Abstract
Riboswitches are ligand-binding elements located in 5′ untranslated regions of messenger RNAs, which regulate expression of downstream genes. In Listeria monocytogenes, a vitamin B12-binding (B12) riboswitch was identified, not upstream of a gene but downstream, and antisense to the adjacent gene, pocR, suggesting it might regulate pocR in a nonclassical manner. In Salmonella enterica, PocR is a transcription factor that is activated by 1,2-propanediol, and subsequently activates expression of the pdu genes. The pdu genes mediate propanediol catabolism and are implicated in pathogenesis. As enzymes involved in propanediol catabolism require B12 as a cofactor, we hypothesized that the Listeria B12 riboswitch might be involved in pocR regulation. Here we demonstrate that the B12 riboswitch is transcribed as part of a noncoding antisense RNA, herein named AspocR. In the presence of B12, the riboswitch induces transcriptional termination, causing aspocR to be transcribed as a short transcript. In contrast, in the absence of B12, aspocR is transcribed as a long antisense RNA, which inhibits pocR expression. Regulation by AspocR ensures that pocR, and consequently the pdu genes, are maximally expressed only when both propanediol and B12 are present. Strikingly, AspocR can inhibit pocR expression in trans, suggesting it acts through a direct interaction with pocR mRNA. Together, this study demonstrates how pocR and the pdu genes can be regulated by B12 in bacteria and extends the classical definition of riboswitches from elements governing solely the expression of mRNAs to a wider role in controlling transcription of noncoding RNAs.
Keywords: asRNA, cobalamin, infection
The Gram-positive bacterium Listeria monocytogenes is a food borne pathogen which causes gastroenteritis as well as meningitis and encephalitis in immunocompromised individuals and abortions in pregnant women (1). The ability of the bacterium to invade diverse tissues while proliferating intracellularly in phagocytic and nonphagocytic cells has led to its adoption as a model organism in both infection biology and cell biology (2, 3). More recently, L. monocytogenes has also become a focus of studies of RNA-based regulation. Multiple transcriptomics studies in L. monocytogenes have now identified 134 putative small trans-acting RNAs (sRNAs), 86 antisense RNAs (asRNAs), and 42 cis-acting riboswitches (4).
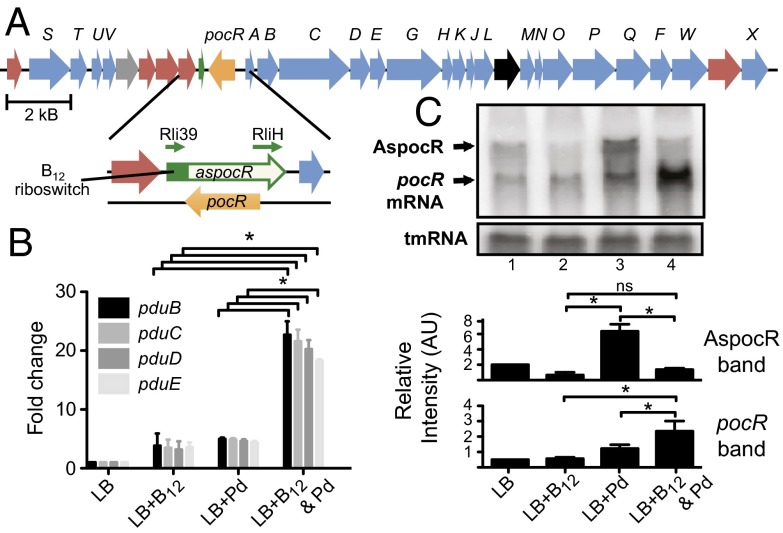
In Gram-positive bacteria, the majority of cis-acting riboswitches affect transcription termination, although some affect translation, Rho-mediated termination, and mRNA cleavage (5–7). Transcribed as part of the 5′ UTR of mRNAs, transcription termination riboswitches adopt structured conformations able to bind specific ligands. Ligand-binding induces formation of a terminator in an elongating mRNA, causing RNA polymerase to cease transcription. As such, transcription termination riboswitches are positioned upstream of ORFs and regulate elongation of transcription into the downstream genes. Intriguingly, in L. monocytogenes we previously noticed one such B12 riboswitch positioned downstream of the lmo1149 gene and in the opposite and convergent orientation to the next adjacent gene, pocR (lmo1150) (8). As this riboswitch lacked a downstream ORF, we hypothesized it might play a nonclassical role. Three previous studies supported the hypothesis that the riboswitch might be transcribed as part of, and regulate transcription of, an asRNA. In the first study, a transcript encompassing the riboswitch, Rli39, was detected by a tiling array analysis (Fig. 1A) (8). In the second study, a noncoding RNA (ncRNA), RliH, was identified by strand-specific Northern blot, antisense to the 5′ end of pocR (Fig. 1A) (9). Finally, in the third study, whole-genome differential RNA-seq analysis identified a transcription start site (TSS) for the Rli39 transcript, but no TSS was detected for RliH (10). These findings suggested that Rli39 and RliH might represent two parts of a long riboswitch-regulated asRNA.
Fig. 1.
Regulation of the pdu locus by B12 and propanediol. (A) Schematic representation of the L. monocytogenes propanediol utilization (pdu) locus: pocR gene (orange), pdu genes (blue), B12 biosynthesis genes (red), unknown gene (gray), ethanolamine utilization gene (black), and B12 riboswitch (green). Enlargement shows the aspocR/pocR locus. Green arrows denote the Rli39 and RliH transcripts. (B) Expression levels of the pduBCDE genes evaluated by qRT-PCR. RNA was isolated from bacteria grown to an OD600 of 0.3–0.5 in LB, LB+B12 (20 nM), LB+Pd (50 mM), or LB+both. Results represent three biological replicates. Error bars show SE. (C) Northern blot of the pocR and AspocR transcripts. Blots were probed with a radioactively labeled PCR probe spanning the aspocR/pocR locus. Band intensities were calculated from four experiments. Units are arbitrary. Statistically significant differences (P < 0.05) are marked with asterisks. RNA was isolated from bacteria grown as described above. tmRNA was used as a loading control.
The rationale for a B12-regulated asRNA opposite to pocR became clearer upon a survey of the literature regarding the role of PocR. In Salmonella enterica, PocR is a transcription factor that, in the presence of propanediol, activates transcription of the pdu and cob genes, which mediate propanediol catabolism and vitamin B12 biosynthesis, respectively (11). Propanediol is a molecule produced in the intestine by commensal bacteria as a byproduct of the fermentation of rhamnose and fucose, and propanediol catabolism requires a B12-dependent diol dehydratase encoded by the pduCDE genes (12). We hypothesized that in L. monocytogenes a B12-regulated asRNA opposite pocR might fine tune the expression of pocR in response to B12. This could inhibit PocR-mediated activation of the B12-dependent pdu genes when propanediol is present but B12 has yet to be synthesized. At present, however, it is unknown how pocR or the pdu genes are regulated in response to B12 in any bacterium.
In this study we show that a B12 riboswitch in L. monocytogenes regulates a cis-encoded asRNA opposite to pocR. This transcript, named AspocR, encompasses the previously identified Rli39 and RliH transcripts. Binding of B12 to the riboswitch leads to premature termination of aspocR transcription, whereas a longer AspocR transcript is produced in the absence of B12. Expression of the long form of AspocR inhibits pocR expression and consequently the activation of the pdu genes, when B12 is unavailable. As propanediol catabolism is thought to be important for the pathogenesis of many intestinal pathogens, our study has implications for both L. monocytogenes and enterobacterial pathogenesis.
Results
Maximal Expression of the pdu Genes Requires both B12 and Propanediol.
In L. monocytogenes, the pdu genes are located in two cassettes of 8 and 20 genes, which surround the pocR gene (Fig. 1A). Additionally, a B12 riboswitch lies 3′ of pocR, but positioned in the antisense orientation (Fig. 1A). Little is known about the regulation of pocR or the pdu genes in response to either propanediol or B12. We thus investigated the regulation of pdu genes in response to the presence of propanediol or B12 by quantitative reverse transcriptase PCR (qRT-PCR). To this end we first evaluated the transcript levels of four pdu genes encoding an accessory protein of propanediol catabolic reactions (pduB), and the three subunits of the B12-dependent diol dehydratase (pduCDE). When either propanediol or B12 alone was present in the medium, expression of the pduB, pduC, pduD, and pduE genes increased, albeit mildly (∼5-fold), compared with the control condition (LB). In contrast, activation of all four genes was significantly higher (∼25-fold) when both propanediol and B12 were present together (Fig. 1B), indicating that maximal expression requires the presence of both propanediol and B12.
The B12 Riboswitch Regulates a Long Transcript.
The demonstration that the pduBCDE genes are regulated in response to propanediol and B12 together is consistent with the hypothesis that the B12 riboswitch regulates an asRNA opposite to pocR. The asRNA could in turn regulate pocR, and subsequently pdu gene expression, in response to B12. To analyze whether such an asRNA is produced, we examined transcription from both strands of the pocR locus by Northern blot using a double-stranded DNA probe to identify the putative antisense RNA and the pocR transcript simultaneously (Fig. 1C). When bacteria were cultured in LB medium alone, both the pocR transcript (∼1,000 nt) and a putative antisense RNA (∼1,400 nt, herein named AspocR) were detected at similar levels (Fig. 1C, lane 1). When B12 was added to the medium, the level of AspocR decreased (Fig. 1C, lane 2), supporting the hypothesis that transcription of aspocR is controlled by a B12 riboswitch. Surprisingly, the level of pocR transcript remained relatively unchanged despite the reduction in AspocR levels, suggesting the cell maintains a basal level of pocR transcript even in the presence of low levels of AspocR, although the mechanism of this regulation is unknown. In the presence of propanediol, however, we observed a significant increase in the levels of both AspocR as well as pocR mRNA (Fig. 1C, lane 3). This increase is likely due to propanediol-dependent activation by PocR itself. PocR has been shown to autoregulate its expression in S. enterica, and both aspocR and pocR contain two nearly identically spaced sites in their promoters, which closely resemble the S. enterica consensus PocR binding site (Fig. S1A) (8, 13, 14). In contrast, in the presence of both propanediol and B12 we observed an even greater increase in pocR mRNA, but the AspocR transcript was no longer detected (Fig. 1C, lane 4), in agreement with our original hypothesis. We also examined expression of both pocR and aspocR by Northern blot using single-stranded RNA probes to detect each transcript individually (Fig. S1 B and C). Using a probe complementary to the riboswitch we detected a short transcript (Fig. S1B, thin arrow), corresponding to the previously reported Rli39 transcript under all conditions (8). However, we also detected a long AspocR transcript when bacteria were grown in presence of propanediol alone (Fig. S1A, thick arrow), and this transcript was detected in LB alone, if blots were overexposed. Expression of pocR increased in the presence of propanediol, but was most strongly activated in the presence of both B12 and propanediol. Together, these results suggest that, in the presence of B12, the riboswitch causes aspocR to be transcribed as a short truncated transcript, whereas in the absence of B12, aspocR is transcribed as a long transcript, which could act as an asRNA to inhibit pocR expression.
Expression of pocR Is Regulated by B12 via AspocR.
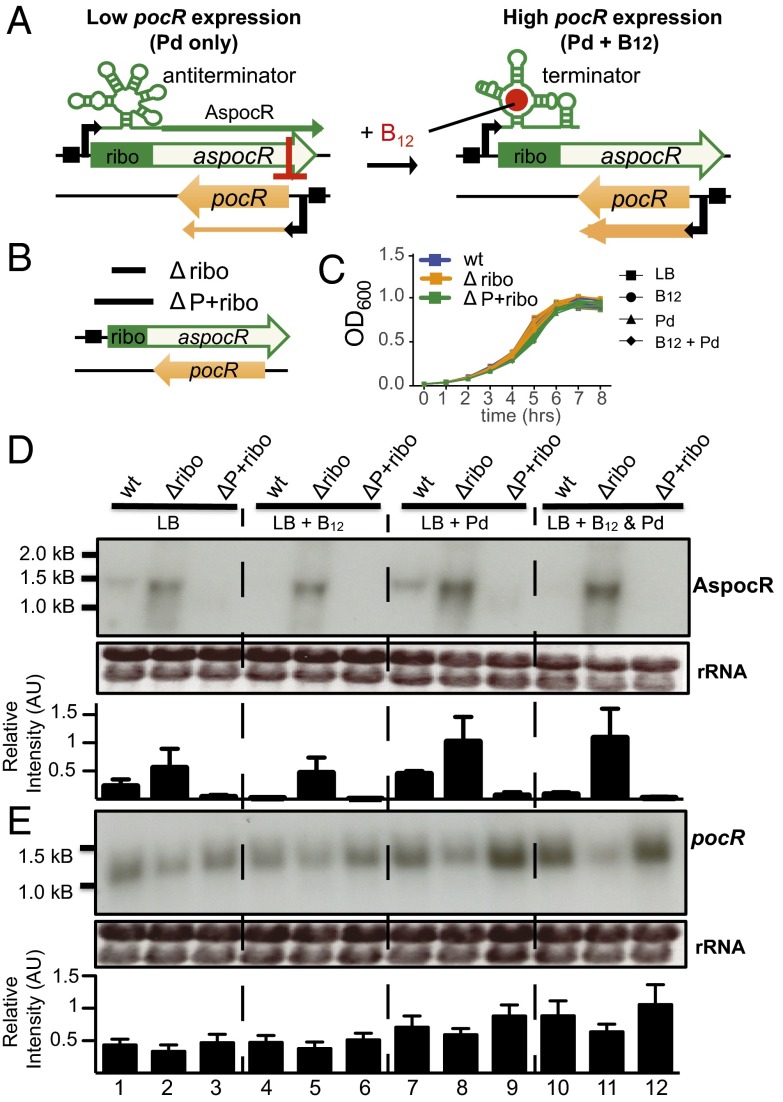
Our data fit well with a model whereby the B12 riboswitch controls expression of an asRNA, which can inhibit pocR expression when B12 levels are low (Fig. 2A). To determine if the long transcript acts as a cis-encoded asRNA regulating pocR, we constructed two mutant strains. In the first mutant (Δribo), we deleted a 229-nt region encompassing the B12 riboswitch, while leaving intact the predicted promoter upstream of the riboswitch (Fig. 2B). We expected that the Δribo mutant would be unable to terminate aspocR transcription, even in the presence of B12. In a second mutant (ΔP+ribo), a 263-nt region encompassing both the riboswitch and the −10 promoter region was deleted (Fig. 2B). We expected that, as it lacked a promoter, the ΔP+ribo mutant would never express the AspocR transcript. Both mutants grew similarly to the wild-type strain under all conditions (Fig. 2C), and RNA was isolated from all strains grown in LB medium alone, LB plus propanediol or B12, or LB plus both in combination. Strand-specific Northern blotting was carried out to analyze levels of the long AspocR transcript and pocR mRNA. In the wild-type strain, low levels of AspocR were produced when bacteria were grown in LB medium (Fig. 2D, lane 1), and aspocR expression increased when bacteria were grown in the presence of propanediol (Fig. 2D, lane 7). However, when only B12, or B12 in combination with propanediol was present in the medium, no AspocR was detected (Fig. 2D, lanes 4 and 10). In agreement with the expected expression patterns of the mutants, the Δribo mutant showed a constitutive expression of aspocR, and aspocR expression increased when bacteria were grown in the presence of propanediol (Fig. 2D, lanes 2, 5, 8, and 11). The absence of the riboswitch results in the observed band migrating slightly faster than that of the wild-type strain and likely accounts for the higher levels of AspocR observed in the Δribo mutant as the riboswitch also leads to the production of truncated transcripts in the wild-type strain. In contrast, no AspocR transcript was detected in the ΔP+ribo mutant under any conditions (Fig. 2D, lanes 3, 6, 9, and 12).
Fig. 2.
Expression of pocR is regulated by B12 via AspocR. (A) Model showing proposed regulation of pocR by AspocR. (B) Schematic representation of the wild-type aspocR/pocR locus with the B12 riboswitch (green) and aspocR promoter (black box). Regions deleted in the Δribo and ΔP+ribo mutants are indicated by black bars. (C) Growth curves of the three strains under conditions indicated. Results represent three experiments. (D and E) Northern blots were carried out using single-stranded RNA probes to detect the long AspocR and pocR transcripts. Band intensities were quantified from three separate experiments. Units are arbitrary. RNA was isolated from bacteria grown to an OD600 of 0.3–0.5 in LB, LB+B12 (20 nM), LB+Pd (50 mM), or LB+both. Ethidium bromide staining of rRNA is shown as a loading control.
We also examined expression of pocR and found that, in all cases, pocR mRNA levels inversely correlated with levels of AspocR. In particular, in the Δribo mutant, which constitutively expresses aspocR, pocR mRNA levels were low under all conditions (Fig. 2E, lanes 2, 5, 8, and 11). In contrast, in the ΔP+ribo mutant, which never expresses aspocR, levels of pocR mRNA were elevated compared with the wild-type strain when bacteria were grown in the presence of propanediol alone (Fig. 2E, lane 9 vs. 7). This indicates that in the ΔP+ribo mutant, pocR was highly expressed when only propanediol was present, whereas in the wild-type strain, pocR mRNA levels were lower when bacteria were grown in the presence of propanediol alone and increased significantly only when bacteria were grown in the presence of both propanediol and B12. Together our data imply a model wherein pocR expression is activated in the presence of propanediol, but is simultaneously inhibited by transcription of the AspocR asRNA. Only when B12 is also present will the riboswitch terminate transcription of the long AspocR transcript, allowing maximal pocR expression.
Effects of aspocR Expression on PocR-Regulated Genes.
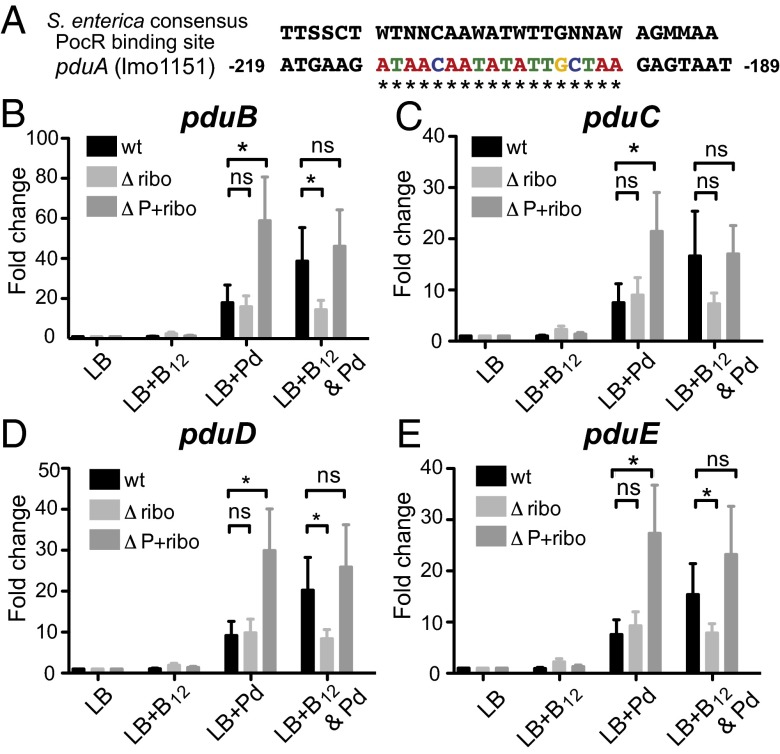
We identified putative PocR binding sites upstream of the pduA (Fig. 3A), pduU, and pduS genes (Fig. S2A) suggesting, as in S. enterica, the pdu genes are regulated by PocR. To determine if AspocR-mediated control of pocR expression affects regulation of the pdu genes, we evaluated the transcript levels of pduBCDE, pduU, and pduS genes by qRT-PCR in the wild type, Δribo, and ΔP+ribo mutants (Fig. 3 and Fig. S2). Bacteria were grown to midlog phase in LB medium in the presence of either propanediol or B12 alone or in combination, and RNA was extracted. When cells were grown in propanediol alone, the pduBCDE genes were strongly up-regulated in the ΔP+ribo mutant compared with the wild type or the Δribo mutant (Fig. 3). Conversely, the pduBCDE genes were minimally expressed in the Δribo mutant, which constitutively expresses aspocR, even in the presence of both propanediol and B12. We observed the same pattern of expression for the pduS and pduU genes as for the pduBCDE genes (Fig. S2), however induction of these genes was lower than for the pduBCDE genes. These data suggest that the regulation of pocR via AspocR is the main switch controlling pdu gene expression in response to B12.
Fig. 3.
Effects of aspocR expression on PocR-regulated genes. (A) Potential PocR-binding site upstream of the pduA gene is shown. Numbering denotes the position relative to the translation start site. Asterisks denote matches to the S. enterica consensus PocR-binding sequence. Levels of the (B) pduB, (C) pduC, (D) pduD, and (E) pduE genes were evaluated by qRT-PCR in the wild-type (WT), Δribo, or ΔP+ribo strains. Statistically significant differences (P < 0.05) are indicated by asterisks. RNA was isolated from bacteria grown to an OD600 of 0.3–0.5 in LB, LB+B12 (20 nM), LB+Pd (50 mM), or LB+both. Results represent the average of four biological replicates. Error bars show SE.
As the vitamin B12 biosynthesis operon has been shown to be activated by PocR in S. enterica (11), we also examined expression of the cblT (lmo1190) and cbiA (lmo1191) genes. cblT and cbiA are the first two genes of the vitamin B12 biosynthesis operon in L. monocytogenes (Fig. S3A) and we identified a PocR-binding site upstream of cblT (Fig. S3B). cblT encodes a putative transporter of a B12 precursor and cbiA encodes a cobyrinic acid synthase involved in B12 synthesis. Interestingly, whereas L. monocytogenes appears to carry only the genes for anaerobic synthesis of B12 (15), both genes are nonetheless activated under aerobic conditions in response to propanediol. This activation is PocR dependent as levels of both cblT and cbiA remained low in the Δribo mutant. We also noted that the intergenic region between cblT and cbiA contains a B12 riboswitch (8), and expression of cbiA was repressed in all strains in the presence of B12 (Fig S3 C and D). Thus, whereas expression of the B12 biosynthesis operon is activated by PocR in response to propanediol, repression of cbiA and potentially the downstream B12 biosynthesis genes is likely mediated by the B12 riboswitch between cblT and cbiA in response to B12.
AspocR Inhibits pocR Expression in Trans and in Vitro.
To date, only a few studies have examined how asRNAs mediate their effects on target genes (16, 17). Owing to their perfect complementarity with the sense-encoded gene, it has been proposed that asRNAs might act through a base-pairing mechanism. However, it has also been proposed that transcription of an asRNA might interfere with transcription of the sense transcript by affecting transcription elongation on the opposite strand. Such a transcriptional interference mechanism requires the asRNA to be transcribed from the same locus as the sense gene. To examine these two possible mechanisms of AspocR-mediated regulation, we tested whether AspocR could ectopically affect expression of pocR. A fragment containing the aspocR/pocR locus, but lacking the promoters for either gene, was cloned into the pAD vector, which stably integrates into the chromosomal tRNAARG locus. A constitutive promoter was cloned in front of aspocR, and a Rho-independent terminator was incorporated at the end of aspocR. A control plasmid carrying only the promoter and terminator was also constructed. The plasmids were transformed into the ΔP+ribo background, which does not express aspocR to generate the ΔP+ribo::aspocR and ΔP+ribo::Ø strains. Bacteria were subsequently grown in LB medium alone, LB in the presence of either propanediol or B12 alone, or in combination, RNA was extracted, and Northern blotting was carried out against the long form of AspocR and the pocR transcript.
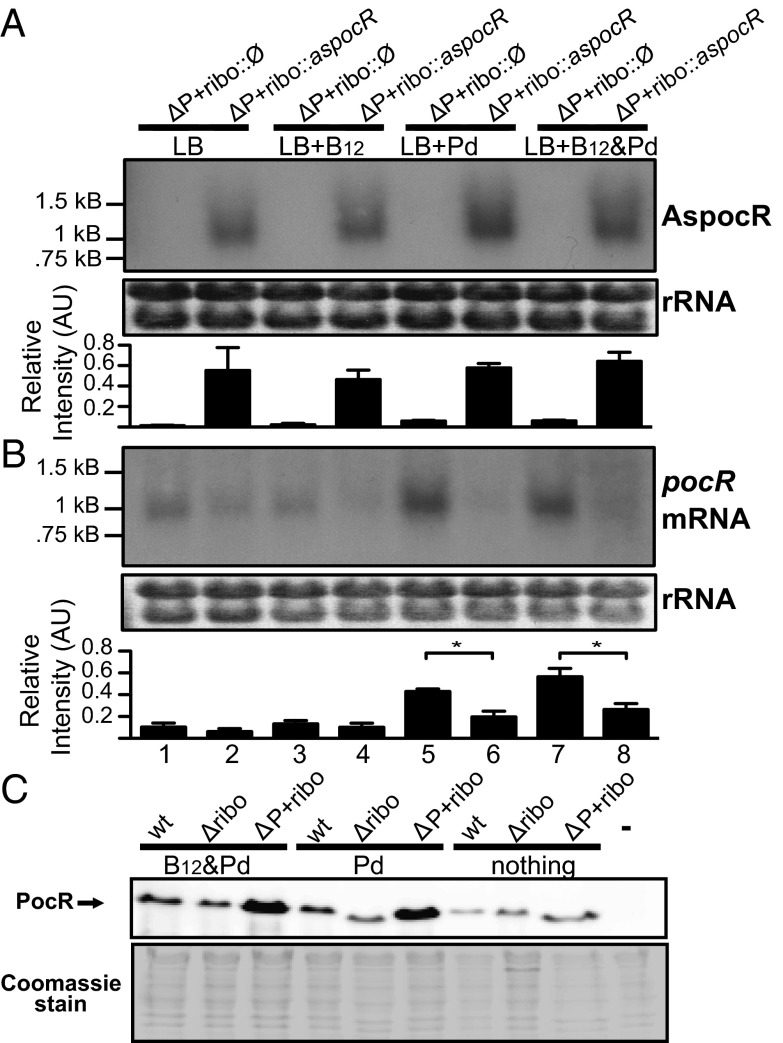
In the ΔP+ribo::Ø strain no AspocR transcript was detected under any condition (Fig. 4, lanes 1, 3, 5, and 7), whereas in the ΔP+ribo::aspocR strain, an AspocR transcript was detected under all conditions, indicating it was constitutively expressed (Fig. 4, lanes 2, 4, 6, and 8). In the ΔP+ribo::Ø strain, pocR transcript levels were unaffected by the presence of the control construct and showed the same pattern of induction as observed previously for the ΔP+ribo mutant. Namely, pocR mRNA levels were elevated in the presence of propanediol alone or propanediol plus B12. In contrast, in the ΔP+ribo::aspocR strain, which constitutively expresses aspocR, levels of pocR transcript were significantly reduced under all conditions. This indicates that AspocR can act in trans and does not need to be transcribed from the same locus to inhibit expression of pocR. While this does not exclude the possibility that AspocR also mediates transcriptional interference when transcribed from its native locus, it suggests that the main effects of AspocR on pocR expression are mediated by base pairing to pocR mRNA.
Fig. 4.
AspocR represses pocR expression in trans and in vitro. Strains were constructed in the ΔP+ribo background with the aspocR gene preceded by a constitutive promoter and followed by a Rho-independent terminator (ΔP+ribo::aspocR) or with a constitutive promoter and terminator but lacking the aspocR gene (ΔP+ribo::Ø). RNA was isolated from bacteria grown to an OD600 of 0.3–0.5 in LB, LB+B12 (20 nM), LB+Pd (50 mM), or LB+both. Northern blots were carried out using single-stranded RNA probes to detect the (A) long AspocR and (B) pocR transcripts. Statistically significant differences (P < 0.05) are indicated by asterisks. Ethidium bromide staining of rRNA is shown as a loading control. Positions of molecular weight markers are indicated. Band intensities were quantified from three separate experiments. Units are arbitrary. (C) Western blot showing PocR protein levels from in vitro transcription/translation experiments. Fragments corresponding to the aspocR/pocR locus from the wild-type, Δribo, or ΔP+ribo strains were used as templates for reactions in the presence of Pd+B12, Pd, or nothing. A control reaction with Pd+B12 but no template is also shown (−). Equal loading was verified by Coomassie staining (shown below).
To further examine the mechanism of AspocR mediated regulation, we measured PocR expression using an in vitro transcription/translation system. In this system we first amplified fragments corresponding to the aspocR/pocR locus from the wild-type, Δribo, or ΔP+ribo strains and cloned them into plasmids. Plasmids were linearized and used as templates for in vitro transcription/translation experiments. When wild-type or Δribo templates were used in transcription/translation reactions, we observed low levels of PocR protein (Fig. 4C), probably due to constitutive AspocR expression from these templates. This conclusion was supported by the findings that a B12 riboswitch was unable to mediate transcription termination in the presence of B12 in another in vitro transcription system (18). In contrast, reactions with the ΔP+ribo template, which lacks the aspocR promoter, showed significant PocR expression in the presence of propanediol and B12 together (Fig. 4C), or propanediol (Fig. 4C) and B12 alone (Fig. S4), presumably due to the absence of AspocR-mediated inhibition.
Finally, we performed experiments to examine the stability of pocR mRNA in the presence or absence of AspocR. Wild-type, Δribo, and ΔP+ribo strains were cultured in LB medium in the presence of both propanediol and B12 to an OD600 of ∼0.4 before rifampicin was added to the medium to stop transcription. RNA was extracted just before rifampicin treatment, and at various times thereafter, and Northern blotting was carried out to determine the levels of pocR mRNA. Consistent with our previous observations, we observed a significant reduction in the amount of pocR mRNA at time 0 in the Δribo mutant compared with the wild type or ΔP+ribo mutants (Fig. S5A, lanes 1, 6, and 11), indicating that the steady-state level of pocR transcript is indeed inhibited in the presence of AspocR. After rifampicin treatment, we observed a slight decrease in the half-life of pocR mRNA in the ΔP+ribo mutant; however, these differences were not significant (Fig. S5B) and suggested AspocR does not promote the degradation of pocR mRNA.
Antisense Oriented Riboswitches in Other Bacteria.
As pocR and the pdu genes are widely conserved in enteropathogens, we examined the pocR genes of other bacteria for the presence of an associated antisense B12 riboswitch. This analysis identified B12 riboswitches antisense to the pocR genes in the closely related species of Listeria welshimeri, Listeria innocua, Listeria seeligeri, and Listeria ivanovii as well as other strains of L. monocytogenes (Dataset S1), suggesting that pocR expression in these bacteria is likely governed by orthologs of aspocR. We also noted a B12 riboswitch adjacent to the pocR gene in other species including S. enterica, Citrobacter koseri, Escherichia coli, Shigella spp., and Yersinia enterocolitica (Dataset S1); however, these B12 riboswitches were not in an antisense orientation to pocR. Thus, regulation of pocR by a B12 riboswitch-regulated asRNA appears limited to the Listeria genus.
Riboswitch-regulated asRNAs are so far limited to the example of the AspocR transcript described here and to another asRNA opposite the ubiG operon in Clostridium acetobutylicum, which is regulated by an S-adenosylmethionine (SAM) riboswitch (19). We were interested in examining whether riboswitch-regulated asRNAs might have been overlooked in bacteria. We designed an algorithm to identify antisense oriented riboswitches in 834 bacterial genomes using gene annotations derived from National Center for Biotechnology Information Refseq and riboswitch annotations from the RNA families (Rfam) database. We examined 15 classes of ligand-binding riboswitches. In total we identified 144 occurrences of riboswitches antisense to the adjacent downstream open-reading frame (Table 1), including the SAM riboswitch previously shown to regulate an asRNA in C. acetobutylicum. Surprisingly, antisense riboswitches were mostly represented in the B12, S-adenosylmethionine (SAM), cyclic-di-GMP, Mg2+ and flouride-binding riboswitch families (2.1–9.1% antisense riboswitches/total), and very few or no antisense riboswitches were identified in the other riboswitch classes examined (detailed in Dataset S2). The reasons accounting for these differences are unknown. However, the presence of antisense-oriented riboswitches in a range of bacterial species suggests that riboswitch-regulated noncoding RNAs likely represent an overlooked mechanism bacteria use to regulate gene expression.
Table 1.
Antisense-oriented riboswitches
| Riboswitch type | # antisense* | % (total analyzed) |
| Vitamin B12 | 45 (27) | 3.8 (1,171) |
| SAM | 18 (9) | 2.1 (844) |
| Cyclic-di-GMP | 16 (3) | 4.5 (355) |
| T-box | 15 (11) | 0.7 (2,300) |
| Mg2+ | 15 (12) | 9.1 (165) |
| TPP | 10 (4) | 0.7 (1,403) |
| FMN | 9 (7) | 1.6 (557) |
| Fluoride | 6 (1) | 2.9 (207) |
| Glycine | 5 (3) | 0.8 (666) |
| Lysine | 4 (2) | 1.1 (369) |
| Purine | 1 (1) | 0.3 (380) |
| Pre-queosine | 0 | 0.0 (113) |
| Pyrimidine | 0 | 0.0 (253) |
| SAH | 0 | 0.0 (85) |
| THF | 0 | 0.0 (38) |
FMN, flavin mononucleotide; SAH, S-adenosylhomocysteine; SAM, S-adenosylmethionine; THF, tetrahydrofolate; TPP, thiamine pyrophosphate.
Number of antisense riboswitches that partially overlap an adjacent ORF are presented in parentheses in this column.
Discussion
In this study, we identied a B12 riboswitch that is transcribed as part of, and regulates transcription of, a noncoding asRNA, AspocR. Regulation by AspocR ensures that pocR, and consequently genes of the propanediol utilization pathway, are maximally expressed only when the cofactor essential for propanediol catabolism, B12, is present. Regulation of pocR by a riboswitch-regulated asRNA represents a unique mechanism for controlling expression of a transcriptional regulatory protein and our analysis of over 800 bacterial genomes suggests that AspocR represents an emerging family of riboswitch-regulated asRNAs.
Surprisingly, like rare cases of other long asRNAs (20), AspocR was able to act in trans, whereas the riboswitch-regulated asRNA from C. acetobutylicum was not (19). The ability of AspocR to act in trans suggests that it can diffuse and bind to the pocR mRNA leading to a number of outcomes. Studies have suggested that base pairing between an asRNA and an mRNA can lead to the degradation of the mRNA, interfere with translation initiation, or repress mRNA transcription through transcriptional attenuation (16). Our data suggest AspocR may work through the latter two mechanisms. Our in vitro transcription/translation experiments showed that significantly higher quantities of PocR protein were produced with a template lacking the aspocR promoter, suggesting that aspocR expression inhibits PocR translation, at least in vitro. Furthermore our in vivo experiments examining the effects of ectopic aspocR expression on pocR mRNA levels showed that the expression of aspocR results in lower steady-state levels of pocR transcript. However, the stability of the pocR mRNA itself was largely unaffected by aspocR expression, suggesting that AspocR may repress pocR expression at the level of transcription rather than by affecting pocR transcript stability. As only a few asRNAs have been studied in detail at the mechanistic level, AspocR, and asRNA-mediated regulation in general, thus deserve further study.
Interestingly, we noted that the AspocR transcript never completely represses pocR expression. Even in the Δribo mutant, which constitutively expresses aspocR, some pocR transcript remains detectable. AspocR seems to act as a rheostat, modulating pocR mRNA levels rather than acting as a complete inhibitor of pocR expression. This modulation makes sense in light of PocR’s dual role in activating the expression of the propanediol catabolic genes and also the genes responsible for anaerobic, de novo synthesis of vitamin B12. If AspocR strongly repressed pocR in the absence of B12, PocR levels would likely be insufficient to activate expression of the B12 biosynthetic genes, locking the cell in a state where it would be unable to activate B12 synthesis. Instead, expression of aspocR only partially represses pocR at low B12 conditions. In the presence of propanediol, this level of PocR appears sufficient to initiate expression of the B12 biosynthesis genes, which under anaerobic conditions will produce B12 essential for propanediol catabolism, but it is insufficient to maximally activate expression of the pdu genes. Presumably, only when sufficient levels of B12 accumulate, either through uptake or de novo synthesis, does B12 bind the riboswitch, terminating transcription of aspocR, and leading to maximal expression of pocR and the pdu genes. Thus, AspocR seems to serve as a fine-tuning element, rather than an all-or-nothing switch.
To our knowledge, our study is unique in showing how pocR and the pdu genes are regulated by B12 in any bacterium. Regulation of propanediol catabolism has recently garnered attention as propanediol, along with the closely related molecule ethanolamine (which is also degraded in a B12-dependent manner), are increasingly considered important nutrient sources for bacterial pathogens during infection (21). An elegant set of studies recently showed that, in the case of S. enterica, growth on ethanolamine in the intestine depends upon the inflammatory immune response induced by the bacterium (22, 23). Inflammation leads to the recruitment of neutrophils to the intestine, whose generation of reactive oxygen species result in the production of an electron acceptor, tetrathionate. S. enterica can then use tetrathionate to anaerobically respire on ethanolamine and outgrow commensal bacteria. The tetrathionate reductase system also supports anaerobic respiration on propanediol in S. enterica (24), but L. monocytogenes lacks an obvious ortholog of tetrathionate reductase. Furthermore, we were unable to grow L. monocytogenes in minimal medium supplemented with propanediol and B12, under aerobic, microaerobic, or anaerobic conditions, even when we included tetrathionate, nitrate, dimethyl sulfoxide, or fumarate as possible electron acceptors. Nonetheless, the observations that L. monocytogenes up-regulates the pdu and ethanolamine utilization (eut) genes during both intracellular growth in intestinal epithelial cells (25), as well as in the intestine of germ-free mice pretreated with lactobacilli (26), suggest that propanediol and ethanolamine both play a role in L. monocytogenes pathogenesis. We suspect that L. monocytogenes may require an electron acceptor specific to its in vivo niche to use propanediol, and it will be of considerable interest to determine how L. monocytogenes and other pathogens use these carbon, nitrogen and energy sources differently during infection.
Finally, our bioinformatics analysis suggests that riboswitch-regulated asRNAs are not uncommon in bacteria. Up to 15 different types of riboswitches have now been identified in prokaryotic genomes, and the position of many transcription termination riboswitches, either in an antisense orientation, or far from the adjacent ORF start, suggests they would be unable to regulate these ORFs in a classical manner. These “marooned” riboswitches are strong candidates to regulate asRNAs, and plausibly sRNAs, suggesting many riboswitch-regulated noncoding RNAs have been thus far overlooked.
Materials and Methods
For greater detail, see Datasets S3 and S4.
Bacterial Strains, Plasmids, Primers, and Growth Conditions.
Strains and plasmids used are listed in Dataset S3. Primers used are listed in Dataset S4. L. monocytogenes and E. coli were grown in LB broth with shaking at 200 rpm at 30 °C and 37 °C, respectively. LB was supplemented with 50 mM 1,2 propanediol and/or 20 nm vitamin B12. For all experiments, bacteria were grown to an OD600 of 0.3–0.5.
Northern Blots.
Blots were carried out according to the Northernmax-Gly kit protocol. Blots were probed with double-stranded DNA probes or single-stranded RNA probes labeled with α-[32P]ATP or α-32P-UTP, respectively.
Quantitative Real-Time PCR.
One microgram of DNaseI-treated total RNA was used for cDNA synthesis with an iScript cDNA Synthesis kit. qRT-PCR reactions were prepared with SYBR Green master mix. Reaction cycling and quantification was carried out in an ABI Prism 7900HT. Expression levels were normalized to the rpoB gene. Samples were evaluated in triplicate and results represent at least three independent experiments. Statistically significant differences were calculated using log2 transformed values evaluated by paired t test.
In Vitro Transcription/Translation and Western Blots.
Fragments encompassing the aspocR/pocR locus of either wild type, ΔP+ribo, or ΔP+ribo strains were used as templates for in vitro transcription/translation reactions with E. coli S30 extracts. Reactions were precipitated, washed, resolubilized, and run on 12% (vol/vol) SDS/PAGE gels. Gels were transferred to Hybond-P membranes and blotted with affinity-purified α-PocR rabbit sera.
Supplementary Material
Acknowledgments
We thank Cristel Archambaud and Marie-Anne Nahori for helpful discussions. This work was supported by grants (to P.C.), European Research Council Advanced Grant 233348, Association Nationale de la Recherche (ANR) (BACNET 09-BLAN- 0024-02), ANR Investissement d’Avenir Programme (10-LABX-62), Fondation Le Roch, and Fondation Jeantet. J.J. was supported by Umeå University, the Swedish Research Council Grants K2011-56X-15144-08-6 and 621-2012-2451, and an ERC Starting Grant (260764–RNAntibiotics). P.C. is an International Senior Research Scholar of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1304795110/-/DCSupplemental.
References
- 1.Cossart P. Illuminating the landscape of host-pathogen interactions with the bacterium Listeria monocytogenes. Proc Natl Acad Sci USA. 2011;108(49):19484–19491. doi: 10.1073/pnas.1112371108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lecuit M. Understanding how Listeria monocytogenes targets and crosses host barriers. Clin Microbiol Infect. 2005;11(6):430–436. doi: 10.1111/j.1469-0691.2005.01146.x. [DOI] [PubMed] [Google Scholar]
- 3.Cossart P, Sansonetti PJ. Bacterial invasion: The paradigms of enteroinvasive pathogens. Science. 2004;304(5668):242–248. doi: 10.1126/science.1090124. [DOI] [PubMed] [Google Scholar]
- 4.Mellin JR, Cossart P. The non-coding RNA world of the bacterial pathogen Listeria monocytogenes. RNA Biol. 2012;9(4):372–378. doi: 10.4161/rna.19235. [DOI] [PubMed] [Google Scholar]
- 5.Winkler WC, Breaker RR. Regulation of bacterial gene expression by riboswitches. Annu Rev Microbiol. 2005;59:487–517. doi: 10.1146/annurev.micro.59.030804.121336. [DOI] [PubMed] [Google Scholar]
- 6.Romby P, Charpentier E. An overview of RNAs with regulatory functions in gram-positive bacteria. Cell Mol Life Sci. 2010;67(2):217–237. doi: 10.1007/s00018-009-0162-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hollands K, et al. Riboswitch control of Rho-dependent transcription termination. Proc Natl Acad Sci USA. 2012;109(14):5376–5381. doi: 10.1073/pnas.1112211109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Toledo-Arana A, et al. The Listeria transcriptional landscape from saprophytism to virulence. Nature. 2009;459(7249):950–956. doi: 10.1038/nature08080. [DOI] [PubMed] [Google Scholar]
- 9.Mandin P, Repoila F, Vergassola M, Geissmann T, Cossart P. Identification of new noncoding RNAs in Listeria monocytogenes and prediction of mRNA targets. Nucleic Acids Res. 2007;35(3):962–974. doi: 10.1093/nar/gkl1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wurtzel O, et al. Comparative transcriptomics of pathogenic and non-pathogenic Listeria species. Mol Syst Biol. 2012;8:583. doi: 10.1038/msb.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rondon MR, Escalante-Semerena JC. The poc locus is required for 1,2-propanediol-dependent transcription of the cobalamin biosynthetic (cob) and propanediol utilization (pdu) genes of Salmonella typhimurium. J Bacteriol. 1992;174(7):2267–2272. doi: 10.1128/jb.174.7.2267-2272.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bobik TA, Xu Y, Jeter RM, Otto KE, Roth JR. Propanediol utilization genes (pdu) of Salmonella typhimurium: Three genes for the propanediol dehydratase. J Bacteriol. 1997;179(21):6633–6639. doi: 10.1128/jb.179.21.6633-6639.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen P, Andersson DI, Roth JR. The control region of the pdu/cob regulon in Salmonella typhimurium. J Bacteriol. 1994;176(17):5474–5482. doi: 10.1128/jb.176.17.5474-5482.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rondon MR, Escalante-Semerena JC. In vitro analysis of the interactions between the PocR regulatory protein and the promoter region of the cobalamin biosynthetic (cob) operon of Salmonella typhimurium LT2. J Bacteriol. 1996;178(8):2196–2203. doi: 10.1128/jb.178.8.2196-2203.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. Comparative genomics of the vitamin B12 metabolism and regulation in prokaryotes. J Biol Chem. 2003;278(42):41148–41159. doi: 10.1074/jbc.M305837200. [DOI] [PubMed] [Google Scholar]
- 16.Thomason MK, Storz G. Bacterial antisense RNAs: How many are there, and what are they doing? Annu Rev Genet. 2010;44:167–188. doi: 10.1146/annurev-genet-102209-163523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sesto N, Wurtzel O, Archambaud C, Sorek R, Cossart P. The excludon: A new concept in bacterial antisense RNA-mediated gene regulation. Nat Rev Microbiol. 2013;11(2):75–82. doi: 10.1038/nrmicro2934. [DOI] [PubMed] [Google Scholar]
- 18.Fox KA, et al. Multiple posttranscriptional regulatory mechanisms partner to control ethanolamine utilization in Enterococcus faecalis. Proc Natl Acad Sci USA. 2009;106(11):4435–4440. doi: 10.1073/pnas.0812194106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.André G, et al. S-box and T-box riboswitches and antisense RNA control a sulfur metabolic operon of Clostridium acetobutylicum. Nucleic Acids Res. 2008;36(18):5955–5969. doi: 10.1093/nar/gkn601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee E-J, Groisman EA. An antisense RNA that governs the expression kinetics of a multifunctional virulence gene. Mol Microbiol. 2010;76(4):1020–1033. doi: 10.1111/j.1365-2958.2010.07161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buchrieser C, Rusniok C, Kunst F, Cossart P, Glaser P. Listeria Consortium Comparison of the genome sequences of Listeria monocytogenes and Listeria innocua: Clues for evolution and pathogenicity. FEMS Immunol Med Microbiol. 2003;35(3):207–213. doi: 10.1016/S0928-8244(02)00448-0. [DOI] [PubMed] [Google Scholar]
- 22.Winter SE, Bäumler AJ. A breathtaking feat: To compete with the gut microbiota, Salmonella drives its host to provide a respiratory electron acceptor. Gut Microbes. 2011;2(1):58–60. doi: 10.4161/gmic.2.1.14911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thiennimitr P, et al. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc Natl Acad Sci USA. 2011;108(42):17480–17485. doi: 10.1073/pnas.1107857108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Price-Carter M, Tingey J, Bobik TA, Roth JR. The alternative electron acceptor tetrathionate supports B12-dependent anaerobic growth of Salmonella enterica serovar typhimurium on ethanolamine or 1,2-propanediol. J Bacteriol. 2001;183(8):2463–2475. doi: 10.1128/JB.183.8.2463-2475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joseph B, et al. Identification of Listeria monocytogenes genes contributing to intracellular replication by expression profiling and mutant screening. J Bacteriol. 2006;188(2):556–568. doi: 10.1128/JB.188.2.556-568.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Archambaud C, et al. Impact of lactobacilli on orally acquired listeriosis. Proc Natl Acad Sci USA. 2012;109(41):16684–16689. doi: 10.1073/pnas.1212809109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.