Abstract
Genome-wide association studies (GWAS) of complex traits have generated many association signals for single nucleotide polymorphisms (SNPs). To understand the underlying causal genetic variant(s), focused DNA resequencing of targeted genomic regions is commonly used, yet the current cost of resequencing limits sample sizes for resequencing studies. Information from the large GWAS can be used to guide choice of samples for resequencing, such as the SNP genotypes in the targeted genomic region. Viewing the GWAS tag-SNPs as imperfect surrogates for the underlying causal variants, yet expecting that the tag-SNPs are correlated with the causal variants, a reasonable approach is a two-phase case-control design, with the GWAS serving as the first-phase and the resequencing study serving as the second-phase. Using stratified sampling based on both tag-SNP genotypes and case-control status, we explore the gains in power of a two-phase design relative to randomly sampling cases and controls for resequencing (i.e., ignoring tag-SNP genotypes). Simulation results show that stratified sampling based on both tag-SNP genotypes and case-control status is not likely to have lower power than stratified sampling based only on case-control status, and can sometimes have substantially greater power. The gain in power depends on the amount of linkage disequilibrium between the tag-SNP and causal variant alleles, as well as the effect size of the causal variant. Hence, the two-phase design provides an efficient approach to follow-up GWAS signals with DNA resequencing.
Keywords: DNA resequencing, Horwitz-Thompson estimate, inverse sampling fraction weights, two-phase sampling
Introduction
Despite the many successes of genome-wide association studies (GWAS) that have identified a large number of genomic regions associated with a wide variety of complex traits, it has been difficult to identify the underlying causal variants. Many of the single nucleotide polymorphisms (SNPs) on commercial genotyping arrays were chosen to “tag” genomic regions. That is, tag SNPs capture the population-level variation of a genomic region, with the size of the representative region depending on the amount of linkage disequilibrium (LD) between the tag-SNPs and other unmeasured variants. In most cases, the tag SNPs do not directly influence complex traits, but rather are correlated with the unmeasured causal variants. Hence, a strategy to find the underlying causal variant(s) is to resequence the DNA of chosen samples to search for variants that have a stronger association with disease than the tag SNPs, as well as supporting evidence of their likely function based on annotation. Furthermore, conditional regression is often used to evaluate the role of newly discovered variants after adjusting out the effects of the tag-SNP caused by LD. That is, we expect that any newly discovered variants in a targeted region that are associated with disease to be also associated with the tag SNP through LD. Without this expectation, then any newly discovered variants would be independent of the tag SNP, making it questionable why a tag SNP that is not causal of disease is associated with disease, independent of causal variants! We use this perspective, of controlling for the association of a tag SNP with disease, when considering designs for sampling subjects for DNA resequencing studies.
Although the cost of DNA resequencing of targeted regions continues to decrease, it is nonetheless cost prohibitive to resequence the large number of samples typically used in GWAS (e.g., 2,000 subjects). Currently, the cost at Mayo Clinic to resequence a region of 500 kb by next-generation sequencing technology is approximately $300 per sample. However, a relatively large number of samples is required in order to have sufficient power to detect variants that are less common than the minor alleles of the tag SNPs. For these reasons, efficient study designs are needed to optimally choose the most informative samples for resequencing studies.
For a fixed budget, an obvious design is to stratify on case/control status and randomly sample an equal number of cases and controls. We refer to this design as a case-control random sample. If there is prior information that covariate information could be used to enrich for genetic causation, a more powerful strategy would be to use covariate data when sampling cases and controls. Unusual cases and controls are those not well explained by their covariate profiles, suggesting that other factors, such as genetic, have a strong role in their phenotypes. For example, over-sampling cases with covariate profiles of low risk, and over-sampling controls with covariate profiles of high risk, can increase power over random sampling. By choosing these extremes, the cases and controls might be enriched for genetic extremes, enhancing power. A limitation of the case-control random sample, however, is that the information from GWAS is ignored, which can result in having too few of the high-risk genotypes suggested from the most significant GWAS SNPs.
An alternative design is to stratify on both case/control status and categories based on one or more tag-SNP genotypes measured in the GWAS sample. This strategy is referred to as a two-phase design, more specifically outcome-dependent two-phase stratified sampling design. The phase-1 sample is the entire sample used for a GWAS, and the phase-2 sample is obtained from the phase-1 sample by stratifying on phase-1 information, and then randomly sampling within strata. These types of two-phase designs have been well developed in survey research [Fuller, 2009], and in epidemiologic studies. Breslow et al. have published extensively on two-phase study designs in epidemiology, advocating it for situations in which a covariate is too costly to measure on everyone (for recent developments and critical references, see [Breslow et al., 2009a]). In this situation, a relatively inexpensive surrogate for the target covariate of interest is measured in the phase-1 sample, subjects are stratified based on this inexpensive covariate, and then the costly target covariate is measured on a subset of subjects, based on a stratified random sample. If the surrogate and target covariates are highly correlated, then there is little loss of statistical efficiency; it is almost as if the expensive target covariates were measured on everyone. Although there are statistical refinements that can improve analyses, for example, using auxiliary information to improve how subjects in the phase-2 sample are weighted, a key to the success of this two-phase study design is the magnitude of correlation between the surrogate and target covariates. In our context, the surrogate covariates are the GWAS tag-SNPs measured in phase-1, and the target covariates are the variants measured by resequencing samples in phase-2.
Because two-stage, or more generally multistage, designs have been widely used in GWAS, it is important to appreciate the difference between a two-phase design as we have described, and a two-stage design often advocated for genetic studies. For a two-stage design, a large number of genetic markers are tested for their association with disease among cases and controls at the first stage, and then a small fraction of the most significant markers are measured in an independent set of cases and controls at the second stage, an even larger sample of subjects than the first stage, a situation which is often not possible in pharmacogenomics studies. The association of the markers carried forward to the second stage is then evaluated in the entire set of cases and controls [Elston et al., 2007]. In contrast, the two-phase design obtains measures on a relatively cheap surrogate marker on all cases and controls, stratifies this entire sample according to the surrogate marker genotype and case-controls status, randomly samples subjects from within each stratum to obtain a phase-2 sample, and then measures an expensive marker on the phase-2 sample. Note that the two-stage design uses independent subjects between the two stages. In contrast, for the two-phase design, the subjects in the phase-2 portion are a subset of those from the phase-1 portion. In addition, the two-stage design analyzes all subjects at the second stage, in contrast to the two-phase design that analyzes only subjects sampled for the phase-2 portion.
To solidify our ideas with a practical example, a GWAS of breast cancer risk among women treated with either tamoxifen or raloxifene to prevent breast cancer identified two regions of interest, on chromosomes 4 and 16. The strongest associated SNPs from each of these two regions had P-values of 8.5 × 10−7 and 1.1 × 10−6, respectively. Furthermore, when these two SNPs were modeled jointly in logistic regression, there was a hint of interaction (modeling additive effects of alleles on the log odds ratio [OR]), with P-value of 5.1 × 10−3. The phase-1 sample of this GWAS had 592 cases and 1,171 controls. Because there were two regions of interest, and the sample size was too costly to resequence all subjects, a two-phase design was used. For this design, there were nine two-SNP genotype categories. Cross-classifying these nine categorieswith case/control status gives 18 strata. With a budget to resequence 400 samples in the two regions of interest, we opted for a design that approximately equally allocated the 400 samples across the 18 strata. As we will show, the analysis of this phase-2 sample can be performed by traditional logistic regression, for which the balanced design adjusts out the effects of the two tag SNPs. Alternatively, sampling weights can be used that allow us to take advantage of the correlation of the tag-SNPs with variants discovered in the resequencing studies, providing more flexible modeling opportunities and improved efficiency of parameter estimation.
In the following sections, we review design and analysis issues for outcome-dependent two-phase stratified sampling, particularly from the viewpoint of a GWAS forming phase-1 and DNA resequencing forming phase-2. Because theamount of LD between the tag-SNP in phase-1 and variants detected in phase-2 is critical, we performed simulations to compare the strengths and weaknesses of two main study designs: (1) stratification on only case/control status; (2) stratification on both case/control status and the three genotype categories of a tag-SNP. For these simulations, we evaluated the impact of LD and other parameters on the relative performance of these two study designs. Finally, we illustrate the application of different analytic methods to a two-phase study of breast cancer risk.
Methods
Design Issues
When designing a two-phase study, critical aspects are the variables used to define the strata, and the balance of the strata sizes between cases and controls. Sampling theory has shown that a stratified sample is optimal when the cell sizes are proportional to the standard deviations of the score statistics on the cells [Fuller, 2009; Reilly, 1996; Reilly and Pepe, 1995]. In our situation, the score statistics are for the variants detected in the phase-2 reseqeuncing study, so the standard deviation of a stratum-specific score statistic will depend on the genotype frequency of a tested variant. Hence, optimal allocation for one variant may not be optimal for another variant. Rather than attempting optimal allocation, a nearly optimal strategy is a balanced design. This is motivated by several features. In epidemiologic studies, frequency matching cases and controls is typically used to control for an important confounding covariate. This suggests that a way to control for the effect of a tag-SNP when designing two-phase resequencing studies is to balance the phase-2 strata such that each of the strata defined by tag-SNP genotypes have an equal number of cases and controls, or at least as equal as possible given the available strata in phase-1 and the total phase-2 sample size. Breslow and Chatterjee [1999] found that this balanced design approach achieves near-optimality, making its simplicity appealing for our objectives.
Theoretical Issues
Three types of estimation procedures have been developed for two-phase designs: weighted likelihood (WL), pseudo-likelihood, and maximum likelihood (ML). See Breslow et al. [Breslow and Chatterjee, 1999; Breslow and Holubkov, 1997] for reviews of these procedures and their relative strengths and weaknesses. Although the ML procedure can be more efficient than the other procedures, depending on the relationship of covariates with response, it can experience convergence failures, and results can be biased if the wrong model is used (e.g., missing covariates or improper functional form of covariates). In contrast, the WL procedure is easy to implement and it is robust to modelmisspecification. Robustness might be particularly important when using two-phase designs to follow-up GWAS with DNA resequencing, because there are opportunities for unknown gene-gene and genecovariate interactions to influence results. For these reasons, we describe only the WL procedure and evaluate two-stage designs based on it.
Following the notation of Breslow and Chatterjee [1999], we consider the GWAS to be the phase-1 sample, and then randomly sample subjects for phase-2 DNA resequencing, using both the case-control status and tag-SNP(s) from phase-1. Suppose that there is a total of N subjects in the phase-1 sample, and these are classified according to a binary case-control status (Y) and genotypes based on tag-SNP(s). For J genotype categories, there will be 2J stratification categories based on this disease-genotype cross-classification. Let Nij denote the number of phase-1 subjects with Y = i (i = 0 for control and i = 1 for case) and genotype stratum S = j. The phase-2 sample is obtained by randomly sampling, without replacement, nij subjects from the Nij that are available in each of the stratification categories. The subjects sampled for phase-2 will have the phase-1 covariate data, possibly including the tag-SNPs as covariates, as well as additional variants detected in the phase-2 DNA resequencing. By assembling the desired covariates into a p-dimensional covariate vector xijk (k = 1, …, nij), we wish to fit the usual logistic regression model, P(y|x) = μ(x′ β) = exp(x′ β)/[1 + exp(x′ β)], but efficiently using both the phase-1 (Nij) and phase-2 (xij k) data to estimate the regression coefficients β. Keep in mind that any covariates representing the tag-SNP(s) will be available for all N subjects, but the variants detected by DNA resequencing will be available for only the subset of n•• subjects in phase-2. This can be viewed as a missing-data problem— subjects not selected for phase-2 are missing variants detected by DNA resequencing—yet the missing data can be overcome through strong correlation of the tag-SNP(s) and variants in the phase-2 subjects.
The WL approach, originating in sampling theory, is based on the usual score statistic
But, to account for the phase-2 sampling, based on sampling fractions fij = nij/Nij, inverse probability-weighted score equations are used. Solving the following estimating equation,
| (1) |
provides the WL estimate, β̂, known as the Horwitz-Thompson estimate. As described elsewhere [Breslow and Holubkov, 1997], β̂ is a consistent estimate with an asymptotic normal distribution and a covariance matrix that can be estimated by the “information sandwich” estimate. For this, the model-based information matrix, Im, and the empirical covariance matrix of the score vectors, D, are computed as illustrated below,
These are then combined into the sandwich estimate
| (2) |
Note that the inverse sampling fractions are used in both the observed information matrix and the empirical covariance matrix. The empirical covariance matrix is a weighted empirical estimator, which computes the within-stratum empirical variance of the scores (Uij• is a sum over k), and then sums over all strata using weight . This latter point is important, when considering the impact of weights on V(β̂).
As we shall see, there are two opposing factors that influence the efficiency of the two-phase design: (1) strong correlations of the tag-SNP(s) with the phase-2 variants can make the two-phase design more powerful than a random case-control design; (2) widely differing sampling fractions, which translate to widely differing values of in the calculation of V(β̂), can increase V(β̂), reducing power. It is also important to realize that the weights, , are 1 or larger, so that some subjects are up-weighted more than others. If a variant measured in phase-2 is perfectly correlated with the phase-1 tag-SNP(s) used to define the strata, then this up-weighting means that the effective sample size for the smaller phase-2 sample, in terms of V(β̂), is the same as the effective sample size for the larger phase-1 sample. This means that variants nearly perfectly correlated with the tag-SNP used to create strata will have P-values that are nearly the same as those found for the tag-SNP in the complete GWAS phase-1 sample. If, however, a variant measured in phase-2 is independent of the phase-1 SNP(s) used to define the strata, then the effective sample size for phase-2 is the same as the actual phase-2 sample size. Recognizing that P-values are influenced by both the size of the genetic effect on the trait and the sample size, this means that P-values for testing whether β = 0 for a variant independent of the tag-SNP will typically be less extreme than that for the tag-SNP, particularly when the phase-2 sample size is much smaller than the phase-1 sample size. In summary, the correlation of the variants measured in phase-2 with the tag-SNP used for stratification influences not only V(β̂), but also the interpretation of P-values, when comparing results from the phase-2 sequence data with the phase-1 tag-SNPs.
Hypothesis Testing
To test the null hypothesis that a particular set of variants is not associated with disease status, a Wald test can be constructed as QWald = β̂′ V (β̂)−1 β̂, which has an asymptotic chi-square distribution whose df is the rank V(β̂). However, we have found the Wald statistic to be numerically unstable, with β̂ too extreme for sparse variants, particularly when only the cases carry a variant and none of the controls, or vice versa. To overcome this problem, we advocate using a robust generalized score statistic, described by Boos [1992]. The generalized score statistic allows for possible inadequate modeling of covariates by using a robust empirical covariance for the score vectors, and does not require iterative fitting of parameters to be tested in the null hypothesis. Suppose the vector β of coefficients, of length m, is partitioned into , where β1 is of length r and β2 is of length (m – r). To test Ho : β1 = 0, treating β2 as nuisance parameters for adjusting covariates, the robust score statistic is = , where is a generalized inverse. The score vector U1 is for the first r terms of U(β) (expression 1) evaluated under Ho : β1 = 0, so that . Because β̂ solves the score equations for the last (m – r)terms, U2 = 0. The robust covariance matrix of vector U1 is based on partitioning the model information matrix Iminto submatrices that correspond with the partition of Let Im,ij denote one of these submatrices (i, j = 1, 2). Similarly, partition the empirical covariance matrix D into analogous submatrices, denoted by Dij. From these submatrices, the covariance of the score vector U1, conditional on the adjusting covariates, is
For large samples, Qscore has chi-square distribution with df the rank of Vs. Note that for a single variant, without any adjusting covariates, the statistic simplifies to Qscore = U2/D.
Analytic Issues
Implementation of the WL procedure is straightforward, taking advantage of existing software for logistic regression. Below we describe use of the R software package. Assuming that y is a vector for the response variable in the phase-2 data, v is a vector of subject-specific codes for the number of minor alleles for a variant measured in the phase-2 data, and f is a vector of subject-specific sampling fractions, the logistic model can be fit by
Note that glm assumes that weights (w) are frequency weights, so that a weight, typically an integer, means that the observations (y and covariates) represent w subjects. For binomial data, this means that y is the fraction of cases among w subjects. This type of weight, however, is not the same as the sampling weights we are using, w = 1/f. Consequently, the above use of glm results in the correct estimate, β̂, but the variance V(β̂) output from glm, as well as P-values, are not correct. Rather, the variance needs to be calculated by expression 2, which can be easily achieved by using the fit of glm, the above fit along with subject-specific indicators of the phase-2 strata (indexed by subscripts i and j), and sampling fractions f. Similar calculations can be used to compute the generalized robust score statistic.
Simulations
To evaluate the strengths and weaknesses of the two-phase design, and compare with randomly sampling based only on case-control status, we simulated a representative phase-1 data set based on a tag-SNP. From this phase-1 data, a phase-2 sample was obtained by stratifying on both the tag-SNP genotype and case-control status. In addition, an alternative phase-2 sample was obtained by stratifying on only case-control status (ignoring the tag-SNP genotype), and randomly sampling a smaller set of cases and controls for the phase-2 sample. Various analyses were conducted for the phase-1 and phase-2 data sets, described below, and the entire process was repeated 1,000 times.
To simulate the genotype data in phase-1, conditional on case-control status, we simulated two-locus genotypes, treating one as a tag-SNP and the other as a causal variant. To generate the distribution of the two-locus genotypes, we specified the minor allele frequency at each locus, the magnitude of LD between the minor alleles of the two loci (in terms of the fraction of the maximum LD, D′), the odds ratio (OR) for the tag-SNP, and the disease prevalence. Because we assumed that the effect of the tag-SNP is only due to LD with the causal variant, the specified parameters determined the OR of the causal SNP. Using this set of parameters, we can determine the frequency of two-locus haplotypes, then in turn the frequency of two-locus genotypes (pairs of haplotypes). Assuming a prevalence (of 0.1), and a logistic model that includes only the causal variant as a risk factor, we used Bayes formula to compute the distribution of two-locus genotypes conditional on case-control status. These conditional distributions were used to simulate two-locus genotypes for cases and controls in the phase-1 data, assuming 1,000 cases and 1,000 controls, as in typical GWAS. For the phase-1 sample, we performed two separate analyses: (1) the tag-SNP alone, mimicking what would be possible in a GWAS, and (2) the causal variant alone, in order to contrast the power for testing the effect of a causal variant in the largest available sample size vs. the sample sizes used in the phase-2 analyses.
Phase-2 Stratified on Tag-SNP and Case-Control Status
For the phase-2 portion, the number of cases and controls were each set to 100. The simulated phase-1 data were stratified according to both the case-control status and the three genotypes of the tag-SNP, and the phase-2 sample was obtained by randomly sampling within each of the strata to achieve as much balance (equal size) as possible across the six strata. If the number of available subjects within a stratum was less than required for a balanced design, all subjects were sampled, and a nearly balanced design was accepted. For this phase-2 data, two different types of analyses were conducted: weighted and unweighted. The weighted approach is the Horwitz-Thompson estimate using inverse probability sampling weights, as outlined above. The unweighted used the typical unweighted logistic regression model, ignoring the sampling weights. We performed this unweighted analysis in order to compare the power of the weighted and unweighted analyses, since it is known that variability in samplingweights can reduce power.
Phase-2 Stratified on Case-Control Status Only
To evaluate the strengths and weaknesses of stratifying on tag-SNP genotypes, we performed a simple stratification on case-control status (ignoring the tag-SNP genotypes), and randomly sampled 100 cases and 100 controls, from the phase-1 sample of 1,000 cases and 1,000 controls. For this data, we performed an unweighted logistic regression analysis.
For reporting our simulation results, we present the OR for the underlying causal variant, as well as the expected r2 for the Pearson correlation of the minor alleles at the tag-SNP and causal variant. For all simulations, we used a nominal Type-I error of 0.05, and 1,000 simulated data sets.
Phase-2 Study Application
As described elsewhere [Ingle et al., 2010], a case-control GWAS was conducted among women treated with either ta-moxifen or raloxifene in randomized clinical trials of women at high risk for breast cancer: cases were women who experienced an invasive breast cancer or ductal carcinoma in situ (DCIS) and controls were women who did not experience an invasive breast cancer or DCIS. A total of 592 cases and 1,172 controls were genotyped by the Illumina Human610-Quad platform, and 547,356 SNPs were analyzed for their association with case-control status. The most significant SNPs from the GWAS were on chromosome 16 (P-value = 8.5 × 10−7), and on chromosome 4 (P-value = 1.1 × 10−6). From a logistic regression analysis of the most significant SNP from each of these two regions, the test for interaction resulted in a P-value of 5.1 × 10−3. To choose a phase-2 sample for DNA resequencing of the two target regions, we stratified the GWAS sample according to case-control status and the joint genotypes of the SNPs on chromosomes 4 and 16. For these 18 strata, we randomly sampled subjects to achieve as much balance as possible. The resulting counts of subjects for the phase-1 and phase-2 samples are presented in Table 1, along with the inverse sampling probability weights. The resequencing effort focused on regions ±250 kb from the tag-SNPs, with approximately 4,000 new variants detected in each of the regions on chromosomes 4 and 16.
Table 1.
Phase-1 and Phase-2 sample counts, stratified on case-control status and the joint genotypes for SNPs on chromosomes 4 and 16
| Genotypesa |
Phase-1 sample |
Phase-2 sample |
Phase-2 weightsb |
||||
|---|---|---|---|---|---|---|---|
| SNP-4 | SNP-16 | Controls | Cases | Controls | Cases | Controls | Cases |
| 0 | 0 | 117 | 79 | 22 | 22 | 5.32 | 3.59 |
| 0 | 1 | 244 | 78 | 22 | 22 | 11.09 | 3.55 |
| 0 | 2 | 112 | 19 | 22 | 19 | 5.09 | 1.00 |
| 1 | 0 | 158 | 105 | 22 | 22 | 7.18 | 4.77 |
| 1 | 1 | 277 | 150 | 22 | 22 | 12.59 | 6.82 |
| 1 | 2 | 113 | 48 | 22 | 23 | 5.14 | 2.09 |
| 2 | 0 | 41 | 42 | 23 | 23 | 1.78 | 1.83 |
| 2 | 1 | 74 | 45 | 23 | 23 | 3.22 | 1.96 |
| 2 | 2 | 31 | 25 | 23 | 23 | 1.35 | 1.09 |
Genotypes coded as count of number of minor alleles; SNP-4 for chromosome 4 strongest SNP; SNP-16 for chromosome 16 strongest SNP.
Weight = 1 / (sampling fraction) = Phase-1 cell size/Phase-2 cell size.
Results
Simulation Results
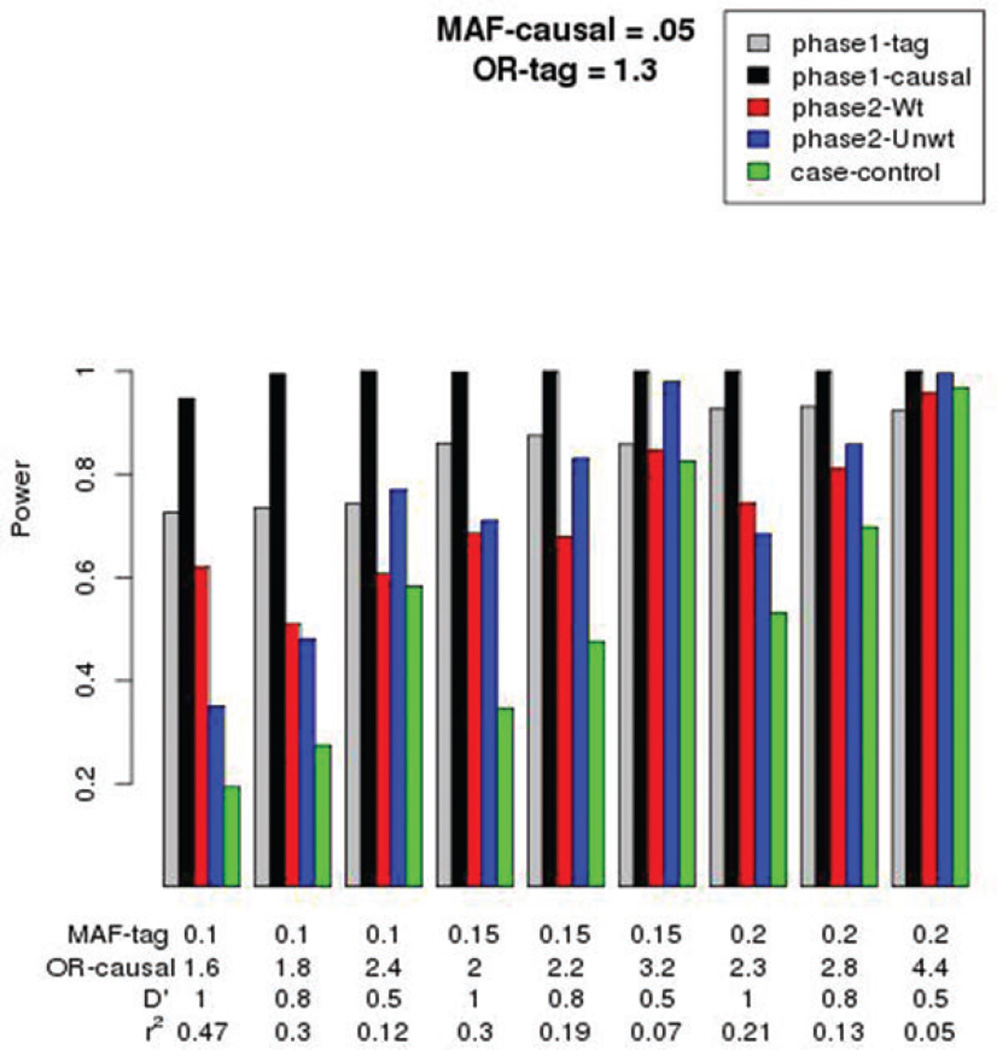
The power to detect an association with the tag-SNP or the causal variant in the phase-1 data set of 1,000 cases and 1,000 controls is presented in Figure 1, for when the OR for the tag-SNP is 1.3 and the minor allele frequency (MAF) of the causal variant is 0.05. The gray bar is for the phase-1 data testing the tag-SNP, and the black bar that for the causal variant. For these comparisons, we increased the MAF of the tag-SNP from 0.1 to 0.15 to 0.2, and for each level of the tag-SNP MAF, we altered the LD with the causal variant from D′ of 1.0, to 0.8, to 0.5. By decreasing the LD while keeping the tag-SNP OR fixed at 1.3, the OR for the causal variant increased, as illustrated along the x-axis of Figure 1. As expected, the power to detect associationwith the causal variant was always greater than that for the tag-SNP in the phase-1 data. The red and blue bars in this plot show the power for the phase-2 data of 100 cases and 100 controls, when stratification was on both case-control status and the tag-SNP genotype. The weighted analysis (red bar) had greater power than the unweighted analysis (blue bar) to detect the causal variant when the OR for the causal variant was relatively small. But, as the OR for the causal variant increased, the power for the unweighted analysis became greater than that for the weighted analysis, suggesting that the variability in the weights reduces power for causal variants with larger effects sizes. The case-control sampling (green bar), which ignored the tag-SNP genotypes when sampling the phase-2 subjects, had lower power than the two-phase design that stratified on both case-control status and phase-1 tag-SNP genotypes, except for the more extreme situation of a common tag-SNP (MAF = 0.2) having weak LD (D′ = 0.5) with a causal variant having large risk (OR causal = 4.4). In this case, the power of the case-control sampling was similar to that of the two-phase design.
Figure 1.
Power for phase-1 GWAS sample (tag and causal variants), for phase-2 sample obtained by stratifying on both case-control status and tag-SNP genotype (weighted =Wt; unweighted = Unwt), and for a phase-2 sample obtained by stratifying only on case-control status (case-control). The OR for the tag-SNP was fixed at 1.3, and the MAF of the causal variant fixed at 0.05. The MAF of the tag-SNP was allowed to vary (see below x-axis), as was the fraction of maximum LD between the tag-SNP and causal variant (D′). These parameters in turn determined the OR of the causal variant, and the expected r2, correlation, between the causal and tag-SNP alleles.
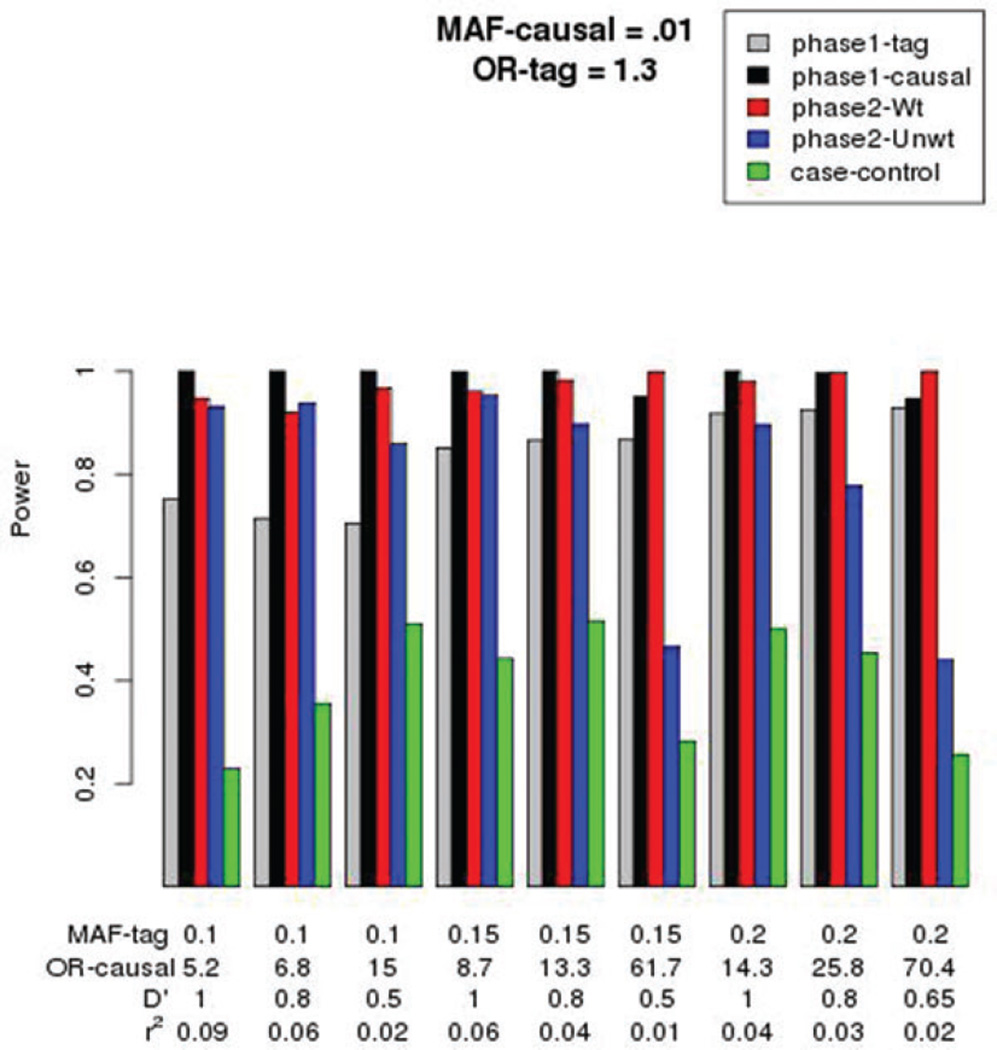
In parallel to Figure 1 simulations, we performed simulations with similar parameters, but decreased the MAF of the causal variant from 0.05 in Figure 1 to an MAF of 0.01 in Figure 2. The general patterns observed in Figure 1 appear in Figure 2, but with a few critical differences. First, as the MAF of the tag-SNP increased (keeping MAF of causal variant fixed at 0.01), and as the LD between tag-SNP and causal variant decreased, the relative power of the phase-2 weighted analysis (red bars) increased over the unweighted analysis (blue bars). This suggests that a weighted analysis is beneficial for detecting rare causal variants with large effect sizes, despite the variation in the weights. Second, the case-control design, which ignores the phase-1 tag-SNP genotypes to obtain the phase-2 sample, has much lower power to detect the rare causal variants.
Figure 2.
Power for phase-1 GWAS sample (tag and causal variants), for phase-2 sample obtained by stratifying on both case-control status and tag-SNP genotype (weighted = Wt; unweighted = Unwt), and for a phase-2 sample obtained by stratifying only on case-control status (case-control). The OR for the tag-SNP was fixed at 1.3, and the MAF of the causal variant fixed at 0.01. Other parameters are as defined in Figure 1.
Phase-2 Study Application
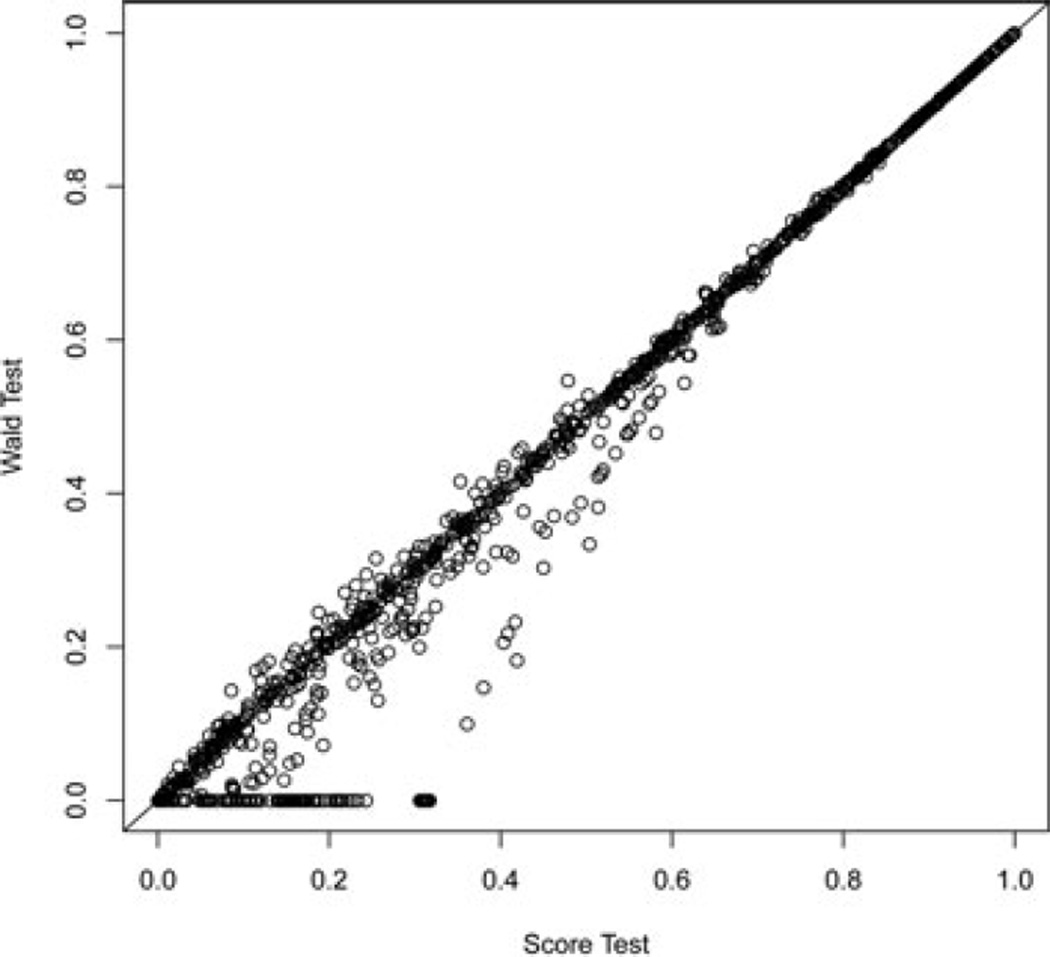
The results from analyzing 3,876 variants detected on chromosome 4 are presented in Figures 3 and 4. Figure 3 compares the P-values computed by the Wald and score tests. A large number of Wald tests resulted in P-value near 0 (n = 1,698), while none of the score tests had P-value near 0. These anomalies occurred when only one of the groups (cases vs. controls) had carriers of a variant, which caused the estimate β̂ to be wildly extreme. This occurred when the MAF of the variant was less than approximately 0.02 in either of the groups. These results suggest that the Wald test should not be used, particularly for less common variants.
Figure 3.
Wald vs. score statistics, P-values, for chromosome 4 region.
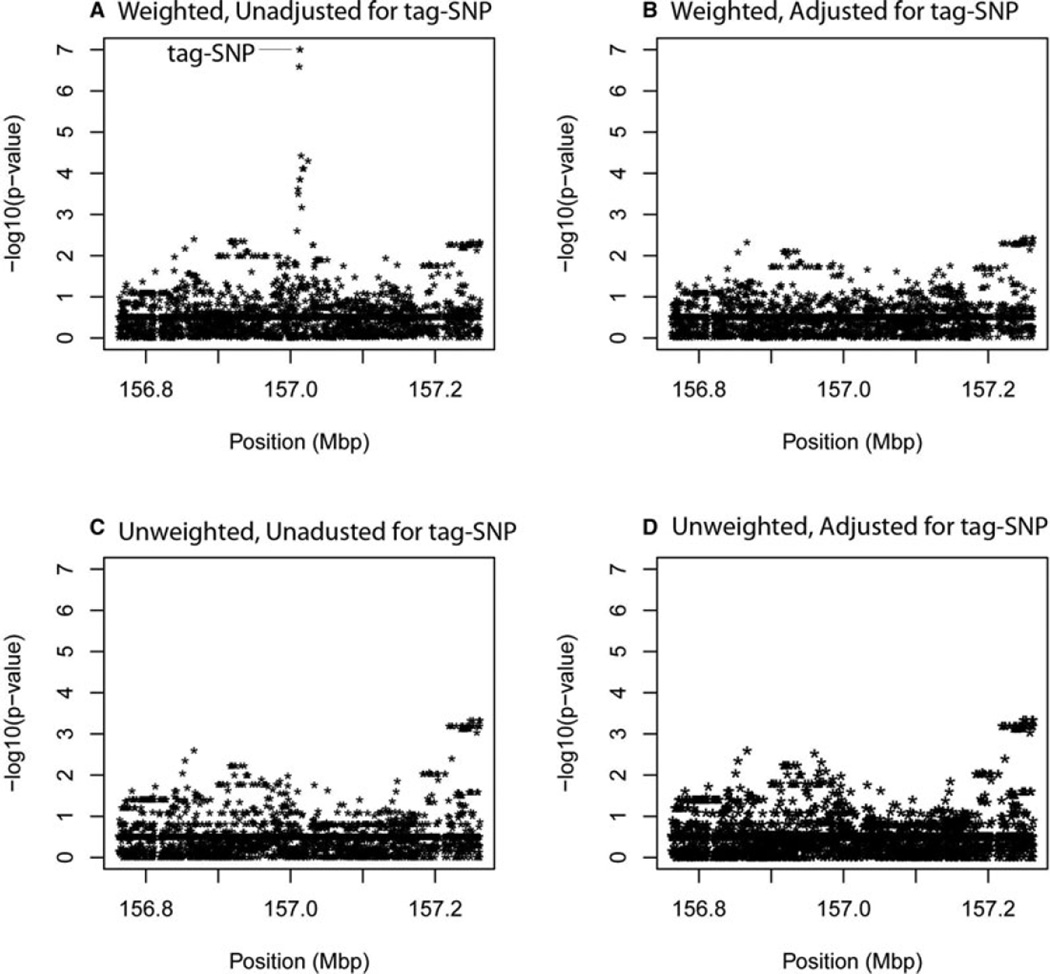
Figure 4.
Score-statistic P-values based on inverse sampling fraction weights (panels A, B), and based on traditional unweighted logistic regression (panels C, D), for chromosome 4 region. The tag-SNP from the phase-1 GWAS is indicated in panel A. This panel illustrates a vertical line of variants that follow the tag-SNP, due to strong correlations with the tag-SNP. Panel B illustrates results from adjusting for the tag-SNP modeled as a covariate. The bottom panels (C, D), illustrate P-values from score-statistics based on logistic regression without inverse sampling fraction weights. Because the cases and controls are perfectly balanced for the tag-SNP genotypes, the results implicitly control for the tag-SNP genotypes. For this reason, the unweighted results are nearly identical between the unadjusted and adjusted analyses. Panels B and D can be used to contrast the model-based adjustment for the tag-SNP (B) with the design-based adjustment for the tag-SNP (D). For most of the variants they are quite similar, although a few differences stand out, such as the cluster to the far right that has somewhat more extreme P-values for the design-based adjustment compared to the model-based adjustment.
Using the score statistics, we computed P-values using the inverse sampling fraction weights, without or with adjustment for the phase-1 GWAS tag-SNP. We repeated these analyses, yet without weights, using traditional logistic regression score statistics. Because the cases and controls were balanced according to the tag-SNP genotypes, ignoringweights implicitly adjusts for the tag-SNP. The results in Figure 4 show that the inverse sampling fraction weights reproduce the P-values for the tag-SNP obtained in the full data set of phase-1, and that any SNPs highly correlated with the tag-SNP are also pulled up to the level of the tag-SNP (panel A). Adjusting for the tag-SNP removes its effect, and any variants highly correlated with it, illustrated in panel B. The unweighted results, panels C and D of Figure 4, illustrate the impact of perfectly matching the cases and controls according to the tag-SNP genotypes. That is, the results are close to what is obtained by weighted analysis that includes the tag-SNP as a covariate. We can view the weighted analysis as a model-based adjustment for the tag-SNP, because assumptions are made on how to model the effect of the tag-SNP. In contrast, we can view the unweighted analysis as a design-based adjustment for the tag-SNP, because the cases and controls are balanced for the tag-SNP genotypes. For most of the variants, the model-based adjustments (panel B) are quite similar to the design-based adjustments (panel D), although a few differences stand out, such as the cluster to the far right that has somewhat more extreme P-values for the design-based adjustment compared to the model-based adjustment.
Although it is difficult to draw broad conclusions on the application of our methods to a single data set, results for chromosome 16 (not shown) showed similar patterns, with weighted analyses reproducing P-values for the tag-SNP according to the larger phase-1 sample results, and any variants highly correlated with the tag-SNP to produce P-values comparable to the tag-SNP, while uncorrelated variants had P-values much less extreme, partially due to the smaller effective sample size for the phase-2 sample (N = 400) relative to the phase-1 sample (N = 1,764).
Discussion
Deciding on how best to follow-up GWAS associations with DNA resequencing studies is challenging, because current cost constraints limit sample sizes, relative to the large samples typically used in GWAS, and because little is known about the underlying genetic architecture and its relationship to the SNPs used in GWAS. By simulations, we illustrate that using the information from tag-SNP genotypes to select cases and controls for subsequent DNA resequencing studies can dramatically increase power to detect causal variants, particularly when the causal variant is less common with a large effect size. Although our simulations are limited, there were no suggestions that the two-phase sampling design would do worse than stratifying only on case-control status to obtain a random sample, and often the power gains were substantial over the case-control stratified sampling. Furthermore, as illustrated by our example application, the two-phase sample design can be analyzed with or without the inverse sampling fraction weights. Using weights allows one to capitalize on the correlation of variants detected by DNA sequencing with the original tag-SNP, to view the patterns of associations local to the tag-SNP. To search for variants associated with disease that are independent of the original tag-SNP, the weighted analysis can included the tag-SNP as an adjusting covariate, performing a model-based adjustment. Furthermore, because cases and controls in the phase-2 sample are balanced for the tag-SNP, they are implicitly balanced for other variants highly correlated with the tag-SNP. This suggests that ignoring weights in the phase-2 analysis is a way to perform a design-based adjustment for the tag-SNP. Our application results nicely illustrate this feature.
Although we have discussed weights based only on the information from a single tag-SNP, additional SNPs from the initial phase-1 GWAS could be used to improve the phase-2 analyses. As clearly described by Breslow et al. [2009a], the variance of the phase-2 parameter estimates depends on two terms:the usualmodel-based variability and the design-based variance. The design-based variance depends on inverse sampling-probability weights. By use of auxiliary variables and information on the total phase-1 cohort, the sampling weights can be adjusted to reduce the contribution from the design-based variance, with adjustment based on either calibrating the weights to phase-1 totals of auxiliary variables, or by estimating improved weights using the auxiliary variables. Auxiliary variables are those available in the phase-1 sample and are highly correlated with the target variable. In our situation, we can view the underlying causal variant as the target variable, and the phase-1 GWAS tag-SNPs—those not used for stratified sampling of the phase-2 sample—as the auxiliary variables. Other covariates, such as disease severity, could be auxiliary variables, but their correlation with the underlying causal variant will likely be unknown, and numerical model-fitting problems can occur when attempting to use too many auxiliary variables. When observed tag-SNP genotypes, other than those used for phase-2 sampling, are used as auxiliary variables, the discrete genotype categories result in the calibration and estimation methods having the same adjusted weights. These adjusted weights are based on a finer level of stratification than that actually used for phase-2 sampling, and it is this finer stratification (called poststratification) that reduces the contribution from the design-based variance. From this perspective, it is possible to improve the phase-2 design we have described by using one, or a few, tag-SNPs from the GWAS to define strata for sampling, and then use the remaining tag-SNPs as auxiliary information. When there are many observed auxiliary tag-SNPs, iterative refinement of the weights is achieved through a “raking” process [Lumley, 2010]. Further research on the strengths and weaknesses of using additional tag-SNPs as auxiliary information is warranted.
An important consideration onwhether to use the tag-SNP information for a two-phase stratified sampling design is the potential amount of correlation of the tag-SNP alleles with the underlying causal variant allele(s). If one believed that the underlying causal variants are independent of the GWAS tag- SNP, then the tag-SNP carries no information on the causal variants, and so stratifying only on case-control status would be best. Yet, it seems contradictory to think that a tag-SNP from GWAS would lead interest to a specific region of the genome, yet not be correlated with underlying causal variants. This could occur, however, if the tag-SNP were in fact a causal variant, and the gene within which it resides has additional independent causal variants. Hence, an open question is what magnitude of correlation between a tag-SNP and an underlying causal variantmakes the two-phase design worth-while, recognizing that nonstandard software is required for analyses. Breslow et al. [2009b], in a different nongenetic context, give some helpful hints. They found that imputation of partially missing data improves precision of the corresponding regression coefficient when the R2 for prediction is at least 0.5. In our context, this would suggest that the squared correlation coefficient between the tag-SNP used for stratification of the phase-2 sample design and underlying causal variants should be at least 0.5 to gain substantial improvements of the two-phase design over a case-control random sample design. Yet, our simulations suggest that even with much lower correlations, the two-phase design has substantial power over a case-control random sample design when the effect size of the less-common causal variant is large. Clearly, too little is known about the genetic architecture of common diseases to suggest general guidelines, but our limited simulations and application to real data suggest that the two-phase design is not likely to perform worse than a case-control random sample design, and in fact can perform substantially better.
Acknowledgments
This research was supported by the US Public Health Service, National Institutes of Health (NIH), contract grant number GM065450 (DJS, JPS). The NSABP trials were supported by NIH contract grant numbers U10-CA-37377 and U10-CA-69974. The administrative coordination and data analyses were supported by NIH grants U19 GM6138 (Mayo PGRN) and P50 CA116201 (Mayo Clinic Breast Cancer SPORE).
References
- Boos DD. On generalized score tests. Am Stat. 1992;46:327–333. [Google Scholar]
- Breslow N, Chatterjee N. Design and analysis of two-phase studies with binary outcome applied to Wilms tumour prognosis. Appl Stat. 1999;48:457–468. [Google Scholar]
- Breslow NE, Holubkov R. Weighted likelihood, pseudo-likelihood and maximum likelihood methods for logistic regression analysis of two-stage data. Stat Med. 1997;16(1–3):103–116. doi: 10.1002/(sici)1097-0258(19970115)16:1<103::aid-sim474>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Breslow N, Lumley T, Ballantyne C, Chambless L, Kulich M. Improved Horvitz-Thompston estimation of model parameters from two-phase stratified samples: applications in epidemiology. Stat Biosc. 2009a;1:1–19. doi: 10.1007/s12561-009-9001-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslow NE, Lumley T, Ballantyne CM, Chambless LE, Kulich M. Using the whole cohort in the analysis of case-cohort data. Am J Epidemiol. 2009b;169(11):1398–1405. doi: 10.1093/aje/kwp055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elston RC, Lin D, Zheng G. Multistage sampling for genetic studies. Ann Rev Genomics Hum Genet. 2007;8:327–342. doi: 10.1146/annurev.genom.8.080706.092357. [DOI] [PubMed] [Google Scholar]
- Fuller W. Sampling Statisics. Hoboken: John Wiley & Sons, Inc; 2009. [Google Scholar]
- Ingle J, Liu M, Wickerham D, Schaid D, Mushiroda T, Kubo M, Costantino J, Goetz M, Ames M, Wang L. Genome-wide assocations of breast events and functional genomic studies in high-risk women receiving tamoxifen or raloxifene on NSABP P1 and P2 prevention trials. A Pharmacogenomics Research Network-RIKEN-NSABP Collaboration. Cancer Res. 2010;70(24 Suppl) Abstract PD05-02. [Google Scholar]
- Lumley T. Complex Surveys: A Guide to Analysis Using R. Hoboken: JohnWiley & Sons, Inc; 2010. [Google Scholar]
- Reilly M. Optimal sampling strategies for two-stage studies. Am J Epidemiol. 1996;143:92–100. doi: 10.1093/oxfordjournals.aje.a008662. [DOI] [PubMed] [Google Scholar]
- Reilly M, Pepe M. A mean score method for missing and auxiliary covariate data in regression models. Biometrika. 1995;82:299–314. [Google Scholar]