Coomassie blue: lane1, 10 μl; lane 2, 30X concentrated supernatant. (C) Purified recombinant gSG6 protein (5 μg) stained by Coomassie blue. M, Molecular weight marker. Supernatant and purified recombinant gSG6 were electrophoresed on 15% polyacrylammide gels.
In the effort to understand the physiological role of gSG6, the Pichia-expressed recombinant protein was used in functional tests to evaluate possible anti-hemostatic (PT, APTT, aorta vasodilation, platelet aggregation), immuno-modulatory (TNF-alpha binding, maturation/differentiation of human dendritic cells) and anti-microbial activities. All these assays gave negative results and did not help elucidating gSG6 function; however, we should point out that an adverse effect of the C-terminal c-myc and 6X-His tags cannot be ruled out.
gSG6 is expressed in distal lateral lobes
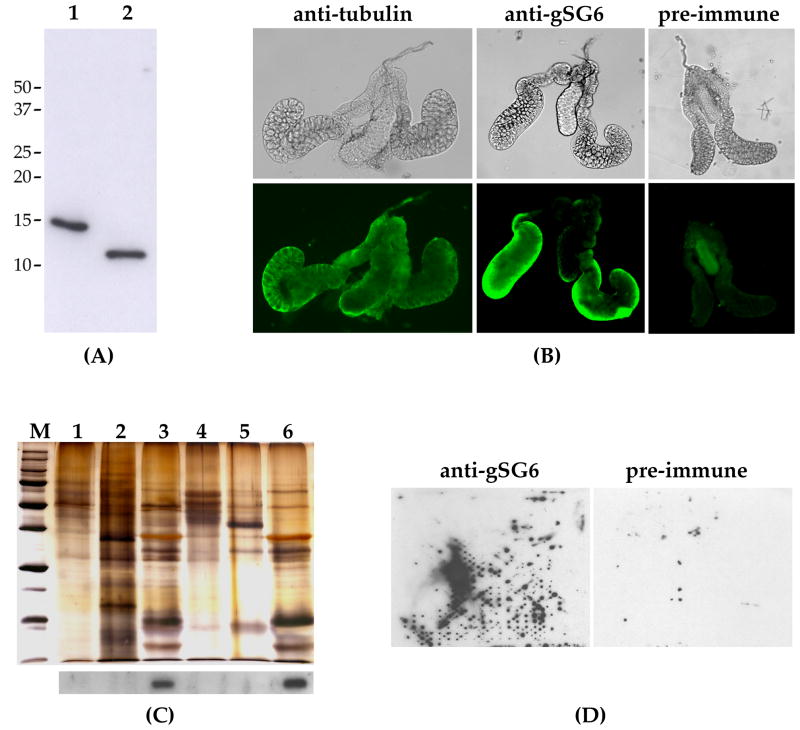
While its functional analysis was in progress, the recombinant gSG6 was used to produce mouse polyclonal antibodies. The immune serum recognizes a single band of approximately 10 kDa in female salivary gland extracts as well as the recombinant protein, which shows a higher molecular mass because of the C-terminal c-myc and 6X-His tags (Figure 2A). Using known amounts of recombinant protein we estimated that a single pair of female glands may carry approximately 10 to 50 nanograms of protein. This represents ~1–5% of the total content, which is assumed to be around 1 microgram {Ribeiro, 1999 #527}, and it is consistent with previous SDS-PAGE analysis performed on total An. gambiae salivary extracts (Francischetti et al. 2002). In vivo, the gSG6 protein was mainly detected by immuno-localization in the distal-lateral lobes of female salivary glands (Figure 2B). In comparison, using a pre-immune serum only shows a background staining in the medial lobe whereas diffuse staining was obtained with an anti-α-tubulin control antibody. The lobe-specific expression was further confirmed by western blot analysis as shown in Figure 2C. Indeed, gSG6 was only detected in protein extracts from dissected whole female salivary glands and from distal-lateral lobes but was absent in protein preparations from proximal-lateral and medial lobes as well as from male glands. This pattern of expression is very similar to that of other female salivary gland-specific proteins in both Ae. aegypti (James 1994) and An. gambiae (Arcà et al. 1999a).
Figure 2. gSG6 protein analysis and localization by an anti-gSG6 antibody.
(A) Western blot analysis with the anti-gSG6 polyclonal serum. 1, recombinant gSG6, ~30 ng; 2, An. gambiae female salivary gland extracts, 1 pair. Molecular weight markers are shown on the left. (B) Immuno-staining of An. gambiae female salivary glands with the anti-tubulin and the anti-gSG6 antibodies. Staining with the pre-immune serum is shown as control. (C) Silver-stained polyacrylammide gel (upper panel) and corresponding western blot with the anti-gSG6 polyclonal serum (lower panel). Protein extracts from: 1, male glands, 10 pairs; 2, female carcass, 1/10; 3, female salivary glands, 5 pairs; 4, proximal-lateral lobes, 6 pairs; 5, medial lobes, 6 pairs; 6, distal-lateral lobes, 6 pairs. Note that one fifth of the total protein extract was used for western analysis and the remaining for the silver staining. (D) Detection of gSG6 on a membrane probed by An. gambiae females: staining with the anti-gSG6 (left) and with a pre immune serum (right) are shown.
Developmental expression of gSG6, a component of the An. gambiae saliva
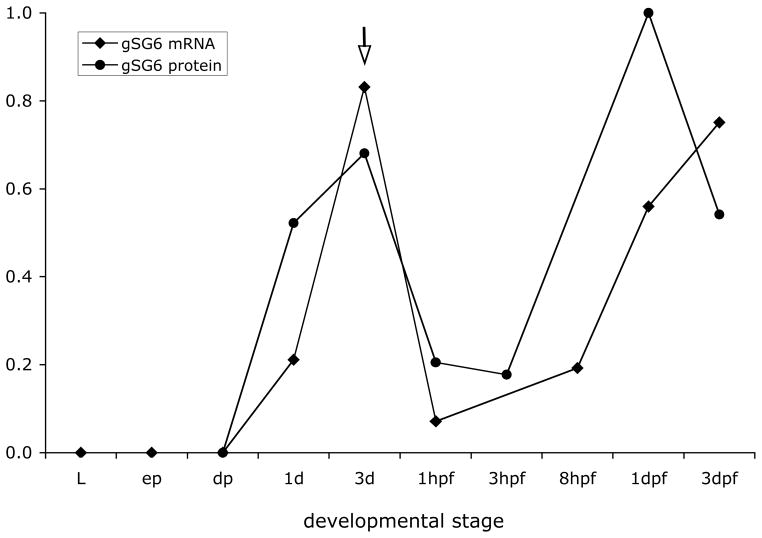
Using quantitative RT-PCR expression analysis we measured gSG6 mRNA levels at different developmental stages. As expected for a sex-specific salivary gland gene, expression is absent during pre-adult stages, starting in young adult females and reaching a maximum three to five days after emergence (Figure 3). A slight progressive decrease of mRNA level was observed with mosquito aging in the absence of blood-feeding (supplementary Figure S2), whereas a substantial drop in the mRNA level was found in 3-days-old females shortly after getting their blood meal (1–3 hours). This observation is fully in agreement with the results obtained by microarray analysis and showing a decrease in salivary transcript abundance after a blood meal (Marinotti et al. 2005). Later on the amount of gSG6 transcript starts to increase going back to high levels in a couple of days. As also shown in Figure 3, the gSG6 protein level exhibits a developmental pattern that is fully consistent with the mRNA expression profile. The protein amount reaches a maximum three to five days after emergence and then decreases with aging. The gSG6 gland content drops dramatically as a consequence of the salivation associated with mosquito probing and feeding, but the salivary glands are replenished in a couple of days. The pattern of expression of gSG6, the presence of a putative signal peptide, its cleavage during maturation (Francischetti et al. 2002) and the absence of other predictable anchoring signals suggest that gSG6 is a secretory protein. We provide here an additional indication that gSG6 is secreted into the mosquito saliva. Indeed, the anti-gSG6 polyclonal serum specifically recognize the protein in small drops of saliva that An. gambiae mosquitoes deposited onto nitrocellulose filters while probing in the attempt to feed (Figure 2D).
Figure 3. gSG6 protein and mRNA levels post emergence and after blood feeding.
gSG6 protein amounts were determined by western blot analysis of salivary gland extracts using silver-staining to normalize the protein content. Relative protein levels are expressed using as reference the maximum amount of protein, which was found 24 hours after blood feeding. gSG6 relative mRNA levels were determined by quantitative RT-PCR on RNA extracted from dissected salivary glands. The maximum mRNA level was found around 5–6 days after emergence (see spplemental Figure S2) and was set arbitrarily as the reference. The arrow points at day 3, when mosquitoes were allowed to feed. Data reported here are from two independent sets of experiments (gSG6 protein) and from three replicas of one RNA experimental set (gSG6 mRNA). L, third and fourth instar larvae; ep, early pupae; dp, dark/late pupae; 1d, 1 day-old; 3d, 3 days-old; hpf, hours post blood feeding; dpf, days post blood feeding.
gSG6 silencing: effect on protein and mRNA level
As using the recombinant protein for in vitro assays did not provide any further insight into the gSG6 function, we decided to use RNAi to knock-down gSG6 and evaluate the possible effects on An. gambiae feeding and probing behaviour. Gene silencing by RNAi in the mosquito An. gambiae (Blandin et al. 2002) is a well established technique widely used in several laboratories for functional analysis. Mosquito salivary glands appeared initially refractory to this methodology until it was shown that higher concentrations of dsRNA were needed to get RNAi working on mosquito salivary genes (Boisson et al. 2006).
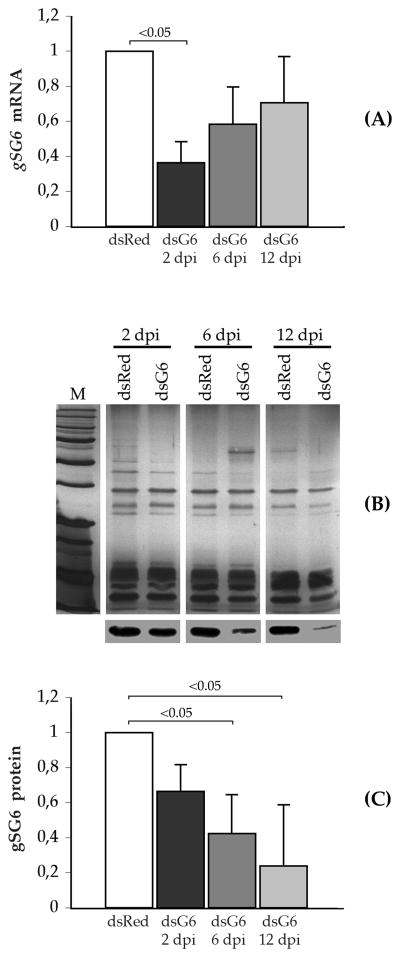
As a first step we assessed if, and to what extent, injection of dsRNA targeting gSG6 (gSG6-dsRNA) into the mosquito was able to affect gSG6 mRNA and protein levels; dsRNA targeting the red fluorescent protein gene (DsRed-dsRNA) was injected as control. gSG6 mRNA levels were measured by qRT-PCR and they appeared lower in gSG6-dsRNA- as compared to DsRed-dsRNA-injected mosquitoes at all time points (Figure 4A). The lowest mRNA amount was found at the earlier time after injection (0.36 ± 0.12, 2 dpi, days post injection) with a progressive increase at the later time points (0.58 ± 0.21, 6 dpi; 0.70 ± 0.26, 12 dpi). The reduction of gSG6 mRNA was weakly significant only at the earlier time point as indicated in Figure 4A (Mann-Whitney test, p=0.049). Similarly, gSG6 protein levels were measured in salivary gland extracts by western blot followed by immunostaining with the anti-gSG6 antibody, whereas silver stained polyacrylamide gels were used for the normalization (Figure 4B). The results of three independent sets of injections are summarized in Figure 4C where the gSG6 protein level appears to progressively decrease from 2 to 12 days post injection (dpi). In comparison to DsRed-dsRNA injected mosquitoes we found that the gSG6 levels were 0.66 ± 0.15 at day 2, 0.42 ± 0.22 at day 6, and 0.24 ± 0.35 at day 12 after the injection of gSG6-dsRNA. This corresponds approximately to a 34%, 58% and 76% relative decrease at two, six and twelve days post injection, respectively. Mann-Whitney test indicated weak significance for the decrease in protein level observed at the later time points, i.e. at 6 days (p=0.049) and 12 days (p=0.046) after injection, as also shown in Figure 4C.
Figure 4. Effect of gSG6-dsRNA injection on gSG6 mRNA and protein levels.
(A) The relative amount of gSG6 transcript was measured by quantitative RT-PCR and normalized using ribosomal S7 mRNA. mRNA levels in gSG6-dsRNA-injected mosquitoes are expressed as a percentage of the amounts found in DsRed-dsRNA-injected mosquitoes at the same time point. (B) Salivary protein extracts at the indicated time points after injection were fractionated by SDS-PAGE and silver stained (top) or analyzed by western blot with the anti-gSG6 polyclonal serum (bottom). (C) Relative levels of gSG6 protein in gSG6-dsRNA-injected mosquitoes are expressed as percentage of the gSG6 amount measured in salivary extracts of DsRed-dsRNA-injected mosquitoes at the corresponding time point. Results reported in (A) and (C) represent the mean of three independent experiments; whisker=1 standard deviation; dsRed, DsRed-dsRNA-injected mosquitoes; dsG6, gSG6-dsRNA-injected mosquitoes; dpi, days-post-injection. Statistical significance as determined by Mann-Whitney test is indicated in 4A (dsRed vs. dsG6 2 dpi, p=0.049, z=1.96) and in 4C (dsRed vs. dsG6 6 dpi, p=0.049, z=1.96; dsRed vs. dsG6 12 dpi, p=0.046, z=1.99).
The different trend observed for gSG6 mRNA and protein accumulation is most probably the result of a slow kinetics of protein degradation; however, it is likely that some other effects, perhaps on translation, give an additional contribution. Anyhow, from this analysis we can conclude that the injection of gSG6-dsRNA into the mosquito hemocoel affects both mRNA and protein levels with a reduction after six days of approximately 60% of the protein and 40% of the mRNA. For this reason this time point was chosen for investigating the effect of gSG6 silencing on mosquito feeding.
gSG6 silencing: effect on feeding and probing behaviour
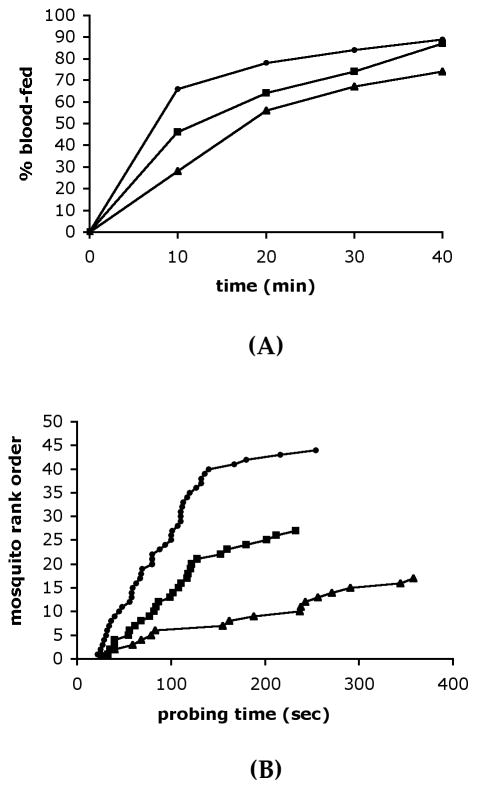
We next asked if the reduced gSG6 protein levels in mosquito saliva had any effect on the An. gambiae blood-feeding ability. To this end mosquitoes were injected with gSG6-dsRNA, or with DsRed-dsRNA as a control, and 6 days later they were given the opportunity to feed. The number of fed individuals was then determined at different time points. After 10 minutes of exposure to guinea pigs, 28% (17/60) of the gSG6-dsRNA injected mosquitoes had fed as compared to 46% (27/61) of those who received DsRed-dsRNA (Figure 5A). During the same time 66% (44/70) of the non-injected mosquitoes had successfully blood-fed. This difference became progressively smaller over time and, after 40 minutes of exposure, 74% (40/60), 87% (48/61) and 89% (58/70) of the mosquitoes had fed, respectively. The difference in blood-feeding capacity between gSG6- and DsRed-dsRNA injected mosquitoes was not significant. However, the class of the gSG6-dsRNA injected mosquitoes constantly shows a lower proportion of successful blood-feeding at all time points (Figure 5A).
Figure 5. Effect of gSG6-dsRNA injection on the mosquito blood feeding ability.
(A) Percentage of non injected (●, n=70), DsRed- (■,n=61) and gSG6-dsRNA-injected mosquitoes (▲, n=60) that blood-fed after exposure to guinea pigs for the indicated time points. (B) Probing time in non injected (●), DsRed- (■) and gSG6-dsRNA-injected mosquitoes (▲). Mosquitoes were exposed to guinea pig for 10 minutes. Probing time was measured in those mosquitoes that successfully fed within this time interval (non injected n=44, DsRed-injected n=27, gSG6-injected n=17). Cumulative results from four independent sets of experiments are reported.
An alternative assay to evaluate the blood feeding ability is the analysis of probing behaviour and the measure of the time that the mosquito needs for its intradermal search for blood. More specifically the probing time is defined as the interval between the insertion of mouthparts into the host skin and the beginning of blood ingestion. This approach has been already successfully used to study the probing behaviour of different mosquito species on different hosts (Ribeiro 2000), as well as to assess the effect of the AgApy gene knock-down by dsRNA injection in An. gambiae (Boisson et al. 2006). Therefore, we measured the probing time of non injected-type, gSG6- and DsRed-dsRNA-injected mosquitoes which successfully fed during the first 10 minutes of exposure to a guinea pig. The results of four independent sets of experiments are summarized in Figure 5B. Mosquitoes injected with gSG6-dsRNA needed on average twice or almost twice the time to locate blood and start feeding as compared, respectively, to non injected (182 vs 90 seconds) or DsRed-dsRNA injected (182 vs 107 seconds) mosquitoes. This difference is significant (Mann-Withney test: non injected vs DsRed, z=1.28, p=0.20; non injected vs gSG6, z=2.90 p=0.0037; DsRed vs gSG6, z=2.19, p=0.028).
gSG6 is conserved among members of the Anopheles gambiae complex
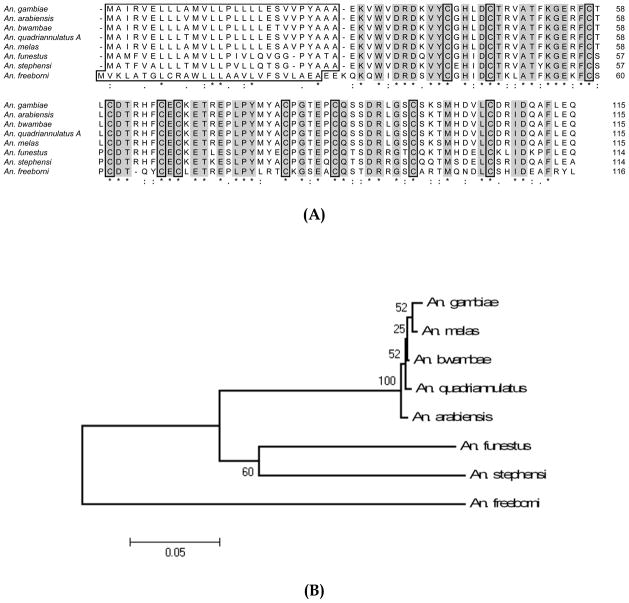
To evaluate the degree of gSG6 variation within closely related species we cloned and sequenced the genomic region encoding the gSG6 protein in other members of the Anopheles gambiae species complex, namely Anopheles arabiensis, Anopheles melas, Anopheles bwambae and Anopheles quadriannulatus A. In addition, the sequence of the more distantly related Anopheles freeborni gSG6 protein was retrieved following a salivary transcriptome analysis from this mosquito (Ribeiro JMC et al., in preparation). The alignment of the An. gambiae gSG6 with the other anopheline deduced proteins identified so far highlights the high degree of conservation of this protein family (Figure 6A). In particular, the predicted mature gSG6 homologs share 99–100% identity within the An. gambiae species complex, whereas among members of the Cellia subgenus, which also includes An. stephensi and An. funestus, the identity is of 80–85%. Comparison of the protein from species of the Cellia subgenus with the deduced gSG6 from An. freeborni, which belongs to the subgenus Anopheles, shows, as expected, a lower degree of conservation (67–71 % identity). The robust separation and clustering of the An. gambiae species complex, as well as the divergence of all the Cellia species from An. freeborni are well summarized by the phylogram obtained from the alignment of the nucleotide sequence encoding the mature polypeptides (Figure 6B).
Figure 6. Alignment of the anopheline gSG6 proteins.
(A) Clustal alignment of anopheline gSG6 proteins. Signal peptides and conserved Cysteines are boxed. Conserved sites are shaded. Accession numbers are as follow: An. gambiae (CAC35522); An. arabiensis (FJ800835); An. bwambae (FJ800837); An. quadriannulatus A (FJ800836); An. melas (FJ800838); An. funestus (ABI83741); An. stephensi (AAO74842); An. freeborni (EZ114340). (B) Phylogram tree (NJ algorithm, bootstrapped 10000 times) constructed from the alignment of the nucleotide sequence encoding the mature gSG6 polypeptides.
DISCUSSION
Although a relatively large number of An. gambiae salivary proteins has been identified so far (Arcà et al. 2005), a function is known (or can be postulated) only for a small fraction of them. This is the case of apyrase/5′-nucleotidase (Lombardo et al. 2000), D7 family members (Calvo et al. 2006), maltase and amylase (Grossman and James 1993; James et al. 1989), cE5/anophelin (Valenzuela et al. 1999), salivary peroxidase (Ribeiro and Valenzuela 1999), gSG7 (Isawa et al. 2002) and the 30 kDa protein (Calvo et al. 2007b; Yoshida et al. 2007). Salivary proteins of blood-feeding arthropods, besides their anti-hemostatic action, have the potential to be exploited for vaccine development (Donovan et al. 2007; Gomes et al. 2008; Titus et al. 2006) and may represent useful markers of exposure to Anopheles bites (Orlandi-Pradines et al. 2007; Remoue et al. 2006; Waitayakul et al. 2006). Thus, both the biological interest in evolution and function of mosquito salivary proteins and their potential applications in malariology encouraged us to approach functional analysis using expression of recombinant proteins and reverse genetics.
The gSG6 protein was selected as an attractive target mainly because of its weak similarity to an anticoagulant from a distant species and the female gland-specific expression. Membrane probing assay and immunostaining with an anti-gSG6 polyclonal serum provided solid evidence in support of the prediction that gSG6 is secreted in mosquito saliva. The secretory nature of the protein, as well as its injection into the host during blood-feeding, is further confirmed by the presence of anti-gSG6 IgG in the sera of individuals from malaria endemic regions (unpublished observations and (Poinsignon et al. 2008)). In addition, immunoblot and immunolocalization showed gSG6 specific expression in distal-lateral lobes. This is in agreement with previous observations indicating that genes specifically expressed in female mosquito salivary glands, i.e. mainly involved in blood-feeding, are transcribed in distal-lateral lobes while proteins found in both male and female glands, i.e. involved in sugar feeding, are expressed in the proximal portion of lateral lobes (Arcà et al. 1999b; James 1994).
We used the Pichia-expressed recombinant protein to address the question of its function but we could not confirm the predicted potential anti-hemostatic action or reveal any other possible activity. In this respect, although mass spectrometry suggested correct folding and absence of post-translational modifications of the recombinant protein (Figure S1), we cannot rule out the possibility of disruption of the functional activity by the C-terminal tags. Using RNAi as a functional genomic tool, we showed that decreased gSG6 protein levels correspond to an impaired blood feeding ability for the mosquito, as indicated by the reduced capacity of getting blood and the increased probing time. Indeed, the time needed by gSG6-dsRNA injected mosquitoes to locate blood and start feeding was significantly higher as compared to non injected (p=0.0037) or control-dsRNA injected (p=0.028) mosquitoes. Similar effects were also observed after silencing the An. gambiae platelet inhibitor apyrase by dsRNA injection (Boisson et al. 2006). These observations strongly support an involvement of the gSG6 protein in the blood feeding process. Since gSG6 is a relatively abundant salivary protein, which can be revealed in salivary extracts by Coomassie-blue staining (Francischetti et al. 2002), it is possible that it may act by binding some important mediator of hemostasis and/or inflammation as shown for D7 family members (Calvo et al. 2006). Nevertheless, its biological properties have yet to be demonstrated. Perhaps expression in other systems (Escherichia coli, insect cells) and/or without tags may help with activity preservation and functional assays.
gSG6 appears to be restricted to anopheline species, since homologs were not found so far in sialotranscriptomes from culicine mosquitoes (Calvo et al. 2007a; Lombardo et al. 2006). However, gSG6 is also absent in the data set coming from the analysis of the salivary transcriptome of Anopheles darlingi (Calvo et al. 2004; Calvo et al. 2009), a member of the Nyssorhynchus subgenus, which includes New World malaria vectors such as An. darlingi and Anopheles albimanus (Harbach 2004; Krzywinski and Besansky 2003). A possible scenario is that the gSG6 protein evolved in the lineage that originated the subgenera Cellia and Anopheles after its separation from the clade that subsequently evolved in the subgenera Lophopodomyia, Kerteszia and Nyssorhynchus. Further insights into the sialotranscriptomes of An. albimanus and/or other mosquitoes belonging to these subgenera may allow to confirm this hypothesis and to get a deeper understanding of the evolutionary history of the gSG6 gene.
The observation that gSG6 is restricted to some anopheline species supports the idea of its possible use as serological marker of exposure to important malaria vectors. Indeed, preliminary analyses indicate that gSG6 is immunogenic and that anti-gSG6 IgG response can be detected in exposed human populations (not shown). Further studies will be needed to confirm its possible use for indirect measures of vector density, for epidemiological studies and to monitor vector control campaigns. In this respect some initial encouraging data have also been obtained with a peptide designed on the gSG6 protein sequence (Poinsignon et al. 2008).
An additional aspect that we started approaching during this study is the apparently fast evolutionary rate of salivary genes and the degree of their variation. Indeed, comparison of salivary and housekeeping protein sets in An. gambiae and An. stephensi showed an average identity of 93 ± 6% when considering housekeeping (H) and 62 ± 15% when comparing salivary (S) proteins (Valenzuela et al. 2003). The same holds for the culicine mosquitoes Ae. aegypti and Ae. albopictus (H=94 ± 13%, S=71.5 ± 11%) (Arcà et al. 2007) and a similar situation was earlier described for the sand fly peptide maxadilan (Lanzaro et al. 1999). A possible explanation of this observations may be the selective pressure exerted by the host immune system on genes of crucial relevance for blood feeding.
The region encoding the mature gSG6 polypeptide shows 79–81% nucleotide identity in the three members of the Cellia subgenus An. gambiae, An. funestus and An. stephensi. To get an insight into the degree of variation between closely related species, we obtained the gSG6 coding sequence from some members of the An. gambiae complex and found very high degree of conservation (98–99% identity). It will probably be more informative looking at salivary genes with longer coding regions and also investigating the degree of intra-specific salivary gene polymorphism in natural mosquito populations. The phylogenetic tree constructed using the nucleotide sequence encoding the mature gSG6 orthologs available so far (Figure 6B) shows a solid separation between species of the An. gambiae complex, which are closely clustered, and the other Cellia. It is worth to point out that the relationships among the different species of the complex, as represented in the tree, are only partly in agreement with the cytogenetic data obtained by polytene chromosome analysis, which indicated An. melas and An. merus as the distant species from An. gambiae s.s (Coluzzi et al. 2002). Most likely the clustering of An. melas with An. gambiae is the result of the very high degree of conservation joined to the very short size of gSG6. Bootstrap analysis also underlies the low degree of reliability of the clustering within the An. gambiae species complex.
In conclusion we started a functional analysis of the An. gambiae salivary transcriptome and provided convincing evidence of the involvement of gSG6 in blood-feeding. We confirmed and extended previous reports (Boisson et al. 2006) showing that RNA interference by dsRNA injection can be reliably employed to target proteins that are present at high levels in mosquito saliva. Furthermore, as recently reported, genes encoding proteins expressed on the salivary gland surface are also amenable to silencing by dsRNA injection (Ghosh et al. 2009), indicating that we have now sufficient tools and information to get deeper insights into the role of salivary glands and saliva in hematophagy, parasite transmission and parasite-vector-host interactions.
MATERIALS & METHODS
Mosquito colony and injections
Mosquito were reared under standard insectary conditions (28°C, 70% humidity) and fed either on 5% glucose or on guinea pigs. Larvae were bred at 25°C and fed on dry cat food. The mosquitoes used in this study belonged to the laboratory An. gambiae M-form population GACAM (Xag, 2R+, 2L+, 3R+, 3L+ from Cameroon, colonized at Rome). Salivary glands and other tissues were dissected in PBS, rapidly frozen by liquid nitrogen and stored at −80°C until needed. When tissues were needed for protein extraction, Complete protease inhibitor cocktail (Roche Applied Science, Mannheim, Germany) was added before freezing or homogenization.
Microinjections were performed using a Nanoject II (Drummond Scientific, PA, USA). Approximately 200 nl of dsRNA (~7 μg/μl) were injected into the thorax of ice cold anesthetized mosquitoes. gSG6-dsRNA or, as a control, dsRNA targeting the red fluorescent protein gene from the coral Discosoma (DsRed-dsRNA) (Matz et al. 1999) were injected into the thorax of 3-days-old females. Salivary glands were dissected at different time points after injection and used to evaluate both mRNA and protein amounts. The gSG6 mRNA and protein levels found at each time point in DsRed-dsRNA injected mosquitoes were used as reference.
An. arabiensis and An. quadriannulatus A were collected in 2005 at the Rekomitjie Tsetse Research Station, Zimbabwe [16°08′ S, 29°24′ E] (Torr et al. 2008), An. melas in 2002 at Mateba, Angola [8°45′ S, 13°23′ E] (Calzetta et al. 2008) and An. bwambae in 1994 at sites of the Semliki Forest, Uganda (30°08′ E, 0°48′ N: VP personal communication). The An. freeborni colony originated in Marysville, California in the 1940s and it has been maitained at NIH.
Protein extraction, analysis and quantitation
If not otherwise specified, all the experimental procedures followed standard protocols as described in (Ausubel et al. 1991; Sambrook et al. 1989). For protein analysis, salivary glands were stored at −80°C. Before use they were subjected to three cycles of freezing/thawing in liquid nitrogen, boiled 10 minutes after addition of 1V of 2x SDS sample buffer, briefly centrifuged and used for SDS-PAGE. Protein extracts from larvae, pupae and carcasses (whole female body after salivary glands removal) were obtained by homogenization followed by centrifugation; supernatants were recovered and stored at −80°C. Samples were boiled for 10 minutes after addition of 1V of 2x SDS sample buffer just before SDS-PAGE analysis. Normalization of western blots was carried out by silver staining of a protein fraction after SDS-PAGE electrophoresis. Typically, for western blot analysis was used one fourth the amount utilized for silver staining. A Gel Doc XR Documentation System and the Quantity One software (BioRad, CA, USA) were used for visualization and quantification according to manufacturer instructions. An unknown protein band of approximately 35 kDa that appears unaffected by blood feeding (i.e. a not secreted salivary gland protein) was arbitrarely selected as control load, quantified and employed for western blot normalization. Linearity of the values with increasing protein amounts was not verified.
RNA extraction and quantitative expression analysis
Total RNA was extracted using the Trizol reagent (Invitrogen, CA, USA). Aliquots of approximately 1 μg were treated with RNAse-free DNAseI (Invitrogen) and retro-transcribed using oligo(dT)12–18 and the Superscript II Reverse Transcriptase (Invitrogen). Quantitative real-time Reverse Transcription-Polymerase Chain Reactions (qRT-PCRs) were performed using an Applied Biosystems 7700 Real-Time PCR system with the S7 ribosomal protein gene as internal control for normalization. A cDNA amount obtained by reverse transcription of 10 ng of total RNA was used in 25 ul reactions containing 1x SYBR green mix (Applied Biosystems, CA, USA) and the following gene-specific primers at the indicated concentrations:
RT_G6F1 (900 nM) GGTACGGTAGTTGAACAAAAAATCAC
RT_G6R (300 nM) TCTCTCTCTCAACCAGAACCTCTT
AgS7_qRT-PCR_F (300 nM) GTGCGCGAGTTGGAGAAGA
AgS7_qRT-PCR_R (900 nM) ATCGGTTTGGGCAGAATGC
Standard curves were prepared both for reference and target genes using 0.08 ng, 0.4 ng, 2.0, 10 ng and 50 ng of templetate. Amplification efficiencies were 100% (S7) and 95.3% (gSG6). The relative quantification method was used to assess gene expression levels.
Recombinant gSG6 clone and Pichia pastoris transformation
Recombinant gSG6 protein was expressed in Pichia pastoris under control of the methanol-inducible alchool oxidase promoter. The 264 bp cDNA fragment encoding the mature gSG6 protein, i.e. not including the endogenous signal sequence, was amplified by PCR using as template the full-length cDNA (Lanfrancotti et al. 2002) and the primers aFG6X (5′-CTGAGCTCGAGAAAAGAGAAAGGTGTGGGTCGACC-3′) and aFG6N (5′-CTGAGGCGGCCGCCTGCTCCAGGAAGGCCTGAT-3′). These primers carry XhoI and NotI restriction sites for directional cloning into the pPICZαA (Invitrogen), a vector that allows for the secretion in the culture medium of a recombinant protein carrying the c-myc epitope and a six-histidine tag at the C-terminus. After sequencing the obtained pPICZαA-G6 construct thus obtained was linearized by SacI and used to electroporate the P. pastoris X-33 strain according to manufacturer instructions (Invitrogen). Transformed colonies were selected on YPD-zeocin plates (1% yeast extract, 2% peptone, 2% glucose, 2% agar, zeocin 100 μg/ml) by incubation at 30°C for 3–10 days. Zeocin-resistant colonies were confirmed as true gSG6 transformants (X-33G6) by PCR amplification using combinations of gene-specific primers aFG6X and aFG6N and vector-specific primers 5′AOX1 (5′-GAGTGGTTCCAATTGACAAGC) and 3′AOX1 (5′-GCAAATGGCATTCTGACATCC-3′).
Protein expression and purification
Optimal expression conditions were determined by small-scale growth of X-33G6 colonies according to manifacturer instruction. Culture supernatants were analyzed by SDS-PAGE and immunoblot with an anti-c-myc antibody (Roche Applied Science, Mannheim, Germany). For large-scale expression a single X-33G6 colony was grown (28°C, 220–250 rpm) in 250 ml of BMGY Medium (1% yeast extract, 2% peptone, 100mM potassium phosphate pH 6.0, 1.34% yeast nitrogen base, 4 × 10−5% biotin, 1% glycerol) until culture reached 2–6 OD600 (i.e. ~16–18 hours). Cells were harvested by centrifugation (3000 g, 5 minutes, room temperature), resuspended at a density of 1.0 OD600 in BMMY Medium (1% yeast extract, 2% peptone, 100mM potassium phosphate pH 6.0, 1.34% yeast nitrogen base, 4 × 10−5% biotin, 0.5% methanol) and grown as above for 36 hours adding methanol at a 0.5% final concentration after 24 hours. Thirty-six hours after starting the induction, the culture was centrifuged (3000 g, 20 minutes, 4°C) and the supernatant was filtered (0.45 μm, Nalgene), concentrated by ultra-filtration (Ultracell 400, BioMax 10, Millipore) and affinity purified according to manufacturer instructions onto an HisTrap FF column on an AKTÄ explorer chromatography system (GE Healthcare). Elution was carried out with an imidazole gradient (20–500 mM). gSG6 containing fractions were pooled and further purified by anion exchange chromatography onto an HiTrapQ HP column (GE Healthcare). Bound proteins were eluted with a linear gradient 0–1M NaCl. Fractions containing the gSG6 protein were analyzed by SDS-PAGE and Coomassie- and/or silver-stained to assess purity of the preparation. Homogeneity of the purified protein was further confirmed by mass-spectrometry (CEINGE Biotecnologie Avanzate, Naples). Protein fractions were concentrated, dialyzed and stored at −20°C.
Antibody production
Purified recombinant gSG6 protein was used to produce specific immune serum in BALB/c mice. Specifically, 50 μg of the recombinant protein and complete Freund’s adjuvant were mixed to form a stable emulsion and injected intraperitoneally. The immunization was repeated after 28, 42, 56 days using 25 μg of antigen in incomplete Freund’s adjuvant. At day 70 the immunized mice were bled to obtain immune serum. Before the immunization cycle, blood (100 μl) was collected from the submandibular vein of each mouse to obtain pre-immune serum.
Immunostainings
For immunolocalization female salivary glands were dissected from 3- to 5-days-old mosquitoes, transferred into a 8-well multitest slide, gently immobilized on the glass surface (leaving the tissue to partially dry), fixed (4% formaldehyde, 30 minutes) and permeabilized (0.05% Triton X-100, 20 minutes). Glands were then washed with PBS, blocked in 1% BSA for 1 hour and washed again. After incubation with the mouse anti-gSG6 polyclonal antibody (1:200, 1 hour) the glands were washed twice with 1% BSA and stained with a goat anti-mouse FITC-conjugated secondary antibody (1:2000, 30 minutes; Sigma-Aldrich, MO, USA). After washing with 1% BSA and mounting by addition of a few drops of VECTASHIELD (Vector, CA, USA) the glands were observed by optical fluorescence microscopy at 40x and 100 x magnifications.
For western blot analysis protein extracts were fractionated by SDS-PAGE on 15% polyacrylamide gels, electro-blotted on to nitrocellulose filters and immunostained according to standard procedures with the mouse anti-gSG6 polyclonal antibody (1:1000), a rabbit anti-mouse horseradish peroxidase-conjugated secondary antibody (1:20000, Sigma) using the chemiluminescent peroxidase substrate-1 (Sigma) according to manufacturer instructions.
Membrane probing
Membrane probing was performed as described (Billingsley et al. 1991). Twenty-five female mosquitoes were placed into a cage, starved for at least five hours, and then allowed to probe a nitrocellulose filter (Hybond C-extra, Hybond Inc., CA, USA) placed on the net with on top a cotton wool previously soaked in a sugar solution (5% glucose, 0.2 μm filtered). After 30 minutes, during which all or most of the mosquitoes probed the filter trying to feed, the membrane was removed, washed and subjected to immunoblot according to standard procedures. The filter was immunostained either with the anti-gSG6 (1:1000) or with the pre-immune serum (1:1000) followed by detection with the rabbit horseradish peroxidase-conjugated anti-mouse antibody (Sigma, 1:20000) as above.
dsRNA preparation
Fragments spanning the coding region of the Discosoma DsRed (Discosoma sp. enhanced red fluorescent protein) and An. gambiae gSG6 genes were cloned as EcoRI-HindIII fragments into the pLL10 vector (Blandin et al. 2002). The 679 bp DsRed fragment was obtained by PCR amplification using the pBac[3xP3-DsRed] vector as template (Horn et al. 2002) and the oligonucleotide primers Red-FE (5′-CATGGAATTCTGGTGCGCTCCTCCAAGAAC-3′) and Red-RH (5′-CATGAAGCTTTACAGGAACAGGTGGTGGCG-3′). The 490 bp gSG6 fragment was amplified by RT-PCR using total RNA from adult female salivary glands and the primers G6F1E (5′-CATGGAATTCAGTCTGCGCTCATTCGCTCC-3′) and G6R1H (5′-CATGAAGCTTGTTCAACTACCGTACCACAG-3′) with the SuperScript one-step RT-PCR system (Invitrogen). Reverse transcription (50°C 30 min) and heat inactivation of the reverse transcriptase (94°C 2 min) were followed by 35 PCR cycles (94°C 30 sec, 55°C 30 sec, 72°C 1 min). The amplified products were cloned into the pLL10 plasmid vector and the resulting constructs were linearized either with EcoRI or with HindIII and used for sense and antisense ssRNA synthesis by the T7 MEGAscript Kit (Ambion, TX, USA). ssRNAs were DNAse I treated, phenol/chlorophorm extracted, ethanol precipitated, resuspended in injection buffer (5 mM KCl, 0.1 mM phosphate buffer pH 6.8) and annealed. dsRNAs were quantified and the concentration adjusted to 7 μg/μl. A nano-injector was used to introduce 3× 69 nl of dsRNA (approx. 1,5 μg per mosquito) into the thorax of ice cold anesthetized mosquitoes.
Probing time measurement and feeding assay
Two days-old adult female mosquitoes were injected or not with gSG6- or DsRed-dsRNAs as described above and five days later their feeding capability was tested. Before the probing time measurements, mosquitoes were starved from sugar for at least 4–5 hours. Three mosquitoes were transferred in cages (one per cage) and allowed to rest for 5 minutes before being offered shaved, immobilized guinea pigs. Probing time is defined as the time taken from the initial insertion of the mouthparts in the skin until the initial engorgement of blood (Ribeiro 2000). If the mosquito terminates the probing unsuccessfully and tries again elsewhere, the second probing time is progressively added to the first one until blood is found; on the contrary, the interprobing time, i.e. the time in between two probings, is not added. For this assay experimental observation was truncated at 600 seconds. In addition, mosquitoes (injected or not with dsRNAs) were allowed to feed on blood for an extended time and fed/unfed mosquitoes were counted at 10, 20, 30 and 40 minutes. Four independent sets of injections for probing time tests and feeding assays were performed. The results were pooled together after validation through Kruskall-Wallis test and then analyzed by the Mann-Withney U test.
Cloning of gSG6 from other species of the An. gambiae complex
The gSG6 coding region of An. arabiensis, An. bwambae, An quadriannulatus and An. melas was amplified by PCR using as template genomic DNA extracted from single mosquitoes of the different species, and the following oligonucleotide primers:
G6_A (5′-GCTCGCATTTATTCAGCATG–3′OH)
G6_B (5′-CGGTAGCTTCTCCACCCTTA–3′OH)
G6_C (5′-ATGGCCATTCGTGTGGAGTTGC–3′OH)
G6_D (5′-TTACTGCTCCAGGAAGGCCTG–3′OH)
The initial denaturation step (94°C 5 min) was followed by 30 PCR cycles (94°C 1 min, 50°C 1 min, 72°C 40 sec) followed by a final elongation step of 7 minutes at 72°C. The amplification products were purified using the QIAquick PCR Purification Kit (QIAGEN) and cloned into the PCR®2.1 plasmid vector (Invitrogen). At least four different clones per each species were isolated and sequenced. After the alignment a consensus sequence was extracted and used both for the clustal analysis and for database submission. The An. freeborni gSG6 sequence was obtained as part of a salivary transcriptome analysis performed as previously described (Calvo et al. 2007a) (Ribeiro, in preparation). Sequence alignment was done using the program ClustalW (Larkin et al. 2007) at the EBI website (http://www.ebi.ac.uk/Tools/webservices/). Percentage of identity/similarity were determined by pairwise alignment of the predicted mature gSG6 polypeptides from different species using the specialized Blast search Align at the NCBI website (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Nucleotide sequences encoding mature gSG6 polypeptides were used for the phylogenetic analysis that was conducted in MEGA4 (Tamura et al. 2007) using the Neighbor-Joining method and bootstrapped 10,000 times. Evolutionary distances were computed using the Kimura 2-parameter method and gaps were treated using the pairwise deletion option.
Supplementary Material
Acknowledgments
We wish to thank R. De Cristofaro (Catholic University, Rome), I. Francischetti (NIAID-NIH, Rockville MD), E. Ricca (University Federico II, Naples) and V. Barnaba (La Sapienza University, Rome) for their help with the functional assays; M. Ruzzi (La Tuscia University, Viterbo) and D. Soldati (Imperial College, London) for the initial help with the Pichia pastoris expression system; G. Lycett (Liverpool School of Tropical Medicine, Liverpool) for kindly providing the pLL10 plasmid; A. della Torre (La Sapienza University, Rome) for making available genomic DNA from An. arabiensis, An. bwambae, An. melas and An. quadriannulatus A. FL was in part supported by the Fondazione “Istituto Pasteur-Cenci Bolognetti”. RR was supported by Fondazione “Compagnia di San Paolo” (Torino) through the Italian Malaria Network. This work is part of the activities of the BioMalPar European Network of Excellence supported by a European grant (LSHP-CT-2004-503578) from the Priority 1 “Life Sciences, Genomics and Biotechnology for Health” in the 6th Framework Programme.
References
- Arcà B, Lombardo F, Capurro M, della Torre A, Spanos L, Dimopoulos G, Louis C, James AA, Coluzzi M. Salivary gland-specific gene expression in the malaria vector Anopheles gambiae. Parassitologia. 1999a;41:483–7. [PubMed] [Google Scholar]
- Arcà B, Lombardo F, de Lara Capurro M, della Torre A, Dimopoulos G, James AA, Coluzzi M. Trapping cDNAs encoding secreted proteins from the salivary glands of the malaria vector Anopheles gambiae. Proc Natl Acad Sci U S A. 1999b;96:1516–21. doi: 10.1073/pnas.96.4.1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcà B, Lombardo F, Francischetti IM, Pham VM, Mestres-Simon M, Andersen JF, Ribeiro JM. An insight into the sialome of the adult female mosquito Aedes albopictus. Insect Biochem Mol Biol. 2007;37:107–27. doi: 10.1016/j.ibmb.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Arcà B, Lombardo F, Valenzuela JG, Francischetti IM, Marinotti O, Coluzzi M, Ribeiro JMC. An updated catalogue of salivary gland transcripts in the adult female mosquito, Anopheles gambiae. J Exp Biol. 2005;208:3971–86. doi: 10.1242/jeb.01849. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RF, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. John Wiley & Sons Inc; New York: 1991. [Google Scholar]
- Billingsley PF, Hodivala KJ, Winger LA, Sinden RE. Detection of mature malaria infections in live mosquitoes. Trans R Soc Trop Med Hyg. 1991;85:450–3. doi: 10.1016/0035-9203(91)90215-k. [DOI] [PubMed] [Google Scholar]
- Blandin S, Moita LF, Kocher T, Wilm M, Kafatos FC, Levashina EA. Reverse genetics in the mosquito Anopheles gambiae: targeted disruption of the Defensin gene. EMBO Rep. 2002;3:852–6. doi: 10.1093/embo-reports/kvf180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisson B, Jacques JC, Choumet V, Martin E, Xu J, Vernick K, Bourgouin C. Gene silencing in mosquito salivary glands by RNAi. FEBS Lett. 2006;580:1988–92. doi: 10.1016/j.febslet.2006.02.069. [DOI] [PubMed] [Google Scholar]
- Calvo E, Andersen J, Francischetti IM, de LCM, deBianchi AG, James AA, Ribeiro JMC, Marinotti O. The transcriptome of adult female Anopheles darlingi salivary glands. Insect Mol Biol. 2004;13:73–88. doi: 10.1111/j.1365-2583.2004.00463.x. [DOI] [PubMed] [Google Scholar]
- Calvo E, Dao A, Pham VM, Ribeiro JM. An insight into the sialome of Anopheles funestus reveals an emerging pattern in anopheline salivary protein families. Insect Biochem Mol Biol. 2007a;37:164–75. doi: 10.1016/j.ibmb.2006.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo E, Mans BJ, Andersen JF, Ribeiro JMC. Function and evolution of a mosquito salivary protein family. J Biol Chem. 2006;281:1935–42. doi: 10.1074/jbc.M510359200. [DOI] [PubMed] [Google Scholar]
- Calvo E, Pham VM, Marinotti O, Andersen JF, Ribeiro JM. The salivary gland transcriptome of the neotropical malaria vector Anopheles darlingi reveals accelerated evolution of genes relevant to hematophagy. BMC Genomics. 2009;10:57. doi: 10.1186/1471-2164-10-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo E, Tokumasu F, Marinotti O, Villeval JL, Ribeiro JM, Francischetti IM. Aegyptin, a novel mosquito salivary gland protein, specifically binds to collagen and prevents its interaction with platelet glycoprotein VI, integrin alpha2beta1, and von Willebrand factor. J Biol Chem. 2007b;282:26928–38. doi: 10.1074/jbc.M705669200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calzetta M, Santolamazza F, Carrara GC, Cani PJ, Fortes F, Di Deco MA, della Torre A, Petrarca V. Distribution and chromosomal characterization of the Anopheles gambiae complex in Angola. Am J Trop Med Hyg. 2008;78:169–75. [PubMed] [Google Scholar]
- Coluzzi M, Sabatini A, della Torre A, Di Deco MA, Petrarca V. A polytene chromosome analysis of the Anopheles gambiae species complex. Science. 2002;298:1415–8. doi: 10.1126/science.1077769. [DOI] [PubMed] [Google Scholar]
- Donovan MJ, Messmore AS, Scrafford DA, Sacks DL, Kamhawi S, McDowell MA. Uninfected mosquito bites confer protection against infection with malaria parasites. Infect Immun. 2007;75:2523–30. doi: 10.1128/IAI.01928-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca L, Seixas E, Butcher G, Langhorne J. Cytokine responses of CD4+ T cells during a Plasmodium chabaudi chabaudi (ER) blood-stage infection in mice initiated by the natural route of infection. Malar J. 2007;6:77. doi: 10.1186/1475-2875-6-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francischetti IM, Valenzuela JG, Pham VM, Garfield MK, Ribeiro JMC. Toward a catalog for the transcripts and proteins (sialome) from the salivary gland of the malaria vector Anopheles gambiae. J Exp Biol. 2002;205:2429–51. doi: 10.1242/jeb.205.16.2429. [DOI] [PubMed] [Google Scholar]
- Ghosh AK, Devenport M, Jethwaney D, Kalume DE, Pandey A, Anderson VE, Sultan AA, Kumar N, Jacobs-Lorena M. Malaria parasite invasion of the mosquito salivary gland requires interaction between the plasmodium TRAP and the anopheles saglin proteins. PLoS Pathog. 2009;5:e1000265. doi: 10.1371/journal.ppat.1000265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes R, Teixeira C, Teixeira MJ, Oliveira F, Menezes MJ, Silva C, de Oliveira CI, Miranda JC, Elnaiem DE, Kamhawi S, Valenzuela JG, Brodskyn CI. Immunity to a salivary protein of a sand fly vector protects against the fatal outcome of visceral leishmaniasis in a hamster model. Proc Natl Acad Sci U S A. 2008;105:7845–50. doi: 10.1073/pnas.0712153105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman GL, James AA. The salivary glands of the vector mosquito, Aedes aegypti, express a novel member of the amylase gene family. Insect Mol Biol. 1993;1:223–32. doi: 10.1111/j.1365-2583.1993.tb00095.x. [DOI] [PubMed] [Google Scholar]
- Harbach RE. The classification of genus Anopheles (Diptera: Culicidae): a working hypothesis of phylogenetic relationships. Bull Entomol Res. 2004;94:537–53. doi: 10.1079/ber2004321. [DOI] [PubMed] [Google Scholar]
- Horn C, Schmid BG, Pogoda FS, Wimmer EA. Fluorescent transformation markers for insect transgenesis. Insect Biochem Mol Biol. 2002;32:1221–35. doi: 10.1016/s0965-1748(02)00085-1. [DOI] [PubMed] [Google Scholar]
- Isawa H, Yuda M, Orito Y, Chinzei Y. A mosquito salivary protein inhibits activation of the plasma contact system by binding to factor XII and high molecular weight kininogen. J Biol Chem. 2002;277:27651–8. doi: 10.1074/jbc.M203505200. [DOI] [PubMed] [Google Scholar]
- James AA. Molecular and biochemical analyses of the salivary glands of vector mosquitoes. Bull Inst Pasteur. 1994;92:133–150. [Google Scholar]
- James AA, Blackmer K, Racioppi JV. A salivary gland-specific, maltase-like gene of the vector mosquito Aedes aegypti. Gene. 1989;75:73–83. doi: 10.1016/0378-1119(89)90384-3. [DOI] [PubMed] [Google Scholar]
- Kamhawi S, Belkaid Y, Modi G, Rowton E, Sacks D. Protection against cutaneous leishmaniasis resulting from bites of uninfected sand flies. Science. 2000;290:1351–4. doi: 10.1126/science.290.5495.1351. [DOI] [PubMed] [Google Scholar]
- Krzywinski J, Besansky NJ. Molecular systematics of Anopheles: from subgenera to subpopulations. Annu Rev Entomol. 2003;48:111–39. doi: 10.1146/annurev.ento.48.091801.112647. [DOI] [PubMed] [Google Scholar]
- Lanfrancotti A, Lombardo F, Santolamazza F, Veneri M, Castrignano T, Coluzzi M, Arcà B. Novel cDNAs encoding salivary proteins from the malaria vector Anopheles gambiae. FEBS Lett. 2002;517:67–71. doi: 10.1016/s0014-5793(02)02578-4. [DOI] [PubMed] [Google Scholar]
- Lanzaro GC, Lopes AH, Ribeiro JM, Shoemaker CB, Warburg A, Soares M, Titus RG. Variation in the salivary peptide, maxadilan, from species in the Lutzomyia longipalpis complex. Insect Mol Biol. 1999;8:267–75. doi: 10.1046/j.1365-2583.1999.820267.x. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lombardo F, Di Cristina M, Spanos L, Louis C, Coluzzi M, Arcà B. Promoter sequences of the putative Anopheles gambiae apyrase confer salivary gland expression in Drosophila melanogaster. J Biol Chem. 2000;275:23861–8. doi: 10.1074/jbc.M909547199. [DOI] [PubMed] [Google Scholar]
- Lombardo F, Lanfrancotti A, Mestres-Simon M, Rizzo C, Coluzzi M, Arcà B. At the interface between parasite and host: the salivary glands of the African malaria vector Anopheles gambiae. Parassitologia. 2006;48:573–80. [PubMed] [Google Scholar]
- Marinotti O, Nguyen QK, Calvo E, James AA, Ribeiro JMC. Microarray analysis of genes showing variable expression following a blood meal in Anopheles gambiae. Insect Mol Biol. 2005;14:365–73. doi: 10.1111/j.1365-2583.2005.00567.x. [DOI] [PubMed] [Google Scholar]
- Matz MV, Fradkov AF, Labas YA, Savitsky AP, Zaraisky AG, Markelov ML, Lukyanov SA. Fluorescent proteins from nonbioluminescent Anthozoa species. Nat Biotechnol. 1999;17:969–73. doi: 10.1038/13657. [DOI] [PubMed] [Google Scholar]
- Orlandi-Pradines E, Almeras L, Denis de Senneville L, Barbe S, Remoue F, Villard C, Cornelie S, Penhoat K, Pascual A, Bourgouin C, Fontenille D, Bonnet J, Corre-Catelin N, Reiter P, Pages F, Laffite D, Boulanger D, Simondon F, Pradines B, Fusai T, Rogier C. Antibody response against saliva antigens of Anopheles gambiae and Aedes aegypti in travellers in tropical Africa. Microbes Infect. 2007;9:1454–62. doi: 10.1016/j.micinf.2007.07.012. [DOI] [PubMed] [Google Scholar]
- Poinsignon A, Cornelie S, Mestres-Simon M, Lanfrancotti A, Rossignol M, Boulanger D, Cisse B, Sokhna C, Arcà B, Simondon F, Remoue F. Novel peptide marker corresponding to salivary protein gSG6 potentially identifies exposure to Anopheles bites. PLoS ONE. 2008;3:e2472. doi: 10.1371/journal.pone.0002472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remoue F, Cisse B, Ba F, Sokhna C, Herve JP, Boulanger D, Simondon F. Evaluation of the antibody response to Anopheles salivary antigens as a potential marker of risk of malaria. Trans R Soc Trop Med Hyg. 2006;100:363–70. doi: 10.1016/j.trstmh.2005.06.032. [DOI] [PubMed] [Google Scholar]
- Ribeiro JM. Blood-feeding in mosquitoes: probing time and salivary gland anti-haemostatic activities in representatives of three genera (Aedes, Anopheles, Culex) Med Vet Entomol. 2000;14:142–8. doi: 10.1046/j.1365-2915.2000.00227.x. [DOI] [PubMed] [Google Scholar]
- Ribeiro JM, Arca B, Lombardo F, Calvo E, Phan VM, Chandra PK, Wikel SK. An annotated catalogue of salivary gland transcripts in the adult female mosquito, Aedes aegypti. BMC Genomics. 2007;8:6. doi: 10.1186/1471-2164-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro JM, Valenzuela JG. Purification and cloning of the salivary peroxidase/catechol oxidase of the mosquito Anopheles albimanus. J Exp Biol. 1999;202:809–16. doi: 10.1242/jeb.202.7.809. [DOI] [PubMed] [Google Scholar]
- Ribeiro JMC. Blood-feeding arthropods: live syringes or invertebrate pharmacologists? Infect Agents Dis. 1995;4:143–52. [PubMed] [Google Scholar]
- Ribeiro JMC, Charlab R, Pham VM, Garfield M, Valenzuela JG. An insight into the salivary transcriptome and proteome of the adult female mosquito Culex pipiens quinquefasciatus. Insect Biochem Mol Biol. 2004;34:543–63. doi: 10.1016/j.ibmb.2004.02.008. [DOI] [PubMed] [Google Scholar]
- Ribeiro JMC, Francischetti IM. Role of arthropod saliva in blood feeding: sialome and post-sialome perspectives. Annu Rev Entomol. 2003;48:73–88. doi: 10.1146/annurev.ento.48.060402.102812. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsh EF, Maniatis Te. Book Molecular Cloning: A Laboratory Manual. 2. Cold Spring Harbor Press; Cold Spring Harbor, NY, City: 1989. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–9. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Titus RG, Bishop JV, Mejia JS. The immunomodulatory factors of arthropod saliva and the potential for these factors to serve as vaccine targets to prevent pathogen transmission. Parasite Immunol. 2006;28:131–41. doi: 10.1111/j.1365-3024.2006.00807.x. [DOI] [PubMed] [Google Scholar]
- Titus RG, Ribeiro JMC. Salivary gland lysates from the sand fly Lutzomyia longipalpis enhance Leishmania infectivity. Science. 1988;239:1306–8. doi: 10.1126/science.3344436. [DOI] [PubMed] [Google Scholar]
- Torr SJ, Della Torre A, Calzetta M, Costantini C, Vale GA. Towards a fuller understanding of mosquito behaviour: use of electrocuting grids to compare the odour-orientated responses of Anopheles arabiensis and An. quadriannulatus in the field. Med Vet Entomol. 2008;22:93–108. doi: 10.1111/j.1365-2915.2008.00723.x. [DOI] [PubMed] [Google Scholar]
- Valenzuela JG, Belkaid Y, Garfield MK, Mendez S, Kamhawi S, Rowton ED, Sacks DL, Ribeiro JMC. Toward a defined anti-Leishmania vaccine targeting vector antigens: characterization of a protective salivary protein. J Exp Med. 2001;194:331–42. doi: 10.1084/jem.194.3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela JG, Francischetti IM, Pham VM, Garfield MK, Ribeiro JMC. Exploring the salivary gland transcriptome and proteome of the Anopheles stephensi mosquito. Insect Biochem Mol Biol. 2003;33:717–32. doi: 10.1016/s0965-1748(03)00067-5. [DOI] [PubMed] [Google Scholar]
- Valenzuela JG, Francischetti IM, Ribeiro JMC. Purification, cloning, and synthesis of a novel salivary anti-thrombin from the mosquito Anopheles albimanus. Biochemistry. 1999;38:11209–15. doi: 10.1021/bi990761i. [DOI] [PubMed] [Google Scholar]
- Waitayakul A, Somsri S, Sattabongkot J, Looareesuwan S, Cui L, Udomsangpetch R. Natural human humoral response to salivary gland proteins of Anopheles mosquitoes in Thailand. Acta Trop. 2006;98:66–73. doi: 10.1016/j.actatropica.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Sudo T, Niimi M, Tao L, Sun B, Kambayashi J, Watanabe H, Luo E, Matsuoka H. Inhibition of collagen-induced platelet aggregation by anopheline anti-platelet protein, a saliva protein from a malaria vector mosquito. Blood. 2007 doi: 10.1182/blood-2007-06-097824. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.