Abstract
Interferon regulatory factor (IRF) 4 is a hematopoietic cell-specific transcription factor that regulates the maturation and differentiation of immune cells. Using an inducible expression system, we found that IRF4 directly induced a specific subset of interferon-stimulated genes (ISG) in a type I interferon (IFN)-independent manner in both epithelial and B cell lines. Moreover, Kaposi sarcoma-associated herpesvirus (KSHV)-encoded viral FLICE inhibitory protein (vFLIP) enhances IRF4-mediated gene induction. Co-expression of IRF4 with vFLIP significantly increased ISG60 (IFIT3) and Cig5 (RSAD2) transcription that was dependent on the ability of vFLIP to activate NF-κB. A vFLIP mutant (A57L) – defective in NF-κB activation, failed to enhance IRF4-mediated ISG induction. Thus, we provide a physiologically relevant mechanism where viral protein mediated NF-κB activation modulates specific ISG induction by IRF4. In contrast, IRF4 also acted as a negative regulator of KSHV replication and transcription activator (RTA) expression after induction of KSHV lytic reactivation in KSHV positive primary effusion lymphoma (PEL) cells. Taken together, these results suggest a dual role for IRF4 in regulating ISG induction and KSHV lytic reactivation in PEL cells.
INTRODUCTION
The interferon regulatory factor (IRF) family of transcription factors are mainly involved in the regulation of innate immune response genes, type I interferons (IFN), and the maturation of immune cells (1, 2). IRF4, a member of the IRF family, is required for proper maturation and differentiation of immune cells (3); as well as acts as both positive (4, 5) and negative (6, 7) regulator of gene transcription. IRF4 was first identified in multiple myeloma cells, where its overexpression caused deregulation of cell cycle regulatory proteins (8, 9), highlighting the diverse functions of IRF4 in regulation of transcription and the importance of balanced IRF4 activity in maintaining homeostasis. IRF4 has also been found to have transformation potential that contributes to several lymophoproliferative diseases (10, 11). It is overexpressed in human T-lymphotropic virus 1 (HTLV-1) infected adult T-cell leukemia (ATL) cells and contributes to their transformed phenotype (12, 13). High IRF4 levels are associated to the transformation of B cells by Epstein-Barr Virus (EBV) LMP1 oncoprotein, resulting in increased cellular growth and proliferation (14, 15). However, in primary effusion lymphoma (PEL), a Kaposi’s sarcoma-associated herpesvirus (KSHV, also called human herpesvirus 8)-associated B cell neoplasm (16, 17), the role of IRF4 has not been defined.
PEL most commonly occurs amongst immunocompromised individuals (16, 17). It has an immunoblastic or plasmablastic appearance and is both IRF4- and CD138-positive (10, 18). PEL cells are characterized by latent infection with KSHV (19), where the virus persists in cells as a naked episome and express only a limited subset viral genes (latent genes) (20–23). These include genes encoding viral FLICE inhibitory protein (vFLIP), viral cyclin (vCYC), latency-associated nuclear antigen LANA, LANA2 (also known as vIRF3), and miRNA encoding genes (24), which modulate antiviral immune responses through various mechanisms. The transition from latency to lytic replication is controlled by the KSHV replication transactivator (RTA) protein which initiates viral lytic gene transcription, leading to virion formation, and death of the host cell.
The vFLIP protein, encoded by the KSHV gene K13/ORF71, was first identified as a viral FLICE-inhibitory protein (25) and led to the subsequent discovery of cellular FLIP proteins (26). More recent studies reveal that the primary function of vFLIP is activation of NF-κB through interactions with IκB Kinase (IKK) complex (27, 28). Constitutive activation of NF-κB by vFLIP is required for Rat-1 cell transformation (29), lymphomagenesis in transgenic mice (30), and survival of PEL cells (31). Furthermore, vFLIP suppresses full lytic viral gene expression through an NF-κB targeting mechanism that is essential for the maintenance of viral latency in PEL (32, 33).
Here, using an inducible IRF4 expression system, we examined the role of IRF4 as a regulator of ISG induction. Our results suggest that IRF4 directly targets ISG60 and Cig5 to positively regulate their expression. IRF4 mediated ISG induction was enhanced by KSHV vFLIP in an NF-κB dependent manner, highlighting the importance of NF-κB on the transcriptional regulation of ISGs. In contrast, we observed a negative regulatory effect of IRF4 on KSHV RTA-mediated transcription and lytic gene expression following viral reactivation. Taken together, these results show that IRF4 plays an important role in shaping innate immune responses in PEL cells and may be essential for maintaining KSHV latency in PEL.
MATERIALS AND METHODS
Cells and reagents
HEK293 cells, 293T, and HEK293 derived cell lines were cultured in DMEM (Lonza) containing 10% fetal bovine serum (Atlanta Biologicals) and 100 I.U./ml penicillin and 100 μg/ml streptomycin (Lonza). BCBL-1, BC-1 and BCP-1, and BJAB cells were cultured in RPMI medium supplemented with 10 to 20% fetal bovine serum. 293iIRF4 cells were transfected with Fugene-6 (Roche) following manufacturer’s protocol. Cells were stimulated for 48 h with varying doses of doxycycline (Dox, Clontech). TNFα was from PreproTech and 12-O-tetradecanoylphorbol-13-acetate (TPA) was from Sigma–Aldrich. The following primary antibodies were used in this study: anti-IRF4 (Cell Signaling), anti-V5 (Invitrogen), anti-RSAD2, anti-Actin, and anti-Tubulin (Santa Cruz), anti-OASL (Abgent), anti- ISG60 and anti-DRBP76 (34), anti-LANA2 (CM-A807), anti-LANA (35), and anti-ORF50 (36).
Plasmids
IRF4, transcript variant 1 (NM_002460), was PCR amplified with N-Terminal V5 tag from pCMV6-IRF4 (Origene). The PCR product was then cloned into pENTR-D/TOPO (Invitrogen) following manufacturer’s guidelines. The expression vector pcDNA/IRF4-V5 was generated by recombination between pENTR/D-TOPO IRF4-V5 and pcDNA/DEST47 using Gateway LR Clonase II enzyme mix (Invitrogen) according to manufacturer’s guidelines. Phosphomimetic mutant of IRF4 S446D was generated by sire-directed mutagenesis of pENTR/D-TOPO IRF4-V5 using QuikChange II site-directed mutagenesis kit (Stratagene) following manufacturer’s protocols using primers: 5′-CCACAGATCTATCCGCCATGACTCTATTCAAGAATGACTC-3′ and 5′-GAGTCATTCTTGAATAGAGTCATGGCGGATAGATCTGTGG-3′. pcDNA/K13-HA was generated by PCR amplification of vFLIP cDNA with a C-terminal HA tag from pMSCV/K13 and cloned into pENTR/D-TOPO (Invitrogen). K13-HA was then subcloned into the EcoRI-XbaI sites in pcDNA3.1(+)/Hygro (Invitrogen). vFLIP mutant, A57L, was generated from pcDNA/K13-HA using the previously described method and the following primers: 5′-CGTTT CCCCTGTTACTGGAATGTCTGTTTCGTG-3′ and 5′-CACGAAACAGACATTCCAGTAACA GGGGAAACG-3′. pcDNA/LANA, pcDNA/LANA2, pcDNA/ORF50 have been previously described (37–39). The IκB-super repressor mutant S32A/S36A has been previously described (40). The reporter plasmids pGl3-Nut-1 and NF-κB firefly luciferase have been previously described (41, 42). pRL-Null vector expressing Renilla luciferase was obtained from Promega.
Lentiviral vectors
Doxycycline inducible lentiviral vectors were generated by performing LR recombination between pENTR/D-TOPO IRF4-V5 or pENTR/D-TOPO IRF4-S446D-V5 with pInducer 20 destination vector (43). Constitutive IRF4 expressing lentiviral vector was generated by LR recombination of pENTR/IRF4-V5 with pLenti CMV Puro DEST (W118-1) (Addgene). Control pInducer20 and pLenti CMV Puro vectors were generated by recombination with pENTR-V5 plasmid (Addgene). Lentiviruses were packaged in 293T and pseudotyped with VSV G protein. Transduction of HEK293, BJAB, and BCBL-1 cells were carried out for 1–5 h at 37°C in the presence of 1μg/ml polybrene. Cells were selected with 500ng/ml G418 or 1μg/ml Puromycin to establish stable cell lines.
Reporter assays
293T (1.5 × 105 cells/well) in 24-well plate were transfected using Fugene 6 as indicated. Twenty four hours later, the cells from each well were collected by trypsin-EDTA digestion and seeded into 6 wells in 96-well plate. Forty eight hours post transfection, luciferase activity was measured using the Dual-Glo luciferase assay system (Promega). Firefly luciferase activity was normalized to Renilla luciferase activity and expressed as fold changes as indicated.
Quantitative PCR analysis of gene expression
Total RNA was extracted using Trizol reagent (Invitrogen) and treated with DNase I at 37°C for 1hr (DNA Free kit, Ambion). 1μg total RNA was used for reverse transcription using iScript cDNA synthesis kit (Bio-Rad) and subjected to real-time PCR using a CFX96 real time system (Bio-Rad) according to manufacturer’s instructions. Primers used for target gene amplification can be found in Supplementary table 1. Samples were normalized to RPL32 and expressed as fold change with respect to untreated vector control cells (value 1), marked with (#).
Chromatin immunoprecipitation (ChIP) assay
ChIP was performed using the ChIP-IT Express kit from Active Motif according to the manufacturer’s protocols. Briefly, 1 × 107 HEK293 cells stably expressing pLenti vector control or pLenti/IRF4-V5 were crosslinked with 1% formaldehyde for 10 min. Cells were then lysed and chromatin was sheared into 200–600 bp fragments by sonication. The cross-linked chromatin was incubated with protein G magnetic beads and anti-V5, anti-Pol II, or control IgG antibody to immunoprecipitate the target protein. DNA was purified after reversing protein/DNA cross-linking; equal amounts of the purified ChIP DNA were subjected to quantitative PCR analysis using primers ISG60 ISRE (5′-GGTCTCAAGCCGTTAGGTTTCATTT-3′; 5′-GAAGTCTTCCTGTCTGCCTCAAGTA-3′) and Cig 5 ISRE (5′-CCCGATCTCTAGTCTTCAGTCTTGG -3′; 5′-GCAGGACACACCTTCTTTGACTAAC-3′). Each sample was normalized to the negative control and expressed as fold change with respect to vector expressing cells. Similarly, × 107 BCBL-1 cells were crosslinked with 1% formaldehyde for 10 min. Cells were then lysed and chromatin was sheared into 200–600 bp fragments by sonication. The cross-linked chromatin was incubated with protein G magnetic beads and anti-IRF4, anti-Pol II, or control IgG antibody to immunoprecipitate the target protein. DNA was purified after reversing protein/DNA cross-linking; equal amounts of the purified ChIP DNA were subjected to quantitative PCR analysis using primers ISG60 ISRE (5′-GGTCTCAAGCCGTTAGGTTTCATTT-3′; 5′-GAAGTCTTCCTGTCTGCCTCAAGTA-3′) and ORF57 RRE/ISRE 5′-ACACTTATGAGTCAGTGTTTTGCCAG-3′; 5′-GGCAGCCAGGTTATATAGTGGGATTA-3′). Each sample was normalized relative to isotype control.
Sub-cellular fractionations
Cells were washed and cell pellets were suspended in hypotonic buffer (20 mM HEPES pH8.0, 10 mM KCl, 1 mM MgCl2, 20% glycerol, 0.1% Triton-X 100) with protease inhibitors. The cell suspensions (100μl) were vortexed for 30s, incubated on ice for 15 min, and centrifuged (16,000g for 10 at 4 °C). The supernatants were collected as soluble cytoplasmic fractions. The remaining nuclear pellets were thoroughly washed in 10 volumes of hypotonic buffer and then resuspended in 100μl RIPA buffer (50mM Tris-HCl [pH7.4], 150mM NaCl, 1% NP-40, 0.25% sodium deoxycholate, 1mM EDTA, 1mM PMSF, 1x Protease inhibitor cocktail) and incubated in ice for 30 minutes prior to SDS-PAGE.
Statistical analysis
Data were analyzed using two-tailed paired Student’s t-test. Values were considered significant at p < 0.05.
RESULTS
IRF4 upregulation leads to ISG induction in PEL cells
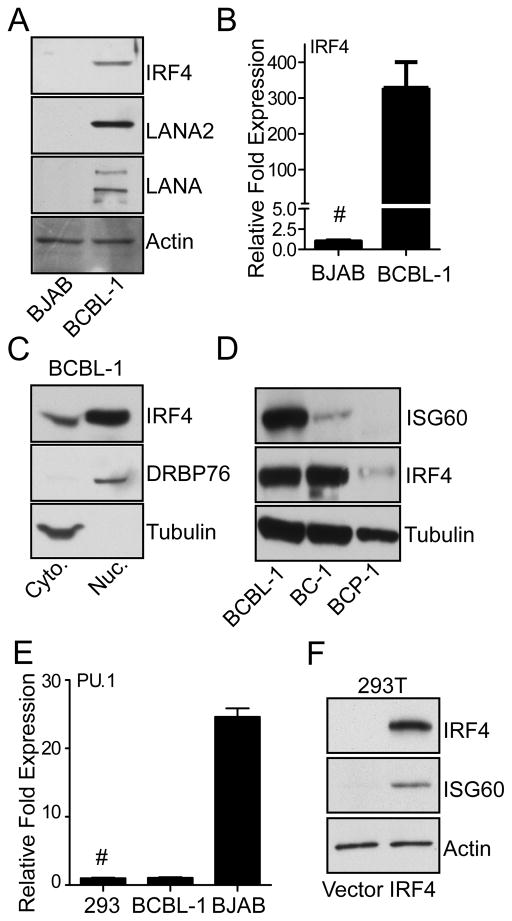
PEL cells are characterized by their plasma cell-like phenotype and express high levels of IRF4 (10, 18, 44). As shown in Fig. 1A and 1B, PEL derived BCBL-1 cells showed expression of IRF4, as well as latency-associated KSHV proteins LANA and LANA2 compared to non-PEL B-cell line BJAB. Transcriptional activities of IRF are usually associated with their activation and nuclear translocation (45), followed by their binding to interferon stimulated regulatory elements (ISRE) located in the promoter regions of their target genes (46, 47). As expected for an activated IRF, in BCBL-1 cells, a major portion of IRF4 protein was present in the DRBP76-positive nuclear fraction (Fig. 1C). To examine whether activated IRF4 in PEL cells induced ISGs, PEL cell lines BCBL-1, BC-1, and BCP-1 were tested for IRF4 and ISG60 expression. As shown in Fig. 1D, both BCBL-1 and BC-1 cells expressed high levels of IRF4 accompanied with various levels of ISG60 expression, while BCP-1 showed lower levels of IRF4 and no detectable ISG60 induction. BCBL-1 and BCP-1 cells are both KSHV positive and EBV negative, while BC-1 cells are both KSHV and EBV positive (19, 48, 49). Previous studies have shown, that PEL cells display an incomplete B cell phenotype and lack the expression of lymphocyte specific transcription factors PU.1 and IRF8 (44). Taking this into consideration, transcript level of PU.1 in BCBL-1 and control 293T cells were analyzed to show that these cells express minimal levels of PU.1 mRNA relative to BJAB cells. Furthermore, IRF4 is an immune cell factor and not expressed in 293T cells at appreciable levels (Fig. 1F). Thus, to examine whether IRF4 expression induces ISG60 in the absence of lymphocyte specific transcription factors, IRF4 was transfected into 293T cells. Exogenous IRF4 expression induced ISG60 protein expression in these cells (Fig. 1F) as well as in HT1080 cells (data not shown). Taken together, these results suggest that IRF4 is capable of inducing the expression of certain ISGs in a variety of cells, including PEL cells.
Figure 1. IRF4 and ISG60 are upregulated in PEL cell lines.
(A) Analysis of IRF4 protein levels in BJAB and BCBL-1 cells. Lysates were prepared from 1×106 cells and immunoblotted with anti-IRF4, anti-LANA anti-LANA2 and anti-β actin antibodies.
(B) IRF4 mRNA analysis in BJAB and BCBL-1 cells. Total RNA was harvested from 1×106 cells and subjected to qRT-PCR using primers against IRF4 and RPL32 as described in Materials and Methods. Expression of IRF4 was normalized to RPL32 and expressed as fold change with respect to BJAB cells (#).
(C) Sub-cellular localization of IRF4 in PEL cell lines. Cytoplasmic and nuclear fractions were prepared from 1×107 BCBL-1 cells and immunoblotted with IRF4, Tubulin, and DRBP76 antibodies.
(D) Analysis of ISG60 and IRF4 protein expression in PEL cell lines BCBL-1, BC-1, and BCP-1. Lysates were prepared from 1×106 cells and immunoblotted with antibodies against IRF4, ISG60, and Tubulin.
(E) PU.1 mRNA analysis in 293, BCBL-1 and BJAB cells. Total RNA was harvested from 1×106 cells and subjected to qRT-PCR using primers against PU.1 and RPL32 as described in Materials and Methods. Expression of PU.1 was normalized to RPL32 and expressed as fold change with respect to 293 cells (#).
(F) Induction of ISG60 by IRF4 in 293Tcells. Cells were transfected with 1μg of pcDNA/IRF4-V5 or vector control plasmids, 48 h post transfection lysates were prepared and immunoblotted with anti-IRF4 and anti-ISG60 antibodies.
IRF4 induces the expression of a specific subset of interferon-stimulated genes
To further characterize the transcriptional activity of IRF4 and identify other potential target ISG, stable HEK293 cells with doxycycline (Dox) inducible V5-tagged IRF4 expression (293iIRF4) were generated. Stimulation of 293iIRF4 cells with increasing doses of Dox for 48 h resulted in a dose-dependent increase in expression of IRF4 mRNA (Fig. 2A) and protein levels (Fig. 2B). Similar to PEL cells, immunoblot analysis in 293iIRF4 cells showed cytoplasmic, but predominantly nuclear localization of IRF4 after treatment with Dox (Fig. 2C), confirming its potential role in direct transcriptional regulation. To determine if IRF4 specifically regulated the endogenous expression of ISG in 293iIRF4 cells, the induction of several well-known ISG transcripts after Dox treatment was examined. Among the ISGs tested, three distinct phenotypes were observed upon IRF4 expression. Several ISG mRNAs showed strong IRF4 dose dependent induction, such as ISG60, Cig5, and OASL (Fig. 2D). This correlated with their protein expression (Fig. 2E). In contrast, ISG15, PKR and IRF7 showed modest to insignificant levels of increase (Fig. 2F). IFNβ which is not an ISG, but is induced via IRF3/IRF7 binding to ISRE, also followed similar pattern showing a very modest increase after IRF4 expression (Fig. 2F). In the third category was MxA, which showed a dose dependent, significant inhibition after Dox treatment (Fig. 2G). IFNα (primers used for the common regions of all human subtypes) also showed similar, albeit modest, pattern (Fig. 2G). Furthermore, the effect of IRF4 on ISG induction was independent of other relevant IRF protein induction, as expression of IRF4 in 293iIRF4, did not change the protein or mRNA expression levels of other IRF family members (Supplementary Fig. 1A and B). These data suggest that IRF4 stimulates the transcription of only a subset of ISGs in an interferon-independent manner, while it can act as possible repressor on other ISGs.
Figure 2. IRF4 induces the expression of a specific subset of interferon-stimulated genes.
(A) Analysis of IRF4 mRNA induction in 293iIRF4 cells. Following 48 h Dox stimulation with indicated doses, total RNA was harvested from 1×106 cells/sample and subjected to qRT-PCR to detect expression of IRF4. Samples were normalized to RPL32 and expressed as fold change with respect to untreated cells (value 1), marked with (#).
(B) Analysis of IRF4 protein induction following Dox stimulation of 293iIRF4 cells. Cell lysates were prepared from Dox stimulated cells (48 h) and immunoblotted with anti-V5 and Actin antibodies.
(C) Sub-cellular localization of IRF4 in 293iIRF4 cells. 1×106 cells were stimulated 0.05μg/ml Dox for 48hrs. Cytoplasmic and nuclear fractions were prepared as described in Materials and Methods, and immunoblotted with indicated antibodies.
(D) In a similar experiment as in (A) ISG60, Cig5, and OASL mRNA induction were analyzed following Dox stimulation.
(E) Analysis of ISG60, Cig5, and OASL protein induction following 48 h Dox stimulation of 293iIRF4 cells. Cell lysates were probed with ISG60, Cig5, OASL, and Actin antibodies.
(F) and (G) Analysis of ISG15, PKR, IRF7, and IFNβ(F) and pan-IFNα and MxA (G) mRNA induction by IRF4 expression following 48 h Dox stimulation.
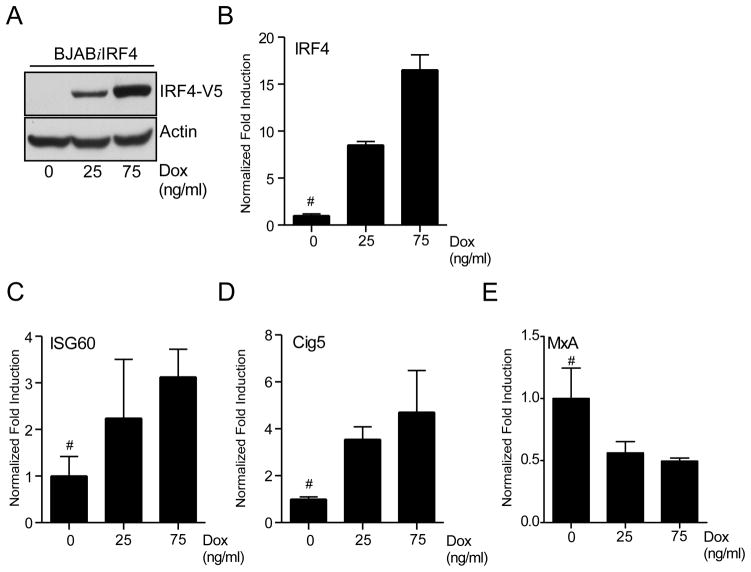
To further address the positive modulation of ISG expression by IRF4 in B cells, a BJAB derived stable cell line, BJABiIRF4 was generated using the Dox inducible IRF4 vector (Fig. 3A and 3B). Similar to 293iIRF4 cells, these cells also showed an increase in ISG60 and Cig5 transcripts and a decrease in MxA transcripts in Dox-dependent manner (Fig. 3C, 3D, and 3E). Taken together, these results indicate that IRF4 activates expression of some ISGs in B cells and may be responsible for the high levels of ISG60 protein observed in most PEL cells.
Figure 3. IRF4 induces expression of ISGs in B-cells.
(A) Analysis of IRF4 protein induction following Dox stimulation in BJABiIRF4 cells. 2×106 cells were stimulated with increasing doses of Dox as indicated for 48. Lysates were prepared after stimulation and immunoblotted with anti-V5 and Actin antibodies.
(B), (C), (D), and (E) qRT-PCR analysis of IRF4 (B), ISG60 (C), Cig5 (D), and MxA (E) expression in Dox treated BJABiIRF4 cells. Cells were stimulated as previously described, total RNA was harvested and subjected to qRT-PCR. Samples were normalized to RPL32 and expressed as fold change with respect to untreated cells (value 1), marked with (#).
Transcriptional activation of ISG60 and Cig5 is directly mediated by IRF4
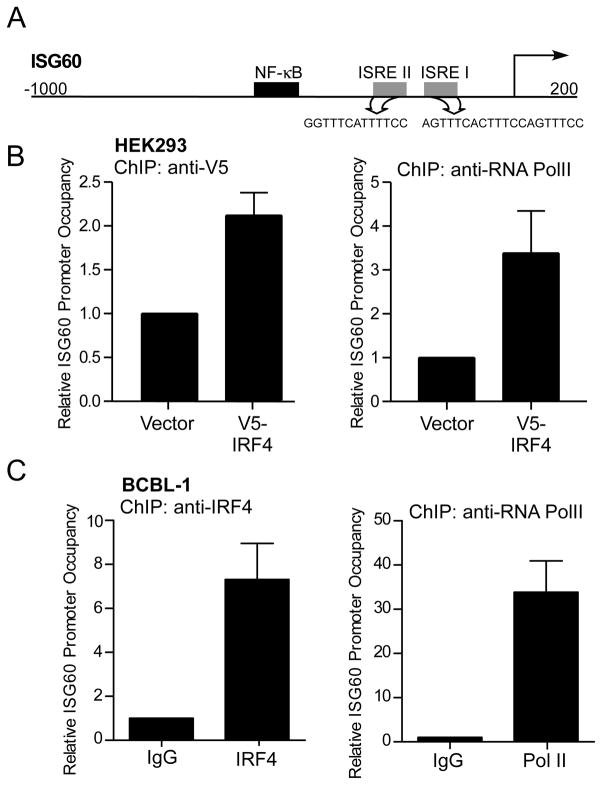
To establish that IRF4 is responsible for direct induction of a subset of ISGs, the mechanism of IRF4 mediated upregulation of ISG60 and Cig5 was examined. Sequence analysis of the 5′ regulatory region of ISG60 and Cig5 genes showed putative ISRE elements (Fig. 4A and Supplementary Fig. 2A) (46, 50). To confirm the ability of IRF4 to specifically bind to the ISRE elements and drive transcription, chromatin immunoprecipitation (ChIP) assays were performed on HEK293 cells constitutively expressing V5-tagged IRF4. ChIP with anti-V5 antibody showed that IRF4 bound to the ISG60 promoter in the region encompassing both the ISREII/I elements of ISG60 (Fig. 4B) and to the region containing the ISRE element on the Cig5 promoter (Supplementary Fig. 2B). To establish IRF4 binding to the ISRE element on the ISG60 promoter in PEL cells, ChIP with anti-IRF4 antibody was carried out in BCBL-1 cells, which showed increased promoter occupancy by IRF4 as compared to isotype control (Fig. 4C). These results suggest that direct binding of IRF4 to the ISRE elements in the promoter regulatory regions of the ISG60 and Cig5 genes results in the their transcriptional activation.
Figure 4. IRF4 binds to ISG60 promoter in 293 and BCBL-1 cells.
(A) Schematic representation of the ISG60 promoter-regulatory region depicting position of two ISRE sites (gray) and their sequences as predicted using the Transcriptional Regulatory Element Database (TRED).
(B) Chromatin-immunoprecipitation of IRF4 bound to ISG60 promoter. Chromatin was prepared from 1×107 HEK293 cells expressing IRF4-V5 (HEK293/pLenti-IRF4-V5) or vector control (HEK293/pLenti). IRF4 and Pol II binding to the promoters were assayed by ChIP assay using anti-V5 and anti-Pol II antibodies for immunoprecipitation. Relative promoter occupancy was determined relative to vector control cells as indicated in materials and methods.
(C) Chromatin-immunoprecipitation of endogenous IRF4 bound to ISG60 promoter in BCBL-1 cells. IRF4 (left) and Pol II (right) binding to the ISG60 promoter was analyzed by ChIP assay as previously described. Relative promoter occupancy was determined relative to isotype control antibody (value 1).
Modulation of IRF4-mediated ISG induction by KSHV latency associated proteins
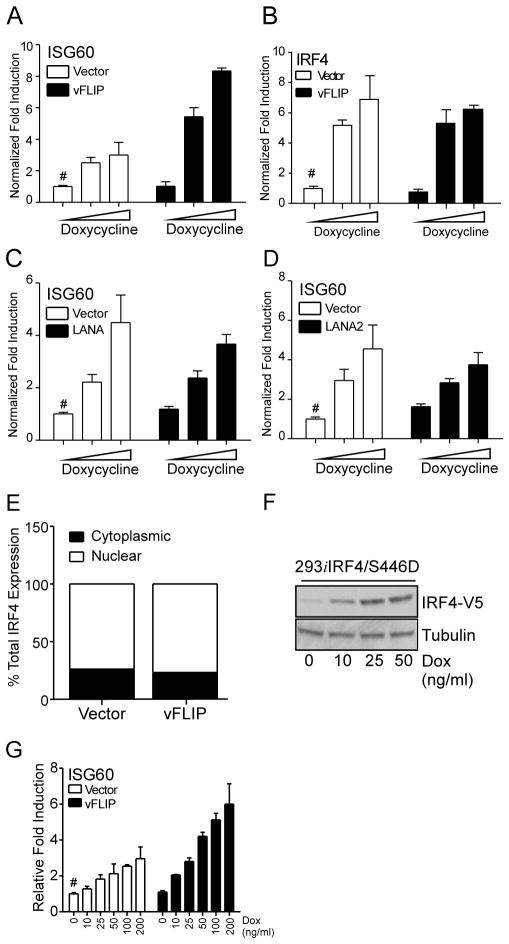
KSHV latency associated proteins have been previously shown to modulate IRF-mediated signaling (51–55). Therefore, to examine the effects of these latency associated viral genes on IRF4 mediated ISG induction we expressed vFLIP, LANA, and LANA2 in 293iIRF4 cells. Among them vFLIP caused a synergistic enhancement in ISG60 transcription in presence of IRF4 (Fig. 5A), whereas LANA or LANA2 did not (Fig. 5C and 5D, respectively). Expression of vFLIP alone did not significantly activate ISG60 (Fig. 5A). Similarly, neither LANA nor LANA2 expressed in the absence of IRF4 had any effect on ISG60 transcript levels (Fig. 5C and 5D). Moreover, expression of the viral latency proteins did not affect the expression of IRF4 (Fig. 5B and data not shown) indicating that KSHV latency associated protein vFLIP enhances IRF4 mediated ISG transcription independently from the modulation of IRF4 expression.
Figure 5. vFLIP enhances IRF4-mediated ISG induction.
Quantitation of ISG60 (A), (C) and (D); and IRF4 (B) mRNA induction in 293iIRF4 cells transfected with KSHV latency-associated viral proteins. 8×105 cells were transfected in a 6-well plate with 800ng of vFLIP (A), LANA (B), LANA2 (C) expression vectors or their respective empty vector controls. Eight hours post transfection, cells were transferred to 24-well plates and stimulated with increasing doses of Dox. Total RNA was harvested 48 h post stimulation. ISG60 and IRF4 mRNA induction was quantified by qRT-PCR. Samples were normalized to RPL32 and expressed as fold change with respect to untreated vector control cells (value 1), marked with (#).
(E) Effect of vFLIP on IRF4 subcellular localization. 293iIRF4 cells were transfected with pcDNA-K13 and stimulated with 0.5 μg/ml Dox for 48 h. Cells lysates were fractioned and subjected to immunoblotting with antibodies against V5. Immunoblots were quantified by densitometry and plotted as total IRF4 fractionated between cytoplasmic and nuclear fractions.
(F) Western blot analysis of IRF4 protein induction following Dox stimulation in 293iIRF4/S446D cells. Cells were stimulated for 48 h with Dox as indicated. Lysates were prepared and subjected to immunoblotting using antibodies against V5 and Tubulin.
(G) Effect of IRF4-S446D on vFLIP-mediated enhancement of ISG60 mRNA induction. 293iIRF4 and 293iIRF4/S446D were transfected and stimulated as previously described. ISG60 mRNA induction was quantified by qRT-PCR. Samples were normalized to RPL32 and expressed as fold change with respect to untreated vector control cells (value 1), marked with (#).
To define the mechanism of synergistic enhancement of IRF4-mediated transcription by vFLIP two approaches were taken. First, to determine if vFLIP increased nuclear translocation of IRF4, 293iIRF4 cells were transfected with vFLIP and stimulated with Dox. As previously observed (Fig. 2C), over 70% of total IRF4 protein expression was detected in the nuclear fraction, and no changes in cellular localization of IRF4 were detected following vFLIP expression (Fig. 5E). Second, to address whether vFLIP could affect the phosphorylation of IRF4, the effect of vFLIP on a constitutively active phosphomimetic mutant of IRF4, S446D, was evaluated (56). Using a Dox-inducible cell line expressing IRF4 S446D, 293iIRF4/S446D (Fig. 5F), the effect of vFLIP co-expression on ISG mRNA induction was examined. Stimulation of 293iIRF4/S446D cells with Dox showed expected increase in ISG60 mRNA, which was further enhanced by vFLIP expression (Fig. 5G) without affecting IRF4 levels. This data suggests that the effect of vFLIP on IRF4-mediated transcription is independent from its phosphorylation of serine 446, and vFLIP does not increase the IRF4 activation.
NF-κB activation is required for enhancement of IRF4-mediated ISG induction by vFLIP
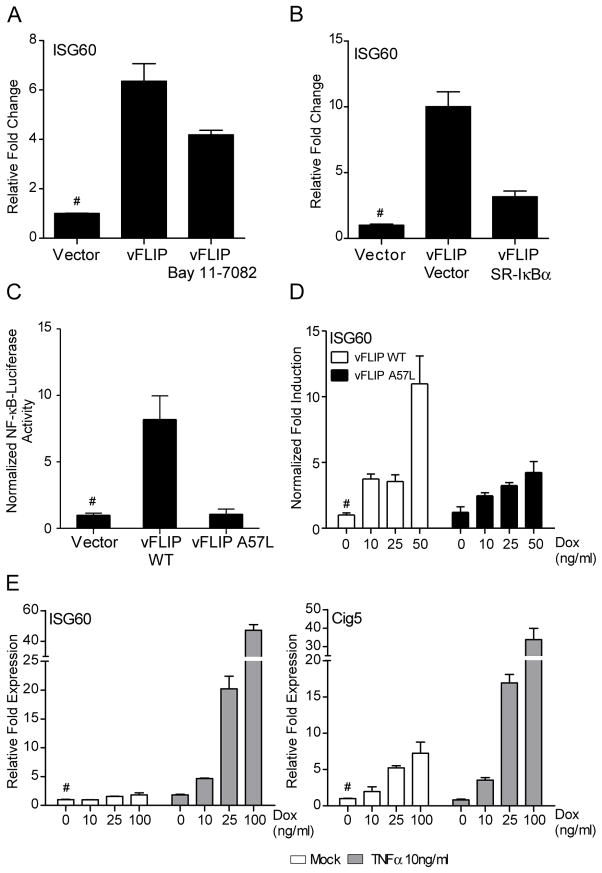
Unlike other viral FLIPs, KSHV vFLIP activates NF-κB signaling by interacting with the IKK complex, resulting in the phosphorylation and subsequent degradation of the NF-κB inhibitor, IκBα (27, 28). To determine if vFLIP affects ISG induction by IRF4 as a result of NF-κB activation, vFLIP-transfected 293iIRF4 cells were treated with the IKK inhibitor Bay 11-7082. The addition of Bay 11-7082 resulted in a 2-fold reduction of ISG60 induction by IRF4 and vFLIP compared to untreated cells (Fig 6A). To exclude non-specific kinase inhibition effects, vFLIP was co-expressed in 293iIRF4 cells with the IκB-super repressor mutant S32A/S36A (SR-IκBα, which prevents NF-κB activation. This resulted in a nearly 7-fold decrease in the vFLIP mediated enhancement of ISG60 induction (Fig. 6B), confirming the requirement for NF-κB activation. Furthermore, a mutant vFLIP that is impaired in its ability to interact with TRAF proteins and activate NF-κB (A57L) (57) was generated and tested for its ability to synergize with IRF4. As expected, vFLIP A57L did not activate NF-κB (Fig. 6C), and had no effect on IRF4 expression (data not shown). Synergistic enhancement of IRF4-mediated ISG60 induction, however, was absent in A57L compared to wild-type vFLIP transfected cells (Fig. 6D). Again, no induction of ISG60 was observed in cells transfected with either WT or mutant vFLIP expression vectors in the absence of Dox treatment. These results suggest that vFLIP upregulates IRF4 mediated ISG60 induction through the activation of NF-κB.
Figure 6. Enhancement of ISG induction by vFLIP requires NF-κB activation.
(A) Effect of Bay 11-7082 on ISG induction by IRF4 and vFLIP. 293iIRF4 were transfected with vector control or vFLIP for 8 h. After transfection, cells were stimulated with Dox in the presence or absence of 5μM Bay 11-7082 for an additional 48 h. ISG60 mRNA induction was analyzed by qRT-PCR as described before. Samples were normalized to RPL32 and expressed as fold change with respect to untreated vector control cells (value 1), marked with (#).
(B) Effect of SR-IκBα expression on ISG induction by IRF4 and vFLIP. 293iIRF4 were co-transfected with vFLIP and SR-IκBα or the respective vector controls for 24 h. After transfection, cells were stimulated with Dox for an additional 48 h. ISG60 mRNA induction was analyzed by qRT-PCR as described before. Samples were normalized to RPL32 and expressed as fold change with respect to untreated vector control cells (value 1), marked with (#).
(C) NF-κB luciferase reporter assays from 293T cells transfected with wild-type vFLIP, vFLIP A57L, or empty vector control. 293T cells were co-transfected with 1μg of cDNA, 0.4 μg of NF-κB luciferase reporter construct and 12 ng of pRL-null. Firefly luciferase activity was measured 48 h post transcription and normalized to renilla luciferase.
(D) qRT-PCR analysis of ISG60 induction in 293iIRF4 cells transfected with wild-type or A57L mutant vFLIP. RNA was extracted after transfection of WT or mutant vFLIP cDNA for 8 h followed by 48 h of Dox stimulation.
(E) Analysis of ISG60 induction in IRF4 expressing 293iIRF4 cells stimulated with TNFα RNA was harvested from 293iIRF4 cells that were stimulated for a total 48 h with Dox in the presence of 100ng/ml TNFα for the last 12 h. Samples were normalized to RPL32 and expressed as fold change with respect to untreated vector control cells (value 1), marked with (#).
Finally, to confirm the involvement of NF-κB in IRF4 mediated ISG60 upregulation, cells were treated with NF-κB activating cytokine, tumor necrosis factor α (TNFα), and examined for ISG60 and Cig5 transcript levels in presence of IRF4. 293iIRF4 cells were stimulated with increasing doses of Dox for 48 h followed by 12 h stimulation with 10ng/ml TNFα. Co-stimulation with TNFα in presence of IRF4 resulted in markedly increased ISG60 and Cig5 transcription (Fig 6E). As observed previously after co-expression of vFLIP, treatment with TNFα alone did not result in a significant induction of either ISG60 or Cig5 mRNA (Fig 6E). Taken together, these results suggest that vFLIP enhances IRF4-dependent ISG induction through NF-κB activation.
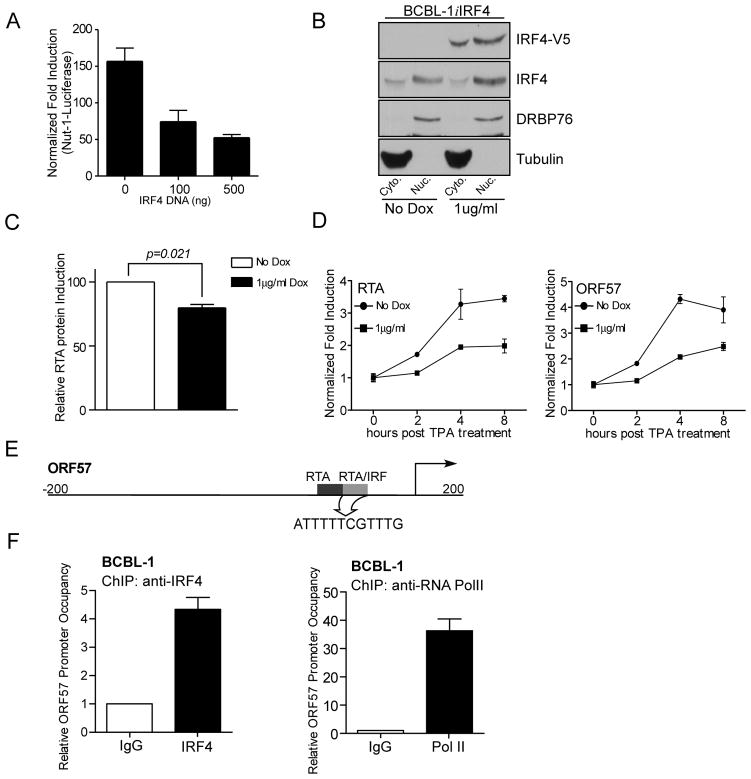
IRF4 inhibits KSHV reactivation from latency
Although, IRF4 has been shown to negatively regulate host gene transcription in specific situations (Fig. 2G and (6, 7)), its role in regulating viral latency has not been described. In the context of KSHV, the viral replication and transcription activator (RTA), encoded by ORF50, activates the expression of viral immediate-early (IE) and early genes, as well as its own expression by binding to RTA-responsive elements (RRE) (58). RTA can also bind to ISRE found in cellular ISGs as well as ISRE-like sequences contained in the promoter regulatory regions of viral genes (59). While IRF4 binds ISRE and modulates ISG expression (Fig. 2 and 4), the ability of regulating KSHV reactivation and gene expression through a similar mechanism by IRF4, has not been elucidated. For this purpose, the effect of IRF4 expression on RTA-mediated transcription was examined using a Nut-1 (PAN) promoter luciferase reporter construct (41). IRF4 expression inhibited RTA mediated luciferase activity in a dose dependent manner (Fig. 7A). Next, the effect of IRF4 on endogenous RTA expression was examined following TPA stimulation to induce lytic gene transcription (20). For this purpose, a stable cell line, BCBL-1iIRF4 was generated, which expressed Dox-inducible V5-tagged IRF4 (Fig. 7B and Supplementary Fig. 3). Stimulation of these cells with TPA in the presence or absence of Dox showed about a 20% reduction in RTA protein expression upon IRF4 expression (Fig. 7C). Finally, to determine whether the inhibitory effect of IRF4 on RTA expression was due to changes in transcriptional induction of RTA, the induction kinetics of RTA mRNA, and ORF57, an RTA-responsive gene known to be negatively regulated by IRF7 (59), mRNA following TPA treatment were examined. Again, IRF4 expression resulted in a repression of RTA and ORF57 mRNA after TPA treatment (Fig. 7D). These data suggest that IRF4 inhibits the expression of RTA-mediated viral gene expression required for lytic KSHV replication. This observation was in accordance to the previous findings showing that IRF4 can act as a negative regulator of ISRE containing, MxA (Fig. 2G). To further understand the mechanism of this inhibition, direct binding of IRF4 to the defined RTA/IRF target element in the ORF57 promoter (Fig. 7E and (59)) was examined by ChIP assay. Using antibodies against endogenous IRF4, it was found that IRF4 was able to occupy the RTA/IRF region in BCBL-1 cells (Fig. 7F), suggesting that the observed inhibition of ORF57 transcription is potentially due to IRF4 competing with RTA for promoter binding. Taken together, these data suggest that IRF4 acts as a dual regulator of cellular and viral gene transcription in PEL cells likely contributing to the maintenance of viral latency.
Figure 7. IRF4 inhibits KSHV viral reactivation.
(A) Effect of IRF4 on Nut-1 luciferase in 293T cells. 293T cells were co-transfected with 100 or 500 ng of IRF4 expression plasmid, 0.4 μg Nut-1 luciferase reporter construct, 24ng pRL-Null and either 75 ng of RTA cDNA or empty vector. Total transfected DNA levels were kept equal with empty vector. 48 h after transfection, luciferase activity was measured. Fold induction was normalized to both Renilla luciferase activity and non-RTA transfected cells.
(B) Analysis of IRF4 protein expression levels in BCBL-1iIRF4 cells. 2×105 cells/ml BCBL-1i4 cells were treated with 1 μg/ml Dox or left untreated for 48 hrs. Subcellular fractions were harvested as previously described and subjected to immunoblot with anti-RTA antibody.
(C) Ectopic IRF4 expression leads to reduced RTA protein expression. 2×105 cells/ml BCBL-1iIRF4 cells were treated with 1 μg/ml Dox or left untreated for 48 h, followed by stimulation with 15ng/ml TPA or DMSO for 12 h prior to harvesting. Cell lysates were subject to immunoblot with anti-RTA antibody. RTA expression levels were quantified for three independent experiments and induction was calculated relative to cells without Dox stimulation.
(D) Inhibition of RTA and ORF57 mRNA induction in BCBL-1 cells by IRF4. BCBL-1iIRF4 cells were stimulated with doxycycline for 48 h followed by stimulation with 15ng/ml TPA for 2, 4, and 8 h. Fold induction of RTA mRNA levels were normalized to RPL32 and non-TPA stimulated cells.
(E) Schematic representation of the ORF57 promoter-regulatory region depicting position of RTA binding sites (black) and the RTA/IRF binding (gray). The RTA/IRF target sequence has been highlighted.
(F) Chromatin-immunoprecipitation of endogenous IRF4 bound to ORF57promoter in BCBL-1 cells. IRF4 (left) and Pol II (right) binding to the ORF57 promoter was analyzed by ChIP assay as previously described. Relative promoter occupancy was determined relative to isotype control antibody (value 1).
DISCUSSION
IRF4 is an immune cell specific transcription factor which plays a role in lymphomagenesis. These functions of IRF4 are defined by its ability to bind various transcription factors and regulate gene expression, such as PU.1in B cells. In this study, we found that in the context of PEL cells, which do not express PU.1, IRF4 can differentially regulate ISG expression. Modulation of ISG expression was also observed in epithelial cells which, like PEL cells, lack the expression of B cell-specific transcription factors. Interestingly, ectopic expression of IRF4 also resulted in similar ISG modulation in B cell lines. Our data showing that IRF4 binds to the cis-elements of specific ISG promoters to induce their transcription expands the current paradigm of IRF4 function, where it may play important role in ISG function in a cell-type specific manner. Next, having identified some direct IRF4 target ISGs, we asked if this activity can be modulated by KSHV latency-associated proteins. As shown in Fig. 5, modulation of IRF4 mediated ISG induction by KSHV-latency associated protein, vFLIP in the context of PEL, provides additional evidence for its biological importance in this malignancy. Among the three latency-associated proteins tested, co-expression of vFLIP resulted in a synergistic activation of ISG60. This effect was specific to vFLIP as neither LANA nor LANA2, which have previously been shown to bind and modulate IRF-mediated transcriptional activation (52–54), modulated IRF4-mediated ISG60 induction. This indicates differential modulation of transcriptional activity of IRFs by KSHV encoded proteins.
Although the role of NF-κB in the induction of IFNβ is well established (60), the requirement for NF-κB in transcriptional control of ISGs has not been well understood. Recent work has shown that a subset of interferon-stimulated genes can be further regulated by NF-κB (61). However, in the absence of a robust NF-κB activation by type I IFNs, physiological relevance of these results has remained unclear. Using both chemical/genetic inhibitors of NF-κB and a vFLIP mutant devoid of NF-κB activity, we showed that NF-κB activation is required for enhancement of ISG60 transcription by vFLIP (Fig. 6). This observation showed that both ISG60 and Cig5 gene expression can be controlled by both IRF4 and NF-κB in a physiologically relevant setting where NF-κB activation is mediated by a viral protein. Furthermore, stimulation with TNFα also resulted in an enhancement of ISG induction, supporting the role of NF-κB in specific ISG regulation. Interestingly, vFLIP expression, or TNFα stimulation alone was not sufficient for the induction of ISGs or IRF4 expression (Fig. 5B and Fig. 6E). Lastly, these results suggest that in B-cells ISG60 or Cig5 enhancement by vFLIP is independent of STAT1 or STAT2 and IFN signaling, which was the case in endothelial cells (62). Thus, IRF proteins primarily drive the induction of ISG60 and Cig5, while NF-κB functions as a secondary enhancer of their transcription.
RTA-mediated gene transcription initiates KSHV lytic replication (58). RTA has also been found to bind ISRE elements in host genes and induce the expression of ISG (59). Recently, IRF7 has been shown to suppress viral reactivation by competing with RTA for binding to RTA-response elements on the ORF57 promoter (63). Results presented in Fig. 7 showing inhibition of RTA-mediated Nut-1 reporter following IRF4 expression supports this model. Considering our finding that IRF4 can bind to ISRE elements in cellular genes (Fig. 4) as well as ISRE-like RRE element found in ORF57 (Fig. 7), it is possible that inhibitory effects of IRF4 on RTA-mediated transcriptional activation are due to competitive inhibition of RTA binding to its target sites, thereby promoting the maintenance of KSHV latency in PEL cells. Interestingly though, ectopic expression IRF4 in BCBL-1 cells resulted in a modest, yet significant decrease in RTA protein expression after 12 hrs after TPA treatment (Fig. 7C). Out of all the lytic genes ORF57 has the best characterized ISRE-like RRE. Thus, we followed the effect of IRF4 expression on mRNA induction of both RTA and ORF57 showing a 2-fold decrease in transcript levels. This is significant because, previous studies have shown that KSHV encoded Nut-1 RNA, a non-coding RNA and the most abundant viral RNA during lytic infection, can bind to IRF4 and inhibit DNA binding (26). This interaction between Nut-1 transcript and IRF4 highlights the importance of IRF4 as a negative regulator of viral gene transcription and shows the virus has developed multiple strategies to circumvent the transcriptional block imposed by IRF4 on RRE/ISRE regulated promoters.
The mechanisms controlling the maintenance of latency and the switch to lytic gene expression of KSHV are complex. Several studies have shown the contribution of NF-κB activation in the negative regulation of RTA mediated transcription (64). Recruitment of RTA to the promoters of lytic genes ORF57 and K-bZIP is inhibited by NF-κB, while the K12 promoter was not affected by NF-κB activation (15). In PEL cells, vFLIP induced NF-κB activation inhibits both ORF50 and ORF57 gene expression contributing to the establishment of latency in an AP-1 dependant manner (65). Furthermore, inhibition NF-κB with Bay 110782 results in spontaneous reactivation (33, 66). On the other hand, NF-κB activation also functions as a positive modulator of lytic replication as shown by Grossman et al (33). Thus, it is likely that both IRF4 and NF-κB function together as negative regulators of KSHV lytic gene expression in the context of PEL. However, further studies will be necessary for a detail understanding of their specific contributions in viral reactivation and maintenance of latency.
Our studies establish an important role of IRF4 in controlling specific ISG induction and its enhancement by vFLIP through NF-κB activation. Concomitantly, IRF4 can act as a negative regulator of KSHV lytic gene expression. Thus, we describe the complex functions of IRF4 that modulate innate immune responses and contribute to the maintenance of KSHV latency in PEL cells.
Supplementary Material
Acknowledgments
We thank Drs. Preet M. Chaudhary and David Lukac, for reagents and Drs. Yuan Chang and Hyun Jin Kwun for reagents, helpful discussions and critical suggestions.
This work was supported in part by AI082673 from National Institute of Allergy and Infectious Diseases Grant (SNS). This project used the UPCI core facilities and was supported in part by award P30CA047904.
Abbreviations
- IFN
Interferon
- NF-κB
Nuclear Factor κB
- IRF
Interferon Regulatory Factor
- ISG
Interferon Stimulated Gene
- ISRE
Interferon Stimulated Regulatory Element
- KSHV
Kaposi Sarcoma-associated Herpesvirus
- vFLIP
viral FLICE Inhibitory Protein
- RTA
Replication and Transcription Activator
- RRE
RTA-Responsive Element
- PEL
Primary Effusion Lymphoma
- Dox
Doxycycline
- TPA
12-O-tetradecanoylphorbol-13-acetate
- TNFα
Tumor necrosis factor α
Footnotes
Author contribution
AF designed and performed the experiments; AF, PSM and SNS designed experiments and wrote the manuscript.
References
- 1.Tamura T, Yanai H, Savitsky D, Taniguchi T. The IRF family transcription factors in immunity and oncogenesis. Annu Rev Immunol. 2008;26:535–584. doi: 10.1146/annurev.immunol.26.021607.090400. [DOI] [PubMed] [Google Scholar]
- 2.Gabriele L, Ozato K. The role of the interferon regulatory factor (IRF) family in dendritic cell development and function. Cytokine Growth Factor Rev. 2007;18:503–510. doi: 10.1016/j.cytogfr.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 3.Chang PC, Fitzgerald LD, Van Geelen A, Izumiya Y, Ellison TJ, Wang DH, Ann DK, Luciw PA, Kung HJ. Kruppel-associated box domain-associated protein-1 as a latency regulator for Kaposi’s sarcoma-associated herpesvirus and its modulation by the viral protein kinase. Cancer Res. 2009;69:5681–5689. doi: 10.1158/0008-5472.CAN-08-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meraro D, Gleit-Kielmanowicz M, Hauser H, Levi BZ. IFN-stimulated gene 15 is synergistically activated through interactions between the myelocyte/lymphocyte-specific transcription factors, PU.1, IFN regulatory factor-8/IFN consensus sequence binding protein, and IFN regulatory factor-4: characterization of a new subtype of IFN-stimulated response element. J Immunol. 2002;168:6224–6231. doi: 10.4049/jimmunol.168.12.6224. [DOI] [PubMed] [Google Scholar]
- 5.Kanno Y, Levi BZ, Tamura T, Ozato K. Immune cell-specific amplification of interferon signaling by the IRF-4/8-PU.1 complex. J Interferon Cytokine Res. 2005;25:770–779. doi: 10.1089/jir.2005.25.770. [DOI] [PubMed] [Google Scholar]
- 6.Yamagata T, Nishida J, Tanaka S, Sakai R, Mitani K, Yoshida M, Taniguchi T, Yazaki Y, Hirai H. A novel interferon regulatory factor family transcription factor, ICSAT/Pip/LSIRF, that negatively regulates the activity of interferon-regulated genes. Mol Cell Biol. 1996;16:1283–1294. doi: 10.1128/mcb.16.4.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosenbauer F, Waring JF, Foerster J, Wietstruk M, Philipp D, Horak I. Interferon consensus sequence binding protein and interferon regulatory factor-4/Pip form a complex that represses the expression of the interferon-stimulated gene-15 in macrophages. Blood. 1999;94:4274–4281. [PubMed] [Google Scholar]
- 8.Iida S, Rao PH, Butler M, Corradini P, Boccadoro M, Klein B, Chaganti RS, Dalla-Favera R. Deregulation of MUM1/IRF4 by chromosomal translocation in multiple myeloma. Nat Genet. 1997;17:226–230. doi: 10.1038/ng1097-226. [DOI] [PubMed] [Google Scholar]
- 9.Shaffer AL, Emre NCT, Lamy L, Ngo VN, Wright G, Xiao W, Powell J, Dave S, Yu X, Zhao H, Zeng Y, Chen B, Epstein J, Staudt LM. IRF4 addiction in multiple myeloma. Nature. 2008;454:226–231. doi: 10.1038/nature07064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carbone A, Gloghini A, Larocca LM, Capello D, Pierconti F, Canzonieri V, Tirelli U, Dalla-Favera R, Gaidano G. Expression profile of MUM1/IRF4, BCL-6, and CD138/syndecan-1 defines novel histogenetic subsets of human immunodeficiency virus-related lymphomas. Blood. 2001;97:744–751. doi: 10.1182/blood.v97.3.744. [DOI] [PubMed] [Google Scholar]
- 11.Mamane Y, Grandvaux N, Hernandez E, Sharma S, Innocente SA, Lee JM, Azimi N, Lin R, Hiscott J. Repression of IRF-4 target genes in human T cell leukemia virus-1 infection. Oncogene. 2002;21:6751–6765. doi: 10.1038/sj.onc.1205843. [DOI] [PubMed] [Google Scholar]
- 12.Sharma S, Grandvaux N, Mamane Y, Genin P, Azimi N, Waldmann T, Hiscott J. Regulation of IFN regulatory factor 4 expression in human T cell leukemia virus-I-transformed T cells. J Immunol. 2002;169:3120–3130. doi: 10.4049/jimmunol.169.6.3120. [DOI] [PubMed] [Google Scholar]
- 13.Mamane Y, Sharma S, Grandvaux N, Hernandez E, Hiscott J. IRF-4 activities in HTLV-I-induced T cell leukemogenesis. J Interferon Cytokine Res. 2002;22:135143. doi: 10.1089/107999002753452746. [DOI] [PubMed] [Google Scholar]
- 14.Xu D, Zhao L, Del Valle L, Miklossy J, Zhang L. Interferon regulatory factor 4 is involved in Epstein-Barr virus-mediated transformation of human B lymphocytes. J Virol. 2008;82:6251–6258. doi: 10.1128/JVI.00163-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Izumiya Y, Izumiya C, Hsia D, Ellison TJ, Luciw PA, Kung HJ. NF-kappaB serves as a cellular sensor of Kaposi’s sarcoma-associated herpesvirus latency and negatively regulates K-Rta by antagonizing the RBP-Jkappa coactivator. J Virol. 2009;83:4435–4446. doi: 10.1128/JVI.01999-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaffe ES. Primary body cavity-based AIDS-related lymphomas. Evolution of a new disease entity. Am J Clin Pathol. 1996;105:141–143. doi: 10.1093/ajcp/105.2.141. [DOI] [PubMed] [Google Scholar]
- 17.Dotti G, Fiocchi R, Motta T, Facchinetti B, Chiodini B, Borleri GM, Gavazzeni G, Barbui T, Rambaldi A. Primary effusion lymphoma after heart transplantation: a new entity associated with human herpesvirus-8. Leukemia. 1999;13:664–670. doi: 10.1038/sj.leu.2401390. [DOI] [PubMed] [Google Scholar]
- 18.Carbone A, Gloghini A, Cozzi MR, Capello D, Steffan A, Monini P, De Marco L, Gaidano G. Expression of MUM1/IRF4 selectively clusters with primary effusion lymphoma among lymphomatous effusions: implications for disease histogenesis and pathogenesis. Br J Haematol. 2000;111:247–257. doi: 10.1046/j.1365-2141.2000.02329.x. [DOI] [PubMed] [Google Scholar]
- 19.Cesarman E, Moore PS, Rao PH, Inghirami G, Knowles DM, Chang Y. In vitro establishment and characterization of two acquired immunodeficiency syndrome-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi’s sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood. 1995;86:2708–2714. [PubMed] [Google Scholar]
- 20.Sarid R, Flore O, Bohenzky RA, Chang Y, Moore PS. Transcription mapping of the Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) genome in a body cavity-based lymphoma cell line (BC-1) J Virol. 1998;72:1005–1012. doi: 10.1128/jvi.72.2.1005-1012.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dresang LR, Teuton JR, Feng H, Jacobs JM, Camp DG, 2nd, Purvine SO, Gritsenko MA, Li Z, Smith RD, Sugden B, Moore PS, Chang Y. Coupled transcriptome and proteome analysis of human lymphotropic tumor viruses: insights on the detection and discovery of viral genes. BMC Genomics. 12:625. doi: 10.1186/1471-2164-12-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhong W, Wang H, Herndier B, Ganem D. Restricted expression of Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genes in Kaposi sarcoma. Proc Natl Acad Sci U S A. 1996;93:6641–6646. doi: 10.1073/pnas.93.13.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wen KW, Damania B. Kaposi sarcoma-associated herpesvirus (KSHV): molecular biology and oncogenesis. Cancer Lett. 2010;289:140–150. doi: 10.1016/j.canlet.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muller JM, Moore H, Myers MM, Shair HN. Dopamine’s role in social modulation of infant isolation-induced vocalization: II. Maternally modulated infant separation responses are regulated by D1- and D2-family dopamine receptors. Dev Psychobiol. 2009;51:158–172. doi: 10.1002/dev.20355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thome M, Schneider P, Hofmann K, Fickenscher H, Meinl E, Neipel F, Mattmann C, Burns K, Bodmer JL, Schroter M, Scaffidi C, Krammer PH, Peter ME, Tschopp J. Viral FLICE-inhibitory proteins (FLIPs) prevent apoptosis induced by death receptors. Nature. 1997;386:517–521. doi: 10.1038/386517a0. [DOI] [PubMed] [Google Scholar]
- 26.Rossetto CC, Pari GS. Kaposi’s sarcoma-associated herpesvirus noncoding polyadenylated nuclear RNA interacts with virus- and host cell-encoded proteins and suppresses expression of genes involved in immune modulation. J Virol. 2011;85:13290–13297. doi: 10.1128/JVI.05886-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chaudhary PM, Jasmin A, Eby MT, Hood L. Modulation of the NF-kappa B pathway by virally encoded death effector domains-containing proteins. Oncogene. 1999;18:5738–5746. doi: 10.1038/sj.onc.1202976. [DOI] [PubMed] [Google Scholar]
- 28.Liu L, Eby MT, Rathore N, Sinha SK, Kumar A, Chaudhary PM. The human herpes virus 8-encoded viral FLICE inhibitory protein physically associates with and persistently activates the Ikappa B kinase complex. J Biol Chem. 2002;277:13745–13751. doi: 10.1074/jbc.M110480200. [DOI] [PubMed] [Google Scholar]
- 29.Sun Q, Zachariah S, Chaudhary PM. The human herpes virus 8-encoded viral FLICE-inhibitory protein induces cellular transformation via NF-kappaB activation. J Biol Chem. 2003;278:52437–52445. doi: 10.1074/jbc.M304199200. [DOI] [PubMed] [Google Scholar]
- 30.Chugh P, Matta H, Schamus S, Zachariah S, Kumar A, Richardson JA, Smith AL, Chaudhary PM. Constitutive NF-kappaB activation, normal Fas-induced apoptosis, and increased incidence of lymphoma in human herpes virus 8 K13 transgenic mice. Proc Natl Acad Sci U S A. 2005;102:12885–12890. doi: 10.1073/pnas.0408577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guasparri I, Keller SA, Cesarman E. KSHV vFLIP is essential for the survival of infected lymphoma cells. J Exp Med. 2004;199:993–1003. doi: 10.1084/jem.20031467. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Rossetto CC, Susilarini NK, Pari GS. Interaction of Kaposi’s sarcoma-associated herpesvirus ORF59 with oriLyt is dependent on binding with K-Rta. J Virol. 2011;85:3833–3841. doi: 10.1128/JVI.02361-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grossmann C, Ganem D. Effects of NFkappaB activation on KSHV latency and lytic reactivation are complex and context-dependent. Virology. 2008;375:94–102. doi: 10.1016/j.virol.2007.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sarkar SN, Peters KL, Elco CP, Sakamoto S, Pal S, Sen GC. Novel roles of TLR3 tyrosine phosphorylation and PI3 kinase in double-stranded RNA signaling. Nat Struct Mol Biol. 2004;11:1060–1067. doi: 10.1038/nsmb847. [DOI] [PubMed] [Google Scholar]
- 35.Corte-Real S, Collins C, Aires da Silva F, Simas JP, Barbas CF, 3rd, Chang Y, Moore P, Goncalves J. Intrabodies targeting the Kaposi sarcoma-associated herpesvirus latency antigen inhibit viral persistence in lymphoma cells. Blood. 2005;106:3797–3802. doi: 10.1182/blood-2005-04-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Toptan T, Fonseca L, Kwun HJ, Chang Y, Moore PS. Complex alternative cytoplasmic protein isoforms of the Kaposi’s sarcoma-associated herpesvirus latency-associated nuclear antigen 1 generated through noncanonical translation initiation. J Virol. 2013;87:2744–2755. doi: 10.1128/JVI.03061-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seaman WT, Ye D, Wang RX, Hale EE, Weisse M, Quinlivan EB. Gene expression from the ORF50/K8 region of Kaposi’s sarcoma-associated herpesvirus. Virology. 1999;263:436–449. doi: 10.1006/viro.1999.9963. [DOI] [PubMed] [Google Scholar]
- 38.Kwun HJ, da Silva SR, Shah IM, Blake N, Moore PS, Chang Y. Kaposi’s sarcoma-associated herpesvirus latency-associated nuclear antigen 1 mimics Epstein-Barr virus EBNA1 immune evasion through central repeat domain effects on protein processing. J Virol. 2007;81:8225–8235. doi: 10.1128/JVI.00411-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rivas C, Thlick AE, Parravicini C, Moore PS, Chang Y. Kaposi’s sarcoma-associated herpesvirus LANA2 is a B-cell-specific latent viral protein that inhibits p53. J Virol. 2001;75:429–438. doi: 10.1128/JVI.75.1.429-438.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shair KH, Schnegg CI, Raab-Traub N. EBV latent membrane protein 1 effects on plakoglobin, cell growth, and migration. Cancer Res. 2008;68:6997–7005. doi: 10.1158/0008-5472.CAN-08-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lukac DM, Renne R, Kirshner JR, Ganem D. Reactivation of Kaposi’s sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology. 1998;252:304–312. doi: 10.1006/viro.1998.9486. [DOI] [PubMed] [Google Scholar]
- 42.Zhu J, Smith K, Hsieh PN, Mburu YK, Chattopadhyay S, Sen GC, Sarkar SN. High-throughput screening for TLR3-IFN regulatory factor 3 signaling pathway modulators identifies several antipsychotic drugs as TLR inhibitors. J Immunol. 184:5768–5776. doi: 10.4049/jimmunol.0903559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meerbrey KL, Hu G, Kessler JD, Roarty K, Li MZ, Fang JE, Herschkowitz JI, Burrows AE, Ciccia A, Sun T, Schmitt EM, Bernardi RJ, Fu X, Bland CS, Cooper TA, Schiff R, Rosen JM, Westbrook TF, Elledge SJ. The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proc Natl Acad Sci U S A. 108:3665–3670. doi: 10.1073/pnas.1019736108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arguello M, Sgarbanti M, Hernandez E, Mamane Y, Sharma S, Servant M, Lin R, Hiscott J. Disruption of the B-cell specific transcriptional program in HHV-8 associated primary effusion lymphoma cell lines. Oncogene. 2003;22:964–973. doi: 10.1038/sj.onc.1206270. [DOI] [PubMed] [Google Scholar]
- 45.Honda K, Taniguchi T. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat Rev Immunol. 2006;6:644–658. doi: 10.1038/nri1900. [DOI] [PubMed] [Google Scholar]
- 46.de Veer MJ, Sim H, Whisstock JC, Devenish RJ, Ralph SJ. IFI60/ISG60/IFIT4, a new member of the human IFI54/IFIT2 family of interferon-stimulated genes. Genomics. 1998;54:267–277. doi: 10.1006/geno.1998.5555. [DOI] [PubMed] [Google Scholar]
- 47.Lou YJ, Pan XR, Jia PM, Li D, Xiao S, Zhang ZL, Chen SJ, Chen Z, Tong JH. IRF-9/STAT2 [corrected] functional interaction drives retinoic acid-induced gene G expression independently of STAT1. Cancer Res. 2009;69:3673–3680. doi: 10.1158/0008-5472.CAN-08-4922. [DOI] [PubMed] [Google Scholar]
- 48.Renne R, Zhong W, Herndier B, McGrath M, Abbey N, Kedes D, Ganem D. Lytic growth of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat Med. 1996;2:342–346. doi: 10.1038/nm0396-342. [DOI] [PubMed] [Google Scholar]
- 49.Boshoff C, Gao SJ, Healy LE, Matthews S, Thomas AJ, Coignet L, Warnke RA, Strauchen JA, Matutes E, Kamel OW, Moore PS, Weiss RA, Chang Y. Establishing a KSHV+ cell line (BCP-1) from peripheral blood and characterizing its growth in Nod/SCID mice. Blood. 1998;91:1671–1679. [PubMed] [Google Scholar]
- 50.Severa M, Coccia EM, Fitzgerald KA. Toll-like receptor-dependent and -independent viperin gene expression and counter-regulation by PRDI-binding factor-1/BLIMP1. J Biol Chem. 2006;281:26188–26195. doi: 10.1074/jbc.M604516200. [DOI] [PubMed] [Google Scholar]
- 51.Cloutier N, Grandvaux N, Flamand L. Synergistic activation of interferon-beta gene transcription by the viral FLICE inhibitory protein of Kaposi’s sarcoma-associated herpesvirus and type I IFN activators. Eur J Immunol. 2007;37:2772–2778. doi: 10.1002/eji.200737181. [DOI] [PubMed] [Google Scholar]
- 52.Cloutier N, Flamand L. Kaposi sarcoma-associated herpesvirus latency-associated nuclear antigen inhibits interferon (IFN) beta expression by competing with IFN regulatory factor-3 for binding to IFNB promoter. J Biol Chem. 285:7208–7221. doi: 10.1074/jbc.M109.018838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bi X, Yang L, Mancl ME, Barnes BJ. Modulation of interferon regulatory factor 5 activities by the kaposi sarcoma-associated herpesvirus-encoded viral interferon regulatory factor 3 contributes to immune evasion and lytic induction. J Interferon Cytokine Res. 31:373–382. doi: 10.1089/jir.2010.0084. [DOI] [PubMed] [Google Scholar]
- 54.Joo CH, Shin YC, Gack M, Wu L, Levy D, Jung JU. Inhibition of interferon regulatory factor 7 (IRF7)-mediated interferon signal transduction by the Kaposi’s sarcoma-associated herpesvirus viral IRF homolog vIRF3. J Virol. 2007;81:8282–8292. doi: 10.1128/JVI.00235-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lubyova B, Pitha PM. Characterization of a novel human herpesvirus 8-encoded protein, vIRF-3, that shows homology to viral and cellular interferon regulatory factors. J Virol. 2000;74:8194–8201. doi: 10.1128/jvi.74.17.8194-8201.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Biswas PS, Gupta S, Chang E, Song L, Stirzaker RA, Liao JK, Bhagat G, Pernis AB. Phosphorylation of IRF4 by ROCK2 regulates IL-17 and IL-21 production and the development of autoimmunity in mice. J Clin Invest. 120:3280–3295. doi: 10.1172/JCI42856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sakakibara S, Pise-Masison CA, Brady JN, Tosato G. Gene regulation and functional alterations induced by Kaposi’s sarcoma-associated herpesvirus-encoded ORFK13/vFLIP in endothelial cells. J Virol. 2009;83:2140–2153. doi: 10.1128/JVI.01871-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guito J, Lukac DM. KSHV Rta Promoter Specification and Viral Reactivation. Front Microbiol. 3:30. doi: 10.3389/fmicb.2012.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang J, Wang J, Wood C, Xu D, Zhang L. Kaposi’s sarcoma-associated herpesvirus/human herpesvirus 8 replication and transcription activator regulates viral and cellular genes via interferon-stimulated response elements. J Virol. 2005;79:5640–5652. doi: 10.1128/JVI.79.9.5640-5652.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chang PC, Izumiya Y, Wu CY, Fitzgerald LD, Campbell M, Ellison TJ, Lam KS, Luciw PA, Kung HJ. Kaposi’s sarcoma-associated herpesvirus (KSHV) encodes a SUMO E3 ligase that is SIM-dependent and SUMO-2/3-specific. J Biol Chem. 2010;285:5266–5273. doi: 10.1074/jbc.M109.088088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pfeffer LM, Kim JG, Pfeffer SR, Carrigan DJ, Baker DP, Wei L, Homayouni R. Role of nuclear factor-kappaB in the antiviral action of interferon and interferon-regulated gene expression. J Biol Chem. 2004;279:31304–31311. doi: 10.1074/jbc.M308975200. [DOI] [PubMed] [Google Scholar]
- 62.Alkharsah KR, Singh VV, Bosco R, Santag S, Grundhoff A, Konrad A, Sturzl M, Wirth D, Dittrich-Breiholz O, Kracht M, Schulz TF. Deletion of Kaposi’s sarcoma-associated herpesvirus FLICE inhibitory protein, vFLIP, from the viral genome compromises the activation of STAT1-responsive cellular genes and spindle cell formation in endothelial cells. J Virol. 85:10375–10388. doi: 10.1128/JVI.00226-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang J, Zhang J, Zhang L, Harrington W, Jr, West JT, Wood C. Modulation of human herpesvirus 8/Kaposi’s sarcoma-associated herpesvirus replication and transcription activator transactivation by interferon regulatory factor 7. J Virol. 2005;79:2420–2431. doi: 10.1128/JVI.79.4.2420-2431.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.de Oliveira DE, Ballon G, Cesarman E. NF-kappaB signaling modulation by EBV and KSHV. Trends Microbiol. 2010;18:248–257. doi: 10.1016/j.tim.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 65.Ye FC, Zhou FC, Xie JP, Kang T, Greene W, Kuhne K, Lei XF, Li QH, Gao SJ. Kaposi’s sarcoma-associated herpesvirus latent gene vFLIP inhibits viral lytic replication through NF-kappaB-mediated suppression of the AP-1 pathway: a novel mechanism of virus control of latency. J Virol. 2008;82:4235–4249. doi: 10.1128/JVI.02370-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brown HJ, Song MJ, Deng H, Wu TT, Cheng G, Sun R. NF-kappaB inhibits gammaherpesvirus lytic replication. J Virol. 2003;77:8532–8540. doi: 10.1128/JVI.77.15.8532-8540.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.