Abstract
Understanding the functional roles of all the molecules in cells is an ultimate goal of modern biology. An important facet is to understand the functional contributions from intermolecular interactions, both within a class of molecules (e.g. protein–protein) or between classes (e.g. protein-DNA). While the technologies for analyzing protein–protein and protein–DNA interactions are well established, the field of protein–lipid interactions is still relatively nascent. Here, we review the current status of the experimental and computational approaches for detecting and analyzing protein–lipid interactions. Experimental technologies fall into two principal categories, namely solution-based and array-based methods. Computational methods include large–scale data-driven analyses and predictions/dynamic simulations based on prior knowledge of experimentally identified interactions. Advances in the experimental technologies have led to improved computational analyses and vice versa, thereby furthering our understanding of protein–lipid interactions and their importance in biological systems.
Keywords: Computational, Experimental, Protein–lipid interactions, Technology
1 Introduction
The proper functioning of cells relies on biomolecular interactions within and between cells. Characterizing these molecular interactions has led to much of our understanding of the molecular mechanisms of cellular regulation, biological processes, and human diseases. Over the last decade, studies in biological science have moved from analysis of single molecules and their individual interactions to broadly parallel, simultaneous analyses of multiple molecules and their interactions. Concurrently with this explosion of data generation from these “interactomics” studies has been a resurgence in the computational approaches applied to extract meaningful information from the data, ultimately providing a better understanding of the regulatory systems that underlie biological processes.
Studying the variety of cellular interactions, e.g. protein–protein interactions (PPI), protein–DNA interactions (PDI), and protein–lipid interactions (PLI), raises unique challenges. Thus far, PPI and PDI have been extensively studied due to the availability of technologies that can detect global interactions and analyze their roles in cellular processes [1]. PPI has been traditionally identified by standard techniques such as spectroscopy and calorimetry [2]. Although these techniques have been critical for establishing our current understanding of the PPI network, they are limited by the number of proteins profiled in each assay. Development of high-throughput experimental techniques (e.g. yeast two hybrid screening and affinity purification MS) and related computational techniques for mapping the experimentally identified interactions have enhanced our understanding of the PPI network [3].
Parallel technologies for PDI can be generalized into two categories: binding site characterization and gene activation [4]. Binding site characterization studies are based on analyzing the binding preferences of a given protein for a variety of DNA sequences in vitro, with microarrays having been applied to good effect for these studies [5]. While these interactions may occur in vitro, they may not be relevant to biological processes. As such, gene activation studies examine the unique sequences bound by proteins, particularly transcription factors, in the cellular context. Both the binding sequences and the downstream sequences can be recovered, e.g. ChIP-chip [6] and ChIP-seq [7,8], providing critical information about the actual function of PDI in controlling cellular responses.
In contrast, our knowledge of PLI has lagged, despite lipids being one of the most abundant classes of cellular metabolites and data showing that the binding by lipids affects the functions of proteins. This is due both to technical difficulties in identification of each member of the diverse class of cellular lipids and to the relative dearth of screening techniques for detecting global PLI. Nevertheless, advances in lipidomic research [9], paired with techniques adapted from PPI and PDI studies, have facilitated the recent development of novel approaches to characterize PLI. In this review, we describe the main experimental approaches, namely solution-based and array-based approaches, for analyzing PLI and introduce recent approaches in computational analysis of PLI. The findings from the experimental studies of PLI could be capitalized upon to expand the current framework of biological network analyses beyond PPI and PDI. Computational approaches developed from network biology or structural biology of PPI and PDI can be further adopted to analyze and integrate PLI to obtain a better understanding of their roles in mediating cellular signaling and metabolic processes.
2 Solution-based methods for studying protein–lipid interactions
Solution-based methods allow one to estimate the equilibrium binding and binding kinetics for PLI in complex solutions that mimic biological environments. In this section, we discuss current experimental techniques to detect PLI in solution. The basic principles and recent applications of the methods are introduced. In addition, the merits and pitfalls of each method are compared (Table 1), providing a guide on which approaches might be of use in the study of PLI under different contexts.
Table 1.
Solution-based techniques to detect PLI
| Methods | Advantages | Disadvantages |
|---|---|---|
| Liposome sedimentation assay | Applicable for proteomics analyses (in vitro and in vivo); available as both label-free and label-based methods. | Difficult to quantify the equilibrium constants. |
| Photoactivated cross-linking assay | Applicable for proteomics analyses (in vitro and in vivo). | Low sensitivity (provides low yield of the cross-linked lipid-binding proteins). |
| Isothermal titration calorimetry (ITC) | Provides thermodynamic parameters; is a label-free method. | Low sensitivity (requires higher protein concentrations). |
| Electron spin resonance (ESR) spectroscopy | Provides stoichiometry, selectivity, and geometric information of PLI; can be applied on opaque samples. | Requires post-hoc molecular modification that limits the applicability of the approach in biological membranes. |
| Fluorescence-based assays | Higher sensitivity. | Problems with light scattering and auto-fluorescence. |
The liposome sedimentation assay is most frequently employed for measuring PLI in solution. Similar to sedimentation velocity analytical ultracentrifugation used for studying PPI [10], the sedimentation efficiency of the liposomes depends, in this case, on their size. Typically, liposomes are mixed with a target protein and the lipid–protein complex is separated from the unbound proteins through high-speed centrifugation (> 20,000 g). The protein-bound liposome (higher molecular size) is contained in the pellet while the unbound proteins (lower molecular size) remain in the supernatant. This method does not require the attachment of labels, and each protein fraction can be readily quantified by SDS-PAGE and coomassie staining [11–13]. In some studies, labels (e.g. GST, MBP, radioactivity) are attached to the proteins to improve detection sensitivity [14–16]. Depending on the detection methods, liposome sedimentation can be used for protein concentrations in the range of 1–20 μM. Alone, this assay is qualitative, useful for determining whether a protein interacts with the lipids of the liposome. However, it was recently combined with proteomics analysis, using Nano-LC-MS/MS, to identify approximately 300 potential acidic phospholipid-binding proteins [17]. This novel combination provides a powerful tool to identify the variety of proteins that bind to a particular lipid.
Combined with the sedimentation assay, photoactivatable groups incorporated into lipids have been successfully used to study PLI [18, 19]. Upon UV activation, photoactivatable groups such as benzophenones, aryl azides, 3-trifluorophenyl diazirines, and alkyl diazirines yield highly reactive species that cross-link to form covalent bonds with proteins in contact with the activated site. The covalently labeled protein can be subsequently separated from unbound proteins through high-speed centrifugation and then detected by SDS-PAGE or MS-based proteomic analyses. Protein interactions with cholesterol, sphingolipids, and phosphatidylcholine have been successfully identified using the photo-cross-linking approach and their applications of photo-cross-linking with lipids have been extensively reviewed [20, 21]. In addition, recent developments of click chemistry coupled with the use of photoactivatable groups have enabled proteome-wide detection of PLI in the mitochondria, contributing to the realization of high throughput PLI proteomics [21].
Isothermal titration calorimetry (ITC) is one of the popular label-free, solution-based tools for detecting PLI. In this case, ITC measures the heat generated (exothermic) or absorbed (endothermic) during formation of protein–lipid complexes [22]. From ITC data, thermodynamic parameters of the interaction (i.e. binding enthalpy, Gibbs free energy, dissociation constant) can be determined. Although this method requires relatively higher protein concentrations than the sedimentation assay, ranging from tens of micromolar to several millimolar, it is increasingly being applied in conjunction with structure-based techniques, such as NMR, Fourier transform infrared, and circular dichroism [23–26], to provide structural details of the PLI coupled with thermodynamic information (Table 2).
Table 2.
Coupled solution-based and structure-based techniques to analyze PLI
| First author/reference | Description | Year |
|---|---|---|
| Alegre-Cebollada [49] | Binding of Sticholysin II Toxin and its mutants to sphingomyelin/DOPC/Cholesterol-containing liposomes studied by ITC and structural changes upon lipid binding by FTIR | 2008 |
| Shao [50] | Binding of Lactadherin C2 Domain to liposomes containing phosphatidylserine by FRET and flow cytometry and structural determination of lipid binding sites by X-ray crystallography | 2008 |
| Erlmann [51] | Binding of Deleted in Liver Cancer 1 (DLC1) to liposomes containing phosphatidylinositol-4,5-bisphosphate by liposome sedimentation assay and FRET | 2009 |
| Hughes [23] | Binding of Phospholamban (PLM1–23) to phospholipids by ITC and NMR | 2009 |
| Jobichen [52] | Binding of GrIR (global regulator of LEE repressor) to phospholipids by ITC and structural characterization of lipid binding domain by X-ray crystallography | 2009 |
| Marsh [53] | Orientation and Peptide-Lipid Interactions of Alamethicin Incorporated in Phospholipid Membranes by ESR and IR | 2009 |
| Mustafa [44] | Binding of serine racemase to liposomes containing phosphatidylinositol (4,5)-bisphosphate (PIP2) by liposome sedimentation assay and fluorescence anisotropy | 2009 |
| Qiu [43] | Interaction of β-amyloid (Aβ) peptide with liposomes by FRET and fluorescence anisotropy | 2009 |
| Guillén [45] | Binding of Hepatitis C virus protein NS4B to liposomes by fluorescence anisotropy and structural changes upon lipid binding by IR | 2010 |
| Tsujita [17] | Identification of acidic phospholipid-binding protein candidates by Nano-LC-MS/MS and confirmation the interactions between acidic phospholipid-binding proteins with lipids by liposome sedimentation assay | 2010 |
| D'Errico [29] | Binding of bovine seminal ribonuclease (BS-RNase) to liposomes by ESR and SPR and structural changes by CD | 2011 |
| Hughes [54] | Binding of the cytosolic domain of Phospholamban (PLM38–72) to phospholipids by ITC and NMR and structural determination by CD | 2011 |
| Hundertmark [26] | Binding of LEA18 (Late embryogenesis abundant 18) protein to liposomes by ITC and structural determination by CD | 2011 |
| Kobashigawa [42] | Binding of EEA1 (Early endosome antigen 1) FYVE domain to liposomes containing phosphoinositides by fluorescence anisotropy and NMR | 2011 |
| Bhunia [25] | Binding of scaffold protein Ste5 to phospholipids by ITC and structural determination by NMR and CD | 2012 |
| Dixon [39] | Binding of IQ motif containing GTPase activating proteins to phosphoinositides by TR-FRET and X-ray crystallography | 2012 |
| Hoernke [24] | Binding of cationic pentapeptides to liposomes by ITC and FTIR | 2012 |
| Krishnakumari [55] | Binding of peptides corresponding to the carboxyl-terminal region of human-β-defensins-1–3 to liposomes by ITC and fluorescence anisotropy | 2012 |
Another widely used and label-based biophysical methodology for PLI is ESR (electronic spin resonance) spectroscopy, which discriminates between immobilized lipids (near the protein interface) and free lipids in solution [27]. By attaching the spin-label nitroxyl ring to the lipid hydrocarbon chain, ESR is able to determine both the stoichiometry of the PLI and the selectivity of the protein for different lipid species [28]. Spin–spin interactions can also be observed between different lipid species or between lipids and spin-labeled proteins. In addition, by changing the location of the spin label on the lipids, ESR can provide geometrical information on the PLI, i.e. the depth to which the membrane protein penetrates the lipid bilayer [29,30]
Fluorescence-based methods, such as FRET (fluorescence resonance energy transfer) and fluorescence anisotropy (see reference [31] for a detail review of the theory), can also be applied to the characterization of PLI. Each of these methods is based on the sensitivity of fluorophores to their local environment. For FRET, once a protein containing a donor fluorophore and a lipid labeled with an acceptor fluorophore are in close proximity (typically 10–100 Å), acceptor fluorescence signals will increase [32–34]. In cases where sensitivity is not an issue, the use of tryptophan, with intrinsic fluorescence, as a donor bypasses the need for post-hoc labeling of the protein. Several paired acceptors for the tryptophan donor (e.g. dansyl [35], pyrene [36], and NBD (nitrobenzoxadiazole) [37]) have been employed to characterize PLI. Unfortunately, light scattering and autofluorescence can be challenges when using conventional FRET. Therefore, time-resolved FRET (TR-FRET) can overcome this issue by employing europium, which has a long fluorescence lifetime, in the range of milliseconds [38,39]. TR-FRET has been applied successfully, due to its high signal-to-noise ratio [40], in 96-well plates for screening purposes [38].
In contrast to FRET, fluorescence anisotropy is determined by measuring the emission intensities parallel and perpendicular to the excitation plane. When a small molecule interacts with a larger one, the hydrodynamic volume of the complex increases, resulting in higher fluorescence anisotropy values. The fluorophore can be labeled either on the protein [41,42] or lipid [43,44]. Intrinsic tryptophan fluorescence has also been used in anisotropy measurements for quantifying the binding affinity of a PLI [45]. In addition to FRET and fluorescence anisotropy, other fluorescence-based methods employed to quantify the interactions between proteins and lipids include two-photon microscopy [46], fluorescence correlation spectroscopy [47], and flow cytometry [48]. As with the sedimentation assay and ITC, fluorescence-based methods have also been combined with structural-based methods for improved characterization of the protein–lipid complexes (Table 2).
3 Array-based methods for analysis of protein–lipid interactions
3.1 Immobilization techniques
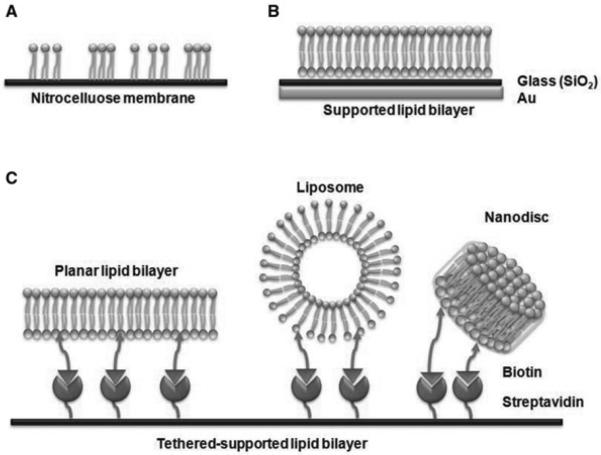
High-throughput screening of PLI have been performed using both protein [56] and lipid [57] microarrays. Here, we will focus on the application of lipid microarrays to probe PLI, with particular emphasis on the most recent developments in this technology. Lipid microarrays typically require the lipid molecules to be immobilized onto a planar surface. Several types of lipid systems can be immobilized onto surfaces (Fig. 1), including single lipids, liposomes, and supported lipid bilayers. In addition to these lipid systems, nanodiscs consisting of a discoidal lipid bilayer enclosed within an amphipathic protein belt were recently used to immobilize lipid bilayers on sensor chips for PLI studies [58]. Although the nanodisc approach requires additional steps to incorporate the lipids within the supporting protein, they provide a more soluble, uniform, and stable environment than the single lipid, liposome, and supported lipid bilayer approaches.
Figure 1.
Immobilization techniques. (A) Protein lipid overlay assay. Lipids can be noncovalently spotted on hydrophobic surfaces such as nitrocellulose membrane. (B) Supported lipid bilayer. The lipid bilayers can be generated from lipid vesicle fusion on silica surface supported by gold. (C) Tethered-supported lipid bilayer. Planar lipid bilayers, liposomes, and nanodiscs containing biotinylated lipids can be anchored onto streptavidin-coated surface.
Techniques for immobilizing liposomes and supported lipid bilayers have been recently reviewed in [59, 60]. The most common and conventional method used for PLI studies is to spot the lipids on nitrocellulose membranes (Fig. 1A), which has been employed in protein lipid overlay assays [61,62]. This method is the simplest one in that it does not require any chemical modification of the lipids or fabrication steps on the surfaces. Immobilization techniques are well developed, and have been adapted into commercial products for screening. These immobilization methods are incorporated in commercial lipid arrays, such as PIP Strips™, PIP MicroStrips™, SphingoStrips™, and PIP Array™. In addition, these membrane surfaces are typically fragile, making them infeasible for high-throughput screening systems for detecting PLI. However, recent studies used the nitrocellulose or PVDF membranes attached onto glass slides for automated spotting systems [63,64]. To obtain more uniform lipid bilayer structures, supported lipid bilayers can be generated from lipid vesicle fused onto hydrated surfaces made of silica or polydimethylsiloxane (PDMS) (Fig. 1B). By applying a thin silicate layer to the gold substrate, without any surface chemistry modifications, the lipid vesicles fused onto the silicate surface to produce a single lipid bilayer [65,66]. In addition, PDMS is an ideal alternative for hydrophilic surfaces to generate supported lipid bilayers [67]. However, it is known that the planar supported lipid bilayers have stability issues, by causing vesicle deformation that induces stress into the lipid bilayer [60]. There are two ways to improve the stability of the supported lipid bilayers. One approach is to introduce poly(ethylene glycol) (PEG)-derivative-lipids (PEG-brush configuration), which helps to retain a functional air-stable bilayer membrane [68, 69]. Alternatively, to achieve gentler immobilization, the irreversible interaction between biotinstreptavidin can be capitalized to generate tethered-supported lipid bilayers [70] (Fig. 1C). In addition, liposomes and nanodiscs also have been immobilized using biotin-streptavidin interactions in the tethered lipid systems [58,71].
3.2 Detection techniques
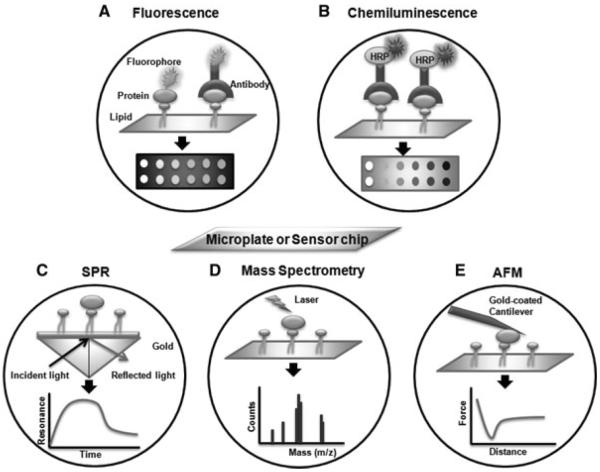
While techniques for immobilization have been extensively studied, methods to detect PLI are not well developed. Similar to protein microarray [72, 73], detection techniques employed in lipid microarray analyses can be categorized into two classes: label-based and label-free methods (Fig. 2). Label-based detection methods require labeling of the target proteins with fluorescent dyes, epitope tags, radioisotopes, or signal-generating enzymes (e.g. horseradish peroxidase (HRP)). Label-free methods (e.g. surface plasmon resonance (SPR), MS, atomic force microscopy (AFM), and interferometry) measure the inherent physical properties of the target proteins such as mass or dielectric properties. Label-based methods are more widely used, because the associated instruments are readily available. However, they are more laborious, requiring the attachment of tags on the proteins that could interfere with protein function. Although label-free techniques avoid these issues, they are typically less sensitive than the label-based approaches. However, the sensitivity of label-free techniques (especially SPR) has dramatically increased in recent years using approaches that can amplify the signals.
Figure 2.
Detection techniques for lipid microarray systems. (A) Fluorescence-based detection uses either fluorophore-tagged proteins or fluorophore-tagged antibodies against target proteins to detect protein–lipid interactions. (B) Chemiluminescence generates signals from HRP-conjugated antibody targeted for lipid-bound proteins. (C) SPR measures changes in the refractive index near a sensor surface. The interaction between the surface immobilized lipid bilayer and the proteins results in a change in the refractive index. (D) TOF-MS method uses laser energy to generate ions, displaying the mass-to-charge values and signal intensities of the proteins that interact with the lipids. (E) AFM equipped with a gold-coated cantilever sensor measures changes in the forces when the proteins interact with the lipids.
3.2.1 Label-based techniques
As with the solution phase analyses discussed above, fluorescent labels are commonly applied in label-based methods (Fig. 2A). Fluorophore-tagged proteins can be detected on lipid-coated surfaces using fluorescence microscopy. Often, these studies are piloted using the model Cholera toxin protein, which binds to GM1 gangliosides and is clinically relevant. In such studies, GM1 lipids are incorporated into phospholipid bilayers immobilized onto a surface, and the Cholera toxin protein is labeled with either FITC [69, 70] or Alexa dyes [74–76]. It was demonstrated that the tethered supported bilayer system has relatively high sensitivity and can detect protein at nanomolar concentrations [70]. In addition, total internal reflection fluorescence microscopy (TIRFM), one of the preferred techniques used to observe single molecules attached to planar surfaces, has been employed to measure the Cholera toxin-GM1 interaction [74]. In TIRFM, since only the membrane-bound proteins can be excited by the evanescent wave of an internally reflected laser beam, the binding kinetics between the proteins and lipids can be quantified. Thus, TIRFM was able to detect the binding of GM1 in planar supported bilayers to Alexa 595 labeled-Cholera toxin protein at concentrations as low as 100 pM [74]. The fluorescence method has also been coupled with antibodies to detect PLI through an ELISA-like approach [67,77]. Thus the fluorescence-based immunodetection method provides a fast, simple, and sensitive detect for high-throughput screening of PLI.
Chemiluminescence detection also can be used with label-based detection methods (Fig. 2B). Protein lipid overlay assay has adapted the chemiluminescence detection method for semiquantitative measurements of PLI. Typically, nitrocellulose membranes immobilized with lipids are incubated with a protein possessing an epitope tag [62, 64, 78]. The lipid-bound protein is then detected by immonoblotting with an HRP-conjugated antibody against the epitope tag. The protein lipid overlay assay has been successfully applied to screen PLI in yeast [64]. In addition, chemiluminescence has been used as a readout for a microplate-based approach to characterize PLI [77, 79]. Chemiluminescence methods typically have a detection limit in the range of 1 nM protein, roughly tenfold lower than fluorescence-based methods.
3.2.2 Label-free techniques
SPR is an optical technique that measures changes in refractive index at a metal-coated surface, and is a powerful analytical method for studying biomolecular interactions. SPR allows for rapid, label-free characterization of PLI, with direct measurement (Fig. 2C) and sensitivities of approximately 3 nM protein having been achieved [80]. Using an amplification approach involving functionalized gold nanoparticles in combination with an in situ atom transfer radical polymerization reaction, the detection limit was lowered to 160 aM (1.6 × 10−16 M) [81], achieving a higher sensitivity than previously reported detection methods. A recent study integrated SPR with neutron reflectivity and electrical impedance spectroscopy to investigate different aspects of a PLI, providing information about the lipid composition as well as the structural properties of β-lactoglobulin regulated by the PLI [82].
Single sample detection of PLI by SPR was successfully measured over a decade ago [83]. Recent publications have focused on using SPR as a screening tool for real-time analyses of multiple samples [58, 65, 80–82, 84, 85]. Further, SPRi (SPR imaging), an advanced format of SPR in which an image of the light reflected from the SPR substrate can be obtained, allows visualization of a whole array in real-time. SPRi was used to detect PLI, using both microfluidic and etched glass array formats [66, 68, 86, 87]. Current efforts are focused on achieving sensitivities with SPRi for parallel measurements that match those of fluorescence-based approaches.
MS technology has recently become one of the most popular methods to detect PLI in parallel, due to its capacity to identify the variety of lipids present in biological samples [88]. Many studies have focused on the identification of phosphoinositide (PI)–protein interactions using the lipid affinity capture method combined with MS [89]. In this approach, either proteins or lipids are immobilized on the affinity matrices. The immobilized molecules then capture their binding partners from cell extracts. The interactions are then detected by LC-MS/MS. Thus far, MS has rarely been applied on lipid microarray to detect PLI. Two types of time of flight MS (TOF-MS), which display mass-to-charge values and signal intensities of individual proteins, have been adapted to measure PLI [76, 90] (Fig. 2D). MALDI-TOF (matrix-assisted laser desorption/ionization time of flight MS) and SELDI-TOF (surface enhanced laser desorption/ionisation-time of flight MS) have been applied to single PLI [76] and array-based measurements [90], respectively. Both MS applications were able to identify oligomeric lipid-binding proteins [76,90].
Other techniques such as AFM, and interferometry are less frequently used to detect PLI. AFM has been used to probe PLI (AFM probing) or generate surface images (AFM imaging) of protein–lipid complexes [90, 91] (Fig. 2E). The interaction force between phospholipids immobilized on a sensor chip and proteins conjugated on a gold-coated cantilever was measured using AFM probing to show that lipids can interact with the C2A domain of synaptotagmin I [91]. AFM imaging showed that the α-synuclein protein penetrated a lipid monolayer and that the dimer form of the protein had higher affinity for the lipid bilayer relative to the monomer [90]. In addition, backscattering interferometry (BSI) was used to measure changes in refractive index on microfluidic chips that result from intermolecular associations, which provides the binding affinity between the proteins and lipids [92].
4 Computational analysis of protein–lipid interactions
In this section, we introduce the available computational approaches for analyzing and predicting PLI. Much of the analysis depends on the characteristics of the data generated by the various approaches that tackle different biological questions. Large-scale interaction data are used to systematically catalog protein–lipid associations, and to identify potentially novel interactions. In contrast, small-scale data with detailed kinetics are beneficial for studying molecular mechanisms of protein–lipid binding and identifying essential domains and binding sites. Since this is a new research area that has not been fully explored, we also provide a perspective of this field.
4.1 Large-scale analysis
Large-scale screening and computational analysis of PLI network in a model organism had not been fully explored until recently. A pioneering study in yeast [64] demonstrated the feasibility and potential benefits of such a systematic study. Using miniaturized nitrocellulose arrays (protein lipid overlay assay), they measured in parallel the binding profiles of 172 soluble proteins to 51 lipids and their metabolic intermediates. As a simple eukaryotic model organism, yeast has the advantages of a smaller number of genes and a well-studied metabolic system. The 51 lipids and their metabolic intermediates cover the major lipid classes and the 172 protein-set contains the majority of proteins with LBDs (lipid-binding domains) and the known lipid-regulated proteins and enzymes involved in lipid metabolism. Thus this study established a framework to systematically catalog PLI in yeast.
To validate the findings from their systematic screening in yeast both computational analyses and experimental techniques were applied to evaluate the PLI identified by the methodology [64], and they include:
-
(i)
(computational) Statistical analysis based on literature-derived reference dataset. The literature-derived reference dataset provides a collection of known PLI. Comparing the large-scale lipid array data with the reference dataset allowed for the estimation of the accuracy, sensitivity, and bias of the methodology based on its recovery of the known interactions.
-
(ii)
(computational) Use the yeast genetic network to provide (indirect) functional relationships in support of the novel interactions. This is based on the assumption that proteins and lipids that physically interact may be functioning in a same pathway, or in different pathways but of similar function. Under this assumption, if the known metabolic pathways involving a lipid X contain enzymes that are functionally related to a protein Y, then the lipid X and protein Y could be functionally related. The yeast genetic network provides such functional relationships between different proteins.
-
(iii)
(experimental) Physiological assays, including protein recruitment to liposomal and biological membranes, to determine whether the protein-lipid pairs measured in vitro could represent actual interactions in vivo.
-
(iv)
(experimental) Perturb the lipid metabolism, (e.g. sphingolipid synthesis) with antibiotics, and use live-cell imaging to determine the effect of the perturbations on the subcellular localization of the proteins that are identified to interact with the lipid.
Further computational analyses, e.g. sequence comparison and protein structure modeling, were applied on the novel interactions that were identified to help uncover potential mechanisms including novel protein–lipid binding sites as well as their functional role in cell regulation [64]. For example, there were proteins identified to interact with lipid classes that are not known to have LBDs, however sequence alignments helped to identify cryptic domains, e.g. CRAL/TRIO CRAL/TRIO (a protein structural domain that binds small lipophilic molecules, it is named after cellular retinaldehyde-binding protein and TRIO guanine exchange factor), in some proteins, which are remote homologs of LBDs that have previously gone undetected in these proteins. Further testing using physiological assays confirmed the prediction that these domains were responsible for lipid binding. Furthermore, they collected proteins confirmed by live-cell imaging to function in sphingolipid metabolism, and subsequently found enriched PH (Pleckstrin homology) domains on these proteins, suggesting the PH domain could be an unanticipated LBD that could function in sphingolipid recognition. Structural analysis of the proteins through X-ray crystallography and protein-lipid docking studies supported the cooperative lipid binding to the PH domain, and suggested the PH domain of the Slm1 protein played an important role in integrating the phosphatidylinositol phosphates (PtdInsP) and the sphingolipid signaling pathway.
The study in yeast demonstrates the first systematic screening of PLI in a model organism, and large-scale protein–lipid data analysis. Computational analyses can be performed to provide initial checks on the data quality and to filter the data using statistical approaches based on current knowledge of PLI. Other types of interaction data, e.g. the known genetic interactions in yeast, could be useful in such cross validation studies. Since gene and protein interaction data are widely available in other organisms, one could consider PPI (signaling pathways) or PDI (gene expression) data to filter and interpret the novel PLI that are identified. Comparing the novel interactions with current PLI databases or LBD domains could facilitate the discovery of novel LBDs, and when coupled with experimental validation, can identify novel functional mechanisms of protein–lipid binding, i.e. their functional role in cell regulation.
4.2 Molecular mechanism
In contrast to the large-scale screening that categorizes PLI and analyzes their functional role, other computational efforts have focused on predicting protein-LBDs in silico to explore the molecular mechanisms of such interactions. These approaches can be categorized into two canonical types: (A) machine learning (classification) approaches based on a training set; (B) abinitio prediction based on dynamic simulations.
In type A, the setting is as follows, given the amino acid sequence of known LBDs, predict which amino acid residues in a protein could be located at the protein–lipid interface. This is a classification problem, i.e. each amino acid in the protein sequence could be classified as being either at the interface or distant from the interface. Machine learning approaches (i.e. to learn a pattern from the current interaction data and use the pattern to predict new interfaces) can be applied to such classification problem, which is essentially similar to the methods used for identifying the interfaces in protein-DNA or protein-RNA interactions [93]. Although the problem setting focuses on the sequence level, the training dataset usually is collected from structural data (in PDB, http://www.rcsb.org/pdb) wherein one could reliably determine if an amino acid residue is involved in an interaction. A commonly used standard is to consider all amino acid residues within 3.5 ångströms of any lipid molecule to be the interaction interface of the protein. An early study by Irausquin and Wang [94] applied Support Vector Machine on sequence data to predict which amino acid on the protein can be bound by the lipid. They achieved around 52% sensitivity (i.e. of all the amino acids in the protein–lipid interface, 52% was identified by this approach) and 71% specificity (i.e. 71% of the predictions made by the approach are correct, which includes both the true positive predictions for which the amino acids actually are located on the protein–lipid interface and the true negative predictions) in a training dataset of 470 lipid-binding proteins. A recent study [95] improved the approach by incorporating Position Specific Scoring Matrix (PSSM) into the feature set, i.e., instead of predicting each amino acid individually. They also consider the flanking sequence around the amino acid and the conservation of the flanking sequence, because the LBDs as the functional part of a protein should have similar patterns and be more conserved than the other sequences. The approach increase the accuracy to around 80% (63% sensitivity, 90% specificity) thereby bolstering the applicability of learning approaches for predicting protein–lipid-binding interfaces. A limitation of this type of approach is that they do not necessarily provide detailed information on the molecular mechanism of how the proteins interact with the lipids. Nevertheless some of the methods were able to learn or infer the amino acid that were likely lipid-interacting residues [94], and to identify hydrophobic amino acids that appeared more frequently at the interface.
More mechanistic information can be obtained with the type B approach, which applies molecular dynamic (MD) simulation to compute protein–lipid-binding structures in silico. MD simulation is a powerful technique for studying the interaction interface and relating protein structure to function [96]. To explore a protein–lipid interaction necessitates the structures of both the protein and lipid bilayer. MD simulation could predict the potential positioning of the protein within the lipid bilayer and thereby provide a mechanistic understanding of the interaction.
Given that a biological system consists of a large number of molecules, conventional MD approach (i.e. atomistic MD simulation) that simulates all the atoms in a biomolecular system is computationally very expensive and infeasible. Therefore limiting its application, i.e. to the protein–lipid interface, and simplifying the model makes it more tractable. One approach taken to simplify the atomistic model was the development of a low-resolution hydrophobic slab model that treated the entire lipid-bilayer as one hydrophobic slab for positioning the membrane proteins, which reduced the computation time, [97]. However, the slab model ignored all possible conformational changes of the membrane that could occur during an interaction, thus making it difficult to study the specificity and functional mechanism of the interaction.
In contrast to assuming a slab configuration, coarse-grained (CG) simulation [98–101] applies a coarse-grained representation of the constituent molecules in a biomolecular system to achieve simplification of the system without losing all the details on the PDI interface. In CG models, the atoms are grouped together to form particles, e.g. by grouping four nonhydrogen atoms into one particle [100], or larger groupings are used to map amino acids into only two interacting CG particles, one for the backbone and one for the side chain [101]. The level of detail of the CG model for exploring the interaction mechanism correlated inversely with the computation time, with more mechanistic details requiring more simulation time. For a given CG model, one could derive the interaction parameters (the force field) for all the CG particles and compute the contribution of each (bonded and nonbonded) interaction to the potential energy of the simulated system [101]. With the CG particles and the force field, the CG-MD simulation begins by placing the membrane protein in a space along with randomly positioned CG lipid molecules to form a bilayer. The simulation is run (for example) for approximately 0.25 microseconds (approximately a day of CPU time, and may vary depending on the complexity of the CG model and the size of the protein) [100] to observe whether the bilayer self-assembles around the protein.
CG-MD results could predict the interaction structure and identify specific features of the PLI. For example, the KvAP (voltage-gated potassium channels) protein is a voltage sensor; however it was not known how the protein sensed voltage. CG-MD simulation of the voltage sensor (VS) domain on the KvAP protein [100] showed significant local distortion of the bilayer in response to changes in the voltage across the cell membrane. This suggested a novel mechanism by which the protein senses voltage—through local structural changes at the interaction surface.
Currently CG-MD is being applied to explore functional specificity of PLI, e.g. whether different amino acid compositions preferentially locate in different regions of the membrane, and if there are differences in the bilayer distortion with different types of transmembrane domains [99]. A structure database that is specific to protein–lipid interactions (http://sbcb.bioch.ox.ac.uk/cgdb) [102] has been constructed using CG-MD self-assembly simulations, which collected 91 proteins and their interaction structures with the DPPC lipid bilayer. Future studies could focus on improving the CG models and CG force fields to account for interactions of membrane proteins with different lipid bilayers [99], e.g. palmitoyloleoyl phosphatidylcholine (POPC), palmitoyl-oleoyl phosphatidylglycerol (POPG), etc., and developing multi-scale approaches that combine CG and atomistic MD simulation to enable simulations with more mechanistic details.
In summary, LBDs on proteins could be predicted based on sequence information using machine-learning approaches, or based on 3D structural information obtained through molecular dynamic simulations. The predictions generate hypotheses that could be tested using experimental methods described in the above sections. Thus integration of experimental and computational techniques would enhance our understanding of the molecular mechanisms of PLIs. For example, dynamic simulations could identify potential regions of interactions between a protein and lipid, and when coupled with experiments could discover novel lipid-binding regions or domains on the protein.
5 Conclusion
Reliable methodologies that characterize the interactions of biomolecules offer tremendous value in advancing our systematic understanding of biological systems. Current challenges still exist in the field of PLI. Solution-based methods are relatively well established as compared to array-based methods and when integrated with structure-based methods provide more in depth information on PLI. Although array-based methods for characterizing PLI have progressed, challenges remain and further improvements in detection are needed to capitalize upon parallel, high-throughput analyses.
Computational approaches are being developed to analyze large-scale data and predict novel LBDs based on current knowledge (Table 3). These approaches, including the statistical analysis of high-throughput systems and the learning and dynamic simulation of molecular interactions, are in their early stages and thus far have been limited by the quality and scale of current PLI data. With the development of detection strategies and accumulation of large-scale PLI data, more computational approaches developed from network biology (i.e. pathway analysis and functional annotation) or structural biology (i.e. conservation analysis and building domain databases) of PPI and PDI could be adopted to analyze PLI. Finally, integrating PLI data with other data, i.e. gene expression and metabolic data, would provide a more complete depiction of cellular signaling and metabolic processes.
Table 3.
Computational-based approaches for studying PLI
| Large-scale analysis | Molecular mechanism | ||
|---|---|---|---|
| Data required | High-throughput PLI screening (protein or lipid microarrays). | Structural and kinetic details of PLI (NMR, FRET, etc.). | |
| Objective | Systematic catalog and analysis of PLIs in an organism/phenotype. | Prediction of PLI, PLI-interface, and dynamics. Exploring the molecular mechanism of PLI. | |
| Techniques | Sequence comparison; statistical analysis; network analysis/ inference [64]. | Machine learning (classification) [93–95]. | Molecular dynamic simulation [96–102]. |
| Usage | Estimation of the accuracy, sensitivity, and bias of high-throughput screening; inference of PLI function [64]. | Prediction of binding interface/motif based on sequence [94,95] | Study the structural basis of PLI [102]; explore functional specificity of PLI [100]. |
| Future perspective | Incorporation of PPI and PDI to achieve comprehensive understanding of the functional mechanisms of PLIs. | Multi-scale approaches that model and combine different levels of molecular information to achieve better understanding of the molecular mechanisms of PLI. | |
Acknowledgments
We would like to acknowledge support from the NIH (R01GM079688 and R01GM089866), and the NSF (CBET 0941055).
Abbreviations
- AFM
atomic force microscopy
- CG
coarse grained
- ESR
electronic spin resonance spectroscopy
- FRET
fluorescence resonance energy transfer
- ITC
isothermal titration calorimetry
- LBD
lipid-binding domain
- MD
molecular dynamic
- PDI
protein-DNA interactions
- PLI
protein–lipid interactions
- PPI
protein–protein interactions
- SPR
surface plasmon resonance
- SPRi
surface plasmon resonance imaging
Footnotes
The authors have declared no conflict of interest.
6 References
- [1].Charbonnier S, Gallego O, Gavin AC. The social network of a cell: recent advances in interactome mapping. Biotechnol. Annu. Rev. 2008;14:1–28. doi: 10.1016/S1387-2656(08)00001-X. [DOI] [PubMed] [Google Scholar]
- [2].Williamson MP, Sutcliffe MJ. Protein-protein interactions. Biochem. Soc. Trans. 2010;38:875–878. doi: 10.1042/BST0380875. [DOI] [PubMed] [Google Scholar]
- [3].Sardiu ME, Washburn MP. Building protein-protein interaction networks with proteomics and informatics tools. J. Biol. Chem. 2011;286:23645–23651. doi: 10.1074/jbc.R110.174052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Xie Z, Hu S, Qian J, Blackshaw S, Zhu H. Systematic characterization of protein-DNA interactions. Cell Mol. Life Sci. 2011;68:1657–1668. doi: 10.1007/s00018-010-0617-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Bulyk ML. Protein binding microarrays for the characterization of DNA-protein interactions. Adv. Biochem. Eng. Biotechnol. 2007;104:65–85. doi: 10.1007/10_025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Wu J, Smith LT, Plass C, Huang TH. ChIP-chip comes of age for genome-wide functional analysis. Cancer Res. 2006;66:6899–6902. doi: 10.1158/0008-5472.CAN-06-0276. [DOI] [PubMed] [Google Scholar]
- [7].Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- [8].Mardis ER. ChIP-seq: welcome to the new frontier. Nat. Methods. 2007;4:613–614. doi: 10.1038/nmeth0807-613. [DOI] [PubMed] [Google Scholar]
- [9].Wenk MR. Lipidomics: new tools and applications. Cell. 2010;143:888–895. doi: 10.1016/j.cell.2010.11.033. [DOI] [PubMed] [Google Scholar]
- [10].Brown PH, Balbo A, Schuck P. Characterizing protein-protein interactions by sedimentation velocity analytical ultracentrifugation. Curr. Protoc. Immunol. 2008;Chapter 18(Unit 18.15.1–18.15.39) doi: 10.1002/0471142735.im1815s81. [DOI] [PubMed] [Google Scholar]
- [11].Ford MG, Pearse BM, Higgins MK, Vallis Y, et al. Simultaneous binding of PtdIns(4,5)P2 and clathrin by AP180 in the nucleation of clathrin lattices on membranes. Science. 2001;291:1051–1055. doi: 10.1126/science.291.5506.1051. [DOI] [PubMed] [Google Scholar]
- [12].Gorman PM, Kim S, Guo M, Melnyk RA, et al. Dimerization of the transmembrane domain of amyloid precursor proteins and familial Alzheimer's disease mutants. BMC Neurosci. 2008;9:17. doi: 10.1186/1471-2202-9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Tsujita K, Suetsugu S, Sasaki N, Furutani M, et al. Coordination between the actin cytoskeleton and membrane deformation by a novel membrane tubulation domain of PCH proteins is involved in endocytosis. J. Cell Biol. 2006;172:269–279. doi: 10.1083/jcb.200508091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ishii M, Fujita S, Yamada M, Hosaka Y, Kurachi Y. Phosphatidylinositol 3,4,5-trisphosphate and Ca2+/calmodulin competitively bind to the regulators of G-protein-signalling (RGS) domain of RGS4 and reciprocally regulate its action. Biochem. J. 2005;385:65–73. doi: 10.1042/BJ20040404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lee SH, Jin JB, Song J, Min MK, et al. The intermolecular interaction between the PH domain and the C-terminal domain of Arabidopsis dynamin-like 6 determines lipid binding specificity. J. Biol. Chem. 2002;277:31842–31849. doi: 10.1074/jbc.M204770200. [DOI] [PubMed] [Google Scholar]
- [16].Dalton AK, Murray PS, Murray D, Vogt VM. Biochemical characterization of rous sarcoma virus MA protein interaction with membranes. J. Virol. 2005;79:6227–6238. doi: 10.1128/JVI.79.10.6227-6238.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Tsujita K, Itoh T, Kondo A, Oyama M, et al. Proteome of acidic phospholipid-binding proteins: spatial and temporal regulation of Coronin 1A by phosphoinositides. J. Biol. Chem. 2010;285:6781–6789. doi: 10.1074/jbc.M109.057018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Schroit AJ, Madsen J, Ruoho AE. Radioiodinated, photoactivatable phosphatidylcholine and phosphatidylserine: transfer properties and differential photoreactive interaction with human erythrocyte membrane proteins. Biochemistry. 1987;26:1812–1819. doi: 10.1021/bi00381a004. [DOI] [PubMed] [Google Scholar]
- [19].Benfenati F, Greengard P, Brunner J, Bähler M. Electrostatic and hydrophobic interactions of synapsin I and synapsin I fragments with phospholipid bilayers. J. Cell Biol. 1989;108:1851–1862. doi: 10.1083/jcb.108.5.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Haberkant P, van Meer G. Protein-lipid interactions: paparazzi hunting for snap-shots. Biol. Chem. 2009;390:795–803. doi: 10.1515/BC.2009.074. [DOI] [PubMed] [Google Scholar]
- [21].Gubbens J, de Kroon AI. Proteome-wide detection of phospholipid-protein interactions in mitochondria by photo cross-linking and click chemistry. Mol. Biosyst. 2010;6:1751–1759. doi: 10.1039/c003064n. [DOI] [PubMed] [Google Scholar]
- [22].Wiseman T, Williston S, Brandts JF, Lin LN. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal. Biochem. 1989;179:131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- [23].Hughes E, Clayton JC, Middleton DA. Cytoplasmic residues of phospholamban interact with membrane surfaces in the presence of SERCA: a new role for phospholipids in the regulation of cardiac calcium cycling? Biochim. Biophys. Acta. 2009;1788:559–566. doi: 10.1016/j.bbamem.2008.10.029. [DOI] [PubMed] [Google Scholar]
- [24].Hoernke M, Schwieger C, Kerth A, Blume A. Binding of cationic pentapeptides with modified side chain lengths to negatively charged lipid membranes: complex interplay of electrostatic and hydrophobic interactions. Biochim. Biophys. Acta. 2012;1818:1663–1672. doi: 10.1016/j.bbamem.2012.03.001. [DOI] [PubMed] [Google Scholar]
- [25].Bhunia A, Mohanram H, Bhattacharjya S. Structural determinants of the specificity of a membrane binding domain of the scaffold protein Ste5 of budding yeast: implications in signaling by the scaffold protein in MAPK pathway. Biochim. Biophys. Acta. 2012;1818:1250–1260. doi: 10.1016/j.bbamem.2012.01.008. [DOI] [PubMed] [Google Scholar]
- [26].Hundertmark M, Dimova R, Lengefeld J, Seckler R, Hincha DK. The intrinsically disordered late embryogenesis abundant protein LEA18 from Arabidopsis thaliana modulates membrane stability through binding and folding. Biochim. Biophys. Acta. 2011;1808:446–453. doi: 10.1016/j.bbamem.2010.09.010. [DOI] [PubMed] [Google Scholar]
- [27].Marsh D, Horváth LI. Structure, dynamics and composition of the lipid-protein interface. Perspectives from spin-labelling. Biochim. Biophys. Acta. 1998;1376:267–296. doi: 10.1016/s0304-4157(98)00009-4. [DOI] [PubMed] [Google Scholar]
- [28].Marsh D. Electron spin resonance in membrane research: protein-lipid interactions from challenging beginnings to state of the art. Eur. Biophys. J. 2010;39:513–525. doi: 10.1007/s00249-009-0512-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].D'Errico G, Ercole C, Lista M, Pizzo E, et al. Enforcing the positive charge of N-termini enhances membrane interaction and antitumor activity of bovine seminal ribonuclease. Biochim. Biophys. Acta. 2011;1808:3007–3015. doi: 10.1016/j.bbamem.2011.08.009. [DOI] [PubMed] [Google Scholar]
- [30].Vitiello G, Grimaldi M, Ramunno A, Ortona O. Interaction of a beta-sheet breaker peptide with lipid membranes. J. Pept. Sci. 2010;16:115–122. doi: 10.1002/psc.1207. [DOI] [PubMed] [Google Scholar]
- [31].Mateo CR, Gómez J, Villalaín J, Gonzálaz Ros JM, editors. Protein-lipid interactions. Springer; New York: 2005. pp. 1–33. [Google Scholar]
- [32].Romoser V, Ball R, Smrcka AV. Phospholipase C beta2 association with phospholipid interfaces assessed by fluorescence resonance energy transfer. G protein betagamma subunit-mediated translocation is not required for enzyme activation. J. Biol. Chem. 1996;271:25071–25078. doi: 10.1074/jbc.271.41.25071. [DOI] [PubMed] [Google Scholar]
- [33].Nomikos M, Mulgrew-Nesbitt A, Pallavi P, Mihalyne G, et al. Binding of phosphoinositide-specific phospholipase C-zeta (PLC-zeta) to phospholipid membranes: potential role of an unstructured cluster of basic residues. J. Biol. Chem. 2007;282:16644–16653. doi: 10.1074/jbc.M701072200. [DOI] [PubMed] [Google Scholar]
- [34].Radhakrishnan A, Stein A, Jahn R, Fasshauer D. The Ca2+ affinity of synaptotagmin 1 is markedly increased by a specific interaction of its C2B domain with phosphatidylinositol 4,5-bisphosphate. J. Biol. Chem. 2009;284:25749–25760. doi: 10.1074/jbc.M109.042499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Gilbert GE, Furie BC, Furie B. Binding of human factor VIII to phospholipid vesicles. J. Biol. Chem. 1990;265:815–822. [PubMed] [Google Scholar]
- [36].Pap EH, Bastiaens PI, Borst JW, van den Berg PA, et al. Quantitation of the interaction of protein kinase C with diacylglycerol and phosphoinositides by time-resolved detection of resonance energy transfer. Biochemistry. 1993;32:13310–13317. doi: 10.1021/bi00211a044. [DOI] [PubMed] [Google Scholar]
- [37].Petrescu AD, Gallegos AM, Okamura Y, Strauss JF, Schroeder F. Steroidogenic acute regulatory protein binds cholesterol and modulates mitochondrial membrane sterol domain dynamics. J. Biol. Chem. 2001;276:36970–36982. doi: 10.1074/jbc.M101939200. [DOI] [PubMed] [Google Scholar]
- [38].Gray A, Olsson H, Batty IH, Priganica L, Peter Downes C. Nonradioactive methods for the assay of phosphoinositide 3-kinases and phosphoinositide phosphatases and selective detection of signaling lipids in cell and tissue extracts. Anal. Biochem. 2003;313:234–245. doi: 10.1016/s0003-2697(02)00607-3. [DOI] [PubMed] [Google Scholar]
- [39].Dixon MJ, Gray A, Schenning M, Agacan M, et al. IQGAP proteins reveal an atypical phosphoinositide (aPI) binding domain with a pseudo C2 domain fold. J. Biol. Chem. 2012;287:22483–22496. doi: 10.1074/jbc.M112.352773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Bazin H, Trinquet E, Mathis G. Time resolved amplification of cryptate emission: a versatile technology to trace biomolecular interactions. J. Biotechnol. 2002;82:233–250. doi: 10.1016/s1389-0352(01)00040-x. [DOI] [PubMed] [Google Scholar]
- [41].Zigoneanu IG, Yang YJ, Krois AS, Haque E, Pielak GJ. Interaction of α-synuclein with vesicles that mimic mitochondrial membranes. Biochim. Biophys. Acta. 2012;1818:512–519. doi: 10.1016/j.bbamem.2011.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Kobashigawa Y, Harada K, Yoshida N, Ogura K, Inagaki F. Phosphoinositide-incorporated lipid-protein nanodiscs: a tool for studying protein-lipid interactions. Anal. Biochem. 2011;410:77–83. doi: 10.1016/j.ab.2010.11.021. [DOI] [PubMed] [Google Scholar]
- [43].Qiu L, Lewis A, Como J, Vaughn MW, et al. Cholesterol modulates the interaction of beta-amyloid peptide with lipid bilayers. Biophys. J. 2009;96:4299–4307. doi: 10.1016/j.bpj.2009.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Mustafa AK, van Rossum DB, Patterson RL, Maag D, et al. Glutamatergic regulation of serine racemase via reversal of PIP2 inhibition. Proc. Natl. Acad. Sci. USA. 2009;106:2921–2926. doi: 10.1073/pnas.0813105106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Guillén J, González-Alvarez A, Villalaín J. A membranotropic region in the C-terminal domain of hepatitis C virus protein NS4B interaction with membranes. Biochim. Biophys. Acta. 2010;1798:327–337. doi: 10.1016/j.bbamem.2009.07.011. [DOI] [PubMed] [Google Scholar]
- [46].Sanchez SA, Bagatolli LA, Gratton E, Hazlett TL. A two-photon view of an enzyme at work: Crotalus atrox venom PLA2 interaction with single-lipid and mixed-lipid giant unilamellar vesicles. Biophys. J. 2002;82:2232–2243. doi: 10.1016/S0006-3495(02)75569-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Melo AM, Prieto M, Coutinho A. The effect of variable liposome brightness on quantifying lipid-protein interactions using fluorescence correlation spectroscopy. Biochim. Biophys. Acta. 2011;1808:2559–2568. doi: 10.1016/j.bbamem.2011.06.001. [DOI] [PubMed] [Google Scholar]
- [48].Temmerman K, Nickel W. A novel flow cytometric assay to quantify interactions between proteins and membrane lipids. J. Lipid. Res. 2009;50:1245–1254. doi: 10.1194/jlr.D800043-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Alegre-Cebollada J, Cunietti M, Herrero-Gálan E, Gavi-lanes JG, Martínez-del-Pozo A. Calorimetric scrutiny of lipid binding by sticholysin II toxin mutants. J. Mol. Biol. 2008;382:920–930. doi: 10.1016/j.jmb.2008.07.053. [DOI] [PubMed] [Google Scholar]
- [50].Shao C, Novakovic VA, Head JF, Seaton BA, Gilbert GE. Crystal structure of lactadherin C2 domain at 1.7A resolution with mutational and computational analyses of its membrane-binding motif. J. Biol. Chem. 2008;283:7230–7241. doi: 10.1074/jbc.M705195200. [DOI] [PubMed] [Google Scholar]
- [51].Erlmann P, Schmid S, Horenkamp FA, Geyer M, et al. DLC1 activation requires lipid interaction through a polybasic region preceding the RhoGAP domain. Mol. Biol. Cell. 2009;20:4400–4411. doi: 10.1091/mbc.E09-03-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Jobichen C, Fernandis AZ, Velazquez-Campoy A, Leung KY, et al. Identification and characterization of the lipid-binding property of GrlR, a locus of enterocyte effacement regulator. Biochem. J. 2009;420:191–199. doi: 10.1042/BJ20081588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Marsh D. Orientation and peptide-lipid interactions of alamethicin incorporated in phospholipid membranes: polarized infrared and spin-label EPR spectroscopy. Biochemistry. 2009;48:729–737. doi: 10.1021/bi801279n. [DOI] [PubMed] [Google Scholar]
- [54].Hughes E, Whittaker CA, Barsukov IL, Esmann M, Middleton DA. A study of the membrane association and regulatory effect of the phospholemman cytoplasmic domain. Biochim. Biophys. Acta. 2011;1808:1021–1031. doi: 10.1016/j.bbamem.2010.11.024. [DOI] [PubMed] [Google Scholar]
- [55].Krishnakumari V, Nagaraj R. Binding of peptides corresponding to the carboxy-terminal region of human-β-defensins-1–3 with model membranes investigated by isothermal titration calorimetry. Biochim. Biophys. Acta. 2012;1818:1386–1394. doi: 10.1016/j.bbamem.2012.02.016. [DOI] [PubMed] [Google Scholar]
- [56].Zhu H, Bilgin M, Bangham R, Hall D, et al. Global analysis of protein activities using proteome chips. Science. 2001;293:2101–2105. doi: 10.1126/science.1062191. [DOI] [PubMed] [Google Scholar]
- [57].Feng L. Probing lipid-protein interactions using lipid microarrays. Prostaglandins Other Lipid Mediat. 2005;77:158–167. doi: 10.1016/j.prostaglandins.2004.09.003. [DOI] [PubMed] [Google Scholar]
- [58].Borch J, Torta F, Sligar SG, Roepstorff P. Nanodiscs for immobilization of lipid bilayers and membrane receptors: kinetic analysis of cholera toxin binding to a glycolipid receptor. Anal. Chem. 2008;80:6245–6252. doi: 10.1021/ac8000644. [DOI] [PubMed] [Google Scholar]
- [59].Bally M, Bailey K, Sugihara K, Grieshaber D, et al. Liposome and lipid bilayer arrays towards biosensing applications. Small. 2010;6:2481–2497. doi: 10.1002/smll.201000644. [DOI] [PubMed] [Google Scholar]
- [60].Christensen SM, Stamou D. Biomimetic Membranes for Sensor and Separation Applications. Springer; New York: 2012. pp. 87–112. [Google Scholar]
- [61].Stevenson JM, Perera IY, Boss WF. A phosphatidylinositol 4-kinase pleckstrin homology domain that binds phosphatidylinositol 4-monophosphate. J. Biol. Chem. 1998;273:22761–22767. doi: 10.1074/jbc.273.35.22761. [DOI] [PubMed] [Google Scholar]
- [62].Dowler S, Kular G, Alessi DR. Protein lipid overlay assay. Sci. STKE. 2002;2002:l6. doi: 10.1126/stke.2002.129.pl6. [DOI] [PubMed] [Google Scholar]
- [63].Kanter JL, Narayana S, Ho PP, Catz I, et al. Lipid microarrays identify key mediators of autoimmune brain inflammation. Nat. Med. 2006;12:138–143. doi: 10.1038/nm1344. [DOI] [PubMed] [Google Scholar]
- [64].Gallego O, Betts MJ, Gvozdenovic-Jeremic J, Maeda K, et al. A systematic screen for protein-lipid interactions in Saccharomyces cerevisiae. Mol. Syst. Biol. 2010;6:430. doi: 10.1038/msb.2010.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Linman MJ, Culver SP, Cheng Q. Fabrication of fracture-free nanoglassified substrates by layer-by-layer deposition with a paint gun technique for real-time monitoring of protein-lipid interactions. Langmuir. 2009;25:3075–3082. doi: 10.1021/la803835a. [DOI] [PubMed] [Google Scholar]
- [66].Linman MJ, Abbas A, Roberts CC, Cheng Q. Etched glass microarrays with differential resonance for enhanced contrast and sensitivity of surface plasmon resonance imaging analysis. Anal. Chem. 2011;83:5936–5943. doi: 10.1021/ac200881q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Phillips KS, Cheng Q. Microfluidic immunoassay for bacterial toxins with supported phospholipid bilayer membranes on poly(dimethylsiloxane) Anal. Chem. 2005;77:327–334. doi: 10.1021/ac049356+. [DOI] [PubMed] [Google Scholar]
- [68].Wang Z, Wilkop T, Han JH, Dong Y, et al. Development of air-stable, supported membrane arrays with photolithography for study of phosphoinositide-protein interactions using surface plasmon resonance imaging. Anal. Chem. 2008;80:6397–6404. doi: 10.1021/ac800845w. [DOI] [PubMed] [Google Scholar]
- [69].Deng Y, Wang Y, Holtz B, Li J, et al. Fluidic and air-stable supported lipid bilayer and cell-mimicking microarrays. J. Am. Chem. Soc. 2008;130:6267–6271. doi: 10.1021/ja800049f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Taylor JD, Phillips KS, Cheng Q. Microfluidic fabrication of addressable tethered lipid bilayer arrays and optimization using SPR with silane-derivatized nanoglassy substrates. Lab. Chip. 2007;7:927–930. doi: 10.1039/b618940g. [DOI] [PubMed] [Google Scholar]
- [71].Losey EA, Smith MD, Meng M, Best MD. Microplate-based analysis of protein-membrane binding interactions via immobilization of whole liposomes containing a biotinylated anchor. Bioconjug. Chem. 2009;20:376–383. doi: 10.1021/bc800414k. [DOI] [PubMed] [Google Scholar]
- [72].Chandra H, Reddy PJ, Srivastava S. Protein microarrays and novel detection platforms. Expert Rev. Proteomics. 2011;8:61–79. doi: 10.1586/epr.10.99. [DOI] [PubMed] [Google Scholar]
- [73].Ray S, Mehta G, Srivastava S. Label-free detection techniques for protein microarrays: prospects, merits and challenges. Proteomics. 2010;10:731–748. doi: 10.1002/pmic.200900458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Moran-Mirabal JM, Edel JB, Meyer GD, Throckmorton D, et al. Micrometer-sized supported lipid bilayer arrays for bacterial toxin binding studies through total internal reflection fluorescence microscopy. Biophys. J. 2005;89:296–305. doi: 10.1529/biophysj.104.054346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Smith KA, Gale BK, Conboy JC. Micropatterned fluid lipid bilayer arrays created using a continuous flow microspotter. Anal. Chem. 2008;80:7980–7987. doi: 10.1021/ac800860u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Joubert JR, Smith KA, Johnson E, Keogh JP, et al. Stable, ligand-doped, poly(bis-SorbPC) lipid bilayer arrays for protein binding and detection. ACS Appl. Mater. Interfaces. 2009;1:1310–1315. doi: 10.1021/am900177p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Rowland MM, Gong D, Bostic HE, Lucas N. et al. Microarray analysis of Akt PH domain binding employing synthetic biotinylated analogs of all seven phosphoinositide headgroup isomers. Chem. Phys. Lipids. 2012;165:207–215. doi: 10.1016/j.chemphyslip.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Consonni SV, Gloerich M, Spanjaard E, Bos JL. cAMP regulates DEP domain-mediated binding of the guanine nucleotide exchange factor Epac1 to phosphatidic acid at the plasma membrane. Proc. Natl. Acad. Sci. USA. 2012;109:3814–3819. doi: 10.1073/pnas.1117599109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Gong D, Smith MD, Manna D, Bostic HE, et al. Microplate-based characterization of protein-phosphoinositide binding interactions using a synthetic biotinylated headgroup analogue. Bioconjug. Chem. 2009;20:310–316. doi: 10.1021/bc8004107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Phillips KS, Han JH, Martinez M, Wang Z, et al. Nanoscale glassification of gold substrates for surface plasmon resonance analysis of protein toxins with supported lipid membranes. Anal. Chem. 2006;78:596–603. doi: 10.1021/ac051644y. [DOI] [PubMed] [Google Scholar]
- [81].Liu Y, Cheng Q. Detection of membrane-binding proteins by surface plasmon resonance with an all-aqueous amplification scheme. Anal. Chem. 2012;84:3179–3186. doi: 10.1021/ac203142n. [DOI] [PubMed] [Google Scholar]
- [82].Junghans A, Champagne C, Cayot P, Loupiac C, Köper I. Probing protein-membrane interactions using solid supported membranes. Langmuir. 2011;27:2709–2716. doi: 10.1021/la103200k. [DOI] [PubMed] [Google Scholar]
- [83].Terrettaz S, Stora Y, Duschl C, Vogel H. Protein binding to supported lipid membranes: investigation of the cholera toxin-ganglioside interaction by simultaneous impedance spectroscopy and surface plasmon resonance. Langmuir. 1993;9:1361–1369. [Google Scholar]
- [84].Das A, Base C, Manna D, Cho W, Dubreuil RR. Unexpected complexity in the mechanisms that target assembly of the spectrin cytoskeleton. J. Biol. Chem. 2008;283:12643–12653. doi: 10.1074/jbc.M800094200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Gheorghiu M, Olaru A, Tar A, Polonschii C, Gheorghiu E. Sensing based on assessment of non-monotonous effect determined by target analyte: case study on pore-forming compounds. Biosens. Bioelectron. 2009;24:3517–3523. doi: 10.1016/j.bios.2009.05.007. [DOI] [PubMed] [Google Scholar]
- [86].Phillips KS, Wilkop T, Wu JJ, Al-Kaysi RO, Cheng Q. Surface plasmon resonance imaging analysis of protein-receptor binding in supported membrane arrays on gold substrates with calcinated silicate films. J. Am. Chem. Soc. 2006;128:9590–9591. doi: 10.1021/ja0628102. [DOI] [PubMed] [Google Scholar]
- [87].Taylor JD, Linman MJ, Wilkop T, Cheng Q. Regenerable tethered bilayer lipid membrane arrays for multiplexed label-free analysis of lipid-protein interactions on poly(dimethylsiloxane) microchips using SPR imaging. Anal. Chem. 2009;81:1146–1153. doi: 10.1021/ac8023137. [DOI] [PubMed] [Google Scholar]
- [88].Harkewicz R, Dennis EA. Applications of mass spectrometry to lipids and membranes. Annu. Rev. Biochem. 2011;80:301–325. doi: 10.1146/annurev-biochem-060409-092612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].D'Santos C, Lewis EA. Integrative Proteomics. InTech; Croatia: 2012. pp. 363–378. [Google Scholar]
- [90].Giannakis E, Pacífico J, Smith DP, Hung LW, et al. Dimeric structures of alpha-synuclein bind preferentially to lipid membranes. Biochim. Biophys. Acta. 2008;1778:1112–1119. doi: 10.1016/j.bbamem.2008.01.012. [DOI] [PubMed] [Google Scholar]
- [91].Park JH, Kwon EY, Jung HI, Kim DE. Direct force measurement of the interaction between liposome and the C2A domain of synaptotagmin I using atomic force microscopy. Biotechnol. Lett. 2006;28:505–509. doi: 10.1007/s10529-006-0010-y. [DOI] [PubMed] [Google Scholar]
- [92].Baksh MM, Kussrow AK, Mileni M, Finn MG, Born-hop DJ. Label-free quantification of membrane-ligand interactions using backscattering interferometry. Nat. Biotechnol. 2011;29:357–360. doi: 10.1038/nbt.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Wang L, Brown SJ. BindN: a web-based tool for efficient prediction of DNA and RNA binding sites in amino acid sequences. Nucleic. Acids Res. 2006;34:W243–W248. doi: 10.1093/nar/gkl298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Irausquin SJ, Wang L. Machine learning approach for prediction of lipid-interacting residues in amino acid sequences. IEEE. 2007:315–319. [Google Scholar]
- [95].Xiong W, Guo Y, Li M. Prediction of lipid-binding sites based on support vector machine and position specific scoring matrix. Protein J. 2010;29:427–431. doi: 10.1007/s10930-010-9269-x. [DOI] [PubMed] [Google Scholar]
- [96].Ash WL, Zlomislic MR, Oloo EO, Tieleman DP. Computer simulations of membrane proteins. Biochim. Biophys. Acta. 2004;1666:158–189. doi: 10.1016/j.bbamem.2004.04.012. [DOI] [PubMed] [Google Scholar]
- [97].Lomize AL, Pogozheva ID, Lomize MA, Mosberg HI. Positioning of proteins in membranes: a computational approach. Protein Sci. 2006;15:1318–1333. doi: 10.1110/ps.062126106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Shih AY, Arkhipov A, Freddolino PL, Schulten K. Coarse grained protein-lipid model with application to lipoprotein particles. J. Phys. Chem. B. 2006;110:3674–3684. doi: 10.1021/jp0550816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Scott KA, Bond PJ, Ivetac A, Chetwynd AP, et al. Coarse-grained MD simulations of membrane protein-bilayer self-assembly. Structure. 2008;16:621–630. doi: 10.1016/j.str.2008.01.014. [DOI] [PubMed] [Google Scholar]
- [100].Sansom MS, Scott KA, Bond PJ. Coarse-grained simulation: a high-throughput computational approach to membrane proteins. Biochem. Soc. Trans. 2008;36:27–32. doi: 10.1042/BST0360027. [DOI] [PubMed] [Google Scholar]
- [101].Spijker P, van Hoof B, Debertrand M, Markvoort AJ, et al. Coarse grained molecular dynamics simulations of transmembrane protein-lipid systems. Int. J. Mol. Sci. 2010;11:2393–2420. doi: 10.3390/ijms11062393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Chetwynd AP, Scott KA, Mokrab Y, Sansom MS. CGDB: a database of membrane protein/lipid interactions by coarse-grained molecular dynamics simulations. Mol. Membr. Biol. 2008;25:662–669. doi: 10.1080/09687680802446534. [DOI] [PubMed] [Google Scholar]