Abstract
DNA end resection is critical for chromosome break repair by homologous recombination and influences the efficiency of repair by nonhomologous DNA end joining. An elegant study by Sinha and colleagues (pp. 1423–1437) published in the June 15, 2009, issue of Genes & Development identified a novel mycobacterial DNA end resection protein complex, AdnAB, that harbors dual DNA motor and dual nuclease functions. Sinha and colleagues also demonstrated that the DNA end-binding protein complex Ku regulates the activity of AdnAB.
Keywords: DNA repair, helicase, double-strand breaks, ATP-dependent nuclease, molecular ruler
DNA double-strand break (DSB) and its repair
DSBs are induced in cells by agents such as ionizing radiation (IR) and genotoxic chemicals, and can also arise during DNA replication, such as when the DNA polymerase ensemble encounters a secondary DNA structure or a pre-existing lesion in the template. If misrepaired or left unrepaired, DSBs can cause prolonged cell cycle arrest, induction of apoptosis, chromosome loss, and chromosomal rearrangements. As such, DSBs pose a particularly dangerous threat to cell viability and genome integrity. In fact, there is ample evidence that mishandling of DSBs can lead to carcinogenesis through activation of oncogenes or inactivation of tumor suppressor genes.
As one might expect, both prokaryotic and eukaryotic cells have evolved elaborate systems with conserved mechanisms for the efficient detection, processing, and repair of DSBs. Nonhomologous DNA end joining (NHEJ) and homologous recombination (HR) are the two major pathways used by cells to repair DSBs. The NHEJ pathway uses the DNA end-binding protein Ku, a specialized DNA ligase (LigD in bacteria, DNA Ligase IV in eukaryotes) and various ancillary factors in eukaryotes to join the broken DNA ends in a manner that is facilitated by DNA microhomology but otherwise has no specific DNA sequence homology requirement. It should be noted that repair by NHEJ is intrinsically error-prone, as it often generates small deletions and, less frequently, insertions at the site of the DSB (Daley et al. 2005). In contrast, HR is considered a more accurate mechanism because homologous sequences are used to direct faithful repair of the DSB (Krogh and Symington 2004; Daley et al. 2005).
A conserved class of DNA-pairing enzymes known as general recombinases, including the bacterial RecA and eukaryotic Rad51 and Dmc1 proteins, mediate the HR reaction. A key feature of HR is the ATP-dependent polymerization of the recombinase protein on ssDNA, derived from DSB processing (see below), to form a right-handed nucleoprotein complex commonly referred to as the presynaptic filament (San Filippo et al. 2008). Once made, the presynaptic filament, in conjunction with its cohort of accessory factors, conducts a genome-wide search for homology and catalyzes the formation of a DNA joint between the recombining DNA molecules. Subsequent steps include DNA synthesis, resolution of the recombination intermediates through one of several pathways, and DNA ligation to complete the reaction (Krogh and Symington 2004; San Filippo et al. 2008).
The first step in repairing a DSB by NHEJ or HR is processing of the broken DNA ends. For NHEJ, only limited processing is necessary to expose microhomology in the two DNA ends to produce a structure suitable for ligation, whereas HR requires much more extensive 5′-end resection to generate the 3′ ssDNA tails for recombinase presynaptic filament formation. Hence, the DNA end-processing step is a crucial stage in determining which pathway is used to repair a DSB. As will be reviewed below, studies done mainly in bacteria thus far have shown DNA end resection to be a surprisingly complex process performed by multisubunit enzymes with combined helicase and nuclease activities. Interestingly, many organisms possess functionally redundant DNA end resection systems that contribute independently to DSB processing.
DNA end resection in prokaryotes
The RecBCD pathway
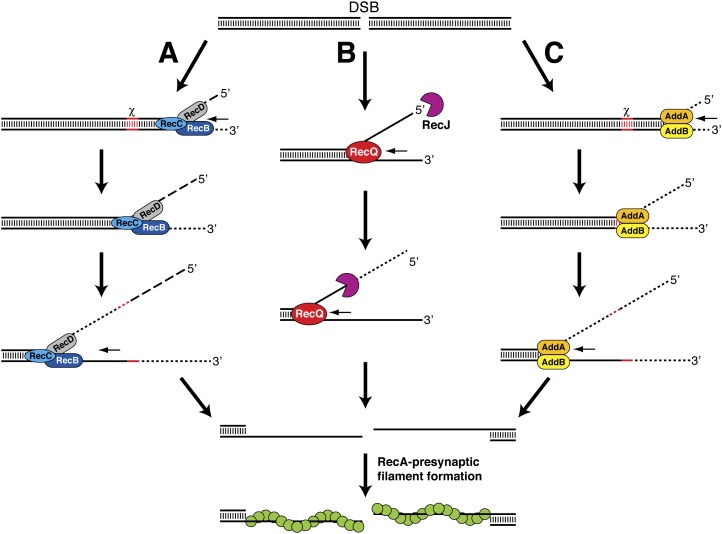
A heterotrimeric complex composed of the RecB, RecC, and RecD proteins mediates DNA end resection in Escherichia coli and many other Gram-negative bacterial species. A large body of biochemical, electron microscopic, single-molecule, and crystallographic studies have helped elucidate the mechanism of action of the RecBCD complex (Dillingham and Kowalczykowski 2008). The RecB and RecD subunits of this complex both harbor an ATP hydrolysis-driven DNA motor (helicase) activity, and a nuclease activity resides within RecB. The RecBCD complex engages blunt or nearly blunt DNA ends and rapidly separates the strands in the DNA through its motor activity (Fig. 1A). As unwinding proceeds, the nascent strands are differentially degraded through nucleolytic cleavage by the RecB subunit until a specific DNA sequence (5′-GCTGGTGG-3′) called χ is encountered. The enzyme ensemble briefly pauses upon χ recognition by the RecC subunit. When translocation resumes, the vigorous 3′-to-5′ exonuclease activity is attenuated, whereas the 5′-to-3′ exonuclease is up-regulated, resulting in the production of a 3′ ssDNA tail capped by the χ sequence. Interaction with the χ sequence also activates RecB to facilitate loading of the RecA recombinase onto the exposed ssDNA to seed the assembly of the presynaptic filament (Dillingham and Kowalczykowski 2008).
Figure 1.
DSB end resection in prokaryotes by the RecBCD (A), RecQ–RecJ (B), and AddAB (C) helicase–nuclease ensembles. Short DNA fragments released by nucleolytic cleavage are indicated by dashed lines. See the text for details.
The RecQ–RecJ pathway
A distinct pathway that is mediated by the RecQ and RecJ proteins performs the DNA end resection when the RecBCD complex has been rendered nonfunctional because of mutations (Spies and Kowalczykowski 2005). RecQ is a DNA helicase capable of unwinding both blunt-ended and 3′-tailed dsDNA. At a DSB, RecQ unwinds the duplex DNA from the broken end to yield a flayed structure with both 5′ and 3′ single-stranded regions (Fig. 1B). RecJ, a 5′-to-3′-ssDNA exonuclease, then digests the 5′-terminated strand, resulting in a 3′-tail that is bound by the ssDNA-binding protein, SSB, which further stimulates RecQ activity (Shereda et al. 2007). Unlike RecBCD, assembly of the presynaptic filament in the RecQ/RecJ-dependent pathway requires a recombination mediator function provided by the heterotrimeric RecFOR complex. RecFOR recognizes and promotes the nucleation and extension of RecA onto the SSB-coated ssDNA to assemble the presynaptic filament (Kowalczykowski 2000).
The AddAB pathway
The Bacillus subtilis AddAB heterodimer is the founding member of a growing class of enzymes that are functionally analogous to RecBCD. Inactivation of addA or addB leads to a phenotype that closely mimics E. coli recBCD− cells (Kooistra et al. 1997; el Karoui et al. 1998). Likewise, AddAB can fully complement E. coli recBCD-null mutants for cell viability, sensitivity to DNA-damaging agents, and HR frequency (Kooistra et al. 1993; el Karoui et al. 1998). The AddA subunit shows regions of homology with RecB, containing an N-terminal helicase domain and a C-terminal nuclease domain. The AddB subunit also harbors a conserved nuclease domain at its C terminus (Haijema et al. 1996; Yeeles and Dillingham 2007). Despite the apparent differences in complex architecture, the DNA end resection reaction mediated by AddAB bears a close resemblance to that catalyzed by RecBCD. AddAB binds to dsDNA ends and, through the coordinated action of the helicase and nuclease domains, catalyzes rapid unwinding and degradation of both DNA strands until a χ sequence is encountered (Fig. 1C; Chédin et al. 2000). Recognition of χ by AddAB extinguishes the 3′ strand cleavage activity; however, degradation of the opposite strand in the 5′-to-3′ direction remains unaffected. The end result is the generation of a 3′ ssDNA tail that abuts the χ sequence (Chédin et al. 2000). It is not currently known whether AddAB actively loads RecA onto ssDNA, as has been demonstrated for RecBCD.
DNA end resection in eukaryotes
Like the situation in bacteria, DNA end resection in eukaryotic cells is associated with multiple helicase and nuclease activities. Since no reconstituted system is as yet available, much of what we know about the possible mechanisms of DNA resection has been inferred from cell-based studies that examined the fate of DSBs in cells—Saccharomyces cerevisiae cells in particular. Based on these studies, it has been surmised that DNA ends are engaged initially by the protein complex that harbors the Mre11, Rad50, and Xrs2 (Nbs1 in humans) proteins. In conjunction with Sae2 (CtIP in humans), the MRX(N) complex initiates limited resection of the broken DNA end (Lengsfeld et al. 2007; Sartori et al. 2007) in increments of 100 or so nucleotides (Mimitou and Symington 2008; Zhu et al. 2008). It should be noted that both Mre11 and Sae2 possess a nuclease activity that could be used in this initial resection stage (Paull and Gellert 1998; Trujillo and Sung 2001; Lengsfeld et al. 2007). The 3′-tailed DNA ends generated by the MRX(N) and Sae2 (CtIP) ensemble are further processed through the coordinated action of the Sgs1 helicase (BLM in humans) and the Dna2 helicase/nuclease, or by the 5′-to-3′ exonucleolytic activity of Exo1, to yield long 3′ ssDNA tails (Gravel et al. 2008; Mimitou and Symington 2008; Nimonkar et al. 2008; Zhu et al. 2008). Interestingly, the ability of human Exo1 protein to resect DNA ends is directly stimulated by the BLM helicase in a manner that does not require the DNA motor activity of BLM (Nimonkar et al. 2008). Once generated, the ssDNA tail becomes bound by the ssDNA-binding protein RPA. For HR to occur, RPA must be displaced to allow for the loading and polymerization of Rad51. This function is performed by accessory proteins called recombination mediators, such as the breast tumor suppressor BRCA2 and S. cerevisiae Rad52 (San Filippo et al. 2008). Importantly, substantial evidence has accrued that the ssDNA tails generated by DNA end resection serve to activate the DNA damage checkpoints, to halt cell cycle progression, and allow time for repair to occur (Raynard et al. 2008).
DNA end resection in archaea
In archaea, HR appears to be the major pathway of DSB repair, since a RecA ortholog, RadA, is present but Ku-like factors are absent in most species. From biochemical studies done with purified Pyrococcus furiosus proteins, we know that DNA end resection by the bipolar helicase HerA and the 5′-to-3′ exonuclease NurA is enhanced by the pfMre11–pfRad50 complex, and that the 3′ ssDNA can be used as substrate for RadA-mediated DNA joint formation (Hopkins and Paull 2008). In the reconstituted end resection reaction, dsDNA ends are first processed by pfMre11-mediated endonucleolytic cleavage to produce a short 3′ ssDNA overhang, which can be engaged by HerA–NurA for further processing in a manner that is dependent on the DNA motor activity of HerA and the nucleolytic function of NurA. Both HerA and NurA are located within the same operon as pfMre11 and pfRad50 (Constantinesco et al. 2004; Hopkins and Paull 2008). Moreover, the Sulfolobus acidocaldarius HerA protein can be coimmunoprecipitated with saMre11 and saRad50, which are enriched in the chromosomal fraction together with RadA upon γ irradiation (Quaiser et al. 2008). These results further support the role of Mre11/Rad50/HerA/NurA in DSB end resection and repair in archaea.
AdnAB, a novel end resection helicase–nuclease complex in mycobacteria
In mycobacterial species, such as Mycobacterium smegmatis, DSBs are repaired through either HR or NHEJ. NHEJ is of particular importance for eliminating DSBs during the latency stage when there is no sister chromatid available to direct HR repair (Shuman and Glickman 2007). NHEJ in mycobacteria is highly mutagenic, being accompanied by insertion or deletion of nucleotides (Stephanou et al. 2007). The deletion type of NHEJ events is indicative of DNA end resection prior to break joining. Interestingly, even though M. smegmatis possesses a canonical RecBCD end-processing pathway, its ablation gives no discernible phenotype with regard to DNA repair and little change in the deletion type of NHEJ events (Stephanou et al. 2007; Aniukwu et al. 2008). Together, these observations provide strong evidence for the presence of another DNA resection machinery in M. smegmatis. However, neither RecQ/RecJ homologs nor AddAB homologs have been identified in this mycobacterium. In the June 15, 2009, issue of Genes & Development, Sinha et al. (2009) applied a proteomic approach to uncover the founding member of a new dimeric DNA motor-driven nuclease complex from M. smegmatis, termed AdnAB (ATP-dependent nuclease), with distinctive DSB end resection properties.
Both AdnA and AdnB harbor an N-terminal helicase-like domain adjoining a C-terminal nuclease domain, thus distinguishing AdnAB from RecBCD, which has two motor and one nuclease entities, and AddAB, which has one motor and two nuclease entities. Linear but not circular dsDNA is an effective cofactor of AdnAB ATPase activity, indicating that it is activated by free DNA ends. AdnAB exhibits both ATP-dependent and ATP-independent nuclease activities. In the absence of ATP, AdnAB releases predominantly 5-mer and 6-mer species by making incisions measured from the 5′ end of the ssDNA. In the presence of ATP, nuclease activity is stimulated and the incision step is increased to 16 or 17 nucleotides from the 5′ end. The AdnA subunit is responsible for the ATP-independent incision of ssDNA, whereas AdnB mediates the ATP-dependent reaction. When both nuclease domains are inactivated, the AdnAB helicase activity is revealed. Sinha et al. (2009) surmised that DSB end processing by AdnAB is coupled to the NTPase motor that unwinds the DNA duplex from free ends and thereby exposes the displaced single strands to the associated single-strand nuclease activity, which degrades the 5′-terminated strand, thereby producing 3′ ssDNA tails (Fig. 2).
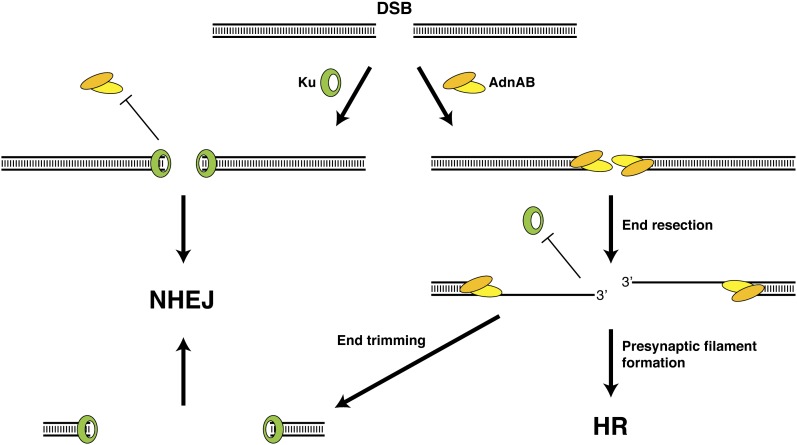
Figure 2.
Interplay between AdnAB and Ku during DSB processing in mycobacteria. Ku and AdnAB compete for binding to DSB ends. If Ku binds the DNA ends first, access to AdnAB is limited, thus favoring repair by NHEJ. If AdnAB is allowed to engage the DNA ends, 5′ resection occurs to expose 3′ ssDNA tails that are used to assemble the RecA presynaptic filament, and repair of the DSB occurs by HR. If the 3′ ssDNA tails are trimmed, subsequent repair by NHEJ will result in deletions at the DSB site.
Regulation of DNA end resection: lessons from bacteria and yeast
Another interesting aspect of the Sinha et al. (2009) study was the demonstration that Ku, the DNA end-binding subunit of the NHEJ machinery, is able to protect dsDNA ends from extensive resection by AdnAB (Fig. 2). It is likely that Ku suppresses AdnAB by physically blocking access to the free DNA ends that are required for activation of its motor activity. The notion of Ku acting as a negative regulator of end resection has been suggested previously from genetic studies in eukaryotes (Tomita et al. 2003; Barlow et al. 2008; Clerici et al. 2008; Wasko et al. 2009). Ku is required to prevent Exo1-dependent ssDNA formation at telomeres (Maringele and Lydall 2002). Moreover, the MMS sensitivity of rad50 mutants but not rad50 exo1 mutants is suppressed by deleting Ku, suggesting that Ku regulates DNA end resection by Exo1 (Tomita et al. 2003; Wasko et al. 2009).
It is interesting to note that in S. cerevisiae the cellular capacity for DNA end resection varies throughout the cell cycle, being the highest during the S and G2 phases when sister chromatids are available for use as HR repair templates. This is in large part due to a tight regulation of DSB repair by the cyclin-dependent kinase (CDK) (Aylon et al. 2004; Ira et al. 2004). In the G1 phase, the balance of DSB repair is shifted toward NHEJ, which remains active throughout the cell cycle. During G1, DNA ends that are suitable for ligation are protected from extensive resection in a Ku-dependent manner (Barlow et al. 2008; Clerici et al. 2008). There is evidence that Ku limits the DSB recruitment of the MRX complex (Clerici et al. 2008). Consistent with this, overexpression of Ku in G2 cells delays DSB processing by impeding MRX loading (Clerici et al. 2008). The dependence on CDK activity for DNA resection in S/G2 is lifted in the absence of Ku, suggesting that CDK might directly target Ku (Clerici et al. 2008). Interestingly, Sae2 is an in vivo CDK target, and a phosphomimic sae2 mutant is able to initiate DSB end resection in G2 cells even in CDK's absence (Huertas et al. 2008). How the CDK-mediated phosphorylation of Sae2 helps overcome the Ku-dependent block of DNA end resection remains to be determined.
Important questions
As discussed earlier, an important aspect of the HR pathway is the coupling of end resection with the loading of the recombinase onto the exposed ssDNA. This can be accomplished directly by the end resection machinery (e.g., RecB), or through the action of accessory proteins (e.g., RecFOR). In eukaryotes, recombination mediators such as BRCA2 and Rad52 facilitate the replacement of RPA with Rad51 (Spies and Kowalczykowski 2005; San Filippo et al. 2008). It remains to be determined how mycobacterial RecA is loaded onto AdnAB-processed DNA ends, although the presence of RecF and RecR homologs in mycobacteria suggest that they might perform this function. Interestingly, the mycobacterial RecA protein interacts directly with SSB, although the biological significance of this interaction is not clear (Reddy et al. 2001).
There appears to be a multiplicity of DSB end-processing pathways in both prokaryotes and eukaryotes. It will be of particular interest to learn whether these pathways indeed operate independently of one another, or whether they normally operate in concert to process DSBs but become genetically separable when selected components are ablated by mutations. We still do not understand the molecular basis for the CDK-dependent alleviation of the block on DSB processing in S and G2 cells. Also, since eukaryotic DNA is packaged into chromatin, how the DNA end resection machinery negotiates this barrier remains to be delineated.
Acknowledgments
The research in our laboratory has been supported by grants from the U.S. National Institutes of Health, the U.S. Department of Defense, and the Susan G. Komen for the Cure Foundation.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1824209.
References
- Aniukwu J, Glickman MS, Shuman S. The pathways and outcomes of mycobacterial NHEJ depend on the structure of the broken DNA ends. Genes & Dev. 2008;22:512–527. doi: 10.1101/gad.1631908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylon Y, Liefshitz B, Kupiec M. The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. EMBO J. 2004;23:4868–4875. doi: 10.1038/sj.emboj.7600469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow JH, Lisby M, Rothstein R. Differential regulation of the cellular response to DNA double-strand breaks in G1. Mol Cell. 2008;30:73–85. doi: 10.1016/j.molcel.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chédin F, Ehrlich SD, Kowalczykowski SC. The Bacillus subtilis AddAB helicase/nuclease is regulated by its cognate Chi sequence in vitro. J Mol Biol. 2000;298:7–20. doi: 10.1006/jmbi.2000.3556. [DOI] [PubMed] [Google Scholar]
- Clerici M, Mantiero D, Guerini I, Lucchini G, Longhese MP. The Yku70–Yku80 complex contributes to regulate double-strand break processing and checkpoint activation during the cell cycle. EMBO Rep. 2008;9:810–818. doi: 10.1038/embor.2008.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinesco F, Forterre P, Koonin EV, Aravind L, Elie C. A bipolar DNA helicase gene, herA, clusters with rad50, mre11 and nurA genes in thermophilic archaea. Nucleic Acids Res. 2004;32:1439–1447. doi: 10.1093/nar/gkh283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley JM, Palmbos PL, Wu D, Wilson TE. Nonhomologous end joining in yeast. Annu Rev Genet. 2005;39:431–451. doi: 10.1146/annurev.genet.39.073003.113340. [DOI] [PubMed] [Google Scholar]
- Dillingham MS, Kowalczykowski SC. RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol Mol Biol Rev. 2008;72:642–671. doi: 10.1128/MMBR.00020-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el Karoui M, Ehrlich D, Gruss A. Identification of the lactococcal exonuclease/recombinase and its modulation by the putative Chi sequence. Proc Natl Acad Sci. 1998;95:626–631. doi: 10.1073/pnas.95.2.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel S, Chapman JR, Magill C, Jackson SP. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes & Dev. 2008;22:2767–2772. doi: 10.1101/gad.503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haijema BJ, Venema G, Kooistra J. The C terminus of the AddA subunit of the Bacillus subtilis ATP-dependent DNase is required for the ATP-dependent exonuclease activity but not for the helicase activity. J Bacteriol. 1996;178:5086–5091. doi: 10.1128/jb.178.17.5086-5091.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins BB, Paull TT. The P. furiosus mre11/rad50 complex promotes 5′ strand resection at a DNA double-strand break. Cell. 2008;135:250–260. doi: 10.1016/j.cell.2008.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P, Cortés-Ledesma F, Sartori AA, Aguilera A, Jackson SP. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature. 2008;455:689–692. doi: 10.1038/nature07215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ira G, Pellicioli A, Balijja A, Wang X, Fiorani S, Carotenuto W, Liberi G, Bressan D, Wan L, Hollingsworth NM, et al. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 2004;431:1011–1017. doi: 10.1038/nature02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooistra J, Haijema BJ, Venema G. The Bacillus subtilis addAB genes are fully functional in Escherichia coli . Mol Microbiol. 1993;7:915–923. doi: 10.1111/j.1365-2958.1993.tb01182.x. [DOI] [PubMed] [Google Scholar]
- Kooistra J, Haijema BJ, Hesseling-Meinders A, Venema G. A conserved helicase motif of the AddA subunit of the Bacillus subtilis ATP-dependent nuclease (AddAB) is essential for DNA repair and recombination. Mol Microbiol. 1997;23:137–149. doi: 10.1046/j.1365-2958.1997.1991570.x. [DOI] [PubMed] [Google Scholar]
- Kowalczykowski SC. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem Sci. 2000;25:156–165. doi: 10.1016/s0968-0004(00)01569-3. [DOI] [PubMed] [Google Scholar]
- Krogh BO, Symington LS. Recombination proteins in yeast. Annu Rev Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- Lengsfeld BM, Rattray AJ, Bhaskara V, Ghirlando R, Paull TT. Sae2 is an endonuclease that processes hairpin DNA cooperatively with the Mre11/Rad50/Xrs2 complex. Mol Cell. 2007;28:638–651. doi: 10.1016/j.molcel.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maringele L, Lydall D. EXO1-dependent single-stranded DNA at telomeres activates subsets of DNA damage and spindle checkpoint pathways in budding yeast yku70Δ mutants. Genes & Dev. 2002;16:1919–1933. doi: 10.1101/gad.225102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimonkar AV, Ozsoy AZ, Genschel J, Modrich P, Kowalczykowski SC. Human exonuclease 1 and BLM helicase interact to resect DNA and initiate DNA repair. Proc Natl Acad Sci. 2008;105:16906–16911. doi: 10.1073/pnas.0809380105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull TT, Gellert M. The 3′ to 5′ exonuclease activity of Mre11 facilitates repair of DNA double-strand breaks. Mol Cell. 1998;1:969–979. doi: 10.1016/s1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- Quaiser A, Constantinesco F, White MF, Forterre P, Elie C. The Mre11 protein interacts with both Rad50 and the HerA bipolar helicase and is recruited to DNA following γ irradiation in the archaeon Sulfolobus acidocaldarius . BMC Mol Biol. 2008;9:25. doi: 10.1186/1471-2199-9-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynard S, Niu H, Sung P. DNA double-strand break processing: The beginning of the end. Genes & Dev. 2008;22:2903–2907. doi: 10.1101/gad.1742408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy MS, Guhan N, Muniyappa K. Characterization of single-stranded DNA-binding proteins from Mycobacteria. The carboxyl-terminal of domain of SSB is essential for stable association with its cognate RecA protein. J Biol Chem. 2001;276:45959–45968. doi: 10.1074/jbc.M103523200. [DOI] [PubMed] [Google Scholar]
- San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shereda RD, Bernstein DA, Keck JL. A central role for SSB in Escherichia coli RecQ DNA helicase function. J Biol Chem. 2007;282:19247–19258. doi: 10.1074/jbc.M608011200. [DOI] [PubMed] [Google Scholar]
- Shuman S, Glickman MS. Bacterial DNA repair by non-homologous end joining. Nat Rev Microbiol. 2007;5:852–861. doi: 10.1038/nrmicro1768. [DOI] [PubMed] [Google Scholar]
- Sinha KM, Unciuleac M-C, Glickman MS, Shuman S. AdnAB: A new DSB-resecting motor–nuclease from mycobacteria. Genes & Dev. 2009;23:1423–1437. doi: 10.1101/gad.1805709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spies M, Kowalczykowski SC. Homologous recombination by RecBCD and RecF pathways. In: Higgins NP, editor. The bacterial chromosome. ASM Press; Washington, DC: 2005. pp. 389–403. [Google Scholar]
- Stephanou NC, Gao F, Bongiorno P, Ehrt S, Schnappinger D, Shuman S, Glickman MS. Mycobacterial nonhomologous end joining mediates mutagenic repair of chromosomal double-strand DNA breaks. J Bacteriol. 2007;189:5237–5246. doi: 10.1128/JB.00332-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita K, Matsuura A, Caspari T, Carr AM, Akamatsu Y, Iwasaki H, Mizuno K, Ohta K, Uritani M, Ushimaru T, et al. Competition between the Rad50 complex and the Ku heterodimer reveals a role for Exo1 in processing double-strand breaks but not telomeres. Mol Cell Biol. 2003;23:5186–5197. doi: 10.1128/MCB.23.15.5186-5197.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo KM, Sung P. DNA structure-specific nuclease activities in the Saccharomyces cerevisiae Rad50*Mre11 complex. J Biol Chem. 2001;276:35458–35464. doi: 10.1074/jbc.M105482200. [DOI] [PubMed] [Google Scholar]
- Wasko BM, Holland CL, Resnick MA, Lewis LK. Inhibition of DNA double-strand break repair by the Ku heterodimer in mrx mutants of Saccharomyces cerevisiae . DNA Repair (Amst) 2009;8:162–169. doi: 10.1016/j.dnarep.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeeles JT, Dillingham MS. A dual-nuclease mechanism for DNA break processing by AddAB-type helicase-nucleases. J Mol Biol. 2007;371:66–78. doi: 10.1016/j.jmb.2007.05.053. [DOI] [PubMed] [Google Scholar]
- Zhu Z, Chung W, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]