Abstract
The gut immune system has a key role in the development of autoimmune diabetes, and factors that control the gut immune system are also regulators of beta-cell autoimmunity. Gut microbiota modulate the function of the gut immune system by their effect on the innate immune system, such as the intestinal epithelial cells and dendritic cells, and on the adaptive immune system, in particular intestinal T cells. Due to the immunological link between gut and pancreas, e.g. the shared lymphocyte homing receptors, the immunological changes in the gut are reflected in the pancreas. According to animal studies, changes in gut microbiota alter the development of autoimmune diabetes. This has been demonstrated by antibiotics that induce changes in the gut microbiota. Furthermore, gut-colonizing microbes may modify the incidence of autoimmune diabetes in animal models. Deficient toll-like receptor (TLR) signaling, mediating microbial stimulus in immune cells, prevents autoimmune diabetes, which appears to be dependent on alterations in the intestinal microbiota. Although few studies have been conducted in humans, recent studies suggest that the abundance of Bacteroides and lack of butyrate-producing bacteria in fecal microbiota are associated with beta-cell autoimmunity and type 1 diabetes. It is possible that altered gut microbiota are associated with immunological aberrancies in type 1 diabetes. The changes in gut microbiota could lead to alterations in the gut immune system, such as increased gut permeability, small intestinal inflammation, and impaired tolerance to food antigens, all of which are observed in type 1 diabetes. Poor fitness of gut microbiota could explain why children who develop type 1 diabetes are prone to enterovirus infections, and do not develop tolerance to cow milk antigens. These candidate risk factors of type 1 diabetes may imply an increased risk of type 1 diabetes due to the presence of gut microbiota that do not support health. Despite the complex interaction of microbiota, host, environment, and disease mechanisms, gut microbiota are promising novel targets in the prevention of type 1 diabetes.
Keywords: Bacteroides genus, beta-cell autoantibodies, butyrate, gut, microbiota
Abbreviations: BB – BioBreeding ; CCR2 – C-C chemokine receptor type 2; DP – diabetes-prone; DR – diabetes-resistant; FINDIA - Finnish Dietary Intervention Trial; FOXP3 – forkhead box P3; HLA – human leukocyte antigen; IgE – immunoglobulin E; IL – interleukin; IFN-gamma – interferon gamma; KRV – Kilhan rat virus; mDC – myeloid dendritic cell; MyD88 – myeloid differentiation primary response gene 88; NF-κB – nuclear factor 'kappa-light-chain-enhancer' of activated B-cells; NOD - non-obese diabetic; OTU – operational taxonomic unit; rRNA – ribosomal ribonucleic acid; TGF-beta – transforming growth factor beta; Th – T helper; TLR – toll-like receptor; TRIGR – Trial to Reduce IDDM in the Genetically at Risk (study)
1. Introduction
During the last decade, our interest in the role of gut microbiota as a regulator of health and disease has increased tremendously, but the understanding of the gut microbiota function is still limited. The composition of gut microbiota has been associated with immune functions, immune-mediated diseases of the host, energy output, and obesity [1]. The development of better techniques for the identification of the species and functional composition of the gut microbiota has enabled this particular research field to develop quickly. It has been shown by using 16S rRNA gene sequencing that non-culturable populations represent a significant part of the human gut microbiota. Thus, genomic information of the gut microbiota is essential. In addition to taxonomic data, modern genome sequencing technologies are able to reveal the functional repertoire, which may not necessarily correlate with the phylogenetic profile of the microbiome. Metagenomics provide information on the functional properties of the gut microbiota, and may reveal important functions of the low-abundance microbes in the gut.
In germ-free mice, lack of microbial signal in the gut mucosa is associated with impaired oral tolerance [2] and impaired regulatory T cell function [3]. The role of gut microbiota in the regulation of immunity and tolerance is well-known, but understanding of the mechanisms involved is hampered by the complex interaction between biology of microbiota and host. Several commensals with key roles in the development of mucosal tolerance have been identified. Colonization of mice with human commensals, i.e. Bacteroides fragilis, facilitates differentiation of regulatory T cells and interleukin-10 (IL-10) production [4]. An immunomodulatory molecule, polysaccharide A of Bacteroides fragilis mediates the conversion of CD4 cells into Foxp3-expressing regulatory T cells during commensal colonization. Also, polysaccharide A is able to prevent and cure experimental colitis in animals. Oral colonization with clostridial species induces mucosal tolerance, characterized by the accumulation of regulatory T cells and activation of TGF-beta in the colon. These changes result in resistance to colitis and IgE response [5]. Such findings support further research into potential therapeutic options, based on the use of tolerance-supporting bacterial strains or their beneficial metabolites in the prevention and treatment of immune-mediated diseases such as autoimmune diabetes. Recently, the search for diabetogenic or type 1 diabetes-protecting microbiota has been intensified. Beside bacteria, other micro-organisms may be identified as regulators for immune homeostasis in type 1 diabetes.
2. Gut microbiota and autoimmune diabetes in animal models
In 1987, it was first suggested that the development of autoimmune diabetes may be dependent on microbiota [6]. Transferring non-obese diabetic (NOD) mice from pathogen-free to germ-free conditions resulted in a marked increase in the incidence of diabetes. In a recent study, the incidence of autoimmune diabetes in NOD mice did not increase in germ-free conditions, but the development of insulitis was accelerated [7]. These studies confirmed the role of gut microbiota as a regulator of inflammation in the islets of Langerhans. Decreased Foxp3 expression was observed in the ileum and colon of germ-free NOD mice. IL-17 expression was lower in the ileum, but higher in the colon in germ-free than in pathogen-free NOD mice. In mesenteric and pancreatic lymph nodes of germ-free NOD mice, elevated levels of IL-17 and interferon gamma (IFN-gamma) expressing CD4 cells were found, compared with pathogen-free NOD mice. Whereas, decreased levels of Foxp3-positive CD4 cells were found in germ-free NOD mice. Despite accelerated insulitis, the number of Foxp3-positive T cells infiltrating the islets was higher in germ-free NOD mice. The results indicated that a lack of microbiota clearly affects the intestinal regulatory T cell compartment and the progression of insulitis. The link between gut microbiota and the development of insulitis can be explained by the shared lymphocyte homing receptors in the gut and inflamed pancreas [8]. It has been shown that oral administration of antigen is able to activate antigen-specific T cells in pancreatic lymph nodes [9]. Thus, the microbiota-induced changes in the gut immune system may also be seen in immune cells of pancreatic lymph nodes and in insulitis.
Several studies in animal models indicate that alterations in the intestinal microbiota are associated with the development of autoimmune diabetes. At an early age, differences in gut bacterial composition were observed between rats that develop and those that did not develop diabetes; diabetic rats had a lower amount of Bacteroides species [10]. Furthermore, modulation of the intestinal flora through antibiotic treatment decreased the incidence and delayed the onset of diabetes (Table 1). It has also been reported that stool from BioBreeding (BB) diabetes-resistant (DR) rats contained more probiotic-like bacteria, whereas Bacteroides, Eubacterium, and Ruminococcus were more prevalent in diabetes-prone rats [11]. Lactobacillus johnsonii, which was isolated from BB-DR rats, prevented diabetes when administered to BB diabetes-prone (DP) rats [12]. Induction of IL-17 immunity in the mesenteric lymph nodes and spleen was seen in the rats who received Lactobacillus johnsonii [13]. These data suggest an interesting paradigm whereby the development of autoimmune diabetes can be circumvented by gut flora-mediated Th17 differentiation. In BB-DP rats, gut permeability and inflammation are associated with the development of diabetes [14]. Diabetes prevention, in association with the upregulation of IL-17 immunity, could be explained by the ability of IL-17 to activate the anti-microbial response in combination with mucosal repair mechanisms [15]. It is also possible that other mechanisms are involved because Lactobacillus johnsonii affects epithelial integrity directly [16]. However, in NOD-mice, increased expression of colonic IL-17 was associated with the development of diabetes, and dietary prevention of diabetes was associated with down-regulation of colonic IL-17 [17]. The upregulation of Th17 immunity has been reported in peripheral blood in patients with type 1 diabetes [18-21], but the role of Th17 immunity in autoimmune diabetes and in human type 1 diabetes is not fully understood. It seems that Th17 cells show some plasticity and share characteristics with Th1 cells. It is critical to determine their phenotype and their effects on the target tissue.
Table 1. Evidence for the gut microbiota as a regulator of autoimmune diabetes in animal models and for altered gut microbiota in human type 1 diabetes.
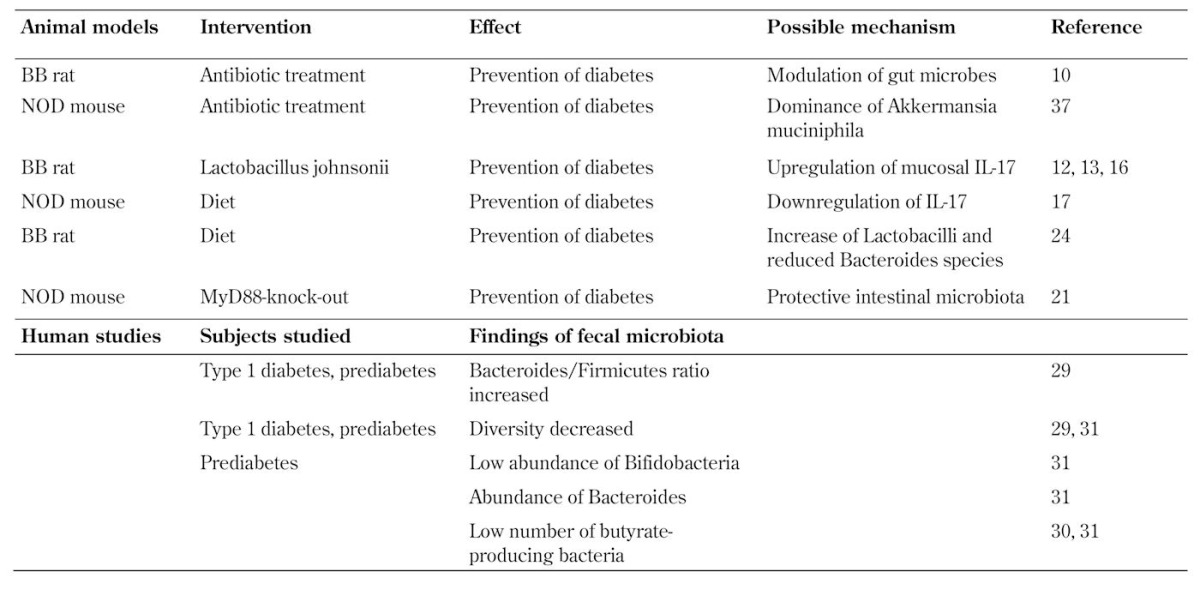
The role of intestinal microbiota as a regulator of autoimmune diabetes is strongly supported by evidence from NOD mice that lack the myeloid differentiation primary response gene 88 (MyD88), an essential signal transducer in toll-like receptor signaling. These MyD88-deficient mice do not develop diabetes [22]. The composition of their microflora differs when compared with wild-type NOD mice. More importantly, the transfer of the intestinal microflora from MyD88-deficient mice protects NOD mice from diabetes, which suggests the activation of tolerogenic mechanisms. See Table 1 for an overview of the gut microbiota-related interventions to treat autoimmune diabetes.
3. Interaction between diet, enteral infections, and bacterial microflora in the gut
The intestinal microbiota includes not only bacteria but also fungi and viruses. The interaction between these microorganisms regulates their growth and function. In a model of virus-induced diabetes, Kilhan rat virus (KRV) infection resulted in a transient increase in the abundance of Bifidobacterium spp. and Clostridium spp. in fecal samples of infected animals from day 5, but not day 12 [23]. Uninfected animals had no such increase. Treatment with a combination of trimethoprim and sulfamethoxazole, beginning on the day of infection, protected the rats from insulitis and autoimmune diabetes. The virus-induced inflammation in the Peyer's patches and pancreatic lymph nodes was reduced with antibiotic treatment. This result provides evidence that the interaction of enteral viruses and bacteria in the gut is of importance in the development of autoimmune diabetes. It is thus possible that altered composition of intestinal bacteria and subsequent immunological changes predispose children with beta-cell autoimmunity to enterovirus infections which are linked to type 1 diabetes [24] (Figure 1).
Figure 1. The complex interaction between the gut microbiota, host, environment, and disease mechanisms in the development of beta-cell autoimmunity and type 1 diabetes.
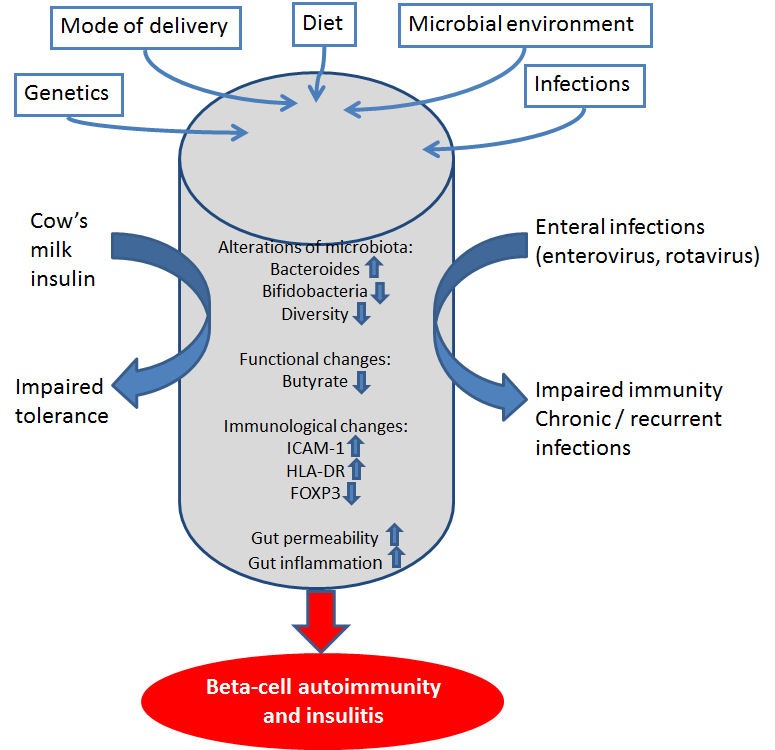
Both genetic and environmental factors affect the development of the gut microbiota, including mode of delivery and diet. In type 1 diabetes, an abundance of Bacteroides and low numbers of butyrate-producing bacteria have been reported. These changes may affect the immunological homeostasis in the intestine and the gut permeability. In the inflamed gut, oral tolerance is not supported, and enhanced immune responses to food antigens, such as cow’s milk, is developed. Altered microbiota could also affect the immune protection against enterovirus infections associated with type 1 diabetes.
Viral infection may also modulate microbiota, as mentioned above [22]. In animal models, it has been demonstrated that dietary interventions modify the composition of the gut microbiota [25]. A hydrolyzed casein-based diet beginning at weaning prevented diabetes in BB-DP rats. Higher IL-10 levels were measured in ileum tissue explants from hydrolyzed casein-fed rats, and beneficial gut microbiota changes were found, i.e. increased Lactobacilli and reduced Bacteroides spp. levels.
4. Less diversity and abundance of Bacter-oidetes in human type 1 diabetes
Recent results from the Human Microbiome Project included follow-up samples from 242 healthy adults. It was demonstrated that within-individual variation over time was consistently lower than between-individual variation, both in organismal composition and in metabolic function [26]. This suggests that in healthy adults the stability of the microbial community at different body sites, such as oral cavity, gut, and skin, is likely associated with health. Low diversity of gut microbes, i.e. number, abundance, and distribution of distinct types of organisms, has been linked to obesity, inflammatory bowel disease [27-29], and type 1 diabetes including beta-cell autoimmunity [30-33].
The published data on gut microbiota in patients with type 1 diabetes or in prediabetic individuals is very limited. In a pilot study, the composition of the intestinal microbiota and metagenomic data was analyzed in serial fecal samples from four Finnish children who developed beta-cell autoimmunity and type 1 diabetes and from four control children who were matched for the age and HLA-DQ risk genotype of type 1 diabetes [30, 31]. In the follow-up samples, collected at three time points during the first years of life, gut microbiota development was more stable in control children, whereas in children who developed beta-cell autoimmunity the microbiome became less diverse and less stable during the follow-up [30]. In combination with findings in other immune-mediated diseases, this suggests that immunological aberrancies are associated with loss of diversity of microflora.
Over time, an increased ratio of Bacteroidetes versus Firmicutes developed in the four children who became beta-cell autoantibody positive during the follow-up [29]. Bacteroidetes sequences increased from 53.27% of all sequences in the first sample to 69.17% in the third sample in the children who developed type 1 diabetes. In contrast, Bacteroidetes sequences decreased from 76.13% to 54.65% of all sequences in control children. The sequences of the second most abundant phylum, Firmicutes, declined from 43.1% to 20.66% in the diabetic children, but increased in controls. Three Firmicutes families were significantly more abundant in controls, namely Ruminococcaceae, Lachnospiraceae, and Eubacteriaceae. At the genus level, Bacteroides increased over time in children who developed type 1 diabetes, whereas Eubacterium and Faecalibacterium (part of the Firmicutes family) increased in controls. At the species level, 16-fold more Bacteroides ovatus sequences were found at the time of autoimmunity in the diabetic children than in controls, and several Bacteroides species were more abundant in these children. However, Bacteroides vulgatus and Bacteroides fragilis were more abundant in controls.
Altered ratios of Bacteroidetes and Firmicutes have also been reported in obesity and Crohn's diseaes, but these changes were different to those observed in type 1 diabetes. An abundance of Firmicutes was seen in obesity, while depletion of both Firmicutes and Bacteroidetes was reported in Crohn’s disease [27-29].
In our recent study, comprising 18 prediabetic children with beta-cell autoimmunity and 18 matched controls, we used 16S rRNA pyrosequencing to examine the composition of bacterial microbiota in feces samples [32]. Children with beta-cell autoimmunity and children negative for beta-cell autoantibodies were matched for age, sex, and HLA risk genotype of type 1 diabetes. Fecal samples were collected from 18 children, who had developed at least two beta-cell autoantibodies during the follow-up in the TRIGR or FINDIA pilot studies, and from matched control children negative for beta-cell autoimmunity. In the TRIGR pilot study, the children were randomized to receive ordinary cow’s milk formula or hydrolyzed casein-based formula [33]. In the FINDIA pilot study, the children received hydrolyzed whey-based formula or insulin-free whey-based formula, as test formula during the first 6 to 8 months of age [34]. We matched cases and controls for formula feeding history to avoid the well-known factors that modulate the composition of intestinal microbiota, i.e. host HLA genotype, gender, or early diet. The most interesting finding was that the Bacteroidetes phylum, the Bacteroidaceae family, and the Bacteroides genus were more common in autoantibody-positive than in autoantibody-negative children (4.6 vs. 2.2%, 3.5 vs. 1.5%, 4.3 vs. 2.0%, p = 0.035, 0.022, and 0.031, Mann-Whitney U test, respectively). Although the Bacteroides genus was associated with autoantibody positivity, only a few of its many members were related to autoantibody positivity on the species level.
We found that the diversity per sample was higher in autoantibody-negative than in autoantibody-positive children, as measured by the number of observed operational taxonomic units (OTUs) and the Chao index (p = 0.028 and p = 0.034, respectively, Mann-Whitney U test). The measurement was carried out in the TRIGR cohort, which comprised older children than the FINDIA cohort. In general, the diversity was higher in the TRIGR cohort than in the FINDIA cohort [32]. Table 1 provides an overview of the human studies related to gut microbiota and type 1 diabetes.
5. Low abundance of butyrate-producing bacteria in children with beta-cell autoimmunity
We also used principal component analysis to compare gut microbiota variation between children with and those without beta-cell autoimmunity. The analysis showed that a low abundance of lactate- and butyrate-producing species was seen in children with beta-cell autoimmunity [32]. These bacteria were B. adolescentis, R. faecis (a member of Clostridium cluster XIVa), and F. prausnitzii (a member of Clostridium cluster IV). B. adolescentes is an acetate- and lactate-utilizing bacterium, whereas the members of Clostridium clusters XIVa and IV are acetate-utilizing butyrate-producing bacteria [35, 36] with anti-inflammatory properties [37]. In children from the FINDIA study, E. hallii, an acetate- and lactate-utilizing and butyrate-producing bacterium from the Clostridium cluster XIVa [36], was inversely related to the number of beta-cell autoantibodies. Our findings of low abundance of butyrate-producing bacteria in children with beta-cell autoimmunity correspond with the observations made in the four children with beta-cell autoimmunity and their controls [30, 31]. The higher proportion of butyrate-producing bacteria in controls could contribute to the maintenance of a healthy gut because of its beneficial effects on gut integrity. Butyrate is an anti-inflammatory factor; it induces mucin synthesis and increases the barrier mechanisms of tight junctions. It also decreases bacterial transport across the gut epithelium.
Brown and co-authors reported that mucin-degrading bacteria of the genera Prevotella and Akkermansia were 20- and 140- fold higher in the four control children compared with the four diabetic children [31]. In NOD mice, the dominance of Akkermansia muciniphila, caused by vancomycin treatment, was associated with protection from autoimmune diabetes [38]. Interestingly, lactate producers, Lactobacillus, Lactococcus, Bifidobacterium, and Streptococcus were more abundant in diabetes cases, although some strains of these genera, such as lactobacilli and bifidobacteria, were introduced as health-promoting probiotics in the treatment and prevention of allergic diseases in children [39]. In contrast, we observed a low abundance of certain lactate-producing bacteria in children with beta-cell autoimmunity [32]. It should be noted that lactate can be further metabolized to butyrate or other short chain fatty acids with less beneficial effects. Therefore, the balance between different bacteria using lactate as substrate for short chain fatty acid metabolisms may be of importance for maintaining health. Producers of lactate (e.g. bifidobacteria) may contribute to the net production of butyrate [36]. We also found that a low abundance (< 12%) of the two most common bifidobacteria, Bifidobacterium adolescentis and Bifidobacterium pseudocatenulatum, was associated with beta-cell autoimmunity.
In metagenomic analyses, the functional changes in the gut microbiota were analyzed in the same eight Finnish children that were included in the study examining gut microbiota composition [31]. Samples from those children who finally developed type 1 diabetes were taken after seroconversion positive for at least two beta-cell autoantibodies. In these samples, the functional diversity in diabetic children was lower than in controls. The metagenomic approach revealed that gut microbiota of the children who developed type 1 diabetes was more abundant in functions related to stress responses, virulence factors, phages, and motility genes. Whereas, central metabolism and respiratory functions were more abundant in the microbiota of the controls.
6. Causality and functional effects of altered microbiota in type 1 diabetes
Potentially, altered microbiota in type 1 diabetes can be associated with alterations in the gut immune system, such as increased gut permeability [40, 41], small intestinal inflammation [42, 43], and impaired tolerance to food antigens [44], observed in type 1 diabetes. However, similar to the relationship between altered gut microbiota and the development of type 1 diabetes, studies on causality for the development of detrimental functional gut effects are lacking. Therefore, it is quite possible that intestinal inflammation in type 1 diabetes modulates the composition of the microbiota and not vice versa, although there is evidence for the first direction of causality. Butyrate regulates epithelial integrity. Therefore, the low abundance of butyrate-producing bacteria in type 1 diabetes could contribute to the increased gut permeability [45, 46]. Butyrate also acts as an anti-inflammatory factor. Therefore, deficient production of butyrate may be associated with the intestinal immune activation in type 1 diabetes.
Other evidence for the development of diabetes and impairment in gut function induced by gut microbiota alterations is given by studies investigating bacteria-based effects on the immune system. The diabetic gut appears to be underequipped with bacteria that promote protective immune mechanisms. As aforementioned, certain members of Clostridium clusters XIV and IV were observed infrequently in the gut of children with beta-cell autoimmunity. Interestingly, these clusters were shown to promote the accumulation of regulatory T cells in the intestine, and to induce mucosal tolerance in germ-free mice [5]. Low expression of FOXP3 and impaired induction of FOXP3-positive regulatory T cells by small intestinal dendritic cells, from patients with type 1 diabetes, have been reported [47, 48]. Despite the upregulation of Th17 immunity in the peripheral blood, we could not demonstrate enhanced IL-17 expression in the small intestinal biopsy samples taken from children with type 1 diabetes [49]. This could be due to changes in commensals, which appear to be important for the regulation of intestinal Th17 response and the balance between Th17 and regulatory T cells [50, 51].
Poor fitness of gut microbiota could explain why children who develop type 1 diabetes do not develop tolerance to cow's milk proteins [44] or are prone to enterovirus infections [23]. These candidate risk factors for type 1 diabetes may imply an increased risk of type 1 diabetes because the presence of gut microbiota does not support health (Figure 1). The interaction of microbiota, host, environment, and disease mechanisms is complex, and the understanding of the role of microbiota in the development of type 1 diabetes is a real challenge.
7. Outlook on future research
In future studies, metagenomics will be of great importance for the assessment of functional changes in the microbiota. These studies should include immunological evaluations of functional effects of the disease-associated gut microbiota on the immune system. This kind of simultaneous monitoring of the phylogenetic and functional composition of the microbiota and the immunological stage of the host is challenging, especially because there are few reliable markers for type 1 diabetes-related immune aberrancies other than beta-cell autoantibodies. In the pathogenesis of human type 1 diabetes, upregulated IL-17 immunity [18-21] and aberrant function and plasticity of regulatory T cells [52, 53] have been of special interest. Also, alterations of dendritic cells, such as decreased expression of CCR2 and enhanced NF-κB signaling in mDCs, may play a role in microbiota-induced diseases [54, 55]. These immunological changes may be modulated by the composition of intestinal microbiota, but their importance in the development of type 1 diabetes has not yet been clarified.
It is of particular interest that the role of butyrate-producing bacteria has been emphasized in the two available studies in children with beta-cell autoimmunity. These studies showed that a deficiency in butyrate-producing bacteria is associated with an increased risk of type 1 diabetes. These results were found independently, but both studies examined Finnish children. Environmental factors are important modulators of gut microbiota, and it is possible that these changes are specifically related to beta-cell autoimmunity in children living in Finland. Dietary habits affect the composition of the intestinal microbiota. According to our unpublished data, there is remarkable variation in the composition of gut microbiota in children living in different Western European countries. Pronounced differences in bacterial assemblages and functional gene repertoires were noted between US residents and subjects living in the Amazonas region of Venezuela or rural Malawi [56].
It is also possible that in immune-mediated diseases, such as in type 1 diabetes, changes in gut microbiota reflect the immunological alterations leading to disease development, and the findings of altered microbiota are secondary to beta-cell autoimmunity. We did not study samples from the children before the development of beta-cell autoimmunity. Therefore, we are not able to evaluate whether these changes in the gut microbiota precede beta-cell autoimmunity [32]. However, we did observe the most significant alterations in gut microbiota in individuals with three or four autoantibodies. Whereas, the difference between individuals with two autoantibodies and control subjects was less pronounced. The progression of beta-cell autoimmunity and immune attack against beta-cells could thus have effects on intestinal microbiota. Alternatively, an altered microbiota could affect the progression of autoimmunity. Thus, innate immunity could be an important regulator of progression from beta-cell autoimmunity to beta-cell destruction and type 1 diabetes.
The causality between altered microbiota and beta-cell autoimmunity needs to be clarified in future studies, including follow-up samples from a large series of healthy children who develop beta-cell autoimmunity and type 1 diabetes. Even though the diversity and abundance of microbes vary widely among healthy individuals, studies from the Human Microbiome Project have shown that there is considerable stability of the intestinal microbial composition in healthy people [26]. This is in contrast to subjects with autoimmunity. It has been argued that follow-up samples are not necessary in studies on the association of microbiota and diseases. However, follow-up samples are essential for the understanding of the role and causality of altered microbiota in disease development, and future microbiome studies will thus need to make use of longer follow-ups of people at risk of developing type 1 diabetes.
Disclosure: The author reports no conflict of interests.
Acknowledgments
Support from the Academy of Finland, Sigrid Juselius Foundation, and Päivikki and Sakari Sohlberg Foundation is acknowledged.
References
- 1.Vaarala O, Atkinson MA, Neu J. The "perfect storm" for type 1 diabetes: the complex interplay between intestinal microbiota, gut permeability, and mucosal immunity. Diabetes. 2008;57(10):2555–2562. doi: 10.2337/db08-0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sudo N, Sawamura S, Tanaka K, Aiba Y, Kubo C, Koga Y. The requirement of intestinal bacterial flora for the development of an IgE production system fully susceptible to oral tolerance induction. J Immunol. 1997;159(4):1739–1745. [PubMed] [Google Scholar]
- 3.Ostman S, Rask C, Wold AE, Hultkrantz S, Telemo E. Impaired regulatory T cell function in germ-free mice. Eur J Immunol. 2006;36(9):2336–2346. doi: 10.1002/eji.200535244. [DOI] [PubMed] [Google Scholar]
- 4.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci USA. 2010;107(27):12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y. et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331(6015):337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suzuki T, Yamada T, Takao T, Fujimura T, Kawamura E, Shimizu ZM, Tamashita R, Nomoto K. Diabetogenic effects of lymphocyte transfusion on the NOD or NOD nude mouse. In: Rygaard J, Sprang-Thomsen M, editors. Immune-deficient animals in biomedical research. Karger; Basel: 1987. [Google Scholar]
- 7.Alam C, Bittoun E, Bhagwat D, Valkonen S, Saari A, Jaakkola U, Eerola E, Huovinen P, Hänninen A. Effects of a germ-free environment on gut immune regulation and diabetes progression in non-obese diabetic (NOD) mice. Diabetologia. 2011;54(6):1398–1406. doi: 10.1007/s00125-011-2097-5. [DOI] [PubMed] [Google Scholar]
- 8.Hanninen A, Nurmela R, Maksimow M, Heino J, Jalkanen S, Kurts C. Islet beta-cell-specific T cells can use different homing mechanisms to infiltrate and destroy pancreatic islets. Am J Pathol. 2007;170:240–250. doi: 10.2353/ajpath.2007.060142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turley SJ, Lee JW, Dutton-Swain N, Mathis D, Benoist C. Endocrine self and gut non-self intersect in the pancreatic lymph nodes. Proc Natl Acad Sci USA. 2005;102:17729–17733. doi: 10.1073/pnas.0509006102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brugman S, Klatter FA, Visser JT, Wildeboer-Veloo AC, Harmsen HJ, Rozing J, Bos NA. Antibiotic treatment partially protects against type 1 diabetes in the Bio-Breeding diabetes-prone rat. Is the gut flora involved in the development of type 1 diabetes? Diabetologia. 2006;49:2105–2108. doi: 10.1007/s00125-006-0334-0. [DOI] [PubMed] [Google Scholar]
- 11.Roesch LF, Lorca GL, Casella G, Giongo A, Naranjo A, Pionzio AM, Li N, Mai V, Wasserfall CH, Schatz D. et al. Culture-independent identification of gut bacteria correlated with the onset of diabetes in a rat model. ISME J. 2009;3(5):536–548. doi: 10.1038/ismej.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lau K, Benitez P, Ardissone A, Wilson TD, Collins EL, Lorca G, Li N, Sankar D, Wasserfall C, Neu J. et al. Lactobacillus johnsonii N6.2 mitigates the development of type 1 diabetes in BB-DP rats. J Immunol. 2011;186(6):3538–3546. doi: 10.4049/jimmunol.1001864. [DOI] [PubMed] [Google Scholar]
- 13.Valladares R, Sankar D, Li N, Williams E, Lai KK, Abdelgeliel AS, Gonzalez CF, Wasserfall CH, Larkin J, Schatz D. et al. Inhibition of type 1 diabetes correlated to a Lactobacillus johnsonii N6.2-mediated Th17 bias. Plos One. 2010;5(5):e10507. doi: 10.1371/journal.pone.0010507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neu J, Reverte CM, Mackey AD, Liboni K, Tuhacek-Tenace LM, Hatch M, Li N, Caicedo RA, Schatz DA, Atkinson M. Changes in intestinal morphology and permeability in the biobreeding rat before the onset of type 1 diabetes. J Pediatr Gastroenterol Nutr. 2005;40:589–595. doi: 10.1097/01.mpg.0000159636.19346.c1. [DOI] [PubMed] [Google Scholar]
- 15.Onishi RM, Gaffen SL. Interleukin-17 and its target genes: mechanisms of interleukin-17 function in disease. Immunology. 2010;129(3):311–321. doi: 10.1111/j.1365-2567.2009.03240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kingma SD, Li N, Sun F, Valladares RB, Neu J, Lorca GL. Lactobacillus johnsonii N6.2 stimulates the innate immune response through Toll-like receptor 9 in Caco-2 cells and increases intestinal crypt Paneth cell number in biobreeding diabetes-prone rats. J Nutr. 2011;141(6):1023–1028. doi: 10.3945/jn.110.135517. [DOI] [PubMed] [Google Scholar]
- 17.Alam C, Valkonen S, Palagani V, Jalava J, Eerola E, Hänninen A. Inflammatory tendencies and overproduction of IL-17 in the colon of young NOD mice are counteracted with diet change. Diabetes. 2010;59(9):2237–2246. doi: 10.2337/db10-0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cooke A. Th17 cells in inflammatory conditions. Rev Diabet Stud. 2006;3(2):72–75. doi: 10.1900/RDS.2006.3.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Honkanen J, Nieminen JK, Gao R, Luopajarvi K, Salo HM, Ilonen J, Knip M, Otonkoski T, Vaarala O. IL-17 immunity in human type 1 diabetes. J Immunol. 2010;185:1959–1967. doi: 10.4049/jimmunol.1000788. [DOI] [PubMed] [Google Scholar]
- 20.Marwaha AK, Crome SQ, Panagiotopoulos C, Berg KB, Qin H, Ouyang Q, Xu L, Priatel JJ, Levings MK, Tan R. Cutting edge: Increased IL-17-secreting T cells in children with new-onset type 1 diabetes. J Immunol. 2010;185:3814–3818. doi: 10.4049/jimmunol.1001860. [DOI] [PubMed] [Google Scholar]
- 21.Arif S, Moore F, Marks K, Bouckenooghe T, Dayan CM, Planas R, Vives-Pi M, Powrie J, Tree T, Marchetti P. et al. Peripheral and islet interleukin-17 pathway activation characterizes human autoimmune diabetes and promotes cytokine-mediated beta-cell death. Diabetes. 2011;60(8):2112–2119. doi: 10.2337/db10-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, Hu C, Wong FS, Szot GL, Bluestone JA, Gordon JI, Chervonsky AV. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hara N, Alkanani AK, Ir D, Robertson CE, Wagner BD, Frank DN, Zipris D. Prevention of virus-induced type 1 diabetes with antibiotic therapy. J Immunol. 2012;189(8):3805–3814. doi: 10.4049/jimmunol.1201257. [DOI] [PubMed] [Google Scholar]
- 24.Oikarinen M, Tauriainen S, Oikarinen S, Honkanen T, Collin P, Rantala I, Mäki M, Kaukinen K, Hyöty H. Type 1 diabetes is associated with enterovirus infection in gut mucosa. Diabetes. 2012;61(3):687–691. doi: 10.2337/db11-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Visser JT, Bos NA, Harthoorn LF, Stellaard F, Beijer-Liefers S, Rozing J, van Tol EA. Potential mechanisms explaining why hydrolyzed casein-based diets outclass single amino acid-based diets in the prevention of autoimmune diabetes in diabetes-prone BB rats. Diabetes Metab Res Rev. 2012;28(6):505–513. doi: 10.1002/dmrr.2311. [DOI] [PubMed] [Google Scholar]
- 26.Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP. et al. A core gut microbiome in obese and lean twins. Nature. 2009;457(7228):480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L, Nalin R, Jarrin C, Chardon P, Marteau P, Roca J, Dore J. Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut. 2006;55(2):205–211. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134(2):577–594. doi: 10.1053/j.gastro.2007.11.059. [DOI] [PubMed] [Google Scholar]
- 30.Giongo A, Gano KA, Crabb DB, Mukherjee N, Novelo LL, Casella G, Drew JC, Ilonen J, Knip M, Hyöty H. et al. Toward defining the autoimmune microbiome for type 1 diabetes. ISME J. 2011;5:82–91. doi: 10.1038/ismej.2010.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brown CT, Davis-Richardson AG, Giongo A, Gano KA, Crabb DB, Mukherjee N, Casella G, Drew JC, Ilonen J, Knip M. et al. Gut microbiome metagenomics analysis suggests a functional model for the development of autoimmunity for type 1 diabetes. Plos One. 2011;6(10):e25792. doi: 10.1371/journal.pone.0025792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goffau M, Luopajärvi K, Knip M, Ilonen J, Ruohtula T, Härkönen T, Orivuori L, Hakala S, Welling G, Harmsen H, Vaarala O. Fecal microbiota composition differs between children with beta-cell autoimmunity and those without. Diabetes. 2013 doi: 10.2337/db12-0526. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Knip M, Virtanen SM, Seppä K, Ilonen J, Savilahti E, Vaarala O, Reunanen A, Teramo K, Hämäläinen AM, Paronen J. et al. Dietary intervention in infancy and later signs of beta-cell autoimmunity. N Engl J Med. 2010;363:1900–1908. doi: 10.1056/NEJMoa1004809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vaarala O, Ilonen J, Ruohtula T, Pesola J, Virtanen SM, Härkönen T, Koski M, Kallioinen H, Tossavainen O, Poussa T. et al. Removal of bovine insulin from cow's milk formula and early initiation of beta-cell autoimmunity in the FINDIA Pilot Study. Arch Pediatr Adolesc Med. 2012 doi: 10.1001/archpediatrics.2011.1559. In press. [DOI] [PubMed] [Google Scholar]
- 35.Duncan SH, Barcenilla A, Stewart CS, Pryde SE, Flint HJ. Acetate utilization and butyryl coenzyme A (CoA):acetate-CoA transferase in butyrate-producing bacteria from the human large intestine. Appl Environ Microbiol. 2002;68:5186–5190. doi: 10.1128/AEM.68.10.5186-5190.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duncan SH, Louis P, Flint HJ. Lactate-utilizing bacteria, isolated from human feces, that produce butyrate as a major fermentation product. Appl Environ Microbiol. 2004;70:5810–5817. doi: 10.1128/AEM.70.10.5810-5817.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermidez-Humaran LG, Gratadoux JJ, Blugeon S, Bridonneau C, Furet JP, Corthier G. et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA. 2008;105(43):16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hansen CH, Krych L, Nielsen DS, Vogensen FK, Hansen LH, Sorensen SJ, Buschard K, Hansen AK. Early life treatment with vancomycin propagates Akkermansia muciniphila and reduces diabetes incidence in the NOD mouse. Diabetologia. 2012;55(8):2285–2294. doi: 10.1007/s00125-012-2564-7. [DOI] [PubMed] [Google Scholar]
- 39.Savilahti E, Kuitunen M, Vaarala O. Pre and probiotics in the prevention and treatment of food allergy. Curr Opin Allergy Clin Immunol. 2008;8(3):243–248. doi: 10.1097/ACI.0b013e3282ffb134. [DOI] [PubMed] [Google Scholar]
- 40.Vaarala O. Leaking gut in type 1 diabetes. Curr Opin Gastroenterology. 2008;24:701–706. doi: 10.1097/MOG.0b013e32830e6d98. [DOI] [PubMed] [Google Scholar]
- 41.Bosi E, Molteni L, Radaelli MG, Folini L, Fermo I, Bazzigaluppi E, Piemonti L, Pastore MR, Paroni R. Increased intestinal permeability precedes clinical onset of type 1 diabetes. Diabetologia. 2006;49:2824–2827. doi: 10.1007/s00125-006-0465-3. [DOI] [PubMed] [Google Scholar]
- 42.Westerholm-Ormio M, Vaarala O, Pihkala P, Ilonen J, Savilahti E. Immunologic activity in the small intestinal mucosa of pediatric patients with type 1 diabetes. Diabetes. 2003;52:2287–2295. doi: 10.2337/diabetes.52.9.2287. [DOI] [PubMed] [Google Scholar]
- 43.Auricchio R, Paparo F, Maglio M, Franzese A, Lombardi F, Valerio G, Nardone G, Percopo S, Greco L, Troncone R. In vitro-deranged intestinal immune response to gliadin in type 1 diabetes. Diabetes. 2004;53:1680–1683. doi: 10.2337/diabetes.53.7.1680. [DOI] [PubMed] [Google Scholar]
- 44.Luopajarvi K, Savilahti E, Virtanen SM, Ilonen J, Knip M, Akerblom HK, Vaarala O. Enhanced levels of cow's milk antibodies in infancy in children who develop type 1 diabetes later in childhood. Pediatr Diabetes. 2008;9(5):434–441. doi: 10.1111/j.1399-5448.2008.00413.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peng L, Li ZR, Green RS, Holzman IR, Lin J. Butyrate enhances the intestinal barrier by facilitating tight junction assembly via activation of AMP-activated protein kinase in Caco-2 cell monolayers. J Nutr. 2009;139:1619–1625. doi: 10.3945/jn.109.104638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li N, Hatch M, Wasserfall CH, Douglas-Escobar M, Atkinson MA, Schatz DA, Neu J. Butyrate and type 1 diabetes mellitus: can we fix the intestinal leak? J Pediatr Gastroenterol Nutr. 2010;51:414–417. doi: 10.1097/MPG.0b013e3181dd913a. [DOI] [PubMed] [Google Scholar]
- 47.Tiittanen M, Westerholm-Ormio M, Verkasalo M, Savilahti E, Vaarala O. Infiltration of forkhead box P3-expressing cells in small intestinal mucosa in coeliac disease but not in type 1 diabetes. Clin Exp Immunol. 2008;152:498–507. doi: 10.1111/j.1365-2249.2008.03662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Badami E, Sorini C, Coccia M, Usuelli V, Molteni L, Bolla AM, Scavini M, Mariani A, King C, Bosi E, Falcone M. Defective differentiation of regulatory FoxP3+ T cells by small-intestinal dendritic cells in patients with type 1 diabetes. Diabetes. 2011;60(8):2120–2124. doi: 10.2337/db10-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lahdenpera AI, Hölttä V, Ruohtula T, Salo HM, Orivuori L, Westerholm-Ormio M, Savilahti E, Fälth-Magnusson K, Högberg L, Ludvigsson J, Vaarala O. Up-regulation of small intestinal interleukin-17 immunity in untreated coeliac disease but not in potential coeliac disease or in type 1 diabetes. Clin Exp Immunol. 2012;167(2):226–234. doi: 10.1111/j.1365-2249.2011.04510.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gaboriau-Routhiau V, Rakotobe S, Lecuyer E, Mulder I, Lan A, Bridonneau C, Rochet V, Pisi A, De Paepe M, Brandi G. et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 51.Lochner M, Berard M, Sawa S, Hauer S, Gaboriau-Routhiau V, Fernandez TD, Snel J, Bousso P, Cerf-Bensussan N, Eberl G. Restricted microbiota and absence of cognate TCR antigen leads to an unbalanced generation of Th17 cells. J Immunol. 2011;186(3):1531–1537. doi: 10.4049/jimmunol.1001723. [DOI] [PubMed] [Google Scholar]
- 52.McClymont SA, Putnam AL, Lee MR, Esensten JH, Liu W, Hulme MA, Hoffmüller U, Baron U, Olek S, Bluestone JA, Brusko TM. Plasticity of human regulatory T cells in healthy subjects and patients with type 1 diabetes. J Immunol. 2011;186(7):3918–3926. doi: 10.4049/jimmunol.1003099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bailey-Bucktrout SL, Bluestone JA. Regulatory T cells: stability revisited. Trends Immunol. 2011;32(7):301–306. doi: 10.1016/j.it.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nieminen JK, Vakkila J, Salo HM, Ekström N, Härkönen T, Ilonen J, Knip M, Vaarala O. Altered phenotype of peripheral blood dendritic cells in pediatric type 1 diabetes. Diabetes Care. 2012;35(11):2303–2310. doi: 10.2337/dc11-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alkanani AK, Rewers M, Dong F, Waugh K, Gottlieb PA, Zipris D. Dysregulated Toll-like receptor-induced interleukin-1beta and interleukin-6 responses in subjects at risk for the development of type 1 diabetes. Diabetes. 2012;61(10):2525–2533. doi: 10.2337/db12-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP. et al. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]


