Abstract
In cell division, cytokinesis is tightly coupled with mitosis to maintain genomic integrity. Failed cytokinesis in humans can result in tetraploid cells that can become aneuploid and promote cancer. However, the likelihood of aneuploidy and cancer after a failed cytokinesis event is unknown. Here we evaluated cell fate after failed cytokinesis. We interrupted cytokinesis by brief chemical treatments in cell populations of human epithelial lines. Surprisingly, up to 50% of the resulting binucleate cells generated colonies. In RPE1 cells, 90% of colonies obtained from binucleate founders had a karyotype that matched the parental cell type. Time-lapse videomicroscopy demonstrated that binucleate cells are delayed in the first growth phase of the cell cycle (G1) and undergo interphase cellular fission (cytofission) that distributes nuclei into separate daughters. The fission is not compatible with delayed cytokinesis because events occur in the absence of polymerized microtubules and without canonical components of the cytokinetic machinery. However, the cytofission can be interrupted by inhibiting function of actin or myosin II. Fission events occur in both two- and three-dimensional culture. Our data demonstrate that cytofission can preserve genomic integrity after failed cytokinesis. Thus, traction-mediated cytofission, originally observed in Dictyostelium, is relevant to human biology—where it seems to be an evolutionarily conserved mechanism that can preserve genomic integrity.
A century ago, Theodor Boveri generated tetrapolar cell divisions in sea urchin eggs and observed that aberrant chromosome distribution led to distinct fates of daughter cells (1). This led to his influential hypothesis that suppression of human cell division in midstream (i.e., failed cytokinesis) generates aneuploidy and promotes cancer. Today there is considerable evidence to support the idea that aneuploidy can be oncogenic (2–5). Cytokinesis can fail by any of several mechanisms: by prolonged activation of the spindle checkpoint (6, 7), by defects in the cytokinetic apparatus (8), or by mechanical obstruction of furrow ingression by chromosomes or engulfed cells (9–11). Interruption of cytokinesis can yield tetraploid progeny that exhibit chromosomal instability and have increased propensity for malignant transformation (12, 13). However, it is unclear how often a single failed cytokinesis event results in lost genomic integrity. To evaluate this, we have directly assessed the fate of individual human epithelial cells after failed cytokinesis.
Here we report the surprising discovery that human cells commonly recover the original chromosome complement after failed cytokinesis by interphase cytofission. Our results demonstrate that human epithelial cells have a fail-safe mechanism that allows recovery of genomic integrity after failed cytokinesis. This unexpected mechanism may be crucial to safeguard the genome during the ∼1013 cell divisions required for human development.
Results and Discussion
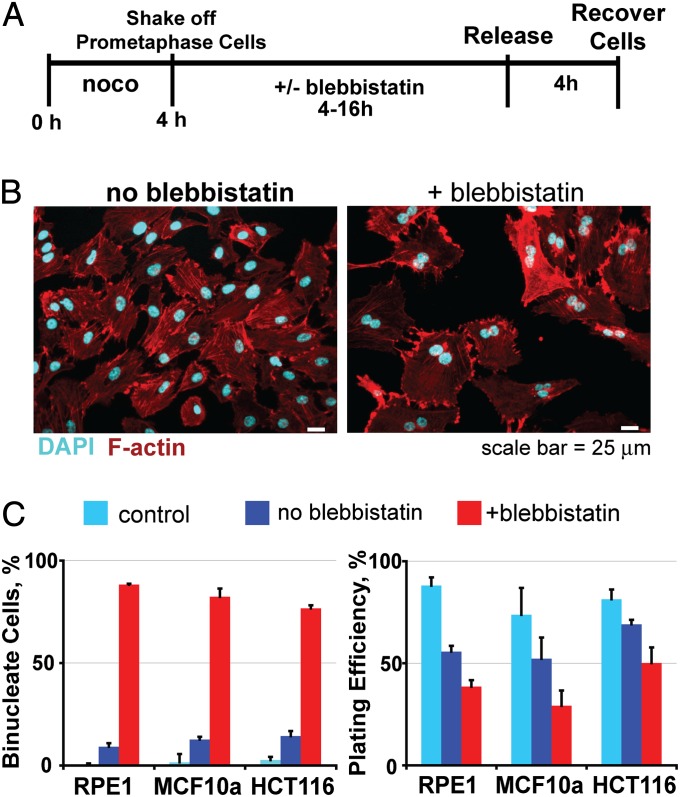
To determine the fate of human epithelial cells after failed cytokinesis, we selected three diverse, near-diploid human cell types: hTERT-immortalized retinal pigment epithelial cells (RPE1), MCF10a breast epithelial cells, and HCT116 colorectal cancer cells. We generated homogenous populations of cells that had executed karyokinesis but failed cytokinesis (Fig. 1 A–C). To do this, cells were synchronized by shake-off of loosely adherent mitotic cells after 4 h of nocodazole treatment. This brief treatment was selected to minimize DNA damage that can occur because of prolonged mitotic arrest (14, 15). After collection, mitotic cells were split into two fractions, and cytokinesis was blocked in one fraction by treatment with blebbistatin, an inhibitor of myosin II (16). Shortly after treatment, more than 80% of blebbistatin-treated cells were binucleate, whereas most control cells not exposed to blebbistatin each had one nucleus (Fig. 1 B and C). Thus, failed cytokinesis produced a population of cells highly enriched for binucleates.
Fig. 1.
Human cells are viable after failed cytokinesis. (A) Procedure to derive a homogenous population of human cells that failed cytokinesis. (B) Representative images of cells after mitotic exit in the absence or presence of the myosin II inhibitor, blebbistatin. After failed cytokinesis, most cells have two nuclei. (C) (Left) Quantification of mononucleate and binucleate cells in each condition before subcloning (±SEM, n = 3 of 100 cells each). (Right) Fraction of plated cells that generated colonies after 21 d (±SEM, n = 3). Control cells were plated without any nocodazole or blebbistatin treatments.
We anticipated that binucleate cells often would be inviable owing to expected aneuploidy in some cells (1, 12) and a p53-dependent G1 arrest or apoptosis in others (17–19). To test this, we subcloned binucleate cells and compared the numbers of colonies recovered with cells plated (Fig. 1C, Right). Untreated cells (control) had a high plating efficiency in these lines, demonstrating their ability to survive subcloning. As expected, a 4-h treatment of nocodazole modestly impaired viability compared with untreated controls (dark blue bars in Fig. 1C). Surprisingly, induction of failed cytokinesis only produced a small additional reduction in plating efficiency. For RPE1 cells, binucleates maintained viability close to that of their matched counterparts, 39% ± 3% (SEM) for binucleate (+blebbistatin) vs. 56 ± 3% for mononucleate cells (no blebbistatin). We observed similar results with MCF10a and HCT116 cells (Fig. 1C) and obtained a similar 36% plating efficiency of RPE1 cells after cytokinesis was blocked by 5 μM cytochalasin D, an inhibitor of actin (20).
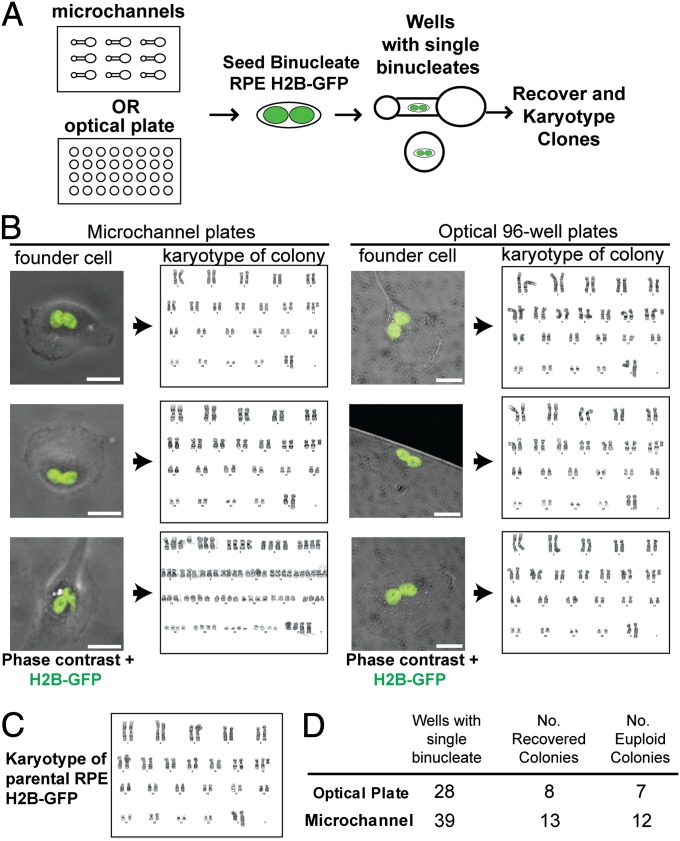
A limitation of the experiments above is that a small number of residual mononucleate cells in the subcloned populations could account for a fraction of the colonies recovered. To address this, we repeated the experiment but directly visualized binucleate cells immediately after subcloning and before allowing colonies to form (Fig. 2A). Using RPE1 cells expressing fluorescent Histone 2B (H2B-GFP), we generated binucleate cells as before and subcloned into plates by limiting dilution. We selected both microchannel plates and optical 96-well plates in parallel experiments to ensure the validity of the findings (microchannel plates allow easy visualization of entire wells; optical plates have high walls between wells, which virtually eliminate risk of well-to-well migration). After marking wells that harbored single binucleate cells, plates were incubated for 3 wk and colonies recovered from 21 of 67 binucleates (31%). To determine whether aneuploidy had developed, karyotype analysis was performed on cells from each colony (Fig. 2 B–D). Strikingly, 19 of the 21 recovered colonies had a karyotype that precisely matched that of the parental RPE1 population. Collectively the results demonstrate that diverse human epithelial cells not only can survive cytokinesis failure but also maintain a stable chromosome complement.
Fig. 2.
Binucleate cells generate euploid daughter cells. (A) Experimental design to identify cell of origin for clonal colonies. After failed cytokinesis, H2B-GFP–labeled RPE1 cells were plated into microchannels or optical-bottom plates by limiting dilution. Channels or wells that contained single binucleate cells were selected for analysis. Colonies were identified after incubation of plates for 3 wk, and karyotype analyses were performed. (B) Images of representative binucleate parental cells that grew into colonies and associated karyograms of cells derived from each clonal colony. (Scale bars, 25 μm.) One of six cells shown yielded near-tetraploid progeny; the others matched the parental. (C) Reference karyotype of H2B-GFP RPE1 cells. (D) Colony formation and number of clones that match the parental karyotype.
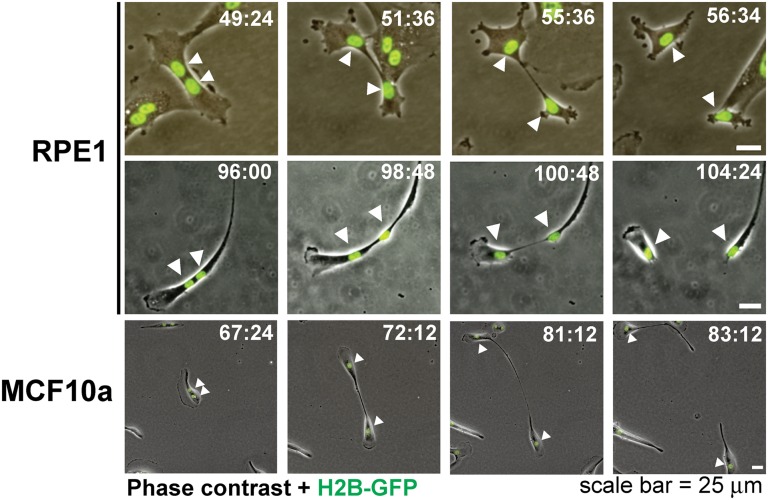
To elucidate the recovery mechanism, we performed time-lapse videomicroscopy of binucleate RPE1 and MCF10a cells expressing H2B-GFP. To eliminate the possibility of unobserved mitoses, 12-min time intervals were selected; this is much shorter than the ∼50 min typically required for mitosis in RPE1 cells (21). Initially, most cells were binucleate with closely adjacent nuclei. We observed a prolonged cell-cycle delay in these cells, as evident from low BrdU incorporation and occasional mitoses, cell stretching, and cytoplast formation (Fig. S1). Strikingly, we observed a number of cell fission events uncoupled to mitosis: cells exhibited nuclear segregation followed by cytoplasmic pinching and division into two daughters. In these cases each daughter inherited an intact nucleus (Fig. 3A and Movies S1–S3). This was observed in both cell types tested and occurred without evidence of intervening mitosis. Fission was observed in up to 2% of cells per day—sufficient to allow recovery in ∼30% of cells in 14 d. Thus, the separate nuclei can successfully segregate into daughter cells in a cytoplasmic fission uncoupled to mitosis. The intermingling of chromosomes is apparently prevented by their enclosure within separate nuclei, an elegant mechanism that immediately accounts for maintained genomic integrity after failed cytokinesis.
Fig. 3.
Cytofission separates nuclei into daughter cells. Cytofission as observed by time-lapse videomicroscopy. Still images of merged phase contrast and fluorescent H2B-GFP are shown for RPE1 and MCF10a cells exhibiting cytofission after prior failed cytokinesis. Time in hours:minutes from the Fig. 1A treatment is shown for each frame. Arrowheads indicate nuclei of cells undergoing fission.
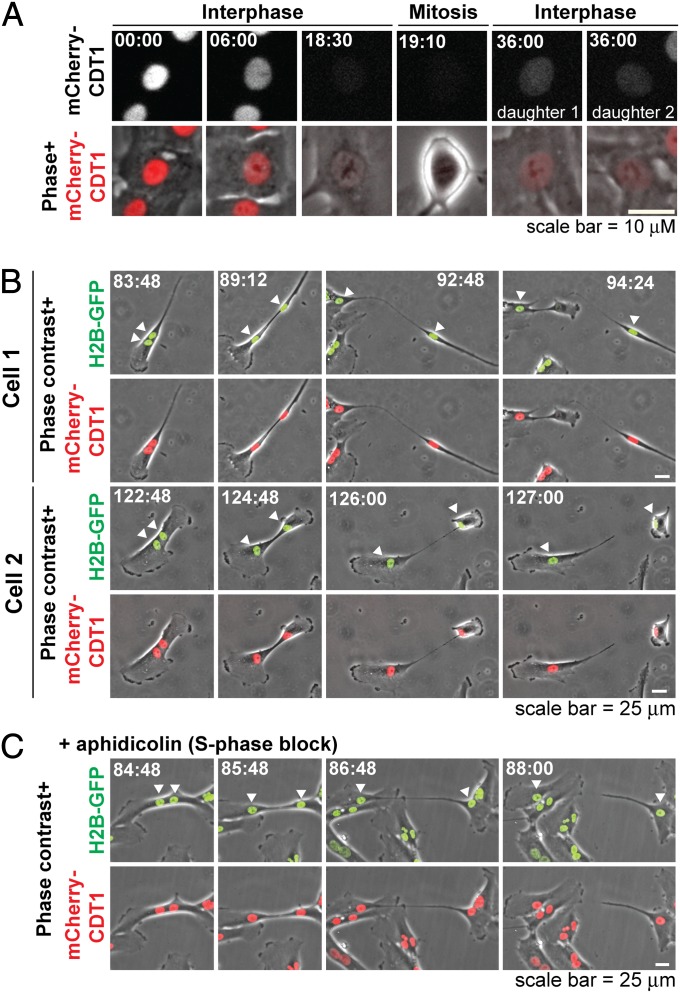
To determine when cytofission was occurring in the cell cycle, we used fluorescent mCherry fused to residues 30–120 of Chromatin licensing and DNA replication factor 1 (CDT1), an established cell-cycle indicator that appears in early G1 and is destroyed in S phase (22). In control cells, mCherry-CDT1 is lost before mitosis and reexpressed in G1 as expected (Fig. 4A). We monitored cytofission in binucleate cells labeled with both H2B-GFP and mCherry-CDT1 by time-lapse videomicroscopy (Fig. 4B and Movies S4 and S5). High mCherry-CDT1 fluorescence was observed in nine of nine cells undergoing fission, demonstrating that cytofission occurs in G1. To confirm this the experiment was repeated in the presence of aphidicolin, which blocks DNA synthesis and precludes transit through S phase (23). Consistent with the above findings, aphidicolin did not preclude cytofission (n = 5 events) or affect robust mCherry-CDT1 expression during fission (n = 2 events; Fig. 4C and Movie S6). We conclude that cytofission occurs during G1 of the cell cycle.
Fig. 4.
Cytofission occurs in G1 of the cell cycle. (A) Control cell demonstrating that mCherry-CDT1 fluorescence is lost before mitosis as expected from this cell-cycle marker. (B) Representative cytofission events in RPE1 cells demonstrating sustained mCherry-CDT1 expression during fission, indicating G1 state. (C) An example of cytofission that occurred in the presence of 5 μM aphidicolin, which precludes S-phase DNA replication. All times are in hours:minutes from blebbistatin treatment. Arrowheads indicate nuclei of cells undergoing fission.
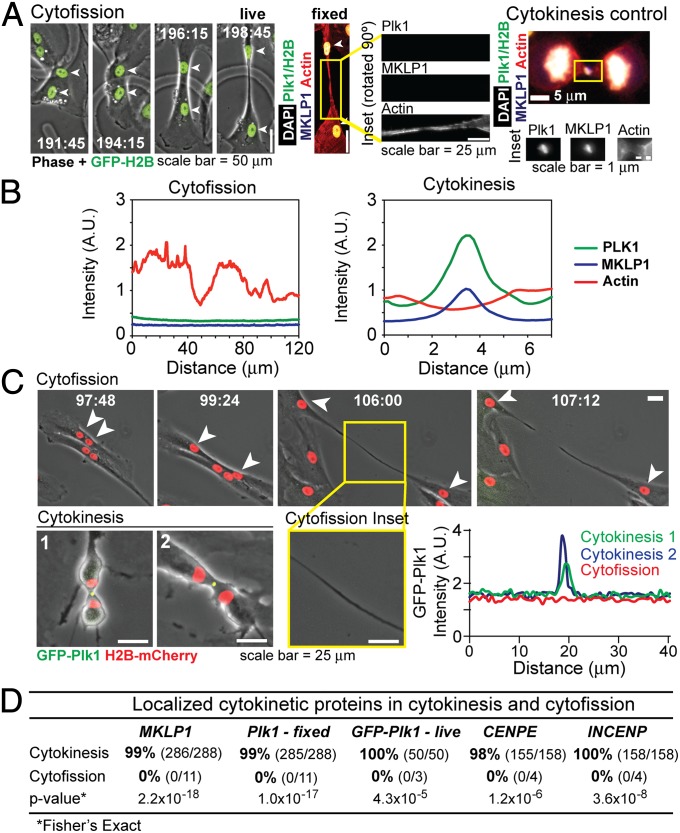
Because G1 follows mitosis, we considered that cytofission could be a manifestation of delayed cytokinesis. To test this we assessed the localization of canonical cytokinesis markers in cells undergoing fission. During cytokinesis, a central-spindle apparatus assembles at midzone microtubules to assemble an equatorial contractile actinomyosin ring (24). Upstream elements of this process include mitotic kinesin-like protein 1 (MKLP1), part of the core central spindle apparatus, and Polo-like kinase 1 (Plk1), which triggers RhoA activity through recruitment of its guanine nucleotide exchange factor (RhoGEF), Ect2 (25, 26). To evaluate localization of these proteins during cytofission, we generated binucleate cells and performed time-lapse videomicroscopy. When cells exhibited cytoplasmic bridging, cells were fixed and stained for MKLP1, Plk1, and actin (Fig. 5A, Fig. S2 and Movie S7). In control cells undergoing canonical mitotic-coupled cytokinesis, we readily observe enrichment of MKLP1 and Plk1 in early and late anaphase. However, in binucleate cells we did not observe any commensurate enrichment of Plk1 or MKLP1 along the cytoplasmic bridge (Fig. 5 A and B). Actin staining, in contrast, was variable across the bridge without a single focus of enhancement. Similarly, inner centromere protein (INCENP) and centromere-associated protein E (CENP-E), other markers of the cytokinesis spindle midzone, were absent in cells undergoing cytofission (n = 4) but present in >98% of control cells (Fig. 5D and Fig. S2).
Fig. 5.
Cells undergoing cytofission lack classic components of the cytokinetic apparatus. (A) Binucleate cell imaged with time-lapse microscopy was fixed during cytofission and stained for actin, PLK1, and MKLP1. (Insets) Indirect immunofluorescence signals for Plk1, MKLP1, and actin. (Right) Control cell undergoing cytokinesis. (B) Line-scans demonstrating intensity of signal across cytoplasmic bridge during cytofission and cytokinesis. Intensity is displayed in arbitrary units (A.U.). (C) Live cell imaging of cells labeled with GFP-Plk1 and H2B-mCherry confirms the lack of Plk1 signal across the cytoplasmic bridge, although it is readily observed in control cells undergoing cytokinesis. Line-scans show signal intensities in arbitrary units across cell bridges. (D) Quantification of observed events. Percentage of observed protein localization is shown, with number of observed localizations (numerator) and cells observed (denominator) in parentheses. Fisher’s exact test was performed to estimate the probability that the absence of markers in cytokinesis occurred by chance.
Fixed assays preclude direct observation of whether a given cell completes fission or exhibits bridge regression. We overcame this limitation with two independent validation experiments. First, live-cell imaging was performed to test whether fluorescent Plk1 can localize during cytofission before final scission into daughters. To do this, we observed binucleate RPE1 cells that stably express both GFP-Plk1 and H2B-mCherry. As expected, control cells undergoing canonical cytokinesis exhibited a robust midzone GFP-Plk1 signal (Fig. 5C and Movie S8). However, no localized signal was observed during cytofission (Fig. 5 C and D and Movie S9). As a second validation we challenged cells with nocodazole, which destabilizes polymerized microtubules; such polymerized microtubules are required for cytokinesis (24). However, we observed that fission occurs even when microtubules are depolymerized by nocodazole (n = 3; Fig. S3 and Movie S10). Thus, we conclude that cytofission does not use the mechanisms of classic cytokinesis.
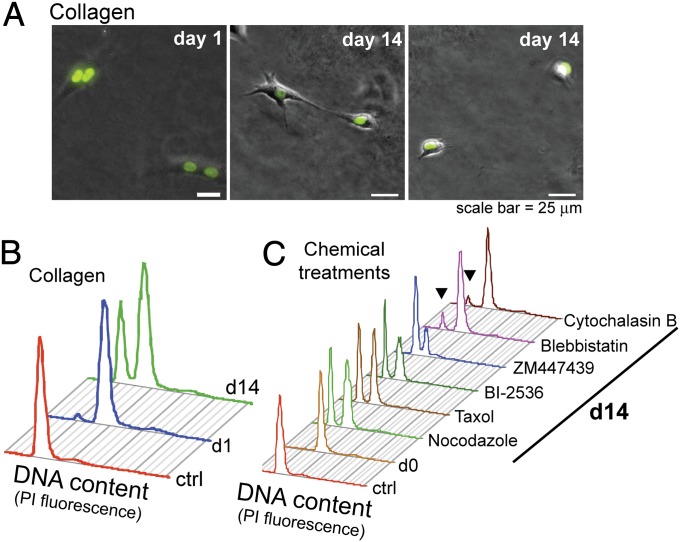
The experiments heretofore have demonstrated that cytofission can maintain genomic integrity after failed cytokinesis in 2D culture. To evaluate whether this could occur in a 3D environment, we developed a flow cytometry assay. In this assay, binucleated cells are plated in the presence of aphidicolin to preclude progression through the cell cycles. As expected, RPE1 cells do not proliferate in the presence of aphidicolin (Fig. S4A); however, the cell number in the population that began as binucleated cells increased consistent with cytofission, which does not require cell-cycle progression. The increase in diploid (2N) cell population is readily measured by flow cytometry (Fig. S4B). Using this assay, we tested whether cytofission could occur in 3D culture by embedding cells in collagen. After 14-d incubation, we observed stretched binucleate and mononucleate cells (Fig. 6A). Moreover, flow cytometry revealed an increased number of 2N cells over time due to cytofission (Fig. 6B). We conclude that human cytofission can occur in 3D contexts resembling intact human tissues.
Fig. 6.
Interphase cytofission occurs in 3D culture and requires myosin and actin function. (A) Cytofission events in RPE1 cells embedded in collagen. Binucleate cells were embedded into collagen (day 1) and imaged on the next day. All experiments were performed in the presence of 5 μM aphidicolin to prevent cell-cycle progression. On day 14, cell stretching and mononucleate cells were observed. (B) PI-FACS quantization of recovered cells on day 14. Cells were removed from collagen gel, stained with PI, and analyzed for DNA content. Most cells have tetraploid (4N) DNA content on day 1, but a large fraction have 2N content by day 14. (C) PI-FACS analysis of cells in 2D culture in the presence of chemical inhibitors for 14 d. Inhibitors included nocodazole 100 ng/mL, Taxol 50 nM, BI2536 50 nM, ZM447439 1 μM, blebbistatin(+/−) 50 μM, and cytochalasin 0.5 μM. Recovery of large 2N populations is observed in the presence of most chemicals, with the exception of blebbistatin and cytochalasin. The control in C is identical to that in B.
To clarify the mechanism of human cytofission, we challenged cells with specific chemical inhibitors followed by DNA content analysis (Fig. 6C). We observed a large increase in 2N cells, indicating intact cytofission, in the presence of chemicals that disrupt microtubule function, as described above. Moreover, we saw no disruption of the appearance of the 2N population after treatment with specific inhibitors of mitotic kinases, including BI-2536 (Plk1) and ZM44739 (Aurora kinases) (27, 28). However, we found markedly reduced 2N cells with blebbistatin and cytochalasin B (arrowheads, Fig. 6C), which inhibit myosin II and actin polymerization, respectively (16, 20). These findings support an active role of actin and myosin filaments in effecting cytofission.
This work demonstrates that cytofission can maintain genomic integrity of human cells in the face of cytokinesis failure. Sporadic cytokinesis failure in epithelial compartments may lead to cells that harbor two nuclei. It is commonly thought that such cells are obligated to die, to remain in a senescent-like state, or to enter mitosis with tetraploid and aneuploid progeny (12, 17, 19, 29, 30). Our findings demonstrate that this is not necessarily so. Instead, accurately segregated chromosomes can remain enveloped in separate nuclei and be distributed into healthy daughters to maintain genomic integrity. This may represent an important innate process that limits the risk of oncogenic transformation in proliferating cell compartments. It will be important to learn whether binucleate cells can similarly resolve to euploid progeny in intact human organs, and whether this could be stimulated to prevent aneuploidy and transformation to cancer.
Contractile-ring independent cell fission has been observed in model organisms such as Dictyostelium and has been termed traction-mediated cytofission or cytokinesis C (31–33). Similarly, contractile-ring independent processes are thought to contribute to mammalian cytokinesis (34, 35). As in Dictyostelium, human cytofission is distinct from cytokinesis; it requires neither polymerized microtubules nor classic components of the cytokinetic apparatus. Intriguingly, we find that blebbistatin inhibits human cytofission, whereas this myosin function is not required for cytofission in Dictyostelium (31). This finding suggests an active role of myosin and actin in human cytofission. In interphase cells, adhesive stress is the major driver of cytofission, and contractile forces are less important (36). If eukaryotic cells are more dependent on myosin II for establishing adhesive force than Dictyostelium, this could resolve the apparent difference. Indeed, a careful balance of adhesion and myosin II function is required for optimal cell motility (37). Thus, it is possible that in other contexts, abrogating myosin II function would enhance cell motility and increase human cytofission.
The cytofission demonstrated here is not expected to resolve polyploidy in all cases of failed cytokinesis. If, for example, cytokinesis is impaired by improper chromosome segregation or “slippage,” aneuploidy is expected to result even with cytofission (17). Moreover, we have observed cytofission during a prolonged G1 arrest in these cells, although this delay in cell-cycle progression is not a requisite effect of failed cytokinesis (19, 30). In contrast, cell-cycle progression and mitosis after failed cytokinesis can promote tetraploidy, aneuploidy, and oncogenesis (1, 12, 13). Nevertheless, we find that human cells harbor an innate fail-safe mechanism that allows euploidy to be maintained in the face of failed cytokinesis.
In conclusion, human cells can maintain coupled karyokinesis and cytofission to maintain genomic integrity without cytokinesis. This demonstrates the relevance of phenomena observed previously in Dictyostelium to human biology. Moreover, it reveals the redundant mechanisms that exist to maintain genomic integrity of human cells for the ∼1013 successful divisions required for development and life. By preventing tetraploidy and aneuploidy, cytofission may play a critical role in restraining human oncogenesis.
Methods
Cell Cultivation.
Cell lines were obtained from American Type Culture Collection. RPE1 and MCF10a cells were cultured in Dulbecco’s modified Eagle’s medium mixed with Ham’s F-12 modified medium (HyClone), and HCT116 were cultured in McCoy’s 5A modified medium (HyClone). Media was supplemented with 100 U/mL of penicillin/streptomycin and 10% (vol/vol) FBS (RPE1 and HCT116) or 5% (vol/vol) horse serum (MCF10a). For MCF10a, media also included 20 ng/mL EGF, 0.5 mL/mL hydrocortisone, 100 ng/mL cholera toxin, and 10 μg/mL insulin (38). Cultures were performed on tissue culture-treated plates, optical plates, or glass coverslips except where noted. To induce cytokinesis failure, exponentially growing cells were synchronized by a 4-h treatment with 0.2 µg/mL nocodazole followed by collection of loosely adherent mitotic cells by shake-off. Collected cells were split into two fractions and treated with DMSO or with 100 µM racemic blebbistatin for 4 h (HCT116) or 16 h (MCF10a and RPE1) and allowed to recover in fresh medium for 4 h. After completion of this procedure, a sample of cells was collected, fixed with formaldehyde in phospho-buffered saline, stained with Hoechst 33258, and mounted onto slides for direct counting of nuclei per cell by fluorescence microscopy.
When necessary, fluorescent markers, including H2B-GFP, H2B-mCherry, and mCherry-Cdt1 (30-120) (Clontech), were introduced into cells with retroviral transduction followed by selection and subcloning and selection of high-expressing clones. GFP-Plk1 cells were derived previously and additionally had both copies of the endogenous PLK1 locus deleted (25).
Clonogenic Assays.
To assess plating efficiency, cells were collected with trypsin treatment, counted, and distributed into 96-well plates in fresh media at 0.5 cells per well followed by incubation for 3 wk. For colony formation assays, the wells harboring colonies were counted, and a Poisson correction was applied assuming a random chance of multiple cells distributed in a given well: plating efficiency was calculated as: γ = −ln(1−γo)/ρ, where γo = observed colonies per well and ρ = plated cells per well.
To directly observe cells in clonogenic assays (Fig. 2), cells were synchronized as above and trypsinized 6 h after removing blebbistatin. Cells were plated into optical 96-well plates or iuvo Microchannel 5250 plates (BellBrook Labs), allowed to adhere overnight, then visualized by fluorescent microscopy. For optical plates, 2-μL droplets were placed at well centers before microscope visualization to eliminate risk of difficult-to-observe contaminating cells at the well periphery. Each well containing a single binucleate cell was imaged and marked for analysis. Next, additional fresh complete medium was added to plates, followed by incubation at 37 °C incubator with 5% CO2 for 21 d. Colonies from wells observed to initially contain one binucleate cell were counted and recovered with trypsin. All clones were expanded, cryopreserved, and subjected to cytogenetic analysis.
Cytogenetics.
Cytogenetic analysis was performed directly on cultured cells. Cells were treated with colcemid for synchronization in metaphase followed by treatment with a 0.075 M KCl hypotonic solution and fixation in 3:1 methanol:acetic acid. G-banding was accomplished using trypsin and Giemsa-Wright stain. Metaphase chromosomes were captured using a GSL slide scanner (Leica Microsystems) and analyzed using CytoVision 4.5.4 (Leica Microsystems). Ten cells from each culture were fully analyzed for chromosome number, identification, and structural abnormalities.
Microscope Image Acquisition.
Microscopy (other than cytogenetics) was performed with a Nikon Eclipse Ti inverted epifluorescence microscope equipped with 10×, 20×, 40×, and 100× objectives and a temperature-controlled stage with 5% CO2 support (In Vivo Scientific). Phase-contrast and wide-field fluorescence images were acquired with a CoolSnap HQ2 camera (Photometrics). For time-lapse videomicroscopy, cells were maintained with 5% CO2 at 37 °C and imaged every 10–15 min (12 min used for most assays). Fixed cells were stained as described below with Alexa-Fluor 488-, 594-, or 647-conjugated secondary antibodies (Invitrogen), and with DAPI with or without rhodamine-phalloidin and imaged with wide-field fluorescence at room temperature. Nikon Elements AR 3.2 was used for image acquisition, analysis, and image threshold adjustments for optimal display. Figures were cropped in Adobe Photoshop CS5.1 and assembled into figures in Adobe Illustrator CS5.1. Line-scans were generated with Nikon Elements using a 16 pixel-width maximal intensity projection of the internuclear bridge identified by actin staining. For videos, files were exported to ImageJ for timestamp overlay.
Flow Cytometry and 3D Culture.
Flow cytometry for DNA content was performed by fixing cells in ethanol and staining with propidium iodide (PI). Cells were analyzed on the 585/42 channel with a BD FACScalibur flow cytometer (BD Biosciences). For 3D culture, cells were embedded in 1.625 mg/mL collagen I gel (Invitrogen). Binucleate H2B-GFP labeled cells were suspended and mixed with 1.6 mg/mL collagen solution in complete medium supplemented with 5 μM aphidicolin at pH 7.4. A collagen-cell suspension was poured into 24-well cell culture plates, allowed to polymerize, and covered with complete medium with aphidicolin. Cells were visualized by epifluorescence microscopy. After 14 d, cells were collected with 2 mg/mL collagenase and processed for flow cytometry.
Immunofluorescence.
Cells prepared as above were cultured on coverslips and fixed with ice-cold methanol or 4% paraformaldehyde for 15 min. Coverslips were washed with phospho-buffered saline and blocked with 3% BSA with 0.1% Triton-X detergent in this same buffer. Primary antibody incubations were performed at room temperature for 60 min in blocking buffer, washed thrice, then incubated with secondary Alexa-Fluor conjugated antibodies for 30 min. Finally coverslips were counterstained with DAPI and, if appropriate rhodamine-phalloidin (actin stain), mounted with ProLong Gold (Invitrogen), and allowed to cure overnight. Primary antibodies (titers) were anti-Plk1 mouse IgG2a (1:500; Santa Cruz Biotechnology, sc-17783, F-8), anti-αTubulin Rat IgG2a (1:1,000; Millipore, MAB1864), Rabbit anti-CENPE (1:500) (39), mouse anti-INCENP (1:500; Upstate 05-940), and rabbit anti-MKLP1 (1:350; Santa Cruz Biotechnology, sc-867). For fluorescence-activated cell sorting, cells were fixed in 100% ethanol and stained with PI followed by analysis of 10,000 cells using 488-nm excitation.
Chemicals used in this study include aphidicolin, racemic blebbistatin (EMD Biosciences), Taxol (Acros), BI2536 (Selleck), ZM447439 (R&D Systems), cytochalasin B (Acros), and nocodazole (EMD Biosciences).
Online Supplemental Material.
Movies are available corresponding to time-lapse videomicroscopy images shown in this article, as follows. Movies S1–S3 correspond to cells illustrated in Fig. 3A; Movies S4 and S5 to Fig. 4B; Movie S6 to Fig. 4C; Movie S7 to Fig. 5A; Movies S8 and S9 to Fig. 5C; and Movie S10 to Fig. S3.
Supplementary Material
Acknowledgments
We thank W. Bement for critical review of the manuscript, A. Kane for performing chromosome analyses, G. Raca for initiating cytogenetic analyses, and Dr. William Brockliss (University of Wisconsin Classics Department) for expert advice. This work was supported by National Institutes of Health Grant R01 GM097245 (to M.E.B.), University of Wisconsin Carbone Cancer Center Grant P30 CA14520, American Cancer Society Grant IRG-58-011-48, Mary Kay Ash Foundation Clinical, Translational Science Award 1UL1RR025011, and the Flight Attendant Medical Research Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1308203110/-/DCSupplemental.
References
- 1. Boveri T (1914) Zur Frage der Entstehung maligner Tumoren [Concerning the Origin of Malignant Tumors] (Fischer Verlag, Jena); trans Harris H (2008). J Cell Sci 121(Suppl 1):1–84. [DOI] [PubMed]
- 2.Hanks S, et al. Constitutional aneuploidy and cancer predisposition caused by biallelic mutations in BUB1B. Nat Genet. 2004;36(11):1159–1161. doi: 10.1038/ng1449. [DOI] [PubMed] [Google Scholar]
- 3.Michel LS, et al. MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature. 2001;409(6818):355–359. doi: 10.1038/35053094. [DOI] [PubMed] [Google Scholar]
- 4.Weaver BA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell. 2007;11(1):25–36. doi: 10.1016/j.ccr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 5.Jeganathan K, Malureanu L, Baker DJ, Abraham SC, van Deursen JM. Bub1 mediates cell death in response to chromosome missegregation and acts to suppress spontaneous tumorigenesis. J Cell Biol. 2007;179(2):255–267. doi: 10.1083/jcb.200706015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brito DA, Rieder CL. Mitotic checkpoint slippage in humans occurs via cyclin B destruction in the presence of an active checkpoint. Curr Biol. 2006;16(12):1194–1200. doi: 10.1016/j.cub.2006.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lanni JS, Jacks T. Characterization of the p53-dependent postmitotic checkpoint following spindle disruption. Mol Cell Biol. 1998;18(2):1055–1064. doi: 10.1128/mcb.18.2.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Straight AF, Field CM. Microtubules, membranes and cytokinesis. Curr Biol. 2000;10(20):R760–R770. doi: 10.1016/s0960-9822(00)00746-6. [DOI] [PubMed] [Google Scholar]
- 9.Krajcovic M, et al. A non-genetic route to aneuploidy in human cancers. Nat Cell Biol. 2011;13(3):324–330. doi: 10.1038/ncb2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mullins JM, Biesele JJ. Terminal phase of cytokinesis in D-98s cells. J Cell Biol. 1977;73(3):672–684. doi: 10.1083/jcb.73.3.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weaver BA, Silk AD, Cleveland DW. Cell biology: Nondisjunction, aneuploidy and tetraploidy. Nature. 2006;449(7104):E9–E10. doi: 10.1038/nature05139. [DOI] [PubMed] [Google Scholar]
- 12.Fujiwara T, et al. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature. 2005;437(7061):1043–1047. doi: 10.1038/nature04217. [DOI] [PubMed] [Google Scholar]
- 13.Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460(7252):278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dalton WB, Nandan MO, Moore RT, Yang VW. Human cancer cells commonly acquire DNA damage during mitotic arrest. Cancer Res. 2007;67(24):11487–11492. doi: 10.1158/0008-5472.CAN-07-5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Orth JD, Loewer A, Lahav G, Mitchison TJ. Prolonged mitotic arrest triggers partial activation of apoptosis, resulting in DNA damage and p53 induction. Mol Biol Cell. 2012;23(4):567–576. doi: 10.1091/mbc.E11-09-0781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Straight AF, et al. Dissecting temporal and spatial control of cytokinesis with a myosin II inhibitor. Science. 2003;299(5613):1743–1747. doi: 10.1126/science.1081412. [DOI] [PubMed] [Google Scholar]
- 17.Ganem NJ, Storchova Z, Pellman D. Tetraploidy, aneuploidy and cancer. Curr Opin Genet Dev. 2007;17(2):157–162. doi: 10.1016/j.gde.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 18.Stukenberg PT. Triggering p53 after cytokinesis failure. J Cell Biol. 2004;165(5):607–608. doi: 10.1083/jcb.200405089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uetake Y, Sluder G. Cell cycle progression after cleavage failure: Mammalian somatic cells do not possess a “tetraploidy checkpoint”. J Cell Biol. 2004;165(5):609–615. doi: 10.1083/jcb.200403014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown SS, Spudich JA. Mechanism of action of cytochalasin: Evidence that it binds to actin filament ends. J Cell Biol. 1981;88(3):487–491. doi: 10.1083/jcb.88.3.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maciejowski J, et al. Mps1 directs the assembly of Cdc20 inhibitory complexes during interphase and mitosis to control M phase timing and spindle checkpoint signaling. J Cell Biol. 2010;190(1):89–100. doi: 10.1083/jcb.201001050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sakaue-Sawano A, et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 2008;132(3):487–498. doi: 10.1016/j.cell.2007.12.033. [DOI] [PubMed] [Google Scholar]
- 23.Wist E, Prydz H. The effect of aphidicolin on DNA synthesis in isolated HeLa cell nuclei. Nucleic Acids Res. 1979;6(4):1583–1590. doi: 10.1093/nar/6.4.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glotzer M. The molecular requirements for cytokinesis. Science. 2005;307(5716):1735–1739. doi: 10.1126/science.1096896. [DOI] [PubMed] [Google Scholar]
- 25.Burkard ME, et al. Chemical genetics reveals the requirement for Polo-like kinase 1 activity in positioning RhoA and triggering cytokinesis in human cells. Proc Natl Acad Sci USA. 2007;104(11):4383–4388. doi: 10.1073/pnas.0701140104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petronczki M, Glotzer M, Kraut N, Peters JM. Polo-like kinase 1 triggers the initiation of cytokinesis in human cells by promoting recruitment of the RhoGEF Ect2 to the central spindle. Dev Cell. 2007;12(5):713–725. doi: 10.1016/j.devcel.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 27.Ditchfield C, et al. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J Cell Biol. 2003;161(2):267–280. doi: 10.1083/jcb.200208091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lénárt P, et al. The small-molecule inhibitor BI 2536 reveals novel insights into mitotic roles of polo-like kinase 1. Curr Biol. 2007;17(4):304–315. doi: 10.1016/j.cub.2006.12.046. [DOI] [PubMed] [Google Scholar]
- 29.Duncan AW, et al. The ploidy conveyor of mature hepatocytes as a source of genetic variation. Nature. 2010;467(7316):707–710. doi: 10.1038/nature09414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wong C, Stearns T. Mammalian cells lack checkpoints for tetraploidy, aberrant centrosome number, and cytokinesis failure. BMC Cell Biol. 2005;6(1):6. doi: 10.1186/1471-2121-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Lozanne A, Spudich JA. Disruption of the Dictyostelium myosin heavy chain gene by homologous recombination. Science. 1987;236(4805):1086–1091. doi: 10.1126/science.3576222. [DOI] [PubMed] [Google Scholar]
- 32.O’Connell CB, Wheatley SP, Ahmed S, Wang YL. The small GTP-binding protein rho regulates cortical activities in cultured cells during division. J Cell Biol. 1999;144(2):305–313. doi: 10.1083/jcb.144.2.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uyeda TQ, Nagasaki A. Variations on a theme: The many modes of cytokinesis. Curr Opin Cell Biol. 2004;16(1):55–60. doi: 10.1016/j.ceb.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 34.Burton K, Taylor DL. Traction forces of cytokinesis measured with optically modified elastic substrata. Nature. 1997;385(6615):450–454. doi: 10.1038/385450a0. [DOI] [PubMed] [Google Scholar]
- 35.Kanada M, Nagasaki A, Uyeda TQ. Adhesion-dependent and contractile ring-independent equatorial furrowing during cytokinesis in mammalian cells. Mol Biol Cell. 2005;16(8):3865–3872. doi: 10.1091/mbc.E05-03-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poirier CC, Ng WP, Robinson DN, Iglesias PA. Deconvolution of the cellular force-generating subsystems that govern cytokinesis furrow ingression. PLOS Comput Biol. 2012;8(4):e1002467. doi: 10.1371/journal.pcbi.1002467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ng MR, Besser A, Danuser G, Brugge JS. Substrate stiffness regulates cadherin-dependent collective migration through myosin-II contractility. J Cell Biol. 2012;199(3):545–563. doi: 10.1083/jcb.201207148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30(3):256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- 39.Brown KD, Wood KW, Cleveland DW. The kinesin-like protein CENP-E is kinetochore-associated throughout poleward chromosome segregation during anaphase-A. J Cell Sci. 1996;109(Pt 5):961–969. doi: 10.1242/jcs.109.5.961. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.