Significance
The transcription factor SOX2 plays a critical role in self-renewal and neuronal differentiation of neural precursors (NPCs); however, the molecular mechanisms underlying its functions are poorly understood. We found that SOX2 regulates the expression of LIN28, a suppressor of let-7 microRNA biogenesis. Exogenous LIN28 rescued the NPC proliferation and some neurogenic deficits in the absence of SOX2. We identified let-7i as a novel and potent inhibitor of neuronal differentiation that represses proneural genes. The discovery of SOX2–LIN28/let-7 pathway that maintains both NPC proliferation and neurogenic potential will enhance our understanding and therapeutic development relevant to neurodegeneration and brain tumors.
Keywords: neural stem cells, mechanisms of pluripotency
Abstract
The transcription factor SRY (sex-determining region)-box 2 (SOX2) is an important functional marker of neural precursor cells (NPCs) and plays a critical role in self-renewal and neuronal differentiation; however, the molecular mechanisms underlying its functions are poorly understood. Using human embryonic stem cell-derived NPCs to model neurogenesis, we found that SOX2 is required to maintain optimal levels of LIN28, a well-characterized suppressor of let-7 microRNA biogenesis. Exogenous LIN28 expression rescued the NPC proliferation deficit, as well as the early but not the late stages of the neurogenic deficit associated with the loss of SOX2. We found that SOX2 binds to a proximal site in the LIN28 promoter region and regulates LIN28 promoter acetylation, likely through interactions with the histone acetyltransferase complex. Misexpression of let-7 microRNAs in NPCs reduced proliferation and inhibited neuronal differentiation, phenocopying the loss of SOX2. In particular, we identified let-7i as a novel and potent inhibitor of neuronal differentiation that targets MASH1 and NGN1, two well-characterized proneural genes. In conclusion, we discovered the SOX2–LIN28/let-7 pathway as a unique molecular mechanism governing NPC proliferation and neurogenic potential.
Self-renewing, multipotent neural precursor cells (NPCs) are capable of terminally differentiating into neuronal and glial lineages during development and in the adult nervous system (1, 2). Disruption of the pathways controlling NPC biology has been implicated in various pathologies, including autism (3), Treacher Collins syndrome (4), and neural tube defects (5), emphasizing the importance of gaining a better understanding of the underlying molecular events and how they may be manipulated to treat and prevent such pathologies. The HMG-box transcription factor SOX2 is ubiquitously expressed in NPCs and supports their self-renewal (6). SOX2 is also required for neurogenesis in the central nervous system (7–10). Recently, we used human embryonic stem cells (hESCs) and mouse models to demonstrate a critical requirement for SOX2 for sensory neurogenesis in dorsal root ganglia (11). However, the mechanisms by which SOX2 functions in self-renewal and neuronal differentiation remain poorly understood.
Small, noncoding microRNAs (miRNAs) are transcribed as long precursors (pri-miRNAs) that are sequentially processed by the RNases Drosha/Pasha to form pri-miRNAs and by Dicer to form the mature miRNAs of ∼20–25 nucleotides. The miRNAs function through imperfect base-pairing with hundreds of target mRNAs to trigger their degradation (or block their translation) by the RNA-induced silencing complex, RISC (12). In some cases, miRNA maturation is tightly controlled; for example, the RNA-binding protein LIN28 regulates the biogenesis of the let-7 miRNA family by inhibiting their maturation at both the pri-miRNA (13, 14) and premiRNA (15, 16) processing steps. Intriguingly, LIN28 protein was found to be associated with SOX2 protein in mouse ESC, but the functional consequences of the interaction are unclear (17, 18). Beyond its role in let-7 biogenesis, LIN28 was recently reported to regulate splicing of a large proportion mRNAs and to bind numerous other mRNAs, many of which play roles in growth and survival (19, 20).
MicroRNAs have been shown to control gene expression in many important cellular functions such as stress signaling and differentiation, and in diseases such as cancer (21, 22). Several miRNAs have been reported to regulate mammalian neurogenesis by promoting neuronal differentiation (miR-124) (23) or by supporting NPC maintenance and opposing their differentiation (miR-184 and miR-137) (24, 25). The let-7 family has been shown to attenuate proliferation by inhibiting cell cycle regulators such as RAS, high mobility group AT-hook 2 (HMGA2), Cyclin D, and CDC25 (26–28). In particular, the let-7b miR was found to inhibit self-renewal and promote neuronal differentiation of mouse NPCs by targeting the critical stem cell transcription factor TLX and the cell cycle regulator CCND1 (29). Here, we present evidence for an unsuspected link between SOX2 and the LIN28/let-7 pathway and provide a molecular mechanism for the dual role of SOX2 in maintaining NPC proliferation and neurogenic potential.
Results
SOX2 Positively Regulates LIN28 Expression.
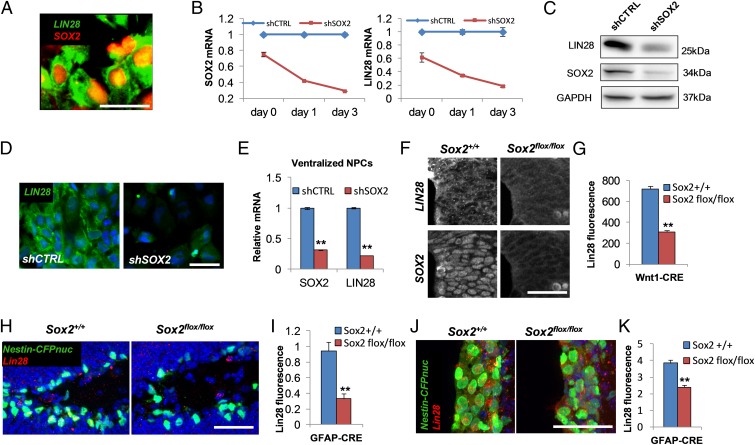
Our previous microarray gene expression analysis of hESC-derived multipotent NPCs (11, 30, 31) suggested that SOX2 regulates the expression of LIN28, a well-studied repressor of let-7 family biosynthesis (13–16). Immunofluorescence analysis confirmed that SOX2 and LIN28 were coexpressed in all NPCs (Fig. 1A). We used NPCs derived from a hESC line carrying a doxycycline (dox)-inducible SOX2 shRNA (11) to knock down SOX2 and monitor the expression of LIN28 transcripts. Following dox treatment, we observed loss of SOX2 expression with concomitant and proportional loss of LIN28, as measured by quantitative PCR (qPCR) analysis (Fig. 1B). Down-regulation of LIN28 (60–80% decrease) was confirmed at the protein level by Western blot and immunocytochemistry (Fig. 1 C and D). The reintroduction of exogenous SOX2 in SOX2 knockdown NPCs rescued LIN28 expression (Fig. S1 A and B; note that the shRNA targets SOX2 3′ UTR and therefore does not affect the exogenous SOX2 coding sequence). SOX2 knockdown also triggered a less pronounced decrease (∼45%) in expression of LIN28B, a LIN28 homolog that also suppresses let-7 maturation (Fig. S1C). Our conventional neuralization protocol (see Materials and Methods for details) resulted in neural precursors uniformly positive for PAX3 and SOX9, and up to 80% positive for SOX10 (Fig. S2 A–K). These neural precursors are biased toward a dorsal identity and are hereafter referred to as NPCs. To investigate whether SOX2 regulation of LIN28 depends on the regional identity of the NPCs, we used ventralized NPCs (vNPCs) obtained by patterning hESC during neuralization with the Sonic Hedgehog (SHH) agonist purmorphamine. vNPCs expressed high levels of the ventral markers OLIG2 and FOXA2 and displayed markedly reduced levels of the dorsal markers PAX3 and GDF7 (Fig. S2 I–K). Knockdown of SOX2 in vNPCs resulted in robust down-regulation of LIN28 (Fig. 1E), suggesting that SOX2 regulation of LIN28 expression is independent of the NPC regional identity.
Fig. 1.
SOX2 regulates the expression of LIN28. (A) Immunostaining for SOX2 and LIN28 in hESC-derived NPCs. (B) qPCR analysis of SOX2 (Left) and LIN28 (Right) expression following SOX2 knockdown in hESC-derived NPCs. shRNA was induced by dox on day 0. Data are normalized to HPRT. shSOX2, SOX2 shRNA; shCTRL, scrambled shRNA. (C) Western blot for SOX2 and LIN28 in SOX2 knockdown cells 3 d after dox induction. (D) Immunostaining for LIN28 in SOX2 knockdown and control NPCs 3 d after dox induction. (E) qPCR analysis of SOX2 and LIN28 expression in ventralized NPCs 3 d after dox induction. (F) Immunostaining for SOX2 and LIN28 in fetal neural tubes (E11) of wild-type (Sox2+/+) and SOX2 knockout (Sox2flox/flox) mice with conditional deletion in the dorsal neural tube (Wnt1-Cre::Sox2flox). (G) Quantification of LIN28 fluorescence shown in F. (H) Immunostaining for LIN28 in the subgranular layer of the adult hippocampal dentate gyrus in wild-type (Sox2+/+) and SOX2-deleted (Sox2flox/flox) mice. (I) Quantification of LIN28 fluorescence in CFP+ cells shown in H. (J) Immunostaining for LIN28 in the subventricular zone of wild-type (Sox2+/+) and SOX2-deleted (Sox2flox/flox) mice. (K) Quantification of LIN28 fluorescence in CFP+ cells shown in J. **P < 0.005. Nestin-CFPnuc transgene labels neuronal precursors in H and J. Nuclei are in blue (DAPI). (Scale bars: 50 µm.) Error bars ± SE.
Next, we used mouse genetic models to investigate the dependence of LIN28 expression on SOX2 in vivo. LIN28 was highly expressed in the developing neural tube (E11) of wild-type animals (Fig. 1F). In contrast, ablation of SOX2 expression in Sox2flox::Wnt1-Cre mutant mice [Sox2flox allele (8) crossed with Wnt1-Cre mice (32)] reduced LIN28 expression by 60% in the dorsal region, where the Wnt1-Cre transgene is most active (Fig. 1 F and G). These results demonstrate that SOX2 is required for expression of physiological levels of LIN28 in the developing neural tube. We also analyzed LIN28 expression in the subgranular layer of the dentate gyrus, the area of adult hippocampal neurogenesis (33). LIN28 was readily detected in NPCs (CFPnuc+ cells; Fig. 1H) of Nestin-CFPnuc transgenic mice (34). We engineered Sox2flox::mGFAP-Cre::Nestin-CFPnuc mice in which SOX2 is ablated in adult NPCs (35) (Fig. S2 L and M). In these mice, levels of LIN28 were reduced by 65% in the neural precursors (CFPnuc+ cells) within the subgranular layer of dentate gyrus (Fig. 1 H and I and Fig. S2 N and O). Finally, we used the same transgenic mouse model to assess the effect of SOX2 knockout on LIN28 expression in the adult NPCs of the subventricular zone (SVZ), another region of the adult mouse brain displaying active neurogenesis. Similarly to that observed in the subgranular layer of dentate gyrus, we found significant reduction of LIN28 levels in the neural precursors (CFPnuc+ cells) of SOX2 conditional knockout mice (Fig. 1 J and K and Fig. S2 P and Q).
Collectively, our in vitro data from human NPCs with different regional identities, and the in vivo data in the developing spinal cord and neurogenic niches of the adult brain, indicate a general requirement of SOX2 for optimal levels of LIN28 expression in neural stem/precursor cells.
SOX2 Modulates Histone Acetyltransferase Activity at the LIN28 Promoter.
A bioinformatic analysis of the LIN28 promoter in several species revealed the presence of putative SOX binding sites, including a conserved site proximal to the transcription initiation site (distance <150 bp; Fig. S3A). We assayed human LIN28 2-kb proximal promoter (containing either a wild-type or a mutated SOX binding site) in wild-type or SOX2 knockdown NPCs (Fig. S3 B and C). The mutated promoter was significantly less active compared with wild type in the presence Sox2; however, no differences were observed in the absence of Sox2 (Fig. S3 B and C), suggesting that SOX2 directly modulate the activity of LIN28 promoter at least in part through the conserved proximal SOX binding site. We next directly assessed SOX2 binding at the human LIN28 promoter in hESC-derived NPCs by ChIP-qPCR, and we found that SOX2 binds in proximity of the transcription start site (Fig. 2 A and B). Overall, these data suggest that SOX2 directly interacts with LIN28 genomic regulatory elements. In a previous study (11), we identified SOX2-interacting proteins in hESC-derived NPCs by immunoprecipitation followed by mass spectrometry (IP-MS). Intriguingly, we found that SOX2 interacts with several protein components of histone acetyltransferase (HAT) complexes (Fig. 2C), and among these were DNMT1-associated protein 1 (DMAP1), TBP-associated factor (TAF), TAF5, TAF6, and the HAT adaptor protein TRRAP (36), which was recently found to interact with SOX2 in mouse NPCs (37). We confirmed that TRRAP is expressed and colocalized with SOX2 in the nuclei of NPCs (Fig. 2D), hinting at the possibility that SOX2 might be associated with a HAT adaptor complex that increases HAT activity on the LIN28 promoter. Indeed, ChIP experiments with a TRRAP-specific antibody demonstrated the presence of TRRAP-containing complexes associated with the LIN28 promoter in the proximity of the SOX2 binding site (Fig. 2E). Furthermore, down-regulation of SOX2 resulted in a significant reduction in TRRAP-containing complexes at the LIN28 promoter (Fig. 2F) and reduced the binding of HATs known to interact with TRRAP [i.e., GCN5 (38) and PCAF (39); Fig. 2F]. These results suggest that the interaction of TRRAP and HATs with LIN28 regulatory elements requires SOX2.
Fig. 2.
SOX2 affects the epigenetic state of human LIN28 promoter. (A) Schematic of the putative SOX2 binding sites on the human LIN28 promoter. (B) ChIP-qPCR showing SOX2 enrichment at the LIN28 promoter. (C) SOX2 was shown to interact with TRRAP and other proteins found in HAT complexes using a SOX2 IP-MS approach. (D) NESTIN/TRRAP (Upper) and SOX2/TRRAP (Lower) costaining showing TRRAP expression in NPCs. (E) ChIP-qPCR analysis of TRRAP binding to LIN28 promoter in proximity to the SOX2 binding site. (F) ChIP-qPCR analysis demonstrated reduced binding of TRRAP and HATs (GCN5 and PCAF) at the LIN28 promoter in SOX2 knockdown (shSOX2) vs. control (shCTRL) cells. (G) ChIP-qPCR with antibodies specific for H3K9Ac, H3K9/18Ac, and control IgG demonstrated reduced levels of H3 acetylation at the LIN28 promoter in shSOX2 vs. shCTRL cells. Analyses were performed 3 d after dox-mediated shRNA induction. *P < 0.05, **P < 0.005. Error bars ± SE. (Scale bars: 50 µm.)
Consistent with the reduced binding of TRRAP and HATs at the LIN28 promoter, SOX2 knockdown reduced the acetylation of histone H3 at lysine 9 (H3K9Ac) and lysine 18 (H3K18Ac), epigenetic markers for open, actively transcribed chromatin (Fig. 2G). A 24-h treatment of NPCs with the HAT inhibitor MB3 significantly reduced the levels of LIN28 mRNA (Fig. S3D), suggesting that histone acetylation is functionally required to maintain high levels of LIN28 expression. To evaluate whether the reduced assembly of the HAT machinery at LIN28 promoter was specific for LIN28 or due to a global loss of acetylation and HAT functional components, we quantified TRRAP, GCN5, and global H3K9/18ac in SOX2 knockdown NPCs. Loss of SOX2 did not alter TRRAP expression (Fig. S4 A and B), and GCN5 was slightly down-regulated (53% and 39% decrease by qPCR and Western blot, respectively; Fig. S4 A and B). The global levels of H3K9/18Ac were not altered in SOX2 knockdown NPCs (Fig. S4 C and D). These results suggest that SOX2 knockdown induces a local reduction of H3K9/18 acetylation at the LIN28 promoter. The loss of histone acetylation in SOX2 knockdown cells could also be explained if SOX2 inhibits histone deacetylases (HDACs) at the LIN28 promoter; if so, we reasoned that chemical inhibition of HDACs should mimic SOX2 function and increase LIN28 expression, even in the absence of SOX2. However, we found that HDAC inhibition in shSOX2-treated cells did not increase LIN28 expression (Fig. S3E), suggesting that SOX2 regulation of LIN28 promoter acetylation is likely mediated only by HAT activity. Considering that TRRAP-containing complexes regulate different types of histone acetylation, it is possible that additional epigenetic mechanisms may be operating. Collectively, our results suggest that SOX2 regulates the epigenetic state (H2K9/18Ac) of LIN28 promoter and that in the absence of SOX2, LIN28 promoter exhibits a decreased H3K9/18 acetylation associated with a more closed chromatin configuration.
LIN28 Modulates NPC Proliferation and Neuronal Commitment.
SOX2 knockdown in proliferating NPCs (i.e., cultured under self-renewal conditions) decreased the cell number compared with control cultures; however, immunostaining for activated caspase 3 (AC3) showed no change in apoptosis, suggesting that the rate of cell death is unchanged under these conditions. Instead, SOX2 knockdown dramatically decreased NPC proliferation, as assessed by immunostaining of the cell cycle marker Ki67 (52% reduction; Fig. 3 A and B) and BrdU incorporation (30.9 ± 1.9% BrdU+ cells in control NPCs vs. 16.9 ± 2.9% in SOX2 knockdown cells). Because LIN28 has been shown to promote proliferation in both let-7–dependent (28) and –independent (19) manners, we asked if exogenous LIN28 would rescue NPC proliferation in the absence of SOX2. Inducible expression of exogenous LIN28 in SOX2 knockdown NPCs restored LIN28 to 63% of its endogenous level (while SOX2 levels remained low; Fig. 3H). Reexpression of LIN28 efficiently rescued NPC proliferation, as assessed by Ki67 staining (89% of control; Fig. 3 A and B) and BrdU incorporation (16.9 ± 2.9% BrdU+ cells in SOX2 knockdown NPCs vs. 35.6 ± 1.2% in SOX2 knockdown cells expressing exogenous LIN28). These results suggest that LIN28 can fully rescue the proliferative deficits associated with SOX2 knockdown and therefore establish LIN28 as a major downstream mediator of SOX2-dependent proliferation in NPCs.
Fig. 3.
LIN28 rescues SOX2-dependent proliferation and an early but not late stage of neurogenesis. (A) Immunostaining for Ki67 in NPCs expressing dox-inducible scrambled shRNA (Left, shCTRL), inducible SOX2 shRNA (Center, shSOX2/RFP), and inducible SOX2 shRNA in conjunction with inducible LIN28 (Right, shSOX2/LIN28). We overexpressed red fluorescent protein (RFP) in shSOX2/RFP cells to control for LIN28 overexpression in shSOX2/LIN28 NPCs. In all of the assays described in this figure, the shSOX2/RFP line performed similarly to the shSOX2 line. (B) Quantification of the Ki67 staining shown in A. (C) Immunostaining for TUJ1 in shCTRL, shSOX2/RFP, and shSOX2/LIN28 lines cultured under neuronal differentiation conditions. (D) Quantification of the TUJ1 staining shown in C. (E) Immunostaining for AC3 in shCTRL, shSOX2/RFP, and shSOX2/LIN28 lines cultured under neuronal differentiation conditions. (F) Quantification of AC3 staining shown in E. (G) MAP2 staining in shCTRL, shSOX2/RFP, and shSOX2/LIN28 lines cultured under neuronal differentiation conditions. (H) qPCR analysis of SOX2 and LIN28 expression in shSOX2/RFP, shSOX2/LIN28, and shCTRL NPC lines was performed to confirm exogenous expression of LIN28 in the shSOX2/LIN28 line. Data are normalized to HPRT. *P < 0.05, **P < 0.005. Nuclei are in blue (DAPI). Error bars ± SE. (Scale bars: 50 µm.)
We previously found that knocking down SOX2 precludes NPC differentiation into neurons without affecting glial fates (11). Indeed, SOX2 knockdown in NPCs cultured under conditions favoring neuronal differentiation strongly reduced the appearance of young TUJ1+ neurons (Fig. 3 C and D) and completely inhibited the more mature MAP2+ cells (Fig. 3G). The loss of neurons was likely due to extensive apoptosis, as evaluated by immunostaining for AC3 (Fig. 3 E and F). Expression of exogenous LIN28 on the background of SOX2 knockdown rescued the early stages of neuronal differentiation (TUJ1 immunostaining; Fig. 3 C and D) and reduced apoptosis by 57% (AC3 immunostaining; Fig. 3 E and F), suggesting that prevention of cell death might contribute to the observed LIN28-dependent rescue. However, LIN28 overexpression was unable to rescue neuronal maturation (MAP2 staining; Fig. 3G), suggesting that other functions of SOX2 are required to achieve neuronal maturation.
Taken together, these results suggest that LIN28 plays a major role in controlling SOX2-dependent NPC proliferation and contributes to SOX2-dependent neurogenesis.
Role of let-7 in NPC Proliferation and Neuronal Differentiation.
LIN28 is well known to be a suppressor of let-7 miR biogenesis. Therefore, we investigated the effect of SOX2 knockdown (and subsequent LIN28 down-regulation) on expression of the let-7 miR family in NPCs. Quantification of mature miRNA expression upon SOX2 knockdown using TaqMan miRNA arrays showed that 50% of total let-7 family members were up-regulated, 25% were not affected, and 25% were down-regulated (Fig. S5A). Subsequent qPCR analysis confirmed the up-regulation of mature let-7b miR (albeit at lower levels, suggesting that TaqMan array analysis may overestimate the up-regulation of let-7 miRs) and identified an increase in mature let-7i miR, which was not detected on the TaqMan array (Fig. 4A). In agreement with the observed up-regulation of let-7b, qPCR analysis of SOX2 knockdown NPCs revealed significant down-regulation of the reported let-7b targets TLX, CCND1 (29), CDC25A (28) (Fig. 4 B–D), and confirmed targets of other let-7, such as E2F transcription factor 2 (E2F2) (26) and Kirsten rat sarcoma viral oncogene homolog (KRAS) (27, 40, 41) (Fig. S5B). The let-7 targets were also down-regulated in vNPCs following SOX2 knockdown (Fig. S5C). Reexpression of LIN28 in SOX2 knockdown NPCs inhibited let-7b and let-7i maturation (Fig. 4E) and derepressed let-7 target genes (Fig. 4F), suggesting a canonical pathway for LIN28 regulation of let-7b and let-7i biogenesis.
Fig. 4.
SOX2 regulates the LIN28/let-7 pathway in NPCs. (A) TaqMan qPCR analysis of mature let-7b and let-7i miRs 3 d after shSOX2 or shCTRL induction by dox, normalized to U6 small RNA. (B–D) qPCR analysis of known let-7b targets (normalized to HPRT) over 3 d following SOX2 knockdown in NPCs. Dox was added on day 0. (E) TaqMan qPCR analysis of mature let-7b and let-7i miRs in control NPCs (shCTRL), cells with inducible SOX2 shRNA (shSOX2/RFP), and cells with inducible SOX2 shRNA and inducible LIN28 (shSOX2/LIN28). Inducible RFP expression in shSOX2/RFP served as a control for LIN28 expression. (F) qPCR analysis of the let-7b target genes CCND1, TLX, and CDC25A in shCTRL, shSOX2/RFP, and shSOX2/LIN28 lines. Cells were analyzed 3 d after dox-mediated induction of shRNA and LIN28/RFP overexpression. (G–J) Monitoring of let-7 activity in NPCs and mature neurons with let-7 sensors. (G) No differences in GFP fluorescence were seen for control and let-7 sensors in nestin-positive NPCs, suggesting low let-7 activity (quantified in H). MAP2+ neurons (21 d of neuronal differentiation) transduced with let-7 sensors (I) uniformly lack GFP expression compared with control (quantified in J), suggesting high let-7 activity in mature neurons. *P < 0.05, **P < 0.005. Error bars ± SE. (Scale bar: 50 µm.)
To monitor let-7 activity during neuronal differentiation, we engineered lentiviral reporters encoding GFP with 3′ UTR containing five perfect let-7b or let-7i target sequences (let-7b/i sensors; Fig. S6). Let-7 activity was not detected in NPCs, consistent with the high levels of SOX2 and LIN28 expression in these cells (Fig. 4 G and H). In contrast, let-7 miRNAs were highly active in mature MAP2+ neurons (Fig. 4 I and J), suggesting that let-7 miRNA accumulates in these cells.
To investigate the consequence of let-7 misexpression in NPCs, we generated hESC lines expressing dox-inducible let-7b or let-7i and differentiated the lines into NPCs. Under self-renewal conditions, overexpression of let-7b reduced NPC proliferation (Fig. 5 A and B), but let-7i had no apparent effect (Fig. 5 A and B). Under neurogenic conditions, let-7b overexpression reduced the number of MAP2+ neurons by ∼35% (Fig. 5 C and D), whereas let-7i overexpression almost completely abolished MAP2 expression and cells with neuronal morphologies (Fig. 5 C and D). Furthermore, let-7i overexpression induced a dramatic increase in apoptosis, similar to the effect of SOX2 knockdown under neurogenic conditions (Fig. S7 A and B). These results indicate that different let-7 family members might have selective effects on NPC proliferation and neurogenesis, although it is possible that the observed functional differences may be diminished at higher expression levels (Fig. S6).
Fig. 5.
Let-7 miRs antagonize NPC self-renewal and neurogenesis. (A and B) Effect of let-7b and let-7i overexpression on NPC proliferation. Immunostaining for Ki67 and pHH3 and quantification of Ki67+ and pHH3+ cells. (C and D) Effect of let-7b and let-7i overexpression on neuronal differentiation of NPCs. Immunostaining and quantification of MAP2 after 21 d. (E) qPCR analysis of proneural genes (MASH1, NGN1) in control (shCTRL), shSOX2, and let-7i–overexpressing NPCs. Data are normalized to HPRT expression. (F) Schematic of the GFP reporters for let-7 activity at MASH1 3′ UTR. Black boxes, wild-type; red crossed boxes, mutated let-7 binding sites from MASH1 3′ UTR. (G and H) GFP fluorescence of wild-type and mutant GFP-MASH1 3′ UTR reporters in the presence of scrambled shRNA (shCTRL) or let-7i miR. (I) Schematic of the SOX2-LIN28/let-7 pathway activity regulating proliferation and neurogenesis in NPCs. *P < 0.05, **P < 0.005. Error bars ± SE. (Scale bars: 50 µm.)
Because the role of let-7 in regulating proliferation is well established, we focused on their involvement in neurogenesis—in particular, let-7i, which showed the strongest antagonistic activity (Fig. 5 C and D). Previously, we suggested that SOX2 may be required in NPCs for expression of the proneural genes MASH1and NGN1 (11). Indeed, overexpression of let-7i strongly reduced expression of both MASH1 and NGN1, mimicking the effects of SOX2 knockdown (Fig. 5E). A bioinformatic analysis predicted the existence of two let-7 binding sites in the human MASH1 3′ UTR and one site in the human NGN1 3′ UTR (Fig. S8). These binding sites were also conserved in the murine Mash1 and Ngn1 mRNAs (Fig. S8 A–C). To determine whether MASH1 is a direct target of let-7i, we fused the GFP coding sequence with a 200-bp sequence of the MASH1 3′ UTR containing the two predicted let-7 binding sites (Fig. 5F). Cotransfection of HEK293T cells with a let-7i expression vector and the GFP-MASH1 3′ UTR reporter resulted in a significant reduction in GFP expression (Fig. 5G), which was reversed by introducing mutations into both let-7 binding sites (Fig. 5H). These data suggest that the proneural gene MASH1 is a direct target of let-7i.
Collectively, these results demonstrate that individual members of the let-7 miR family may preferentially affect NPC proliferation or neuronal differentiation and suggest that the combined activity of let-7 miRs phenocopies the loss of SOX2 in NPCs with respect to proliferation and neurogenesis. We identify LIN28 as a direct SOX2 effector that inhibits activation of the let-7 family, thus enabling the proliferation and subsequent neuronal differentiation of NPCs (Fig. 5I). Our findings provide a mechanism for the previously observed dual requirement for SOX2 in proliferation and neuronal differentiation of NPCs.
Discussion
The molecular mechanisms underlying SOX2 function in NPCs are not well understood. Here, we discovered that SOX2 is required to maintain endogenous levels of LIN28, a master regulator of let-7 miR biogenesis that was recently implicated in global modulation of mRNA splicing (20). The knockdown studies in hESC-derived NPCs and genetic ablation of SOX2 in mouse models established that SOX2 is required for optimal LIN28 expression in cultures and in vivo. We found that the function of SOX2 in NPC proliferation is largely mediated by LIN28, because LIN28 overexpression is sufficient to rescue the proliferation deficits associated with the loss of SOX2. Under neurogenic conditions, LIN28 was able to rescue apoptosis and the early (βIII-tubulin+ cells) but not late (MAP2+ cells) stages of neurogenesis after SOX2 knockdown. Based on our in vitro findings, we speculate that SOX2 regulates LIN28 expression in part by recruiting HATs through the TRRAP adaptor to the LIN28 promoter to maintain a high level of H3K9/18 acetylation and an open chromatin state of LIN28 promoter in NPCs. It is possible, however, that other mechanisms downstream of SOX2, such as a classical transcriptional transactivation (42) or a global regulation of HATs levels (43), contribute to the regulation of LIN28 expression in NPCs.
Recently, LIN28 was shown to regulate the abundance of splicing factors and to interact with numerous mRNAs (19, 20), several of which play roles in growth and survival (19, 20), thus providing one potential mechanism by which LIN28 may promote NPC proliferation independently of let-7. However, the role of LIN28 in the repression of let-7 miR family biosynthesis is better understood (13–16). The let-7 family plays well-established roles in controlling proliferation (26–28), such as the role of let-7b in targeting TLX and CCND1 in neural stem cells and in promoting neuronal differentiation (29). However, let-7 miRNAs can act as inhibitors of neuronal differentiation (44); for example, let-7 regulates neuronal differentiation in zebrafish through targeting Mash1/Ascl1 (45). We found that let-7b overexpression inhibited NPC proliferation, whereas let-7i overexpression was significantly less active under the same conditions. Although both let-7b and let-7i inhibited NPC neurogenesis, let-7i overexpression produced a much stronger effect. Modeling the mRNA degradation with the let-7 sensors suggest that the effect of a specific let-7 miR on a particular target mRNA depends on the degree of miR–mRNA complementarity and on the ratio of particular let-7 miR and its mRNA target. Although the let-7 family members are often considered to have redundant functions (21), studies in squamous cell carcinomas of the head suggest that let-7b inhibits proliferation, let-7d represses epithelial to mesenchymal transition, and let-7i mainly inhibits mesenchymal cell movement (46–48). Our observations support the possibility that the let-7 family members exhibit target selectivity and thus might promote different cell fates even when expressed in the same cell type.
In conclusion, we identified LIN28 as a direct mediator of SOX2-dependent proliferation in neural precursors. We showed that LIN28 is required in NPCs, at least in part, to suppress the precocious biogenesis of active let-7 family miRNAs. Indeed, let-7 activity was detected in mature MAP2+ neurons but not in NPCs. The let-7 activity would be deleterious for self-renewing multipotent NPCs because it inhibits both proliferation (e.g., TLX, CCND1, CDC25A) and neuronal commitment (e.g., NGN1, MASH1). It is tempting to speculate that the down-regulation of SOX2 and LIN28 followed by the induction of let-7 expression that occurs during the final stages of neurogenesis is required to orchestrate cell cycle exit and the extinction of proneural gene transcripts that are only transiently required during neurogenesis and are absent in mature neurons (49). This hypothesis is consistent with the evidence that LIN28 negatively regulates gliogenesis independently of let-7 miRs (44), as well as our previous observation that gliogenesis is not affected by the loss of SOX2 from neural precursors (11). In humans, SOX2 haploinsufficiency results in abnormal development of the hippocampus, and epilepsy (50); anophthalmia (51); retarded growth and sensorineural deafness (52); and genital anomalies (53). Our findings suggest that some of these phenotypes could be mediated through the LIN28/let-7 pathway, thus identifying potential new targets for therapeutic intervention.
Materials and Methods
Maintenance of hESCs.
H9 hESCs were maintained on a feeder layer of mouse embryonic fibroblasts and in Matrigel-coated plates [BD Biosciences; final Matrigel dilution = 1:30 in PBS, 2-h coating at room temperature (RT)] in Knockout DMEM (Gibco) containing 20% serum replacement (Gibco), 1× nonessential amino acids (Gibco), 2 mM l-glutamine (Gibco), 0.1 mM β-mercaptoethanol (Gibco), 1× antibiotics/antimycotics (Omega), and 8 ng/mL bFGF (Sigma). hESCs were passaged every 5–7 d by manual removal of morphologically identifiable differentiated colonies followed by treatment with 1 mg/mL collagenase type IV (Gibco) diluted in knockout DMEM. The medium was changed daily.
Derivation, Maintenance, and Differentiation of Human NPCs.
Neurospheres were generated from hESCs as previously described (30, 54). The protocol yields NPCs with a clear dorsal identity (Fig. S2 A–K). To ventralize the cells, NPCs at the rosette stage (5–7 d after neural induction) were treated with the SHH agonist purmorphamine (0.5 µM) for 7 d. Monolayer cultures of hESC-derived NPCs were propagated on Matrigel-coated plates in base medium [1:1 ratio of DMEM/F12 GlutaMAX-neurobasal medium (Gibco), 2% B27 supplement without vitamin A (Gibco), 10% BIT 9500 (StemCell Technologies), and 1 mM glutamine (Gibco)] supplemented with 20 ng/mL EGF (Chemicon), 20 ng/mL bFGF, 5 µg/mL insulin (Sigma), 10 ng/mL LIF (Millipore), and 5 mM nicotinamide (Sigma). hESC-derived NPCs were subcultured enzymatically with Accutase (Chemicon) at a 1:2 or 1:3 ratio approximately once a week. To induce neuronal differentiation, hESC-derived NPCs were seeded onto fibronectin-coated plates (2 µg/mL, overnight coating) at high density (3 × 105 cells/cm2) and cultured for 3 wk with base medium supplemented with 40 ng/mL basic FGF and 40 ng/mL brain-derived neurotrophic factor. Unless otherwise stated, all data were obtained by using the hESC-derived NPCs with a default dorsal identity.
Immunocytochemistry and Immunohistochemistry.
Immunocytochemistry.
Cells were rinsed with PBS and fixed in 4% paraformaldehyde (PFA) in PBS for 15 min at RT. Cells were blocked with 3% BSA and 0.5% Triton X-100 in PBS (PBSAT) for 1 h at RT and then incubated overnight at 4 °C with primary antibodies (Table S1) diluted in PBSAT. The appropriate fluorochrome-conjugated secondary antibodies were used at 1:1,000 dilution. Nuclei were stained with DAPI.
Immunohistochemistry.
PFA-fixed mouse sections (12-µm thickness) in low pH Antigen Unmasking Solution (Vector Laboratories) were heated in a microwave until boiling and then cooled to RT. This process was repeated 3–4 times and sections were then washed with PBS and blocked with 5% BSA, 5% goat serum, 0.5% Triton X-100 in PBS (PBSGT) for 1 h at RT. Sections were incubated with primary antibodies overnight at 4 °C, washed with PBSGT, and incubated for 1 h at RT with the appropriate fluorochrome-conjugated secondary antibody.
Quantitative PCR.
Quantification of mRNA levels.
Total RNA was extracted using the RNeasy kit (Qiagen) and 1 µg was reverse-transcribed using the QuantiTect kit (Qiagen) according to the manufacturer’s instructions. Purified cDNA (2 μL) diluted 1:8 was used as a template. qPCR was performed with SYBR Green PCR Master Mix (Invitrogen) according to the manufacturer’s recommendations. Hypoxanthine phosphoribosyltransferase (HPRT) or GAPDH was used for normalization. Data were analyzed using the ∆(∆CT) method. Primers are listed in Table S2.
Quantification of mature miRNA levels—TaqMan qPCR.
The small RNA fraction containing the miRNA pool was purified with the NucleoSpin miRNA kit (Macherey-Nagel) according to the manufacturer’s instructions. miRNA-specific cDNA preparation and qPCR quantification was performed using the TaqMan small RNA assay (Applied Biosystems). An aliquot of 10 ng of purified small RNA pool was used in the retrotranscription reactions. qPCR was performed with TaqMan universal PCR master mix according to the manufacturer’s guidelines. Data were analyzed using the ∆(∆CT) method and normalized to U6 snRNA.
Quantification of mature miRNA levels—TaqMan arrays.
RNA was purified with the mirVANA miRNA Isolation Kit (Ambion). The purity and integrity of the total RNA were analyzed on RNA NanoChip (Agilent Technologies) using the Eukaryote Total RNA Nano Assay protocol. Total RNA samples with an RNA integrity number of 10 were used for miRNA analysis. The miRNA profiling was performed using TaqMan MicroRNA assays according to the manufacturer’s protocol (Applied Biosystems). For each sample, two RT reactions (Megaplex Pool A and Pool B) and two TaqMan MicroRNA Arrays (A and B Cards) were performed for a full miRNA profile. Briefly, 1 μg of total RNA was used for each Megaplex reverse transcription reaction in a total reaction volume of 7.5 μL using MicroRNA Reverse Transcription Kit (Applied Biosystems). The cDNA synthesis was performed on ABI’s 96-well Thermal Cycler (Veriti) for 40 cycles of 16 °C for 2 min, 42 °C for 1 min, 50 °C for 1 s, and 85 °C for 5 min. Each RT product was mixed with 2× TaqMan Universal PCR Master Mix II, with no uracil-N-glycosylase (part no. 4440040; Applied Biosystems) and loaded into the two (A and B) TaqMan low-density array (TLDA) human miRNA V3.0 384-well cards. PCR mix (100 μL) was dispensed into each port of the 384-well TLDA card. The cards were centrifuged twice for 1 min each at 331 × g in a Sorvall centrifuge (Thermo Scientific) and sealed with a microfluidic card sealer. The TLDA cards were run on a 7900HT Fast Real-Time PCR system with robotics (Applied Biosystems) using Sequence Detection Systems (SDS) software V2.3. Conditions were 10 min at 95 °C followed by 40 cycles of 95 °C for 15 s and 60 °C for 60 s. Raw Ct values were calculated using SDS RQ Manager V1.2 with automatic baseline threshold setting, and all samples were normalized using miRNA mammalian endogenous control gene, U6. The miRNA arrays were performed by the functional genomics core of the Sanford–Burnham Medical Research Institute (Lake Nona, FL).
Quantification of total miRNA levels.
The small RNA fraction containing the miRNA pool was purified with the NucleoSpin miRNA kit (Macherey–Nagel) according to the manufacturer’s instructions. The miRNAs were polyadenylated and reverse-transcribed with the Mir-X miRNA First-Strand Synthesis Kit (Clontech). qPCR quantification was performed with SYBR Green PCR Master Mix (Invitrogen). The universal reverse primers included in the Mir-X kit were used in conjunction with miRNA-specific forward primers consisting of the full sequence of the mature miRNA being analyzed. Because the mature miRNA sequence is also contained in pri- and premiRNAs, this technique quantifies the total level of a particular miRNA.
ChIP-qPCR.
ChIP was performed using the EZ-ChIP kit (Millipore) according to the manufacturer’s recommendations with the following modifications: 2 ×106 cell equivalents were used for each immunoprecipitation; cells were sonicated to obtain chromatin fragments of 200–500 bp; and 5–10 µg of the immunoprecipitating antibodies were used in each ChIP. Antibodies included normal rabbit IgG (nonspecific control; Millipore, PP64), rabbit anti-SOX2 (Millipore; AB5603), rabbit anti-TRRAP (Santa Cruz; sc-1141), rabbit anti-PCAF (Santa Cruz; sc-8999), rabbit anti-GCN5 (Santa Cruz; sc-20698), mouse anti-H3K9Ac (Cell Signaling; 9671), and rabbit anti-H3K9/18Ac (Millipore; 07-593). To evaluate factor-specific enrichment at different promoter sites, qPCR was performed using the purified chromatin as a template. Amplification was performed with site-specific primers designed to flank the genomic region of interest (i.e., the SOX binding site at position −32 on the LIN28 promoter). Primer sequences were as follows: forward, GGGTTGGGTCATTGTCTTTTAG; reverse, AAAGGGTTGGTTCGGAGAAG. qPCR data were normalized to the values obtained with normal rabbit IgG. To compare factor-specific enrichment at particular sites across different cell lines [i.e., control shRNA-expressing cells (shCTRL) vs. SOX2 knockdown (shSOX2)], qPCR data were normalized to 1% of the purified input DNA, which was used as a measure of the total amount of chromatin present in the sample.
Identification of SOX2-Interacting Proteins.
IP-MS was performed as previously described (11). Briefly, hESC-derived NPCs were cultured as described above, lysed, and subjected to IP using the Pierce Crosslink Immunoprecipitation Kit with rabbit anti-SOX2 or normal rabbit IgG control antibodies. Samples were reduced, alkylated, and digested with sequencing grade modified trypsin (Promega) using standard procedures. The resulting peptides were desalted with a peptide microtrap (Michrom Bioresources), dried in a SpeedVac, and resuspended in 0.1% formic acid/5.0% (vol/vol) acetonitrile. Each sample was run in triplicate. LC-MS/MS analyses were performed using an HTC-PAL Autosampler/Paradigm MS2 HPLC connected to a 0.2 × 150-mm Magic C18 column/CaptiveSpray (Michrom) coupled to an LTQ Orbitrap Velos mass spectrometer equipped with electron transfer dissociation (Thermo Fisher), using a decision tree (55) top-20 data-dependent method and a 15-min HPLC gradient. Spectra were searched against an ipi.v.3.73 human protein database using a Sorcerer-SEQUEST Enterprise (SageN Research), and results were filtered to a false discovery rate of 0.005–0.008 using ProteinProphet (Trans-Proteomic Pipeline).
3′ UTR Reporters and let-7 Sensor/Sponges.
3′ UTR reporters.
To study the interaction between let-7i and MASH1 mRNA, a 200-bp fragment of MASH1 3′ UTR containing two predicted let-7 binding sites was cloned downstream of the GFP coding sequence. Reporters containing mutated versions of the let-7 binding sites were also engineered. In these constructs, the wild-type binding site at position 2091 (AACATGTAATGCTATTACCTCT) was mutated to AAGCAGTAATGTAGCATAGATG, whereas the binding site at position 2197 (AGAGGCCACCAGTTGTACTTCA) was mutated to TTGCTGAACCTCAACTGCGGAA. The mutations were designed to completely disrupt the binding of let-7 as predicted by the RNA22 algorithm (Fig. S8D). HEK293T cells were transfected with the reporter and let-7i expressing plasmid. Cotransfection with a vector expressing nontargeting shRNA was used as a control. To compare fluorescence across different samples, pictures were taken with the same exposure time and contrast/brightness parameters. For each experiment, a minimum of 75 cells was analyzed. GFP fluorescence was quantified under the different experimental conditions using ImageJ software (http://rsb.info.nih.gov/ij/).
Let-7 sensors/sponges.
The GFP coding sequence was cloned downstream of the doxycycline-inducible promoter of the pTRIPZ vector (Open Biosystems). The resulting pTRIPZ-GFP vector was used as a control in all experiments using the sensors/sponges. The let-7b and let-7i sensors/sponges were constructed by inserting five perfectly complementary let-7b or let-7i sites in the 3′ UTR of GFP of the pTRIPZ-GFP vector. To monitor the activity of let-7 in NPCs, cells were transduced with lentiviruses packaged with the appropriate plasmids. GFP expression from the control construct and sensors was induced with 0.5 µg/mL doxycycline. Cells were fixed and analyzed by immunocytochemistry 4 d later. To monitor the activity of let-7 in mature (MAP2+) neurons, NPCs were differentiated into neurons for 17 d, and cells were then transduced with control and sensor lentivectors. Doxycycline (0.5 µg/mL) was added to the neutralizing medium, and the cells were fixed and analyzed 4 d later, for a total of 21 d under neuronal differentiation conditions.
Let-7 sensors/sponges were also used to block let-7 activity in SOX2 knockdown NPCs; for this, shSOX2 NPCs were transduced with control and sponge lentivectors. SOX2 shRNA and GFP sponges were induced with 3 µg/mL doxycycline until cells were fixed and analyzed. To examine the sponge effects on proliferation, the cells were cultured under self-renewal conditions for 4 d in doxycycline-containing media. To assess the sponge effects on neurogenic differentiation, NPCs were cultured in neuronal differentiation media containing doxycycline for 21 d. Note that GFP expression from sponges could not be used to assess transduction efficiency because it depends on the endogenous let-7 levels. The efficiency of transduction was therefore tested with the control pTRIPZ-GFP vector under both culture conditions. NPCs were transduced with nearly 100% efficiency under self-renewal conditions and ∼20% efficiency under neuronal differentiation conditions.
Electroporation and Luciferase Assay in hES-Derived NPCs.
The 2-kb LIN28 proximal promoter was cloned into a firefly-luciferase reporter along with the mutant LIN28 promoter containing a mutated SOX binding site (wild-type site: 5′ CTTTGAA 3′; mutant site: 5′ CTCCTCA 3′, position −32 from transcription start site human genomic sequence). The constructs were electroporated into human NPC lines expressing either control or SOX2 doxycycline-inducible shRNAs. Electroporation was performed with the Neon system (Invitrogen) as follows: 105 cells were resuspended in NPC media (without antibiotics/antimycotics and with the addition of 0.5% FBS) in the presence of 800 ng LIN28 reporters and 500 ng of control vector expressing Renilla luciferase. Cells were electroporated with three pulses of 10 ms at 1,400 V and plated onto Matrigel-coated plates; the next day, media was changed to standard NPC media with the addition of 1 µg/mL doxycycline, to induce shRNA expression. Luciferase activity was monitored 3 d after with the Dual Glow Luciferase Assay System (Promega) according to manufacturer instructions.
Mutant Mice.
We achieved conditional SOX2 deletion in vivo by exploiting two distinct genetic models. Mice carrying Sox2loxP alleles (8) were crossed with mice expressing Cre recombinase under the control of either the Wnt1 promoter (Wnt1-Cre) (32) or the mGFAP promoter (mGFAP-Cre) (35). Crossing of Sox2loxP::Wnt1-Cre mice resulted in conditional deletion of SOX2 in embryonic dorsal neural tubes and neural crest derivatives. This model was used to study SOX2-dependent LIN28 expression at embryonic stages (E11) in the mouse spinal cord. Five consecutive sections at the level of the forelimb were analyzed in both wild-type and knockout mice. Sox2loxP::mGFAP:Cre crosses resulted instead in conditional ablation of SOX2 in adult NPCs of the dentate gyrus. Sox2loxP::mGFAP:Cre mice were crossed onto the background of Nestin-CFPnuc (34) to allow visualization of neural stem/precursor cells within the adult dentate gyrus. Two-month-old mice were used to study SOX2-dependent LIN28 expression in the context of adult hippocampal and subventricular neurogenesis. In both models, mice homozygous for Sox2 deletion (Sox2LoxP/LoxP) were compared with the wild-type littermates.
Generation of Stable hESC Lines Carrying Doxycycline-Inducible Lentivectors for shRNA and Overexpression.
Inducible SOX2 shRNA-expressing hESCs.
hESC lines stably expressing shSOX2 and shCTRL were established as previously described (11).
Inducible let-7 overexpression.
Sequences corresponding to mature let-7b or let-7i were cloned between the mir-30 regulatory regions of the inducible pTRIPZ lentivector (Open Biosystems), which makes exogenous let-7 processing independent of LIN28. Vectors were packaged into lentiviral particles by the Viral Vectors Core at the Sanford–Burnham Medical Research Institute (La Jolla, CA) and used to generate stable hESC lines. To eliminate nontransduced hESCs, the puromycin resistance gene present in the pTRIPZ vector was exploited. Cells were cultured in the presence of 2.5 µg/mL puromycin for at least 10 d before being used in experiments. TaqMan qPCR was used to confirm doxycycline-dependent let-7 expression in these lines.
Inducible LIN28 overexpression and SOX2 shRNA.
The red fluorescent protein (RFP) and shRNA coding sequences downstream of the doxycycline-inducible promoter of the pTRIPZ vector were replaced with the LIN28 coding sequence. The resulting vector encoding doxycycline-inducible LIN28 was stably integrated in the genome of the shSOX2 hESCs described above by means of lentiviral transduction. Selection of transduced cells was performed by exploiting the puromycin resistance gene present in the pTRIPZ vector. The resulting hESC line expressed both SOX2 shRNA and LIN28 in a doxycycline-inducible fashion (shSOX2/LIN28 line). A cell line expressing inducible SOX2 shRNA and inducible RFP was used as a control (shSOX2/RFP). qPCR was used to confirm the concomitant SOX2 down-regulation and LIN28 expression in the shSOX2/LIN28 line.
Digital Image Analysis.
Pictures of cells cultured under different experimental conditions were taken with the same exposure time and contrast/brightness parameters. For quantification of nuclear markers (such as Ki67), positive cells were counted and expressed as a percentage of total (DAPI+) cells. Measurements were further normalized to the values obtained under the experimental condition that served as a control.
Quantification of cytoplasmic markers (such as TUJ1 and MAP2) was difficult to perform manually due to the dense cultures required to induce neuronal differentiation. We therefore used a semiautomated approach to evaluate neuronal marker expression using immunocytochemistry. Pictures were taken with the same exposure time and contrast/brightness parameters. The total area showing immunoreactivity for a particular marker was determined using ImageJ and normalized to the total area positive for DAPI, which provides an estimate of the total number of cells present in a given field. A minimum of three random fields containing at least 100 cells was analyzed for each condition.
miRNA Target Prediction and Statistical Analysis.
miRNA target sites were predicted using the software RNA22, available online (https://cm.jefferson.edu/rna22v1.0/). All experiments were performed at least in triplicate. Statistical significance was assessed using the Student t test. P < 0.05 was considered significant. In all graphs, error bars represent SE values.
Supplementary Material
Acknowledgments
We thank Dr. Sonia Albini and Prof. Lorenzo Puri for reagents and for sharing their expertise in chromatin immunoprecipitation techniques. We are grateful to Prof. Ranjan Perera and Subramaniam Shyamala Govindarajan for their help with TaqMan miRNA array analysis. We thank Dr. Laurence M. Brill for his technical support with the IP-MS experiments. This work was supported by a California Institute for Regenerative Medicine fellowship (to F.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. K.H. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1220176110/-/DCSupplemental.
References
- 1.Lui JH, Hansen DV, Kriegstein AR. Development and evolution of the human neocortex. Cell. 2011;146(1):18–36. doi: 10.1016/j.cell.2011.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhao C, Deng W, Gage FH. Mechanisms and functional implications of adult neurogenesis. Cell. 2008;132(4):645–660. doi: 10.1016/j.cell.2008.01.033. [DOI] [PubMed] [Google Scholar]
- 3.Li H, et al. Transcription factor MEF2C influences neural stem/progenitor cell differentiation and maturation in vivo. Proc Natl Acad Sci USA. 2008;105(27):9397–9402. doi: 10.1073/pnas.0802876105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dixon J, et al. Tcof1/Treacle is required for neural crest cell formation and proliferation deficiencies that cause craniofacial abnormalities. Proc Natl Acad Sci USA. 2006;103(36):13403–13408. doi: 10.1073/pnas.0603730103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Marco P, Merello E, Cama A, Kibar Z, Capra V. Human neural tube defects: Genetic causes and prevention. Biofactors. 2011;37(4):261–268. doi: 10.1002/biof.170. [DOI] [PubMed] [Google Scholar]
- 6.Graham V, Khudyakov J, Ellis P, Pevny L. SOX2 functions to maintain neural progenitor identity. Neuron. 2003;39(5):749–765. doi: 10.1016/s0896-6273(03)00497-5. [DOI] [PubMed] [Google Scholar]
- 7.Cavallaro M, et al. Impaired generation of mature neurons by neural stem cells from hypomorphic Sox2 mutants. Development. 2008;135(3):541–557. doi: 10.1242/dev.010801. [DOI] [PubMed] [Google Scholar]
- 8.Favaro R, et al. Hippocampal development and neural stem cell maintenance require Sox2-dependent regulation of Shh. Nat Neurosci. 2009;12(10):1248–1256. doi: 10.1038/nn.2397. [DOI] [PubMed] [Google Scholar]
- 9.Puligilla C, Dabdoub A, Brenowitz SD, Kelley MW. Sox2 induces neuronal formation in the developing mammalian cochlea. J Neurosci. 2010;30(2):714–722. doi: 10.1523/JNEUROSCI.3852-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taranova OV, et al. SOX2 is a dose-dependent regulator of retinal neural progenitor competence. Genes Dev. 2006;20(9):1187–1202. doi: 10.1101/gad.1407906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cimadamore F, et al. Human ESC-derived neural crest model reveals a key role for SOX2 in sensory neurogenesis. Cell Stem Cell. 2011;8(5):538–551. doi: 10.1016/j.stem.2011.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11(9):597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 13.Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14(8):1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320(5872):97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heo I, et al. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol Cell. 2008;32(2):276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 16.Rybak A, et al. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol. 2008;10(8):987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 17.Cox JL, Mallanna SK, Luo X, Rizzino A. Sox2 uses multiple domains to associate with proteins present in Sox2-protein complexes. PLoS ONE. 2010;5(11):e15486. doi: 10.1371/journal.pone.0015486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mallanna SK, et al. Proteomic analysis of Sox2-associated proteins during early stages of mouse embryonic stem cell differentiation identifies Sox21 as a novel regulator of stem cell fate. Stem Cells. 2010;28(10):1715–1727. doi: 10.1002/stem.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peng S, et al. Genome-wide studies reveal that Lin28 enhances the translation of genes important for growth and survival of human embryonic stem cells. Stem Cells. 2011;29(3):496–504. doi: 10.1002/stem.591. [DOI] [PubMed] [Google Scholar]
- 20. Wilbert ML, et al. (2012) LIN28 binds messenger RNAs at GGAGA motifs and regulates splicing factor abundance. Mol Cell 48(2):195–206. [DOI] [PMC free article] [PubMed]
- 21.Boyerinas B, Park SM, Hau A, Murmann AE, Peter ME. The role of let-7 in cell differentiation and cancer. Endocr Relat Cancer. 2010;17(1):F19–F36. doi: 10.1677/ERC-09-0184. [DOI] [PubMed] [Google Scholar]
- 22.Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell. 2012;148(6):1172–1187. doi: 10.1016/j.cell.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng LC, Pastrana E, Tavazoie M, Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci. 2009;12(4):399–408. doi: 10.1038/nn.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu C, et al. Epigenetic regulation of miR-184 by MBD1 governs neural stem cell proliferation and differentiation. Cell Stem Cell. 2010;6(5):433–444. doi: 10.1016/j.stem.2010.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szulwach KE, et al. Cross talk between microRNA and epigenetic regulation in adult neurogenesis. J Cell Biol. 2010;189(1):127–141. doi: 10.1083/jcb.200908151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dong Q, et al. MicroRNA let-7a inhibits proliferation of human prostate cancer cells in vitro and in vivo by targeting E2F2 and CCND2. PLoS ONE. 2010;5(4):e10147. doi: 10.1371/journal.pone.0010147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar MS, et al. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci USA. 2008;105(10):3903–3908. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson CD, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67(16):7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- 29.Zhao C, et al. MicroRNA let-7b regulates neural stem cell proliferation and differentiation by targeting nuclear receptor TLX signaling. Proc Natl Acad Sci USA. 2010;107(5):1876–1881. doi: 10.1073/pnas.0908750107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bajpai R, et al. Molecular stages of rapid and uniform neuralization of human embryonic stem cells. Cell Death Differ. 2009;16(6):807–825. doi: 10.1038/cdd.2009.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Curchoe CL, et al. Early acquisition of neural crest competence during hESCs neuralization. PLoS ONE. 2010;5(11):e13890. doi: 10.1371/journal.pone.0013890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Danielian PS, Muccino D, Rowitch DH, Michael SK, McMahon AP. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr Biol. 1998;8(24):1323–1326. doi: 10.1016/s0960-9822(07)00562-3. [DOI] [PubMed] [Google Scholar]
- 33.Suh H, et al. In vivo fate analysis reveals the multipotent and self-renewal capacities of Sox2+ neural stem cells in the adult hippocampus. Cell Stem Cell. 2007;1(5):515–528. doi: 10.1016/j.stem.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Encinas JM, Vaahtokari A, Enikolopov G. Fluoxetine targets early progenitor cells in the adult brain. Proc Natl Acad Sci USA. 2006;103(21):8233–8238. doi: 10.1073/pnas.0601992103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garcia AD, Doan NB, Imura T, Bush TG, Sofroniew MV. GFAP-expressing progenitors are the principal source of constitutive neurogenesis in adult mouse forebrain. Nat Neurosci. 2004;7(11):1233–1241. doi: 10.1038/nn1340. [DOI] [PubMed] [Google Scholar]
- 36.Lee KK, Workman JL. Histone acetyltransferase complexes: One size doesn’t fit all. Nat Rev Mol Cell Biol. 2007;8(4):284–295. doi: 10.1038/nrm2145. [DOI] [PubMed] [Google Scholar]
- 37.Engelen E, et al. Sox2 cooperates with Chd7 to regulate genes that are mutated in human syndromes. Nat Genet. 2011;43(6):607–611. doi: 10.1038/ng.825. [DOI] [PubMed] [Google Scholar]
- 38.Martinez E, et al. Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol Cell Biol. 2001;21(20):6782–6795. doi: 10.1128/MCB.21.20.6782-6795.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vassilev A, et al. The 400 kDa subunit of the PCAF histone acetylase complex belongs to the ATM superfamily. Mol Cell. 1998;2(6):869–875. doi: 10.1016/s1097-2765(00)80301-9. [DOI] [PubMed] [Google Scholar]
- 40.Johnson SM, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120(5):635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 41.Yu ML, et al. Vascular smooth muscle cell proliferation is influenced by let-7d microRNA and its interaction with KRAS. Circ J. 2011;75(3):703–709. doi: 10.1253/circj.cj-10-0393. [DOI] [PubMed] [Google Scholar]
- 42.Tanaka S, et al. Interplay of SOX and POU factors in regulation of the Nestin gene in neural primordial cells. Mol Cell Biol. 2004;24(20):8834–8846. doi: 10.1128/MCB.24.20.8834-8846.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ura H, et al. Eed/Sox2 regulatory loop controls ES cell self-renewal through histone methylation and acetylation. EMBO J. 2011;30(11):2190–2204. doi: 10.1038/emboj.2011.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Balzer E, Heine C, Jiang Q, Lee VM, Moss EG. LIN28 alters cell fate succession and acts independently of the let-7 microRNA during neurogliogenesis in vitro. Development. 2010;137(6):891–900. doi: 10.1242/dev.042895. [DOI] [PubMed] [Google Scholar]
- 45.Ramachandran R, Fausett BV, Goldman D. Ascl1a regulates Müller glia dedifferentiation and retinal regeneration through a Lin-28-dependent, let-7 microRNA signalling pathway. Nat Cell Biol. 2010;12(11):1101–1107. doi: 10.1038/ncb2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jakymiw A, et al. Overexpression of dicer as a result of reduced let-7 microRNA levels contributes to increased cell proliferation of oral cancer cells. Genes Chromosomes Cancer. 2010;49(6):549–559. doi: 10.1002/gcc.20765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chang CJ, et al. Let-7d functions as novel regulator of epithelial-mesenchymal transition and chemoresistant property in oral cancer. Oncol Rep. 2011;26(4):1003–1010. doi: 10.3892/or.2011.1360. [DOI] [PubMed] [Google Scholar]
- 48.Yang WH, et al. RAC1 activation mediates Twist1-induced cancer cell migration. Nat Cell Biol. 2012;14(4):366–374. doi: 10.1038/ncb2455. [DOI] [PubMed] [Google Scholar]
- 49.Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat Rev Neurosci. 2002;3(7):517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- 50.Sisodiya SM, et al. Role of SOX2 mutations in human hippocampal malformations and epilepsy. Epilepsia. 2006;47(3):534–542. doi: 10.1111/j.1528-1167.2006.00464.x. [DOI] [PubMed] [Google Scholar]
- 51.Fantes J, et al. Mutations in SOX2 cause anophthalmia. Nat Genet. 2003;33(4):461–463. doi: 10.1038/ng1120. [DOI] [PubMed] [Google Scholar]
- 52.Hagstrom SA, et al. SOX2 mutation causes anophthalmia, hearing loss, and brain anomalies. Am J Med Genet A. 2005;138A(2):95–98. doi: 10.1002/ajmg.a.30803. [DOI] [PubMed] [Google Scholar]
- 53.Williamson KA, et al. Mutations in SOX2 cause anophthalmia-esophageal-genital (AEG) syndrome. Hum Mol Genet. 2006;15(9):1413–1422. doi: 10.1093/hmg/ddl064. [DOI] [PubMed] [Google Scholar]
- 54.Cimadamore F, et al. Nicotinamide rescues human embryonic stem cell-derived neuroectoderm from parthanatic cell death. Stem Cells. 2009;27(8):1772–1781. doi: 10.1002/stem.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Swaney DL, McAlister GC, Coon JJ. Decision tree-driven tandem mass spectrometry for shotgun proteomics. Nat Methods. 2008;5(11):959–964. doi: 10.1038/nmeth.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.