Abstract
Inflorescence architecture of barley (Hordeum vulgare L.) is common among the Triticeae species, which bear one to three single-flowered spikelets at each rachis internode. Triple spikelet meristem is one of the unique features of barley spikes, in which three spikelets (one central and two lateral spikelets) are produced at each rachis internode. Fertility of the lateral spikelets at triple spikelet meristem gives row-type identity to barley spikes. Six-rowed spikes show fertile lateral spikelets and produce increased grain yield per spike, compared with two-rowed spikes with sterile lateral spikelets. Thus, far, two loci governing the row-type phenotype were isolated in barley that include Six-rowed spike1 (Vrs1) and Intermedium-C. In the present study, we isolated Six-rowed spike4 (Vrs4), a barley ortholog of the maize (Zea mays L.) inflorescence architecture gene RAMOSA2 (RA2). Eighteen coding mutations in barley RA2 (HvRA2) were specifically associated with lateral spikelet fertility and loss of spikelet determinacy. Expression analyses through mRNA in situ hybridization and microarray showed that Vrs4 (HvRA2) controls the row-type pathway through Vrs1 (HvHox1), a negative regulator of lateral spikelet fertility in barley. Moreover, Vrs4 may also regulate transcripts of barley SISTER OF RAMOSA3 (HvSRA), a putative trehalose-6-phosphate phosphatase involved in trehalose-6-phosphate homeostasis implicated to control spikelet determinacy. Our expression data illustrated that, although RA2 is conserved among different grass species, its down-stream target genes appear to be modified in barley and possibly other species of tribe Triticeae.
Keywords: cytokinin, EGG APPARATUS1, grain number, yield potential
Inflorescence architecture is highly diverse among members of the grass family (Poaceae), in which spikelets represent the fundamental building blocks, comprising one or more florets enclosed by two glumes. Economically important grass species of the tribe Triticeae, such as wheat (Triticum spp.), barley, triticale (×Triticosecale Wittm. ex A. Camus), and rye (Secale cereale L.), typically possess a branchless spike-shaped inflorescence, whereas inflorescences of tropical species, e.g., maize, Sorghum spp., and rice (Oryza sativa L.), have highly branched tassels or panicles. Inflorescence branching in maize appears to be largely regulated through the RAMOSA gene network, which involves the RAMOSA1 (RA1), RA2, RA3, and RAMOSA1 ENHANCER LOCUS2 (REL2) genes (1). In maize, determinacy of the spikelet pair meristems (SPMs) is mediated through RA2, which encodes a lateral organ boundaries (LOB) domain-containing transcriptional regulator that functions upstream of the RA1 and REL2 determinacy-providing complex (2–4). Expression of RA1 is dependent upon RA3, which encodes a phosphatase involved in trehalose metabolism (5). Orthologs of RA1 and RA3 are unique to the grass tribe Andropogoneae (paired spikelets arise from SPMs), but close paralogs of RA3, with an as of yet unknown function, exist in barley (HvSRA) and other grasses (5).
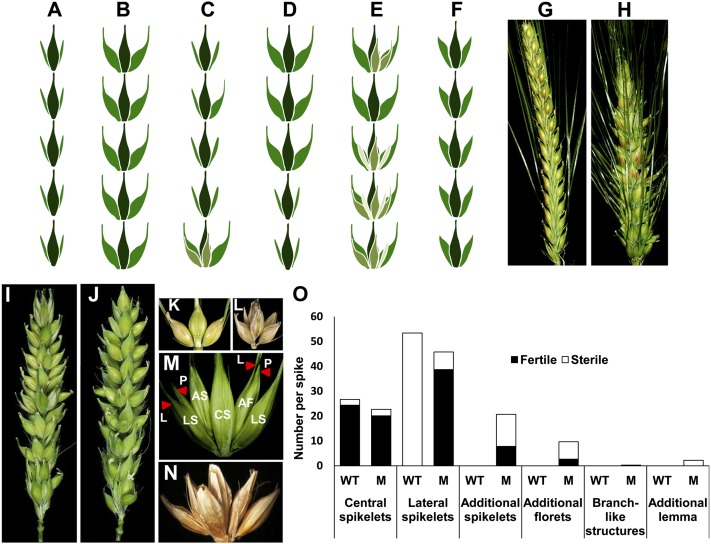
The barley inflorescence is an indeterminate spike that produces three single-flowered spikelets in a distichous manner at each rachis internode that develop into one central and two lateral spikelets (Fig. 1A) (6, 7). Based upon lateral spikelet/floret size and fertility barley is classified into two different row-types; i.e., two-rowed and six-rowed barley (8). In two-rowed barley, the central spikelet is fertile and produces grain, and the two lateral spikelets remain sterile (Fig. 1G). In six-rowed barley, all three spikelets are fertile and develop into grains (Fig. 1B). The six-rowed phenotype is controlled by at least five independent loci that include Six-rowed spike1 (vrs1), vrs2, vrs3, vrs4, and Intermedium-C (Int-c). Vrs1 encodes a homeodomain-leucine zipper class I transcription factor that is a negative regulator of lateral spikelet fertility (9). Mutant vrs1.a promotes lateral spikelet fertility resulting in a complete six-rowed spike (Fig. 1B). Alleles at the locus int-c, which is an ortholog of the maize domestication gene TEOSINTE BRANCHED1 (HvTB1) (10), modify lateral spikelet development with respect to allelic constitution at vrs1 (Fig. 1F). Loss-of-function vrs1.a is generally accompanied by Int-c.a in six-rowed barley and the functional Vrs1.b by int-c.b in two-rowed barley. The remaining vrs loci vrs2, vrs3, and vrs4 show varying levels of lateral spikelet fertility with complete lateral spikelet fertility observed in vrs4 mutants (Fig. 1 C–E). Apart from lateral spikelet fertility, vrs4 mutants show indeterminate triple spikelet meristems (TSMs), thereby producing additional spikelets/florets. The apparent indeterminacy of the TSM in vrs4 mutants suggests that vrs4 is involved in a genetic pathway that regulates the highly conserved determinate nature of the TSM inherent to Hordeum species.
Fig. 1.
Spike morphology of different row-type loci and vrs4 phenotype. (A) Two-rowed spike (Vrs1): fertile central spikelets (CS) and sterile lateral spikelets (LS). (B) Six-rowed spike1 (vrs1): completely fertile CS and LS. (C) Six-rowed spike2 (vrs2): LS fertility observed at the spike base with occasional additional spikelets [additional spikelet (AS) fertile or sterile, AS in light green color]; along the spike, LS are occasionally enlarged and set seed, (D) Six-rowed spike3 (vrs3): spike base appears two-rowed, and the remaining portion appears six-rowed. (E) Six-rowed spike4 (vrs4): spike similar to vrs1, with frequent awn bearing AS (light green colored). (F) Intermedium-C (int-c): LS are enlarged, set seed, which are usually smaller than in vrs1, LS without awns. (G) Two-rowed wild-type spike (Piroline). vrs4 mutant spikes: (H) vrs4.l, (I) BW-NIL(mul1.a). (J) BW-NIL(vrs4.k): six-rowed-like appearance and formation of AS and additional florets (AF). (K) Spikelet triplet at one rachis internode. (L and M) BW-NIL(vrs4.k) (L) and vrs4.l (M) showing AS and AF; initial triplet: CS, LS with lemma (L) and palea (P). AF developed on the rachilla of LS. (N) Seed formation involving AS and AF. (O) Classes of axillary structures produced by vrs4 mutant vrs4.l and wild-type Piroline spikes. Total number of fertile or sterile axillary structures from autumn grown plants is shown. Data were recorded on five plants with on average two spikes per plant. (see SI Dataset S1A for SE and significance of t test).
In the present study, we isolated the vrs4 locus through genetic mapping and extensive mutant analysis. Our findings showed that Vrs4 underlies HvRA2, an ortholog of maize transcription factor RAMOSA2, which is important for inflorescence development in grasses. Tissue localization through mRNA in situ hybridization in immature spikes showed that HvRA2 is expressed very early during inflorescence development and most probably confines barley’s TSM meristem to three spikelets. Gene expression analysis in combination with microarray experiments demonstrated that HvRA2 may likely function in two distinct pathways, thereby establishing spike architecture through regulation of TSM determinacy and controlling the Hordeum-specific row-type pathway.
Results
vrs4 Mutants Display Indeterminate TSM and Six-Rowed Phenotype.
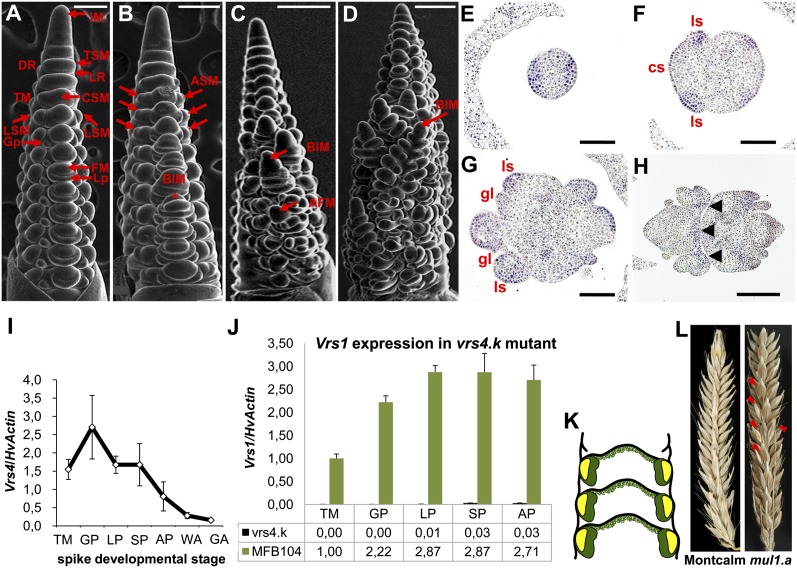
Unlike maize or sorghum, in which spikelets develop from SPMs, immature barley spikes develop a TSM that arises from the first axillary meristem right after double ridge stage (first reproductive stage). TSMs are determinate and form three distinct mounds producing three secondary axillary meristems (AMs), one central spikelet meristem (CSM) and two lateral spikelet meristems (LSMs) (Fig. 2A and SI Appendix, Fig. S1A). Each spikelet meristem initially differentiates into a pair of glume primordia (Fig. 2A), and then into one floral meristem (FM). Scanning electron microscopy (SEM) analysis of wild type and vrs4 mutants at double ridge stage revealed no morphological differences, suggesting that there is no change in determinacy of AM that gives rise to triple spikelet primordia (Fig. 2 A and B). The appearance of two additional mounds on either side of the LSMs was the first visible deviation observed in vrs4 mutant inflorescences (Fig. 2B). The additional spikelets/florets emerging at rachis internodes (SI Appendix, Fig. S1 B and C) were frequently fertile and developed into grains (Fig. 1 L and N). Additional spikelets had the same orientation (lemma primordia on abaxial side) as normal spikelets (Fig. 1M and SI Appendix, Fig. S1 A, B, D, and E). However, additional florets were oriented opposite to the normal spikelet (lemma primordia on the adaxial side showing the alternate orientation of floret formation on the rachilla) and lacked glumes (Fig. 1M). Apart from the additional spikelets/florets (Fig. 1O and SI Appendix, Fig. S1G) we rarely found additional pistils (SI Appendix, Fig. S1I), suggesting that FM determinacy was also affected in vrs4 mutants. Loss of determinacy became even more apparent when CSMs frequently produced branch-like inflorescence meristems (BIM) with ridges (resembling predouble ridge stage of primary inflorescence) (Fig. 2C and SI Appendix, Fig. S1C), which later differentiated into triple mounds (Fig. 2D) and eventually produced a vrs4-like inflorescence meristem (SI Appendix, Fig. S1J). The majority of BIMs aborted as growth continued (SI Appendix, Fig. S1 B and C), and only few meristems at the base developed into additional spikelets. Similar developmental abnormalities were observed in two other vrs4 Bowman near isogenic alleles BW-NIL(vrs4.k) and BW-NIL(mul1.a) (SI Appendix, Fig. S2 A–E). SEM analysis revealed that vrs4 mutants lost determinacy of the TSMs, and subsequently SMs, thus producing supernumerary spikelets and florets. The spring-grown mutant allele vrs4.l showed an enhanced vrs4 phenotype with a significantly higher number of spikelets and branch-like structures than autumn grown plants (SI Appendix, Fig. S1 B and C and SI Dataset S1A).
Fig. 2.
Scanning electron microscopy and transcript localization of Vrs4 mRNA in immature barley spikes. (A) Wild-type inflorescence at lemma primordium (LP) stage showing inflorescence meristem (IM) forming double ridge (DR), upper ridge containing triple spikelet meristem (TSM), and lower leaf ridge (LR). TSM forms a triple mound (TM), which transitions into one central spikelet meristem (CSM) and two lateral spikelet meristems (LSMs). A pair of glume primordia (GP) and a single floral meristem (FM) are produced by each SM. (B) vrs4 at LP initiating additional spikelet meristems (ASMs) (red arrows). CSMs develop into branch-like inflorescence meristem (BIM). Occasionally, CSMs initiate additional florets on rachilla (asterisk). (C) vrs4 at stamen primordium (SP) stage showing BIM at DR and a developing additional floret meristem (AFM). (D) vrs4 at awn primordium (AP) stage showing BIM at TM. (E–H) In situ RNA hybridization of HvRA2 in two-rowed barley cv. Bonus. Transverse sections at DR (E), TM (F), GP (G), and SP (H). cs, central spikelet; ls, lateral spikelet; gl, glume. (I) Transcript levels of HvRA2 determined by quantitative RT-PCR in cv. Bonus. Constitutively expressed HvActin was used for normalization. X-axis represents spike developmental stages. Mean ± SE of three biological replicates. WA, white anther; GA, green anther. (J) Vrs1 expression in BW-NIL(vrs4.k) and wild-type MFB104. Mean ± SE of three biological replicates. Expression values are given at the bottom of the graph. (K) Vrs1 (yellow) and Vrs4 (green) are expressed in overlapping domains of lateral spikelets. (L) Spikes of cv. Montcalm carrying vrs1.a1 allele and vrs4 mutant allele mul1.a. Additional spikelets in mul1.a are indicated by red triangles. (Scale bars: 200 µm in A and B; 333 µm in C and D; and 100 µm in E–H.)
Apart from the indeterminate nature of TSMs, vrs4 mutants displayed another important feature of complete fertility and development of lateral spikelets resulting in a six-rowed phenotype. The six-rowed phenotype observed in vrs4 mutants was analogous to that of vrs1 mutants. In the present study, all vrs4 mutant alleles, except int-e.4, int-e.20, and int-e.72, showed complete or partial six-rowed phenotype (Fig. 1 H–J and SI Appendix, Fig. S3).
Genetic Mapping and Mutant Analysis Reveals That Vrs4 Underlies the Barley Ortholog of Maize RAMOSA2.
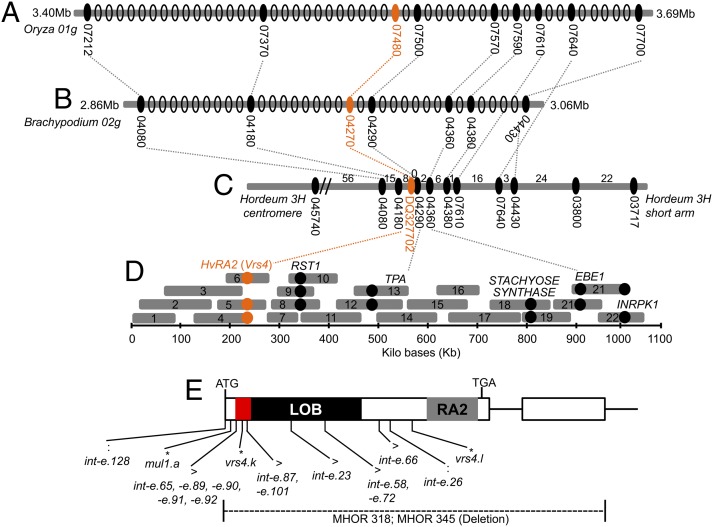
We initially mapped vrs4 to the short arm of chromosome 3H using five F2 mapping populations comprised of 188–214 gametes (SI Appendix, Table S1). Mapped markers were derived from syntenic Brachypodium (chromosome 2) and rice (chromosome 1) gene sequences based on the virtual gene order reported in the barley genome zipper (11) (SI Dataset S1B). In all five mapping populations tested, the vrs4 phenotype cosegregated with a cluster of markers (barley orthologs of Brachypodium/rice genes) derived from the short arm of 3H (37.17–41.68 cM in genome zipper) (SI Appendix, Fig. S4). Flanking markers of the corresponding marker cluster contained 68 predicted Brachypodium genes (Bradi2g03717 to Bradi2g04380) (SI Appendix, Table S2) within the interval. Using recombinant screens in 2,172 gametes (vrs4.k × Golden Promise population), we further refined the interval and mapped vrs4 to a single BAC contig (1,073 kb) containing at least six predicted genes, including barley RAMOSA2 (HvRA2), RESURRECTION1 (RST1), TRYPTOPHAN AMINO TRANSFERASE (TPA), EMBRYO SAC/BASAL ENDOSPERM TRANSFER LAYER/EMBRYO SURROUNDING REGION (EBE), STACHYOSE SYNTHASE, and RECEPTOR LIKE PROTEIN KINASE (INRPK1) (Fig. 3D). Of the six predicted genes, we considered HvRA2 as an interesting candidate gene for the Vrs4 locus, because it was identified as being essential for imposing determinacy on SPM identity in maize (3).
Fig. 3.
High-resolution linkage and physical map of vrs4 and analysis of vrs4 mutants. (A–C) Rice (A) and Brachypodium (B) chromosomal regions syntenic with barley chromosome 3H (C) in the vrs4 region. Predicted genes in rice, Brachypodium and barley chromosomes are indicated by ovals (Gene names start with Os01g (rice) or Bradi2g (Brachypodium) followed by five digit number). Solid black ovals mark the genes from which markers were derived for mapping in the vrs4 region. Numbers of recombinants are indicated between mapped markers on the barley chromosome. (D) Single BAC contig sequenced in the vrs4 region (22 overlapping BAC clones covering 1,073 kb) (see SI Appendix, Table S5 for corresponding BAC names). Predicted genes identified from the BAC sequences are indicated as circles. (E) HvRA2 gene structure showing a distinct grass-specific domain (red box), and RA2 as well as the LOB domains. Lesions in 18 ORF-mutant alleles are indicated below the gene structure (see also SI Appendix, Fig. S5). :, Nucleotide substitution leading to premature stop codon; *, INDELS leading to frame shift; >, nonsynonymous SNP in the coding region.
Because vrs4 mutants in general showed loss of determinacy of TSMs, we sequenced HvRA2 in 20 vrs4 mutants, and found that 18 showed molecular lesions within the HvRA2 ORF (Fig. 3E and SI Appendix, Fig. S5). It had been proposed that the number of meristem primordia giving rise to spikelets is partially unrestricted in vrs4 mutants resulting in formation of additional spikelets/florets (7). From the mutant analysis, we found that irrespective of the type of lesion in various mutant alleles of vrs4, all showed indeterminacy of the TSM to a certain degree (Fig. 1 H–J and SI Appendix, Fig. S3). The molecular nature of the lesions in mutants clearly correlated with the severity of vrs4 phenotype. Five mutants with the strongest spikelet phenotype had either a premature stop codon [BW-NIL(vrs4.k), BW-NIL(mul1.a), vrs4.l (syn. Xc 41.5), int-e.26 and int-e.128 (syn. hex-v.48)] or a putative gene deletion (MHOR 318 and MHOR 345; Figs. 1 H–J and 3E and SI Appendix, Fig. S3). Weaker alleles possessed amino acid substitutions either within the LOB domain [int-e.23, BW-NIL(int-e.58), int-e.72] or in other conserved regions (see below) of the protein (int-e.65, int-e.66, int-e.87, int-e.89, int-e.90, int-e.91, int-e.92, int-e.101) (Fig. 3E and SI Appendix, Fig. S3). The vrs4 mutants int-e.4 and int-e.20 did not show any lesions, suggesting possible transcriptional or posttranscriptional regulation in these alleles (allelism data for int-e.4 and int-e.20 with vrs4 are in SI Appendix). The collection of 18 ORF mutants strongly indicated that HvRA2 underlies the Vrs4 locus.
HvRAMOSA2 Encodes a LOB Domain Transcription Factor with Grass-Specific Domains.
HvRA2 is a class I LOB domain protein consisting of a cysteine-rich repeat (CX2CX6CX3C), a highly conserved GAS block, and a Leucine-zipper coiled-coil motif (LX6LX3LX6L) (SI Appendix, Fig. S6), signature features conserved in all class I LOB domain proteins (12). Apart from the canonical LOB domain, grasses (monocots) posses a unique C-terminal putative activation domain (44 aa) that is not present in Arabidopsis and other eudicots (3) (SI Appendix, Fig. S6). Based on sequence alignments generated from orthologous/homologous RA2 proteins from a range of monocot and eudicot species, we identified another conserved grass specific N-terminal region (16 aa) preceding the LOB domain (Fig. 3E and SI Appendix, Fig. S6). The last four amino acids of the N-terminal domain (PGAG) are strictly conserved across different grass species. Interestingly, seven of the vrs4 mutants possessed amino acid substitutions in the last four amino acids of the conserved N-terminal region, signifying its putative functional role in grasses. Phylogenetic analysis of RA2 homologs/orthologs in eudicots and monocots grouped them into monocot- and eudicot-specific clades (SI Appendix, Fig. S7). Among 23 LOB domain-encoding genes (LBD; 19 class I and four class II) from barley (SI Appendix, Table S3), HvRA2 was unique with conserved N- and C-terminal domains and clearly separated from other barley and eudicot LBD family members (SI Appendix, Fig. S7 and Table S3).
Resequencing of HvRA2 in diverse Barley Genotypes Shows Reduced Nucleotide Diversity.
In an attempt to identify row-type-specific alleles at Vrs4 in natural variation, we conducted sequence analysis of the HvRA2 ORF and parts of the 5′ and 3′ regulatory regions (promoter, UTRs) in a set of 77 diverse two-rowed and six-rowed barley genotypes (SI Dataset S1C). Unlike Vrs1 and Int-c, which showed row-type-specific alleles in natural variation (9, 10) Vrs4 appeared to be conserved across diverse two-rowed and six-rowed germplasm, revealing very low natural nucleotide variation in the coding region (SI Dataset S1C). Haplotype analysis using these sequence data resulted in one major and seven minor nucleotide haplotypes (GenBank accession nos. KC854546 to KC854553) without specificity toward a particular geographic region or row-type (SI Appendix, Fig. S8 and SI Dataset S1C). Among the eight nucleotide haplotypes identified, only one haplotype comprising of the two-rowed genotype, Palmella Blue, coded for an amino acid substitution in an unconserved region of the protein without showing any effect on wild-type phenotype (see SI Dataset S1C for protein sequence alignment of nucleotide haplotypes). We found complete conservation in the 3′UTR, suggesting that it may be a target site for cis-regulatory elements or involved in mRNA metabolism.
HvRA2 Is Expressed Early in Spike Development and Functions Upstream of Vrs1.
In situ hybridization analyses revealed that the first expression of HvRA2 was detected during the double ridge stage, when spikelet primordia begin to differentiate (Fig. 2E), making HvRA2 one of the earliest expressed genes during AM differentiation. At triple-mound stage, when spikelet primordia differentiate into three distinct spikelet meristems, HvRA2 mRNA signals were abundant all over the lateral spikelet primordia with weaker expression in the central spikelet primordia (Fig. 2F), indicating a role in providing determinacy and confinement to the TSM. At glume primordium stage, during which floret primordia initiate, HvRA2 mRNAs were detected in both central and lateral spikelets (Fig. 2G). Transcript levels of HvRA2 in developing spikes at triple mound and glume primordium stages were relatively higher than during later stages (Fig. 2I).
Analyzing different six-rowed mutants, Sakuma et al. noticed that Vrs1 transcripts were particularly down-regulated in the vrs4 mutant background (13). We also found that Vrs1 transcripts in BW-NIL(vrs4.k) were significantly lowered at different spike developmental stages (Fig. 2J). Unlike HvRA2 (Fig. 2E), Vrs1 expression in wild-type plants starts at triple-mound stage with no detectable expression during double ridge stage (9). Thus, HvRA2 expression is temporally ahead of Vrs1. Furthermore, HvRA2 transcripts at triple-mound stage were mainly localized to lateral spikelets (Fig. 2F), suggesting an expression domain overlap between VRS1 and HvRA2 proteins (Fig. 2K). Consistent with this, most of the vrs4 mutants showed a complete six-rowed phenotype similar to vrs1 mutants (Fig. 1 H–J and SI Appendix, Fig. S3), indicating that Vrs1 transcripts require HvRA2 action. Double mutant analysis supported this view, because vrs4 allele BW-NIL(mul1.a), isolated in a six-rowed background (progenitor: Montcalm, vrs1.a1), displayed the six-rowed spike with complete lateral spikelet fertility, as observed in Montcalm, and also indeterminacy of the TSM, diagnostic of vrs4 mutants (Fig. 2L).
Microarray Analysis of vrs4 Reveals HvRA2 as an Important Regulator of Inflorescence Development.
Because Vrs4 encodes a LOB domain transcription factor HvRA2, we performed microarray analysis in two vrs4 deletion mutants, MHOR 318 and MHOR 345, and their respective wild types, Ackermann’s Donaria and Heine’s Haisa, to identify its potential downstream target genes in barley. From the microarray data, we found compelling evidence for Vrs4-mediated regulation of Vrs1 in both mutants analyzed, with highly significant down-regulation of Vrs1 in vrs4 mutants (SI Appendix, Fig. S9A). Importantly, among other genes significantly down-regulated in both mutants, we identified trehalose-6-phosphate (T6P) synthase and HvSRA (a putative T6P phosphatase) (SI Appendix, Fig. S9A), both involved in trehalose biosynthesis, implying possible regulation of T6P homeostasis by Vrs4 during inflorescence development and growth (14, 15). From the microarray expression data, we identified other significant differentially regulated genes in both vrs4 mutants that might likely function in establishing row-type and spikelet determinacy in barley.
The other important differentially regulated gene in both vrs4 mutants was EGG APPARATUS1-LIKE (EA1-LIKE). EA1 encodes a secretary protein that attracts pollen tube growth toward egg apparatus (16). The significant down-regulation of the EA1-LIKE gene in wild-type two-rowed inflorescence (SI Appendix, Fig. S9B) suggests that both Vrs1 and EA1-LIKE may function in a similar pathway. Consistent with this phenomenon, Vrs1 transcripts show localization to pistils of lateral spikelets in barley (13). Alternatively, elevated expression of EA1-LIKE could be due to the proliferating additional spikelets/florets in vrs4 mutants.
The differentially regulated genes likely involved in enhanced meristematic activity of vrs4 mutants include LONELYGUY-LIKE (LOG-LIKE), which encodes cytokinin nucleotide phosphoribohydrolase, a cytokinin biosynthesis enzyme, required to maintain meristem activity (17). Up-regulation of LOG-LIKE genes in vrs4 mutants indicated that higher amounts of LOG-LIKE transcripts promoted meristematic activity in vrs4 mutants (SI Appendix, Fig. S9B). LOG1 is required for the accumulation of KNOTTED1-type homeobox (KNOX) transcripts in rice (17). Thus, overproduction of cytokinin results in increased steady state levels of KNOX gene transcripts (18). Conversely, KNOX proteins were shown to activate cytokinin biosynthesis in the shoot apical meristem of Arabidopsis and rice (19, 20). Thus, KNOX and cytokinin may act in a positive feedback loop, with KNOX genes promoting cytokinin biosynthesis and cytokinins up-regulating KNOX biosynthesis. Our data showed up-regulation of KNAT3 (KNOX) in vrs4 mutants (SI Appendix, Fig. S9B), suggesting a possible coregulation of LOG-LIKE and KNOX proteins in vrs4 mutants. The microarray analysis for these differentially regulated genes is supported by quantitative RT-PCR data (SI Appendix, Fig. S10).
Independent quantitative RT-PCR analyses of a few important genes (not present on the microarray and identified based on extrapolation of vrs4 mutant phenotype with known mutant inflorescence architecture genes characterized in rice or maize) revealed differential regulation for cytokinin oxidase/dehydrogenase (CKX2), which is involved in cytokinin degradation and meristem size (21). HvCKX2 transcript levels in BW-NIL(vrs4.k) and MHOR 318 were lowered (SI Appendix, Fig. S11), consistent with higher meristematic activity in the mutant inflorescence. Transcripts of another gene, PINFORMED1-LIKE (PIN1-LIKE) (22), were also up-regulated in the vrs4 mutants, suggesting that altered auxin transport affected lateral organ development in vrs4 mutants (SI Appendix, Fig. S11).
Discussion
The regulation of inflorescence development has been broadly elucidated in plant species, such as Arabidopsis and maize (23), with the help of a wealth of mutants that display abnormal inflorescence development. However, in barley, functional knowledge of genes and genetic mechanisms involved in spike development has only started to emerge relatively recently, with the cloning of genes such as Vrs1 (9) and Int-c (10). Mutants of vrs4, like other barley row-type mutants, produce a six-rowed phenotype, but the most interesting phenotype is the indeterminate TSM, resulting in the formation of additional spikelets/florets that frequently develop into grains. Thus, actions of vrs4, possibly along with other as of yet unknown genes, may help elucidating how to increase yield potential in barley and other Triticeae species.
Our resequencing data showed very limited natural variation in HvRA2 across diverse barley germplasm with complete conservation in the 3′UTR. A low level of polymorphisms in the 3′ regulatory region was also observed in maize domestication loci RA1 (24) and GRASSY TILLERS1 (25). We hypothesize that naturally occurred variations at this locus might have undergone purifying selection in two- and six-rowed barleys, as severe mutations would disrupt determinacy of the TSM, leading to disordered spike formation. However, weaker natural mutant alleles of vrs4 might still be hidden among intermedium barleys, which represent a large and diverse phenotypic row-type class within cultivated barley.
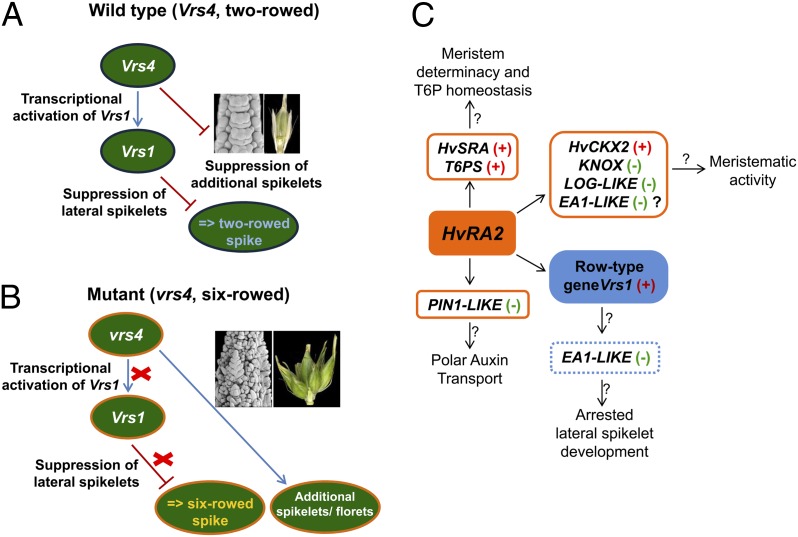
Another important finding from the present study is the transcriptional regulation of Vrs1 by HvRA2. Our microarray analysis confirmed that Vrs1 is significantly down-regulated in vrs4 mutants. The HvRA2 mRNA in situ hybridization results showed that HvRA2 and Vrs1 are expressed in highly overlapping domains of lateral spikelets, suggesting a putative interaction. Circumstantial evidence supporting this view came from the LOB domain consensus DNA recognition motif 5′-CCGGCG-3′ (26) present in the Vrs1 promoter region close to the transcriptional start site (−60 to −65 bp) and also in the 5′UTR. Previous in vitro protein binding assays showed strong binding affinity of LOB domain proteins to this consensus motif (26). However, the physical interaction between HvRA2 and Vrs1 cis elements needs to be established. Clearly, reduction of Vrs1 transcripts in vrs4 mutants indicated that HvRA2 directly or indirectly controls Vrs1 transcripts, and thus the row-type pathway (Fig. 4 A and B).
Fig. 4.
Models of Vrs4 interactions showing HvRA2 putative targets. (A) Functional Vrs4 suppresses additional spikelet formation and activates Vrs1 transcription. (B) Mutant vrs4 cannot control determinacy of the TSM; thus, additional spikelets are formed. Transcriptional activation of Vrs1 expression is omitted (red cross) resulting in lateral spikelet fertility. (C) HvRA2 may regulate transcripts of T6P synthase (T6PS) and HvSRA, a putative T6P phosphatase, thereby maintaining T6P homeostasis and spikelet determinacy. In Vrs4, transcript levels of PIN1-LIKE are maintained at constant level but are up-regulated in vrs4. In wild-type plants, HvRA2 functions upstream of HvHox1, which in turn may down-regulate transcripts of EA1-LIKE resulting in abortion of lateral spikelets, possibly producing a two-rowed spike (hypothetical). However, up-regulation of EA1-LIKE in vrs4 mutants may likely be due to enhanced meristematic activity related to the six-rowed spike phenotype and additional spikelets/florets. Transcript levels of genes involved in meristematic activity [e.g., cytokinin oxidase (CKX2), LONELY GUY-LIKE, and KNOX genes] are also regulated by HvRA2.
In maize, inflorescence morphology is largely governed by the RAMOSA pathway under the coordinated regulation of RA1, RA2, and RA3. Maize RA3 shows a highly localized expression pattern at the base of inflorescence branches and encodes a functional T6P phosphatase, suggesting a role for trehalose signaling in meristem determinacy (5). A clear ortholog of RA3 is not present in barley. However, a homolog of RA3, termed SISTER OF RAMOSA3 (SRA), has been identified in barley (HvSRA) and related grasses (5). The rice homolog of RA3 (OsSRA) also showed highly localized expression pattern at the base of inflorescence branches (5), suggesting a role in inflorescence patterning. In maize, RA1 transcripts are under the control of RA2 and RA3, whereby RA2 controls the fate of SPMs through the RA1–REL2 determinacy complex (4). The SPM is specific to the tribe Andropogoneae, as is RA1 (1, 2). Thus, HvRA2 seemed to have maintained a conserved function (control over meristem determinacy), but diversified the downstream targets for executing this function in barley (as the ortholog/homolog of RA1 is missing in barley). In the large barley mutant collection, several spike mutants for the multifloret or branched spike, such as extra floret (flo)-a, flo-b, flo-c, compositum1 (com1), com2, and multiflorus2 (mul2), were reported, but the underlying genes for these mutants have still not been identified. These loci could be potential candidate targets of Vrs4 for conferring spikelet determinacy. Although RA2 and RA3 act independently in maize, we found that HvRA2 may regulate transcripts of HvSRA and T6P synthase, which in turn may regulate specific growth responses through synthesis of T6P (Fig. 4C) (27). Moreover, HvRA2 coopted a genus-specific function for the row-type pathway through regulation of Vrs1 (control over lateral spikelet fertility), thereby clearly indicating different gene sets and networks for inflorescence development compared with maize (Fig. 4C). In addition, we showed transcript differences for HvCKX2 and auxin transporters in vrs4 mutants, further suggesting a role in inflorescence growth and development (Fig. 4C).
Taken together, our results suggest that Vrs4 is a central player in establishing inflorescence architecture of barley spikes, as well as in determining yield potential and grain number. Thus, a better understanding of the underlying gene regulatory networks during spike formation may help to improve future grain yields of small-grain cereals.
Materials and Methods
Plant Material.
Allelic vrs4 mutants (SI Appendix, Table S4) were obtained from the Nordic Genetic Resource Center, the National Small Grains Collection (US Department of Agriculture), and the IPK gene bank. Seventy-seven diverse barley accessions were obtained from IPK gene bank for haplotype analysis. The mutant alleles vrs4.l (syn. Xc 41.5), its two-rowed progenitor Piroline, and Bowman isolines BW-NIL(vrs4.k) and BW-NIL(mul.1a) were used for phenotypic descriptions and SEM analysis. Plant material used for generating the mapping populations is discussed in SI Appendix.
Vrs4 Phenotype Characterization.
vrs4.l was selected as a reference for comparing inflorescence development with the wild-type Piroline. Piroline and vrs4.l were grown in fields in Tsukuba, Japan, during the autumn (October 2011 to June 2012) and spring (March 2012 to June 2012) seasons. For the analysis shown in Fig. 1O and SI Dataset S1A, data were recorded in five plants (two spikes per plant) from each of the genotypes. Probability values were determined using t test. The number of rachis internodes and number of seeds produced by mutant and wild type were counted. A spikelet triplet consisting of three spikelets was considered a basic set of axillary structures and the remaining axillary structures were considered additional axillary structures. Each triplet or multispikelet was divided into one central and two lateral units, and the number of additional spikelets/florets was counted in each unit. An additional axillary structure was counted as a supernumerary floret when the rachilla of the first axillary structure was not visible. The total number of branch-like structures and additional lemma-like structures produced by a spike were counted. Fertility of axillary structure was scored by counting the number of seeds produced by each type of axillary structure.
Marker Development.
For initial mapping, BOPA SNP markers (28) or SSR and EST based markers (SI Dataset S1B) were selected from the high-density transcript map of barley chromosome 3H (29). For further marker development in the defined vrs4 interval, we relied on the barley genome zipper (11). Gene sequences from the syntenic interval in rice or Brachypodium were extracted from respective genome browser servers (http://rice.plantbiology.msu.edu/cgi-bin/gbrowse/rice/ and http://jbrowse.brachypodium.org/JBrowse.html). The obtained gene sequences were BLASTed against the IPK Barley BLAST server (http://webblast.ipk-gatersleben.de/barley/viroblast.php) to obtain barley sequences. SNP polymorphisms identified from the primers designed were converted to restriction enzyme based CAPS (30) (http://tools.neb.com/NEBcutter2/) or dCAPS (31) markers (http://helix.wustl.edu/dcaps/).
Quantitative RT-PCR.
Total RNA was extracted from immature spike tissues (double ridge, triple mound, glume primordium, lemma primordium, stamen primordium, and awn primordium stages) using the PureLink RNA Mini kit (Invitrogen). RNase-free DNase (Invitrogen) was used to remove genomic DNA contamination. The RNA integrity and quantities were measured using the Agilent Bioanalyzer and NanoDrop (Peq Lab), respectively. Reverse transcription and cDNA synthesis were carried out with 1 µg of RNA using the QuantiTect Reverse Transcription kit (Qiagen). Real-time PCR was performed using the QuantiTect SYBR Green PCR kit (Qiagen) and the ABI Prism 7900HT sequence detection system (Applied Biosystems). RT-PCR results were analyzed using SDS2.2 software (Applied Biosystems). Quantitative RT-PCR primer sequences are listed in SI Dataset S1B.
mRNA in Situ Hybridization.
The Vrs4 gene segment comprising parts of the first and second exons (395 bp) was amplified from cDNA isolated from cv. Bonus immature spikes using specific primers (SI Dataset S1B). The PCR product was cloned into the pBluescript II KS (+) vector (Stratagene). Clones linearized by EcoRI or NotI were used as templates to generate antisense (EcoRI) and sense (NotI) probes using T3 or T7 RNA polymerase. In situ hybridization was conducted as described (9).
BAC Sequencing, Assembly, and Annotation.
A single BAC contig spanning 1,073 kb of the minimum tiling path of barley chromosome 3H was shotgun sequenced using 454 technology (SI Appendix, Table S5) (32) and assembled as described (33). ORFs from the BAC sequences were predicted using the Genescan web server (34). The predicted genes were annotated using the BLASTX function of the National Center for Biotechnology Information (NCBI).
Phylogenetic Analysis.
Barley LBD genes were extracted from the IPK barley BLAST server using HvRA2 as a query sequence. HvRA2 protein was used as NCBI BLASTp query to retrieve RA2 and RA2-LIKE proteins (E-value cutoff 10-30) from monocots and eudicots, respectively (SI Appendix, Table S6). For phylogenetic analysis, the protein sequences were initially aligned using the MUSCLE algorithm implemented in MEGA 5.1 (35). A maximum likelihood (ML) phylogenetic tree was constructed using the ML heuristic method Nearest Neighbor Interchange (NNI) implemented in MEGA 5.1. The bootstrap consensus tree inferred from 1,000 replicates is taken to represent the evolutionary history of the sequences analyzed. Branches corresponding to partitions reproduced in less than 50% bootstrap replicates are collapsed.
Scanning Electron Microscopy.
Immature spike tissues at five stages (triple mound, glume, lemma, stamen, and awn primordium) from field-grown as well as greenhouse-grown plants were used for scanning electron microscopy (SEM). SEM was conducted as described (36).
Supplementary Material
Acknowledgments
We thank Dr. S. R. Palakolanu (International Crops Research Institute for the Semi-Arid Tropics) for help with phylogenetic analysis; Dr. B. Kilian (IPK Gatersleben) for providing germplasm for haplotype analysis; H. Bockelman (US Department of Agriculture–Agricultural Research Service) and IPK gene bank for providing initial mutant germplasm; I. Walde for help with the VeraCode experiment; P. Gawroński for helpful discussions; and H. Ernst, H. Koyama, S. König, K. Lipfert, M. Püffeld, M. Pürschel, C. Trautewig, and C. Weißleder for excellent technical support. This work was supported by grants from the Ministry of Education (IZN), Saxony-Anhalt (to N. Sreenivasulu), the Ministry of Agriculture, Forestry, and Fisheries of Japan (Genomics for Agricultural Innovation Grant TRG1004; to T.K.), the Deutsche Forschungsgemeinschaft (DFG Grant SCHN 768/2-1; to T.S.), and the BMBF (German Federal Ministry of Education and Research, GABI-FUTURE Start_Young Investigator Program Grant 0315071; to T.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos.: KC854546–KC854554).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1221950110/-/DCSupplemental.
References
- 1.Kellogg EA. Floral displays: Genetic control of grass inflorescences. Curr Opin Plant Biol. 2007;10(1):26–31. doi: 10.1016/j.pbi.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 2.Vollbrecht E, Springer PS, Goh L, Buckler ES, Martienssen R. Architecture of floral branch systems in maize and related grasses. Nature. 2005;436(7054):1119–1126. doi: 10.1038/nature03892. [DOI] [PubMed] [Google Scholar]
- 3.Bortiri E, et al. ramosa2 encodes a LATERAL ORGAN BOUNDARY domain protein that determines the fate of stem cells in branch meristems of maize. Plant Cell. 2006;18(3):574–585. doi: 10.1105/tpc.105.039032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gallavotti A, et al. The control of axillary meristem fate in the maize ramosa pathway. Development. 2010;137(17):2849–2856. doi: 10.1242/dev.051748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Satoh-Nagasawa N, Nagasawa N, Malcomber S, Sakai H, Jackson D. A trehalose metabolic enzyme controls inflorescence architecture in maize. Nature. 2006;441(7090):227–230. doi: 10.1038/nature04725. [DOI] [PubMed] [Google Scholar]
- 6.Sreenivasulu N, Schnurbusch T. A genetic playground for enhancing grain number in cereals. Trends Plant Sci. 2012;17(2):91–101. doi: 10.1016/j.tplants.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 7.Forster BP, et al. The barley phytomer. Ann Bot (Lond) 2007;100(4):725–733. doi: 10.1093/aob/mcm183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mansfeld R. Das morphologische System der Saatgerste, Hordeum vulgare L.s.l. Der Züchter. 1950;20:8–24. [Google Scholar]
- 9.Komatsuda T, et al. Six-rowed barley originated from a mutation in a homeodomain-leucine zipper I-class homeobox gene. Proc Natl Acad Sci USA. 2007;104(4):1424–1429. doi: 10.1073/pnas.0608580104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramsay L, et al. INTERMEDIUM-C, a modifier of lateral spikelet fertility in barley, is an ortholog of the maize domestication gene TEOSINTE BRANCHED 1. Nat Genet. 2011;43(2):169–172. doi: 10.1038/ng.745. [DOI] [PubMed] [Google Scholar]
- 11.Mayer KF, et al. Unlocking the barley genome by chromosomal and comparative genomics. Plant Cell. 2011;23(4):1249–1263. doi: 10.1105/tpc.110.082537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shuai B, Reynaga-Peña CG, Springer PS. The lateral organ boundaries gene defines a novel, plant-specific gene family. Plant Physiol. 2002;129(2):747–761. doi: 10.1104/pp.010926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sakuma S, et al. Divergence of expression pattern contributed to neofunctionalization of duplicated HD-Zip I transcription factor in barley. New Phytol. 2013;197(3):939–948. doi: 10.1111/nph.12068. [DOI] [PubMed] [Google Scholar]
- 14.Schluepmann H, Berke L, Sanchez-Perez GF. Metabolism control over growth: A case for trehalose-6-phosphate in plants. J Exp Bot. 2012;63(9):3379–3390. doi: 10.1093/jxb/err311. [DOI] [PubMed] [Google Scholar]
- 15.Eveland AL, Jackson DP. Sugars, signalling, and plant development. J Exp Bot. 2012;63(9):3367–3377. doi: 10.1093/jxb/err379. [DOI] [PubMed] [Google Scholar]
- 16.Márton ML, Cordts S, Broadhvest J, Dresselhaus T. Micropylar pollen tube guidance by egg apparatus 1 of maize. Science. 2005;307(5709):573–576. doi: 10.1126/science.1104954. [DOI] [PubMed] [Google Scholar]
- 17.Kurakawa T, et al. Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature. 2007;445(7128):652–655. doi: 10.1038/nature05504. [DOI] [PubMed] [Google Scholar]
- 18.Rupp H-M, Frank M, Werner T, Strnad M, Schmülling T. Increased steady state mRNA levels of the STM and KNAT1 homeobox genes in cytokinin overproducing Arabidopsis thaliana indicate a role for cytokinins in the shoot apical meristem. Plant J. 1999;18(5):557–563. doi: 10.1046/j.1365-313x.1999.00472.x. [DOI] [PubMed] [Google Scholar]
- 19.Yanai O, et al. Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Curr Biol. 2005;15(17):1566–1571. doi: 10.1016/j.cub.2005.07.060. [DOI] [PubMed] [Google Scholar]
- 20.Sakamoto T, et al. Ectopic expression of KNOTTED1-like homeobox protein induces expression of cytokinin biosynthesis genes in rice. Plant Physiol. 2006;142(1):54–62. doi: 10.1104/pp.106.085811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ashikari M, et al. Cytokinin oxidase regulates rice grain production. Science. 2005;309(5735):741–745. doi: 10.1126/science.1113373. [DOI] [PubMed] [Google Scholar]
- 22.Gälweiler L, et al. Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science. 1998;282(5397):2226–2230. doi: 10.1126/science.282.5397.2226. [DOI] [PubMed] [Google Scholar]
- 23.Malcomber ST, Preston JC, Reinheimer R, Kossuth J, Kellogg EA. Developmental gene evolution and the origin of grass inflorescence diversity. Adv Bot Res. 2006;44:425–481. [Google Scholar]
- 24.Sigmon B, Vollbrecht E. Evidence of selection at the ramosa1 locus during maize domestication. Mol Ecol. 2010;19(7):1296–1311. doi: 10.1111/j.1365-294X.2010.04562.x. [DOI] [PubMed] [Google Scholar]
- 25.Whipple CJ, et al. grassy tillers1 promotes apical dominance in maize and responds to shade signals in the grasses. Proc Natl Acad Sci USA. 2011;108(33):E506–E512. doi: 10.1073/pnas.1102819108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Husbands A, Bell EM, Shuai B, Smith HM, Springer PS. LATERAL ORGAN BOUNDARIES defines a new family of DNA-binding transcription factors and can interact with specific bHLH proteins. Nucleic Acids Res. 2007;35(19):6663–6671. doi: 10.1093/nar/gkm775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eveland AL, et al. Digital gene expression signatures for maize development. Plant Physiol. 2010;154(3):1024–1039. doi: 10.1104/pp.110.159673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Close TJ, et al. Development and implementation of high-throughput SNP genotyping in barley. BMC Genomics. 2009;10:582. doi: 10.1186/1471-2164-10-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sato K, Nankaku N, Takeda K. A high-density transcript linkage map of barley derived from a single population. Heredity (Edinb) 2009;103(2):110–117. doi: 10.1038/hdy.2009.57. [DOI] [PubMed] [Google Scholar]
- 30.Vincze T, Posfai J, Roberts RJ. NEBcutter: A program to cleave DNA with restriction enzymes. Nucleic Acids Res. 2003;31(13):3688–3691. doi: 10.1093/nar/gkg526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neff MM, Turk E, Kalishman M. Web-based primer design for single nucleotide polymorphism analysis. Trends Genet. 2002;18(12):613–615. doi: 10.1016/s0168-9525(02)02820-2. [DOI] [PubMed] [Google Scholar]
- 32.Meyer M, Stenzel U, Hofreiter M. Parallel tagged sequencing on the 454 platform. Nat Protoc. 2008;3(2):267–278. doi: 10.1038/nprot.2007.520. [DOI] [PubMed] [Google Scholar]
- 33.Steuernagel B, et al. De novo 454 sequencing of barcoded BAC pools for comprehensive gene survey and genome analysis in the complex genome of barley. BMC Genomics. 2009;10:547. doi: 10.1186/1471-2164-10-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 1997;268(1):78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- 35.Tamura K, et al. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lolas IB, et al. The transcript elongation factor FACT affects Arabidopsis vegetative and reproductive development and genetically interacts with HUB1/2. Plant J. 2010;61(4):686–697. doi: 10.1111/j.1365-313X.2009.04096.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.