Abstract
The importance of dynamics to biomolecular function is becoming increasingly clear. A description of the structure–function relationship must, therefore, include the role of motion, requiring a shift in paradigm from focus on a single static 3D picture to one where a given biomolecule is considered in terms of an ensemble of interconverting conformers, each with potentially diverse activities. In this Perspective, we describe how recent developments in solution NMR spectroscopy facilitate atomic resolution studies of sparsely populated, transiently formed biomolecular conformations that exchange with the native state. Examples of how this methodology is applied to protein folding and misfolding, ligand binding, and molecular recognition are provided as a means of illustrating both the power of the new techniques and the significant roles that conformationally excited protein states play in biology.
Keywords: energy landscape, transiently and sparsely populated biomolecular states, invisible states, structure–function paradigm
The field of structural biology emerged from seminal X-ray diffraction studies of biomolecules such as DNA (1), myoglobin (2), hemoglobin (3), and lysozyme (4) that were carried out over 50 years ago. Since these landmark investigations, our knowledge of the relation between structure and function has greatly expanded, driven by significant improvements in both experimental and computational approaches and, of course, by the exponential increase in the number of structures that are now available. Despite tremendous advances, structural biology remains largely focused on studies of the lowest-energy conformational states of biomolecules, and although there are notable exceptions (5), the end game often remains the determination of a single static 3D structure that is then used as a starting point for understanding molecular function. It is becoming increasingly clear, however, that this narrow approach is not sufficient and that a description of molecular structure must take into account conformational fluctuations that can occur over a broad range of both time and length scales.
The conformational space that can be explored by a given biomolecule is usually explained by invoking the concept of an energy landscape (6), a multidimensional hypersurface that governs the thermodynamics and kinetics of conformational transitions (7, 8). Because biomolecular stability is dictated by the sum of a large number of attractive and repulsive interactions of similar strength, the resultant free-energy landscape is rugged (9), with the global minimum in the surface thought to correspond to the native state of the biomolecule. In addition, there are often local minima that are separated from the global minimum by free-energy barriers that can be overcome by thermal fluctuations. These low-lying conformationally excited states have remained elusive to quantitative investigation because their sparse population and transient nature complicates their study by conventional structural biology techniques that are so powerful for characterizing highly populated, ground-state conformers.
In this Perspective, we concentrate on the progress that has been made in providing an atomic-level description of low-lying excited conformational states of both proteins and nucleic acids using emerging solution NMR methods that bring invisible conformers into focus, rather than presenting a more comprehensive account of the many different NMR methodologies for studying biomolecular dynamics that are now available. Structures of a number of excited-state conformations, chosen from examples involving protein folding and misfolding, molecular recognition, and ligand binding, illustrate both the impact of these new approaches and the detailed atomic-resolution information that can now be obtained.
NMR Methods for Studying Excited Biomolecular Conformations
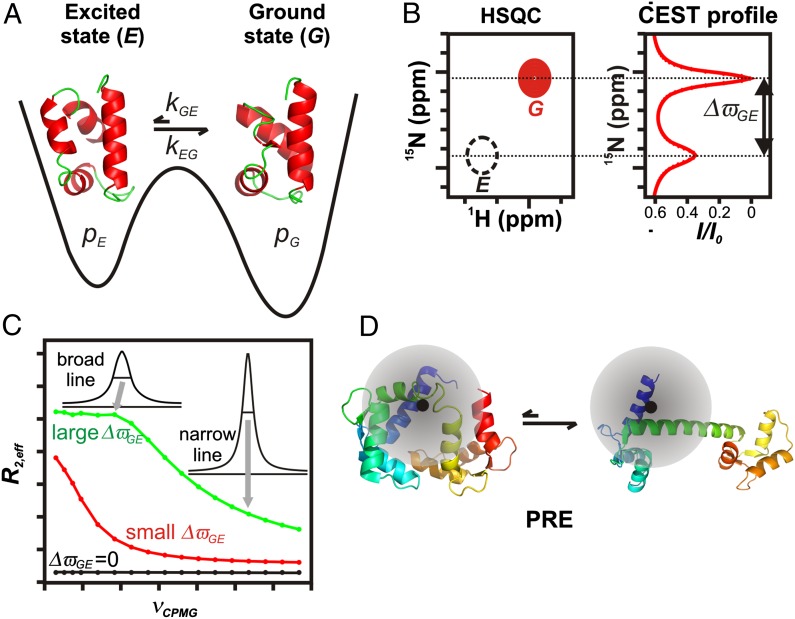
Consider an exchange process in which a highly populated and long-lived ground-state conformer, G, exchanges with a sparsely populated, transiently formed excited state, E (Fig. 1A). The exchange rate constant for the interconversion between the two states is denoted by kex = kGE + kEG and defines a timescale for the exchange process, τex = 1/kex. In this Perspective, we focus on cases where the fractional population of the excited state, pE, is no more than a few percent and where spectra from E cannot be observed directly (Fig. 1B, Left). Detailed structural information on E can still be obtained, however, from the observed spectrum that essentially corresponds to that of the ground state by using a variety of methods, such as chemical-exchange saturation transfer (CEST) (Fig. 1B, Right) (10, 11), Carr–Purcell–Meiboom–Gill relaxation dispersion (CPMG RD) (Fig. 1C) (12, 13), R1ρ (12), or paramagnetic relaxation enhancement (PRE) (Fig. 1D) (14). Each of these methods is sensitive to a different range of exchange timescales, τex, and they can be further separated on the basis of the type of structural data that they generate. For example, CEST and RD provide site-specific chemical shifts of E that are then used in concert with chemical shift based structure prediction algorithms and a database of structures of protein fragments to determine the conformation of the excited state (15, 16). The development of structure calculation methods relying on chemical shifts rather than conventional NOE distance restraints has played a pivotal role in determining excited-state structures (17–19), as described in several examples below. In contrast to CEST and RD approaches, PRE-based measurements provide distances between NMR active nuclei and the unpaired electron of an attached paramagnetic spin label that is used to probe molecular structure. These distances are then used as restraints in molecular dynamics calculations to visualize structures of the transiently populated ensemble (14).
Fig. 1.
NMR methods for studying sparsely populated, transiently formed biomolecular conformers. (A) Schematic 1D energy landscape showing the ground state of a protein in exchange with a thermally accessible excited state. Exchange between G and E is in the microsecond-to-millisecond regime, with pE << pG. (B) Schematic (Left) showing a small region from a standard 15N-1H Heteronuclear Single Quantum Coherence dataset. The peak derived from state E (dashed black) is not visible in a typical spectrum and is shown here only for clarity. The CEST profile (Right) is obtained by varying the 15N frequency of a weak radio frequency (B1) field. Reduction in resonance intensity (I/I0) of the ground-state peak is seen when irradiation frequencies correspond to resonance positions of G or E. Consequently, ΔϖGE can be readily obtained from the CEST profile. (C) CPMG RD profiles, R2,eff vs. νCPMG, for different ΔϖGE values (0 ppm, black; 1.7 ppm, red; 6.8 ppm, green). Values of R2,eff are calculated from intensities of correlations derived from the ground state as a function of pulsing frequency. Higher pulsing frequencies more effectively refocus the dephasing (excess peak line width) arising from exchange, resulting in narrower peaks and smaller R2,eff values. CPMG dispersion profiles can be fit to extract ΔϖGE values and exchange parameters. (D) Cartoon representation of a protein labeled with a paramagnetic spin label (black) that exchanges between a ground-state conformation and a compact excited state. The shaded circle represents the sphere of influence of the spin label. The regions of the protein that are proximal to the unpaired electron, and, hence the pattern of PREs, are clearly different in the two states.
Like other methodologies described in this section, the CEST experiment (Fig. 1B) was developed close to a half-century ago (20) and applied to biomolecular systems shortly thereafter (21, 22). The use of the CEST approach to amplify signals (23, 24) arising from sparsely populated biomolecular states in exchange with a ground conformation is, however, more recent (10, 11). Typically, the method is applicable for systems where kex is between 20–300 s−1 and pE is ≥1% (11). In the CEST experiment, a weak radiofrequency (rf) field is applied at frequencies (one at a time) spanning the entire range of chemical shifts of a particular nucleus (for example, amide 15N). The weak rf perturbation causes a substantial and irreversible loss in signal intensity of the ground-state peak if it is applied proximal in frequency to it. More important, a loss in intensity of the ground-state peak also occurs if the rf field is applied at a frequency near that of the corresponding peak in the excited state because the effect of the perturbation on E is transferred to G by the exchange process connecting the two states. Thus, a CEST profile as in Fig. 1B is obtained, graphing the normalized ground-state peak intensity, I/Io, as a function of the rf irradiation frequency. Large chemical shift differences (ΔϖGE) between the ground (ϖG) and excited states (ϖE) can be read off the CEST profile (Fig. 1B), whereas smaller ΔϖGE values can be obtained by numerical fits of the profile to the appropriate exchange model.
As kex increases to between 200–2,000 s−1, pE ≥ 1%, the CPMG RD experiment becomes the method of choice (12, 13). Here, the effective transverse relaxation rate of a ground-state spin, (R2,eff, proportional to the line width) is recorded as a function of the rate of application of chemical shift refocusing pulses, νCPMG. These pulses systematically modulate R2,eff, producing RD profiles (R2,eff vs. νCPMG) of the sort illustrated in Fig. 1C that depend on kex, pE, and ΔϖGE. As illustrated, the shape of a RD profile can be extremely sensitive to the magnitude of ΔϖGE, with kex, pE, and ΔϖGE extracted by fitting the curves numerically to a model of chemical exchange.
As the exchange rate increases, it is no longer possible to pulse fast enough to significantly affect relaxation rates. In this case, rotating frame relaxation rate (R1ρ) measurements are performed (25). These experiments are conceptually similar to CPMG RD methods, with the variation of R1ρ as a function of rf field strength and offset providing a sensitive measure of exchange rates and, in favorable cases, also of chemical shifts. Recent applications of the R1ρ technique have included studies of rate processes with kex as fast as 40,000 s−1 (26–28).
PRE measurements become powerful in the case of rapidly exchanging conformational states where the effective R2 values of NMR spins are population-weighted averages of rates in the G and E states (14). The PRE effect originates from magnetic interactions between the target NMR active spin that is separated by a distance r from an unpaired electron in a paramagnetic center (Fig. 1D, black circle) attached to the biomolecule of interest. The r-6 dependence of the PRE effect provides distance information that can extend up to 35 Å depending on the electron source (14). In cases where electron–nuclear distances are substantially smaller in E than in G, the corresponding spectra of the G state are only relatively slightly affected by the paramagnet so that accurate R2 values can be measured. Because these rates are related directly to distances in the excited state, they can be used in molecular dynamics computational protocols as distance restraints for structure refinement (14).
Formation of Nonnative Structure Along a Folding Pathway
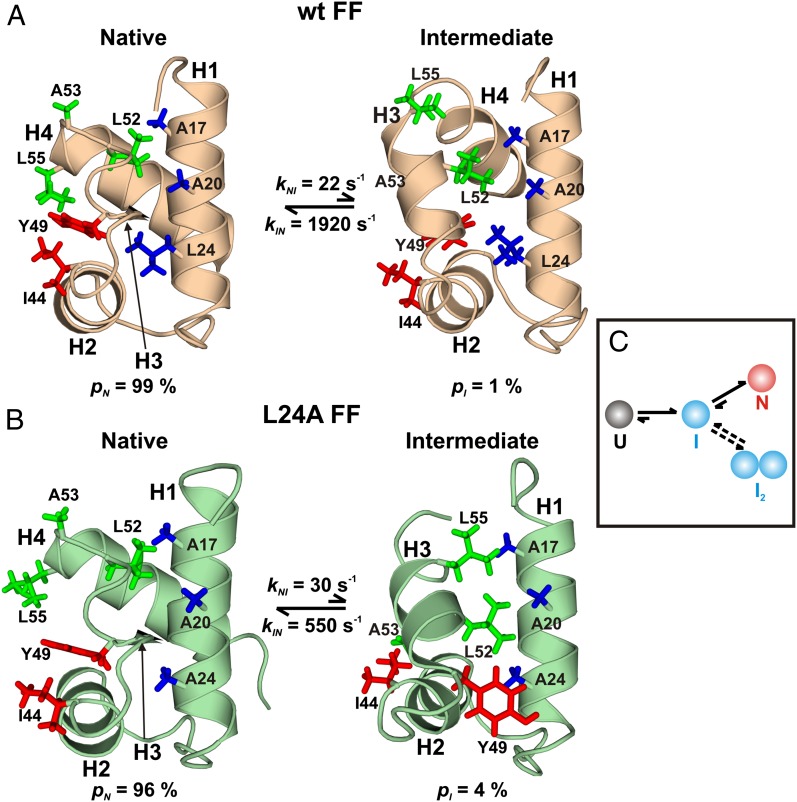
Studies of protein folding have long been of scientific interest (29), but they have assumed even more importance recently with the realization that folding intermediates may play central roles in nucleating the aggregation of proteins implicated in disease (30). Thus, understanding the pathways by which proteins fold can provide insight not only into how the primary amino acid sequence encodes tertiary structure but also, perhaps, about how improperly folded states can be generated that then serve as scaffolds for aggregation and self-assembly. One system that has emerged as a prototype in studies of folding is the FF domain from human HYPA/FBP11 (31), a 71-residue, four-helix bundle that has been shown by stopped-flow fluorescence (32) and RD NMR methods (33) to fold via an on-pathway intermediate (I). Formation of I from the unfolded state (U) occurs on the microsecond timescale (32), whereas the transition between I and N is slower (kIN, ∼2,000 s−1; 25 °C) for the wild type (wt) domain (33) and corresponds to the rate-limiting step for folding. At equilibrium, 25 °C, and in the absence of denaturants, state I is populated at 1%, with an average lifetime (τI = 1/kIN) of 0.5 ms (Fig. 2A), and it cannot be directly observed in NMR spectra (17).
Fig. 2.
Transient intermediates along the FF domain folding pathway. Structures of the N [Protein Data Bank (PDB) ID code 1UZC for wt] and I (PDB ID codes 2KZG and 2L9V for wt and L24A) states of wt (A) and L24A (B) FF domains. The exact values of rate constants and populations differ slightly depending on the isotope-labeling scheme used and sample conditions. Side chains making nonnative hydrophobic contacts in each of the I states are highlighted and colored according to the secondary structure element from which they originate. Y49 flips from inside the core of the protein to the outside during the L24A FF domain I-to-N transition. (C) A model for FF folding depicting both the folding and the dimerization pathways. (A and B are adapted from ref. 35.)
The structure of I was solved recently (17) by CPMG RD methodology using backbone 15N, 13C, and 1H chemical shifts and amide bond-vector orientations (34) in conjunction with CS-Rosetta, a chemical shift-based structure-prediction program that relies on a database of peptide fragments from high-resolution protein structures (15). Whereas helices H1 and H2 are native-like in I there are also nonnative features (Fig. 2A), most notably that helix H3 is longer in I, whereas H4 is shorter and incompletely formed compared with its counterpart in N. The longer H3 helix docks against helices H1 and H2, forming unique patterns of nonnative hydrophobic contacts, as can be seen in Fig. 2A (17). Because the intermediate is on-pathway (32, 33), its structure provides a direct view of the sequence of events occurring during the folding of the wt FF domain, with helices H1 and H2 forming a folding core that serves as a template for the organization of the rest of the structure. Furthermore, the structure of the intermediate reveals why the I to N transition is rate limiting, because a number of nonnative, yet stabilizing interactions must be broken before adopting the native fold.
As a test of the hypothesis that nonnative contacts in I are responsible for slowing folding, an L24A mutant FF domain was studied that folds from an on-pathway intermediate with a rate constant of ∼500 s−1 (30 °C), fourfold slower than for the wt FF domain (35). The structure of the L24A FF folding intermediate determined by RD NMR derived constraints is very similar to that of the wt I state. However, the cavity created by mutating the large hydrophobic L24 side chain to Ala is filled by Y49 (Fig. 2B), a conformation that is not possible in the wt intermediate. Additional nonnative contacts are, thus, formed relative to the wt I state that likely play a role in further slowing the folding rate (35).
Although there is a large body of information about how mutations affect a protein’s native state, much less is known about the plasticity of excited-state conformers. A recent CEST study of the A39G mutant FF domain (36) establishes that the I state 15N chemical shifts are very similar to those of the wt intermediate, indicating that the A39G mutation only minimally perturbs the structure of I.
It is, of course, naïve to assume that the folding pathway of any protein involves only a single intermediate. However, in the types of NMR experiments described above, transitions from the I state of the FF domain to additional intermediate states remain “hidden” by the presence of the native conformer. Therefore, a truncation mutant lacking H4 was designed as a mimic of the intermediate state. Because H4 is poorly formed in the intermediate, the mutant construct destabilizes the N state of the wt FF domain, without introducing significant changes to I (37). In this manner, I becomes the dominant conformation in solution and is now “visible” using standard NMR methods. The structure of the truncated mutant was determined using classical NMR approaches and shown to be the same as that of the invisible I state elucidated as described above (38). The importance of cross-validating structures of excited states via traditional NMR studies of designed, stabilizing mutants cannot be overemphasized, especially given that the development of the RD methodology is still progressing. In this case, the designed mimic had a further advantage in that it could be used to probe additional pathways from I. Notably, the I conformer was shown to dimerize, hinting that the intermediate may serve as branching point between folding and misfolding (Fig. 2C) (37).
On the Edge Between Protein Folding and Aggregation
Although it is understood that partially folded intermediates can play critical roles in misfolding and aggregation into insoluble β-sheet–rich fibrillar aggregates (39), so-called amyloid fibrils, the detailed mechanism by which this occurs is less well appreciated. One such system for which atomic resolution information is now available is the A39V/N53P/V55L Fyn SH3 domain (18), a 66-residue fragment that folds from U via an on-pathway intermediate (40). In its native state, the SH3 domain forms an incomplete five-stranded antiparallel β-barrel (41) (Fig. 3A). Backbone 15N, 13C, and 1H chemical shifts, as well as orientational restraints for the A39V/N53P/V55L Fyn SH3 I state, were obtained using CPMG RD methodology and subsequently used in a CamShift restrained molecular dynamics computational protocol (16) to arrive at a structure for I (Fig. 3A). Residues 57–59 are disordered in the I structure, so that β5 has not yet formed. In the absence of its partner strand β5, β1 associates more closely with β2 by forming a nonnative hydrogen bond between T2 and I28, whereas F4 changes in orientation to occupy the space of β5 in the native state, thereby partly compensating for the loss of packing interactions in I (Fig. 3A) (18). Most importantly, in the absence of a fully formed β5, strand β1 that is rich in hydrophobic residues becomes exposed and serves as a starting point for self-association.
Fig. 3.
An excited state of the Fyn SH3 domain at the interface between folding and misfolding. (A) Three-dimensional structures of the N (PDB ID code 2LP5) and I (PDB ID code 2L2P) states of the A39V/N53P/V55L Fyn SH3 domain, color-coded according to the surface aggregation propensity score (Sagg) (91). F4, which fills the position occupied by β5 in N, is shown in stick representation (gray). (B) A39V/N53P/V55L Fyn SH3 Δ(57–60), a structural mimic of I as seen from chemical shift correlations (Left Inset), aggregates in a time-dependent fashion leading to a loss in intensity of NMR resonances in 1H-15N HSQC datasets with concomitant formation of amyloid fibrils that can be visualized in an electron micrograph (Right Inset). (Adapted from ref. 18. Reprinted with permission from AAAS.)
The structure of I was validated by making the A39V/N53P/V55L Fyn SH3 Δ(57–60) truncation mutant that is predicted to adopt a structure similar to that of I (18). A comparison of 15N chemical shifts of this intermediate state proxy with those of I (Fig. 3B, Inset) establishes that indeed the Δ(57–60) mutant is a good mimic. Furthermore, under NMR conditions, the mimic forms fibrils that have amyloid-like characteristics (Fig. 3B, Inset); notably both wt Fyn SH3 Δ(57–60) and wt Fyn SH3 Δ(56–60) also form similar fibrillar aggregates, whereas other mutants that populate the U state as high or higher than the mimic do not. This suggests that, at least in the case of the Fyn SH3 domain, it is not the globally unfolded state but rather the thermally accessible excited state I that is critical for aggregation and fibril formation.
Evolving Protein Function by Altering the Energy Landscape
Protein function and misfunction often critically depend on the interconversion between different molecular conformations that can have diverse biological activities (42). A detailed characterization of each conformer is, therefore, relevant for understanding the structure–function paradigm that, in turn, can lead to new ways of engineering novel functions through the introduction of a small number of key mutations.
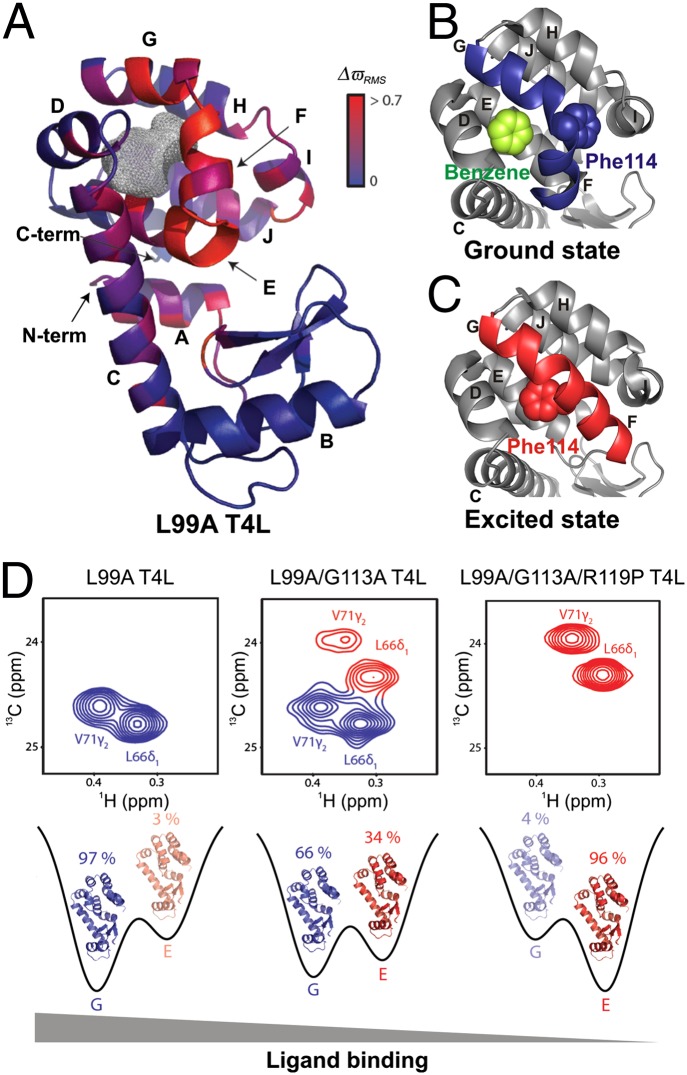
By means of example, consider the substitution of Ala for Leu at position 99 of T4 lysozyme (L99A T4L) that creates an internal cavity of ∼150 Å3 (43). The cavity can accommodate hydrophobic molecules such as substituted benzenes (Fig. 4 A and B), and detailed X-ray diffraction studies have established that the ground-state conformations of wt and L99A T4L are virtually identical (44). However, CPMG RD measurements show that L99A T4L is dynamic on the millisecond timescale, unlike the wt protein (45). These dynamics are the result of exchange between the native state and an alternative conformation that is populated to ∼3% at room temperature. The structure of this excited conformer has been determined from backbone 15N, 13C, and 1H chemical shifts obtained using CPMG RD experiments in concert with a CS-Rosetta structure refinement protocol (19) (Fig. 4C).
Fig. 4.
Probing the structure–function relationship at different points on the energy landscape. (A) Ground-state X-ray structure of L99A T4L (PDB ID code 3DMV), color-coded according to the magnitude of chemical shift differences (ΔϖRMS) between the ground and excited states (19). The gray mesh delineates the cavity that results from the L99A mutation. (B and C) Comparison of the C-terminal domain of L99A T4L in the ground (PDB ID code 3DMX) (B) and excited (PDB ID code 2LCB) (C) states, highlighting the different orientations of the F and G helices in each of the conformers. The F114 side chain that rotates into the cavity in the excited state and the bound benzene in the ground state are shown using space-filling representations. (D) The ground and excited-state populations can be manipulated by a small number of mutations. G (Left) and E states become comparable in population in the L99A/G113A construct (Center), as seen in 13C-1H correlation spectra. The new set of peaks (red) have chemical shift values that are in excellent agreement with those obtained for state E from fits of the L99A CPMG RD profiles. Populations of the ground and excited states are inverted in L99A/G113A/R119P T4L (Right), as the ground-state peaks (blue) disappear from the NMR spectrum. The affinity of T4L for ligand in each of the constructs is indicated. [Adapted by permission from Macmillan Publishers Ltd: Nature (19), copyright 2011.]
A comparison of the native (Fig. 4 A and B) and excited-state (Fig. 4C) structures reveals that conformational changes are localized near the cavity and that helices F and G, which are perpendicular in the ground conformer, form a single long helix in the excited state. Furthermore, in the excited state, the ψ backbone dihedral angle of F114 changes from +49° to −36° and the rotation of the side-chain χ1 torsion angle from gauche to trans positions the F114 side chain into the cavity to occupy the space of L99 in the wt protein (19).
Rosetta structure-based design calculations were used to generate mutants that stabilized the excited state. For example, G113A stabilizes the long contiguous F-G helix in the excited state by replacing a helix destabilizing residue (G) with one that favors helical structure (A). A further mutation, R119P, destabilizes the ground conformer because of steric clashes between Pro119 and Thr115. Notably, the chemical shifts of L99A/G113A/R119P T4L are identical to those of the excited state of L99A T4L, demonstrating that an inversion of populations has been achieved by the G113A, R119P mutations. Further studies establish that only the ground state can bind benzene, as expected, because the positioning of F114 into the cavity in the excited state precludes ligand binding in this conformation (19). The excited-state structure, thus, provides a framework for understanding how to manipulate the delicate balance between states on a rugged protein energy landscape, leading ultimately to a change in function. It also illustrates how this might happen naturally during evolution as a result of mutations, such as the G113A/R119P substitutions described here, to generate protein folds with distinct functionalities (Fig. 4D) (46).
Structural Plasticity of DNA and Implications for Molecular Recognition
In the canonical view of the structure of DNA the two strands of the double helix are linked by Watson–Crick base pairs. The linear nucleotide sequence then forms the genetic code that is used to establish a protein’s primary structure (47). However, it is increasingly clear that recognition of specific DNA sequences by DNA-binding partners involves more than a simple linear code and that the inherent plasticity of certain DNA sequences provides a second type of mechanism that regulates the specificity and affinity of interactions with target molecules (48).
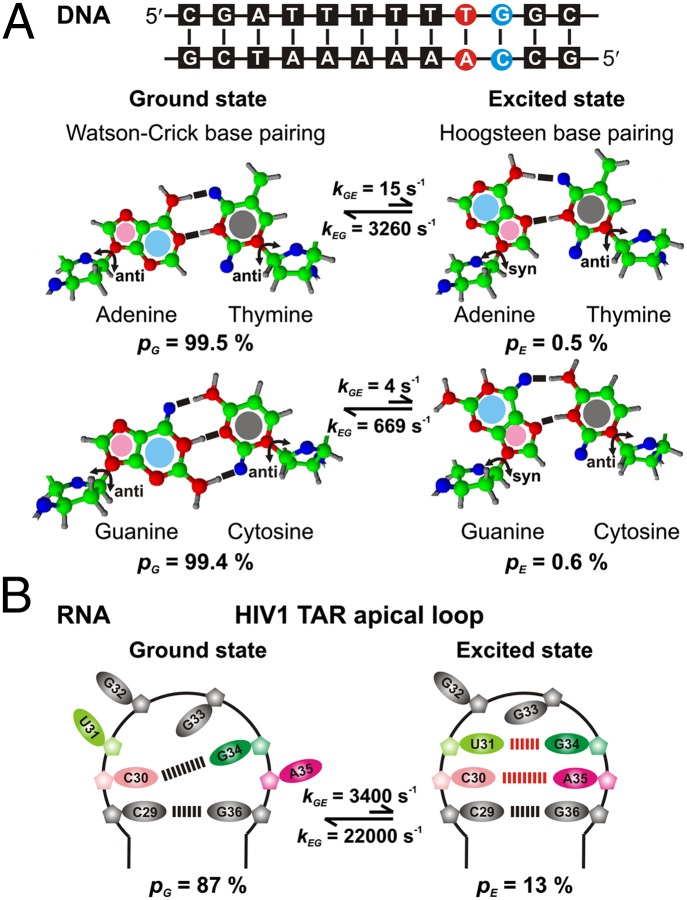
Remarkably, transiently populated DNA conformations have now been detected at single A-T or G-C base pairs occurring at CA/TG steps in naked double-stranded canonical DNA duplexes (49). These conformationally excited states have been characterized using on- and off-resonance 13C R1ρ RD experiments, using DNA sequences such as the one shown in Fig. 5A, for which excited-state populations of ∼0.5% and lifetimes between 0.3–1.5 ms are established (26 °C; pH 5.4). Particularly large chemical shift changes, ΔϖGE = 2–3 ppm, were detected at the sugar C1′ and base C8 (A, G) or C6 (C) carbons adjacent to the glycosidic bond between the sugar and the base, providing strong evidence that the exchange processes involve rotations of ∼180° along the glycosidic bond. This results in a conversion of A or G from an anti (G state) to a syn (E) conformation. The magnitude, direction and location of the changes in chemical shifts, as well as the thermodynamics of the conformational exchange event, led to the assignment of the excited state as one wherein the canonical Watson–Crick base pairing between A-T and G-C was replaced by a Hoogsteen base pair (Fig. 5A) (49).
Fig. 5.
Nucleic acid conformational plasticity can modulate function. (A) Sequence of A6-DNA, which shows significant local conformational exchange at the highlighted A-T (red) and G-C (blue) base pairs. Molecular structures of Watson–Crick (G state) and Hoogsteen (E state) A-T (Upper) and G-C (Lower) base pairs. Purine residues A and G undergo an anti to syn conformational transition from G to E. (B) Secondary structure and hydrogen-bonding partners in G and E states of the HIV1 TAR apical loop. A pair of noncanonical A-C and G-U base pairs is formed in E.
The existence of Hoogsteen base pairs was verified by trapping the noncanonical base paired excited state using modified bases, N1-methylated adenine (A-T) or guanine (G-C) at the CA/TG step. The chemical shifts observed for the modified DNA duplexes were in good agreement with predictions based on analysis of the R1ρ RD data, providing strong evidence that these modified oligonucleotide sequences are faithful structural mimics of the excited state. The presence of Hoogsteen base pairing was further confirmed by analysis of distances in NOE datasets recorded on these mimics (49).
The detection of Hoogsteen base pairing has brought to light the conformational flexibility inherent in free DNA. It appears that the DNA sequence codes for alternate low-lying excited-state conformations that may be specifically recognized by proteins. Hoogsteen base pairs are known to be recognition motifs for several enzymes and transcription factors (50, 51), and they introduce distortions into DNA topology (51, 52) and expose DNA surfaces that are distinct from those in a Watson–Crick structure. The fact that the same linear sequence converts between distinct conformations that may be important in protein-nucleic acid recognition, provides an additional layer of regulation of these biomolecular interactions.
Regulation of RNA Function Through Dynamics
RNA molecules organize into 3D structures, dictated to a large extent by nucleotide sequence. As with DNA, described above, the sequence–structure relationship for RNA is complex, because these molecules can adopt a variety of different conformations that, in turn, dictate biological function. Al-Hashimi and coworkers have recently developed an elegant approach for obtaining atomic level descriptions of sparsely populated, transiently formed RNA conformations using a combined NMR, mutagenesis, and structure-prediction approach (53).
One application of the methodology is provided by studies of the HIV-1 transactivation response element (TAR) using 13C R1ρ RD (53). Fits of the dispersion data were consistent with a two-site conformational exchange process that converts the ground state to a higher-energy conformer that has a fractional population of 13% and a lifetime of ∼45 μs. The structural properties of the excited state could be elucidated on the basis of 13C chemical shifts extracted from analysis of the RD profiles that are sensitive reporters of sugar pucker, base stacking, and syn vs. anti glycosidic bond angles. The organization of this excited state features two noncanonical base pairs between U31-G34 and C30-A35 with the G34 base in a syn conformation (Fig. 5B). Notably, the excited-state structure was predicted to be the second-lowest energy conformation of the RNA fragment studied (after the ground state) using the secondary structure prediction program, MC-Fold (54). The excited state was stabilized by mutating either C30 to U or A35 to G, thereby converting the noncanonical C30-A35 interaction into a canonical Watson–Crick base pair. The stabilized sequence could then be studied by conventional NMR methods, with the observed chemical shifts in good agreement with those of E obtained indirectly via RD, thereby validating the structure of the excited state.
The structure of E has important implications for function. Because key recognition residues (C30, U31, G34, and A35) are sequestered by forming base pairs, the excited state cannot bind viral transactivator protein Tat and human cyclin T1 that activate transcription of the HIV-1 genome (55). Moreover, the G↔E equilibrium can be shifted to the inactive state (E) by a number of proteins that are known to bind to the TAR apical loop or by lowering the pH (53). Understanding conformational dynamics may, therefore, lead to the development of new anti-HIV drugs that down-regulate HIV-1 genome transactivation by stabilizing the excited state of TAR.
Excited states have also been detected for the ribosomal A-site internal loop and the HIV-1 stem loop 1 using analogous methods to those described for TAR (53). Although a quantitative description of transient nucleic acid structure is only just emerging, it is clear that the conformational plasticity that has recently been observed and characterized in fundamental structural elements of RNA may well play an important role in mediating important biological functions including protein recognition, gene expression/regulation, and catalysis (53).
Interplay Between Conformational Selection and Induced Fit
Calmodulin (CaM) is a universal eukaryotic Ca2+-sensing messenger protein that has been studied extensively by a large number of different biophysical methods (56). Ca2+-loaded CaM (CaM-4Ca2+) exists in an extended conformation (57) that transforms into a highly compact structure when bound to target proteins (Fig. 6 A and B) (58). The structural plasticity of this important molecule has been explored by using PREs to detect and visualize transiently populated conformations in different ligated states of the protein (59). Paramagnetic nitroxide spin labels were conjugated to Cys residues substituted in four different positions on CaM, and PREs were measured on the metal-free (apoCaM) and loaded states and in the presence and absence of myosin light chain kinase peptide (MLCK). Interdomain PREs were observed for both apoCaM and CaM-4Ca2+, implying that both forms of CaM transiently sample compact conformations. The measured PREs were used to guide a simulated annealing computation to visualize the structures of these sparsely populated states (Fig. 6C). Notably, the corresponding structures of CaM-4Ca2+ include many conformers that are similar to those adopted by substrate-bound CaM-4Ca2+ (compare Fig. 6 B and C). In contrast, a similar analysis shows that the ensemble of apoCaM structures is distinct and does not overlap with the peptide bound state (59). The direct visualization of conformationally excited states of various forms of CaM suggests a recognition mechanism whereby Ca2+ activates CaM for binding target peptides by modifying both local structure, as well as interdomain interactions, biasing some of the conformations toward those that are primed for peptide binding. This adds yet another degree of complexity to the ways in which CaM recognizes its targets, emphasizing the interplay between induced fit and conformational selection (59).
Fig. 6.
Compact transient conformations in the Ca2+ sensor signaling protein calmodulin. (A) Structures of CaM-4Ca2+ (green) (PDB ID code 1CLL) and CaM-4Ca2+ bound to MLCK peptide (blue) (PDB ID code 1CDL). (B) Superposition of the structures in A best fit to the N-terminal (Left) or C-terminal (Right) domains. Structures of 26 other peptide-bound structures of CaM-4Ca2+ are best fit in similar ways and shown superimposed as atomic probability density maps in gray. (C) Atomic probability density maps of the compact excited-state conformations of CaM-4Ca2+ shown at contour levels ranging from 0.1 (blue) to 0.5 (red) and best fit to the N-terminal (Left) or C-terminal (Right) domains of free CaM-4Ca2+ (dark green). The corresponding density maps of the major state, which lacks significant interdomain PREs, are shown in gray at a contour level of 0.1. [Adapted with permission from ref. 59 (Copyright 2011, American Chemical Society).]
Applications to Other Transient Conformers
The energy landscape of a biomolecule is shaped largely by its primary sequence but also by binding partners, cofactors, and substrates, and there is increasing evidence of the importance of thermally accessible excited states that reside on this landscape in guiding biomolecular function. We have focused on only a few examples above, emphasizing cases where atomic-resolution structures are obtained from NMR relaxation data. Many other important examples of where NMR relaxation methodologies have been used in insightful ways, albeit without obtaining detailed structures of the transiently formed states that are implicated in function, have been described in the literature, and important lessons are emerging.
Detailed NMR studies have shown that at least in one case virtually every stage of enzyme kinetics, including the catalytic step, benefits from sparsely populated states on the energy landscape. Conformational fluctuations of dihydrofolate reductase (DHFR) to low-lying excited states have been shown to channel the enzyme through its functional cycle and to facilitate substrate/cofactor binding, product release, and transition-state formation (60, 61). In this enzyme system, each intermediate in the catalytic cycle populates excited states that resemble preceding and/or succeeding intermediates, suggesting that ligand binding and release is guided via a conformational selection mechanism. In addition, thermal fluctuations in DHFR promote the structural changes necessary for transition state assembly and, thereby, mediate the catalytic hydride transfer step (62). Functionally important excited states have also been identified in various enzymes, such as adenylate kinase (63), cyclophilin A (64), RNaseA (65), HIV-1 protease (66), and triosephosphate isomerase (65), with transition rates matching catalytic turnover numbers, providing evidence that conformational exchange plays an important role in regulating the rate of product formation for many of these enzymes. Some enzymes, such as cyclophilin A, exhibit fluctuations in the absence of substrate, indicating that their dynamics predispose them toward a functional form. In contrast, for other enzymes, such as DHFR (60) and DNA pol β (67), fluctuations appear or disappear in direct response to a modulation of the energy landscape by substrates and cofactors.
The stability of low-lying excited states can be modulated in a number of different ways to achieve downstream regulation of function. Proteins in signal transduction cascades are often activated by a population shift mechanism, whereby a physical or chemical signal switches the position of a preexisting equilibrium in favor of the active state. For instance, the native state of the blue light-sensing Avena sativa LOV2 domain interconverts with a low-populated conformer that resembles the light-activated state of the protein (68). Irradiation of LOV2 with blue light stabilizes the excited state and inverts the populations of interconverting conformations, leading to activation. Similarly, phosphorylation of the N-terminal receiver domain activates the nitrogen regulatory protein (NtrC) from Salmonella typhimurium by initiating a large conformational change that exposes a hydrophobic surface responsible for downstream signal transmission (69). NMR RD measurements, however, show that such an activated state is present even in the absence of phosphorylation, albeit at a low level (69). In a dramatic recent example that has implications for new strategies for drug discovery, it was demonstrated via a combined NMR RD and calorimetry study that allosteric regulation can be effected via transiently populated conformational states (70). In the absence of the nucleotide cGMP, a mutant catabolite activator protein (CAP*) transiently populates an excited state that is active in the sense that it binds DNA, whereas the ground state remains inactive. Thus, CAP* is able to bind DNA (via its DNA-binding domains) despite the fact that the ground-state conformer is DNA binding-incompetent. Addition of cGMP that binds to a separate region of the protein abrogates DNA binding by eliminating the excited state without altering the structure of the ground conformer.
Transiently formed encounter complexes are important intermediates in molecular recognition events. They have been detected in PRE studies, focusing on the binding of the HOXD9 homeodomain to a 24-bp double-stranded DNA with a single protein-specific binding site (71) and in RD measurements where they are on pathway in the coupled binding and folding of the intrinsically disordered pKID domain in the presence of the partner KIX protein (72).
In addition to modulating protein function, low-lying excited states can play important roles in misfunction as well, as suggested by RD studies of the Fyn SH3 domain (18) described above. Excited states are thought to play a role in a number of debilitating diseases by functioning as potential starting points for protein self-assembly. For example, the aggregation of transthyretin (TTR) causes senile systemic amyloidosis and familial amyloid polyneuropathy (73). Dissociation of the tetrameric two-sheet β-sandwich TTR into a monomeric form and subsequent misfolding of the monomer has been postulated as the mechanism leading to its amyloid fibril formation (74). A monomeric variant of TTR has now been shown by RD NMR to transiently populate an intermediate state characterized by structural fluctuations in one of the sheets, suggesting that amyloid formation in this case may occur by partial rather than global unfolding (75). Transiently populated states have also been observed in the cataract-causing Opj variant of γS-crystallin, a structural protein in the eye lens (76).
Future Perspectives
As described above, it is now possible, at least in some cases, to determine the atomic resolution 3D structures of biomolecular excited states. These studies will ultimately benefit from combining NMR with other biophysical methods. For example, central to the success of the described NMR approaches is the continuously growing power of computation, coupled with improved structure determination algorithms (15, 16, 77, 78). In fact, translation of chemical shifts into 3D structure using database approaches and sparse experimental restraints has only become possible in the past few years. The growth in computational power has also led to impressive increases in sampling times in molecular dynamics simulations, so that rare events can be studied by computation (79). Millisecond-timescale molecular dynamics (MD) simulations of small protein domains in explicit solvent are now possible, and these trajectories provide a powerful framework to analyze NMR RD data in terms of the structures that are formed along pathways and that lead to biochemical function (80, 81). Equilibrium simulations of the native state of bovine pancreatic trypsin inhibitor protein (BPTI), establishing the existence of thermally accessible transiently populated states (82), have already been cross-validated with NMR data. Reasonable agreement between calculated amide 15N chemical shifts of the conformers observed in MD and shifts measured from R1ρ measurements has been observed (83), although the experimentally derived populations of the minor states of BPTI are still not reproduced by simulation. It is likely that the synergy between experiment and computation will only become stronger as long-time scale simulations become more common.
Although the majority of NMR and computational applications have focused on relatively small protein systems, with time, size barriers are becoming less problematic. For example, RD NMR studies have been performed on the ClpP protease (84), a 300-kDa double-ring structure, as well as the 670-kDa proteasome, using 13CH3-labeled methyl groups in an otherwise highly deuterated environment (85). These measurements take advantage of experiments that preserve a large fraction of the NMR signal, protecting it from the relaxation losses that are normally catastrophic for large biomolecules. In other studies of supramolecular systems, 15N-based CEST experiments have been used to quantify the mechanism of exchange between Aβ monomers and fibrils (>1 MDa), facilitating the extraction of structural properties of the Aβ peptide in the bound state (10). The continued development of solid-state NMR methodologies will provide additional promising approaches for studies of transiently populated states, alleviating some of the size requirements that can limit solution applications (86, 87).
In addition to the advances in NMR described above, there have also been innovations in other experimental areas that have contributed substantially toward our understanding of the role of biomolecular fluctuations at an atomic level. For instance, ambient-temperature X-ray crystallography (88) has emerged recently as a powerful probe of sparsely populated protein conformations. In this approach, room-temperature X-ray diffraction datasets are analyzed using an algorithm that scans the electron-density specifically for alternate protein side-chain conformations that were not modeled. As a landmark illustration of the power of this technique, minor conformations in regions of cyclophilin A that were observed to be dynamic by solution NMR methods were detected using X-ray data acquired at room temperature, providing a structural rationalization of the NMR data (5). Notably, these additional states could not be observed from analysis of datasets recorded at cryogenic temperatures. Other exciting methodologies beyond the scope of review here include studies of transiently formed states by Laue diffraction methods (89) and single-molecule florescence approaches (90).
It is evident that structural studies of the native state of a biomolecule are merely the starting point in a larger quest to understand function. Further NMR measurements of the sort described here, along with insight from a large spectrum of different biophysical techniques, will lead to a more complete description of how these fascinating molecules carry out the myriad of important tasks that they perform and, importantly, how their activities can be modulated in a rational manner.
Acknowledgments
We thank Alex Bain and Pramodh Vallurupalli for a critical reading of the manuscript. A.S. is a recipient of a Canadian Institutes of Health Research (CIHR) postdoctoral fellowship. L.E.K. holds a Canada Research Chair in Biochemistry and is funded by the CIHR.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Watson JD, Crick FH. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953;171(4356):737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 2.Kendrew JC, et al. A three-dimensional model of the myoglobin molecule obtained by x-ray analysis. Nature. 1958;181(4610):662–666. doi: 10.1038/181662a0. [DOI] [PubMed] [Google Scholar]
- 3.Perutz MF, et al. Structure of haemoglobin: A three-dimensional Fourier synthesis at 5.5-A. resolution, obtained by X-ray analysis. Nature. 1960;185(4711):416–422. doi: 10.1038/185416a0. [DOI] [PubMed] [Google Scholar]
- 4.Blake CC, et al. Structure of hen egg-white lysozyme. A three-dimensional Fourier synthesis at 2 Angstrom resolution. Nature. 1965;206(4986):757–761. doi: 10.1038/206757a0. [DOI] [PubMed] [Google Scholar]
- 5.Fraser JS, et al. Hidden alternative structures of proline isomerase essential for catalysis. Nature. 2009;462(7273):669–673. doi: 10.1038/nature08615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frauenfelder H, Sligar SG, Wolynes PG. The energy landscapes and motions of proteins. Science. 1991;254(5038):1598–1603. doi: 10.1126/science.1749933. [DOI] [PubMed] [Google Scholar]
- 7.Lazaridis T, Karplus M. Thermodynamics of protein folding: A microscopic view. Biophys Chem. 2003;100(1-3):367–395. doi: 10.1016/s0301-4622(02)00293-4. [DOI] [PubMed] [Google Scholar]
- 8.Dill KA, Chan HS. From Levinthal to pathways to funnels. Nat Struct Biol. 1997;4(1):10–19. doi: 10.1038/nsb0197-10. [DOI] [PubMed] [Google Scholar]
- 9.Dill KA. Dominant forces in protein folding. Biochemistry. 1990;29(31):7133–7155. doi: 10.1021/bi00483a001. [DOI] [PubMed] [Google Scholar]
- 10.Fawzi NL, Ying J, Ghirlando R, Torchia DA, Clore GM. Atomic-resolution dynamics on the surface of amyloid-β protofibrils probed by solution NMR. Nature. 2011;480(7376):268–272. doi: 10.1038/nature10577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vallurupalli P, Bouvignies G, Kay LE. Studying “invisible” excited protein states in slow exchange with a major state conformation. J Am Chem Soc. 2012;134(19):8148–8161. doi: 10.1021/ja3001419. [DOI] [PubMed] [Google Scholar]
- 12.Palmer AG, 3rd, Kroenke CD, Loria JP. Nuclear magnetic resonance methods for quantifying microsecond-to-millisecond motions in biological macromolecules. Methods Enzymol. 2001;339:204–238. doi: 10.1016/s0076-6879(01)39315-1. [DOI] [PubMed] [Google Scholar]
- 13.Hansen DF, Vallurupalli P, Kay LE. Using relaxation dispersion NMR spectroscopy to determine structures of excited, invisible protein states. J Biomol NMR. 2008;41(3):113–120. doi: 10.1007/s10858-008-9251-5. [DOI] [PubMed] [Google Scholar]
- 14.Clore GM. Exploring sparsely populated states of macromolecules by diamagnetic and paramagnetic NMR relaxation. Protein Sci. 2011;20(2):229–246. doi: 10.1002/pro.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen Y, et al. Consistent blind protein structure generation from NMR chemical shift data. Proc Natl Acad Sci USA. 2008;105(12):4685–4690. doi: 10.1073/pnas.0800256105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robustelli P, Kohlhoff K, Cavalli A, Vendruscolo M. Using NMR chemical shifts as structural restraints in molecular dynamics simulations of proteins. Structure. 2010;18(8):923–933. doi: 10.1016/j.str.2010.04.016. [DOI] [PubMed] [Google Scholar]
- 17.Korzhnev DM, Religa TL, Banachewicz W, Fersht AR, Kay LE. A transient and low-populated protein-folding intermediate at atomic resolution. Science. 2010;329(5997):1312–1316. doi: 10.1126/science.1191723. [DOI] [PubMed] [Google Scholar]
- 18.Neudecker P, et al. Structure of an intermediate state in protein folding and aggregation. Science. 2012;336(6079):362–366. doi: 10.1126/science.1214203. Available at http://www.sciencemag.org/content/336/6079/362.abstract. [DOI] [PubMed] [Google Scholar]
- 19.Bouvignies G, et al. Solution structure of a minor and transiently formed state of a T4 lysozyme mutant. Nature. 2011;477(7362):111–114. doi: 10.1038/nature10349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forsén S, Hoffman RA. Study of moderately rapid chemical exchange reactions by means of nuclear magnetic double resonance. J Chem Phys. 1963;39(11):2892–2901. [Google Scholar]
- 21.Gupta RK, Redfield AG. Double nuclear magnetic resonance observation of electron exchange between ferri- and ferrocytochrome c. Science. 1970;169(3951):1204–1206. doi: 10.1126/science.169.3951.1204. [DOI] [PubMed] [Google Scholar]
- 22.Cayley PJ, et al. Nuclear magnetic resonance studies of the binding of trimethoprim to dihydrofolate reductase. Biochemistry. 1979;18(18):3886–3895. doi: 10.1021/bi00585a008. [DOI] [PubMed] [Google Scholar]
- 23.Ward KM, Aletras AH, Balaban RS. A new class of contrast agents for MRI based on proton chemical exchange dependent saturation transfer (CEST) J Magn Reson. 2000;143(1):79–87. doi: 10.1006/jmre.1999.1956. [DOI] [PubMed] [Google Scholar]
- 24.van Zijl PC, Yadav NN. Chemical exchange saturation transfer (CEST): What is in a name and what isn’t? Magn Reson Med. 2011;65(4):927–948. doi: 10.1002/mrm.22761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Palmer AG, 3rd, Massi F. Characterization of the dynamics of biomacromolecules using rotating-frame spin relaxation NMR spectroscopy. Chem Rev. 2006;106(5):1700–1719. doi: 10.1021/cr0404287. [DOI] [PubMed] [Google Scholar]
- 26.Ban D, et al. Exceeding the limit of dynamics studies on biomolecules using high spin-lock field strengths with a cryogenically cooled probehead. J Magn Reson. 2012;221:1–4. doi: 10.1016/j.jmr.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 27.Hansen AL, Nikolova EN, Casiano-Negroni A, Al-Hashimi HM. Extending the range of microsecond-to-millisecond chemical exchange detected in labeled and unlabeled nucleic acids by selective carbon R(1ρ) NMR spectroscopy. J Am Chem Soc. 2009;131(11):3818–3819. doi: 10.1021/ja8091399. [DOI] [PubMed] [Google Scholar]
- 28.Lundström P, Akke M. Microsecond protein dynamics measured by 13Calpha rotating-frame spin relaxation. ChemBioChem. 2005;6(9):1685–1692. doi: 10.1002/cbic.200500086. [DOI] [PubMed] [Google Scholar]
- 29.Dill KA, MacCallum JL. The protein-folding problem, 50 years on. Science. 2012;338(6110):1042–1046. doi: 10.1126/science.1219021. [DOI] [PubMed] [Google Scholar]
- 30.Chiti F, Dobson CM. Amyloid formation by globular proteins under native conditions. Nat Chem Biol. 2009;5(1):15–22. doi: 10.1038/nchembio.131. [DOI] [PubMed] [Google Scholar]
- 31.Allen M, Friedler A, Schon O, Bycroft M. The structure of an FF domain from human HYPA/FBP11. J Mol Biol. 2002;323(3):411–416. doi: 10.1016/s0022-2836(02)00968-3. [DOI] [PubMed] [Google Scholar]
- 32.Jemth P, et al. Demonstration of a low-energy on-pathway intermediate in a fast-folding protein by kinetics, protein engineering, and simulation. Proc Natl Acad Sci USA. 2004;101(17):6450–6455. doi: 10.1073/pnas.0401732101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Korzhnev DM, Religa TL, Lundström P, Fersht AR, Kay LE. The folding pathway of an FF domain: Characterization of an on-pathway intermediate state under folding conditions by (15)N, (13)C(α) and (13)C-methyl relaxation dispersion and (1)H/(2)H-exchange NMR spectroscopy. J Mol Biol. 2007;372(2):497–512. doi: 10.1016/j.jmb.2007.06.012. [DOI] [PubMed] [Google Scholar]
- 34.Vallurupalli P, Hansen DF, Stollar E, Meirovitch E, Kay LE. Measurement of bond vector orientations in invisible excited states of proteins. Proc Natl Acad Sci USA. 2007;104(47):18473–18477. doi: 10.1073/pnas.0708296104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Korzhnev DM, et al. Nonnative interactions in the FF domain folding pathway from an atomic resolution structure of a sparsely populated intermediate: An NMR relaxation dispersion study. J Am Chem Soc. 2011;133(28):10974–10982. doi: 10.1021/ja203686t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vallurupalli P, Kay LE. Probing slow chemical exchange at carbonyl sites in proteins by chemical exchange saturation transfer NMR spectroscopy. Angew Chem Int Ed Engl. 2013;52(15):4156–4159. doi: 10.1002/anie.201209118. [DOI] [PubMed] [Google Scholar]
- 37.Korzhnev DM, Religa TL, Kay LE. Transiently populated intermediate functions as a branching point of the FF domain folding pathway. Proc Natl Acad Sci USA. 2012;109(44):17777–17782. doi: 10.1073/pnas.1201799109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barette J, Velyvis A, Religa TL, Korzhnev DM, Kay LE. Cross-validation of the structure of a transiently formed and low populated FF domain folding intermediate determined by relaxation dispersion NMR and CS-Rosetta. J Phys Chem B. 2012;116(23):6637–6644. doi: 10.1021/jp209974f. [DOI] [PubMed] [Google Scholar]
- 39.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 40.Neudecker P, et al. Identification of a collapsed intermediate with non-native long-range interactions on the folding pathway of a pair of Fyn SH3 domain mutants by NMR relaxation dispersion spectroscopy. J Mol Biol. 2006;363(5):958–976. doi: 10.1016/j.jmb.2006.08.047. [DOI] [PubMed] [Google Scholar]
- 41.Noble ME, Musacchio A, Saraste M, Courtneidge SA, Wierenga RK. Crystal structure of the SH3 domain in human Fyn; comparison of the three-dimensional structures of SH3 domains in tyrosine kinases and spectrin. EMBO J. 1993;12(7):2617–2624. doi: 10.2210/pdb1shf/pdb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karplus M, Kuriyan J. Molecular dynamics and protein function. Proc Natl Acad Sci USA. 2005;102(19):6679–6685. doi: 10.1073/pnas.0408930102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eriksson AE, Baase WA, Wozniak JA, Matthews BW. A cavity-containing mutant of T4 lysozyme is stabilized by buried benzene. Nature. 1992;355(6358):371–373. doi: 10.1038/355371a0. [DOI] [PubMed] [Google Scholar]
- 44.Eriksson AE, et al. Response of a protein structure to cavity-creating mutations and its relation to the hydrophobic effect. Science. 1992;255(5041):178–183. doi: 10.1126/science.1553543. [DOI] [PubMed] [Google Scholar]
- 45.Mulder FA, Mittermaier A, Hon B, Dahlquist FW, Kay LE. Studying excited states of proteins by NMR spectroscopy. Nat Struct Biol. 2001;8(11):932–935. doi: 10.1038/nsb1101-932. [DOI] [PubMed] [Google Scholar]
- 46.Tokuriki N, Tawfik DS. Protein dynamism and evolvability. Science. 2009;324(5924):203–207. doi: 10.1126/science.1169375. [DOI] [PubMed] [Google Scholar]
- 47.Nelson DL, Cox MM. Lehninger Principles of Biochemistry. New York, NY: WH Freeman; 2010. [Google Scholar]
- 48.Rohs R, et al. Origins of specificity in protein-DNA recognition. Annu Rev Biochem. 2010;79:233–269. doi: 10.1146/annurev-biochem-060408-091030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nikolova EN, et al. Transient Hoogsteen base pairs in canonical duplex DNA. Nature. 2011;470(7335):498–502. doi: 10.1038/nature09775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Patikoglou GA, et al. TATA element recognition by the TATA box-binding protein has been conserved throughout evolution. Genes Dev. 1999;13(24):3217–3230. doi: 10.1101/gad.13.24.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kitayner M, et al. Diversity in DNA recognition by p53 revealed by crystal structures with Hoogsteen base pairs. Nat Struct Mol Biol. 2010;17(4):423–429. doi: 10.1038/nsmb.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hsu GW, Ober M, Carell T, Beese LS. Error-prone replication of oxidatively damaged DNA by a high-fidelity DNA polymerase. Nature. 2004;431(7005):217–221. doi: 10.1038/nature02908. [DOI] [PubMed] [Google Scholar]
- 53.Dethoff EA, Petzold K, Chugh J, Casiano-Negroni A, Al-Hashimi HM. Visualizing transient low-populated structures of RNA. Nature. 2012;491(7426):724–728. doi: 10.1038/nature11498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Parisien M, Major F. The MC-Fold and MC-Sym pipeline infers RNA structure from sequence data. Nature. 2008;452(7183):51–55. doi: 10.1038/nature06684. [DOI] [PubMed] [Google Scholar]
- 55.Tahirov TH, et al. Crystal structure of HIV-1 Tat complexed with human P-TEFb. Nature. 2010;465(7299):747–751. doi: 10.1038/nature09131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Finn BE, Forsén S. The evolving model of calmodulin structure, function and activation. Structure. 1995;3(1):7–11. doi: 10.1016/s0969-2126(01)00130-7. [DOI] [PubMed] [Google Scholar]
- 57.Chattopadhyaya R, Meador WE, Means AR, Quiocho FA. Calmodulin structure refined at 1.7 A resolution. J Mol Biol. 1992;228(4):1177–1192. doi: 10.1016/0022-2836(92)90324-d. [DOI] [PubMed] [Google Scholar]
- 58.Ikura M, et al. Solution structure of a calmodulin-target peptide complex by multidimensional NMR. Science. 1992;256(5057):632–638. doi: 10.1126/science.1585175. [DOI] [PubMed] [Google Scholar]
- 59.Anthis NJ, Doucleff M, Clore GM. Transient, sparsely populated compact states of apo and calcium-loaded calmodulin probed by paramagnetic relaxation enhancement: Interplay of conformational selection and induced fit. J Am Chem Soc. 2011;133(46):18966–18974. doi: 10.1021/ja2082813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boehr DD, McElheny D, Dyson HJ, Wright PE. The dynamic energy landscape of dihydrofolate reductase catalysis. Science. 2006;313(5793):1638–1642. doi: 10.1126/science.1130258. [DOI] [PubMed] [Google Scholar]
- 61.Boehr DD, McElheny D, Dyson HJ, Wright PE. Millisecond timescale fluctuations in dihydrofolate reductase are exquisitely sensitive to the bound ligands. Proc Natl Acad Sci USA. 2010;107(4):1373–1378. doi: 10.1073/pnas.0914163107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bhabha G, et al. A dynamic knockout reveals that conformational fluctuations influence the chemical step of enzyme catalysis. Science. 2011;332(6026):234–238. doi: 10.1126/science.1198542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wolf-Watz M, et al. Linkage between dynamics and catalysis in a thermophilic-mesophilic enzyme pair. Nat Struct Mol Biol. 2004;11(10):945–949. doi: 10.1038/nsmb821. [DOI] [PubMed] [Google Scholar]
- 64.Eisenmesser EZ, Bosco DA, Akke M, Kern D. Enzyme dynamics during catalysis. Science. 2002;295(5559):1520–1523. doi: 10.1126/science.1066176. [DOI] [PubMed] [Google Scholar]
- 65.Loria JP, Berlow RB, Watt ED. Characterization of enzyme motions by solution NMR relaxation dispersion. Acc Chem Res. 2008;41(2):214–221. doi: 10.1021/ar700132n. [DOI] [PubMed] [Google Scholar]
- 66.Louis JM, Ishima R, Torchia DA, Weber IT. HIV-1 protease: Structure, dynamics, and inhibition. Adv Pharmacol. 2007;55:261–298. doi: 10.1016/S1054-3589(07)55008-8. [DOI] [PubMed] [Google Scholar]
- 67.Berlow RB, Swain M, Dalal S, Sweasy JB, Loria JP. Substrate-dependent millisecond domain motions in DNA polymerase β. J Mol Biol. 2012;419(3-4):171–182. doi: 10.1016/j.jmb.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yao X, Rosen MK, Gardner KH. Estimation of the available free energy in a LOV2-J α photoswitch. Nat Chem Biol. 2008;4(8):491–497. doi: 10.1038/nchembio.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gardino AK, Kern D. Functional dynamics of response regulators using NMR relaxation techniques. Methods Enzymol. 2007;423:149–165. doi: 10.1016/S0076-6879(07)23006-X. [DOI] [PubMed] [Google Scholar]
- 70.Tzeng S-R, Kalodimos CG. Allosteric inhibition through suppression of transient conformational states. Nat Chem Biol. 2013;9(7):462–465. doi: 10.1038/nchembio.1250. [DOI] [PubMed] [Google Scholar]
- 71.Iwahara J, Clore GM. Detecting transient intermediates in macromolecular binding by paramagnetic NMR. Nature. 2006;440(7088):1227–1230. doi: 10.1038/nature04673. [DOI] [PubMed] [Google Scholar]
- 72.Sugase K, Dyson HJ, Wright PE. Mechanism of coupled folding and binding of an intrinsically disordered protein. Nature. 2007;447(7147):1021–1025. doi: 10.1038/nature05858. [DOI] [PubMed] [Google Scholar]
- 73.Hammarström P, Wiseman RL, Powers ET, Kelly JW. Prevention of transthyretin amyloid disease by changing protein misfolding energetics. Science. 2003;299(5607):713–716. doi: 10.1126/science.1079589. [DOI] [PubMed] [Google Scholar]
- 74.Liu K, Cho HS, Lashuel HA, Kelly JW, Wemmer DE. A glimpse of a possible amyloidogenic intermediate of transthyretin. Nat Struct Biol. 2000;7(9):754–757. doi: 10.1038/78980. [DOI] [PubMed] [Google Scholar]
- 75.Lim KH, Dyson HJ, Kelly JW, Wright PE. Localized structural fluctuations promote amyloidogenic conformations in transthyretin. J Mol Biol. 2013;425(6):977–988. doi: 10.1016/j.jmb.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mahler B, et al. Characterization of a transient unfolding intermediate in a core mutant of γS-crystallin. J Mol Biol. 2011;405(3):840–850. doi: 10.1016/j.jmb.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wishart DS, et al. CS23D: A web server for rapid protein structure generation using NMR chemical shifts and sequence data. Nucleic Acids Res. 2008;36(Web Server issue):W496–W502. doi: 10.1093/nar/gkn305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Iwahara J, Schwieters CD, Clore GM. Ensemble approach for NMR structure refinement against (1)H paramagnetic relaxation enhancement data arising from a flexible paramagnetic group attached to a macromolecule. J Am Chem Soc. 2004;126(18):5879–5896. doi: 10.1021/ja031580d. [DOI] [PubMed] [Google Scholar]
- 79.Klepeis JL, Lindorff-Larsen K, Dror RO, Shaw DE. Long-timescale molecular dynamics simulations of protein structure and function. Curr Opin Struct Biol. 2009;19(2):120–127. doi: 10.1016/j.sbi.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 80.Lindorff-Larsen K, Piana S, Dror RO, Shaw DE. How fast-folding proteins fold. Science. 2011;334(6055):517–520. doi: 10.1126/science.1208351. [DOI] [PubMed] [Google Scholar]
- 81.Piana S, Lindorff-Larsen K, Shaw DE. Atomic-level description of ubiquitin folding. Proc Natl Acad Sci USA. 2013;110(15):5915–5920. doi: 10.1073/pnas.1218321110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shaw DE, et al. Atomic-level characterization of the structural dynamics of proteins. Science. 2010;330(6002):341–346. doi: 10.1126/science.1187409. [DOI] [PubMed] [Google Scholar]
- 83.Xue Y, Ward JM, Yuwen T, Podkorytov IS, Skrynnikov NR. Microsecond time-scale conformational exchange in proteins: Using long molecular dynamics trajectory to simulate NMR relaxation dispersion data. J Am Chem Soc. 2012;134(5):2555–2562. doi: 10.1021/ja206442c. [DOI] [PubMed] [Google Scholar]
- 84.Sprangers R, Gribun A, Hwang PM, Houry WA, Kay LE. Quantitative NMR spectroscopy of supramolecular complexes: Dynamic side pores in ClpP are important for product release. Proc Natl Acad Sci USA. 2005;102(46):16678–16683. doi: 10.1073/pnas.0507370102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sprangers R, Kay LE. Quantitative dynamics and binding studies of the 20S proteasome by NMR. Nature. 2007;445(7128):618–622. doi: 10.1038/nature05512. [DOI] [PubMed] [Google Scholar]
- 86.Tollinger M, Sivertsen AC, Meier BH, Ernst M, Schanda P. Site-resolved measurement of microsecond-to-millisecond conformational-exchange processes in proteins by solid-state NMR spectroscopy. J Am Chem Soc. 2012;134(36):14800–14807. doi: 10.1021/ja303591y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hu K-N, Yau W-M, Tycko R. Detection of a transient intermediate in a rapid protein folding process by solid-state nuclear magnetic resonance. J Am Chem Soc. 2010;132(1):24–25. doi: 10.1021/ja908471n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fraser JS, et al. Accessing protein conformational ensembles using room-temperature X-ray crystallography. Proc Natl Acad Sci USA. 2011;108(39):16247–16252. doi: 10.1073/pnas.1111325108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Westenhoff S, et al. Time-resolved structural studies of protein reaction dynamics: A smorgasbord of X-ray approaches. Acta Crystallogr A. 2010;66(Pt 2):207–219. doi: 10.1107/S0108767309054361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cornish PV, Ha T. A survey of single-molecule techniques in chemical biology. ACS Chem Biol. 2007;2(1):53–61. doi: 10.1021/cb600342a. [DOI] [PubMed] [Google Scholar]
- 91.Pechmann S, Levy ED, Tartaglia GG, Vendruscolo M. Physicochemical principles that regulate the competition between functional and dysfunctional association of proteins. Proc Natl Acad Sci USA. 2009;106(25):10159–10164. doi: 10.1073/pnas.0812414106. [DOI] [PMC free article] [PubMed] [Google Scholar]