Abstract
Receptor activator of nuclear factor-kappaB -ligand (RANKL), encoded by the gene TNFSF11, is required for osteoclastogenesis, and its expression is upregulated in pathologic bone loss. Transcript variants of TNFSF11 mRNA have been described that encode a membrane-bound and a putative secreted form of RANKL. We identify a TNFSF11 transcript variant that extends the originally identified transcript encoding secreted RANKL. We demonstrate that this TNFSF11 transcript variant is expressed by the human osteosarcoma cell line, Saos-2, and by both primary human T cells and Jurkat T cells. Of relevance to the production of RANKL in pathologic bone loss, expression of this secreted TNFSF11 transcript is upregulated in Jurkat T cells and primary human T cells upon activation. Furthermore, this transcript can be translated and secreted in Jurkat T cells in vitro and is able to support osteoclast differentiation. Our data highlight the complexity of the TNFSF11 genomic locus and demonstrate the potential for the expression of alternate mRNA transcripts encoding membrane-bound and secreted forms of RANKL. Implications of alternate mRNA transcripts encoding different RANKL protein isoforms should be carefully considered and specifically examined in future studies, particularly those implicating RANKL in pathologic bone loss.
Keywords: RANKL, TNFSF11, secreted RANKL, T cells, inflammatory bone loss
Introduction
Receptor activator of nuclear factor-kappaB (NF-κB) ligand (RANKL) is a multipotent cytokine that is essential for osteoclast differentiation and bone resorption1, 2. It also plays a role in innate 3 and adaptive immune responses, brain and thermal regulation, breast tissue development and lactation 4. Human T cells express RANKL upon activation 5, and it was demonstrated that T cell-derived RANKL interacted with RANK (receptor activator of NF-κB) expressed on dendritic cells (DC) and prevented apoptosis of these cells in vitro, resulting in increased DC survival 6, 7. RANKL was subsequently cloned from osteoblast-lineage cells 8, 9, facilitating the discovery of the critical role for RANKL in the differentiation and function of osteoclasts.
Tumour necrosis factor (TNF) super family member 11 (TNFSF11), the gene encoding RANKL, was originally identified as containing five exons, with Exon 1 encoding the 5′ untranslated region (UTR), intracellular and transmembrane protein domains, and Exons 2–5 encoding the extracellular domain (Genbank Accessions: AF053712 and AB064269)5, 7–9. This transcript encodes a ~ 45kDa type 2 transmembrane protein (UniProtKB/Swiss-Prot O14788, isoform 1; NCBI NP_003692; 317 amino acid protein, designated canonical sequence) belonging to the TNF family of cytokines 5, 8, 9; 7. The extracellular domain shares structural homology with other TNF family members, including TNF-related apoptosis-inducing ligand (TRAIL), Fas ligand, and TNF itself 5, 8, 9. Like TNF, the extracellular domain of RANKL can be cleaved (at amino acids 139-140) from the cell surface in vivo by proteases to produce a 26kDa soluble active protein 10.
An alternate human TNFSF11 mRNA transcript was identified in two different squamous cell carcinoma cell lines that caused clinical hypercalcemia (Genbank Accession: AB037599) 11. This TNFSF11 mRNA transcript contained an alternate 5′UTR, encoded by a novel exon located upstream of the original transcription start site, that spliced directly onto Exon 2 of the original TNFSF11 gene. The longest open reading frame for this TNFSF11 transcript encoded the extracellular domain of the RANKL protein encoded by Exons 2–5 (UniProtKB/Swiss-Prot O14788-2, isoform 2; NP_143026; contains amino acids 74-317 of canonical sequence). Importantly, supernatants from cultures of the parent squamous cell carcinoma cell lines contained a protein corresponding to the predicted molecular weight of the RANKL extracellular domain (~27 kDa) that supported osteoclast differentiation 11. This established plausibility that physiologic and/or pathological expression of a TNFSF11 mRNA transcript encoding a soluble RANKL protein occurs in vivo and can influence osteoclast and/or dendritic cell biology. This was further supported by the demonstration that primary myeloma cells and myeloma cell lines express this alternative TNFSF11 mRNA transcript and promote osteoclast differentiation12, 13. Additionally, in vitro, use of both the membrane-bound form of RANKL and recombinant forms consisting of the extracellular domain have activity in stimulating osteoclast differentiation, function and survival 8, 9, 14–17. Interestingly, transgenic mice over-expressing a soluble form of RANKL (amino acids 71-316) develop an osteoporotic phenotype with significant bone loss by 3–4 months of age, 18 demonstrating that soluble RANKL is sufficient to support osteoclastogenesis in vivo. Expression of soluble RANKL, including soluble RANKL derived from T cells, has been shown to be elevated in pathologic conditions associated with bone resorption, such as periodontal disease 19–22. Whether this soluble RANKL results from cleavage of the membrane-bound form of RANKL, or whether it is a product of an alternate transcript encoding the extracellular RANKL protein, is not known.
The existence of alternate TNFSF11 transcripts encoding distinct RANKL protein isoforms suggests greater complexity in the regulation of RANKL gene and protein expression, than is widely recognized. Since T cells are a source of RANKL in pathologic bone disease and have been shown to release a soluble form of RANKL when activated, we focused our studies on determining expression of the TNFSF11 transcript variants in these cells. Here, we delineate the transcripts from the human TNFSF11 gene that are expressed in the setting of T cell activation, and may therefore contribute to pathologic bone loss.
Results
Identification of a novel mRNA transcript encoding a putative secreted human RANKL isoform
Prior to NCBI annotation of the human genomic locus encoding RANKL, we identified two expressed sequence tagged (EST) sequences (accessions: BC040889, BM564083) from the human Unigene cluster for RANKL/TNFSF11 (HS.333791) that aligned with the originally described secreted TNFSF11 mRNA transcript (accession: AB037599), but had additional sequence at the 5′ end (schematic Figure 1A). This suggested that these ESTs may represent a more complete transcript for the secreted mRNA variant of human RANKL, which has since been annotated (NCBI) as TNFSF11 Variant 2 (RefSeq accession: NM_033012). The transcript variant encoding the membrane-bound form of human RANKL is now recognized as TNFSF11 Variant 1 (RefSeq accession: NM_003701). We will refer to the alternate human TNFSF11 transcripts using this nomenclature. Similar to the originally described secreted TNFSF11 transcript AB037599, the longest open reading frames (start identified by A+1TG) for BC040889 and BM564083 encode the entire extracellular domain of the human RANKL protein (isoform 2).
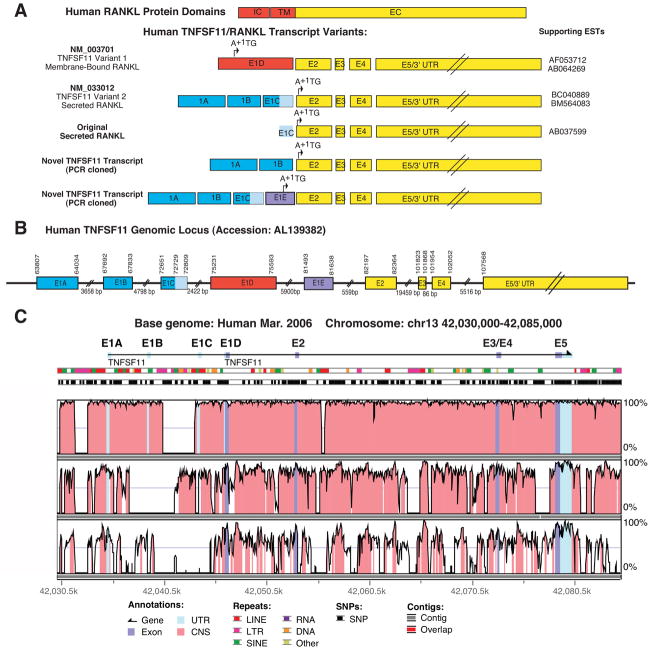
Figure 1. TNFSF11: mRNA transcript variants, genomic loci and sequence conservation.
A. Color coded schematics showing the exon structure for the human TNFSF11 mRNA transcripts aligned with the known RANKL protein domains. Key: E, Exon; EST, expressed sequence tag; IC, intracellular; TM, transmembrane; EC, extracellular; A+1TG, translation start site. B. Schematic showing the genomic structure of the human TNFSF11/RANKL locus. Numbering corresponds to the human genomic contig Accession: AL139382. C. Vista genome alignment of the human TNFSF11 genomic locus (identified by exon-intron schematic at top) to rhesus monkey, dog and mouse genomic loci. Shaded areas identify regions where sequence identity across a 50 bp window is 70% or greater. Key: Light blue shading: conservation of untranslated sequences; dark blue shading: conserved coding sequence; pink shading: conserved intron sequence. Location of repeat elements and single nucleotide polymorphisms (SNPs) are marked as indicated in the figure.
Alignment of TNFSF11 Variant 2 with the genomic contig containing the TNFSF11 genomic locus (accession: AL139382), identified this sequence to contain a 5′ extension of TNFSF11 Variant 1 (NM_003701) with two additional exons located 4.96kb and 8.88kb upstream. To clearly denote the genomic structure of the TNFSF11 locus, we have designated the most 5′ exon common to both TNFSF11 variants as Exon 2, with the more 5′ exons that vary between the transcripts designated according to their location (from distal to proximal: E1A, E1B, E1C and E1D, Figure 1B). The more complete, putative secreted human RANKL transcript, TNFSF11, Variant 2, consists of Exons 1A, 1B, 1C and Exons 2-5, while the membrane-bound human RANKL transcript, TNFSF11 Variant 1, consists of Exon 1D and Exons 2-5 (Figure 1A and B).
Analysis of cross-species alignments of the TNFSF11 genomic loci generated using Vista Genome Browser demonstrated nearly 100% conservation of the sequences surrounding and including Exons 1A, 1B and 1C with the rhesus monkey genome (Figure 1C). In addition, sequence encompassing Exons 1A and 1D show greater than 70% conservation with the dog and mouse genomes (Figure 1C and Supplementary Figure 1 (human and mouse)), suggesting that these regions contain functional genomic elements that may contribute to the regulation of TNFSF11 gene expression. Indeed there are numerous transcription factor binding sites conserved between human and mouse sequences within the ~ 1kb sequence proximal to Exon 1A (Supplemental Figure 1). These include consensus-binding sites for NFAT/STAT, AP-1/STAT, STAT alone, GATA, ikaros-2 and PAX, all of which are important for transduction of activation signals in T cells.
TNFSF11 Variant 2 is expressed and encodes a secreted protein that can support osteoclast differentiation
Since both osteoblast-lineage cells and T cells are important sources of RANKL in physiologic and pathologic bone remodeling 12, 13, 19–21, we sought to verify expression of TNFSF11 Variant 2 in the human osteoblast-like cell line Saos-2, and in the human T cell line, Jurkat Clone E6. RT-PCR using primers specific for TNFSF11 Variant 2 (Exons 1B- 5), and subsequent sequencing of the cloned PCR products, demonstrated that both cell lines expressed this transcript (Figure 2A, upper band). This was confirmed by RT-PCR using an Exon 1A specific forward primer (data not shown). In addition, both cell lines expressed another shorter transcript, which lacked Exon 1C (Figure 2A, lower band, Figure 1A). The absence of Exon 1C does not alter the open-reading frame which again encodes the full-length extracellular domain of RANKL. Interrogation of the genomic structure of the TNFSF11 locus contiguous at Exon1C showed that this region contains ALU repeat sequences known to have cryptic splice sites 23. This may result in alternative splicing at Exon 1C, leading to the production of this shorter TNFSF11 transcript variant. An additional variant of TNFSF11 Variant 2 was also amplified intermittently at low levels: this contained an additional exon between Exons 1C and 2 (Figure 1A), which mapped to the intronic region between Exons 1D and 2 and was subsequently designated as Exon 1E (Figure 1B). The addition of this exon lead to an extended open reading frame with an addition of 19 amino acids to the extracellular domain. These 19 amino acids showed no homology with any signaling domain or protein. Since this sequence was not amplified consistently and was present at very low levels, we did not pursue the characterization of this transcript.
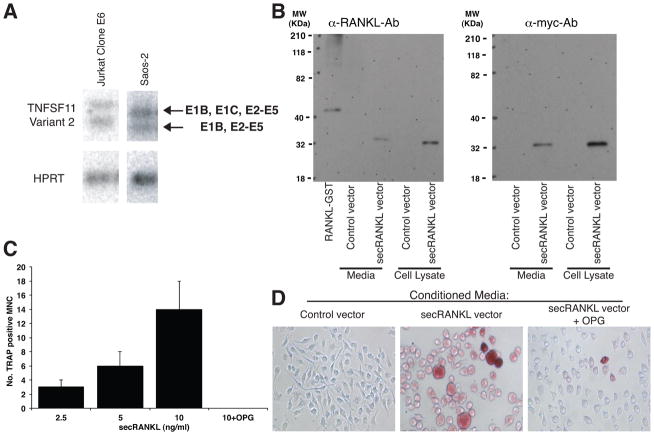
Figure 2. Jurkat T cells and Saos-2 cells express the TNFSF11 Variant 2 which encodes a secreted protein that can support osteoclast differentiation.
A. Semi-quantitative PCR followed by southern hybridization using primers to E1B and E5, shows expression of TNFSF11 Variant 2 in Saos-2 and Jurkat T cells. A second PCR product containing E1B and E2-5 was also amplified. Expression of HPRT is shown as a loading control. B. Western blots showing that the longest open reading frame of TNFSF11 Variant 2 is translated and secreted when expressed as a myc-fusion protein in 293FT cells. Identity of the protein in cell lysates and culture media was confirmed using both an anti-RANKL antibody and anti-myc antibody. C. and D. Conditioned media from 293FT cells transfected with secRANKL vector supports osteoclast differentiation (identified by counting TRAP-positive cells (red staining) with > 3 nuclei which is blocked by inclusion of 200ng/mL OPG. C. Representative results are shown C. Osteoclast quantitation/well (average of 3 wells +/−SD). D. Representative images of each culture condition (the equivalent of 10ng/mL RANKL is shown for secRANKL vector).
To verify whether TNFSF11 Variant 2 encodes a translated and viable secreted protein, the longest open reading frame for this transcript (Exon 2-Exon 5, encoding the entire extracellular domain of human RANKL) was cloned into pcDNA3.1/myc-his B vector and transfected into 293FT cells (secRANKL vector). Western blots of protein from transfected cells, immunoprecipitated with OPG-Fc, were probed with either an anti-RANKL antibody (Figure 2B, left) or anti-myc antibody (Figure 2B, right). These identified a 32 kDa protein that was present in both the cell lysate and culture supernatants (media) from 293FT cells transfected with secRANKL vector and absent in 293FT cells transfected with vector alone (control vector). The molecular weight of this secreted protein is within range of the predicted molecular weight of the putative secreted RANKL protein (28kDa, excluding post-translational modification). Together these data indicate that the open reading frame encoded by TNFSF11 Variant 2 is translated and that the protein can be secreted from cells.
To evaluate if the secreted protein encoded by TNFSF11 Variant 2 is functional, bone marrow macrophages were cultured for 7 days in conditioned medium from 293FT cells transfected with the secRANKL vector which contained the equivalent of 2.5, 5, or 10ng/mL RANKL (quantified by ELISA). A dose dependent increase in osteoclast differentiation (osteoclasts identified as TRAP positive cells >3 nuclei) was observed with the conditioned medium from the 293FT cells transfected with the secRANKL vector (Figure 2C, 2D). The number of osteoclasts formed in these assays was low but importantly this action was blocked by the inclusion of the decoy receptor for RANKL, OPG (200 ng/ml), indicating that this effect was RANKL dependent (Figure 2C, 2D).
Activation of T cells induces expression of TNFSF11 Variant 2 mRNA
Activated T cells are recognized as an important source of RANKL in pathologic bone loss induced by inflammation 19–22,24,28–36 We therefore focused on the regulation of the TNFSF11 Variant 2, encoding a putative secreted RANKL protein, in T cells in response to activation signals. Stimulation of Jurkat T cells with PMA/ionomycin (P/I) for 24 hrs resulted in upregulation of TNFSF11 Variant 2 (detected by RT-PCR, Figure 3A, left panel). A similar increase in total TNFSF11 mRNA expression in response to P/I stimulation was also demonstrated using quantitative-PCR analysis using a primer and probe set located in the common 3′ end of the TNFSF11 transcripts, which identifies all variants (Figure 3A, right panel). Interestingly, no expression of the membrane-bound RANKL transcript, (TNFSF11 Variant 1), by either RT- or quantitative PCR using specific primers or primer plus probe sets, was detected in either unstimulated or stimulated Jurkat T cells (data not shown). The induction of TNFSF11 Variant 2 (Figure 3A, left panel) and total TNFSF11 mRNAs (Figure 3A, right panel) by PMA/ionomycin treatment was inhibited by pre-treatment with the calcineurin inhibitor, cyclosporine A (CsA) suggesting the involvement of the calcineurin signaling pathway in regulating expression of this transcript. Jurkat T cells activated by the more physiologic stimuli of antibodies to CD3 and CD28, also upregulated expression of TNFSF11 Variant 2 (Supplemental Figure 2).
Figure 3. Expression of TNFSF11 Variant 2 is induced by classical T cell activation stimuli.
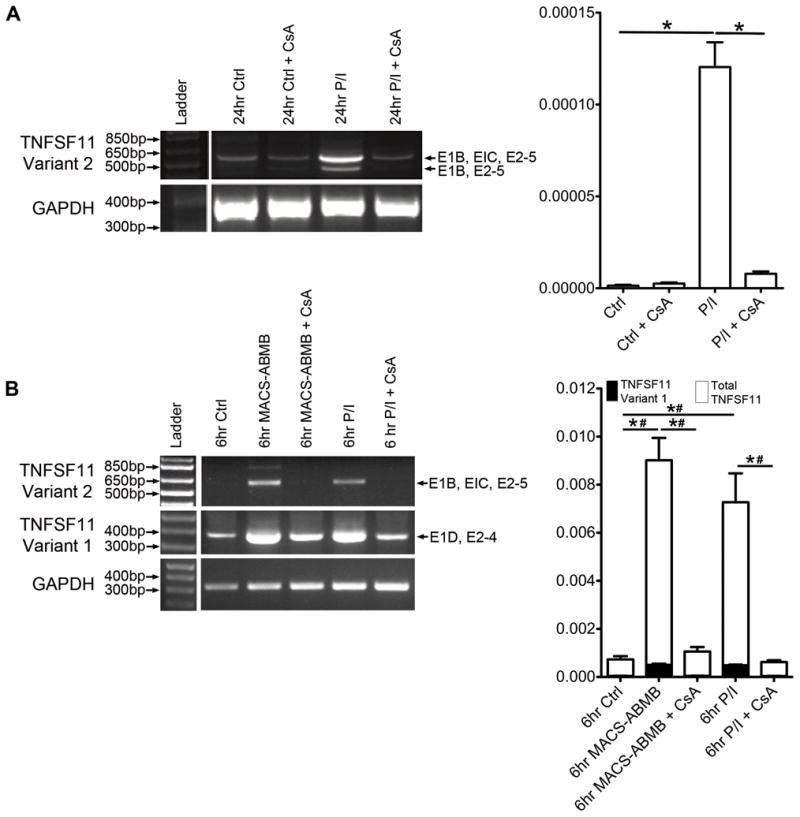
A. TNFSF11 transcript expression in Jurkat T cells stimulated for 24 hrs with PMA/ionomycin (P/I) with and without cyclosporine A (CsA) pre-treatment. Left panel: representative semi-quantitative RT-PCR results using primers specific for TNFSF11 Variant 2 (E1B-E5) (minimum of 3 biologic repeats). GAPDH is shown as loading control. Right panel: quantitative RT-PCR showing relative expression (GAPDH house keeping gene) of total TNFSF11 expression (n = 3). * p ≤ 0.05. B. TNFSF11 mRNA expression in primary human T cells stimulated for 6 hrs with either anti-CD2/anti-CD3/anti-CD28 conjugated to MACS Bead particles (MACS-ABMB) or P/I with and without CsA pre-treatment. Left panel: representative semi-quantitative RT-PCR using primers specific for TNFSF11 Variant 1 (amplified from E1D-E4) and Transcript Variant 2 (amplified from E1B-E5) (minimum of 3 biologic repeats). GAPDH was used as the housekeeping gene. Right panel: quantitative RT-PCR showing relative expression (GAPDH housekeeping gene) of total TNFSF11 and TNFSF11 Variant 1 expression (n = 3). * and # indicate statistically significant differences (p ≤ 0.05) between indicated conditions for TNFSF11 Variant 1 and total TNFSF11 expression respectively.
Primary human T cells from healthy patients isolated from buffy coats via negative selection were also analyzed for TNFSF11 mRNA transcript expression. In contrast to the Jurkat T cell line, RT-PCR indicated that primary T cells express both membrane-bound (Variant 1) and secreted (Variant 2) TNFSF11 transcripts. Expression of both transcripts was upregulated following stimulation with either P/I or activation with the more physiologic stimuli of anti-CD2/CD3/CD28 antibody coated bead particles (Figure 3B). In primary T cells, quantitative PCR indicated a significant increase in total TNFSF11 mRNA expression, but importantly, membrane-bound RANKL (TNFSF11 Variant 1) expression represented only a fraction of the total mRNA (Figure 3B, right panel). Direct quantitative comparison of the expression of the TNFSF11 Variant 2 was not possible due to high GC sequence content within Exons 1A-C, which prevented the development of a suitable quantitative PCR assay.
In primary T cells, we also investigated the involvement of several signaling pathways in the induction of TNFSF11 expression by anti-CD2/CD3/CD28 antibody coated-bead particles. Semi-quantitative PCR analysis demonstrated that induction of both the TNFSF11 transcript variants was significantly attenuated in the presence the inhibitor of calcineurin cyclosporine (Figure 3B, left panel) as well as p38 MAP kinase (p38MAPK, SB239063, data not shown). In contrast, MEK kinase (PD98059) and NF-κB (CAPE) inhibitors did not significantly affect the induction of either of the TNFSF11 transcripts. This attenuation was verified by quantitative PCR showing reduced expression of both total and membrane-bound TNFSF11 mRNA in the presence of CsA (Figure 3B, right panel) or SB239063 (Supplemental Figure 3) but not PD98059 or CAPE inhibitors (Supplemental Figure 3).
Jurkat T cells express the secreted RANKL protein isoform
Having demonstrated that the longest open reading frame of TNFSF11 Variant 2 could be translated into a viable secreted RANKL protein, we sought to determine expression of this RANKL isoform in the Jurkat T cell line. Western blot analysis for RANKL protein expression demonstrated the presence of a ~30 kDa protein in cell lysates but not culture supernatants from Jurkat T cells (Figure 4A). PMA/ionomycin treatment for 24 hrs increased expression of this protein compared to vehicle treated cells (Figure 4A). As expected, based on the absence of TNFSF11 Variant 1 mRNA expression in Jurkat T cells, no expression of the higher molecular weight membrane-bound RANKL protein was detected in the absence or presence of PMA/ionomycin.
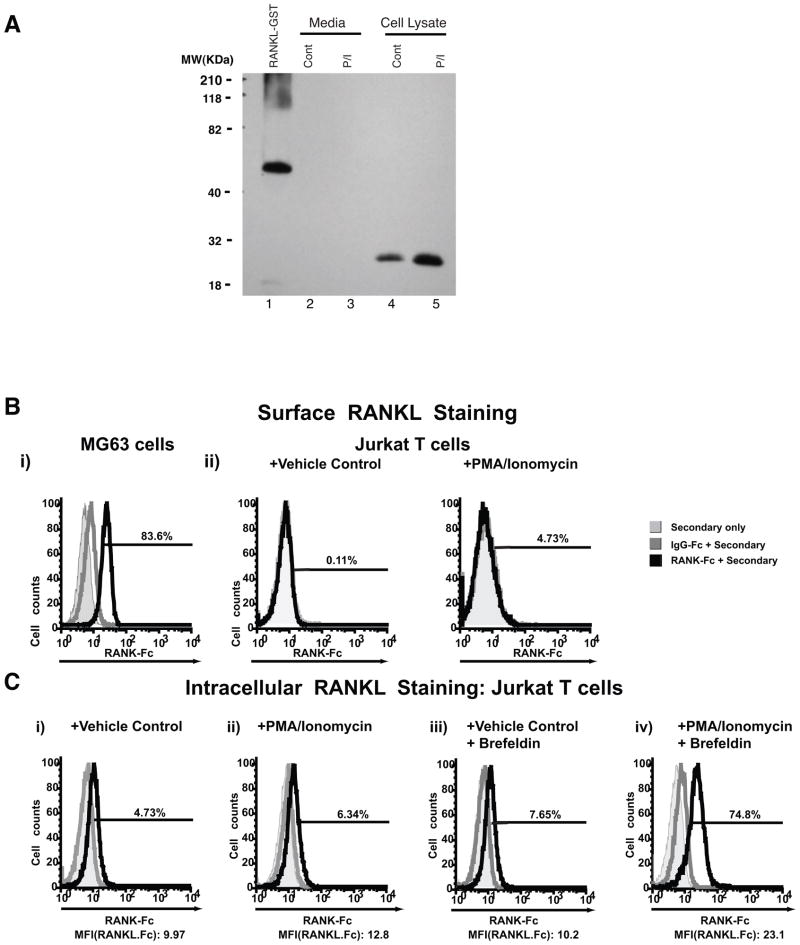
Figure 4. Jurkat T cells only express the secreted form of RANKL and not membrane-bound RANKL protein.
A. Western Bot showing expression of secreted RANKL protein isoform in Jurkat T cell lysates and culture media with or without PMA/ionomycin (P/I) stimulation. RANKL-GST (a recombinant full-length transmembrane RANKL protein) was run as a control. Cont – control. B. Flow cytometry analysis of surface (membrane-bound) RANKL protein expression (i) Unstimulated MG-63 and (ii) Jurkat T cells, vehicle control or P/I stimulated. C. Flow cytometry analysis of intracellular RANKL protein expression in permeabilized Jurkat T cells treated with or without brefeldin before harvest. (i) Control unstimulated or (ii) PMA/ionomycin stimulated Jurkat T cells without brefeldin treatment (iii) Control unstimulated or (iv) P/I stimulated Jurkat T cells treated with brefeldin.
Flow cytometry staining using RANK-Fc was performed to further explore the expression of the human RANKL protein isoforms in Jurkat T cells (Figure 4B and C). To confirm successful detection of membrane-bound (cell surface) expression of RANKL, the MG-63 osteosarcoma cell line was used as positive control. 83.6% of MG-63 cells were found to express cell surface RANKL protein when compared to IgG-Fc treated cells (Figure 4Bi). Consistent with our previous observations, vehicle or PMA/ionomycin stimulated Jurkat T cells did not express membrane-bound RANKL (Figure 4Bii). To determine if Jurkat T cells express the secreted RANKL isoform, intracellular staining and subsequent flow cytometry was performed on Jurkat T cell treated with vehicle or PMA/ionomycin with or without brefeldin pre-treatment, which blocks protein secretion. Unstimulated Jurkat T cells, with and without brefeldin treatment, had low level binding of RANK-Fc with less than 8% of cells positive for expression (Fig. 4Ci and iii). Stimulating Jurkat T cells with PMA/ionomycin in the absence of brefeldin did not significantly alter detectable intracellular RANK-Fc binding compared to unstimulated controls (Figure 4Cii). In contrast, brefeldin treatment of Jurkat T cells stimulated with PMA/ionomycin led to a dramatic increase in both the percentage of RANKL positive cells (74.8%) and an increase in the mean-fluorescence intensity (Figure 4Civ). These results demonstrate that, when protein secretion is inhibited, PMA/ionomycin stimulation of Jurkat T cells results in an increase in the intracellular accumulation of RANKL protein. Combining this with the observation that Jurkat T cells do not express membrane-bound RANKL mRNAs, our data supports that PMA/ionomycin stimulated Jurkat T cells express a secreted protein isoform of RANKL.
Discussion
RANKL, encoded by the gene TNFSF11, has commonly been recognized as a membrane-bound protein that can be cleaved, resulting in a soluble, active product. In addition an alternate mRNA transcript was identified that encoded a putative secreted isoform11. In this study, we identify a full-length transcript that extends the originally described secreted human RANKL mRNA by the addition of two exons upstream of the 5′ end of the TNFSF11 gene. We demonstrate that this novel transcript, TNFSF11 Variant 2, which encodes the extracellular domain of the human RANKL protein (isoform 2), is expressed by the human osteosarcoma cell line Saos-2 and by both primary human T cells and a Jurkat T cell line. Furthermore we show that this transcript can be translated in an in vitro expression system, as well as endogenously in the Jurkat T cells and importantly this secreted RANKL protein isoform could support osteoclastogenesis. Furthermore, bone loss occurs in transgenic mice over-expressing the entire extracellular domain of the RANKL protein 18. These data provide compelling support for in vivo activity of the protein isoform encoded by TNFSF11 Variant 2 can support osteoclastogenesis18.
It should be noted that additional TNFSF11 transcript variants have previously been identified in human peripheral white blood cells 25–27. These are distinguished from TNFSF11 Variants 1 and 2 by the use of alternate transcription start sites and splice sites within the exon we designate as Exon 1D. They encode a putative RANKL protein (UniProtKB/Swiss-Prot reference O14788-3, isoform 3) lacking the intracellular domain, with a truncated transmembrane domain (Accession: AB064270) and the entire extracellular domain of human RANKL (Accession: AB064268). To date, no other studies have confirmed the expression of these TNFSF11 transcripts, and unlike the findings with TNFSF11 Variant 2 in our study, this isoform was not secreted, but accumulated within the golgi when expressed in a fibroblast cell line25, 26.
The current confirmation and clarification of the expression of alternate TNFSF11 transcript variants encoding different RANKL protein isoforms require that careful attention be given to TNFSF11 primer/probe design, with appropriate interpretation of expression data. Most studies use primers located within the region of the TNFSF11 gene common to all transcripts (Exons 2-5), with only one other report directly assessing expression of the secreted RANKL transcript variant in human myeloma cells 12. Future studies using transcript specific primer/probe sets for the alternate TNFSF11 transcripts will inform the relative abundance of membrane-bound RANKL and secreted RANKL proteins, and provide insight into the function of these human RANKL isoforms both in physiologic and pathologic conditions. Furthermore, expression of the alternate TNFSF11 transcripts and extension of the TNFSF11 genomic locus should be considered in studies directed towards understanding the transcriptional regulation of the RANKL gene.
We focused on determining the expression of the extended TNFSF11 transcript Variant 2 in human T cells, since these cells are a known source of RANKL in inflammation and have been shown to play a role in pathologic bone loss 19, 24, 28–36. Our observations indicate that primary T cells express transcripts for both the membrane-bound and soluble forms of RANKL (TNFSF11 Variant 1 and 2 respectively). Importantly, expression of both transcripts was induced following stimulation with anti-CD3/CD28 or PMA/Ionomycin, and the latter stimulation induced endogenous production of a secreted protein in Jurkat T cells. Our observations are consistent with previous reports in which the release of RANKL into culture supernatants was observed following anti-CD3/CD28 stimulation of primary human T cells 2, 34. Furthermore, in a study using primary T cells isolated from patients with ankylosing spondylitis, brefeldin treatment prior to stimulation with PMA/Ionomycin led to an increase in the percentage of cells staining positive for intracellular RANKL compared to membrane-bound RANKL 37. The accumulating data support that the soluble RANKL isoform 2, in addition to the membrane-bound isoform 1, is likely to be produced in inflammatory conditions. Extrapolation from the total RANKL expression in these cells suggests that the secreted product is likely to be the more abundant transcript variant under these conditions.
Understanding of the regulation of the TNFSF11 genomic locus is incomplete, but current knowledge supports complexity at the level of transcriptional regulation (reviewed recently in 38) 39–46. Clear delineation of the mechanisms of RANKL gene regulation and protein expression is vital to full understanding of the diverse biologic activities of this protein. Multiple distal enhancers have been identified that are responsible for regulating RANKL expression and the dominance of these distal control regions appears to vary between species 40, 43–45 and cell type 39. Importantly, the region surrounding and encompassing Exon 1A, 1B or 1C does not match any of the enhancers/regulatory regions already identified for the TNFSF11 locus 43–47. Significant inter-species conservation of the sequence proximal to Exon 1A suggests that this region has a regulatory function. The presence of multiple conserved transcription factor binding sites for factors that mediate T cell responses (eg NFAT, STAT, IK-2) highlights the potential for this region to aid transcriptional activation of this TNFSF11 variant 2 in response to activation signals in both primary and Jurkat T cells. Furthermore, given the location of the distal 5′ exons of the TNFSF11 transcript variants, it is likely that independent proximal promoters orchestrate expression of these alternate transcripts.
Using antagonists to a variety of signaling intermediates, we demonstrated that expression of TNFSF11 Variants 1 and 2 (encoding membrane-bound and secreted proteins respectively) in primary T cells is dependent in part on the activation of the calcineurin-NFAT and potentially the p38 MAPK pathways. These signaling pathways have both been previously implicated in control of TNFSF11 gene expression 42, 48, 49. Recently NFATc3 was implicated in mediating calcium-induced RANKL expression in murine osteoblasts (MC3T3-E1 subclone) by binding to an NFAT binding element residing at −941 to −963 bp within the proximal TNFSF11 promoter 48. Another recent study using activation stimuli in murine T cells demonstrated that the previously identified mRL-D5 enhancer39, 40, 43, plus a novel T cell control region (TCCR, located −123 to −156 bp upstream) were key regulatory elements. c-Fos, and not NFATs (p38 MAPK was not directly examined), was determined to be a key transcription factor involved in TNSF11 gene expression mediated by the TCCR in an activated murine T cell line 39. As has been previously suggested 38, it is not surprising that numerous, redundant and complex signal pathway usage is employed in the regulation of this important gene locus. Further research combining both in vitro and in vivo molecular approaches will be required to fully elucidate these control mechanisms.
In summary we demonstrate the expression of a more extended variant of the originally described secreted human RANKL transcript in Saos-2 osteosarcoma cells, Jurkat T cells and primary human T cells. Compelling evidence is provided that the secreted RANKL transcript variant is expressed in human T cells, is induced by activation and leads to the production of a secreted protein that is functional. Future studies to assess the secretion of RANKL or production and function of soluble forms of RANKL will therefore be important to pursue.
Materials and Methods
Database mining and sequence analysis
Prior to NCBI annotation of the human genomic locus encoding RANKL we aligned the full length mRNA and expressed sequence tagged (EST) sequences from the Unigene Cluster for human TNFSF11/RANKL, (HS.333791, www.ncbi.nlm.nih.gov/UniGene) to the known human RANKL mRNA sequence, AF053712 and the sequence for the putative secreted human RANKL (AB037599). To identify exon boundaries and locate exons within the human RANKL genomic locus, EST sequences were aligned with the genomic contig AL139382 containing the human RANKL gene, TNFSF11, using the Blast2 algorithm (blast.ncbi.nlm.nih.gov). The Vista Genome Browser (http://pipeline.lbl.gov/cgi-bin/GenomeVista) was used to determine cross-species sequence conservation for the TNFSF11 genomic locus using the human TNFSF11 locus as the base sequence and a window size of 50 bp, consensus width of 50 bp and an identity limit of 70%. TRANSFAC analysis (http://www.biobase-international.com) combined with rVista (http://rvista.dcode.org/) was used to identify conserved consensus transcription factor binding sites between mouse and human sequences proximal to and including Exon 1A (Core cutoff: 1.0; Matrix cutoff: 0.90).
Cell culture
Jurkat Clone E6-1 human T cell leukemia cells and MG-63 human osteosarcoma cells were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). Jurkat T cells were grown in RPMI-1640 medium (Mediatech, Manassas, VA, USA) supplemented with 10% fetal bovine serum (FBS, Atlanta), 2 mM L-glutamine (Invitrogen, Grand Island, NY, USA), 10 mM Hepes (Gibco, Grand Island, NY, USA) and 1 mM sodium pyruvate (Gibco). MG-63 cells were maintained in Eagle’s Minimum Essential Medium (EMEM, Mediatech) supplemented with 10% FBS (Atlanta Biologicals, Lawrenceville, GA, USA), 1 mM sodium pyruvate (Gibco) and 0.1 mM non-essential amino acids (Gibco). Primary T cells were prepared from human “buffy coats” (use approved by The University of Queensland Human Ethics Committee). Peripheral blood mononuclear cells (PBMC) were harvested using Ficoll-Paque™ Plus gradient separation and residual red blood cells were lysed using ammonium chloride solution. T cell enrichment was subsequently achieved by magnetic assisted cell sorting using a negative cell selection strategy (Miltenyi Biotec, North Ryde, Australia), which resulted in a highly enriched T cell fraction that was demonstrated to be greater than 90% CD3+ by flow cytometric analysis. Isolated primary T cells were immediately used in activation assays as outlined below.
Determination of expression of TNFSF11 mRNA transcripts in T cells
Stimulation of T cells for RNA expression: Jurkat T cells were serum starved overnight in 0.125% BSA in RPMI medium (Gibco). For PMA/ionomycin stimulation, 16 nM Phorbol 12-myristate 13-acetate (PMA, Sigma-Aldrich, Saint Louis, MO, USA) and 66.7 μM ionomycin (Sigma-Aldrich) for 24 hrs (Jurkat cells) or 6 hrs (primary T cells). Primary T cells were also treated for up to 24 hrs using the MACS T cell activation/expansion kit (Miltenyi BioTech) containing anti-CD2/anti-CD3/anti-CD28 antibodies conjugated to MACSiBead particles (MACS-ABMB). Prior to stimulation with PMA/ionomycin or MACS T cell activation macrobeads, some cells were pretreated for 30 min with cyclosporine A (CsA, Sigma-Aldrich, a calcineurin inhibitor that prevents nuclear factor of activated T cells, NFAT, activation) for 30 min prior to stimulation with PMA/ionomycin or Macs T cell activation macrobeads. Primary T cells were also stimulated in the presence of additional T cell activation signaling pathway inhibitors: Caffeic acid phenyl ester (CAPE, 10 μM, Sigma-Aldrich) is an inhibitor of nuclear factor (NF)-κB, SB239063 (20 μM, Sigma-Aldrich) is an inhibitor of p38 mitogen-activated protein kinases (MAPK, α and β) or PD98059 (25 μM, Sigma-Aldrich) is a mitogen activated protein kinase (MEK)-extracellular signal-regulated kinases (ERK) pathway inhibitor.
Total RNA was prepared using Trizol Reagent (Invitrogen) or Qiagen RNeasy Mini kit (Qiagen, Valencia, CA, USA) (as per manufacturers specification) and DNase-treated using DNA-free (Ambion, Grand Island, NY, USA). 1 μg of RNA was used for cDNA synthesis using Advantage™ RT-for-PCR kit (Clontech, Mountain View, CA, USA) following manufacturer instructions. Primers specific to E1B and E5 (RANKL-EST-F-248 and sRANKL-R-450, 38 PCR cycles) were used to amplify TNFSF11 Variant 2 transcripts (38 PCR cycles). Primers specific to E1D (hRANKL-F-315) and E4 (EC-R-744) were used to amplify TNFSF11 Variant 1. Amplification of GAPDH (21 PCR cycles) or HPRT (26 cycles) was used for normalization purposes. For initial validation purposes, PCR products were run on a 1.2% TAE agarose gel, denatured and transferred to nitrocellulose membrane for probing with 32P labeled oligoprobes for RANKL and HPRT). Hybridized products were visualized using a phosphoimager (refer to Table 1 for primer sequences). Commercially available Taqman assays were purchased from Applied Biosystems (Grand Island, NY, USA) for quantitative total TNFSF11 (Hs00243522), TNFSF11 Variant 1 (Hs00243519) and GAPDH (Hs99999905_m1) with cycling performed as per manufacturer’s specifications using ABI Prism 7000 sequence detection system (Applied Biosystems). Relative expression levels were calculated using experimentally determined primer efficiency and the ΔCT method 50.
Table 1.
Primers
| Primer Name | Sequence | Location | Product |
|---|---|---|---|
| RANKL-EST-F-248 | 5′-AGA AAC TGC TGA AAT ATT GAA CAC A-3′ | E1B | 640bp TNFSF11 Variant 2 E1B-E5 |
| sRANKL-R-450 | 5′-CCC CGA TCA TGG TAC CAA GAG GAC-3′ | E5 | |
| hRANKL-F-315 | 5′-AGC GTC GCC CTG TTC TTC TA- 3′ | E1D | 337bp TNFSF11 Variant 1 E1D-E4 |
| EC-R-744 | 5′-TGT CGG TGG CAT TAA TAG TGA GA-3′ | E4 | |
| RANKL Oligo Probe | 5′-GGT TCC CAT AAA GTG AGT CTG TCC T-3′ | E4-5 | NA |
| huHPRT-F-532 | 5′-GGT CAG GCA GTA TAA TCC AAA GA-3′ | E6 | 406bp hypoxanthine phospho-ribosyltransferase 1 |
| huHPRT-R-937 | 5′-ATA GGC TCA TAG TGC AAA TAA ACA GT-3′ | E9 | |
| HPRT Oligo Probe | 5′-GTT GGA TAT AAG CCA GAC TTT GTT G-3′ | E7-8 | NA |
Abbreviations: bp, base pairs; R, reverse; F, forward; E, exon.
Identification of RANKL protein expression by immunoprecipitation and western analyses
Jurkat T cells at exponential growth phase were washed three times with PBS, cultured in 0.1% FBS in RPMI-1640 media for 24 hrs, treated with 30 nM of PMA and 300 nM of ionomycin for 24 hrs, and then samples for media as well as lysate were prepared.
For media sample preparation, media from the unstimulated or stimulated Jurkat T cells were centrifuged and filtered with 0.2 μm cut-off filter membrane to remove cell debris and membranes and concentrated using a 5 kDa cut-off Microcon centrifugal filter device (Millipore-Amicon, Billerica MA, USA). Cell lysates were prepared by lysing cells in 0.1% Triton-X-100 in PBS (pH7.4) buffer containing 1% of the following cocktail protease inhibitors: 1 mM EDTA, 2 mM 4-(2-Aminoethyl) benzenesulphonyl fluoride (AEBSF), 130 μM Bestatin, 14 μM E-64, 1 μM Leupeptin, and 1 μM Aprotinin (Sigma-Aldrich). The samples from concentrated conditioned media or cell lysate were applied to a 96-well microplate wells coated with OPG-Fc (R&D Systems, Minneapolis, MN, USA). Wells were blocked with 1% BSA (R&D Systems, Minneapolis MN), incubated at room temperature for 2 hrs, and washed with 0.05% Tween 20 in PBS. The bound protein was then eluted from wells with 1X SDS-PAGE sample buffer. The eluted protein was subjected to SDS-PAGE followed by Western blot analysis with a rabbit anti-human RANKL polyclonal antibody (PeproTech Inc, Rocky Hill NJ, USA). Bound antibody was detected using an HRP-conjugated anti-rabbit IgG or HRP-conjugated anti-mouse-IgG (Cell Signaling Technology, Danvers MA, USA) and enhanced chemiluminescence (ECL) blot detection system (PerkinElmer Life Sciences, Boston, MA, USA).
Cloning the longest open reading frame of TNFS11 Variant 2
Total RNA was isolated from Saos-2 cells stimulated with 100 nM PTHrP and cDNA was prepared from 1 μg total RNA by reverse transcription using an Advantage™ RT-for PCR kit. The amplicon corresponding to the protein coding region of secRANKL was amplified by forward primer (5′-AAAGAAGATATCATGGATCCTAATAGAAT-3′) containing EcoRV restriction site (italicized) and reverse primer (5′-CTGGGTCTAGATCTATATCTCGAACTT-3′) containing Xba 1 restriction site (italicized) using Roche Extend High Fidelityplus PCR system (Roche, Branford, CT, USA). RT-PCR was carried out under conditions of 10 cycles (94°C for 20 S, 55°C for 30s, and 72°C for 60s) as first run and then 35 cycles (94°C for 20s, 55°C for 30s, and 72°C for 60s with an incremental increase of 10s at the extension step for each cycle). An A overhang at the end of PCR product was added by applying 1 μl Taq DNA polymerase and incubating at 68°C for 10 min. The PCR products were cloned into PCR II vector (Invitrogen). The plasmids were transformed to dam/dcm deficient competent bacteria for demethylating EcoRV site and then cut by EcoRV and Xba I. The excised fragments were gel purified using Qiagen Qiax Gel Extraction kit and then cloned into pCDNA3.1/myc-his B vector (Invitrogen). 293FT cells (ATTC) were transiently transfected with 5 μg of pcDNA3.1/myc-his B control vector and vector containing the TNFSF11 Variant 2 open reading frame sequence from the beginning of exon 2 (A+1TG) through to the stop codon in Exon 5, using lipofectamine/plus (Invitrogen) in 100 mm diameter plates. After 6 hrs, transfection solution was carefully removed and cells were cultured in 1% FBS media for approximately 40 hrs. Samples of both media and lysates were prepared. Western detection was as described for endogenous RANKL expression except that a mouse anti-myc monoclonal antibody (Invitrogen) was also applied to confirm expression of the recombinant RANKL protein followed by detection with HRP-conjugated anti-mouse-IgG (Cell Signaling) and enhanced chemiluminescence (ECL) blot detection system (PerkinElmer Life Sciences).
Enzyme-linked immunosorbent assay (ELISA)
RANKL protein concentration was measured using a sandwich ELISA developed in our laboratory. Recombinant human RANKL (PeproTech) was used as a standard. 100 μl/well OPG-FC (R&D system) was coated onto 96-well microplates (Corstarcoporation, Cambridge, MA) at final protein concentrations of 0.25 μg/ml in 50 mM carbonate-bicarbonate buffer, pH 9.6, and incubated overnight at 4°C. The plates were blocked using 5% skim milk, 0.1% Tween-20 in PBS (pH 7.4). 100 μl of supernatant from cultured cells was applied to each well and incubated at room temperature for 2 hours. After washing, the plates were incubated with rabbit anti-hRANKL polyclonal antibody (PeproTech), 250 ng/well for 1 hour at room temperature followed by incubation with an anti-rabbit IgG horseradish peroxidase conjugate (Cell Signaling). The bound horseradish peroxidase was assayed with 3, 3′, 5, 5′-tetramethylbenzidine (TMB) (Sigma) as substrate, and the reaction was stopped with 2M H2SO4. ODs were determined at 450 nM. The detection limit for this assay was 35 pg/ml.
Osteoclastogenesis assays
Murine bone marrow macrophages were prepared from femora and tibiae of 4–6 week old male C57BL/6 mice as previously described 51. The bone marrow macrophages (1.5 × 105 cells/well) were seeded in 48 well plates and different volumes of media collected form 293 T cell transfected with the secRANKL expression vector which corresponded to 2.5, 5 or 10ng/mL RANKL protein (measured by ELISA) were applied in total of 300 ul of α-MEM supplemented with 10% fetal bovine serum (Hyclone), 20 ng/ml of M-CSF, 100 unit/ml of penicillin and 100 ug/ml of streptomycin (Invitrogen). In some cases 200ng/mL OPG (R&D) was also added. After 7 days of culture, cells were fixed and stained for tartrate resistant acid phosphatase (TRAP). TRAP-positive cells with >3 nuclei were counted as osteooclasts.
Flow cytometry analyses for membrane-bound RANKL and secreted RANKL protein expression
Jurkat T cells, seeded in 6 well tissue culture plates (Corning, Lowell, MA USA) at a density of 1 X 106 cells/ml, were stimulated with 30 nM PMA and 300 nM ionomycin or DMSO for 24 hrs. Non-stimulated human MG-63 osteosarcoma cells were used as positive control for membrane-bound RANKL protein expression. For intracellular flow cytometry, Brefeldin A (Sigma-Aldrich) was added 4 hrs before harvest to some Jurkat T cells stimulated with PMA/ionomycin as well as control cells. For membrane-bound RANKL protein detection, MG-63 and Jurkat T cells (PMA/ionomycin stimulated or control DMSO treated) were harvested and washed twice in FACS buffer (DPBS supplemented with 1% FBS and 0.1% sodium azide). Cells were resuspended in FACS buffer and then incubated with either IgG1-Fc (110-HG) or RANK-Fc (Cat # 683-RK) chimeras (5 μg/ml, R &D Systems) for 45 min on ice, washed twice for 30 min on ice in FACS buffer containing, phyco-erythin conjugated goat anti-human IgG (Fcγ fragment specific, (eBioscience, Cat # 12-4998, San Diego, CA, USA) for 30 min on ice in FACS buffer. After washing the cells twice with buffer, they were resuspended in FACS buffer and analyzed by flow cytometry. For analysis of intracellular expression of RANKL protein, BD cytofix/cytoperm kit components (BD Biosciences, San Diego, CA, USA) were used. Cells were harvested and washed once with FACS buffer before being incubated with fixation/permeabilization solution, then washed and resuspended in BD Perm/Wash buffer. Fixed/permeabilized cells were incubated with IgG1-Fc, RANK-Fc chimeras as detailed above. All flow cytometric analyses were performed using FACSCalibur (BD Biosciences) and the results were analyzed by Flow Jo software (Tree Star, Ashland, OR, USA).
Supplementary Material
Acknowledgments
This work was supported by: National Institutes of Health R01 AR047665 and 055952 awarded to Dr. Gravallese; Arthritis National Research Foundation (Dr. Walsh); The University of Queensland Early Career Grant, The Clive and Vera Ramaciotti Foundations Establishment Gift and a National Health and Medical Research (NHMRC) Council Career Development Award, awarded to Dr. Pettit; a NHMRC Dora Lush Postgraduate Scholarship (409914) awarded to Dr. Alexander.
Footnotes
Supplementary Information: Supplemental Figures 1–3 and associated information is available at the Genes & Immunity’s website, http://www.nature.com/gene/index.html.
Conflicts of Interest:
The authors declare no conflicts of interest, financial or otherwise.
References
- 1.Yasuda H, Shima N, Nakagawa N, Yamaguchi K, Kinosaki M, Mochizuki S, et al. Osteoclast differentiation factor is a ligand for osteoprotegerin/osteoclastogenesis-inhibitory factor and is identical to TRANCE/RANKL. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(7):3597–602. doi: 10.1073/pnas.95.7.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kong YY, Yoshida H, Sarosi I, Tan HL, Timms E, Capparelli C, et al. OPGL is a key regulator of osteoclastogenesis, lymphocyte development and lymph-node organogenesis. Nature. 1999;397(6717):315–23. doi: 10.1038/16852. [DOI] [PubMed] [Google Scholar]
- 3.Anjubault T, Martin J, Hubert FX, Chauvin C, Heymann D, Josien R. Constitutive expression of TNF-related activation-induced cytokine (TRANCE)/receptor activating NF-kappaB ligand (RANK)-L by rat plasmacytoid dendritic cells. PloS one. 7(3):e33713. doi: 10.1371/journal.pone.0033713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanada R, Hanada T, Sigl V, Schramek D, Penninger JM. RANKL/RANK-beyond bones. J Mol Med (Berl) 2011;89(7):647–56. doi: 10.1007/s00109-011-0749-z. [DOI] [PubMed] [Google Scholar]
- 5.Wong BR, Rho J, Arron J, Robinson E, Orlinick J, Chao M, et al. TRANCE is a novel ligand of the tumor necrosis factor receptor family that activates c-Jun N-terminal kinase in T cells. The Journal of biological chemistry. 1997;272(40):25190–4. doi: 10.1074/jbc.272.40.25190. [DOI] [PubMed] [Google Scholar]
- 6.Wong BR, Josien R, Lee SY, Sauter B, Li HL, Steinman RM, et al. TRANCE (tumor necrosis factor [TNF]-related activation-induced cytokine), a new TNF family member predominantly expressed in T cells, is a dendritic cell-specific survival factor. J Exp Med. 1997;186(12):2075–80. doi: 10.1084/jem.186.12.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anderson DM, Maraskovsky E, Billingsley WL, Dougall WC, Tometsko ME, Roux ER, et al. A homologue of the TNF receptor and its ligand enhance T-cell growth and dendritic-cell function. Nature. 1997;390(6656):175–9. doi: 10.1038/36593. [DOI] [PubMed] [Google Scholar]
- 8.Yasuda H, Shima N, Nakagawa N, Yamaguchi K, Kinosaki M, Goto M, et al. A novel molecular mechanism modulating osteoclast differentiation and function. Bone. 1999;25 (1):109–13. doi: 10.1016/s8756-3282(99)00121-0. [DOI] [PubMed] [Google Scholar]
- 9.Lacey DL, Timms E, Tan HL, Kelley MJ, Dunstan CR, Burgess T, et al. Osteoprotegerin ligand is a cytokine that regulates osteoclast differentiation and activation. Cell. 1998;93 (2):165–76. doi: 10.1016/s0092-8674(00)81569-x. [DOI] [PubMed] [Google Scholar]
- 10.Lum L, Wong BR, Josien R, Becherer JD, Erdjument-Bromage H, Schlondorff J, et al. Evidence for a role of a tumor necrosis factor-alpha (TNF-alpha)-converting enzyme-like protease in shedding of TRANCE, a TNF family member involved in osteoclastogenesis and dendritic cell survival. The Journal of biological chemistry. 1999;274(19):13613–8. doi: 10.1074/jbc.274.19.13613. [DOI] [PubMed] [Google Scholar]
- 11.Nagai M, Kyakumoto S, Sato N. Cancer cells responsible for humoral hypercalcemia express mRNA encoding a secreted form of ODF/TRANCE that induces osteoclast formation. Biochem Biophys Res Commun. 2000;269(2):532–6. doi: 10.1006/bbrc.2000.2314. [DOI] [PubMed] [Google Scholar]
- 12.Lai FP, Cole-Sinclair M, Cheng WJ, Quinn JM, Gillespie MT, Sentry JW, et al. Myeloma cells can directly contribute to the pool of RANKL in bone bypassing the classic stromal and osteoblast pathway of osteoclast stimulation. Br J Haematol. 2004;126(2):192–201. doi: 10.1111/j.1365-2141.2004.05018.x. [DOI] [PubMed] [Google Scholar]
- 13.Farrugia AN, Atkins GJ, To LB, Pan B, Horvath N, Kostakis P, et al. Receptor activator of nuclear factor-kappaB ligand expression by human myeloma cells mediates osteoclast formation in vitro and correlates with bone destruction in vivo. Cancer research. 2003;63 (17):5438–45. [PubMed] [Google Scholar]
- 14.Fuller K, Wong B, Fox S, Choi Y, Chambers TJ. TRANCE is necessary and sufficient for osteoblast-mediated activation of bone resorption in osteoclasts. J Exp Med. 1998;188(5):997–1001. doi: 10.1084/jem.188.5.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsuzaki K, Udagawa N, Takahashi N, Yamaguchi K, Yasuda H, Shima N, et al. Osteoclast differentiation factor (ODF) induces osteoclast-like cell formation in human peripheral blood mononuclear cell cultures. Biochem Biophys Res Commun. 1998;246(1):199–204. doi: 10.1006/bbrc.1998.8586. [DOI] [PubMed] [Google Scholar]
- 16.Tsukii K, Shima N, Mochizuki S, Yamaguchi K, Kinosaki M, Yano K, et al. Osteoclast differentiation factor mediates an essential signal for bone resorption induced by 1 alpha,25-dihydroxyvitamin D3, prostaglandin E2, or parathyroid hormone in the microenvironment of bone. Biochem Biophys Res Commun. 1998;246(2):337–41. doi: 10.1006/bbrc.1998.8610. [DOI] [PubMed] [Google Scholar]
- 17.Takahashi N, Udagawa N, Suda T. A new member of tumor necrosis factor ligand family, ODF/OPGL/TRANCE/RANKL, regulates osteoclast differentiation and function. Biochem Biophys Res Commun. 1999;256(3):449–55. doi: 10.1006/bbrc.1999.0252. [DOI] [PubMed] [Google Scholar]
- 18.Mizuno A, Kanno T, Hoshi M, Shibata O, Yano K, Fujise N, et al. Transgenic mice overexpressing soluble osteoclast differentiation factor (sODF) exhibit severe osteoporosis. J Bone Miner Metab. 2002;20(6):337–44. doi: 10.1007/s007740200049. [DOI] [PubMed] [Google Scholar]
- 19.Vernal R, Dutzan N, Hernandez M, Chandia S, Puente J, Leon R, et al. High expression levels of receptor activator of nuclear factor-kappa B ligand associated with human chronic periodontitis are mainly secreted by CD4+ T lymphocytes. J Periodontol. 2006;77(10):1772–80. doi: 10.1902/jop.2006.050376. [DOI] [PubMed] [Google Scholar]
- 20.Bostanci N, Ilgenli T, Emingil G, Afacan B, Han B, Toz H, et al. Gingival crevicular fluid levels of RANKL and OPG in periodontal diseases: implications of their relative ratio. J Clin Periodontol. 2007;34(5):370–6. doi: 10.1111/j.1600-051X.2007.01061.x. [DOI] [PubMed] [Google Scholar]
- 21.Sakellari D, Menti S, Konstantinidis A. Free soluble receptor activator of nuclear factor-kappab ligand in gingival crevicular fluid correlates with distinct pathogens in periodontitis patients. J Clin Periodontol. 2008;35(11):938–43. doi: 10.1111/j.1600-051x.2008.01314.x. [DOI] [PubMed] [Google Scholar]
- 22.Silva MJ, Kajiya M, AlShwaimi E, Sasaki H, Hong J, Ok P, et al. Bacteria-reactive immune response may induce RANKL-expressing T cells in the mouse periapical bone loss lesion. J Endod. 2012;38(3):346–50. doi: 10.1016/j.joen.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kreahling J, Graveley BR. The origins and implications of Aluternative splicing. Trends Genet. 2004;20(1):1–4. doi: 10.1016/j.tig.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 24.Kong YY, Feige U, Sarosi I, Bolon B, Tafuri A, Morony S, et al. Activated T cells regulate bone loss and joint destruction in adjuvant arthritis through osteoprotegerin ligand. Nature. 1999;402(6759):304–9. doi: 10.1038/46303. [DOI] [PubMed] [Google Scholar]
- 25.Ikeda T, Kasai M, Utsuyama M, Hirokawa K. Determination of three isoforms of the receptor activator of nuclear factor-kappaB ligand and their differential expression in bone and thymus. Endocrinology. 2001;142(4):1419–26. doi: 10.1210/endo.142.4.8070. [DOI] [PubMed] [Google Scholar]
- 26.Ikeda T, Kasai M, Suzuki J, Kuroyama H, Seki S, Utsuyama M, et al. Multimerization of the receptor activator of nuclear factor-kappaB ligand (RANKL) isoforms and regulation of osteoclastogenesis. The Journal of biological chemistry. 2003;278(47):47217–22. doi: 10.1074/jbc.M304636200. [DOI] [PubMed] [Google Scholar]
- 27.Suzuki J, Ikeda T, Kuroyama H, Seki S, Kasai M, Utsuyama M, et al. Regulation of osteoclastogenesis by three human RANKL isoforms expressed in NIH3T3 cells. Biochem Biophys Res Commun. 2004;314(4):1021–7. doi: 10.1016/j.bbrc.2003.12.191. [DOI] [PubMed] [Google Scholar]
- 28.Horwood NJ, Kartsogiannis V, Quinn JM, Romas E, Martin TJ, Gillespie MT. Activated T lymphocytes support osteoclast formation in vitro. Biochem Biophys Res Commun. 1999;265(1):144–50. doi: 10.1006/bbrc.1999.1623. [DOI] [PubMed] [Google Scholar]
- 29.Gravallese EM, Manning C, Tsay A, Naito A, Pan C, Amento E, et al. Synovial tissue in rheumatoid arthritis is a source of osteoclast differentiation factor. Arthritis and rheumatism. 2000;43(2):250–8. doi: 10.1002/1529-0131(200002)43:2<250::AID-ANR3>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 30.Takayanagi H, Iizuka H, Juji T, Nakagawa T, Yamamoto A, Miyazaki T, et al. Involvement of receptor activator of nuclear factor kappaB ligand/osteoclast differentiation factor in osteoclastogenesis from synoviocytes in rheumatoid arthritis. Arthritis and rheumatism. 2000;43(2):259–69. doi: 10.1002/1529-0131(200002)43:2<259::AID-ANR4>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 31.Teng YT, Nguyen H, Gao X, Kong YY, Gorczynski RM, Singh B, et al. Functional human T-cell immunity and osteoprotegerin ligand control alveolar bone destruction in periodontal infection. The Journal of clinical investigation. 2000;106(6):R59–67. doi: 10.1172/jci10763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang Y, Mehta CK, Hsu TY, Alsulaimani FF. Bacteria induce osteoclastogenesis via an osteoblast-independent pathway. Infect Immun. 2002;70(6):3143–8. doi: 10.1128/IAI.70.6.3143-3148.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu D, Xu JK, Figliomeni L, Huang L, Pavlos NJ, Rogers M, et al. Expression of RANKL and OPG mRNA in periodontal disease: possible involvement in bone destruction. Int J Mol Med. 2003;11(1):17–21. doi: 10.3892/ijmm.11.1.17. [DOI] [PubMed] [Google Scholar]
- 34.Kawai T, Matsuyama T, Hosokawa Y, Makihira S, Seki M, Karimbux NY, et al. B and T lymphocytes are the primary sources of RANKL in the bone resorptive lesion of periodontal disease. The American journal of pathology. 2006;169(3):987–98. doi: 10.2353/ajpath.2006.060180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giuliani N, Colla S, Sala R, Moroni M, Lazzaretti M, La Monica S, et al. Human myeloma cells stimulate the receptor activator of nuclear factor-kappa B ligand (RANKL) in T lymphocytes: a potential role in multiple myeloma bone disease. Blood. 2002;100(13):4615–21. doi: 10.1182/blood-2002-04-1121. [DOI] [PubMed] [Google Scholar]
- 36.Colucci S, Brunetti G, Rizzi R, Zonno A, Mori G, Colaianni G, et al. T cells support osteoclastogenesis in an in vitro model derived from human multiple myeloma bone disease: the role of the OPG/TRAIL interaction. Blood. 2004;104(12):3722–30. doi: 10.1182/blood-2004-02-0474. [DOI] [PubMed] [Google Scholar]
- 37.Stupphann D, Rauner M, Krenbek D, Patsch J, Pirker T, Muschitz C, et al. Intracellular and surface RANKL are differentially regulated in patients with ankylosing spondylitis. Rheumatology international. 2008;28(10):987–93. doi: 10.1007/s00296-008-0567-y. [DOI] [PubMed] [Google Scholar]
- 38.O’Brien CA. Control of RANKL gene expression. Bone. 2010;46(4):911–9. doi: 10.1016/j.bone.2009.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bishop KA, Coy HM, Nerenz RD, Meyer MB, Pike JW. Mouse Rankl expression is regulated in T cells by c-Fos through a cluster of distal regulatory enhancers designated the T cell control region. The Journal of biological chemistry. 2011;286(23):20880–91. doi: 10.1074/jbc.M111.231548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bishop KA, Meyer MB, Pike JW. A novel distal enhancer mediates cytokine induction of mouse RANKl gene expression. Molecular endocrinology. 2009;23(12):2095–110. doi: 10.1210/me.2009-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takami M, Cho ES, Lee SY, Kamijo R, Yim M. Phosphodiesterase inhibitors stimulate osteoclast formation via TRANCE/RANKL expression in osteoblasts: possible involvement of ERK and p38 MAPK pathways. FEBS letters. 2005;579(3):832–8. doi: 10.1016/j.febslet.2004.12.066. [DOI] [PubMed] [Google Scholar]
- 42.Takami M, Takahashi N, Udagawa N, Miyaura C, Suda K, Woo JT, et al. Intracellular calcium and protein kinase C mediate expression of receptor activator of nuclear factor-kappaB ligand and osteoprotegerin in osteoblasts. Endocrinology. 2000;141(12):4711–9. doi: 10.1210/endo.141.12.7852. [DOI] [PubMed] [Google Scholar]
- 43.Fu Q, Manolagas SC, O’Brien CA. Parathyroid hormone controls receptor activator of NF-kappaB ligand gene expression via a distant transcriptional enhancer. Molecular and cellular biology. 2006;26(17):6453–68. doi: 10.1128/MCB.00356-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nerenz RD, Martowicz ML, Pike JW. An enhancer 20 kilobases upstream of the human receptor activator of nuclear factor-kappaB ligand gene mediates dominant activation by 1,25-dihydroxyvitamin D3. Molecular endocrinology. 2008;22(5):1044–56. doi: 10.1210/me.2007-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim S, Yamazaki M, Zella LA, Shevde NK, Pike JW. Activation of receptor activator of NF-kappaB ligand gene expression by 1,25-dihydroxyvitamin D3 is mediated through multiple long-range enhancers. Molecular and cellular biology. 2006;26(17):6469–86. doi: 10.1128/MCB.00353-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim S, Yamazaki M, Zella LA, Meyer MB, Fretz JA, Shevde NK, et al. Multiple enhancer regions located at significant distances upstream of the transcriptional start site mediate RANKL gene expression in response to 1,25-dihydroxyvitamin D3. J Steroid Biochem Mol Biol. 2007;103(3–5):430–4. doi: 10.1016/j.jsbmb.2006.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martowicz ML, Meyer MB, Pike JW. The mouse RANKL gene locus is defined by a broad pattern of histone H4 acetylation and regulated through distinct distal enhancers. Journal of cellular biochemistry. 2011;112(8):2030–45. doi: 10.1002/jcb.23123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee HL, Bae OY, Baek KH, Kwon A, Hwang HR, Qadir AS, et al. High extracellular calcium-induced NFATc3 regulates the expression of receptor activator of NF-kappaB ligand in osteoblasts. Bone. 2011;49(2):242–9. doi: 10.1016/j.bone.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 49.Rossa C, Ehmann K, Liu M, Patil C, Kirkwood KL. MKK3/6-p38 MAPK signaling is required for IL-1beta and TNF-alpha-induced RANKL expression in bone marrow stromal cells. Journal of interferon & cytokine research: the official journal of the International Society for Interferon and Cytokine Research. 2006;26(10):719–29. doi: 10.1089/jir.2006.26.719. [DOI] [PubMed] [Google Scholar]
- 50.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic acids research. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Crotti TN, Flannery M, Walsh NC, Fleming JD, Goldring SR, McHugh KP. NFATc1 regulation of the human beta3 integrin promoter in osteoclast differentiation. Gene. 2006;372:92–102. doi: 10.1016/j.gene.2005.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.