Abstract
Plantazolicin (PZN), a polyheterocyclic, Nα,Nα-dimethylarginine–containing antibiotic, harbors remarkably specific bactericidal activity toward strains of Bacillus anthracis, the causative agent of anthrax. Previous studies demonstrated that genetic deletion of the S-adenosyl-l-methionine–dependent methyltransferase from the PZN biosynthetic gene cluster results in the formation of desmethylPZN, which is devoid of antibiotic activity. Here we describe the in vitro reconstitution, mutational analysis, and X-ray crystallographic structure of the PZN methyltransferase. Unlike all other known small molecule methyltransferases, which act upon diverse substrates in vitro, the PZN methyltransferase is uncharacteristically limited in substrate scope and functions only on desmethylPZN and close derivatives. The crystal structures of two related PZN methyltransferases, solved to 1.75 Å (Bacillus amyloliquefaciens) and 2.0 Å (Bacillus pumilus), reveal a deep, narrow cavity, putatively functioning as the binding site for desmethylPZN. The narrowness of this cavity provides a framework for understanding the molecular basis of the extreme substrate selectivity. Analysis of a panel of point mutations to the methyltransferase from B. amyloliquefaciens allowed the identification of residues of structural and catalytic importance. These findings further our understanding of one set of orthologous enzymes involved in thiazole/oxazole-modified microcin biosynthesis, a rapidly growing sector of natural products research.
Keywords: enzymology, mutagenesis, RiPP natural product
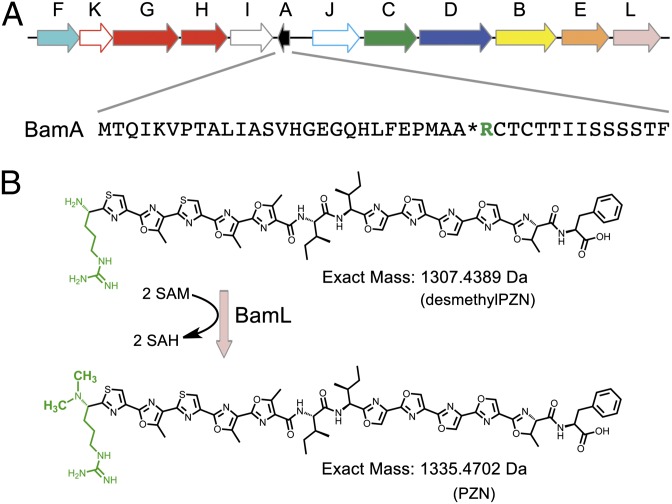
Plantazolicin (PZN) is a poly-azol(in)e-containing molecule of ribosomal origin from the plant-growth promoting bacterium, Bacillus amyloliquefaciens FZB42 (1-3). PZN exhibits selective bactericidal activity toward Bacillus anthracis (3). All of the genes required for PZN production, immunity, and export cluster within a 10-kb region of the FZB42 genome (Fig. 1A). Genome mining has identified highly similar PZN biosynthetic gene clusters in Bacillus pumilus, Clavibacter michiganensis subsp. sepedonicus, Corynebacterium urealyticum, and Brevibacterium linens (3). PZN is biosynthesized from a 41-residue, inactive precursor peptide (Fig. 1A). Distinguishing chemical features of PZN are the two contiguous poly-azol(in)e moieties, which like all thiazole/oxazole-modified microcin (TOMM) natural products, originate from Cys and Ser/Thr residues on the C-terminal region of the precursor peptide (4–6). During heterocycle formation, a cyclodehydratase first converts Cys and Ser/Thr to thiazoline and (methyl)oxazoline, respectively. This ATP-dependent transformation formally removes water from the preceding amide bond (7–10). Subsequent dehydrogenation yields the aromatic thiazole and (methyl)oxazole (11). During PZN maturation, all 10 Cys and Ser/Thr residues within the C-terminal core region are cyclized, yielding 9 azole heterocycles and 1 methyloxazoline (Fig. 1B). Further modification includes leader peptide proteolysis and methylation to yield the final metabolite. The PZN methyltransferase dimethylates the N terminus (Arg) and is S-adenosyl-l-methionine (SAM)-dependent. The N-terminal methylation of ribosomal peptides from bacteria is exceptionally rare. With respect to natural products, the only compounds that we are aware of that undergo N-terminal dimethylation besides PZN are the linaridin antibiotics (e.g., cypemycin, grisemycin), the best studied of which contain Nα,Nα-dimethylAla (12–14).
Fig. 1.
PZN biosynthesis. (A) The PZN biosynthetic gene cluster with single-letter codes above each ORF. The amino acid sequence of the PZN precursor peptide (designated as A) is shown. *, leader peptide cleavage site; B, dehydrogenase; C/D, cyclodehydratase; E, putative leader peptidase; F, putative immunity protein; G/H, ABC transporters; green, site of dimethylation; I/J, unknown function; K, transcriptional regulator; L, methyltransferase. (B) The BamL and SAM-dependent conversion of desmethylPZN to PZN with the N-terminal Arg residues shown in green. Note that the most C-terminal heterocycle is a methyloxazoline, which can be selectively hydrolyzed to yield Thr.
Through genetic manipulation and alteration of cultivation conditions, PZN biosynthetic intermediates have been intercepted, permitting investigation into the details of downstream tailoring reactions and the bioactivity of partially processed substrates (3). After genetic deletion of the methyltransferase from B. amyloliquefaciens, desmethylPZN was isolated in roughly equivalent yield (1). DesmethylPZN was found to be devoid of antibiotic activity, as was also shown for desmethylcypemycin (12). As with PZN, the corresponding methyltransferase for cypemycin (CypM) tailoring was confirmed through deletion studies (12) and has recently been characterized in vitro (14). Although CypM and the PZN methyltransferase catalyze similar reactions, they do not share significant amino acid sequence similarity outside of the predicted SAM-binding sites. In this work, we report the in vitro reconstitution of the PZN methyltransferase, which exhibits a striking selectivity for polyheterocyclized substrates. We have also determined the high-resolution cocrystal structures of the PZN methyltransferases from B. amyloliquefaciens (BamL) and B. pumilus (BpumL), each bound to S-adenosyl-l-homocysteine (SAH), and have carried out detailed characterization of site-specific variants of BamL. These studies provide a molecular rationale for the unexpected selectivity for an otherwise broadly acting superfamily of catalysts.
Results
Enzymatic Conversion of DesmethylPZN to PZN.
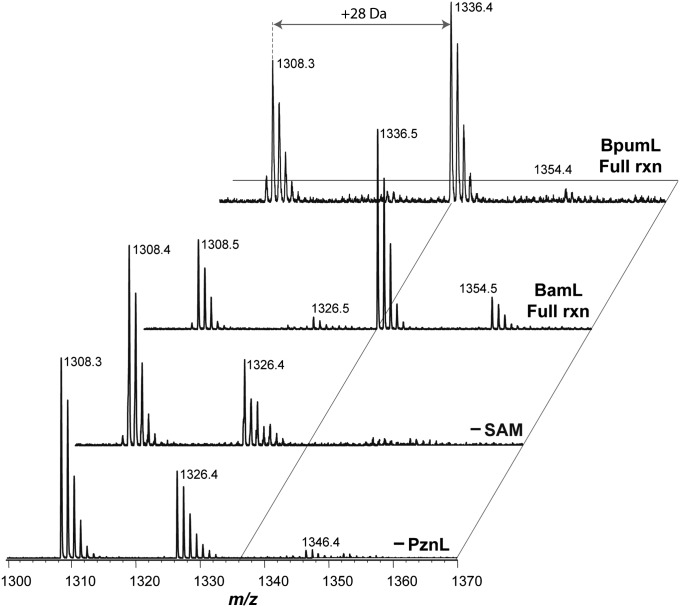
To assess enzymatic activity, we cloned, expressed, and purified BamL as a fusion with maltose-binding protein (MBP). MBP-BamL was then added to reactions containing desmethylPZN, SAM, and a nucleosidase that converts the SAH by-product to adenine and S-ribosyl-l-homocysteine, minimizing potential product inhibition (Pfs) (15). Reactions were initiated by the addition of SAM, quenched at desired time points, and analyzed by MALDI-MS (Fig. 2). In the presence of desmethylPZN and SAM, MBP-BamL catalyzed the formation of PZN (m/z 1336). Omitting either SAM or BamL resulted in no product formation. Identical results were achieved with the BpumL methyltransferase (from B. pumilus), another bona fide producer of PZN (3) (Fig. 2). Removal of the MBP tag from BamL led to a lower level of PZN formation, suggesting that MBP does not interfere with methyltransferase activity (SI Appendix, Table S1). The higher level of PZN formation with MBP was likely due to increased stability conferred by the tag (16). Using the MALDI-MS end point assay, we further evaluated the activity of MBP-BamL by testing the effect of pH, buffer, and a variety of other additives (SI Appendix, Table S1). Omission of Pfs from the reaction led to only a modest decrease (∼25%) in product formation. Using isothermal titration calorimetry (ITC), we obtained a dissociation constant between SAM and BamL of 7.5 ± 0.8 µM (SI Appendix, Fig. S1). In accord with lack of SAH by-product inhibition, identical ITC runs with SAH generated unusual titration curves that could not be mathematically fit to any reasonable binding model. To confirm the site of BamL dimethylation, collision-induced dissociation (CID) spectra were obtained using Fourier transform MS. A previously characterized diagnostic fragment ion, m/z 1277.4291 Da, was observed, indicating that the site of methylation was on the expected N-terminal α amine (SI Appendix, Fig. S2) (3).
Fig. 2.
Activity of purified methyltransferases. MALDI-MS was used to monitor the conversion of desmethylPZN (m/z 1308) to PZN (m/z 1336). The m/z 1326 and 1354 species represent hydrolyzed desmethylPZN and PZN, respectively. Samples containing all reaction components used either the methyltransferase from BamL or BpumL. The SAM cosubstrate and BamL enzyme were omitted from the samples labeled −SAM and −BamL, respectively. PznL, PZN methyltransferase (either BamL or BpumL); rxn, reaction.
Despite the variety of in vitro reconstitution reactions screened (SI Appendix, Table S1), monomethylPZN was never detected (m/z 1322). Even at shorter time points, when consumption of desmethylPZN was incomplete, monomethylPZN was not observed. In an additional effort to detect this species, we performed BamL reactions under pseudosingle turnover conditions using variable concentrations of BamL, desmethylPZN, and SAM (SI Appendix, Fig. S3). Under no condition was monomethylPZN (m/z 1322) detected. However, when all reaction components were supplied at 100 µM, we observed a minor peak consistent with hydrolyzed (+18 Da) monomethylPZN (m/z 1340). Unfortunately, the low signal-to-noise ratio did not allow for structural confirmation.
DesmethylPZN Substrate Analogs.
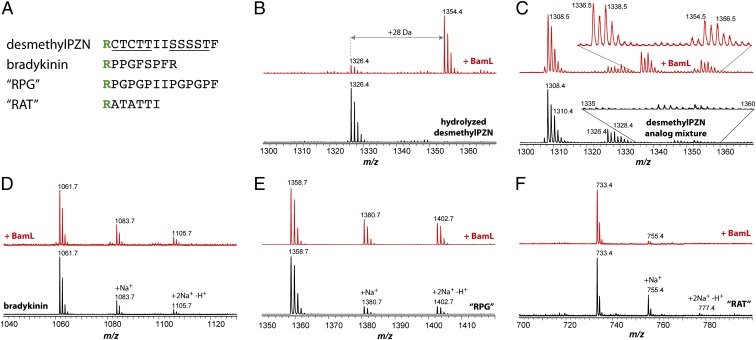
Because of the hydrolytic instability of PZN, we observed by MALDI-MS hydrolyzed desmethylPZN (m/z 1326) and hydrolyzed PZN (m/z 1354) in all in vitro reactions (Fig. 2 and SI Appendix, Fig. S3). These species arise from the hydrolysis of the sole methyloxazoline ring. We have previously reported on the lability of this heterocycle, which reinstates the most C-terminal Thr (Fig. 1) (3). To establish whether BamL could directly accept hydrolyzed desmethylPZN (m/z 1326) as a substrate to produce hydrolyzed PZN (m/z 1354), we subjected desmethylPZN to conditions that yielded quantitative conversion to hydrolyzed desmethylPZN for BamL substrate testing (SI Appendix, Materials and Methods). Analysis of reaction products run under identical conditions showed that BamL processed hydrolyzed desmethylPZN with efficiency approximately equal to that of desmethylPZN (Figs. 2 and 3 A and B). From earlier work, we noted that another desmethylPZN variant harboring two methyloxazolines (dihydrodesmethylPZN, m/z 1310) was readily accessible by using oxygen-saturated cultivation (3). Because of separation difficulties, we tested a mixture of m/z 1308, 1310, 1326, and 1328 (desmethylPZN analog mixture) for processing by BamL. Analysis of reaction products by MALDI-MS showed the presence of PZN (m/z 1336), dihydroPZN (m/z 1338, note isotopic ratio), hydrolyzed PZN (m/z 1354), and hydrolyzed dihydroPZN (m/z 1356, Fig. 3C). These data indicate BamL successfully processed dihydrodesmethylPZN.
Fig. 3.
Substrate tolerance of BamL. MALDI-MS was used to assess the ability of BamL to process substrates other than desmethylPZN. (A) Subset of peptides tested as substrates with the N-terminal Arg shown in bold. Heterocyclized residues of desmethylPZN are underlined. The most C-terminal heterocycle is reverted to Thr in hydrolyzed desmethylPZN. (B–F) Bottom spectra, unreacted peptides; top spectra, after reaction with BamL. (B) Hydrolyzed desmethylPZN (m/z 1326). (C) DesmethylPZN analog mixture that contained desmethylPZN (m/z 1308), dihydrodesmethylPZN (m/z 1310), and the hydrolyzed forms (m/z 1326 and 1328, respectively). (D) Bradykinin (m/z 1061). (E) “RPG” peptide (m/z 1358). (F) “RAT” peptide (m/z 733).
Unusual Substrate Selectivity of BamL.
To further explore substrate permissiveness, we tested bradykinin and a synthetic peptide termed “RPG” (sequence given in Fig. 3) as BamL substrates. To a first approximation, bradykinin and the RPG peptide mimic desmethylPZN. Both harbor an N-terminal Arg, are similar in size to desmethylPZN, and contain Pro where several azole heterocycles are found in desmethylPZN (Fig. 3A). As with Pro, azoles are five-membered, nitrogen-containing heterocycles that serve as peptide conformational restraints. Although not a perfect match for an azole, the ability of Pro to structurally and functionally substitute in TOMM natural products has been documented (17). Upon treatment of bradykinin and the RPG peptide with BamL and SAM, no reaction products were detected, even after increasing the enzyme concentration and allowing for extended reaction times (Fig. 3 D and E). Similarly, adrenocorticotropic hormone peptide 18–39, with an N-terminal amino acid sequence of RPV, was not processed by BamL. Because Pro is an imperfect match for an azole, we also tested whether BamL would process an additional synthetic peptide “RAT” (sequence given in Fig. 3), which contained the left segment of the native PZN precursor sequence except that the Cys residues were replaced with Ala. Again, no reaction product was observed, even after increasing the enzyme concentration to near single turnover levels (Fig. 3F). Two tetrapeptides, RAAA and RGGG, were also evaluated as BamL substrates, but no reaction products were detected. As a final test, BamL accepted Arg amide (carboxylate replaced with a neutral amide) as a substrate, albeit a very inefficient one. After increased reaction time with elevated substrate concentration, high-resolution MS identified a mass consistent with dimethylArg amide (experimental 202.1677 Da, theoretical 202.1688 Da, error 4.5 ppm). The very low intensity of this peak suggested that <5% of the starting material was converted. These results suggested that BamL does not simply recognize peptides with an N-terminal Arg and furthermore is intolerant to substrates not containing desmethylPZN-like (i.e., polyheterocyclic) functionality. In contrast to the specificity exhibited by BamL, other characterized SAM-dependent methyltransferases acting on small molecules and peptides have much broader substrate scopes. For example, the methyltransferases involved in rebeccamycin (18, 19), dehydrophos (20), aminocoumarin (21), and CypM (14) biosynthesis methylate a wide range of substrates.
Structure Determination of Two PZN Methyltransferases.
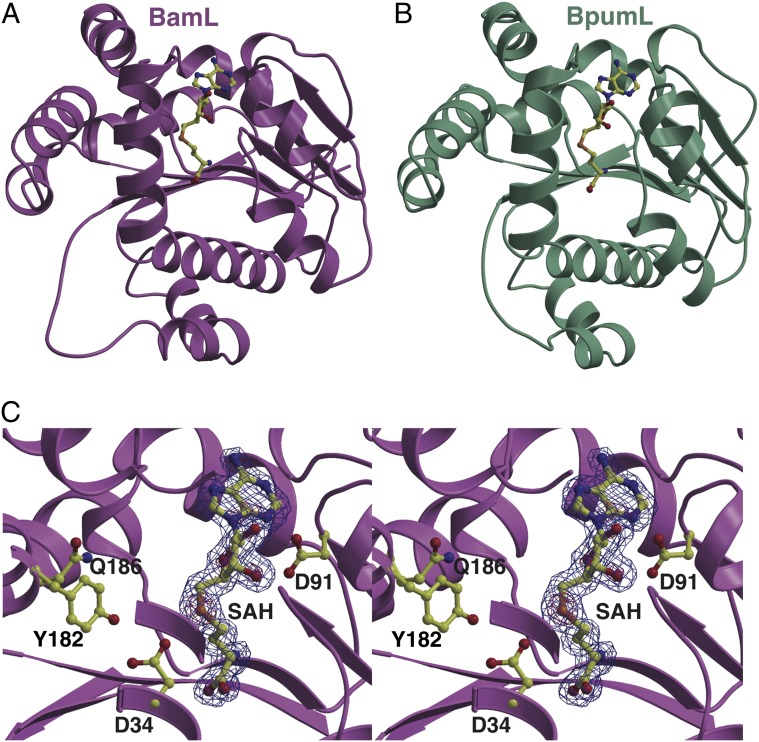
To provide insight into the unexpected lack of activity toward nonheterocyclized peptide substrates, we determined the crystal structures of BamL (1.75 Å resolution) and BpumL (2.0 Å resolution), each in complex with SAH. Initial crystallographic phases were determined at 3.2 Å resolution using SeMet-labeled crystals of BpumL, followed by model building and refinement against higher resolution data until convergence. Phases for BamL were determined by molecular replacement using a partial model of BpumL. Relevant data collection and refinement statistics are given in SI Appendix, Table S2. The BamL and BpumL structures consist of a core Rossman-fold domain composed of seven β-strands surrounded by six α-helices, with an overall architecture similar to other methyltransferases (Fig. 4 A and B). As expected from the 48% sequence identity between BamL and BpumL, the structures are superimposable with an rmsd of 1.1 Å over 256 aligned α carbons. A DALI search against the Protein Data Bank (PDB) identifies the closest structural homologs as the bacterial Hen1 methyltransferase (PDB ID code, 3JWG; Z-score, 16.8; rmsd, 2.5 Å over 174 aligned α carbons) and the hypothetical bacterial YecO protein (PDB ID code, 1IM8; Z-score, 16.7; rmsd, 2.9 Å over 194 aligned α carbons). The conservation in structure is restricted to the core Rossman fold domain and persists despite minimal similarities (roughly 10–13% identity) across the primary structure.
Fig. 4.
Ribbon diagrams derived from the X-ray crystal structures of (A) BamL and (B) BpumL, with the bound SAH ligand shown in yellow ball-and-stick representation. (C) Electron density maps calculated using Fourier coefficients (Fobs – Fcalc) with phases derived from the refined 1.75 Å resolution structure of BamL calculated with the coordinates for SAH omitted before one round of crystallographic refinement. The map is contoured at 2.6σ (blue mesh) and 8σ (red mesh) and the final refined coordinates are superimposed. The coordinates for SAH and polar active site residues that are important in catalysis are shown as yellow (ball-and-stick).
Unlike the structures of typical small molecule methyltransferases, the architectures of BamL and BpumL are minimalistic and lack additional secondary structure decorations. One molecule of SAH is bound across the interior of BamL with the adenine ring enclosed in a cavity defined along the sides by S92, F137, and a loop encompassing residues S112–A114 across the top. The side chain of D91 is within interaction distance with the 2′ hydroxyl of the ribose (Fig. 4C). BamL residues R42 and H131 provide additional contacts with SAH via the homocysteine carboxylate. Notably, the trajectory of the homocysteine moiety of SAH is not collinear with the adenine but is deflected toward the center of the polypeptide by F21 and a loop encompassing G68–Q71. This would position the electrophilic methyl group of SAM at the base of a long tunnel running to the surface of the protein. The walls of this tunnel are defined by a number of hydrophobic residues including F21, Y33, T38, L132, L162, Y182, and L183. These residues are strictly conserved between BamL and BpumL, with the exception of L183, which is I181 in BpumL (SI Appendix, Fig. S4). Because the depth of this tunnel is of sufficient length to accommodate the first four to five residues of desmethylPZN, we presume that it defines the substrate-binding pocket (SI Appendix, Fig. S5). At the base of the tunnel, a shorter, perpendicular cavity is found. This shorter cavity is immediately adjacent to the presumptive position of the electrophilic methyl group of SAM and is of suitable size to accommodate the N-terminal Arg side chain of (desmethyl)PZN. Residues D34, Y182, and Q186 of BamL presumably stabilize the N-terminal amine of the substrate (Fig. 4C). Evidence for the role of these residues in substrate engagement is borne out by the mutational analyses described in the following section.
Selection and Enzymatic Activity of BamL Mutants.
To better understand catalysis, we aligned the amino acid sequence of BamL with a series of well-characterized methyltransferases. With the exception of identifying residues that comprise the SAM-binding pocket, this exercise shed little light on what residues may be important for BamL catalysis because of the poor conservation across the methyltransferase superfamily (22). Fortunately, sequence alignment of the five known PZN methyltransferases (including BpumL) identified in our earlier work (3) revealed a manageable number of conserved residues with which to begin the enzymological dissection of BamL (SI Appendix, Fig. S4). We thus targeted eight residues of BamL to be replaced with Ala (SI Appendix, Table S3). We then analyzed the structures of BamL and BpumL cocrystalized with SAH and mutated seven additional positions hypothesized to be critical either for substrate binding (desmethylPZN and SAM) or catalysis. At four of these locations, the wild-type (WT) residue was replaced with Ala and an additional alternative residue, ultimately yielding a database of 19 mutant proteins (Table 1).
Table 1.
Relative enzymatic activity of BamL mutants
| BamL | Cond. A conv. | Cond. B conv. | Cond. C conv. | BamL | Cond. A conv. | Cond. B conv. | Cond. C conv. |
| WT | 1.00 ± 0.16* | 1.00 ± 0.15* | 1.00 ± 0.08* | L132F | 1.00 ± 0.06 | <0.05† | ND |
| W20A‡ | ND | ND | ND | D161A | 0.16 ± 0.01 | <0.05† | ND |
| F21A‡ | ND | ND | ND | L162A | 0.65 ± 0.11 | <0.05† | 0.24 ± 0.05 |
| D34A | 0.62 ± 0.08 | <0.05† | ND | L162F | 0.76 ± 0.13 | <0.05† | ND |
| T38A | 1.00 ± 0.03 | 0.59 ± 0.10 | 0.37 ± 0.12 | R164A | 0.43 ± 0.01 | <0.05† | ND |
| T38F | 1.00 ± 0.04 | <0.05† | 0.25 ± 0.03 | Y182A | 0.23 ± 0.08 | <0.05† | ND |
| R42A | 0.40 ± 0.07 | <0.05† | <0.05† | Y182F | 0.96 ± 0.03 | <0.05† | 0.37 ± 0.02 |
| D91A‡ | ND | ND | ND | D185A | 0.77 ± 0.06 | <0.05† | ND |
| D126A‡ | ND | ND | ND | Q186A | 0.45 ± 0.07 | <0.05† | <0.05† |
| L132A | 0.92 ± 0.08 | <0.05† | 0.59 ± 0.10 | S190A | 1.00 ± 0.07 | 0.74 ± 0.32 | ND |
Relative activity was determined by comparing the ion intensities of the starting material to product by MALDI-TOF-MS. Results derive from two independent enzyme preparations and at least two independent reactions per prep. Values are normalized to that obtained for WT enzyme. Cond. A: MBP-BamL (10 µM), Pfs (10 µM), desmethylPZN (50 µM), and SAM (3 mM) were added to Tris buffer (50 mM, pH 7.8) and allowed to proceed for 16 h at 37 °C. Cond. B: MBP-BamL (1 µM), Pfs (1 µM), desmethylPZN (5 µM), and SAM (3 mM) were added to Tris buffer (50 mM, pH 7.8) and allowed to proceed for 1 h at 22 °C. Cond. C: MBP-BamL (1 µM), Pfs (1 µM), desmethyl PZN (50 µM), and SAM (500 µM) were added to Tris buffer (50 mM, pH 7.8) and allowed to proceed for 1 h at 22 °C. Cond., condition; conv., fraction converted; ND, not determined.
Error is reported as SD of the mean (n = 4).
The detection limit for product formation is estimated to be ∼5% of that found in the WT reaction.
Several point mutants were extensively degraded during heterologous expression in E. coli (SI Appendix, Materials and Methods); thus, their activities were ND.
To establish if any point mutant reduced enzymatic activity or structural instability, we first expressed and purified all 19 mutant BamL proteins from Escherichia coli under identical conditions (SI Appendix, Materials and Methods). BamL W20A, F21A, D91A, and D126A underwent extensive proteolytic degradation and were low yielding, indicating that these positions play an important role in structural stability (SI Appendix, Fig. S6). In an effort to obtain values for the kinetic constants kcat and Km for the mutant panel, we attempted quantitative liquid chromatography MS, coupled-enzymatic assays, and tritiated SAM assays. Despite many attempts, a number of problems arose that limited our ability to determine the desired kinetic parameters. As an alternative, we used internally calibrated MALDI-MS to compare the ion intensities of the desmethylPZN starting material with the PZN product under three reaction conditions (Table 1). In what is referred to as reaction condition A [10 µM MBP-BamL, 10 µM Pfs (SAH nucleosidase), 50 µM desmethylPZN, and 3 mM SAM for 16 h at 37 °C], all tested mutants except for BamL T38A, T38F, L132A, L132F, Y182F, and S190A showed impaired activity. In a more stringent test of activity, the concentrations of MBP-BamL, Pfs, and desmethylPZN were all lowered 10-fold, and the reaction time was limited to 1 h at 22 °C (referred to as reaction condition B). Under condition B, we only observed product formation from BamL T38A and S190A. Importantly, the SAM concentration was intentionally kept high (3 mM) in reaction condition B. To evaluate if a BamL point mutant affected the ability to properly handle SAM, we examined a subset of the mutants in reaction condition C. Here, the desmethylPZN concentration was returned to 50 µM and the SAM concentration was reduced to 500 µM (other variables were the same as in condition B). Upon comparison with condition B, detectable product formation was resurrected from reactions with BamL T38F, L132A, L162A, and Y182F (Table 1). This tripartite assay allowed for the generalized assessment of which residues were required for proper handling of the desmethylPZN and SAM cosubstrates.
Detection of Monoalkylated PZN Analogs.
Although our earlier attempts to identify monomethylPZN as a BamL reaction intermediate were unsuccessful, we observed a strong indication that this species was formed upon analysis of the BamL Y182F reaction using condition A (SI Appendix, Fig. S7). After supplying WT BamL to the Y182F reaction mixture containing m/z 1308, 1322, and 1336, all species converged to m/z 1336 (PZN). Convergence provided definitive evidence that m/z 1322 was indeed monomethylPZN, an on-pathway reaction intermediate as expected.
A close inspection of the SAM-binding pocket within BamL/BpumL suggested that slightly larger alkyl substituents might be tolerated (Fig. 4 and SI Appendix, Fig. S5). To evaluate if an ethyl group could be transferred from a SAM analog bearing an electrophilic ethyl group, rather than the naturally occurring methyl substituent, we synthesized “ethyl SAM” using an established procedure (23). Enzymatic reactions were carried out as performed previously. Analysis of the reaction product by MALDI-MS revealed the transfer of a single ethyl group to desmethylPZN, yielding monoethylPZN (SI Appendix, Fig. S7). This variant is isobaric with native PZN (m/z 1336), which bears two methyl groups. Subjecting this species to CID gave a diagnostic fragment ion of m/z 1277, which localized the ethyl modification to the N terminus. Intriguingly, despite increased reaction times, addition of fresh enzyme, and the use of either ethyl SAM or natural SAM, the corresponding dialkylated PZN derivative was not observed. This suggests that there are insurmountable steric barriers to the formation of monomethyl-monoethylPZN (m/z 1350) and diethylPZN (m/z 1364).
Discussion
In this work, we successfully reconstituted the in vitro activity of the methyltransferase involved in PZN biosynthesis (Fig. 2). We discovered that, in contrast to other small-molecule methyltransferases, BamL is highly specific for substrates closely related to desmethylPZN. Indeed, a variety of peptidic substrates containing N-terminal Arg residues, with and without conformationally restrictive Pro residues included, all failed to be processed (Fig. 3). The molecular basis for this unusual selectivity is that the PZN-binding cleft is sufficiently narrow that substrates lacking contiguous polyheterocyclic structures are simply too wide to be accommodated (Fig. 4 and SI Appendix, Fig. S5). Heterocyclization substantially reduces the width of a peptide as the carbonyl oxygens are removed and the β-nucleophilic side chains (Cys, Ser/Thr) are backbone-cyclized. Moreover, contiguously heterocyclic peptides can more readily adopt the narrow, planar conformations required to reach the active site (Fig. 1 and SI Appendix, Fig. S5). We posit that the lack of activity toward the tetrapeptide substrates bearing minimal side chains (RAAA and RGGG) could arise from these and related features. Contrary to RAAA/RGGG, desmethylPZN can likely pass through the tunnel “pinch point” without adopting an energetically unfavorable conformation or having the enzyme undergo a major structural rearrangement. DesmethylPZN is also considerably more hydrophobic than the tetrapeptides, as evidenced by reverse-phase C18 HPLC elution (desmethylPZN, 72% MeOH; RAAA/RGGG, <15% MeOH for both). Moreover, desmethylPZN is structurally preorganized in a linear form; thus, the entropic penalty for enzyme binding should be vastly lower than for the more conformationally flexible tetrapeptides.
To establish if PZN methyltransferases harbor a narrow substrate-binding cleft, we solved the high-resolution crystal structures of two family members (BamL and BpumL), each in complex with SAH (Fig. 4). Although the overall fold of BamL/BpumL was characteristic of other known methyltransferases, a remarkable feature was a deep tunnel (14–15 Å) running from the surface of the protein toward the active site. Several generally conserved residues within the PZN methyltransferases define the walls of this tunnel, and an orthogonal cleft that we propose stabilizes the guanidinium group of PZN. Although attempts to crystallize BamL/BpumL with (desmethyl)PZN were unsuccessful, analysis of the available structures prompted us to postulate that the tunnel binds the five N-terminal residues of PZN (SI Appendix, Fig. S5). To experimentally support this model, a series of bioinformatics- and structure-guided point mutations were introduced into BamL. The resultant methyltransferase activities were measured in a three-tiered assay, alerting us to residues of potential importance in substrate binding, enzymatic catalysis, and protein structural stability (Table 1). Four BamL mutants (W20A, F21A, D91A, and D126A) were extensively degraded during E. coli heterologous expression and their activities were not determined (Table 1 and SI Appendix, Fig. S6). For mutants less prone to degradation, we first used an excess of enzyme, an elevated concentration of both substrates (SAM and desmethylPZN), and an extended reaction time to detect any trace of activity (condition A). The mutations tested that compose the putative PZN substrate-binding tunnel were BamL D34A, T38A/F, L132A/F, L162A, Y182A/F, and Q186A. Of these, D34A, L162A, and Q186A exhibited decreased activity, and Y182A activity was severely impaired, likely because of structural instability (Table 1 and SI Appendix, Fig. S6). Under condition B, where the enzyme concentration, reaction time, and desmethylPZN concentration were greatly decreased, the only tunnel mutation that gave detectable product formation was T38A. To support that the tunnel mutants were indeed involved in PZN binding, the desmethylPZN concentration was raised and the SAM concentration was lowered (condition C). Product formation was reinstated for all tested mutations (i.e., T38F, L132A, L162A, and Y182F) except for R42A and Q186A, the latter of which is in close proximity to two structurally stabilizing positions (W20 and F21).
Using ITC, we measured a dissociation constant of 7.5 ± 0.8 µM between BamL and SAM (SI Appendix, Fig. S1). In accord with the crystal structures, the three-tiered enzymatic assay supports critical roles for BamL R42 and D91 in binding SAM. As mentioned previously, enzymatic activity was not resurrected for R42A upon changing from condition B to condition C, indicative of an interaction with SAM (Table 1). The structure reveals that the side chain of BamL R42 is engaged in an ionic interaction with the carboxylate moiety of SAH. Further, the carboxylate side chain of BamL D91 makes several contacts with the SAH ribose hydroxyl groups, which upon mutation to Ala yielded degraded protein.
To be effective catalysts, BamL and other PZN methyltransferases need to precisely position the SAM methyl donor and acceptor (SI Appendix, Fig. S8). Given that amines tend to be protonated at physiological pH, BamL might be expected to include an active site base. Although the pKa of this group could be dramatically perturbed in the interior of an enzyme, negating the need for an active site base, our data suggest that BamL D34 and Y182 may be directly involved in catalysis. Although residual activity remains with the point mutants D34A and Y182F (Table 1), the side chains of these residues are in close proximity to each other and the thioether sulfur of SAH (Fig. 4). Considering our unsuccessful attempts to identify monomethylPZN as a reaction intermediate with WT BamL (SI Appendix, Fig. S3), it was gratifying to observe this intermediate with BamL Y182F (SI Appendix, Fig. S7). This result, taken together with the data in Table 1, suggests severely impaired catalysis. It is notable that two members of the PZN methyltransferase family, from C. urealyticum and B. linens, naturally harbor a Phe at this position (BamL Y182F) but a Tyr at BamL F21 (SI Appendix, Fig. S4). As with position 182, residue 21 is in close proximity to D34 and the thioether sulfur atom of SAH.
The results presented here have revealed that the enzyme responsible for converting desmethylPZN to PZN exhibits an unusually selective substrate scope in vitro. Given that PZN is a highly discriminating antibiotic with known activity only toward B. anthracis, the causative agent of anthrax, the characterization of the responsible biosynthetic enzymes remains an important undertaking. Our study lays a foundation for future work wherein the rational engineering of PZN variants bearing unnatural modifications can be produced for the purposes of establishing structure-activity relationships.
Materials and Methods
Detailed methods are provided the SI Appendix, Materials and Methods. Briefly, these cover the experimental procedures followed for the preparation of starting materials, such as isolation of desmethylPZN, cloning, expression, and purification of methyltransferase enzymes, and the generation of site-directed mutants. Also given are the conditions for methyltransferase reconstitution, MS, protein crystallization, structure determination, protein analysis, and ITC.
Supplementary Material
Acknowledgments
We thank John Cronan for the Pfs (SAH nucleosidase) expression plasmid and Xinyun Cao for her technical assistance. We thank members of the D.A.M. laboratory for critically reviewing this manuscript. The Bruker UltrafleXtreme MALDI mass spectrometer was purchased in part with National Center for Research Resources’ National Institutes of Health (NIH) Grant S10 RR027109. Use of the Advanced Photon Source, operated for the US Department of Energy by the Argonne National Laboratory, was supported by Contract DE-AC02-06CH11357. This work was supported in part by the institutional funds provided by the University of Illinois, the NIH Director’s New Innovator Award Program Grant DP2 OD008463 (to D.A.M.), and the Robert C. and Carolyn J. Springborn Endowment (to B.J.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4KVZ and 4KWC).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1306101110/-/DCSupplemental.
References
- 1.Scholz R, et al. Plantazolicin, a novel microcin B17/streptolysin S-like natural product from Bacillus amyloliquefaciens FZB42. J Bacteriol. 2011;193(1):215–224. doi: 10.1128/JB.00784-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kalyon B, et al. Plantazolicin A and B: Structure elucidation of ribosomally synthesized thiazole/oxazole peptides from Bacillus amyloliquefaciens FZB42. Org Lett. 2011;13(12):2996–2999. doi: 10.1021/ol200809m. [DOI] [PubMed] [Google Scholar]
- 3.Molohon KJ, et al. Structure determination and interception of biosynthetic intermediates for the plantazolicin class of highly discriminating antibiotics. ACS Chem Biol. 2011;6(12):1307–1313. doi: 10.1021/cb200339d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee SW, et al. Discovery of a widely distributed toxin biosynthetic gene cluster. Proc Natl Acad Sci USA. 2008;105(15):5879–5884. doi: 10.1073/pnas.0801338105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arnison PG, et al. Ribosomally synthesized and post-translationally modified peptide natural products: Overview and recommendations for a universal nomenclature. Nat Prod Rep. 2013;30(1):108–160. doi: 10.1039/c2np20085f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Melby JO, Nard NJ, Mitchell DA. Thiazole/oxazole-modified microcins: Complex natural products from ribosomal templates. Curr Opin Chem Biol. 2011;15(3):369–378. doi: 10.1016/j.cbpa.2011.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McIntosh JA, Schmidt EW. Marine molecular machines: Heterocyclization in cyanobactin biosynthesis. ChemBioChem. 2010;11(10):1413–1421. doi: 10.1002/cbic.201000196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Milne JC, Eliot AC, Kelleher NL, Walsh CT. ATP/GTP hydrolysis is required for oxazole and thiazole biosynthesis in the peptide antibiotic microcin B17. Biochemistry. 1998;37(38):13250–13261. doi: 10.1021/bi980996e. [DOI] [PubMed] [Google Scholar]
- 9.Dunbar KL, Melby JO, Mitchell DA. YcaO domains use ATP to activate amide backbones during peptide cyclodehydrations. Nat Chem Biol. 2012;8(6):569–575. doi: 10.1038/nchembio.944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Melby JO, Dunbar KL, Trinh NQ, Mitchell DA. Selectivity, directionality, and promiscuity in peptide processing from a Bacillus sp. Al Hakam cyclodehydratase. J Am Chem Soc. 2012;134(11):5309–5316. doi: 10.1021/ja211675n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Milne JC, et al. Cofactor requirements and reconstitution of microcin B17 synthetase: A multienzyme complex that catalyzes the formation of oxazoles and thiazoles in the antibiotic microcin B17. Biochemistry. 1999;38(15):4768–4781. doi: 10.1021/bi982975q. [DOI] [PubMed] [Google Scholar]
- 12.Claesen J, Bibb M. Genome mining and genetic analysis of cypemycin biosynthesis reveal an unusual class of posttranslationally modified peptides. Proc Natl Acad Sci USA. 2010;107(37):16297–16302. doi: 10.1073/pnas.1008608107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Claesen J, Bibb MJ. Biosynthesis and regulation of grisemycin, a new member of the linaridin family of ribosomally synthesized peptides produced by Streptomyces griseus IFO 13350. J Bacteriol. 2011;193(10):2510–2516. doi: 10.1128/JB.00171-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Q, van der Donk WA. Catalytic promiscuity of a bacterial α-N-methyltransferase. FEBS Lett. 2012;586(19):3391–3397. doi: 10.1016/j.febslet.2012.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hendricks CL, Ross JR, Pichersky E, Noel JP, Zhou ZS. An enzyme-coupled colorimetric assay for S-adenosylmethionine-dependent methyltransferases. Anal Biochem. 2004;326(1):100–105. doi: 10.1016/j.ab.2003.11.014. [DOI] [PubMed] [Google Scholar]
- 16.Kapust RB, Waugh DS. Escherichia coli maltose-binding protein is uncommonly effective at promoting the solubility of polypeptides to which it is fused. Protein Sci. 1999;8(8):1668–1674. doi: 10.1110/ps.8.8.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mitchell DA, et al. Structural and functional dissection of the heterocyclic peptide cytotoxin streptolysin S. J Biol Chem. 2009;284(19):13004–13012. doi: 10.1074/jbc.M900802200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang C, et al. RebG- and RebM-catalyzed indolocarbazole diversification. ChemBioChem. 2006;7(5):795–804. doi: 10.1002/cbic.200500504. [DOI] [PubMed] [Google Scholar]
- 19.Zhang C, Weller RL, Thorson JS, Rajski SR. Natural product diversification using a non-natural cofactor analogue of S-adenosyl-L-methionine. J Am Chem Soc. 2006;128(9):2760–2761. doi: 10.1021/ja056231t. [DOI] [PubMed] [Google Scholar]
- 20.Lee JH, et al. Characterization and structure of DhpI, a phosphonate O-methyltransferase involved in dehydrophos biosynthesis. Proc Natl Acad Sci USA. 2010;107(41):17557–17562. doi: 10.1073/pnas.1006848107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anderle C, et al. Improved mutasynthetic approaches for the production of modified aminocoumarin antibiotics. Chem Biol. 2007;14(8):955–967. doi: 10.1016/j.chembiol.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 22.Martin JL, McMillan FM. SAM (dependent) I AM: The S-adenosylmethionine-dependent methyltransferase fold. Curr Opin Struct Biol. 2002;12(6):783–793. doi: 10.1016/s0959-440x(02)00391-3. [DOI] [PubMed] [Google Scholar]
- 23.Islam K, Zheng W, Yu H, Deng H, Luo M. Expanding cofactor repertoire of protein lysine methyltransferase for substrate labeling. ACS Chem Biol. 2011;6(7):679–684. doi: 10.1021/cb2000567. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.