Abstract
Celiac disease is an intestinal autoimmune disease driven by dietary gluten and gluten-specific CD4+ T-cell responses. In celiac patients on a gluten-free diet, exposure to gluten induces the appearance of gluten-specific CD4+ T cells with gut-homing potential in the peripheral blood. Here we show that gluten exposure also induces the appearance of activated, gut-homing CD8+ αβ and γδ T cells in the peripheral blood. Single-cell T-cell receptor sequence analysis indicates that both of these cell populations have highly focused T-cell receptor repertoires, indicating that their induction is antigen-driven. These results reveal a previously unappreciated role of antigen in the induction of CD8+ αβ and γδ T cells in celiac disease and demonstrate a coordinated response by all three of the major types of T cells. More broadly, these responses may parallel adaptive immune responses to viral pathogens and other systemic autoimmune diseases.
Keywords: autoimmunity, mucosal immunity
Celiac disease (CD) is a common autoimmune disease with an estimated prevalence of 1% among people of European ancestry. It is characterized by small intestinal mucosal injury and nutrient malabsorption in genetically susceptible individuals due to dietary gluten ingestion. CD4+ T cells bearing αβ T-cell receptors (TCRs) are critical in the pathogenesis of the disease, as it occurs almost exclusively in HLA-DQ2– or HLA-DQ8–positive individuals (1, 2). CD-associated gluten peptide CD4+ T-cell epitopes have been discovered, and HLA-DQ2/8–restricted gluten-reactive CD4+ T cells have been identified in individuals with CD (3–5). Nonetheless, no gluten-induced enteropathy is seen in humanized mouse models expressing HLA-DQ2 and a gluten-specific TCR (6, 7), suggesting that CD4+ T cells alone are unable to induce tissue damage in CD (1, 2).
An increase in intestinal intraepithelial lymphocytes (IELs), composed of both CD8+ αβ T cells and γδ T cells, is a hallmark of CD. IELs are responsible for the detrimental consequences of CD, including tissue damage and lymphoma development. CD8+ TCRαβ+ IELs (CD8+ IELs) function as effectors in protective immunity to pathogens (8), and in CD they assume a natural killer (NK)-like phenotype to kill intestinal epithelial cells in a manner independent of TCR specificity (9). In rare instances, IELs in CD may transform into enteropathy-associated T-cell lymphoma (EATL), an aggressive lymphoma with a very poor prognosis (10). EATL cells have been shown to have clonal TCRαβ or TCRγδ rearrangements, indicating that either CD8+ IELs or γδ IELs may give rise to lymphoma (11, 12).
Despite intense efforts, gluten-specific IELs in CD have not been readily identified, and there is no significant genetic association of CD with any HLA class I alleles. Moreover, the cytolytic function of IELs in CD can be induced irrespective of their TCR specificity (9). Thus, although the link between dietary gluten and the CD4+ response is well-established, the link between dietary gluten and the recruitment and activation of CD8+ or γδ IELs in celiac disease is unknown. Furthermore, the role of the antigen specificity of IELs in CD is unclear. Here we find that CD8+ and γδ T cells bearing gut-homing receptors are induced by gluten ingestion in CD patients in parallel with gluten-specific CD4+ T cells, and they bear TCR sequences that indicate an antigen-focused response. This indicates that antigen-specific responses of all three of these major T-cell types play a role in this disease.
Results
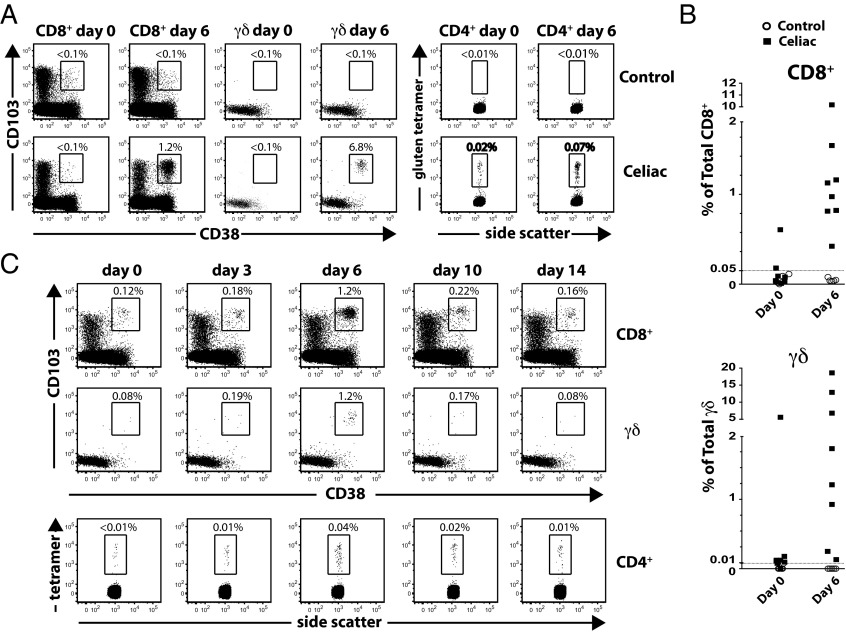
Celiac disease requires the continuous presence of dietary gluten. Reintroducing dietary gluten to celiac patients who are on a gluten-free diet induces large numbers of gluten-specific CD4+ T cells in the peripheral blood 6 d later (4, 5, 13). These cells express the β7 integrin receptor, indicating that they will home to the intestine (5). They also express the activation marker CD38 and lack the expression of CD62L, consistent with an effector phenotype (14). This is generally thought to represent the initiation of an immune response to gluten, and captures activated gluten-reactive CD4+ effector T cells en route from mesenteric lymph nodes or gut-associated lymphoid tissue to the intestine. In an effort to better characterize the context of this immune response, we studied peripheral blood T cells in celiac patients undergoing gluten challenge by time-of-flight mass cytometry (CyTOF) (15), which allows for the independent assessment of many more cellular parameters (currently >40) than fluorescence-based flow cytometry. Indeed, we observed an increase in gluten peptide/HLA-DQ2 tetramer-positive CD4+ T cells in the peripheral blood in all five HLA-DQ2+ celiac patients on day 6 following gluten challenge (Fig. 1 A and C). Unexpectedly, we also observed a large increase in the number of peripheral blood CD8+ αβ and γδ T cells expressing the intestinal epithelial-homing markers αE (CD103) and β7 integrins (16) and the activation marker CD38 (Fig. 1 A and B and Table S1) at this same time point. These cells were not detected in healthy HLA-DQ2+ controls, who underwent oral gluten challenge after at least 1 mo on a gluten-free diet.
Fig. 1.
Induction of activated, gut-homing CD8+ αβ and γδ T cells in peripheral blood of celiac patients following oral gluten challenge. (A) Representative FACS analysis of CD8+ αβ and γδ T-cell (Left) and CD4+ T-cell (Right) response to oral gluten challenge in CD vs. nonceliac control. Expansion of CD103+ (αE integrin), CD38+, and gluten tetramer+ CD4+ T-cell populations is seen on day 6 in CD. Most CD38+CD103+ cells also express β7 integrin; only CD103 staining is depicted here. (B) Relative frequency of αEβ7+CD38+CD8+ T cells as a percentage of total CD8+ cells (Top) and relative frequency of αEβ7+CD38+ γδ cells as a percentage of total γδ T cells (Bottom). (C) Time course showing relative percentage of CD38+CD103+CD8+ (Top), CD38+CD103+ γδ (Middle), and gluten tetramer+ CD4+ (Bottom) in the same patient at the indicated time points following oral gluten challenge. Parallel recruitment of CD38+CD103+ and gluten tetramer+ cells peaks on day 6 before returning to baseline.
The kinetics with which these CD8+ and γδ T cells appear is the same as that of gluten-specific CD4+ T cells, peaking at day 6 after gluten challenge and declining to the baseline by day 14 (Fig. 1C). A similar response was also detected in two celiac patients who underwent rechallenge after returning to a gluten-free diet for at least 1 mo (Fig. 1 A and B and Table S1).
The magnitude of the peripheral blood gluten-specific CD4+ T-cell response is known to be quite variable (4). Similarly, the extent of the αEβ7+CD38+ T-cell response varied between patients, ranging from 0.37% to 10.17% of total peripheral blood CD8+ and from 0.06% to 18.61% of total peripheral blood γδ T cells (Fig. 1B and Table S1). One celiac patient (celiac 2) had αEβ7+CD38+CD8+ and γδ T cells above background levels on day 0, but showed a further increase following gluten challenge. The individual with the lowest detectable response (celiac 6) was an HLA-DQ8+ celiac patient whose disease was diagnosed incidentally by intestinal biopsy, had equivocal antibody test results, and has always been clinically asymptomatic to gluten. Three individuals with active celiac disease, as determined by ongoing symptoms and positive autoantibody titers, were found to have αEβ7+CD38+CD8+ and γδ T-cell proportion below background levels of 0.05% and 0.01%, respectively (Fig. S1). This aspect is similar to the absence of gluten-specific CD4+ T cells in peripheral blood of patients with active celiac disease (4, 5). Also, although plasma cells secreting anti-gluten and autoantibodies are present in celiac intestinal lesions (17–19), we did not detect a similar increase in intestinal-homing B cells (not shown). This is consistent with reports indicating that tissue transglutaminase-specific B cells were undetectable in the peripheral blood of celiac patients (19, 20). In summary, dietary gluten induces the activation and concomitant peripheral blood presence of CD4+ and CD8+ αβ T cells and γδ T cells with gut-homing potential in celiac patients who have been on a gluten-free diet (Fig. 1 and Table S1).
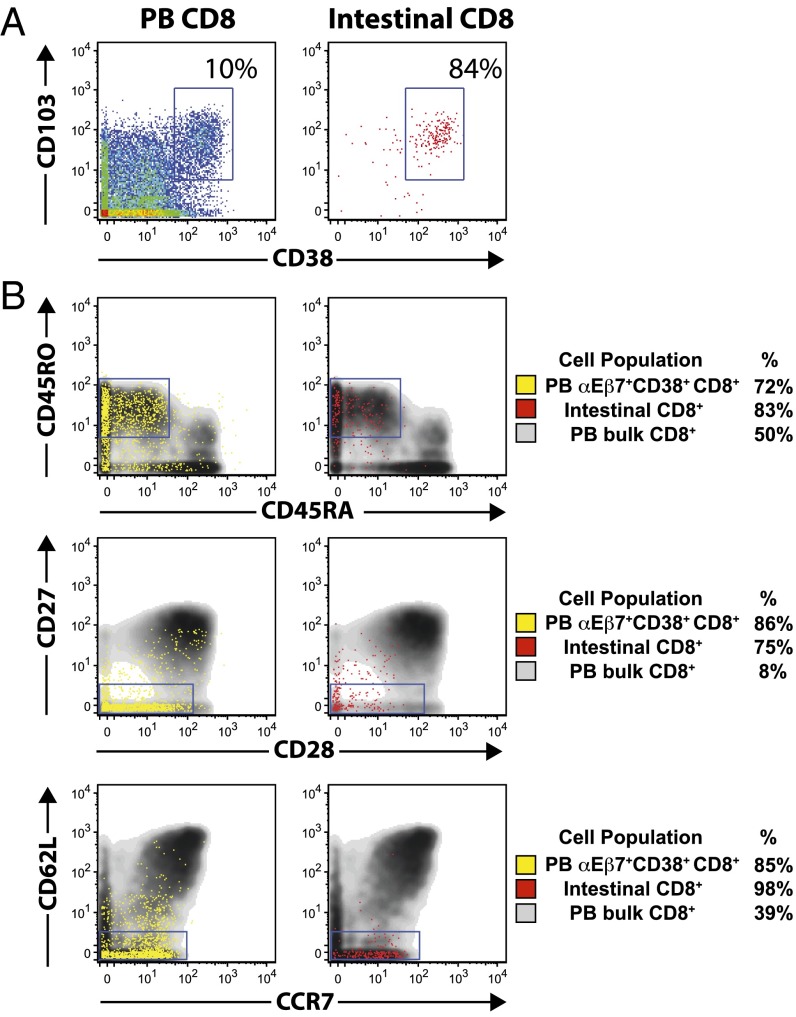
Gluten-reactive CD4+ T cells in the peripheral blood of celiac patients have been shown to be CD38+CD62L−, suggesting that they are gut-bound effector cells (7). CyTOF analysis showed that αEβ7+CD38+CD8+ T cells are CD38+, CD45RO+, CD27−, CD28low, CD62L−, and CCR7low (Fig. 2). This phenotype closely resembles the phenotype of CD8+ T cells isolated from duodenal tissue biopsy specimens of patients with active celiac disease (Fig. 2). CD8+ T cells of this phenotype have been reported to represent differentiated effectors and, accordingly, αEβ7+CD38+CD8+ T cells resemble peripheral blood effector memory CD8+ T cells (Fig. S2) (15, 21, 22). αEβ7+CD38+ γδ cells are predominantly CD45RO+ and CD27−, mirroring intestinal γδ cells from celiac biopsies (Fig. S3). CD45RO+, CD27− γδ T cells are thought to be memory cells (23).
Fig. 2.
Peripheral blood αEβ7+CD38+CD8+ T cells induced by oral gluten challenge express surface markers of effector memory cells and resemble intestinal epithelial CD8+ T lymphocytes from celiac mucosal biopsies. (A) CyTOF analysis of total peripheral blood (PB) CD8+ from a gluten-challenged individual (Left) and total intestinal CD8+ T cells from a celiac patient with active disease (Right) with respect to CD103 and CD38 expression. (B) CyTOF analyses of peripheral blood αEβ7CD38+CD8+ T cells (yellow) and total intestinal CD8+ T cells (red) are overlaid on total peripheral blood CD8+ T cells. Peripheral blood αEβ7+CD38+CD8+ and celiac intestinal CD8+ cells are predominantly CD38+CD45RO+CD45RA−CD27−CD28lowCD62L−CCR7−, consistent with an effector memory phenotype.
The fact that gluten ingestion induces the activation of gluten-specific CD4+ T cells in CD is well-established. However, whether or not the CD8+ and γδ IELs induced in the intestine are responding to specific antigens is unknown. To address this question, we performed single-cell TCR sequencing, which provides a nonbiased means to assess the TCR repertoire without requiring expansion of T-cell clones in culture (24). Single T cells were sorted into 96-well PCR plates from peripheral blood samples of celiac patients following gluten challenge. TCRβ or TCRγ genes were amplified by a series of nested PCRs, and PCR products were directly sequenced.
We were able to perform sequencing on single T cells with high efficiency. We sorted and sequenced 90 single tetramer-positive CD4+ T cells recognizing the gluten epitope DQ2-α-II from the blood of two celiac patients on day 6 after oral gluten challenge (Table S2). Sequences were successfully obtained from 77/90 (86%) of wells into which single T cells were sorted. Consistent with published sequences of DQ2-α-II–reactive T cells from blood and tissue (25), the majority (79%) of unique TCRβ sequences of individual DQ2-α-II–tetramer+ T cells used TRBV7-2 and most (74%) contained the described dominant arginine in position 5 of the CDR3β loop (Table S2), thus validating our methodology.
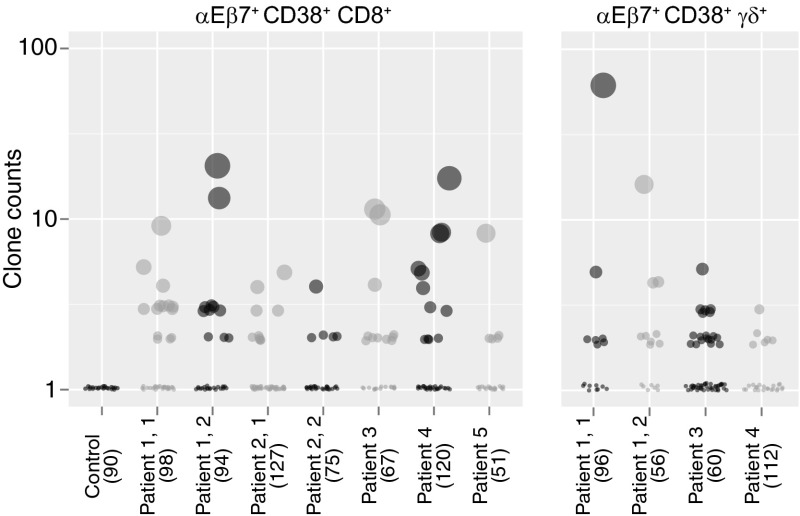
We then sequenced αEβ7+CD38+CD8+ and γδ T cells isolated from celiac patients on day 6 following gluten challenge. αEβ7+CD38+CD8+ T cells, sequenced in five celiac patients, and αEβ7+CD38+ γδ T cells, sequenced in three celiac patients, were found to have a high degree of clonal expansion that was not observed in CD8+CD45RO+ control T cells (Fig. 3). αEβ7 T cells were sequenced in celiac patients who underwent repeat gluten challenge to determine whether both challenges would elicit a similar responding TCR repertoire. Indeed, identical TCRβ and TCRδ clones and similarity in frequency of common clones were found in the two gluten challenges of these patients who underwent repeat challenge after returning to a gluten-free diet for at least 1 mo (Fig. S4).
Fig. 3.
Single-cell TCR sequencing of peripheral blood αEβ7+CD38+CD8+ and αEβ7+CD38+ γδ T cells reveals clonal expansion upon gluten challenge in celiac disease. αEβ7+CD38+CD8+ TCRs were sequenced in five separate patients following gluten challenge, two of whom underwent rechallenge. αEβ7+CD38+ γδ TCRs were sequenced in three patients, one of whom underwent rechallenge. Each individual dot represents a distinct TCR clone. The size of dots and the position along the y axis, plotted on a log scale, indicate the relative frequency of a particular clone. The total number of clones sequenced in each patient is indicated in parentheses.
We next evaluated sequences from αEβ7+CD38+CD8+ and γδ T cells to determine whether we could observe a convergence of TCR features among distinct TCR sequences. To evaluate convergence, we analyzed the nonredundant, unique TCR repertoire of αEβ7+CD38+ T cells. For a particular MHC–peptide, specific CD8+ T-cell responses are often biased toward the use of a particular TCRVβ gene (26). We initially examined TCRVβ gene use. Even individuals of significantly different genetic backgrounds share similar frequency of V gene use in their TCR repertoire, indicating that skewing within a particular population of cells is not attributable to genetic variation in baseline V gene use (27). When assessing the nonredundant TCRβ repertoire of αEβ7+CD38+CD8+ T cells in celiac samples, we found significant overrepresentation of particular V regions in multiple celiac samples compared with unselected healthy controls (Fig. S5 A and B).
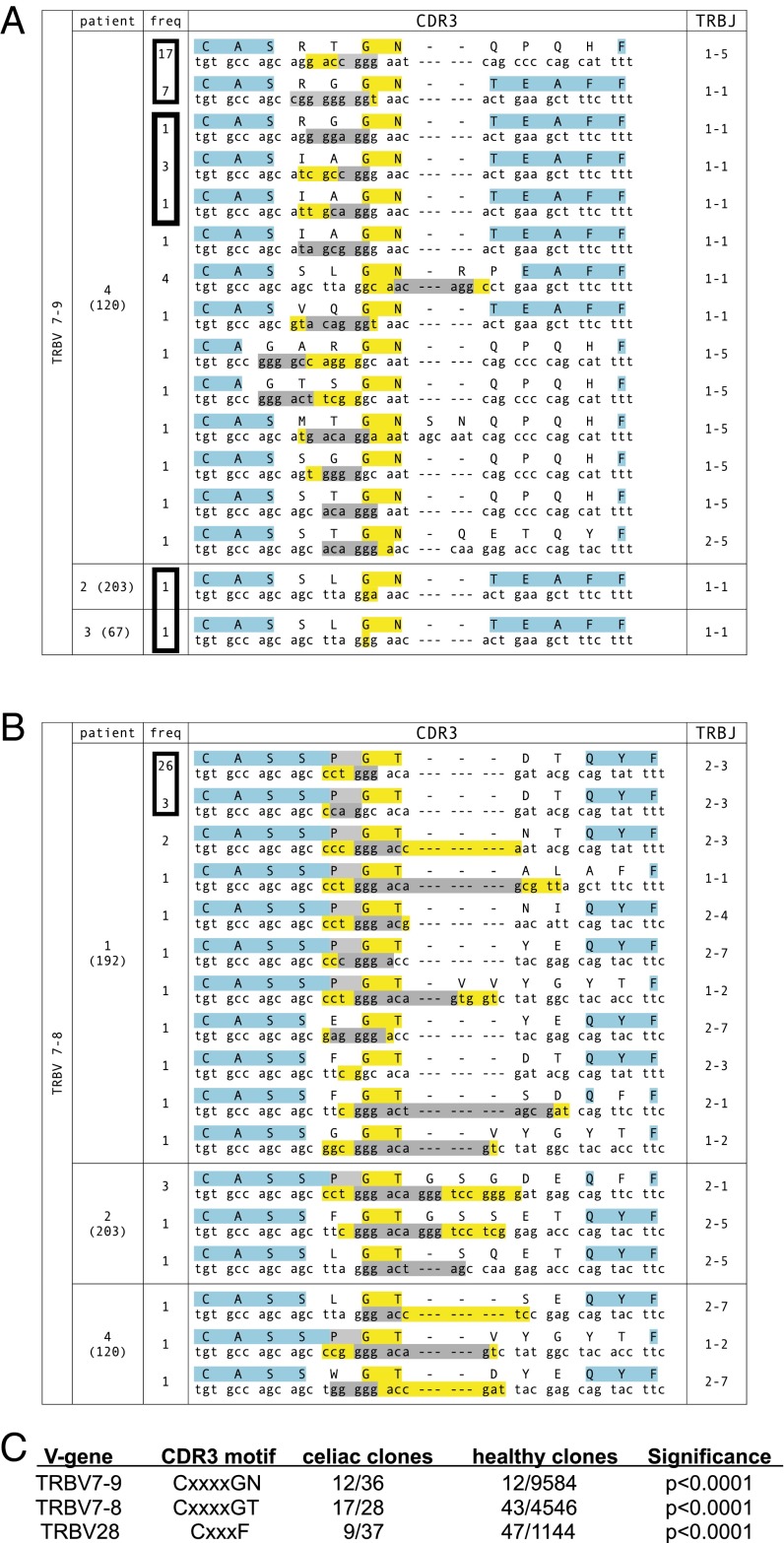
Most of the peptide specificity of TCRβ is determined by the CDR3 loop, which is usually positioned over the antigenic peptide (28, 29). We then determined whether convergence could be observed within CDR3β motifs, focusing on groups using TCRVβ genes that were overrepresented in a nonredundant sampling within a particular individual and had members that were clonally expanded. Strikingly, in αEβ7+CD38+CD8+ T cells, we found four separate examples where identical TCRβ proteins used different DNA sequences (Fig. 4 A and B). In three of these instances, the identically convergent TCRβ occurred in the same patient, and represented a dominantly expressed TCRβ in that individual. In the other instance, the identical TCRβ occurred in different patients (Fig. 4A).
Fig. 4.
Convergent αEβ7+CD38+CD8+TCRβ CDR3 motifs are found among clones within the same celiac patient and across different patients following gluten challenge. (A and B) Convergent motifs CxxxxGN (A) and CxxxxGT (B) are seen in TCRβ clones using TRBV7-9 and TRBV7-8, respectively. The frequency of each clone is indicated and the total number of T cells sequenced in the patient is indicated in parentheses. The protein sequence with the corresponding DNA sequence is shown. Within the protein sequence, yellow indicates absolutely conserved amino acids, gray indicates relatively conserved (≥50%) amino acids, and blue indicates conserved amino acids that are encoded within the V or J genes. Within the DNA sequence, nucleotides in yellow are formed through N or P addition, whereas nucleotides in gray are encoded by D genes. Boxes around frequency numbers highlight distinct clones sharing identical protein sequences. (C) Convergences of motifs seen in TCRβ clones using TRBV7-9, TRBV7-8, and TRBV28 are statistically significant compared with reference control TCRβ sequences.
Additionally, within TCRVβ sequences using TRBV7-8, TRBV7-9, and TRBV28, we could identify characteristic amino acid motifs in the center of the CDR3β that were very common within celiac αEβ7+CD38+CD8+ T cells compared with healthy reference CDR3β sequences (30) (Fig. 4). For instance, the GN motif at positions 6–7 within the CDR3 region of TCRβ clones using TRBV7-9 was highly enriched, occurring in 16 out of 40 unique (nonredundant) TCRβ clones, while occurring in only 12/9,584 of TCRβ clones using TRBV7-9 within the reference database (P < 0.0001) (Fig. 4 A and C). In patient 4, this motif occurred in 14 of 19 unique TCRβ clones, and 5 of these unique clones converged on two identical TCRβs. This motif also occurred in two other patients, who converged upon an identical TCRβ. TCRβ clones using TRBV7-8 similarly converged on a GT motif at position 6–7, which occurred in 17 out of 29 unique TCRβ clones, in contrast to only 43/4,546 TRBV7-8–containing TCRβ clones within the reference database (P < 0.0001) (Fig. 4 B and C). In all instances where the same TCR was formed using distinct VDJ rearrangements within the same patient, there were at least two nucleotide changes within the CDR3, making a PCR or sequencing error improbable.
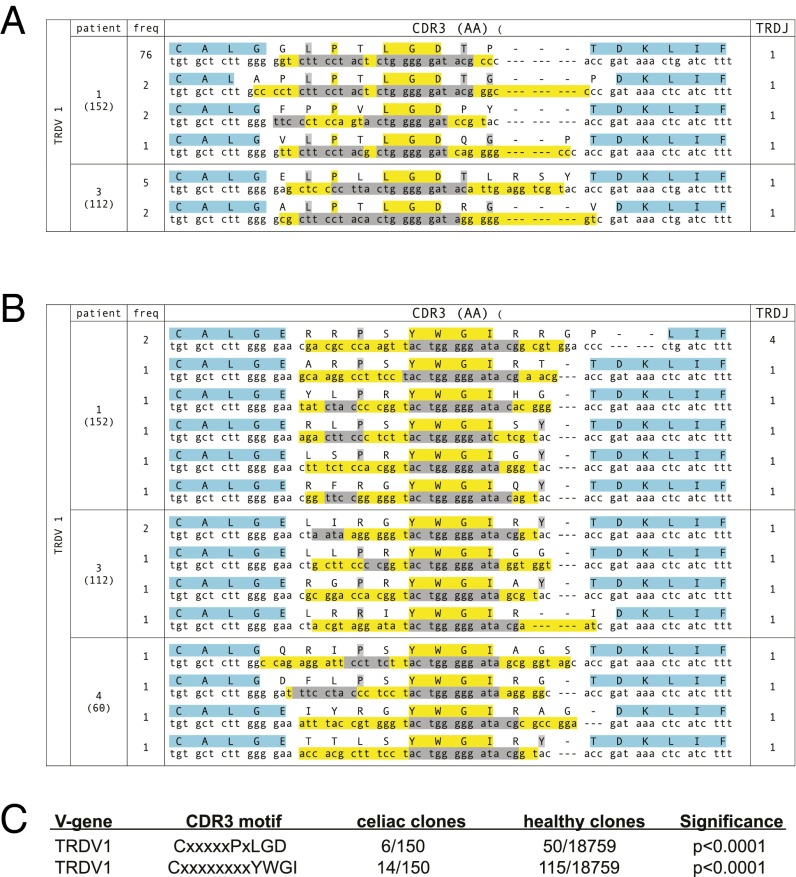
We applied a similar analysis to αEβ7+CD38+ γδ T cells. Intestinal γδ T cells are appreciated to be heavily biased toward TRDV1 use (31). Consistent with this, the majority (80%, 150/188) of unique αEβ7+CD38+ TCRδ sequences from CD patients use TRDV1 (Table S3). We analyzed CDR3δ sequences using TRDV1 to determine whether convergent motifs could be seen in celiac patients. For comparison, we sequenced TCRδ from bulk small intestinal γδ T cells from a person without celiac disease and bulk blood γδ T cells from nine different control patients, obtaining 18,579 unique TCRδ sequences using TRDV1. The most highly expanded sequence, which was present in 76/152 total sequences, shared the CxxxxxPxLGD motif with five other unique CDR3δ sequences across two patients. This motif was rare in reference sequences, occurring in only 50/18,579 unique sequences (P < 0.0001; Fig. 5 A and C). We also found that the amino acid motif CxxxxxxxxYWGI was highly enriched within TCRDV1+ CDR3δ in αEβ7+CD38+ γδ cells compared with reference TCRDV1+ γδ T-cell sequencing, occurring in all three celiac patients at a total frequency of 14/152 unique sequences while only present in 115/18,579 unique reference sequences (P < 0.0001; Fig. 5 B and C).
Fig. 5.
Convergent αEβ7+CD38+TCRδ CDR3 motifs are found among clones within the same celiac patient and across different patients following gluten challenge. (A and B) Convergent motifs CxxxxxPxLGD (A) and CxxxxxxxxYWGI (B) are seen in TCRδ clones using TRBV1. The frequency of each clone is indicated and the total number of T cells sequenced in the patient is indicated in parentheses. The protein sequence with the corresponding DNA sequence is shown. Within the protein sequence, yellow indicates absolutely conserved amino acids, gray indicates relatively conserved (≥50%) amino acids, and blue indicates conserved amino acids that are encoded within the V or J genes. Within the DNA sequence, nucleotides in yellow are formed through N or P addition, whereas nucleotides in gray are encoded by D genes. (C) Convergences of motifs seen in TCRδ clones using TRBV1 are statistically significant compared with reference control TCRδ sequences.
The high clonality of αEβ7+CD38+ T cells, the similarity of TCR repertoire upon a second gluten challenge, and the conservation of CDR3 motifs in different T-cell clones suggest that both CD8+ αβ and γδ T cells are activated in an antigen-specific manner in response to dietary gluten.
Discussion
In CD, dietary gluten induces the infiltration of T cells in the small intestine and the destruction of intestinal epithelial cells. We find that along with the induction of gluten-specific CD4+ cells, the reintroduction of dietary gluten to celiac patients on a gluten-free diet induces the peripheral appearance of large numbers of activated CD8+ and γδ T cells expressing gut-homing markers. These findings are consistent with the supposition that these T cells are activated and imprinted with gut-homing potential in secondary lymphoid organs by dendritic cells presenting gut-derived antigens (32). Like peripheral blood gluten-specific CD4+ T cells, these cells express surface markers consistent with memory or effector cells, indicating that they are programmed as such before gut recruitment. This suggests that at least some of the pathogenic IELs in CD are purposefully activated and recruited to the gut. Importantly, these cells respond with a very focused TCR repertoire, indicating that they are selected in an antigen-specific manner before entering the intestine.
The presence of inflammation has long been postulated to promote the loss of tolerance, and prevailing models of CD pathogenesis propose that IELs are activated as a result of inflammation that is initiated by gluten-specific CD4+ cells. The inflammatory cytokine IL-15 is up-regulated within celiac intestinal mucosa, and has been implicated in promoting inflammation through diverse means, including impairing regulatory T-cell generation promoting NK-like function of CD8+ IELs, and enabling the expansion of IELs (9, 33). CD8+ IELs have been shown to demonstrate cytotoxicity through stimulation by IL-15 and activation through NK receptors including CD94 and NKG2D (9, 34). Whereas αEβ7+CD38+CD8+ T cells clearly show markers of effector cells and are capable of IFN-γ production, they largely do not express perforin, CD57, or higher levels of NKG2D (Fig. S6). Therefore, it is possible that tissue factors, including IL-15, are further required for cytotoxicity.
The function of γδ IELs is more poorly understood. In human CD, both cytotoxic and anti-inflammatory functions have been attributed to subsets of γδ IELs (35, 36). In mice, γδ IELs appear to be constitutively activated with high cytotoxic potential at baseline (37). However, they express both activating and inhibitory NK receptors, and it has been suggested that the combination of these NK receptors can keep the effector functions of γδ IELs in check but enable them to be readily switched on. Thus, both CD8+ and γδ IEL cell populations may ultimately mediate tissue destruction through NK receptors and require tissue-derived factors. However, we find that αEβ7+CD38+ T cells express markers of differentiated effector cells before gut recruitment, and their appearance parallels the appearance of gluten-reactive CD4+ T cells in blood, rather than occurring later. Also, although increased numbers of IELs and mildly increased levels of IL-15 are present in celiac patients on a gluten-free diet (38), the recruitment we describe precedes significant intestinal inflammation and tissue damage, which only reliably occur histologically after 2–4 wk of continuous gluten exposure (39). These findings suggest that IELs in CD are not simply activated as bystanders as a consequence of gut inflammation.
As celiac IEL populations are induced by gluten, a long-standing question has been whether their TCRs recognize gluten. Despite extensive study, gluten-derived peptide epitopes recognized by CD8+ T cells in CD have not been apparent, and there is no significant genetic association of CD with particular HLA class I alleles. Therefore, it is generally thought that IELs do not mediate tissue damage through gluten recognition. Nevertheless, one group has identified a class I gluten epitope recognized by CD8+ T cells isolated from CD intestinal mucosa (40). If the αEβ7+CD38+CD8+ T cells we describe are responding to gluten, this would imply a rapid and efficient cross-presentation of gluten on MHC class I. Besides gluten, other possibilities for IEL ligands include self-antigens or infectious pathogens. The possibility of self-antigen recognition is supported by the very selective destruction of intestinal epithelial cells and the presence of autoantibodies, including antibodies to tissue transglutaminase (10, 41, 42). The role of an infectious cofactor in CD has been proposed based on epidemiologic data showing that neonatal infection seems to predispose individuals to the development of CD (43).
This process through which these three T-cell subsets are synchronously mobilized and recruited to intestinal tissue clearly has implications in immunity to infections. The development of autoimmunity in CD likely represents a misdirected application of processes that are meant to be protective. Due to the well-established dependence of CD on the CD4+ T-cell response, the coordinated T-cell response we describe here presumably depends upon gluten-specific CD4+ T cells. In this context, multiple aspects of the effector CD8+ T-cell responses to viruses have been shown to depend upon CD4+ T-cell help, including the primary effector response, the generation of memory, and recruitment to sites of infection (44–47). This process has been termed “licensing,” referring to the ability of CD4+ T cells to license cognate effector CD8+ T-cell responses. Here we speculate that CD4+ T cells may be “licensing” self-antigen–specific CD8+ T cells to become activated and recruited to the intestine, subsequently leading to tissue damage. This process may share mechanisms with the processes that have been described to coordinate CD4+ and effector T-cell responses to viruses.
Like CD, most autoimmune diseases with HLA associations are associated with MHC class II alleles, including type 1 diabetes, multiple sclerosis, rheumatoid arthritis, and ulcerative colitis (48). Despite the association of these diseases with class II alleles rather than class I alleles, CD8+ effector T cells play an important role in the pathogenesis of these diseases. For instance, although type 1 diabetes is strongly associated with class II alleles, autoreactive CD8+ T cells are extensively found in inflamed diabetic islets and are appreciated to be the primary effectors driving tissue damage (49–51). Thus, the scenario we outline above for celiac disease may be generalizable to other forms of autoimmunity, in that an initial misdirected CD4+ T-cell response may license effector CD8+ and γδ T cells to cause tissue destruction at a particular site.
The mobilization of specific lymphocytes into the peripheral blood 6 d after antigenic challenge, as has been reported in CD (4, 5) and in the context of influenza vaccination (52), has provided an invaluable window into antigen-specific responses in human subjects. It will be interesting to see whether other such migrations are occurring at specific times in other autoimmune diseases. We also suggest that the analysis of activated T cells with gut-homing markers in the peripheral blood on day 6 after gluten challenge may be a superior method to diagnose CD in individuals currently on a gluten-free diet. An estimated 1.6 million Americans follow a gluten-free diet without an established diagnosis of CD (53). Available tests, including antibody levels and intestinal biopsy results, can be completely normal in CD patients on a gluten-free diet. Consequently, such individuals are often asked to continually eat gluten-containing foods for 2–4 wk before testing (39). This is often intolerable and precludes an accurate diagnosis. Our study shows promise in the reliable clinical diagnosis of CD with only short-term gluten exposure.
Methods
Gluten Challenge.
All human sample collection was performed with informed consent under Stanford University Institutional Review Board oversight Volunteers underwent oral gluten challenge as described (4). At time of participation, all volunteers adhered to a strict gluten-free diet for at least 1 mo. After an initial blood draw, volunteers consumed four slices of white bread per day for 3 consecutive days (days 1, 2, and 3) and returned for a second blood draw on day 6. All celiac patient volunteers had a clinical diagnosis of celiac disease established by small intestinal biopsy in addition to serologic antibody testing. Five of six celiac volunteers were HLA-DQ2.5+. One celiac volunteer was HLA-DQ8+ according to clinical testing. Healthy HLA-DQ2.5+ volunteers were either parents of children with celiac disease or individuals who endorsed gluten intolerance. Patients were tested for HLA-DQ2.5 by PCR (SI Methods). All healthy volunteers had a negative clinical diagnostic workup for celiac disease, and were able to comply with a gluten-free diet for at least 1 mo before participation.
Tetramer Analysis and Flow Cytometry.
All FACS experiments were performed on ARIAII or LSRII instruments (Becton Dickinson). Water-soluble MHC–DQ2 molecules with covalently tethered peptides were produced in a baculovirus expression system (54). Two different MHC–DQ2.5 molecules with engineered biotinylation sites were produced with tethered deamidated T-cell epitopes of α-gliadin, DQ2-α-I (QLQPFPQPELPY) and DQ2-α-II (PQPELPYPQPE). Proteins were biotinylated, purified, and stored in PBS, 50% (vol/vol) glycerol at −20 °C. Tetramers were prepared by incubating protein with streptavidin–fluorophore conjugates (eBioscience) at a 4:1 molar ratio. Tetramer staining was performed at room temperature for 1 h using 10 mg/mL tetramer. Antibody clones used for flow cytometry are in SI Methods.
Intestinal Biopsy Preparation.
Small intestinal biopsies were obtained with informed consent from celiac patients undergoing endoscopy at Stanford University Hospital and processed as described (55). See SI Methods.
CyTOF Staining and Data Acquisition.
CyTOF and data acquisition were performed as described (16) on cryopreserved peripheral blood mononuclear cells or freshly isolated intestinal lymphocytes. See SI Methods.
CyTOF Antibody Labeling.
Purified antibodies (lacking carrier proteins) were labeled 100 μg at a time according to instructions provided by DVS Sciences with heavy metal-preloaded maleimide-coupled MAXPAR chelating polymers via Pre-Load Method version 2.1 (16).
Single-Cell Sorting and TCR Sequencing.
Single-cell sorting was performed using an ARIAII cell sorter (Becton Dickinson). TCR sequences from single cells were obtained by a series of three nested PCRs as described (24). The full method and TCR sequence analysis are described in SI Methods.
Supplementary Material
Acknowledgments
We thank members of the M.M.D. and Y.-h.C. laboratory for helpful discussions. We thank Ludvig Sollid for critical reading of the manuscript and helpful suggestions. We are indebted to all volunteers who participated in this study. We thank Jennifer Iscol and the Celiac Community Foundation of Northern California for help with volunteer recruitment. We thank the Human Immune Monitoring Core and the Stanford Shared FACS Facility for the use of equipment. A.H. was supported by a National Institutes of Health (NIH) T32 Gastroenterology Training Grant. E.W.N. was supported by a fellowship through the American Cancer Society. C.K., Y.-h.C., and M.M.D. are funded by NIH grants: DK063158 (C.K.), AI057229-07 (M.M.D.), and AI090019 (M.M.D.). M.M.D. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1311861110/-/DCSupplemental.
References
- 1.Fallang LE, et al. Differences in the risk of celiac disease associated with HLA-DQ2.5 or HLA-DQ2.2 are related to sustained gluten antigen presentation. Nat Immunol. 2009;10(10):1096–1101. doi: 10.1038/ni.1780. [DOI] [PubMed] [Google Scholar]
- 2.Sollid LM, Qiao SW, Anderson RP, Gianfrani C, Koning F. Nomenclature and listing of celiac disease relevant gluten T-cell epitopes restricted by HLA-DQ molecules. Immunogenetics. 2012;64(6):455–460. doi: 10.1007/s00251-012-0599-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lundin KE, et al. Gliadin-specific, HLA-DQ(alpha 1*0501,beta 1*0201) restricted T cells isolated from the small intestinal mucosa of celiac disease patients. J Exp Med. 1993;178(1):187–196. doi: 10.1084/jem.178.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brottveit M, et al. Assessing possible celiac disease by an HLA-DQ2-gliadin tetramer test. Am J Gastroenterol. 2011;106(7):1318–1324. doi: 10.1038/ajg.2011.23. [DOI] [PubMed] [Google Scholar]
- 5.Ráki M, et al. Tetramer visualization of gut-homing gluten-specific T cells in the peripheral blood of celiac disease patients. Proc Natl Acad Sci USA. 2007;104(8):2831–2836. doi: 10.1073/pnas.0608610104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Kauwe AL, et al. Resistance to celiac disease in humanized HLA-DR3-DQ2-transgenic mice expressing specific anti-gliadin CD4+ T cells. J Immunol. 2009;182(12):7440–7450. doi: 10.4049/jimmunol.0900233. [DOI] [PubMed] [Google Scholar]
- 7.Du Pré MF, et al. Tolerance to ingested deamidated gliadin in mice is maintained by splenic, type 1 regulatory T cells. Gastroenterology. 2011;141(2):610–620. doi: 10.1053/j.gastro.2011.04.048. [DOI] [PubMed] [Google Scholar]
- 8.Abadie V, Discepolo V, Jabri B. Intraepithelial lymphocytes in celiac disease immunopathology. Semin Immunopathol. 2012;34(4):551–566. doi: 10.1007/s00281-012-0316-x. [DOI] [PubMed] [Google Scholar]
- 9.Meresse B, et al. Coordinated induction by IL15 of a TCR-independent NKG2D signaling pathway converts CTL into lymphokine-activated killer cells in celiac disease. Immunity. 2004;21(3):357–366. doi: 10.1016/j.immuni.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 10.Jabri B, Sollid LM. Tissue-mediated control of immunopathology in coeliac disease. Nat Rev Immunol. 2009;9(12):858–870. doi: 10.1038/nri2670. [DOI] [PubMed] [Google Scholar]
- 11.Chan JK, et al. Type II enteropathy-associated T-cell lymphoma: A distinct aggressive lymphoma with frequent γδ T-cell receptor expression. Am J Surg Pathol. 2011;35(10):1557–1569. doi: 10.1097/PAS.0b013e318222dfcd. [DOI] [PubMed] [Google Scholar]
- 12.Tack GJ, et al. Origin and immunophenotype of aberrant IEL in RCDII patients. Mol Immunol. 2012;50(4):262–270. doi: 10.1016/j.molimm.2012.01.014. [DOI] [PubMed] [Google Scholar]
- 13.Anderson RP, Degano P, Godkin AJ, Jewell DP, Hill AV. In vivo antigen challenge in celiac disease identifies a single transglutaminase-modified peptide as the dominant A-gliadin T-cell epitope. Nat Med. 2000;6(3):337–342. doi: 10.1038/73200. [DOI] [PubMed] [Google Scholar]
- 14.du Pré MF, et al. CD62L(neg)CD38+ expression on circulating CD4+ T cells identifies mucosally differentiated cells in protein fed mice and in human celiac disease patients and controls. Am J Gastroenterol. 2011;106(6):1147–1159. doi: 10.1038/ajg.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Newell EW, Sigal N, Bendall SC, Nolan GP, Davis MM. Cytometry by time-of-flight shows combinatorial cytokine expression and virus-specific cell niches within a continuum of CD8+ T cell phenotypes. Immunity. 2012;36(1):142–152. doi: 10.1016/j.immuni.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorfu G, Rivera-Nieves J, Ley K. Role of beta7 integrins in intestinal lymphocyte homing and retention. Curr Mol Med. 2009;9(7):836–850. doi: 10.2174/156652409789105525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dieterich W, et al. Identification of tissue transglutaminase as the autoantigen of celiac disease. Nat Med. 1997;3(7):797–801. doi: 10.1038/nm0797-797. [DOI] [PubMed] [Google Scholar]
- 18.Sulkanen S, et al. Tissue transglutaminase autoantibody enzyme-linked immunosorbent assay in detecting celiac disease. Gastroenterology. 1998;115(6):1322–1328. doi: 10.1016/s0016-5085(98)70008-3. [DOI] [PubMed] [Google Scholar]
- 19.Di Niro R, et al. High abundance of plasma cells secreting transglutaminase 2-specific IgA autoantibodies with limited somatic hypermutation in celiac disease intestinal lesions. Nat Med. 2012;18(3):441–445. doi: 10.1038/nm.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marzari R, et al. Molecular dissection of the tissue transglutaminase autoantibody response in celiac disease. J Immunol. 2001;166(6):4170–4176. doi: 10.4049/jimmunol.166.6.4170. [DOI] [PubMed] [Google Scholar]
- 21.Sallusto F, Lenig D, Förster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401(6754):708–712. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- 22.Appay V, et al. Memory CD8+ T cells vary in differentiation phenotype in different persistent virus infections. Nat Med. 2002;8(4):379–385. doi: 10.1038/nm0402-379. [DOI] [PubMed] [Google Scholar]
- 23.De Rosa SC, et al. Ontogeny of gamma delta T cells in humans. J Immunol. 2004;172(3):1637–1645. doi: 10.4049/jimmunol.172.3.1637. [DOI] [PubMed] [Google Scholar]
- 24.Su LF, Kidd BA, Han A, Kotzin JJ, Davis MM. Virus-specific CD4(+) memory-phenotype T cells are abundant in unexposed adults. Immunity. 2013;38(2):373–383. doi: 10.1016/j.immuni.2012.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qiao SW, et al. Posttranslational modification of gluten shapes TCR usage in celiac disease. J Immunol. 2011;187(6):3064–3071. doi: 10.4049/jimmunol.1101526. [DOI] [PubMed] [Google Scholar]
- 26.Kedzierska K, Turner SJ, Doherty PC. Conserved T cell receptor usage in primary and recall responses to an immunodominant influenza virus nucleoprotein epitope. Proc Natl Acad Sci USA. 2004;101(14):4942–4947. doi: 10.1073/pnas.0401279101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramakrishnan NS, Grunewald J, Janson CH, Wigzell H. Nearly identical T-cell receptor V-gene usage at birth in two cohorts of distinctly different ethnic origin: Influence of environment in the final maturation in the adult. Scand J Immunol. 1992;36(1):71–78. doi: 10.1111/j.1365-3083.1992.tb02942.x. [DOI] [PubMed] [Google Scholar]
- 28.Kjer-Nielsen L, et al. A structural basis for the selection of dominant alphabeta T cell receptors in antiviral immunity. Immunity. 2003;18(1):53–64. doi: 10.1016/s1074-7613(02)00513-7. [DOI] [PubMed] [Google Scholar]
- 29.Garboczi DN, et al. Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature. 1996;384(6605):134–141. doi: 10.1038/384134a0. [DOI] [PubMed] [Google Scholar]
- 30.Warren RL, et al. Exhaustive T-cell repertoire sequencing of human peripheral blood samples reveals signatures of antigen selection and a directly measured repertoire size of at least 1 million clonotypes. Genome Res. 2011;21(5):790–797. doi: 10.1101/gr.115428.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chowers Y, Holtmeier W, Harwood J, Morzycka-Wroblewska E, Kagnoff MF. The V delta 1 T cell receptor repertoire in human small intestine and colon. J Exp Med. 1994;180(1):183–190. doi: 10.1084/jem.180.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sigmundsdottir H, Butcher EC. Environmental cues, dendritic cells and the programming of tissue-selective lymphocyte trafficking. Nat Immunol. 2008;9(9):981–987. doi: 10.1038/ni.f.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DePaolo RW, et al. Co-adjuvant effects of retinoic acid and IL-15 induce inflammatory immunity to dietary antigens. Nature. 2011;471(7337):220–224. doi: 10.1038/nature09849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meresse B, et al. Reprogramming of CTLs into natural killer-like cells in celiac disease. J Exp Med. 2006;203(5):1343–1355. doi: 10.1084/jem.20060028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jabri B, et al. Selective expansion of intraepithelial lymphocytes expressing the HLA-E-specific natural killer receptor CD94 in celiac disease. Gastroenterology. 2000;118(5):867–879. doi: 10.1016/S0016-5085(00)70173-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bhagat G, et al. Small intestinal CD8+TCRgammadelta+NKG2A+ intraepithelial lymphocytes have attributes of regulatory cells in patients with celiac disease. J Clin Invest. 2008;118(1):281–293. doi: 10.1172/JCI30989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fahrer AM, et al. Attributes of gammadelta intraepithelial lymphocytes as suggested by their transcriptional profile. Proc Natl Acad Sci USA. 2001;98(18):10261–10266. doi: 10.1073/pnas.171320798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Di Sabatino A, et al. Epithelium derived interleukin 15 regulates intraepithelial lymphocyte Th1 cytokine production, cytotoxicity, and survival in coeliac disease. Gut. 2006;55(4):469–477. doi: 10.1136/gut.2005.068684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leffler D, et al. Kinetics of the histological, serological and symptomatic responses to gluten challenge in adults with coeliac disease. Gut. 2013;62(7):996–1004. doi: 10.1136/gutjnl-2012-302196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mazzarella G, et al. Gliadin activates HLA class I-restricted CD8+ T cells in celiac disease intestinal mucosa and induces the enterocyte apoptosis. Gastroenterology. 2008;134(4):1017–1027. doi: 10.1053/j.gastro.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meresse B, Malamut G, Cerf-Bensussan N. Celiac disease: An immunological jigsaw. Immunity. 2012;36(6):907–919. doi: 10.1016/j.immuni.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 42.Sollid LM, Jabri B. Triggers and drivers of autoimmunity: Lessons from coeliac disease. Nat Rev Immunol. 2013;13(4):294–302. doi: 10.1038/nri3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sandberg-Bennich S, Dahlquist G, Källén B. Coeliac disease is associated with intrauterine growth and neonatal infections. Acta Paediatr. 2002;91(1):30–33. doi: 10.1080/080352502753457905. [DOI] [PubMed] [Google Scholar]
- 44.Nakanishi Y, Lu B, Gerard C, Iwasaki A. CD8(+) T lymphocyte mobilization to virus-infected tissue requires CD4(+) T-cell help. Nature. 2009;462(7272):510–513. doi: 10.1038/nature08511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Janssen EM, et al. CD4+ T cells are required for secondary expansion and memory in CD8+ T lymphocytes. Nature. 2003;421(6925):852–856. doi: 10.1038/nature01441. [DOI] [PubMed] [Google Scholar]
- 46.Shedlock DJ, Shen H. Requirement for CD4 T cell help in generating functional CD8 T cell memory. Science. 2003;300(5617):337–339. doi: 10.1126/science.1082305. [DOI] [PubMed] [Google Scholar]
- 47.Sun JC, Bevan MJ. Defective CD8 T cell memory following acute infection without CD4 T cell help. Science. 2003;300(5617):339–342. doi: 10.1126/science.1083317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trowsdale J. The MHC, disease and selection. Immunol Lett. 2011;137(1-2):1–8. doi: 10.1016/j.imlet.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 49.Coppieters KT, et al. Demonstration of islet-autoreactive CD8 T cells in insulitic lesions from recent onset and long-term type 1 diabetes patients. J Exp Med. 2012;209(1):51–60. doi: 10.1084/jem.20111187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang B, Gonzalez A, Benoist C, Mathis D. The role of CD8+ T cells in the initiation of insulin-dependent diabetes mellitus. Eur J Immunol. 1996;26(8):1762–1769. doi: 10.1002/eji.1830260815. [DOI] [PubMed] [Google Scholar]
- 51.Wong FS, Visintin I, Wen L, Flavell RA, Janeway CA., Jr CD8 T cell clones from young nonobese diabetic (NOD) islets can transfer rapid onset of diabetes in NOD mice in the absence of CD4 cells. J Exp Med. 1996;183(1):67–76. doi: 10.1084/jem.183.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wrammert J, et al. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature. 2008;453(7195):667–671. doi: 10.1038/nature06890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rubio-Tapia A, Ludvigsson JF, Brantner TL, Murray JA, Everhart JE. The prevalence of celiac disease in the United States. Am J Gastroenterol. 2012;107(10):1538–1544. doi: 10.1038/ajg.2012.219. [DOI] [PubMed] [Google Scholar]
- 54.Quarsten H, et al. Staining of celiac disease-relevant T cells by peptide-DQ2 multimers. J Immunol. 2001;167(9):4861–4868. doi: 10.4049/jimmunol.167.9.4861. [DOI] [PubMed] [Google Scholar]
- 55.Shacklett BL, Critchfield JW, Lemongello D. Isolating mucosal lymphocytes from biopsy tissue for cellular immunology assays. Methods Mol Biol. 2009;485:347–356. doi: 10.1007/978-1-59745-170-3_23. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.