Abstract
The PAT family of lipid storage droplet proteins comprised five members, each of which has become an established regulator of cellular neutral lipid metabolism. Perilipin 5 (also known as lsdp-5, MLDP, PAT-1, and OXPAT), the most recently discovered member of the family, has been shown to localize to two distinct intracellular pools: the lipid storage droplet (LD), and a poorly characterized cytosolic fraction. We have characterized the denser of these intracellular pools and find that a population of perilipin 5 not associated with large LDs resides in complexes with a discrete density (~ 1.15 g/ml) and size (~ 575 kDa). Using immunofluorescence, western blotting of isolated sucrose density fractions, native gradient gel electrophoresis, and co-immunoprecipitation, we have shown that these small (~ 15 nm), perilipin 5-encoated structures do not contain the PAT protein perilipin 2 (ADRP), but do contain perilipin 3 and several other as of yet uncharacterized proteins. The size and density of these particles as well as their susceptibility to degradation by lipases suggest that like larger LDs, they have a neutral lipid rich core. When treated with oleic acid to promote neutral lipid deposition, cells ectopically expressing perilipin 5 experienced a reorganization of LDs in the cell, resulting in fewer, larger droplets at the expense of smaller ones. Collectively, these data demonstrate that a portion of cytosolic perilipin 5 resides in high density lipid droplet complexes that participate in cellular neutral lipid accumulation.
Keywords: Perilipin, Lipolysis, Lipid trafficking, Lipid droplets, Muscle, Adipose
1. Introduction
Nearly all cells possess the ability to store neutral lipid, but when excessive this storage is not without consequence. As the obesity epidemic spreads in Western society, a corresponding increase is observed in type II diabetes mellitus (T2DM) [1]. While not exclusively causal, ectopic neutral lipid storage in liver and muscle is emerging as key factors in the etiology of these diseases [2]. Several recent studies have made mechanistic advances into the cause of insulin resistance as it relates to altered neutral lipid storage. Bostrom et al. have shown that sequestration of pro-fusogenic proteins of the SNAP/SNARE pathway becomes localized on LDs preventing their use in fusion of GLUT4 coated vesicles with the plasma membrane, decreasing cellular insulin sensitivity [3] and [4]. Others have shown that overabundant neutral lipid storage in liver blunts signaling responses (AKT) important in the insulin response and may likely contribute to the development of T2DM [5].
At the core of these pathologies is the cell’s ability to sequester neutral lipid. The PAT family consists of five related genes which give rise to at least seven different proteins (perilipin A, B, and C, ADRP [also called Adipophilin or ADFP], TIP47, OXPAT [also called MLDP, perilipin 5, PAT-1, or LSDP5] and s3-12). This family has recently undergone a revision in nomenclature, such that now the proteins are all known as perilipins (to reflect their binding to the periphery of lipid droplets) and numbered in their order of discovery (perilipin, ADRP, TIP-47, s3-12, and OXPAT, respectively) [6]. Some combination of these proteins coats the surface of all LDs. The protein family members can be divided into those proteins solely found on the surface of the lipid storage droplet (perilipins 1 and 2; the constitutive or c-PATs) and those that are found on both the lipid storage droplet and the cytosol (perilipins 3, 4, and 5; the exchangeable or e-PATs) [7]. For reviews, please see [8], [9], [10], [11], [12], [13] and [14].
PAT proteins have been shown to act as cofactors for lipases (perilipin 1A with HSL or ATGL), as binding partners for other proteins (perilipin 1A with CGI-58/abhd-5, a coactivator of ATGL and lyso-PA acyltransferase) [15], [16], [17], [18], [19], [20] and [21] and as interaction partners with trafficking proteins (perilipin 3 with M6PR and Rab 9, and perilipin 2 with SNAP-23, syntaxin 5, and Vamp 4) [3], [4], [22], [23] and [24]. In general, PAT proteins appear to coat LDs in a hierarchical fashion as LDs mature, with perilipin 4 found on the most nascent droplets close to the plasma membrane, followed by perilipin 3, and then perilipin 2 or perilipin 1 found on the most mature LDs [20], [25], [26], [27] and [28]. It is not totally clear where perilipin 5 exists in this hierarchy, although it does not appear to displace perilipin 2 from the droplet surface.
Initial studies on perilipin 5 revealed that the gene is expressed in tissues with high capacity for fatty acid oxidation (such as brown adipose tissue, liver, heart, and skeletal muscle) and regulated by PPAR-α [29], [30] and [31]. Subsequent studies have shown that perilipin 5 also interacts with CGI-58 and HSL [20] and [32]. All of these studies have focused on the roles perilipin 5 play on the lipid storage droplet, although it is exchangeable and may be found on other structures (cytosol or endoplasmic reticulum) as well [33].
In the current work, we present novel evidence suggesting that the cytosolic pool of perilipin 5 is not found free in solution, but rather is bound to structures of discrete density. Furthermore, we have characterized this pool of perilipin 5, and found that it consists, at least partially, of discrete structures of small size (< 20 nm in diameter) with biochemical characteristics reminiscent of circulating high density lipoprotein particles. We propose that these high density lipid droplets (HDLDs) may represent nascent intracellular sites for lipid accumulation. Upon treatment of cells with oleic acid to promote neutral lipid storage, perilipin 5 was found to shift from this cytosolic HDLD pool to more buoyant LDs. We propose that these two distinct pools play key roles in lipid storage and mobilization mediated by perilipin 5.
2. Methods
2.1. Reagents
Unless otherwise noted, all chemical reagents were purchased from Sigma (St. Louis, MO) or Fisher Chemical, and all culture media were obtained from Gibco/Invitrogen. Rabbit polyclonal serum against perilipin 2 and the perilipin-GFP cDNA construct were gifts from Dr. C. Londos (National Institutes of Health). Mouse monoclonal anti-FLAG (M2), anti-FLAG M2 affinity agarose gel, FLAG eluting peptide, and the GPO-Trinder triglyceride kit were purchased from Sigma. Mouse monoclonal anti-perilipin 2 and guinea pig polyclonal anti-perilipin 3 antibodies were obtained from Research Diagnostics. Rabbit polyclonal anti-perilipin 3 antibodies were purchased from Novex. Rabbit anti-calreticulin antibody was purchased from BD Biosciences. Mouse monoclonal antibody against mitochondrial complex IV was purchased from MitoSciences. Rabbit polyclonal anti-FLAG was purchased from Cell Signaling Technology or Sigma. Murine CHO-K1 cells were obtained from the American Type Culture Collection. BODIPY 558/568 C12, ProLong Gold Antifade mounting kits, PS-Speck microscope point source kit, and Alexa Fluor-conjugated secondary antibodies used for immunofluorescence were obtained from Molecular Probes/Invitrogen. Precast Novex Tris–glycine acrylamide gels, Novex Bis–Tris gradient polyacrylamide gels, NativePAGE sample preparation kit, NativeMark protein molecular weight standards, and NativePAGE running buffer kit were obtained from Invitrogen. The high molecular weight native protein marker kit used in native gradient gel electrophoresis was purchased from GE Healthcare, as was the protein silver staining kit. Anti-mouse and anti-rabbit TrueBlot horseradish peroxidase (HRP)-conjugated secondary antibodies (raised against the native forms of IgG molecules) were obtained from eBioscience, and were used in the detection of primary antibodies when probing immunoblots of immunoprecipitated material; all other HRP-conjugated secondary antibodies used in immunoblotting were purchased from Pierce Biotechnology. SuperSignal chemiluminescent substrate and the BCA protein assay kit were also obtained from Pierce Biotechnology. Oleic acid was obtained from EMD Chemicals/Calbiochem. Formvar-carbon coated 300 mesh copper grids were from Ted Pella. Rabbit anti-mouse perilipin 5 polyclonal antisera were generated by Invitrogen against the peptide NH3+-PGSSLGELDQQNVVNRC-COO−. This peptide corresponds to a region of the amino terminus of perilipin 5 from Mus musculus which has a minimal overlap with other PAT family members. No crossreactivity was found with either perilipin 2 or 3 (perilipins 1 and 4 have not been tested but are not expressed in the cell lines employed in these studies). The specificity of the antisera was checked against 3xFLAG-tagged perilipin 5 and an anti-perilipin 5 antibody graciously provided by Dr. Perry Bickel (UT Health Science Center, Houston, Texas).
2.2. Generation of perilipin 5-3x-FLAG fusion protein and derivatives
The polymerase chain reaction was used to introduce a novel BglII site as well as delete the stop codon found in the cDNA coding for murine perilipin 5. This facilitated the in-frame cloning of the c-DNA into the expression vector. The T7 primer was used in the forward direction and the sequence of the mutagenic reverse primer was 5′-GATAGTCAGATCTCCCAGCTCTGGCATC-3′. This PCR product was cloned into a pGEM T-Vector, cut with BglII and EcoRI, and subsequently cloned into the 3x-FLAG expression vector between the BglII and EcoRI sites. To remove the FLAG epitopes, the perilipin 5-3x-FLAG plasmid was cut with BglII, filled in with Klenow fragment to generate a stop codon, and a blunt ended ligation was performed to recircularize the plasmid. Transfection grade plasmid DNA was obtained using a Qiagen endo-free maxiprep kit. The identity of the perilipin 5 cDNA was confirmed by sequencing of the complete coding sequence.
2.3. Cell culture and expression of perilipin 5 in CHO cells
CHO-K1 cells were maintained in Ham’s F-12 medium (Invitrogen) containing 5% Cosmic Calf Serum (HyClone), 10 mM l-glutamine, 14 mM NaHCO3, 100 units/ml penicillin, and 100 units/ml streptomycin at 37 °C in a 5% CO2 atmosphere. Functional studies of perilipin 5 were performed using CHO-K1 cells stably overexpressing epitope-tagged murine perilipin 5, transfected according to the LipofectAMINE protocol (Invitrogen). Where indicated, some CHO cells were transfected with the perilipin 5 expression vector in which the FLAG epitopes were removed. In control experiments, CHO cells were transfected with the 3x-FLAG expression vector in the absence of perilipin 5 cDNA. Selection was performed using 300 μg/ml G418. For lipid loading experiments, cells were either treated for 24 h with fresh medium or medium supplemented with 400 μM oleic acid complexed to fatty acid-free bovine serum albumin (lipid loading media) to promote the storage of triacylglycerols (TAGs).
2.4. Animal studies
8–10 week old male C57Bl6J mice from The Jackson Labs were housed on a 12-hour light/dark cycle and provided food and water ad libitum. The afternoon prior to the experiment food was removed and animals were fasted until 10 AM the next morning to elevate hepatic perilipin 5 expression [29], [30] and [31]. All animal experimentation was conducted in accordance with the guidelines of the Animal Care and Use Committee of Otterbein College (protocol #2010-03-01-01).
At the time of the experiment, animals were euthanized by exsanguination; livers and hearts were removed, rinsed in PBS, and minced in the same buffer. Larger chunks of tissue were removed by a low speed centrifugation (100 ×g, 30 s), and cells in the resulting supernatant were allowed to swell in hypotonic lysis buffer (HLB; 10 mM TRIS, pH 7.4, 1 mM EDTA,) on ice for 20 min prior to shearing through a 28 g needle five passes. This suspension was centrifuged for 15 min at 15,000 ×g to produce a post nuclear supernatant (PNS) used in subsequent analysis.
2.5. Immunofluorescence microscopy, imaging and neutral lipid staining
Immunofluorescence microscopy was conducted as previously described with modifications [25] and [34]. Cells were cultured on sterilized glass coverslips, and neutral lipids were visualized by supplementing lipid loading media or fresh basal growth media with 6 μM BODIPY 558/568 C12 for 24 h. Cells were then fixed with 4% paraformaldehyde in PBS for 1 h and washed three times with PBS before mounting with Prolong Gold for visualization.
In immunolabeling experiments, fixed cells were made permeable by incubation with 0.2% Triton X-100 in PBS for 15 min at room temperature, and blocked with 1% non-fat dry milk/5% normal goat serum in PBS for 1 h. For antigen detection, the following primary antibodies were employed: rabbit polyclonal anti-FLAG (1:100), rabbit polyclonal anti-perilipin 2 serum (1:500), mouse monoclonal anti-FLAG (1:200), mouse monoclonal anti-perilipin 2 (1:5), rabbit anti-perilipin 3 (1:500) and rabbit polyclonal anti-perilipin 5 (1:1000).
After washing three times with PBS containing 0.01% Triton X-100, cells were incubated with secondary Alexa Fluor 488-conjugated goat anti rabbit antibody (1:100). All antibodies were diluted in PBS containing 2% goat serum with 0.01% Triton X-100. Following three additional washes with PBS containing 0.01% Triton X-100, coverslips were mounted using ProLong Gold (Molecular Probes). For double labeling, primary antibodies raised in different species were used simultaneously. Cells were then incubated with species-specific secondary antibodies bearing different fluorophores (all at 1:200 and prepared as described above). Cells were visualized using a Zeiss 510 laser scanning confocal microscope (Carl Zeiss Inc).
2.6. Lipid droplet morphometry
Cells cultured on glass coverslips were incubated for 24 h in lipid loading media supplemented with 6 μM BODIPY 558/568 C12 to label neutral lipids, then fixed and stained as described above. Nuclei were counterstained with 5 μg/ml 4′,6-diamidino-2-phenylindole (DAPI, Sigma). Specimens were then mounted and viewed using a Nikon Eclipse TE200 inverted epifluorescence microscope (Nikon Instruments). Single channel grayscale images (15–25 randomly selected optical fields per treatment condition) were collected using a SPOT RT camera and SPOT 4.0.5 imaging software (Diagnostic Instruments). Prior to analysis, the pixels on the digital images were inverted, such that the BODIPY- stained LDs appeared dark against a white background to facilitate LD analysis. LD morphometric analysis was performed using two methods. To address whether perilipin 5-3X-FLAG expression altered maximal LD size, the diameter of the largest LD per cell was evaluated using SPOT imaging software (method 1). Cells were classified based on the presence or absence of perilipin 5-3X-FLAG expression as determined by anti-FLAG staining. This method was insufficient, however, to allow precise measurements of the numerous, smaller lipid droplets present in each cell. For a more comprehensive analysis, the images were re-analyzed using BioPix Tissue 1.5 software (BioPix Software) in which the size and number of lipid droplets were evaluated by manual pixel counting (method 2) [35]. For all size determinations, only in-focus objects were evaluated; these were classified into size categories based on diameter. Over 100 randomly selected cells per condition were analyzed in each instance.
2.7. Subcellular fractionation
Cells were fractionated based on Weller and Yamaguchi protocols [36] and [37] with the following modifications. Briefly, following overnight treatments specified in each experiment, cells were rinsed in ice-cold PBS, harvested by scraping, and collected by low-speed centrifugation. The cell pellets (~ 120 × 106 cells/pellet) were resuspended in hypotonic lysis buffer (HLB) supplemented with protease inhibitor cocktail (Sigma) and the pellets were allowed to swell on ice for 20 min. Homogenization was achieved using 5 passages though a 28-gage needle, and the homogenates were subjected to centrifugation at 15,000 ×g for 15 min/4 °C to obtain a postnuclear supernatant (PNS). The PNS was adjusted to 30% sucrose. For each homogenate, a discontinuous sucrose gradient was prepared in HLB using one of two protocols. In pilot experiments, conventional gradients (0–30% sucrose) comprised the following (from bottom to top): weighted PNS (30% sucrose, adjusted to 5 ml), 20% sucrose (2.5 ml), 10% sucrose (2.5 ml), and HLB (1 ml). In subsequent experiments, an extended sucrose gradient (0–60% sucrose) was used consisting of: 60% sucrose (2 ml), 40% sucrose (2 ml), weighted PNS (30% sucrose, 2 ml), 20% sucrose (2 ml), 10% sucrose (2 ml), and HLB (1 ml). Gradients were subjected to centrifugation at 100,000 ×g for 16 h/4 °C (SW28.1 Ti swinging bucket rotor, Beckman Coulter). Following centrifugation, either six or eleven (as indicated in the text and figures) equal-volume fractions were collected from the top. Aliquots from each fraction were then subjected to dot blotting or separated either by SDS-PAGE, or NDGGE, and transferred electrophoretically to nitrocellulose membranes. Immunoblots were probed as described in the text and figure legends.
2.8. Immunoprecipitation
CHO-K1 cells expressing perilipin 5-3x-FLAG were rinsed in ice-cold PBS and collected into HLB, and a PNS was prepared as described above. The PNS (from approximately 2 × 107 cells in a 1 ml volume) was added to 40 μl of the washed anti-FLAG M2 affinity gel resin (prepared as indicated in the accompanying literature) and incubated overnight on a rotary mixer. The resin was recovered by brief centrifugation (5000 ×g for 30 s) and washed 3 times in Tris-buffered saline (TBS; 150 mM NaCl, 10 mM Tris–HCl, pH 7.2). The immunoprecipiated material was eluted from the affinity resin using 100 μl of 1 mg/ml FLAG peptide.
2.9. Protein electrophoresis and immunoblot analysis
For SDS-PAGE, cells were harvested as described above, and gel electrophoresis using Novex Tris–glycine 10% acrylamide gels (Invitrogen) was performed to resolve 25 μg of total protein per lane. For non-denaturing gradient gel electrophoresis (NDGGE), proteins were resolved at 150 V for 2 h using Novex 3–12% Bis–Tris gradient gels (Invitrogen) and components of the NativePAGE gel system, according to the instructions of the manufacturer (Invitrogen). Proteins resolved under either native or denaturing conditions were subsequently either transferred electrophoretically to nitrocellulose membranes or subjected to silver staining using a kit, according to the instructions of the manufacturer (GE Healthcare). Alternately, protein dot blots were prepared using the Bio-Rad BioDot apparatus according to the manufacturer’s instructions.
For immunoblotting, membranes were blocked for 1 h using 5% nonfat dry milk in Tris buffered saline/Tween 20 (TBST) (150 mM NaCl, 10 mM Tris–HCl pH 7.2, 0.1% Tween 20) prior to incubation with primary antibodies (1 h at 25 °C or overnight at 4 °C). The following primary antibodies were used: mouse anti-FLAG (1:1000), rabbit anti-ADRP (1:2000), and guinea pig anti-TIP47 (1:2000). After washing, the membranes were exposed to horseradish peroxidase-conjugated secondary antibodies and detected using SuperSignal chemiluminescent substrate (Pierce) and the VersaDoc Imaging System (Bio-Rad).
2.10. Transmission electron microscopy (TEM)
Electron microscopy was performed as previously described [38]. Briefly, a drop of sample was placed on a 300-mesh formvar-carbon coated copper grid (Ted Palla) for 1 min and the excess fluid was wicked away using a laboratory cleaning tissue. For negative staining, a drop of 0.2% phosphotungstic acid (PTA) was then applied to the grid for 30 s and subsequently drawn off from the edge of the grid with filter paper. The specimens were allowed to air-dry prior to observation using a Tecnai G2 Spirit electron microscope (FEI) operated at 80 kV.
2.11. Quantification of total cellular triacylglycerols
Cells were grown to confluence in 100 mm culture dishes. Following two washes with cold PBS, cells were collected in 500 μl cold PBS and then sonicated for 10 s at a 2.0 power setting on a Misonix 3000 Sonicator (Misonix) equipped with a titanium Microtip attachment. A micro-scale version of the Trinder triglyceride assay (Sigma) was performed on cell sonicates and read on an Amersham Biosciences BioTrakII microtiter plate reader. Total triglyceride content was normalized to total cellular protein assessed using a BCA protein assay (Pierce).
2.12. Particle degradation assays
Isolated particles or post nuclear supernatants were treated with 1% Tween-20, 1% Triton-X-100, or 100 units of Candida rugosa lipase (all prepared in 10 mM TRIS, pH 7.4). Following a 1 h incubation particles were analyzed using NDGGE and immunoblotting as previously described. To further validate lipase degradation assays, plasma samples were treated as above and analyzed using a modification of the technique of Papadopoulos and Kintzios [54]. Briefly, plasma samples were electrophoresed through 0.5% agarose gels cast and run in 50 mM TRIS pH 8.6. Gels were fixed by treating for 45 min with 2.5% TCA, rinsed in water for 20 min, dried on glass plates overnight, and stained for 1 h in a freshly prepared saturated solution of oil red O in 80% methanol.
3. Results
3.1. Perilipin 5 exists in two distinct cellular compartments
Previous studies have reported that perilipin 5 localizes to lipid storage droplets [29], [30] and [31]. However, several of these same studies also found that a significant fraction of the protein was not bound to conventional LDs, but rather, was associated with a poorly-defined cytosolic pool. To further probe this observation, Chinese Hamster Ovary fibroblasts (CHO-K1) were transfected with a carboxy-terminal 3-xFLAG labeled perilipin 5 c-DNA construct (perilipin 5-3X-FLAG). CHO cells do not express endogenous perilipin 5, but have been successfully employed in ectopic expression studies by several labs to elucidate multiple functions of PAT proteins [15], [31], [34] and [39].
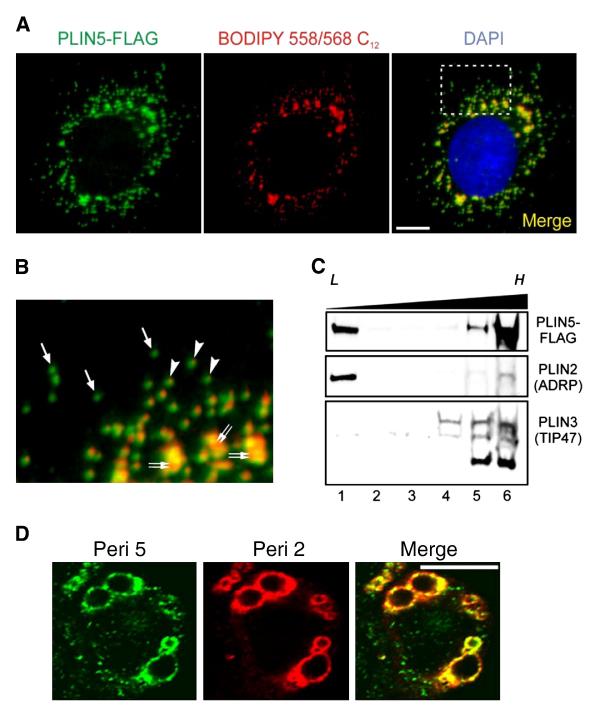
Epitope-tagged perilipin 5 was found to localize to the perimeter of lipid storage droplets (Fig. 1A and B, denoted by double arrows). In the absence of exogenous fatty acids, CHO-K1 cells also formed numerous small lipid droplets in the cytosol, as evidenced by coincident neutral lipid staining (arrowheads, Fig. 1B). However, a significant portion of the perilipin 5 staining did not colocalize with neutral lipid, but instead was found on punctate structures distributed throughout the cytosol (single arrows, Fig. 1B).
Fig. 1.
Perilipin 5 localizes to biochemically distinct intracellular pools. A. Confocal images of a representative CHO cell stably expressing the perilipin 5-3X-FLAG construct (green). The perilipin 5-3X-FLAG protein was detected using an anti-FLAG antibody (green). Neutral lipids were stained through incorporation of BODIPY 558/568 C12 (red), and nuclei were stained with DAPI (blue). Scale bar = 10 μm. B. Inset from merged image (right panel in A). Under basal growth conditions, perilipin 5-3X-FLAG localized to both LDs (double arrows) as well as smaller punctate structures in the cytosol. Of the latter, some were associated with neutral lipid staining (arrowheads), while others were apparently devoid of BODIPY 558/568 C12 labeling (arrows). C. Perilipin 5-3X-FLAG-expressing CHO cell lysates prepared following incubation in lipid-loading medium, were subjected to ultracentrifugation using conventional discontinuous sucrose density gradients (0–30%). Immunoblots were prepared using aliquots of equal volume fractions of the gradients, arranged from low (L) to high (H) density, and probed with anti-FLAG, anti-perilipin 2 (ADRP), and anti-perilipin 3 (TIP47) antibodies. Whereas the majority of perilipin 2 and perilipin 3 partitioned into either low or high-density fractions, respectively, perilipin 5-3X-FLAG was recovered in fractions at both extremes of the density gradient. D. Confocal images of a representative CHO cell stably expressing the perilipin 5-3X-FLAG construct (green) and stained with an anti-perilipin 2 antibody (red). Merged images are shown in yellow. Both perilipin 5 and perilipin 2 localize to lipid storage droplets but only perilipin 5 is found on punctate structures.
To confirm this observation, cells were disrupted and fractionated using ultracentrifugation in a 0–30% sucrose gradient. The protein composition of the gradient fractions was examined using immunoblotting, which revealed that the majority of perilipin 5 partitioned with dense fractions at the bottom of the gradient (migrating with natively expressed perilipin 3, lanes 5 and 6, Fig. 1C). In contrast, the majority of perilipin 2 immunoreactivity was found in the most buoyant fraction at the top of the gradient (lane 1, Fig. 1C). Some of the perilipin 5 partitioned into the buoyant fraction, as would be expected when in association with low-density LDs.
To determine if the non-buoyant fraction of perilipin 5 was associated with other organelles, immunofluorescence microscopy was employed. No colocalization was observed with antibodies directed against LAMP-1 (lysosomes), LAMP-2 (lysosomes), EAA 1 (early endosomes), complex Va protein (mitochondria), or golgin 97 (Golgi) (data not shown).
Immunofluoresence experiments were also conducted to test for the presence of perilipin 2 (Fig. 1D). Perilipin 5 was observed on the surface of structures presumed to be lipid storage droplets and colocalized with perilipin 2 on these structures. Perilipin 2 was not observed in the cytosol or on the punctate perilipin 5 structures.
3.2. Perilipin 5 forms discrete structures in the cytosol
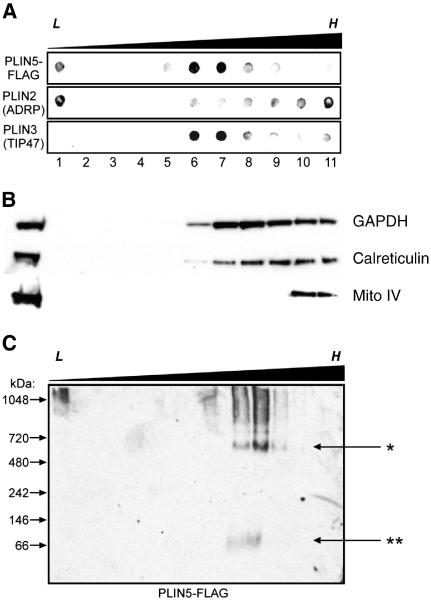
Due to the lack of colocalization with other structures and the punctate nature of the perilipin 5 staining, we hypothesized that perilipin 5 was organized into some type of unique macromolecular structure. To resolve perilipin 5 from other organelles, sucrose gradients were extended from 0–30% to 0–60% (Fig. 2A). If perilipin 5 existed as a free protein in the cytosol, it would be expected to partition at the bottom of these gradients, rather than co-migrating with marker proteins for other organelles. Dot blot analysis revealed that perilipin 5 partitioned into two different density ranges: perilipin 5 bound to low density LDs was found in the least buoyant fraction (< 1.0 g/ml), and perilipin 5 while a second pool of perilipin 5 partitioned in the 1.10 to 1.17 g/ml density range (lanes 6 and 7, Fig. 2A). These fractions were less dense than those in which markers for cytosol (GAPDH), endoplasmic reticulum (calreticulin) or mitochondria (mitochondrial complex IV) migrated (Fig. 2B).
Fig. 2.
A portion of perilipin 5 is associated with high-density structures of a discrete size. A. Perilipin 5-FLAG-expressing CHO cell lysates were subjected to ultracentrifugation using extended discontinuous sucrose density gradients (0–60%). Dot blots were prepared using aliquots of equal volume fractions of the gradients, arranged from low (L) to high (H) density, and probed with anti-FLAG, anti-perilipin 2 (ADRP), and anti-perilipin 3 (TIP47) antibodies. When the gradients were extended over a wider density range, perilipin 5 was found to partition into both low-density (buoyant) fractions (lane 1) as well as fractions of higher density (lanes 6–7), but was essentially absent in the fractions of highest density (lanes 9–11). B. Marker proteins for cytosol (NADPH), ER (calreticulin) and mitochondria (mito complex IV) are shown. C. Non-denaturing gradient gel electrophoresis was performed using sucrose gradient fractions prepared as described in A. Immunoblots were probed with anti-FLAG antibodies. Immunoreactive material that partitioned into the denser fractions of the gradient also migrated into the gel under non-denaturing conditions, revealing an approximate size of 575 kDa (marked by the asterisk, *). A small amount of immunoreactivity was found migrating near the predicted size of perilipin 5 (double asterisk, **). Please note that each panel contains representative data from different experiments.
Due to the discrete density range of the non-buoyant pool of perilipin 5 we hypothesized that this portion of perilipin 5 was bound either to membrane bilayer-invested vesicles, small lipid-filled structures, or both. Since perilipin 5 fails to colocalize with any of the common organelle markers we used, we hypothesized that at least a portion might associate with unique, small, dense, lipidic structures. To further characterize this pool of perilipin 5, NDGGE of sucrose gradient fractions coupled with immunoblotting was employed. The least dense fractions of the sucrose gradient contained particles too large to penetrate the gel (Fig. 2C, leftmost lane); however, perilipin 5 in the higher density range did migrate into the gel, with the majority found as a single discrete band of approximately 575 kDa, consistent with structures having a Stokes diameter of ~ 15 nm (Fig. 2C, asterisk).
3.3. Electron microscopy reveals distinct size of high-density perilipin 5-containing structures
In images obtained using fluorescent microscopy with high numerical aperture objectives, the size of discrete, perilipin 5-containing cytosolic structures was determined to be on the order of ~ 200 nm. Equivalent measurements were obtained using sub-resolution fluorescence microspheres, indicating that the size of these perilipin 5-labeled particles was possibly below the resolution limit of optical microscopy. To independently verify the size of the particles we performed TEM on immunoprecipitated material isolated from lysates of CHO cells stably expressing perilipin 5-3XFLAG. The immunoprecipitated particles were found to be of relatively uniform size with a roughly circular appearance, and exhibited an average diameter of 16.7 nm and a median size of 12.5 nm (Fig. 3A–C). This was in close agreement with the predicted Stokes diameter of ~ 15 nm determined using NDGGE. Quantitative analysis of these structures using manual pixel counting further revealed an apparent periodicity to the sizes of the particles, with minor peaks occurring at 35–40, 60–65, and 85–90 nm (Fig. 3D).
Fig. 3.
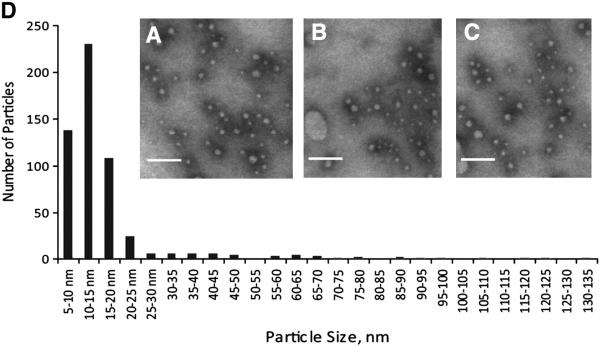
A majority of high-density perilipin 5-containing structures have a size of less than 20 nm. Transmission electron microscopy was performed on material immunoprecipitated from the post-nuclear supernatant isolated from perilipin 5-3X-FLAG-expressing CHO cells grown under basal conditions. A–C Representative images of negative stained immunoprecipitated fractions. Scale bars = 100 nm. D. Histogram showing the size distribution of 500 particles from randomly chosen, non-overlapping microscopic fields, as determined using morphometric analysis.
3.4. High-density perilipin 5-associated structures are devoid of perilipin 2
Co-immunoprecipitation was employed to determine which other proteins might also reside on the particles identified using EM (Fig. 4). Four different fractions from the sucrose gradient of perilipin 5-3x-FLAG expressing cells were analyzed: the least dense fraction (1) and the more dense fractions (6 and 7, combined) of both oleic acid treated and untreated cells. Each of these fractions was precipitated with an anti-FLAG resin and subjected to immunoblotting with the indicated antibodies. Perilipin 5-3x-FLAG was found in the denser fractions of both gradients, but in the least dense fraction only of oleic acid treated cells (Fig. 4A), validating the findings of immunoblots conducted on gradient fractions. Perilipin 2 was only found in association with perilipin 5 on large, buoyant lipid droplets and not on the dense structures, consistent with our immunofluorescent microscopy findings (Fig. 1D).
Fig. 4.
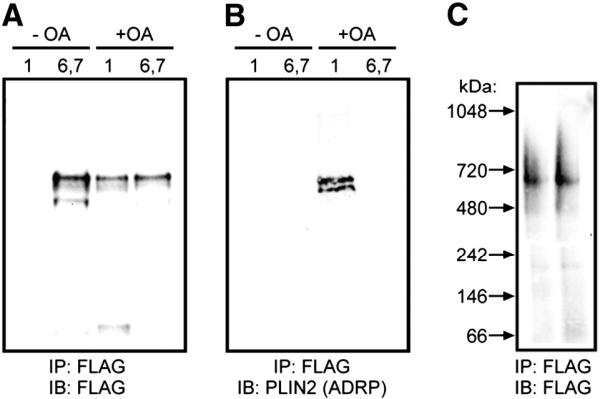
Co-immunoprecipitation of perilipin 5 coated particles confirms a lack of perilipin 2/ADRP in high-density structures. In A–C, cells were either treated with oleic acid (+ OA) or basal growth conditions (− OA). Sucrose gradient (0–60%) ultracentrifugation was performed and fractions 1 (low density) or 6 and 7 (high density) were immunoprecipitated with anti-FLAG beads. The eluted material was subjected to SDS-PAGE and either prepared for immunoblot analysis (A, B), or developed using silver staining, as noted. D. Immunoprecipitated particles from CHO cells were eluted with FLAG peptide and analyzed by non-denaturing gradient gel electrophoresis followed by immunoblotting and detection using anti-FLAG antibodies. No shift in the size of these ~ 575 kDa particles was observed following immunoprecipitation.
When native gradient gel electrophoresis was applied to material immunoprecipitated from dense fractions of PLIN5-expressing CHO cells, discrete bands of ~ 575 kDa were detected in the resulting immunoblots (Fig. 4C). These data suggest that the particles observed by TEM (above) had retained their integrity throughout the immunoprecipitation procedure.
3.5. Expression of perilipin 5 in CHO cells results in altered lipid storage and lipid droplet size
Other groups have found that ectopically expressed perilipin 5 resulted in increased lipid storage [30] and [31]. To confirm that ectopic expression of perilipin 5 in CHO was not resulting in anomalous lipid storage, we quantified TAG levels in these cells. Expression of the perilipin 5-3x-FLAG fusion protein resulted in a 70% increase in cellular TAG levels over control levels (Fig. 5). This increase was also observed when cells were treated with oleic acid to increase lipid accumulation.
Fig. 5.
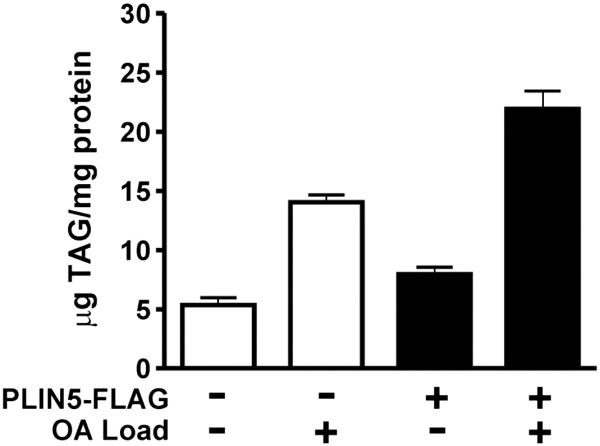
CHO fibroblasts expressing perilipin 5 store more TAG than control cells. CHO cells expressing the perilipin-5 3-X-FLAG fusion protein or transfected with a control plasmid were assayed for cellular TAG in the presence or absence of exogenous oleic acid (OA) treatment for 24 h (n = 6). TAG data were normalized to total protein concentration (mean ± SEM). Relative to control cells, perilipin 5 expressors stored 0.7-fold more TAG under basal and lipid-loaded conditions.
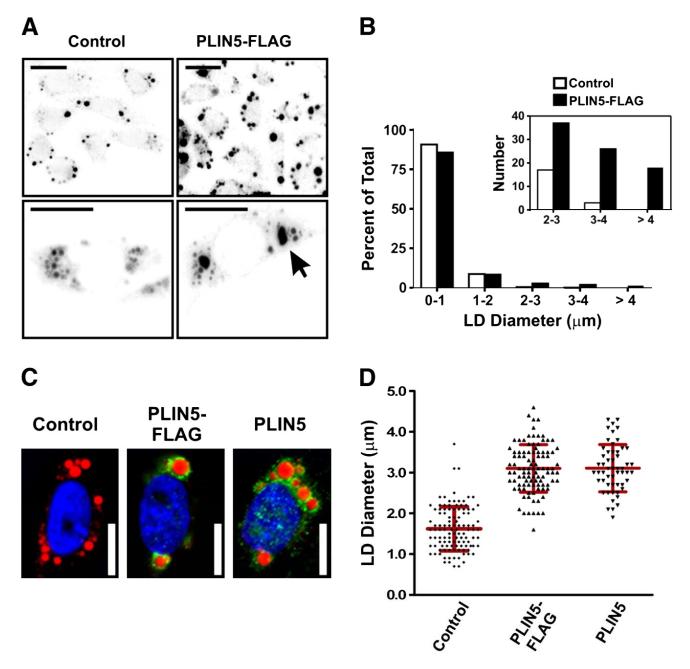
Expression of perilipin 5-3x-FLAG in CHO cells resulted in an increase in LD size (Fig. 6A and B). What was intriguing, however, was the apparent change in LD size distribution. While expression of perilipin 5 only increased neutral lipid stores in these cells by 1.7 fold, the diameter of the largest LD observed in each perilipin 5-expressing cells was increased by over 2.5 fold (Fig. 6B). This increase in the diameter of the largest droplets was associated with a shift in the size of the smaller droplets (Fig. 6B). To confirm that this effect was not an artifact of conjugation with the FLAG peptide, the experiment was repeated using CHO cells transfected with a native perilipin 5 construct. No difference was noted in either the diameter of the largest droplet in each cell or in the distribution of the droplet size (Fig. 6C and D). In this instance our data suggest that perilipin 5 participates in the maintenance of the organization and structure of intracellular LDs.
Fig. 6.
Expression of perilipin 5 mediates a remodeling of lipid droplets. A. Low magnification micrographs (top row) and images of representative cells (bottom row) of lipid-loaded control CHO cells (transfected with control plasmid), and CHO cells expressing perilipin 5-3X-FLAG. Lipid droplets were stained through incorporation of BODIPY 558/568 C12 and appear dark on a light background. Note the increased size of the lipid droplets in the cells expressing perilipin 5-3X-FLAG (arrow). Scale bars = 10 μm. B. The total number and size of LDs in CHO cells stably transfected with control plasmid or perilipin 5-3X-FLAG was determined using quantitative morphometric analysis. Following lipid-loading for 24 h, a significant shift in the percentage of larger LDs was observed with expression of perilipin 5 (inset). C and D. The experiments shown in panels A and B were repeated using a plasmid in which the FLAG epitope was removed (perilipin 5). C. Lipid-loaded CHO cells stably transfected with either control plasmid, or with perilipin 5-3X-FLAG or perilipin 5 expression plasmids, were fixed and immunolabeled with anti-perilipin 5 antibodies (green). Neutral lipids were stained through incorporation of BODIPY 558/568 C12 (red), and nuclei were stained with DAPI (blue). Scale bars = 10 μm. D. Scatterplots of the largest LD diameter per cell in the experiment described in C, as determined using morphometric analysis. There was no discernable effect of the FLAG epitope tag on the accumulation of large lipid droplets.
3.6. Characterization of perilipin 5 in mouse heart and liver validates what is observed in cultured cell models
To ensure that the observed results in CHO were not a result of overexpression or of the model system used, select experiments were repeated using heart and liver from fasted mice. These tissues and conditions were chosen based on previous reports indicating perilipin 5 expression would be elevated in these organs and metabolic states.
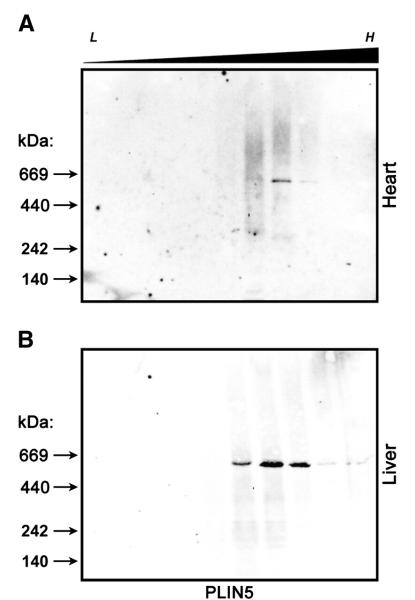
Lysates of heart and liver tissue were subjected to sucrose gradient ultracentrifugation and NDGGE (Fig. 7A and B). Native perilipin 5 in either heart (A) or liver (B) was found to migrate in the sucrose gradient in a similar density range (d = 1.10–1.17 g/cm3) to what was found using CHO cells expressing FLAG tagged perilipin 5. When sucrose gradient fractions were subjected to NDGGE and immunoblot analysis with an anti-perilipin 5 antibody, perilipin 5 was found to migrate in a relatively sharp band with a molecular weight of ~ 575 kDa and diameter of approximately 15 nm, again, indistinguishable from what was observed using the CHO model system. Differences in size or density were not observed between heart and liver samples.
Fig. 7.
Native murine perilipin 5 also forms high-density structures of a discrete size. Heart (A) and liver (B) samples from fasted 8–10 male C57/Bl6 mice were subjected to ultracentrifugation using extended discontinuous sucrose density gradients (0–60%) and examined using non-denaturing gradient gel electrophoresis as described in Fig. 2. Immunoblots were probed with anti-perilipin 5 antibodies. The approximate density at which staining was observed (d = 1.10–1.17 g/ml) and size of the bands (~ 575 kDa) was indistinguishable from those observed in the CHO model system. n = 5 animals, representative blots shown.
Collectively, these data indicate that our observations and predictions made using the transfected CHO model are an accurate reflection of the biology occurring in mouse heart and liver.
3.7. The high density pool of perilipin 5 interacts with perilipin 3 and contains a lipid core
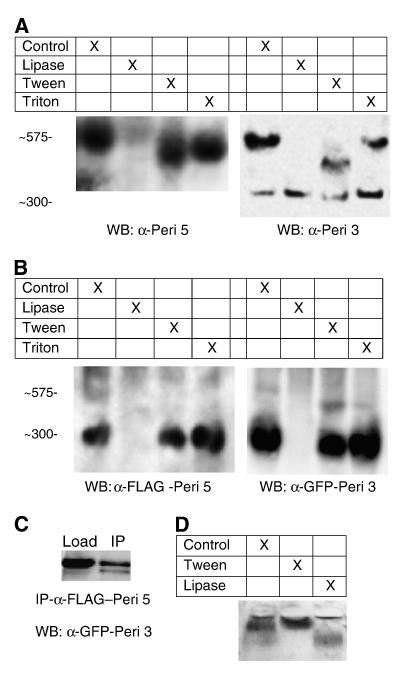
To determine the content of the core of the perilipin-5 particles, we characterized particles isolated from sucrose gradients of liver samples described in the previous experiment. These particles were subjected to one of four treatments: 100 units of C. rugosa lipase, 1% Tween-20, 1% Triton-X-100, or a control incubation. Following treatment, the samples were analyzed using NDGGE and immunoblotting as previously described.
Treatment of the particles with lipase ablated the band migrating at 575 kDa. Similarly, treatment with Tween-20 had a mild effect, decreasing the intensity of the 575 kDa band and causing a shift of staining to smaller sized particles. Triton, a milder detergent than Tween, had at best a weak effect on the band. Based on these observations we conclude that the particle has a core of neutral lipid. In the presence of lipase the core is degraded and the particle loses its integrity. Low concentrations of Tween 20 begin to affect the particle the same way as the lipase, but Triton-X-100 is unable to solubilize the particle under these conditions.
To confirm that the lipase treatment would degrade a particle with a triacylglycerol core, plasma samples were treated with detergent or lipase as described and subjected to agarose electrophoresis and oil red o staining. In untreated samples bands were evident for VLDL/chylomicra, LDL, and HDL. In samples treated with Tween-20 the particles collapsed into a single band which entered the gel with difficulty. In lipase treated samples the triacylglycerol rich bands (VLDL and chylomicra) were ablated but the cholesteryl ester rich bands (LDL and HDL) remained further confirming that lipase treatment was able to degrade triacylglycerol rich particles.
To determine if perilipin 3 resides on the same complex, blots were probed with an anti-perilipin-3 antibody. Bands were observed at 575 kDa, indicating that if the two perilipin proteins were not cohabitating the same droplet, they were at the least on droplets of similar size. In addition to the 575 kDa band we also observed a new band at 300 kDa. This band did not seem to be effected as strongly by the lipase or Tween treatments.
To further probe the interactions of perilipins 5 and 3, CHO cells were cotransfected with cDNA constructs coding for the perilipin 5-3xFLAG fusion protein and a perilipin-3-EGFP fusion protein. Cells were cultured for 16–20 h post transfection and post nuclear supernatants were prepared and treated as previously described. To confirm that the banding pattern seen was the same for both proteins, blots were first probed with an anti-GFP antibody, then stripped and reprobed with an anti-FLAG antibody (Fig. 8B). As was seen in liver samples, a lipase sensitive band was seen that was detectable with both anti-FLAG antibodies (indicating the presence of the perilipin-5-3xFLAG fusion protein) as well as anti-GFP antibodies (indicating the presence of the perilipin-3-EGFP fusion protein). However, it should be noted that this band was observed at 300 kDa and not at the larger 575 kDa band seen in our other studies. A minor band of 575 kDa was observed in several samples.
Fig. 8.
The small, dense perilipin 5 structures are also coated with perilipin 3 and have a lipid core. A. Mouse liver samples from fraction 7 of gradients shown in Fig. 7 were treated with 100 units of Candida rugosa lipase, 1% Tween-20, or 1% Triton-X-100 for 1 h and subjected to non-denaturing gradient gel electrophoresis and immunoblotting as previously described. The panel on the left was probed with an anti-perilipin 5 antibody while the panel on the right was probed with an anti-perilipin 3 antibody. Both sets of samples had banding apparent at 575 kDa. An additional band was seen migrating at approximately 300 kDa in blots probed with anti-perilipin 3 antibody (but not perilipin 5). Treatment with lipase ablated the perilipin 5 coated particles seen migrating at 575 kDa. Treatment with Tween-20 caused a shift in the 575 kDa band, however no change was seen in the Triton treated samples (n = 4, representative blots shown). B. CHO cells were transiently transfected with cDNA fusion constructs coding for both perilipin-5-3xFLAG and perilipin-3-EGFP. Post nuclear supernatants were prepared and experiments were performed as previously described. Blots were first detected using anti-GFP antibody, then stripped and reprobed with anti-FLAG antibody. Under these conditions the predominant band was observed at 300 kDa with a minor band seen at 575. Both proteins migrated to the same region of the gel. Treatment with lipase eliminated these bands (n = 4, representative blots shown). C. To determine if perilipin 3 was found on the same particle as perilipin 5, post nuclear supernatants were immunoprecipitated with an anti-FLAG antibody as previously described. The perilipin 3-GFP fusion protein was detected in the precipitate by probing with either anti-perilipin-3 or anti-GFP (data not shown). D. To confirm that lipase treatment would degrade the core of triacylglycerol rich particles, plasma samples were analyzed following treatment with tween-20 or lipase and analyzed using agarose gel electrophoresis as described in Methods. Treatment of samples with lipase degraded triacylglycerol rich lipoproteins but not cholesteryl ester rich lipoproteins.
To confirm that perilipins 3 and 5 were indeed residing on the same particle, samples of the transient transfections were immunoprecipitated using an anti-FLAG resin (Fig. 8C). The perilipin 3-GFP fusion protein was detected in the precipitate by probing with either anti-perilipin-3 or anti-GFP antibodies (data not shown) indicating that in these samples, some fraction of perilipin 3 was found on the same particle as perilipin 5.
4. Discussion
Perilipin 5 is the most recently identified member of the PAT protein family. Several studies on this protein have shown that it resides in multiple intracellular locations: ER, LDs, and an uncharacterized cytosolic pool [29], [30], [31] and [33]. This work characterizes the cytosolic pool and offers that rather than existing as free protein in the cytosol, perilipin 5 is associated with discrete, high density lipid droplet (HDLD) particles with a lower size limit of approximately 15 nm in diameter. Furthermore, based on the size distribution of LDs, we speculate that this protein is involved in the trafficking of lipid from distal regions of the cell to central LDs.
Of the PAT proteins, perilipin 1A is the best characterized, but it is also one of the most divergent in amino acid sequence and likely has the most specified function. While perilipin 1A is not found in the cytosol under basal conditions, prolonged stimulation by PKA can cause LDs to fragment into thousands of “micro LDs”. It is unclear if they have a lower size limit, and if so, what other proteins may reside on those structures. It has been speculated that this increase in surface area by fragmentation grants lipases greater access to the TAG contents of the LD under conditions of prolonged lipolysis [46].
Immunofluorescence microscopy on the punctate HDLDs failed in many instances to costain for neutral lipid when stained with BODIPY C12. This may be due to the small amount of lipid we propose to be contained in these particles or the dye may not be sensitive enough to stain these structures.
Others have proposed that the e-PATs, specifically perilipins 3 and 4, act to coat developing lipid droplets when the cell is presented with a fatty acid challenge [26], [27], [28] and [47]. Our data support this conclusion, and also suggest that perilipin 5 may perform an analogous, although opposite function to the dispersion phenomenon observed with perilipin 1A. We speculate that, without excess fatty acids for the cell to store, perilipin 5 resides in the cytosol until the cell encounters a fatty acid challenge. When the cell encounters a fatty acid challenge, the excess fatty acids that enter the cell, are packaged into small (10–20 nm diameter) TAG particles coated by perilipins 3 and 5, and then trafficked to the larger LD. There, droplets are coated by perilipins 2, the terminal c-PAT protein commonly found in muscle and liver cells, and perilipin 5.
While perilipin 3 has clearly been demonstrated to be a lipid droplet protein, other functions of perilipin 3 are a subject of controversy in the literature. Previous studies have shown perilipin 3 to be a mannose-6-phosphate receptor trafficking protein, interacting with Rab-9 and Rho BTB3, while other studies have cast doubt on some of these ascribed functions and interaction partners [22], [23], [24], [47], [48] and [49]. Many of these same studies have also shown that perilipin 3 is found in the cytosol as well as other locations in the cell (plasma membrane or Golgi as well as LDs). A recent study has colocalized perilipin 3 with tubular ER [33]. This same study found that loading cells with a soluble diacylglycerol drives all three e-PATs (perilipins 3, 4, and 5) to the ER from the cytosol. Sequence analysis reveals that the closest relative to perilipin 5 is perilipin 3, and our findings fit quite well with what has been observed regarding e-PATs by Skinner et al. The nature of the cytosolic pool of any of the e-PATs has not been explored. We propose that under normal conditions, at least a portion of perilipin 5 resides on small dense particles within the cytosol. When cells are treated with oleic acid to promote lipid deposition, perilipin 5 shifts from this dense pool to the lipid storage droplet in our system, while in that of Skinner et al. it shifts from the cytosol to ER (presumably before transitioning to a mature perilipin 2 coated LD). While we have not observed co-localization of perilipin 5 with ER, these differences may be explained by the differences in model system or experimental treatment. It also may be that differences in these studies indicate that the cell responds differently to different lipid challenges (oleic acid vs. soluble diacylglycerols). Based on our data, it is quite plausible that perilipin 5 serves as an intermediate between perilipins 3 and 2, binding to small, nascent pools of lipid, interacting with trafficking proteins, and effecting movement of lipids to mature LDs.
Our data indicate that perilipin 3 also resides on a discrete HDLD structure with defined lipid and protein components. Perilipin 3 and perilipin 5 (but not perilipin 2) were found to migrate at a similar density in sucrose gradient ultracentrifugation and have a similar banding pattern in NDGGE. Likewise, using an anti-FLAG resin we were able to immunoprecipitate EGFP labeled perilipin 3 in a cotransfected CHO model system.
We currently have no good explanation as to why HDLD particles in cells expressing ectopic perilipin 3-EGFP are smaller in size. This may be due to expression levels or a previously unknown complication of the EGFP tag. However, we speculate that this may provide a tool to further characterize perilipin protein function in future studies. It may be that when perilipin 3 is overexpressed it sequesters perilipin 5 in 300 kDa particles preventing its maturation into larger particles (i.e. 575 kDa particles).
We did not observe significant colocalization of perilipin 3 and perilipin 5 using immunofluorescence; however, that may be due to sensitivity issues related to the small sizes of the particles in question. The other data presented clearly indicate that perilipins 3 and 5 spend some time either directly interacting or bound to the same particle.
It should be noted that the cotransfection studies shown in Fig. 8B and C were conducted with a 3xFLAG tagged perilipin 5 protein and an EGFP tagged perilipin 3 protein. Given the nature of these epitope tags it is possible that the tag is influencing the localization or partitioning of the proteins, however, the comigration of perilipin 5 and perilipin 3 in sucrose density gradients and gradient gels are suggestive that these proteins occupy similar, if not the same pool. Immunofluorescence microscopy of doubly labeled cells failed to show significant colocalization of perilipin 3-EGFP and perilipin 5-3xFLAG, however, the epitope tagged perilipin 3 exhibited reticular staining in those experiments. Using liver lysates we were unable to cleanly coimmunoprecipitate perilipin 5 with an anti perilipin 3 antibody or perilipin 3 with an anti perilipin 5 antibody.
Recent reports by Bulankina et al. propose a hypothetical arrangement of perilipin 3 on a discoidal structure analogous to nascent HDL-3 [47] in which the amphipathic helical regions of perilipin 3 are partially wrapped in a disk of phospholipids. Perilipins 3 and 5 are conserved through this region in the amino terminus. Based on our electron microscopy data, we propose a similar interaction of these amphipathic helical regions of perilipin 5 with a spherical particle.
4.1. Changes in droplet size reflect a role for perilipin 5
We propose that perilipin 5 functions as part of the cellular machinery that transports lipid from distal regions of the cell to mature LDs. In support of this hypothesis, we have observed that an overall change in the distribution of LDs accompanies ectopic perilipin 5 expression, with larger and fewer LDs in lipid-loaded CHO cells expressing perilipin 5 than controls. While the argument can be made that this is an effect of ectopic expression of a PAT protein, ectopic expression of perilipin 1A and a series of mutant perilipins all target to LDs and impact lipid storage without affecting LD size (total mass of lipid is increased without a change in the diameter of LDs compared to control cells), indicating an increase in the total number of droplets [34]. Likewise, we have not observed these effects when perilipin 2 is overexpressed in either CHO or C2C12 cells (B. Colopy and JT Tansey, unpublished data).
4.2. A proposed structure for high-density perilipin5-containing particles
Our observations indicate that perilipin 5 does not exist as a free protein in the cytosol. Based on the diameter and the density of the observed particles, we propose that at least a portion of the cytosolic fraction of perilipin 5 resides on a lipoprotein-like structure in which a neutral lipid core is surrounded by a coating of phospholipids and protein.
We have not excluded the possibility that these small particles have an aqueous core surrounded by a phospholipid bilayer (effectively a small unilamellar vesicle); however, we find this hypothesis lacking in several regards. First, electron microscopy failed to show a phospholipid bilayer on immunoprecipitated particles. Secondly, while density calculations cannot rule out the possibility of an aqueous core, the mass percent protein in these particles would be less than 25%, a low value compared to other biological structures of this size. Finally, the radius of curvature for a particle with a diameter of 15 nm creates unfavorable strains and molecular packing for a bilayer system [40]. Recent studies suggest that this may be independent of the type of lipid forming the bilayer [41]. While synthetic small unilamellar vesicles can be produced in the laboratory with diameters as small as 25 nm (significantly larger than our particles), the synthesis of these particles requires extrusion, organic solvents, strong detergents, non-naturally occurring lipids or high energy sonication [42] and [43]. The nearest sized biological structure referenced in the literature are small caveolae (20–50 nm at the low end of their size range) [44]. Mathematical modeling of organelle formation also indicates the expected minimum size of bilayer structure is larger than our 15 nm particles [45]. The perilipin 5 containing particles we have identified have a similar size and density to high-density lipoprotein particles and as such, we hypothesize that they have a similar structure.
4.3. Lipoproteins: an existing paradigm
Several groups have suggested that the PAT proteins may play analogous roles to apolipoproteins found on plasma lipoproteins [7], [20], [25], [26], [27] and [28]. Plasma apolipoproteins act as ligands for receptors, cofactors for lipases, and amphipathic coatings for the hydrophobic core of the lipoprotein. Through these diverse roles apolipoproteins regulate the trafficking and metabolism of the neutral lipid contents of the lipoprotein. PAT proteins exhibit many analogous properties. Perilipin 1A acts as a regulator of both HSL and ATGL [17], [34], [39], [50], [51] and [52]. Both perilipins 1A and 5 interact with CGI-58 [15], [16], [32] and [53]. Perilipin 2 and 3 have been shown to interact with cellular machinery involved in trafficking and perilipin 2 sequesters proteins involved in vesicular or LD fusion [3] and [35]. All PAT proteins coat LDs and several have been shown to play important regulatory roles in the metabolism of the neutral lipid contents.
Several significant differences separate these two analogous systems. Perilipin 1A appears to be regulated via phosphorylation of PKA sites, but it is the only member of the family recognized to be regulated by this mechanism. In addition, PATs are intracellularly localized while lipoproteins are extracellularly localized. Finally, LDs can reach sizes that are orders of magnitude larger than the largest lipoprotein. We sought to explore the possibility that PATs may be acting as a sort of intracellular lipoprotein by characterizing their structure, composition, and functions. If structures analogous to plasma lipoproteins were found in cells, it could provide an analogous paradigm for the study of lipid traffic and storage in the organism. Here, we describe the cytosolic compartment of perilipin 5 and propose that it is similar in both size and density to plasma lipoproteins. We do not propose that these small structures are the only perilipin 5 containing structures in the cytosol, but that they are clearly one of the most prevalent in the absence of exogenous lipid. Likewise, our data reveal that perilipin 5 is not found free in the cytosol, but rather is always found bound to some lipidic structure. Further identification of the components of this complex will be necessary to decipher the function and mechanism of perilipin 5 in lipid metabolism.
5. Conclusions
Questions have been raised as to why a LD protein has any residence time in the cytosol. These studies now reveal that, at least in the instance of perilipins 3 and 5, a portion of the cytosolic pool containing these proteins is bound to a lipidic structure. Perilipin 2 has been shown to sequester proteins involved in vesicular fusion, and we speculate that the cytosolic pool of perilipin 5 may act in a similar fashion. We propose a model wherein, in the absence of excess fatty acids, perilipin 5 resides in cytosolic HDLDs awaiting a fatty acid challenge. When lipid levels rise, cytosolic perilipin 5 coats these nascent LDs until they can be stabilized by a c-PAT (perilipin 1 or 2).
Highlights.
▶ Perilipin 5 is found in two distinct pools in the cell.
▶ The more dense pool of perilipin 5 also contains perilipin 3 but not perilipin 2.
▶ The dense pool has a lipid rich core and discrete size and density.
▶ Expression of perilipin 5 promotes droplet reorganization.
Acknowledgements
The authors would like to thank Drs. Perry Bickel for anti-perilipin 5 and 3 and Dean Londos for anti-perilipin 2 and 3 antibodies. This work was supported by an Otterbein University Faculty Development Committee award to JTT and an NIH faculty development award K08 HD049628 to WEA. JTT would like to dedicate this work to the memory of his friend and mentor Dean Londos.
Abbreviations
- CHO
Chinese Hamster Ovary fibroblasts
- LD
lipid droplet
- TAG
triacylglycerol
- HDLD
high density lipid droplet
- HSL
hormone sensitive lipase
- ATGL
adipocyte triacylglycerol lipase
- SNAP
synaptosomal associated protein
- SNARE
soluble NSF attachment protein receptors
- Glut4
glucose transporter 4
- T2DM
type 2 diabetes mellitus
- PPAR-α
peroxisome-proliferation activated receptor alpha
References
- [1].Cowie CC, Engelgau MM, Rust KF, Saydah SH, Byrd-Holt DD, Williams DE, Eberhardt MS, Geiss LS, Flegal KM, Gregg EW. Prevalence of diabetes and impaired fasting glucose in adults in the US population—National Health and Nutrition Examination Survey 1999–2002. Diabetes Care. 2006;29:1263–1268. doi: 10.2337/dc06-0062. [DOI] [PubMed] [Google Scholar]
- [2].Petersen KF, Shulman GI. Etiology of insulin resistance. Am. J. Med. 2006;119:10S–16S. doi: 10.1016/j.amjmed.2006.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Bostrom P, Andersson L, Rutberg M, Perman J, Lidberg U, Johansson BR, Fernandez-Rodriguez J, Ericson J, Nilsson T, Boren J, Olofsson SO. SNARE proteins mediate fusion between cytosolic lipid droplets and are implicated in insulin sensitivity. Nat. Cell Biol. 2007;9:1286–1293. doi: 10.1038/ncb1648. [DOI] [PubMed] [Google Scholar]
- [4].Bostrom P, Andersson L, Li L, Perkins R, Hojlund K, Boren J, Olofsson SO. The assembly of lipid droplets and its relation to cellular insulin sensitivity. Biochem. Soc. Trans. 2009;37:981–985. doi: 10.1042/BST0370981. [DOI] [PubMed] [Google Scholar]
- [5].Bell M, Wang H, Chen H, McLenithan JC, Gong DW, Yang RZ, Yu DZ, Fried SK, Quon MJ, Londos C, Sztalryd C. Consequences of lipid droplet coat protein downregulation in liver cells: abnormal lipid droplet metabolism and induction of insulin resistance. Diabetes. 2008;57:2037–2045. doi: 10.2337/db07-1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kimmel AR, Brasaemle DL, McAndrews-Hill M, Sztalryd C, Londos C. Adoption of PERILIPIN as a unifying nomenclature for the mammalian PAT-family of intracellular lipid storage droplet proteins. J. Lipid Res. 2010;51:468–471. doi: 10.1194/jlr.R000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Wolins NE, Brasaemle DL, Bickel PE. A proposed model of fat packaging by exchangeable lipid droplet proteins. FEBS Lett. 2006;580:5484–5491. doi: 10.1016/j.febslet.2006.08.040. [DOI] [PubMed] [Google Scholar]
- [8].Tansey JT, Sztalryd C, Hlavin EM, Kimmel AR, Londos C. The central role of perilipin A in lipid metabolism and adipocyte lipolysis. IUBMB Life. 2004;56:379–385. doi: 10.1080/15216540400009968. [DOI] [PubMed] [Google Scholar]
- [9].Londos C, Sztalryd C, Tansey JT, Kimmel AR. Role of PAT proteins in lipid metabolism. Biochimie. 2005;87:45–49. doi: 10.1016/j.biochi.2004.12.010. [DOI] [PubMed] [Google Scholar]
- [10].Granneman JG, Moore HPH. Location, location: protein trafficking and lipolysis in adipocytes. Trends Endocrinol. Metab. 2008;19:3–9. doi: 10.1016/j.tem.2007.10.006. [DOI] [PubMed] [Google Scholar]
- [11].Brasaemle DL. The perilipin family of structural lipid droplet proteins: stabilization of lipid droplets and control of lipolysis. J. Lipid Res. 2007;48:2547–2559. doi: 10.1194/jlr.R700014-JLR200. [DOI] [PubMed] [Google Scholar]
- [12].Ducharme NA, Bickel PE. Minireview: lipid droplets in lipogenesis and lipolysis. Endocrinology. 2008;149:942–949. doi: 10.1210/en.2007-1713. [DOI] [PubMed] [Google Scholar]
- [13].Bickel PE, Tansey JT, Welte MA. PAT proteins, an ancient family of lipid droplet proteins that regulate cellular lipid stores. Biochim. Biophys. Acta-Mol. Cell Biol. Lipids. 2009;1791:419–440. doi: 10.1016/j.bbalip.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Goodman JM. Demonstrated and inferred metabolism associated with cytosolic lipid droplets. J. Lipid Res. 2009;50:2148–2156. doi: 10.1194/jlr.R001446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Yamaguchi T, Omatsu N, Matsushita S, Osumi T. CGI-58 interacts with perilipin and is localized to lipid droplets—possible involvement of CGI-58 mislocalization in Chanarin–Dorfman syndrome. J. Biol. Chem. 2004;279:30490–30497. doi: 10.1074/jbc.M403920200. [DOI] [PubMed] [Google Scholar]
- [16].Subramanian V, Rothenberg A, Gomez C, Cohen AW, Garcia A, Bhattacharyya S, Shapiro L, Dolios G, Wang R, Lisanti MP, Brasaemle DL. Perilipin A mediates the reversible binding of CGI-58 to lipid droplets in 3T3-L1 adipocytes. J. Biol. Chem. 2004;279:42062–42071. doi: 10.1074/jbc.M407462200. [DOI] [PubMed] [Google Scholar]
- [17].Granneman JG, Moore HPH, Granneman RL, Greenberg AS, Obin MS, Zhu ZX. Analysis of lipolytic protein trafficking and interactions in adipocytes. J. Biol. Chem. 2007;282:5726–5735. doi: 10.1074/jbc.M610580200. [DOI] [PubMed] [Google Scholar]
- [18].Lass A, Zimmermann R, Haemmerle G, Riederer M, Schoiswohl G, Schweiger M, Kienesberger P, Strauss JG, Gorkiewicz G, Zechner R. Adipose triglyceridelipase-mediated lipolysis of cellular fat stores is activated by CGI-58 and defective in Chanarin–Dorfman syndrome. Cell Metab. 2006;3:309–319. doi: 10.1016/j.cmet.2006.03.005. [DOI] [PubMed] [Google Scholar]
- [19].Ghosh AK, Ramakrishnan G, Chandramohan C, Rajasekharan R. CGI-58, the causative gene for Chanarin–Dorfman syndrome, mediates acylation of lysophosphatidic acid. J. Biol. Chem. 2008;283:24507–24515. doi: 10.1074/jbc.M801783200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wang H, Hu L, Dalen K, Dorward H, Marcinkiewicz A, Russell D, Gong D, Londos C, Yamaguchi T, Holm C, Rizzo MA, Brasaemle D, Sztalryd C. Activation of hormone-sensitive lipase requires two steps, protein phosphorylation and binding to the PAT-1 domain of lipid droplet coat proteins. J. Biol. Chem. 2009;284:32116–32125. doi: 10.1074/jbc.M109.006726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Montero-Moran G, Caviglia JM, McMahon D, Rothenberg A, Subramanian V, Xu Z, Lara-Gonzalez S, Storch J, Carman GM, Brasaemle DL. CGI-58/ABHD5 is a coenzyme A-dependent lysophosphatidic acid acyltransferase. J. Lipid Res. 2010;51:709–719. doi: 10.1194/jlr.M001917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Diaz E, Pfeffer SR. TIP47: a cargo selection device for mannose 6-phosphate receptor trafficking. Cell. 1998;93:433–443. doi: 10.1016/s0092-8674(00)81171-x. [DOI] [PubMed] [Google Scholar]
- [23].Orsel JG, Sincock PM, Krise JP, Pfeffer SR. Recognition of the 300-kDa mannose 6-phosphate receptor cytoplasmic domain by 47-kDa tail-interacting protein. Proc. Natl. Acad. Sci. U. S. A. 2000;97:9047–9051. doi: 10.1073/pnas.160251397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Krise JP, Sincock PM, Orsel JG, Pfeffer SR. Quantitative analysis of TIP47-receptor cytoplasmic domain interactions—implications for endosome-to-trans Golgi network trafficking. J. Biol. Chem. 2000;275:25188–25193. doi: 10.1074/jbc.M001138200. [DOI] [PubMed] [Google Scholar]
- [25].Brasaemle DL, Barber T, Wolins NE, Serrero G, BlanchetteMackie EJ, Londos C. Adipose differentiation-related protein is an ubiquitously expressed lipid storage droplet-associated protein. J. Lipid Res. 1997;38:2249–2263. [PubMed] [Google Scholar]
- [26].Wolins NE, Quaynor BK, Skinner JR, Schoenfish MJ, Tzekov A, Bickel P. S3-12, adipophilin, and TIP47 package lipid in adipocytes. J. Biol. Chem. 2005;280:19146–19155. doi: 10.1074/jbc.M500978200. [DOI] [PubMed] [Google Scholar]
- [27].Wolins NE, Skinner JR, Schoenfish MJ, Tzekov A, Bensch KG, Bickel PE. Adipocyte protein S3-12 coats nascent lipid droplets. J. Biol. Chem. 2003;278:37713–37721. doi: 10.1074/jbc.M304025200. [DOI] [PubMed] [Google Scholar]
- [28].Wolins NE, Rubin D, Brasaemle DL. TIP47 associates with lipid droplets. J. Biol. Chem. 2001;276:5101–5108. doi: 10.1074/jbc.M006775200. [DOI] [PubMed] [Google Scholar]
- [29].Yamaguchi T, Matsushita S, Motojima K, Hirose F, Osumi T. MLDP, a novel PAT family protein localized to lipid droplets and enriched in the heart, is regulated by peroxisome proliferator-activated receptor alpha. J. Biol. Chem. 2006;281:14232–14240. doi: 10.1074/jbc.M601682200. [DOI] [PubMed] [Google Scholar]
- [30].Wolins NE, Quaynor BK, Skinner JR, Tzekov A, Croce MA, Gropler MC, Varma V, Yao-Borengasser A, Rasouli N, Kern PA, Finck BN, Bickel PE. OXPAT/PAT-1 is a PPAR-induced lipid droplet protein that promotes fatty acid utilization. Diabetes. 2006;55:3418–3428. doi: 10.2337/db06-0399. [DOI] [PubMed] [Google Scholar]
- [31].Dalen KT, Dahl T, Holter E, Arntsen B, Londos C, Sztalryd C, Nebb HI. LSDP5 is a PAT protein specifically expressed in fatty acid oxidizing tissues. Biochim. Biophys. Acta-Mol. Cell Biol. Lipids. 2007;1771:210–227. doi: 10.1016/j.bbalip.2006.11.011. [DOI] [PubMed] [Google Scholar]
- [32].Granneman JG, Moore HPH, Mottillo EP, Zhu ZX. Functional interactions between Mldp (LSDP5) and Abhd5 in the control of intracellular lipid accumulation. J. Biol. Chem. 2009;284:3049–3057. doi: 10.1074/jbc.M808251200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Skinner JR, Shew TM, Schwartz DM, Tzekov A, Lepus CM, Abumrad NA, Wolins NE. Diacylglycerol enrichment of endoplasmic reticulum or lipid droplets recruits perilipin 3/TIP47 during lipid storage and mobilization. J. Biol. Chem. 2009;284:30941–30948. doi: 10.1074/jbc.M109.013995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Tansey JT, Huml AM, Vogt R, Davis KE, Jones JM, Fraser KA, Brasaemle DL, Kimmel AR, Londos C. Functional studies on native and mutated forms of perilipins—a role in protein kinase A-mediated lipolysis of triacylglycerols in Chinese hamster ovary cells. J. Biol. Chem. 2003;278:8401–8406. doi: 10.1074/jbc.M211005200. [DOI] [PubMed] [Google Scholar]
- [35].Andersson L, Bostrom P, Ericson J, Rutberg M, Magnusson B, Marchesan D, Ruiz M, Asp L, Huang P, Frohman MA, Boren J, Olofsson SO. PLD1 and ERK2 regulate cytosolic lipid droplet formation. J. Cell Sci. 2006;119:2246–2257. doi: 10.1242/jcs.02941. [DOI] [PubMed] [Google Scholar]
- [36].Yamaguchi T, Omatsu N, Omukae A, Osumi T. Analysis of interaction partners for perilipin and ADRP on lipid droplets. Mol. Cell. Biochem. 2006;284:167–173. doi: 10.1007/s11010-005-9045-y. [DOI] [PubMed] [Google Scholar]
- [37].Weller PF, Monahanearley RA, Dvorak HF, Dvorak AM. Cytoplasmic lipid bodies of human eosinophils—subcellular isolation and analysis of arachidonate incorporation. Am. J. Pathol. 1991;138:141–148. [PMC free article] [PubMed] [Google Scholar]
- [38].Tansey JT, Thuren TY, Jerome WG, Hantgan RR, Grant K, Waite M. Hydrolysis of phosphatidylcholine by hepatic lipase in discoidal and spheroidal recombinant high-density lipoprotein. Biochemistry. 1997;36:12227–12234. doi: 10.1021/bi970356w. [DOI] [PubMed] [Google Scholar]
- [39].Wang H, Hu LP, Dalen K, Dorward H, Marcinkiewicz A, Russell D, Gong DW, Londos C, Yamaguchi T, Holm C, Rizzo MA, Brasaemle D, Sztalryd C. Activation of hormone-sensitive lipase requires two steps, protein phosphorylation and binding to the PAT-1 domain of lipid droplet coat proteins. J. Biol. Chem. 2009;284:32116–32125. doi: 10.1074/jbc.M109.006726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Claessens M, Leermakers FAM, Hoekstra FA, Stuart MAC. Entropic stabilization and equilibrium size of lipid vesicles. Langmuir. 2007;23:6315–6320. doi: 10.1021/la0637622. [DOI] [PubMed] [Google Scholar]
- [41].Kamal MM, Mills D, Grzybek M, Howard J. Measurement of the membrane curvature preference of phospholipids reveals only weak coupling between lipid shape and leaflet curvature. Proc. Natl. Acad. Sci. U. S. A. 2009;106:22245–22250. doi: 10.1073/pnas.0907354106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Nieh MP, Kucerka N, Katsaras J. Methods in Enzymology Liposomes, Pt G. Elsevier Academic Press Inc.; San Diego: 2009. Spontaneously formed unilamellar vesicles; pp. 3–20. [DOI] [PubMed] [Google Scholar]
- [43].Genc R, Ortiz M, O’Sullivan CK. Curvature-tuned preparation of nanoliposomes. Langmuir. 2009;25:12604–12613. doi: 10.1021/la901789h. [DOI] [PubMed] [Google Scholar]
- [44].Thorn H, Stenkula KG, Karlsson M, Ortegren U, Nystrom FH, Gustavsson J, Stralfors P. Cell surface orifices of caveolae and localization of caveolin to the necks of caveolae in adipocytes. Mol. Biol. Cell. 2003;14:3967–3976. doi: 10.1091/mbc.E03-01-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Binder B, Goede A, Berndt N, Holzhutter H-G. A conceptual mathematical model of the dynamic self-organisation of distinct cellular organelles. PLoS One. 2009;4:e8295. doi: 10.1371/journal.pone.0008295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Marcinkiewicz A, Gauthier D, Garcia A, Brasaemle DL. The phosphorylation of serine 492 of perilipin A directs lipid droplet fragmentation and dispersion. J. Biol. Chem. 2006;281:11901–11909. doi: 10.1074/jbc.M600171200. [DOI] [PubMed] [Google Scholar]
- [47].Bulankina AV, Deggerich A, Wenzel D, Mutenda K, Wittmann JG, Rudolph MG, Burger KNJ, Honing S. TIP47 functions in the biogenesis of lipid droplets. J. Cell Biol. 2009;185:641–655. doi: 10.1083/jcb.200812042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Espinosa EJ, Calero M, Sridevi K, Pfeffer SR. RhoBTB3: a Rho GTPase-family ATPase required for endosome to Golgi transport. Cell. 2009;137:938–948. doi: 10.1016/j.cell.2009.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Aivazian D, Serrano RL, Pfeffer S. TIP47 is a key effector for Rab9 localization. J. Cell Biol. 2006;173:917–926. doi: 10.1083/jcb.200510010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Sztalryd C, Xu GH, Dorward H, Tansey JT, Contreras JA, Kimmel AR, Londos C. Perilipin A is essential for the translocation of hormone-sensitive lipase during lipolytic activation. J. Cell Biol. 2003;161:1093–1103. doi: 10.1083/jcb.200210169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Miyoshi H, Souza SC, Zhang HH, Strissel KJ, Christoffolete MA, Kovsan J, Rudich A, Kraemer FB, Bianco AC, Obin MS, Greenberg AS. Perilipin promotes hormone-sensitive lipase-mediated adipocyte lipolysis via phosphorylation-dependent and -independent mechanisms. J. Biol. Chem. 2006;281:15837–15844. doi: 10.1074/jbc.M601097200. [DOI] [PubMed] [Google Scholar]
- [52].Shen WJ, Patel S, Miyoshi H, Greenberg AS, Kraemer FB. Functional interaction of hormone-sensitive lipase and perilipin in lipolysis. J. Lipid Res. 2009;50:2306–2313. doi: 10.1194/jlr.M900176-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Granneman JG, Moore HPH, Krishnamoorthy R, Rathod M. Perilipin controls lipolysis by regulating the interactions of AB-hydrolase containing 5 (Abhd5) and adipose triglyceride lipase (Atgl) J. Biol. Chem. 2009;284:34538–34544. doi: 10.1074/jbc.M109.068478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Papadoupoulos NM, Kintzios JA. Determination of human serum lipoprotein patterns by agarose gel electrophoresis. Anal. Biochem. 1969;30:421–426. doi: 10.1016/0003-2697(69)90136-5. [DOI] [PubMed] [Google Scholar]