Abstract
Alterations in the metabolic control of lipid and glucose homeostasis predispose an individual to develop cardiometabolic diseases such as type 2-diabetes and atherosclerosis. Work over the last years has suggested that miRNAs play an important role in regulating these physiological processes. The contribution of miRNAs in regulating metabolism is exemplified by miR-33, an intronic miRNA encoded in the Srebp genes. miR-33 controls cellular cholesterol export and fatty acid degradation while its host genes stimulate cholesterol and fatty acid synthesis. Other miRNAs, such as miR-122, also play a critical role in regulating lipid homeostasis by controlling cholesterol synthesis and lipoprotein secretion in the liver. This review article summarizes the recent findings in the field, highlighting the contribution of miRNAs in regulating lipid and glucose metabolism. We will also discuss how the modulation of specific miRNAs may be a promising strategy to treat metabolic diseases.
MicroRNAs (miRNAs) are small (18–25 nucleotides in length), evolutionarily conserved, non-coding RNAs that have an important function in gene regulation, acting predominantly at the post-transcriptional level1, 2. Mature miRNA products are generated from a longer primary miRNA (pri-miRNA) transcript through sequential processing by the ribonucleases DROSHA and DICER. miRNAs typically control the expression of their target genes by imperfect base pairing to the 3’-untranslated regions (3’UTR) of messenger RNAs (mRNAs), thereby inducing repression of their target mRNAs1, 2. This inhibitory effect can occur by either transcript destabilization, translational inhibition, or both (more detailed information about miRNA biogenesis, function and targeting activity can be found in recent reviews covering these topics). Importantly, a single miRNA can regulate the expression of hundreds of genes and the expression of a single gene can be regulated by multiple miRNAs. The effect of a particular miRNA on gene expression is likely to be dictated by the relative expression of the miRNA and its target genes which can compete for the binding in their 3’UTRs. Of note, one miRNA often regulates multiple genes that are involved in a specific signaling cascade or cellular mechanism, thus making miRNAs potent biological regulators1, 2. Since miRNAs have been described in the early 90’s as regulators of developmental timing in Caenorhabditis elegans, they have been shown to participate in almost every cellular process investigated, including metabolic homeostasis2–5.
microRNAs as regulators of lipid metabolism
Growing evidence suggests that faulty regulation of lipid metabolism promotes metabolic diseases. In addition to the classical transcriptional regulators, SREBPs and LXRs, several miRNAs have been shown to post-transcriptionally regulate the expression of key genes involved in lipid homeostasis, including miR-122, miR-33, miR-106, miR-758, miR-26, miR-370, miR-378/378*, let-7, miR-27, miR-143, miR34a and miR-3356–21. In the present review we will focus our attention on the liver specific miR-122 and the well-characterized intronic miR-33.
miR-122
miR-122 is the most abundant miRNA in the liver, with approximately up to 135,000 copies per human hepatocyte, accounting for ~ 75% of total miRNA expression in this organ 22–25. miR-122 plays important roles in a wide variety of liver functions ranging from cholesterol metabolism, liver cancer, stress responses, and viral infection to circadian regulation of hepatic genes7, 23–27. Two pioneering studies have shown that antisense targeting of miR-122 results in a significant reduction of plasma cholesterol levels7, 22. The first study shows that the effect on plasma cholesterol results most likely from decreased expression of many cholesterol biosynthetic genes, including 3-hydroxy-3-methylglutaryl-CoA reductase (Hmgcr), the rate-limiting enzyme in the cholesterol biosynthesis pathway22. Despite this, the effects of miR-122 on cholesterol biosynthesis are indirect and it is unclear which direct targets of miR-122 mediate them. Interestingly, this study underlines that a miRNA loss-of-function phenotype may be caused by genes that are not directly targeted by the miRNA. The second study implements a similar antisense technology (ASO) against miR-122 in mice and not only confirms the effect on plasma cholesterol, but also reports a significant decrease in plasma triglycerides, as well as decreased hepatic steatosis, in high-fat-diet-fed mice7. Furthermore, hepatocytes isolated from ASO-miR-122-treated mice display decreased hepatic fatty acid and sterol synthesis and increased fatty acid oxidation, likely due to the observed increased levels of AMP-activated kinase (AMPK)7. Subsequent studies using locked nucleic acid (LNA) chemistry in mice and non-human primates corroborate the reduced plasma cholesterol levels without any apparent liver toxicity28. Recently, miR-122 liver-specific knockout and miR-122 germline knockout mice have been shown to have a significant reduction (~30%) in total serum cholesterol and triglyceride (TG) levels and therefore, recapitulate the effects observed with antisense inhibitors of miR-12223, 25. Interestingly, the study of Tsai and colleagues also found a significant down-regulation of the microsomal TG transfer protein (MTTP), which is essential for the assembly of lipoproteins25. Intriguingly, Mttp is not a direct target of miR-122 and the mechanism by which miR-122 regulates its expression is still unknown. Altogether, these results demonstrate that miR-122 plays an important role in regulating serum cholesterol and TG levels by controlling cholesterol biosynthesis and very-low density lipoprotein (VLDL) secretion in the liver.
In addition, a recent report has also shown that the knockdown of miR-122 results in the regulation of hundreds of mRNAs, of which a disproportionately high fraction accumulates in a circadian fashion26. The transcripts associated with these pathways indeed show the strongest time point-specific changes upon miR-122 depletion. The identification of peroxisome proliferator-activated receptor (PPAR) alpha, beta, and gamma and the PPAR alpha coactivator, Smarcd1/Baf60a, as novel targets of miR-122 suggest an involvement of the circadian metabolic regulators of the Ppar family in miR-122-mediated metabolic control26. Taken together these results suggest that inhibition of miR-122 might be a feasible therapeutic approach. In another study, 46 miRNAs were differentially expressed in humans with nonalcoholic fatty liver disease (NAFLD)29. miR-122 was downregulated in NAFLD, and this was correlated with increased expression of lipogenic genes in human livers29. Knockdown of miR-122 in HepG2 cells recapitulated the lipogenic gene expression profile observed in individuals with NAFLD. In this case, it seems likely that miR-122 down-regulation is a compensatory mechanism that counters increasing hepatic lipid levels, rather than a causative agent in the development of NAFLD. Future studies should clarify this apparent discrepancy. In line with these observations, some of the above-mentioned reports have also shown that antagonism of miR-122, in both mice and non-human primates, not only lowers low-density lipoproteins (LDL) levels but the levels of high-density lipoproteins (HDL) as well. These a priori adverse effects, together with the recently reported increased risk of developing hepatocellular carcinoma23, 25, challenge the therapeutic approach of miR-122 inhibition for the treatment of metabolic lipid diseases.
miR-33
miR-33 consists of two intronic microRNAs, miR-33a and miR-33b, which are encoded within the introns of the Srebp2 and Srebp1 genes, respectively6, 15, 16, 19. While miR-33a and miR-33b share their target activity, they differ in their pattern of evolutionary conservation. miR-33a is encoded within intron 16 of the human Srebp2 gene and is conserved in many animal species6, 15, 16, 19. However, the conservation of miR-33b, which is found within intron 17 of the human Srebp1 gene, is lost in many species including rodents and rabbits. miR-33a and miR-33b are co-transcribed with their respective host genes, thereby participating in the regulation of physiological processes related to Srebp2 and -16, 15, 16, 19. Indeed, we and others have found that miR-33a and miR-33b regulate intracellular cholesterol and fatty acid homeostasis in concert with their host genes. Specifically, miR-33a has been shown to target genes involved in cholesterol export such as the adenosine tri-phosphate binding cassette (ABC) transporters Abca1 and Abcg115, 16, 19 and the endolysosomal transport protein Niemann-Pick C1 (Npc1)19. In agreement with the regulation of ABCA1 by miR-33, modulation of miR-33a levels results in encompassing effects in cholesterol efflux in macrophages thus suggesting that miR-33 may participate in the regulation of HDL levels in vivo. Indeed, three independent studies have demonstrated that endogenous inhibition of miR-33 using different strategies leads to a significant increase in hepatic ABCA1 expression and plasma HDL levels15, 16, 19, findings that were later confirmed in the miR-33 knockout mice 30. Most importantly, anti-miR-33 therapy also results in increased plasma HDL levels in non-human primates31. Interestingly, in the well-characterized model for hypercholesterolemia, LDLr knockout mice, anti-miR-33 therapy promotes reverse cholesterol transport (RCT) and atherosclerosis regression32. Despite this, the effect of anti-miR-33 therapy on RCT might not be solely due to ABCA1 up-regulation since it has been recently reported that miR-33 also targets two canalicular transporters, Abcb11 and Atp8b1, which regulate bile secretion33.
In addition to the important role of miR-33a and its host gene, Srebp2, in regulating cholesterol metabolism, the genomic localization of miR-33b in an intron of the Srebp1 gene, led several groups to study the contribution of miR-33 in controlling additional metabolic pathways such as fatty acid metabolism6, 10. Importantly, miR-33a and miR-33b contribute to the regulation of fatty acid metabolism by controlling the expression of carnitine O-octanyl transferase (Crot), carnitine palmitoyltransferase 1A (Cpt1a), and hydroxyacyl-CoA dehydrogenase-3-ketoacyl-CoA thiolase-enoyl-CoA hydratase (trifunctional protein) β-subunit (Hadhb)6, 10. CROT and CPT1A regulate the transport of fatty acids to the mitochondria for their degradation and HADHB is directly involved in mitochondrial fatty acid β-oxidation. Interestingly, endogenous inhibition of miR-33 in human hepatic cells increases the degradation of fatty acids, suggesting that anti-miR-33 therapy may be useful for treating hepatic steatosis by increasing the degradation rate of fatty acids in the liver6, 10. In this regard, non-human primates treated with anti-miR-33 oligonucleotides show a significant reduction of plasma VLDL levels31. These results could be explained by a reduced lipidation and secretion of ApoB-containing lipoproteins due to the increased fatty acid oxidation that might be occurring in the liver of non-human primates treated with anti-miR-33 oligonucleotides. However, this remains to be addressed.
In addition to the regulation of fatty acid oxidation, miR-33a and miR–33b have also been shown to control the expression of Ampkα1 and sirtuin 6 (Sirt6), which are involved in the regulation of lipid and glucose metabolism6. The latter will be discussed in the following section. AMPKα1 regulates key lipogenic enzymes, including HMGCR and ACC. Thus, inhibition of AMPKα1 by miR-33 may increase HMGCR and ACC activity to boost intracellular levels of cholesterol and fatty acids. Altogether, these results suggest a paradigm in which miR-33a and miR-33b act in concert with their host genes, Srebp2 and Srebp1, to increase intracellular cholesterol and fatty acid levels by balancing transcriptional induction and post-transcriptional repression of lipid metabolism genes. Finally, insulin receptor substrate 2 (Irs2), an adaptor protein that controls insulin signaling in the liver, has also been shown to be a miR-33 target, thereby affecting the signaling of a complex downstream network of proteins including protein kinase B (PKB; also known as AKT) phosphorylation and FOXO1 cytoplasmic localization6. Collectively, these data indicate that both isoforms of miR-33 participate in the regulation of relevant pathways that impact three of the primary risk factors of metabolic syndrome, namely insulin resistance, low HDL and high VLDL and suggest that anti-miR-33 therapies may be an attractive approach for treating metabolic diseases.
In contrast to miR-122, miR-33 is less expressed in the liver compared with other tissues such as the brain16, 21. However the presence of multiple binding sites in the 3’UTR of some of the key target genes, including Abca1 and Crot, explain why anti-miR-33 therapy is able to increase their expression in the liver6, 19. The role of miR-33 in the brain is under intensive investigation since ABCA1 also plays an important role in regulating Aβ clearance and its expression has been associated with neurological disorders, including Alzheimer’s disease34, 35. Altogether, these findings show that miR-33 is playing key roles in controlling many physiological processes and much work is necessary to understand the impact of anti-miR-33 therapy in human physiology to rule out possible adverse effects of the chronic treatment with anti-miR-33 oligonucleotides.
Other miRNAs that regulate lipid metabolism
Additional miRNAs (miR-106, miR-758, miR-26, miR-370, miR-378/378*, let-7, miR-27, miR34a and miR-335) have been described to participate in the regulation of lipid metabolism. Among them, miR-758, miR-26 and miR-106b have been shown to regulate cellular cholesterol efflux by targeting ABCA1 in macrophages, hepatocytes and neuronal cell lines, therefore indicating that the post-transcriptional regulation of ABCA1 expression is mediated by multiple miRNAs12, 18, 36. miR-370 has been shown to reduce fatty acid β-oxidation via its targeting activity towards Cpt1a11. In addition, miR-370 appears to participate in the regulation of miR-122 by increasing the expression of lipogenic genes, including Srebp1 and Dgat211. miR-34a targets hepatic sirtuin 1 (Sirt1) and interestingly, the expression of miR-34a was inversely correlated with levels of SIRT1 in fatty livers of diet-induced obese mice37. Both strands of miR-378 have been shown to regulate TG synthesis in 3T3-L1 adipocytes, thereby cooperating in the regulation of lipid accumulation during adipogenesis9. Interestingly, overexpression of miR378/378* in ST2 cells increases the expression of fatty acid binding protein 4 (FABP4), FASN, SCD1, Kruppel-like factor 15 (KLF15), and resistin9. Finally, let-7, miR-143, miR-335, miR-27 and miR-103/107 were also reported to control adipocyte differentiation8, 13, 14, 17, 20, 21.
microRNAs as regulators of glucose metabolism and insulin signaling
Diabetes mellitus is the most common metabolic disorder world-wide and is a major risk factor for cardiovascular disease38. Diabetes mellitus is characterized by elevated blood glucose levels due to a lack of insulin-producing pancreatic β-cells (Type 1 diabetes) or insulin resistance in peripheral tissues (Type 2 diabetes)38. Plasma glucose levels are tightly controlled by insulin and glucagon. Changes in circulating glucose modulate insulin production by the pancreatic β-cells, leading to an increase in glucose uptake in peripheral tissues, including the muscle and adipose tissue39. Moreover, insulin inhibits glucose synthesis and glycogen degradation and stimulates lipid synthesis in the liver39.
Insulin binds to its receptor (INSR) and stimulates an intracellular signaling pathway involving IRS1 and 2, phosphatidylinositol 3-kinase (PI3K) and AKT40. AKT phosphorylates and inactivates forkhead box O1 (FOXO1), a key transcription factor that regulates glucose 6-phosphatase (G6pc) and phosphoenolpyruvate carboxykinase (Pck1) expression, leading to a significant reduction of glucose production in the liver41. Insulin also stimulates the translocation of glucose transporters, such as GLUT-4 in the muscle and adipose tissues, thus promoting glucose clearance42. In addition to hormones, miRNAs have emerged as critical regulators of glucose metabolism by regulating insulin production and secretion, as well as insulin sensitivity. The global impact of miRNAs in glucose production and pancreatic β-cell functions was defined with the generation of pancreas-specific dicer knockout mice43. These mice survive until birth but fail to grow and die by postnatal day 3. The absence of miRNAs during pancreatic β-cell development causes defective Notch signaling, leading to an increase in cell death and several defects in all pancreatic cell lineages43. To solve this problem, Melkman-Zehavi and colleagues developed pancreatic dicer-conditional knockout mice inducible upon treatment with tamoxifen44. Inactivation of pancreatic dicer expression in adult mice results in enhanced blood glucose and reduced plasma insulin levels44. Dicer-deficient β-cells show a significant decrease in insulin synthesis and secretion, which is associated with the up-regulation of basic helix-loop-helix family member e22 (Bhlhe22) and Sox6, two transcriptional repressors of the insulin gene44. Interestingly, four miRNAs, including miR-24, miR-26, miR-182 and miR-148 regulate Bhlhe22 and Sox6 expression at the post-transcriptional level and are significantly down-regulated in dicer-deficient pancreatic β-cells44.
Besides the studies in dicer null mice, several reports have recently shown the critical role of specific miRNAs in regulating insulin production and sensitivity. The importance of some of them will be discussed in the following sections of this review article.
miR-375
miR-375 is one of the most abundant miRNAs in the pancreas and regulates insulin secretion independently of changes in plasma glucose levels45. miR-375 null mice are normoinsulinemic but hyperglycemic and glucose intolerant 46. These mice also have an increase in the number of pancreatic α-cells and fasting and fed plasma glucagon levels46. The increase in plasma glucagon levels results in a significant increase in G6PC and PCK1 expression and glucose production in the liver. miR-375 also regulates the expression of a cluster of genes controlling cellular growth and proliferation, including caveolin-1 (Cav-1), inhibitor of DNA binding 3 (Id3), Ras-dexometasone-induced-1 (Rasd1) and the human antigen D/embryonic lethal abnormal vision-like 4 (HuD/Elavl4)46. HuD/Elavl4 is an RNA-binding protein that regulates preproinsulin (Ins2) translation and insulin production47. This finding suggests that the reduced insulin secretion observed in the pancreatic β-cells from the miR-375 null mice may be due to increased HuD expression in pancreatic β-cells. Moreover, miR-375 targets myotrophin (Mtpn), a gene involved in actin depolymerization and vesicular trafficking, thereby reducing insulin exocitosis46.
Other miRNAs that regulates insulin secretion
In addition to miR-375, other miRNAs have been shown to regulate insulin release including miR-124a, miR-9, miR-96a and miR-3348–51. miR-124a regulates insulin secretion by controlling the expression of Rab27a which is involved together with its effector, granuphilin/Slp4, in the exocytosis of insulin-containing secretory granules in pancreatic β-cells48. Granuphilin/Slp4, a Rab effector known to negatively modulate insulin exocytosis, is also regulated by miR-9, which directly targets Onecut-2 (Oc2)49, 50. OC2 binds to the granuphilin promoter and represses its transcriptional activity. Therefore, over-expression of miR-9 in insulin-secreting cells results in a reduction of insulin exocytosis elicited by glucose or potassium49, 50. Similarly to miR-9, miR-96 also up-regulates the expression of granuphilin, but independently of OC248. miR-96 and miR-124a over-expression also inhibits Noc2, a Rab effector involved in exocytosis, in an insulin-secreting mouse cell line (MIN6B1)48. Lastly, miR-29 also controls insulin secretion by regulating the monocarboxylate transporter 1 (Mct1) expression52. In summary, these reports demonstrate that miR-9, mi-96 and miR-124 control the expression of multiple genes regulating the exocytotic machinery to fine-tune insulin release.
Changes in cellular cholesterol content affect insulin secretion. In this regard, the ABCA1 transporter plays an important role in regulating cholesterol homeostasis in pancreatic β-cells. Indeed, β-cell specific deletion or loss-of function mutations in ABCA1 result in impaired glucose tolerance, insulin secretion and β-cell dysfunction53. Very interestingly, it has been recently shown that miR-33 also modulates the expression of ABC transporters in human and mouse pancreatic islets51. Overexpresion of miR-33 reduces the expression of ABCA1 and insulin secretion in primary islets and MIN6, a mouse insulinoma cell line. Conversely, endogenous inhibition of miR-33 increases ABCA1 expression and insulin secretion in wild-type mouse islets but not in islets isolated from ABCA1-deficient mice51. Altogether, these results suggest that miR-33 also plays an important role in regulating insulin secretion and glucose homeostasis. Further studies are important to elucidate the role of miR-33 in regulating glucose metabolism in vivo.
Regulation of insulin signaling by miRNAs
Other miRNAs regulate insulin sensitivity in the liver and peripheral tissues by controlling the expression of many components of the insulin signaling pathway, including insulin-like growth factor receptor 1 (IGF1R), insulin receptor (INSR), IRS2, phosphatidylinositol 3-kinase regulatory subunit-α (PIK3IP1), AKT2, tuberous sclerosis protein 1 (TSC1), CAV1 and rapamycin-insensitive companion of mTOR (RICTOR).
Two independent groups have recently shown that the Let-7 family of miRNAs regulates glucose homeostasis and insulin sensitivity54, 55. Global and pancreas specific over-expression of Let-7 in mice results in impaired glucose tolerance and reduced glucose-induced pancreatic insulin secretion54, 55. Specific knockdown of Let-7 in Let-7 transgenic mice reverses the phenotype by improving insulin sensitivity in the muscle and adipose tissues. Let-7 directly targets many components of the insulin-signaling pathway such as Igf1r, Insr, Irs2, Pik3ip1, Akt2, Tsc1, and Rictor, thereby reducing insulin sensitivity54, 55. LIN28 tightly controls the expression of Let-7. This RNA-binding protein represses the biogenesis of Let-7 miRNAs and is highly expressed during normal embriogenesis and is up-regulated in some cancers56, 57. Interestingly, Lin28 transgenic mice reduce the expression of Let-7 and improve glucose clearance and insulin sensitivity55. By contrast, skeletal muscle-specific Lin28 knockout mice show impaired glucose tolerance55. Altogether these studies strongly suggest that the Lin28/Let-7 axis regulates glucose metabolism54, 55.
In addition to Let-7, other miRNAs, including miR-33, miR-103, miR-107 and miR-29a/b, also regulate the insulin-signaling pathway6, 52, 58, 59. As described before, miR-33 targets Irs2 and regulates insulin sensitivity in human hepatic cell lines. Moreover, miR-33 also regulates the expression of Ampkα and Sirt6, which are involved in regulating lipid and glucose metabolism6. miR-103 and miR-107 were recently shown to be up-regulated in obese mice59. Moreover the expression of both miRNAs is increased in subjects with NAFLD, a condition often associated with diabetes. miR-103 and miR-107 have similar mature sequences and are thought to target similar genes. Over-expression of miR-107 results in an increase in fasting glucose and insulin levels59. Conversely, silencing of miR-103/miR-107 enhances insulin sensitivity in the liver and in the adipose tissue. Mechanistically, miR-103/107 inhibition increases the expression of CAV-1, a scaffold protein required for caveolae formation, and enhances insulin signaling by increasing insulin receptor stability in the cell membrane59. Indeed, miR-103/107 antagomirs are not able to enhance insulin sensitivity in Cav-1 null mice. miR-29a and miR-29b are also up-regulated in white adipose tissue and in the liver of diabetic rats. In addition, to targeting Cav-2, another caveolae structural component52, 58, the miR-29 family members also regulate phophatidylinositol 3-kinase regulatory subunit alpha (PIK3R1), which regulates insulin signaling.
Other miRNAs that regulates glucose homeostasis
The complexity of miRNAs in regulating physiological processes is exemplified by miR-208a, a heart-specific miRNA that also regulates glucose metabolism and energy homeostasis60. miR-208 regulates the expression of the mediator complex 13 (MED13) which controls the transcription of the thyroid hormone (TH) and other nuclear hormone receptors. TH enhances energy expenditure and regulates body weight. Interestingly, mice administrated with anti-miR-208 oligonucleotides are resistant to obesity and glucose intolerant60. In contrast, Med13 cardiac-specific transgenic mice are resistant to diet-induced obesity with improved glucose tolerance60. This remarkable finding demonstrates how a cardiac-specific miRNA is able to regulate systemic energy homeostasis.
Besides the role of miR-208 in the heart, another interesting possibility that could be explored is that this miRNA maybe secreted in microvesicles by the heart and regulate insulin signaling and glucose metabolism in other peripheral tissues. However, several studies have not be able detect miR-208 in the plasma of subjects with type 2 diabetes (D2M) or dyslipidemia61.
In summary, multiple miRNAs are able to control glucose metabolism by regulating a network of genes in the liver and peripheral tissues. The contribution of specific miRNAs will be determined by the tissue and metabolic state.
Circulating miRNAs
Recently, several studies have highlighted the presence of miRNAs in the plasma. Plasma miRNAs are packaged in microvesicles (including exosomes) that protect them from degradation62. Moreover, recent reports have also identified these small RNAs associated with proteins including the RNA-binding protein Argonaute 2 (AGO2)63. The role of circulating miRNAs is under intense investigation and some studies suggest that they might play important roles in regulating atherogenesis and endothelial cell functions. Some miRNAs are enriched in the plasma under pathological conditions, including myocardial infarction (miR-208, miR-1, miR-133a and miR-21)64, hepatic steatosis and hepatic injury (miR-122)65 and hypertension (Let-7e)66 or reduced such as miR-126 in D2M61; therefore they can be used as disease biomarkers. Finally, Vickers and colleagues have also recently found miRNAs associated with lipoproteins. Interestingly, the HDL-miRNA profile of normal subjects is significantly different from that of familial hypercholesterolemia subjects67. HDL-miRNAs can be delivered to hepatic cells via the scavenger receptor class B type I (SR-BI), however the physiological relevance of this process in regulating gene expression in the liver and peripheral tissues, including atherosclerotic plaque macrophages, remains unknown and warrants further investigation.
Concluding remarks
miRNAs have emerged as key regulators of many physiological processes including lipid and glucose metabolism. Several preclinical studies have pointed out that targeting specific miRNAs, such as miR-33, miR-122, miR-103/107 and let-7 may be a promising strategy to ameliorate cardiometabolic disorders. However, the complexity of gene networks that a single miRNA may control and the potential adverse effects of the total inhibition of a specific miRNA remains to be deeply explored. For example, a global deficiency of miR-122 results in reduced plasma cholesterol levels but increased hepatic steatosis and hepatic cancer23, 25. Contrastingly, miR-122 pharmacological inhibition for few a months leads to a significant reduction of plasma lipid levels and reverses hepatic steatosis in mice. These paradoxical results strongly suggest that much work is necessary to fully understand the role of a single miRNA in regulating animal physiology. New approaches that integrate RNA-sequencing, proteomics and system biology methodologies will help us to elucidate how the modulation of gene networks by miRNAs contribute to the regulation of metabolic processes.
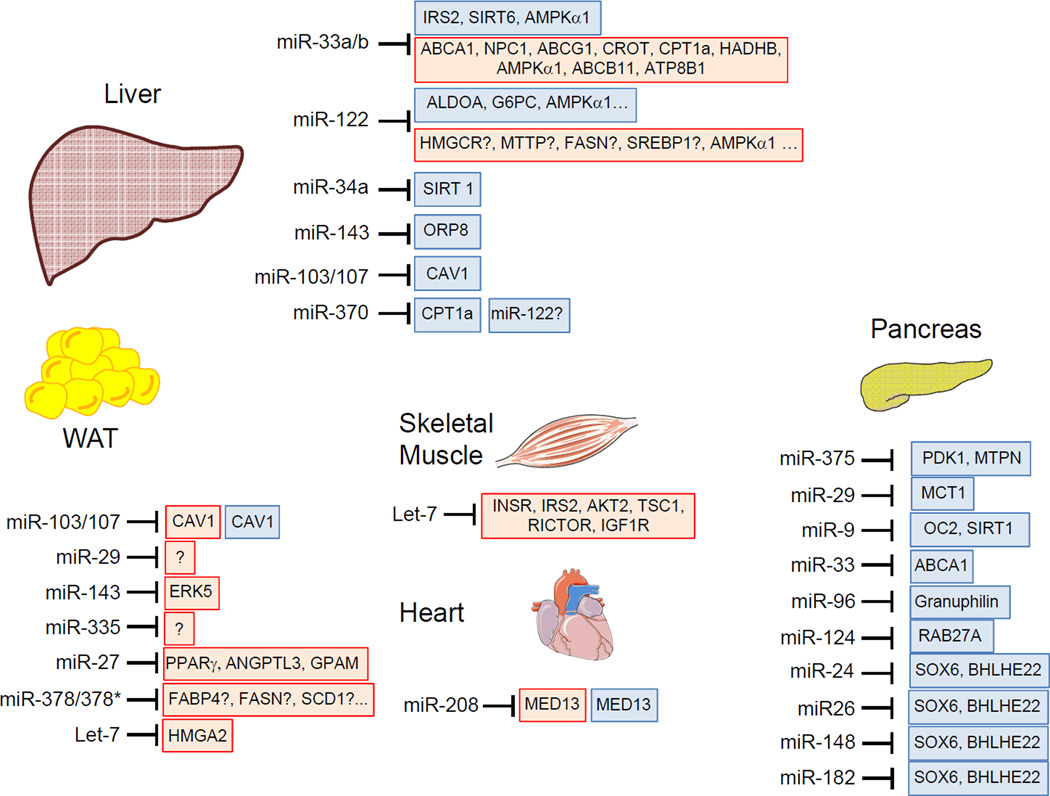
Figure 1. miRNA regulation of lipid metabolism, insulin signaling and glucose homeostasis.
Schematic overview of miRNAs involved in the regulation of glucose and lipid metabolism. Red boxes highlight genes involved in lipid metabolism and blue boxes highlight those genes related to insulin signaling and glucose homeostasis. Unknown direct target genes or molecular mechanisms that regulate the highlighted genes are marked with a question mark. Note that the target genes showed in the figure are those validated experimentally but these genes can be also modulated by other miRNAs and the miRNAs highlighted can regulate other genes that do not appear in the figure.
Table.
MicroRNA involved in lipid and glucose
| Regulator | Tissue/Cell Type | Target genes | Function | Reference |
|---|---|---|---|---|
| Let-7 | Mouse adipose and skeletal muscle | INSR, IGF1R, IR2, TSC1, RICTOR | ↑Insulin Sensitivity,↑ Glucose metabolism | 54,55 |
| 3T3-L1 cells | HMGA2 | ↓Adipogenesis, Adipose differentiation | 20 | |
| miR-9 | NS-1E cells, Mouse pancreatic β islets | OCT, SIRT1 | ↓Insulin secretion | 49,50 |
| miR-24 | Mouse pancreatic β cells | SOX6, BHLHE22 | ↑Insulin biosynthesis | 44 |
| miR-26 | Mouse pancreatic β cells | SOX6, BHLHE23 | ↑Insulin biosynthesis | 45 |
| miR-27 | 3T3-L1 cels | PPARG, ANGTPL3, GPAM | ↓Adipogenesis, Adipose differentiation | 13,14 |
| miR-29 | MIN6 cells, Mouse pancreatic β islets 3T3-L1 cels |
MCT1 ? |
↓Insulin secretion |
52 58 |
| miR-33a/b | Huh-7, HepG2, Mouse liver | ABCA1, ABCG1, ABCB11, ATP8B1 | ↓ Cholesterol Transport/Export |
15, 16, 19, 33 |
| Huh-7, HepG2, Mouse liver | NPC1, CROT, CPT1A, HADHB, AMPKα1 | ↓ β-Oxidation | 10,19,6 | |
| Huh-7, HepG2, Mouse liver | IRS2, SIRT6 | ↓Insulin signaling | 6 | |
| MIN6 cells, Mouse and human pancreatic β islets | ABCA1 | ↓Insulin secretion | 51 | |
| miR-34a | HepG2, Mouse liver | SIRT1 | ↑Insulin secretion | 37 |
| miR-96 | INS-1E cells, MIN6 cells | Granuphilin | ↓Insulin secretion | 48 |
| miR-103/107 | 3T3-L1 cels, Mouse Liver | CAV-1 | ↓Adipose differentiation, ↓Insulin sensitivity | 59 |
| miR-122 | Mouse liver | HMGCR | ↑Cholesterol Synthesis | 22 |
| HepG2 | ALDO, G6PC | Glucose homeostasis | 7 | |
| AMPKα1 | ↑ β-Oxidation | 7 | ||
| miR-124 | MIN6 cells | RAB27A | ↓Insulin secretion | 48 |
| miR-143 | Human adipocytes | ERK5 | ↑Adipose differentiation | 8 |
| miR-148 | MI N6 cells | SOX6, BHLHE22 | ↑Insulin biosynthesis | 44 |
| miR-182 | MI N6 cells | SOX6, BHLHE23 | ↑Insulin biosynthesis | 45 |
| miR-208 | Mouse heart | MED13 | ↑Insulin sensitivity, ↑glucose tolerance | 60 |
| miR-370 | HepG2 | CPT1A | ↓ β-Oxidation | 11 |
| miR-375 | Mouse pancreas | PDK, MTPN | Pancreas homeostasis | 46 |
| miR-378/378* | 3T3-L1 cels | FABP4, FASN, SCD1 | ↑Adipose differentiation | 9 |
Acknowledgment
None
Source of funding
C.F.-H. and Y.S Labs are supported by grants from the National Institute of Health (R01HL106063 and R01HL107953 to C.F.H and R01HL105945 to Y.S).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures
None
References
- 1.Ambros V. The functions of animal micrornas. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. Micrornas: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ambros V. Microrna pathways in flies and worms: Growth, death, fat, stress, and timing. Cell. 2003;113:673–676. doi: 10.1016/s0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- 4.Lee RC, Feinbaum RL, Ambros V. The c. Elegans heterochronic gene lin-4 encodes small rnas with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 5.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in c. Elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 6.Davalos A, Goedeke L, Smibert P, Ramirez CM, Warrier NP, Andreo U, Cirera-Salinas D, Rayner K, Suresh U, Pastor-Pareja JC, Esplugues E, Fisher EA, Penalva LO, Moore KJ, Suarez Y, Lai EC, Fernandez-Hernando C. Mir-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc Natl Acad Sci U S A. 2011;108:9232–9237. doi: 10.1073/pnas.1102281108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, Subramaniam A, Propp S, Lollo BA, Freier S, Bennett CF, Bhanot S, Monia BP. Mir-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 8.Esau C, Kang X, Peralta E, Hanson E, Marcusson EG, Ravichandran LV, Sun Y, Koo S, Perera RJ, Jain R, Dean NM, Freier SM, Bennett CF, Lollo B, Griffey R. Microrna-143 regulates adipocyte differentiation. J Biol Chem. 2004;279:52361–52365. doi: 10.1074/jbc.C400438200. [DOI] [PubMed] [Google Scholar]
- 9.Gerin I, Bommer GT, McCoin CS, Sousa KM, Krishnan V, MacDougald OA. Roles for mirna-378/378* in adipocyte gene expression and lipogenesis. Am J Physiol Endocrinol Metab. 2010;299:E198–E206. doi: 10.1152/ajpendo.00179.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerin I, Clerbaux LA, Haumont O, Lanthier N, Das AK, Burant CF, Leclercq IA, Macdougald OA, Bommer GT. Expression of mir-33 from an srebp2 intron inhibits cholesterol export and fatty acid oxidation. J Biol Chem. 2010;285:33652–33661. doi: 10.1074/jbc.M110.152090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iliopoulos D, Drosatos K, Hiyama Y, Goldberg IJ, Zannis VI. Microrna-370 controls the expression of microrna-122 and cpt1alpha and affects lipid metabolism. J Lipid Res. 2010;51:1513–1523. doi: 10.1194/jlr.M004812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim J, Yoon H, Ramirez CM, Lee SM, Hoe HS, Fernandez-Hernando C. Mir-106b impairs cholesterol efflux and increases abeta levels by repressing abca1 expression. Exp Neurol. 2012;235:476–483. doi: 10.1016/j.expneurol.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim SY, Kim AY, Lee HW, Son YH, Lee GY, Lee JW, Lee YS, Kim JB. Mir-27a is a negative regulator of adipocyte differentiation via suppressing ppargamma expression. Biochem Biophys Res Commun. 2010;392:323–328. doi: 10.1016/j.bbrc.2010.01.012. [DOI] [PubMed] [Google Scholar]
- 14.Lin Q, Gao Z, Alarcon RM, Ye J, Yun Z. A role of mir-27 in the regulation of adipogenesis. FEBS J. 2009;276:2348–2358. doi: 10.1111/j.1742-4658.2009.06967.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marquart TJ, Allen RM, Ory DS, Baldan A. Mir-33 links srebp-2 induction to repression of sterol transporters. Proc Natl Acad Sci U S A. 2010;107:12228–12232. doi: 10.1073/pnas.1005191107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Najafi-Shoushtari SH, Kristo F, Li Y, Shioda T, Cohen DE, Gerszten RE, Naar AM. Microrna-33 and the srebp host genes cooperate to control cholesterol homeostasis. Science. 2010;328:1566–1569. doi: 10.1126/science.1189123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakanishi N, Nakagawa Y, Tokushige N, Aoki N, Matsuzaka T, Ishii K, Yahagi N, Kobayashi K, Yatoh S, Takahashi A, Suzuki H, Urayama O, Yamada N, Shimano H. The up-regulation of microrna-335 is associated with lipid metabolism in liver and white adipose tissue of genetically obese mice. Biochem Biophys Res Commun. 2009;385:492–496. doi: 10.1016/j.bbrc.2009.05.058. [DOI] [PubMed] [Google Scholar]
- 18.Ramirez CM, Davalos A, Goedeke L, Salerno AG, Warrier N, Cirera-Salinas D, Suarez Y, Fernandez-Hernando C. Microrna-758 regulates cholesterol efflux through posttranscriptional repression of atp-binding cassette transporter a1. Arterioscler Thromb Vasc Biol. 2011;31:2707–2714. doi: 10.1161/ATVBAHA.111.232066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rayner KJ, Suarez Y, Davalos A, Parathath S, Fitzgerald ML, Tamehiro N, Fisher EA, Moore KJ, Fernandez-Hernando C. Mir-33 contributes to the regulation of cholesterol homeostasis. Science. 2010;328:1570–1573. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun T, Fu M, Bookout AL, Kliewer SA, Mangelsdorf DJ. Microrna let-7 regulates 3t3-l1 adipogenesis. Mol Endocrinol. 2009;23:925–931. doi: 10.1210/me.2008-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vickers KC, Shoucri BM, Levin MG, Wu H, Pearson DS, Osei-Hwedieh D, Collins FS, Remaley AT, Sethupathy P. Microrna-27b is a regulatory hub in lipid metabolism and is altered in dyslipidemia. Hepatology. doi: 10.1002/hep.25846. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of micrornas in vivo with 'antagomirs'. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 23.Hsu SH, Wang B, Kota J, Yu J, Costinean S, Kutay H, Yu L, Bai S, La Perle K, Chivukula RR, Mao H, Wei M, Clark KR, Mendell JR, Caligiuri MA, Jacob ST, Mendell JT, Ghoshal K. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of mir-122 in liver. J Clin Invest. 2012;122:2871–2883. doi: 10.1172/JCI63539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S, Orum H. Therapeutic silencing of microrna-122 in primates with chronic hepatitis c virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsai WC, Hsu SD, Hsu CS, Lai TC, Chen SJ, Shen R, Huang Y, Chen HC, Lee CH, Tsai TF, Hsu MT, Wu JC, Huang HD, Shiao MS, Hsiao M, Tsou AP. Microrna-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J Clin Invest. 2012;122:2884–2897. doi: 10.1172/JCI63455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gatfield D, Le Martelot G, Vejnar CE, Gerlach D, Schaad O, Fleury-Olela F, Ruskeepaa AL, Oresic M, Esau CC, Zdobnov EM, Schibler U. Integration of microrna mir-122 in hepatic circadian gene expression. Genes Dev. 2009;23:1313–1326. doi: 10.1101/gad.1781009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kojima S, Gatfield D, Esau CC, Green CB. Microrna-122 modulates the rhythmic expression profile of the circadian deadenylase nocturnin in mouse liver. PLoS One. 2010;5:e11264. doi: 10.1371/journal.pone.0011264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjarn M, Hansen HF, Berger U, Gullans S, Kearney P, Sarnow P, Straarup EM, Kauppinen S. Lna-mediated microrna silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 29.Cheung O, Puri P, Eicken C, Contos MJ, Mirshahi F, Maher JW, Kellum JM, Min H, Luketic VA, Sanyal AJ. Nonalcoholic steatohepatitis is associated with altered hepatic microrna expression. Hepatology. 2008;48:1810–1820. doi: 10.1002/hep.22569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horie T, Ono K, Horiguchi M, Nishi H, Nakamura T, Nagao K, Kinoshita M, Kuwabara Y, Marusawa H, Iwanaga Y, Hasegawa K, Yokode M, Kimura T, Kita T. Microrna-33 encoded by an intron of sterol regulatory element-binding protein 2 (srebp2) regulates hdl in vivo. Proc Natl Acad Sci U S A. 2010;107:17321–17326. doi: 10.1073/pnas.1008499107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rayner KJ, Esau CC, Hussain FN, McDaniel AL, Marshall SM, van Gils JM, Ray TD, Sheedy FJ, Goedeke L, Liu X, Khatsenko OG, Kaimal V, Lees CJ, Fernandez-Hernando C, Fisher EA, Temel RE, Moore KJ. Inhibition of mir-33a/b in non-human primates raises plasma hdl and lowers vldl triglycerides. Nature. 2011;478:404–407. doi: 10.1038/nature10486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rayner KJ, Sheedy FJ, Esau CC, Hussain FN, Temel RE, Parathath S, van Gils JM, Rayner AJ, Chang AN, Suarez Y, Fernandez-Hernando C, Fisher EA, Moore KJ. Antagonism of mir-33 in mice promotes reverse cholesterol transport and regression of atherosclerosis. J Clin Invest. 2011;121:2921–2931. doi: 10.1172/JCI57275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Allen RM, Marquart TJ, Albert CJ, Suchy FJ, Wang DQ, Ananthanarayanan M, Ford DA, Baldan A. Mir-33 controls the expression of biliary transporters, and mediates statin- and diet-induced hepatotoxicity. EMBO Mol Med. 2012;4:882–95. doi: 10.1002/emmm.201201228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wahrle SE, Jiang H, Parsadanian M, Hartman RE, Bales KR, Paul SM, Holtzman DM. Deletion of abca1 increases abeta deposition in the pdapp transgenic mouse model of alzheimer disease. The Journal of biological chemistry. 2005;280:43236–43242. doi: 10.1074/jbc.M508780200. [DOI] [PubMed] [Google Scholar]
- 35.Wahrle SE, Jiang H, Parsadanian M, Kim J, Li A, Knoten A, Jain S, Hirsch-Reinshagen V, Wellington CL, Bales KR, Paul SM, Holtzman DM. Overexpression of abca1 reduces amyloid deposition in the pdapp mouse model of alzheimer disease. J Clin Invest. 2008;118:671–682. doi: 10.1172/JCI33622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun D, Zhang J, Xie J, Wei W, Chen M, Zhao X. Mir-26 controls lxr-dependent cholesterol efflux by targeting abca1 and arl7. FEBS Lett. 2012;586:1472–1479. doi: 10.1016/j.febslet.2012.03.068. [DOI] [PubMed] [Google Scholar]
- 37.Lee J, Padhye A, Sharma A, Song G, Miao J, Mo YY, Wang L, Kemper JK. A pathway involving farnesoid x receptor and small heterodimer partner positively regulates hepatic sirtuin 1 levels via microrna-34a inhibition. J Biol Chem. 2010;285:12604–12611. doi: 10.1074/jbc.M109.094524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ashcroft FM, Rorsman P. Diabetes mellitus and the beta cell: The last ten years. Cell. 2012;148:1160–1171. doi: 10.1016/j.cell.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin HV, Accili D. Hormonal regulation of hepatic glucose production in health and disease. Cell Metab. 2011;14:9–19. doi: 10.1016/j.cmet.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sale EM, Sale GJ. Protein kinase b: Signalling roles and therapeutic targeting. Cell Mol Life Sci. 2008;65:113–127. doi: 10.1007/s00018-007-7274-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gross DN, Wan M, Birnbaum MJ. The role of foxo in the regulation of metabolism. Curr Diab Rep. 2009;9:208–214. doi: 10.1007/s11892-009-0034-5. [DOI] [PubMed] [Google Scholar]
- 42.Leto D, Saltiel AR. Regulation of glucose transport by insulin: Traffic control of glut4. Nat Rev Mol Cell Biol. 2012;13:383–396. doi: 10.1038/nrm3351. [DOI] [PubMed] [Google Scholar]
- 43.Lynn FC, Skewes-Cox P, Kosaka Y, McManus MT, Harfe BD, German MS. Microrna expression is required for pancreatic islet cell genesis in the mouse. Diabetes. 2007;56:2938–2945. doi: 10.2337/db07-0175. [DOI] [PubMed] [Google Scholar]
- 44.Melkman-Zehavi T, Oren R, Kredo-Russo S, Shapira T, Mandelbaum AD, Rivkin N, Nir T, Lennox KA, Behlke MA, Dor Y, Hornstein E. Mirnas control insulin content in pancreatic beta-cells via downregulation of transcriptional repressors. EMBO J. 2011;30:835–845. doi: 10.1038/emboj.2010.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P, Stoffel M. A pancreatic islet-specific microrna regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 46.Poy MN, Hausser J, Trajkovski M, Braun M, Collins S, Rorsman P, Zavolan M, Stoffel M. Mir-375 maintains normal pancreatic alpha- and beta-cell mass. Proc Natl Acad Sci U S A. 2009;106:5813–5818. doi: 10.1073/pnas.0810550106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee EK, Kim W, Tominaga K, Martindale JL, Yang X, Subaran SS, Carlson OD, Mercken EM, Kulkarni RN, Akamatsu W, Okano H, Perrone-Bizzozero NI, de Cabo R, Egan JM, Gorospe M. Rna-binding protein hud controls insulin translation. Mol Cell. 2012;45:826–835. doi: 10.1016/j.molcel.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lovis P, Gattesco S, Regazzi R. Regulation of the expression of components of the exocytotic machinery of insulin-secreting cells by micrornas. Biol Chem. 2008;389:305–312. doi: 10.1515/BC.2008.026. [DOI] [PubMed] [Google Scholar]
- 49.Plaisance V, Abderrahmani A, Perret-Menoud V, Jacquemin P, Lemaigre F, Regazzi R. Microrna-9 controls the expression of granuphilin/slp4 and the secretory response of insulin-producing cells. J Biol Chem. 2006;281:26932–26942. doi: 10.1074/jbc.M601225200. [DOI] [PubMed] [Google Scholar]
- 50.Ramachandran D, Roy U, Garg S, Ghosh S, Pathak S, Kolthur-Seetharam U. Sirt1 and mir-9 expression is regulated during glucose-stimulated insulin secretion in pancreatic beta-islets. FEBS J. 2011;278:1167–1174. doi: 10.1111/j.1742-4658.2011.08042.x. [DOI] [PubMed] [Google Scholar]
- 51.Wijesekara N, Zhang LH, Kang MH, Abraham T, Bhattacharjee A, Warnock GL, Verchere CB, Hayden MR. Mir-33a modulates abca1 expression, cholesterol accumulation, and insulin secretion in pancreatic islets. Diabetes. 2012;61:653–658. doi: 10.2337/db11-0944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pullen TJ, da Silva Xavier G, Kelsey G, Rutter GA. Mir-29a and mir-29b contribute to pancreatic beta-cell-specific silencing of monocarboxylate transporter 1 (mct1) Mol Cell Biol. 2011;31:3182–3194. doi: 10.1128/MCB.01433-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brunham LR, Kruit JK, Pape TD, Timmins JM, Reuwer AQ, Vasanji Z, Marsh BJ, Rodrigues B, Johnson JD, Parks JS, Verchere CB, Hayden MR. Beta-cell abca1 influences insulin secretion, glucose homeostasis and response to thiazolidinedione treatment. Nat Med. 2007;13:340–347. doi: 10.1038/nm1546. [DOI] [PubMed] [Google Scholar]
- 54.Frost RJ, Olson EN. Control of glucose homeostasis and insulin sensitivity by the let-7 family of micrornas. Proc Natl Acad Sci U S A. 2011;108:21075–21080. doi: 10.1073/pnas.1118922109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu H, Shyh-Chang N, Segre AV, Shinoda G, Shah SP, Einhorn WS, Takeuchi A, Engreitz JM, Hagan JP, Kharas MG, Urbach A, Thornton JE, Triboulet R, Gregory RI, Altshuler D, Daley GQ. The lin28/let-7 axis regulates glucose metabolism. Cell. 2011;147:81–94. doi: 10.1016/j.cell.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the let-7 precursor loop mediates regulated microrna processing. RNA. 2008;14:1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microrna processing by lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.He A, Zhu L, Gupta N, Chang Y, Fang F. Overexpression of micro ribonucleic acid 29, highly up-regulated in diabetic rats, leads to insulin resistance in 3t3-l1 adipocytes. Mol Endocrinol. 2007;21:2785–2794. doi: 10.1210/me.2007-0167. [DOI] [PubMed] [Google Scholar]
- 59.Trajkovski M, Hausser J, Soutschek J, Bhat B, Akin A, Zavolan M, Heim MH, Stoffel M. Micrornas 103 and 107 regulate insulin sensitivity. Nature. 2011;474:649–653. doi: 10.1038/nature10112. [DOI] [PubMed] [Google Scholar]
- 60.Grueter CE, van Rooij E, Johnson BA, DeLeon SM, Sutherland LB, Qi X, Gautron L, Elmquist JK, Bassel-Duby R, Olson EN. A cardiac microrna governs systemic energy homeostasis by regulation of med13. Cell. 2012;149:671–683. doi: 10.1016/j.cell.2012.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zampetaki A, Kiechl S, Drozdov I, Willeit P, Mayr U, Prokopi M, Mayr A, Weger S, Oberhollenzer F, Bonora E, Shah A, Willeit J, Mayr M. Plasma microrna profiling reveals loss of endothelial mir-126 and other micrornas in type 2 diabetes. Circulation research. 2010;107:810–817. doi: 10.1161/CIRCRESAHA.110.226357. [DOI] [PubMed] [Google Scholar]
- 62.Hunter MP, Ismail N, Zhang X, Aguda BD, Lee EJ, Yu L, Xiao T, Schafer J, Lee ML, Schmittgen TD, Nana-Sinkam SP, Jarjoura D, Marsh CB. Detection of microrna expression in human peripheral blood microvesicles. PloS one. 2008;3:e3694. doi: 10.1371/journal.pone.0003694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL, Tait JF, Tewari M. Argonaute2 complexes carry a population of circulating micrornas independent of vesicles in human plasma. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zile MR, Mehurg SM, Arroyo JE, Stroud RE, DeSantis SM, Spinale FG. Relationship between the temporal profile of plasma microrna and left ventricular remodeling in patients after myocardial infarction. Circulation. Cardiovascular genetics. 2011;4:614–619. doi: 10.1161/CIRCGENETICS.111.959841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cermelli S, Ruggieri A, Marrero JA, Ioannou GN, Beretta L. Circulating micrornas in patients with chronic hepatitis c and non-alcoholic fatty liver disease. PloS one. 2011;6:e23937. doi: 10.1371/journal.pone.0023937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li S, Zhu J, Zhang W, Chen Y, Zhang K, Popescu LM, Ma X, Lau WB, Rong R, Yu X, Wang B, Li Y, Xiao C, Zhang M, Wang S, Yu L, Chen AF, Yang X, Cai J. Signature microrna expression profile of essential hypertension and its novel link to human cytomegalovirus infection. Circulation. 2011;124:175–184. doi: 10.1161/CIRCULATIONAHA.110.012237. [DOI] [PubMed] [Google Scholar]
- 67.Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. Micrornas are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nature cell biology. 2011;13:423–433. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]