Significance
Failure of the host immune system to control infection with Mycobacterium tuberculosis is a major determinant of tuberculosis (TB) disease. In this work, we examined the role of macrophage migration inhibitory factor (MIF), a cytokine that is encoded in a functionally polymorphic locus in humans, in TB. We found genetic low expressers of MIF to be enriched in a population of patients with HIV and disseminated TB. From our work in cellular and mouse models, we propose a key mechanism by which MIF regulates bacterial recognition as the first step in triggering inflammatory pathways to enable mycobacterial control.
Abstract
Macrophage migration inhibitory factor (MIF), an innate cytokine encoded in a functionally polymorphic genetic locus, contributes to detrimental inflammation but may be crucial for controlling infection. We explored the role of variant MIF alleles in tuberculosis. In a Ugandan cohort, genetic low expressers of MIF were 2.4-times more frequently identified among patients with Mycobacterium tuberculosis (TB) bacteremia than those without. We also found mycobacteria-stimulated transcription of MIF and serum MIF levels to be correlated with MIF genotype in human macrophages and in a separate cohort of US TB patients, respectively. To determine mechanisms for MIF’s protective role, we studied both aerosolized and i.v. models of mycobacterial infection and observed MIF-deficient mice to succumb more quickly with higher organism burden, increased lung pathology, and decreased innate cytokine production (TNF-α, IL-12, IL-10). MIF-deficient animals showed increased pulmonary neutrophil accumulation but preserved adaptive immune response. MIF-deficient macrophages demonstrated decreased cytokine and reactive oxygen production and impaired mycobacterial killing. Transcriptional investigation of MIF-deficient macrophages revealed reduced expression of the pattern recognition receptor dectin-1; restoration of dectin-1 expression recovered innate cytokine production and mycobacterial killing. Our data place MIF in a crucial upstream position in the innate immune response to mycobacteria and suggest that commonly occurring low expression MIF alleles confer an increased risk of TB disease in some populations.
Although one third of the world’s population is infected with Mycobacterium tuberculosis (TB), only 5–10% of those infected go on to give rise to the 8.6 million annual incident TB cases and 1.5 million deaths (1). HIV infection increases the risk of TB reactivation 20-fold, and coinfected individuals contribute to 13% of worldwide TB cases but almost 30% of TB-related deaths, further highlighting the importance of host immunity in determining TB outcome (2, 3). The convergence of these epidemics is particularly prominent on the African continent, which has the highest rates of TB cases and deaths per capita, 80% of which occur in people living with HIV (1).
A genetic component to immune control of TB leading to differences in disease susceptibility and outcome has long been recognized (4, 5). Although exquisite mycobacterial susceptibility has been identified in children who have defects in the IFN-γ/IL-12/IL-23 axis, TB genetics in adults has been more difficult to define (6). A number of the candidate genes that have emerged are postulated to affect macrophage handling of mycobacteria. For example, multiple polymorphisms in SLC11A1, involved in phagosome maturation and vesicle trafficking, have been associated with differing degrees of TB risk in different populations (7). Additionally, polymorphisms in the genes for the pattern recognition receptors (PRRs) DC-SIGN (Dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin), TLR (Toll-like receptor) 1/2/8/9, and NOD2 (Nucleotide-binding oligomerization domain-containing protein 2) have emerged in TB as well as leprosy susceptibility (6).
Challenges in establishing candidate genes for TB susceptibility and severity have come from incomplete functionality or mechanistic data for some polymorphisms as well as nonreproducibility among different populations. The different frequencies with which some candidate polymorphisms occur in populations from different ethnicities as well as heterogeneity of clinical phenotyping within cohorts contribute to this challenge (8). Studying cohorts at the extremes of TB diseases (i.e., bacteremia, meningitis) has been suggested as a method for uncovering novel TB susceptibility genes (6). In a study of TB meningitis, this approach has uncovered polymorphisms in the LTA4H promoter that highlight the dual role of the inflammatory response in TB—facilitating mycobacterial control vs. causing inflammatory pathology (9). Additionally, although HIV infection a major risk factor for TB, there is increasing evidence that host genetic factors also influence TB among coinfected patients (10).
In this context, we explored the role of macrophage migration inhibitory factor (MIF), an important mediator of innate immunity and macrophage responses, in TB. MIF was first described in the context of TB disease (11, 12). Encoded on human chromosome 22q11, MIF is released by macrophages, lymphocytes, and pulmonary epithelial cells in response to microbial stimuli (13–15). Several of MIF’s downstream functions support its proinflammatory actions, including overriding glucocorticoid-mediated immune suppression; inducing sustained activation of the ERK 1/2 MAP kinase pathway; inhibiting activation-induced, p53-dependent apoptosis; and up-regulating TLR4 expression on macrophages (16–19). MIF knockout (Mif−/−) mice are protected from pathogenic inflammation in some models (20) but are susceptible to infection in others (21–23). Thus, whether MIF mediates protection from infection or potentiates damaging host response depends on the nature of the pathogen as well as the type of immunity induced.
Two MIF promoter polymorphisms occur commonly across different populations. The number of CATT tetranucleotide repeats at -794 is associated with differential MIF expression: the -794 CATT5 variant is a low-expression allele, and the -794 CATT6,7,8 variants are higher-expression alleles. An SNP at -173 (G/C) of the MIF promoter also has been found in many studies to be in linkage disequilibrium with the high-expression, -794 CATT7 MIF promoter allele and to correlate with disease susceptibility or severity (24). Globally, the CATT5 allele is more frequently identified among African Americans and Africans compared with their Caucasian American or Western European counterparts (25).
Differences in the distribution of high- and low-expression MIF alleles relating to differing phenotypes in clinical disease have been described in a variety of inflammatory conditions, including rheumatoid arthritis, scleroderma, lupus, and asthma (26–28). In infectious diseases, there are reported associations between high-expression MIF alleles and improved outcomes in outpatient pneumonia and meningococcal meningitis, but more severe malaria (29–31). Analogous to the role of MIF defined in mouse models, it seems that the influence of MIF on the severity of infectious diseases (exacerbation or protection) depends on the particular pathogen or disease pathology.
Experimental studies suggest that MIF is necessary for development of PPD (Purified protein derivative)-elicited delayed type hypersensitivity lesions, that it contributes to macrophage control of M. tuberculosis infection in vitro, and that partially backcrossed Mif−/− mice are more susceptible to mycobacterial lethality (32–34). To date, however, there has been no systematic evaluation of the mechanism by which MIF may play a role in the tuberculous immune response. Initial studies of the role of variant MIF alleles in TB have yielded disparate results. Although investigations of the -794 CATT and -173 SNP have suggested associations between the -794 CATT7,8 alleles and TB in a Chinese Han population, and the -173 C allele and TB in Columbian and Moroccan populations, an investigation of MIF levels in HIV/TB-coinfected patients in Tanzania found that patients with low circulating MIF levels have higher mortality (35–38). Notably, MIF genotype investigations have not been undertaken in African HIV/TB-coinfected patients despite the critical role of immune control and the enormity of disease burden in this population.
We report herein an examination of the role of MIF and its variant alleles in human mycobacterial infection and in experimental models. We found the low-expresser MIF genotype, -794 CATT5/5, to be enriched in an HIV-positive population with disseminated TB. Mycobacteria-stimulated transcription in human monocytes was found to be strictly dependent on the number of CATT repeats in the MIF promoter, and MIF serum levels to correlate with MIF genotype in TB infected subjects. We found MIF to be crucial to the control of M. tuberculosis and Mycobacterium bovis disease in mouse models, with innate immune deficits leading to increased neutrophil-mediated tissue damage in genetically MIF-deficient animals. Finally, we uncovered a unique MIF-dependent mechanism for exacerbated mycobacterial disease in which reduced macrophage expression of the innate recognition receptor dectin-1 decreased mycobacteriacidal cytokine and reactive oxygen species (ROS) production, impairing macrophage mycobacterial control.
Results
Increased Frequency of the Low-Expression MIF Genotype CATT5/5 Is Demonstrated in Patients with M. tuberculosis Bacteremia.
The prevalence of the MIF-794 CATT5–8 microsatellite length polymorphism was investigated in a cohort of patients who presented to two hospitals in Uganda with severe sepsis, for whom M. tuberculosis was isolated from blood cultures in 21% of the group (81 of 394) (39). Clinical characteristics and CATT genotyping results were compared in the patients with M. tuberculosis bacteremia vs. those without (Table 1). As expected, a greater proportion of mycobacteremic subjects were HIV positive, had lower CD4 counts, and were less likely to have initiated HAART (Highly active antiretroviral therapy) therapy. Mycobacteremic subjects also were more likely to be genotypic low expressers of MIF [-794 CATT5/5, 33% vs. 18%, odds ratio (OR) 2.2, P = 0.009]. This association remained significant in multivariate analysis adjusting for age, sex, CD4 count, and use of HAART (OR 2.4; Table 2).
Table 1.
Patients with the MIF low-expression genotype, CATT5/5, are enriched among patients with disseminated M. tuberculosis infection
| Variable | Case n = 83 | Control n = 311 | P value |
| Age, y | 34.4 ± 8.8 | 34.2 ± 10.2 | |
| Female | 38 (45.8) | 161 (51.9) | |
| HIV positive | 83 (100) | 256 (82.9) | <0.001 |
| CD4 count (cells/μL) | 36.1 ± 48.4 | 185.9 ± 253.7 | <0.001 |
| Receiving HAART | 7 (8.4) | 73 (23.6) | 0.002 |
| Mortality at 30 d | 41 (52.6) | 75 (26.6) | <0.001 |
| MIF low-expresser, CATT5/5 | 27 (32.5) | 56 (18.0) | 0.009 |
The MIF-794 CATT5–8 microsatellite length polymorphism (rs5844572) was genotyped in a Ugandan population with sepsis, with (case) and without (control) disseminated TB, using DNA isolated from PBMCs. Groups were compared by Student t test or χ2, and corrected P values were calculated as appropriate. Values are number (percentage) or mean ± SD.
Table 2.
Relationship between low-expresser MIF genotype and risk for disseminated TB is preserved in multivariate analysis
| Characteristic | Adjusted odds ratio (95% confidence interval) |
| MIF low-expresser, CATT5/5 | 2.421 (1.300–4.524)* |
| Age | 1.014 (0.985–1.045) |
| Female | 0.683 (0.390–1.196) |
| Higher CD4 count | 0.986 (0.981–0.991)* |
| Use of HAART | 0.212 (0.089–0.508)* |
Adjustment was performed for age, sex, higher CD4 count, and use of HAART. Higher CD4 count (per unit increase) and use of HAART were independently protective against disseminated TB. *P ≤ 0.05.
The -794 CATT Length MIF Polymorphism Is Functional upon Mycobacterial Stimulation, and Serum MIF Levels Correlate with Patient CATT Genotype in TB Disease.
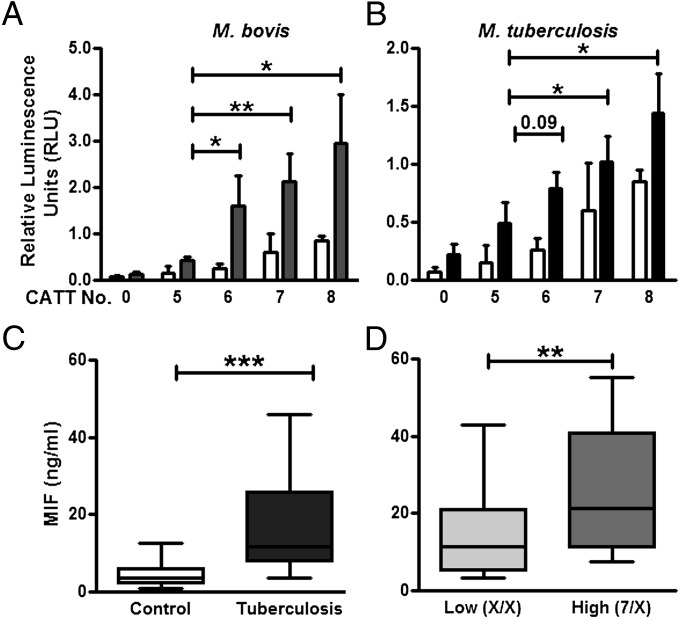
To assess the functionality of the MIF -794 CATT microsattelite repeat in response to mycobacteria, we measured the transcriptional activity of human monocytes transfected with -794 CATT5–8 MIF promoter luciferase fusion plasmids at baseline and upon stimulation with heat-killed and γ-irradiated M. bovis and M. tuberculosis, respectively. Both basal and stimulated transcriptional activity of the MIF promoter increased proportionally as a function of CATT length, with CATT5 showing the lowest and CATT8 the highest gene transcription (Fig. 1 A and B).
Fig. 1.
Human monocytes demonstrate -794 CATT length-dependent MIF transcription upon mycobacterial stimulation, and serum MIF levels correlate with CATT genotype in TB disease. (A and B) Human THP-1 monocytes transfected with MIF promoter/luciferase reporter fusion plasmids bearing 0, 5, 6, 7, and 8 CATT repeats showed a CATT–dependent enhancement of transcription upon stimulation with heat-killed M. bovis (A) or irradiated M. tuberculosis (B). Serum MIF levels were evaluated in a cohort of patients with pulmonary TB (n = 133, TBTC) vs. in a cohort of disease-free controls (n = 143). (C) TB patients had higher circulating MIF than controls. (D) Patients with TB who have the high expression MIF genotype (CATT7/X, n = 36) had higher circulating levels of the cytokine compared with their lower MIF expresser counterparts (CATTX/X, X = the low expresser CATT5 or CATT6 alleles, n = 97). Mean ± SEM, and box and whisker plots with 10th and 90th percentiles reported. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
MIF has been reported to be elevated in both serum and bronchoalveolar lavage fluid in patients with pulmonary TB (35, 40). MIF genotype and serum levels were investigated in 133 HIV-negative patients whose sera were obtained from the US Centers for Disease Control and Prevention (CDC) Tuberculosis Trials Consortium (TBTC) Study 22 (41). The mean serum MIF was elevated threefold in the TB patients, before the initiation of drug therapy, compared with disease-free controls (18.6 ng/mL vs. 5.4 ng/mL; Fig. 1C). In the TB cohort, patients carrying at least one high-MIF expresser allele, -794 CATT7, also had higher serum MIF levels than those with low-MIF expresser alleles, -794 CATT5,6, alone (25.4 ng/mL vs. 16.8 ng/mL; Fig. 1D). We did not find any correlation between MIF genotype and serum MIF levels in disease-free controls, in agreement with prior reports (27).
MIF Is Expressed and Released from Macrophages in Response to Tuberculous Stimuli.
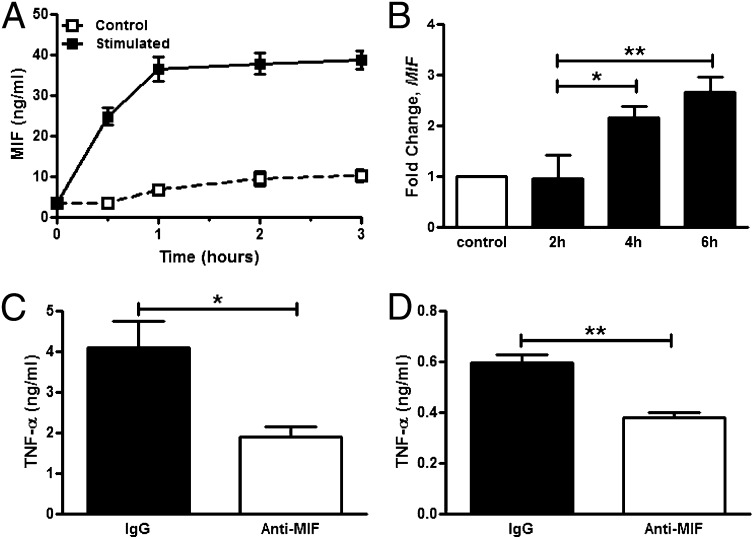
To determine the kinetics of MIF release from macrophages, human THP-1 macrophages were stimulated with γ-irradiated M. tuberculosis, and the conditioned media was sampled. MIF was released quickly after stimulation and plateaued after 1 h (Fig. 2A). Transcriptional up-regulation of MIF was not detected at 2 h after stimulation but occurred thereafter, at 4 and 6 h (Fig. 2B). This temporal pattern of MIF production from macrophages—early release of protein followed by transcriptional up-regulation—reflects initial MIF release from preformed intracellular pools (14, 42). Similar results also were observed upon stimulation of human THP-1 macrophages with PPD, and M. tuberculosis ligands—lipomannan and phosphatidylinositol mannosides-6.
Fig. 2.
MIF is released from human macrophages upon mycobacterial stimulation, and anti-MIF reduces TNF-α production. Human THP-1 monocytes were differentiated and stimulated with irradiated M. tuberculosis. (A and B) MIF release, measured by ELISA of conditioned media, was observed (A), and subsequently MIF transcription, measured by RT-PCR, was up-regulated (B). (C and D) Antibody neutralization of MIF (IgG1, clone IIID.9) reduced production of downstream TNF-α in THP-1 (C) as well as primary human macrophages (D) compared with IgG control treatment. Mean ± SEM reported. *P ≤ 0.05, **P ≤ 0.01.
MIF is known to act in an autocrine/paracrine fashion to promote downstream cytokine production (16, 17, 19). The effect of MIF on macrophage cytokine production was evaluated in the setting of mycobacterial stimulation by incubating THP-1 macrophages with a neutralizing anti-MIF monoclonal antibody before stimulation with irradiated M. tuberculosis. Analysis of the conditioned media showed significantly lower concentrations of TNF-α (Fig. 2C). MIF neutralization also led to decreased IL-1β (1.41 ng/mL vs. 2.01 ng/mL, P = 0.01) and IL-6 (0.41 ng/mL vs. 0.50 ng/mL, P = 0.04) production from M. tuberculosis ligand-stimulated macrophages. When primary macrophages prepared from differentiated human peripheral blood mononuclear cells (PBMCs) were subject to antibody neutralization of MIF before infection with M. bovis, significantly less TNF-α release also was observed (Fig. 2D).
Mif−/− Mice Succumb More Rapidly to Aerosolized M. tuberculosis Infection, with Increased Pulmonary Pathology and Decreased Innate Cytokine Production.
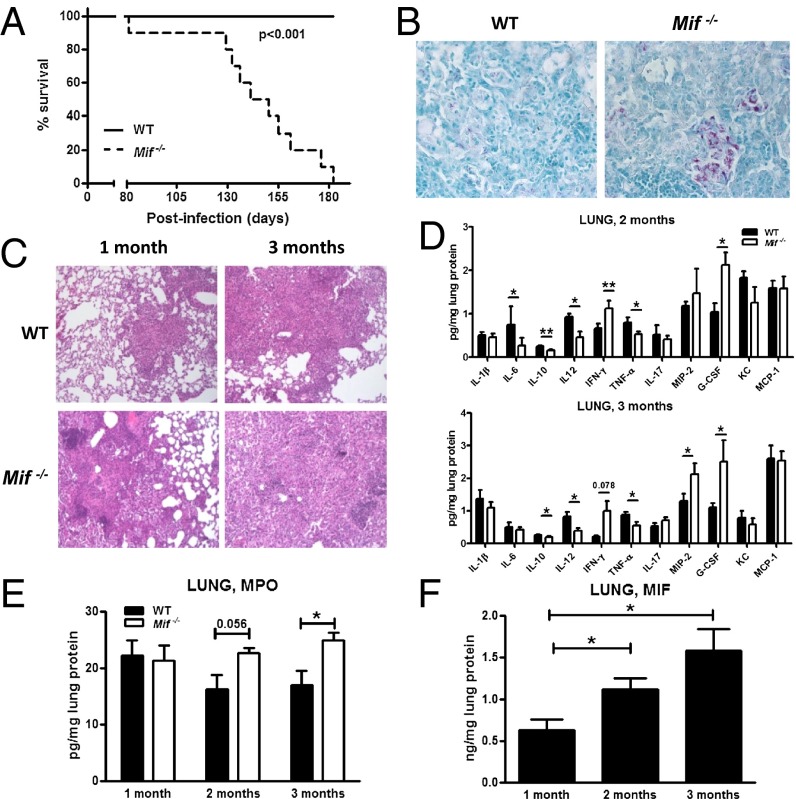
To better understand the genetic association between variant MIF alleles and TB disease, we studied a mouse model of virulent mycobacterial infection. WT and Mif−/− mice were infected with 200 cfu of M. tuberculosis clinical isolate HN878 (W. Beijing Family) by aerosol exposure. Mif−/− animals succumbed more rapidly to TB disease than their WT counterparts (Fig. 3A). Lung bacterial loads, which were similar at 1 mo after infection, were threefold higher in the Mif−/− animals compared with WT later in the course of disease (Fig. S1). Increased bacterial burden also was reflected in the histology of the formalin fixed lungs, in which a greater number of aggregates of acid-fast bacilli were observed in the Mif−/− animals (Fig. 3B).
Fig. 3.
Tuberculous immune response is impaired in the setting of MIF deficiency. Mice were infected with 200 cfu of M. tuberculosis HN878 via aerosol route. Mif−/− mice succumbed to infection with M. tuberculosis more rapidly than their WT counterparts (A) and had increased accumulation of acid-fast bacilli-positive bacteria within the lungs (B), exacerbated lung pathology H&E sections (C), and defective production of innate cytokines (IL-6, IL-12, and TNF-α) (D). The expression of IFN-γ and cytokines promoting neutrophil migration (G-CSF, MIP-2) as well as markers of neutrophil presence in the lung (MPO) were increased in Mif−/− mice (D and E). Lung MIF content was increased over the course of infection (F). n = 10 mice in the survival groups, n ≥ 4 per group otherwise. Experiments repeated at least twice, and mean ± SEM reported. *P ≤ 0.05, **P ≤ 0.01. (Magnification: B, 40×; C, 10×.)
Although the survival curves of the WT and Mif−/− animals in the setting of M. tuberculosis infection did not diverge until 4 mo after infection, histopathologic differences in the lungs were more immediately apparent (Fig. 3C). At 1 mo after infection, WT mice developed small granulomatous lesions occupying ≤20% of the lung parenchyma, as assessed by histologic examination. In contrast, Mif−/− mice developed larger, more diffuse lesions that involved 50–60% of parenchyma. By 3 mo, WT mice had more organized, coalescent granulomas, whereas Mif−/− animals had diffuse mononuclear cell infiltration of the lung parenchyma.
To uncover possible mechanisms by which MIF contributes to the killing of intracellular pathogens or control of pathogen dissemination, we investigated serum and lung lysates after M. tuberculosis infection for cytokine and chemokine production by multiplex array. We noted a pattern of decreased levels of macrophage innate cytokines (IL-6, TNF-α, IL-10, and IL-12), increased IFN-γ, and increased neutrophil chemoattractants [G-CSF, MIP-2 (Macrophage inflammatory protein 2)] in the Mif−/− animals (Fig. 3D). This innate cytokine defect in the Mif−/− animals also was apparent in the serum (Fig. S1). Greater neutrophil accumulation in the Mif−/− lungs over the course of infection was suggested by increased myeloperoxidase (MPO) activity in the lung lysates of these animals (Fig. 3E). Pulmonary MIF production was induced by M. tuberculosis infection and detectable both by ELISA (Fig. 3F) and immunohistochemical staining (Fig. S2). MIF was detected within the granuloma structures, but the density of the inflammatory infiltrate as well as the limitations of staining precluded determination of its precise cellular source.
Mif −/− Mice Have Higher Lung Bacterial Burdens After i.v. M. bovis Infection, with Increased Neutrophil Accumulation and Decreased Innate Cytokine Production, but a Preserved Adaptive Immune Response.
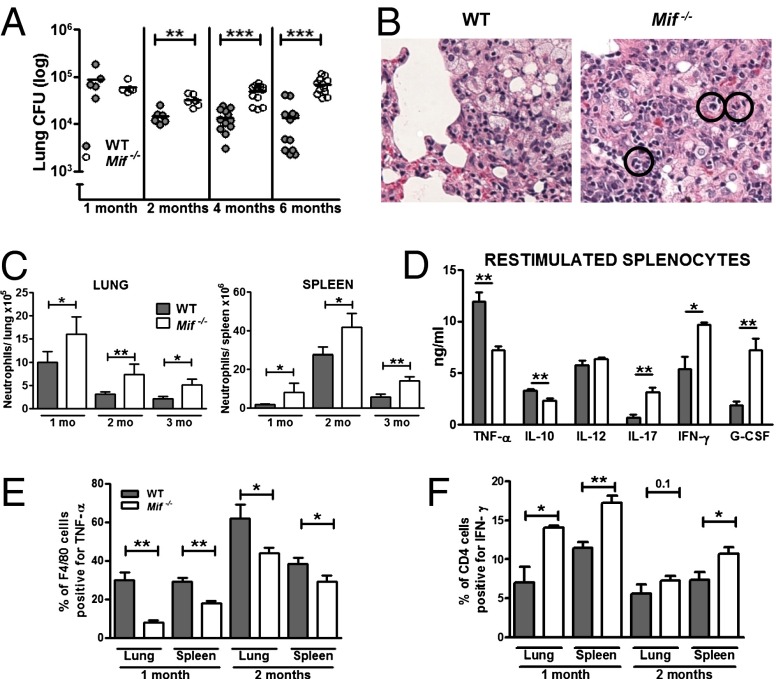
We also studied a murine model of i.v. infection with M. bovis, bacillus Calmette–Guérin, which is amenable to experimental manipulation without the requirement for strict biosafety level 3 (BSL3) isolation. Although WT or Mif−/− mice infected with M. bovis did not demonstrate overt signs of illness for many weeks, earlier mortality in the Mif−/− animals was eventually observed (Fig. S3). The inability of the Mif−/− animals to control mycobacterial infection nevertheless was evident in M. bovis infection, with infected mice demonstrating 10-fold higher lung bacterial burdens after 4 mo (Fig. 4A). Lung histology at 6 mo showed greater inflammatory pathology in the Mif−/− mice, marked by the prominence and persistence of neutrophilic infiltrates (Fig. 4B). Neutrophilia in the lungs and spleens of M. bovis-infected animals was quantified using flow cytometry to identify the Gr1hi population in single-cell suspensions of the organs. Mif−/− animals demonstrated a greater number of neutrophils in both lungs and spleens (Fig. 4C). Interestingly, this difference was observed at 1 mo, when organ bacterial burdens were similar, and persisted over the 6-mo course of the experiment.
Fig. 4.
Mif−/− animals have increased bacterial load, greater neutrophil accumulation, and impaired innate cytokine production during M. bovis infection. Animals were infected with 1 × 107 M. bovis via tail vein injection. (A) Mif−/− animals had higher lung bacterial loads at 2, 4, and 6 mo after infection compared with WT. (B) More severe lung pathology was observed in the Mif−/− animals, marked by increased neutrophil accumulation, H&E sections, neutrophils indicated in black circles. (C) Flow cytometric analysis of single-cell suspensions of lungs and spleens confirmed increased neutrophilia in the Mif−/− mice. (D) When 106 splenocytes from infected animals were plated and restimulated ex vivo with M. bovis, decreased TNF-α and IL-10 were noted in the culture supernatants along with increased IL-17, IFN-γ, and G-CSF. (E and F) Decreased macrophage production of TNF-α and increased CD4 cell production of IFN-γ were confirmed by flow cytometry. n ≥ 4 animals per group, experiments repeated at least twice. Mean ± SEM reported. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001. (Magnification: B, 40×.)
Cell activation and cytokine production responses in infected hosts were additionally investigated by restimulation of splenocytes with M. bovis ex vivo. Multiplex profiling of the conditioned media showed an MIF-related defect in the production of TNF-α and IL-10, augmented production of T-cell cytokines (IFN-γ, IL-17), and a threefold increase in the production of the neutrophil attractant G-CSF (Fig. 4D). Although macrophage numbers (F4/80+CD11b+) were identical in WT and Mif−/− animals, macrophage function, measured by intracellular TNF-α content, was impaired in both lung and spleen (Fig. 4E). The pulmonary and splenic content of CD4 and CD8 T cells was preserved in the Mif−/− animals. However, the ability of the Mif−/− CD4 T cells to produce IFN-γ upon restimulation was increased compared with WT cells, particularly in T cells obtained from the lungs (Fig. 4F).
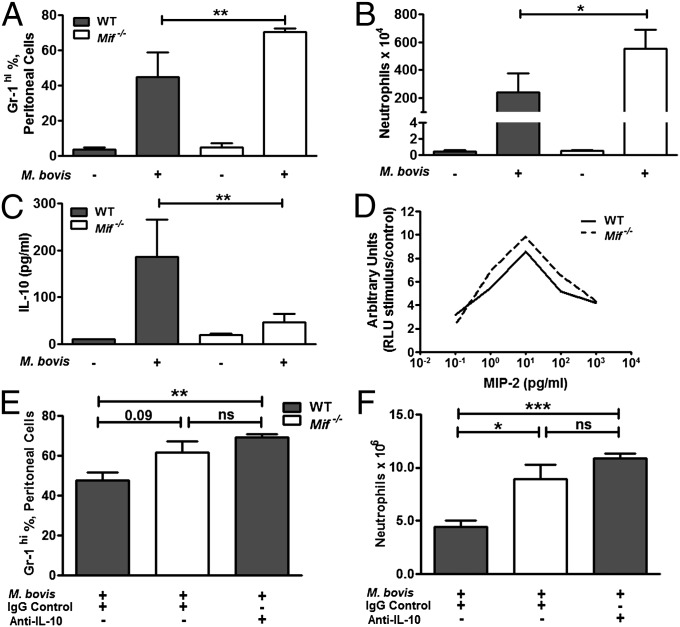
Neutrophils play an important role in the outcome of infection by contributing both to enhanced pathogen clearance as well as to increased inflammatory tissue damage (43). To investigate more closely the dynamics of MIF-dependent neutrophil accumulation, we modeled the innate response to mycobacteria by injection of M. bovis into the peritoneums of WT and Mif−/− mice. Peritoneal lavage cells were collected at 8 h and neutrophils enumerated by flow cytometry. As in lungs, an enhancement in neutrophil accumulation was observed in Mif−/− animals compared with WT; this was reflected both by the proportion and the absolute number of neutrophils (Figs. 5 A and B). No differences between macrophages and T-cell numbers in WT and Mif−/− lavage were evident. Among mediators of neutrophil recruitment and survival, we found reduced expression of IL-10 in Mif−/− vs. WT lavage fluid (Fig. 5C), which was similar to the reduction in IL-10 observed in the M. tuberculosis and M. bovis pulmonary infection models. No differences in G-CSF or MIP-2 levels were detected, and there was no difference in the chemotaxis responses of Mif−/− vs. WT neutrophils in vitro (Fig. 5D). IL-10 mediates the efferocytosis of neutrophils (43), and imunoneutralization of IL-10 in WT mice quantitatively recapitulated the effect of MIF deficiency on M. bovis-induced neutrophil accumulation (Fig. 5 E and F).
Fig. 5.
Neutrophil accumulation and decreased levels of IL-10 are demonstrated in Mif−/− mice upon peritoneal M. bovis injection. Peritoneal exudate was collected by lavage from mice injected i.p. with 1 × 107 cfu M. bovis for 8 h. (A) Proportion of neutrophils in the peritoneal exudate was increased in the Mif−/− compared with WT. (B) Absolute neutrophil numbers were also increased in the Mif−/− compared with WT. (C) IL-10 was found to be decreased in the lavage fluid in Mif −/− compared with WT. (D) Bone marrow neutrophils, isolated from WT and Mif−/− mice by density gradient separation, were found to have identical migration in response to MIP-2 stimulus. (E and F) Antibody neutralization of IL-10 before M. bovis injection resulted in increased peritoneal exudate neutrophils. n = 5 animals per group, experiments repeated at least twice. Mean ± SEM reported. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
These data collectively support an MIF-dependent defect in innate immunity that leads to increased lethality from mycobacterial infection. The enhanced IFN-γ response of CD4 T cells in Mif−/− mice may be in compensation to macrophage failure or to increased antigen exposure because of an accruing mycobacterial burden.
Mif−/− Macrophages Have Impaired Mycobacterial Control Resulting from Reduced Cytokine and ROS Production.
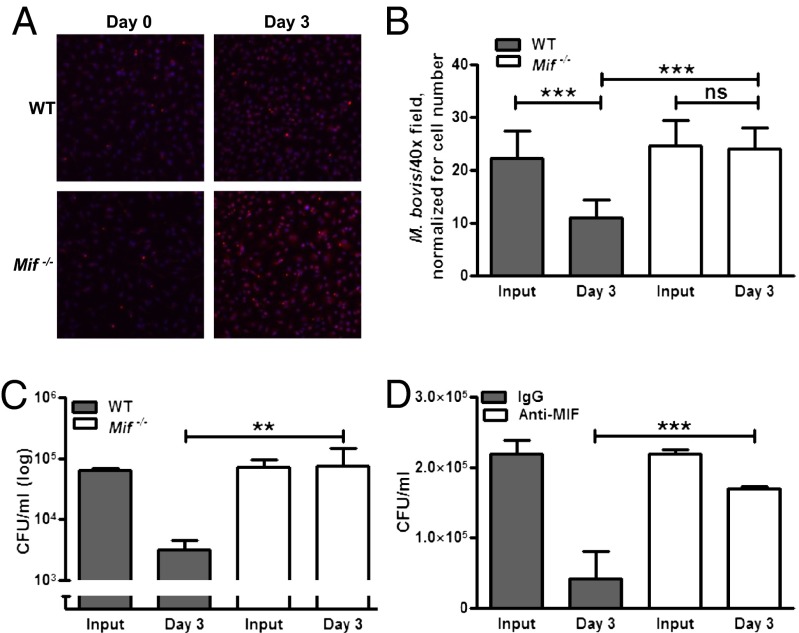
We next infected bone marrow-derived macrophages (BMDMs) from WT and Mif−/− mice with M. bovis and enumerated intracellular mycobacterial content by fluorescent detection. Macrophage mycobacterial uptake was identical in WT and Mif−/− cells, but there were decreased mycobacterial numbers at 3 d in infected WT macrophages, and no reduction was detected in Mif−/− macrophages (Fig. 6 A and B). These results were confirmed, accounting for bacterial viability, by plating the infected macrophage lysates at the indicated times and enumerating bacterial cfus. At 3 d after infection, WT macrophages showed a 1.4 log decrease in intracellular mycobacterial content relative to Mif−/− macrophages (Fig. 6C). Given the antiapoptotic effects of MIF, cell viability in these cultures was assessed by enumeration and by supernatant lactate dehydrogenase measurement. Both were equivalent in WT and Mif−/− macrophages throughout the course of the study.
Fig. 6.
MIF-deficient macrophages have a defect in their ability to control mycobacterial infection. BMDMs from WT and Mif−/− mice were infected with 10:1 MOI M. bovis. (A and B) Mif−/− macrophages demonstrated no reduction in intracellular bacteria at 3 d after infection compared with a significant reduction noted in the WT (detected by immunofluorescent antibody staining; in A, red, M. bovis; blue, DAPI; quantified in B). Subsequently, colony counts (cfus) in infected macrophage lysates were enumerated at 3 d after infection. (C) Although bacterial input (assessed 3 h after invasion) was identical, WT macrophages demonstrated a reduction in intracellular bacteria at 3 d, whereas no reduction was observed in Mif −/− cells. (D) Similar findings were observed with monoclonal antibody neutralization of MIF (clone IIID.9). Mean ± SEM reported. *P ≤ 0.05 **P ≤ 0.01 ***P ≤ 0.001. (Magnification: A, 40×.)
Immunoneutralization of MIF, which has been reported to reduce M. tuberculosis killing by cultured human macrophages (34), also reproduced the mycobactericidal defect of genetic MIF deficiency (Fig. 6D), indicating that MIF’s action on M. bovis-infected macrophage is mediated through an extracellular, autocrine/paracrine pathway.
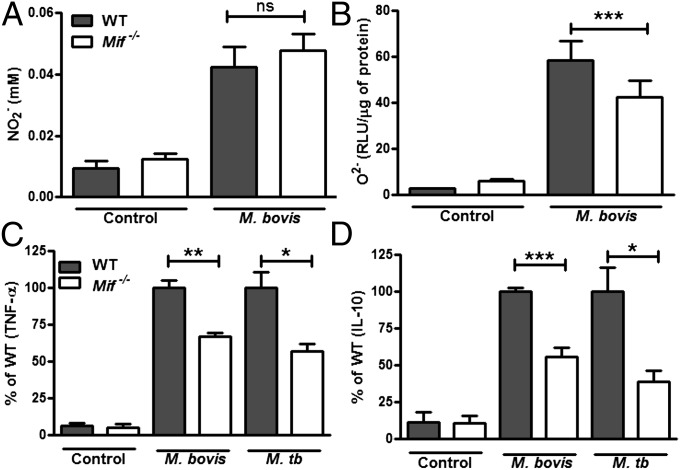
In terms of direct effectors of mycobacterial killing, we found impaired ROS generation but preserved nitric oxide (NO) production in M. bovis-infected Mif−/− macrophages (Fig. 7 A and B). The production of TNF-α and IL-10 from M. bovis-infected Mif−/− BMDMs also was impaired (Fig. 7 C and D), suggesting that the macrophage is responsible for the reduced levels of these cytokines observed in vivo. A similar 50% reduction in TNF-α and IL-10 production was observed in M. tuberculosis-infected Mif−/− BMDMs (Fig. 7 C and D). Although there is evidence that IL-10 may promote mycobacterial infection in certain settings (44, 45), the observations herein of increased mycobacterial burden, increased neutrophil numbers, and inflammatory lung damage, with decreased IL-10 production, suggest an intrinsic failure of Mif−/− macrophages to down-regulate a pathogenic immune response.
Fig. 7.
MIF-deficient macrophages have reduced ROS and cytokine production. BMDMs from WT and Mif−/− were infected with M. bovis and the conditioned media collected for NO (NO2−) quantification, ROS (O2−) determination, and cytokine analysis. (A and B) Although Mif−/− BMDMs were not impaired in their ability to produce NO (A), they generated reduced amounts of ROS in response to M. bovis infection (B). (C and D) Additionally, the Mif−/− BMDMs demonstrated impaired TNF-α and IL-10 production after infection with M. bovis or M. tuberculosis. Mean ± SEM reported. *P ≤ 0.05 **P ≤ 0.01, ***P ≤ 0.001.
Dectin-1 Up-Regulation and Downstream Spleen Tyrosine Kinase Phosphorylation Are Impaired in Mif−/− Macrophages.
Transcriptional microarray analysis of M. bovis-infected WT and Mif−/− BMDMs was performed to identify potential downstream mediators that may account for the MIF-dependent defects in the macrophage response to mycobacteria. Principal component analysis mapping demonstrated grouping of the transcription profiles by genotype and infection status (Fig. S4). A list of differentially regulated genes in infected Mif−/− vs. WT macrophages was generated, and a selection of the most significant genes is shown in Fig. S4. This list identified a number of genes, including the CD74 cell surface receptor for MIF, which may be down-regulated in the absence of MIF stimulation (46). Interestingly, the most significant, Mif-dependent reduction in mRNA expression (P = 0.006) was observed for dectin-1, which encodes a cell surface receptor for fungal β-1,3-glucans and for mycobacterial ligand(s) (47, 48).
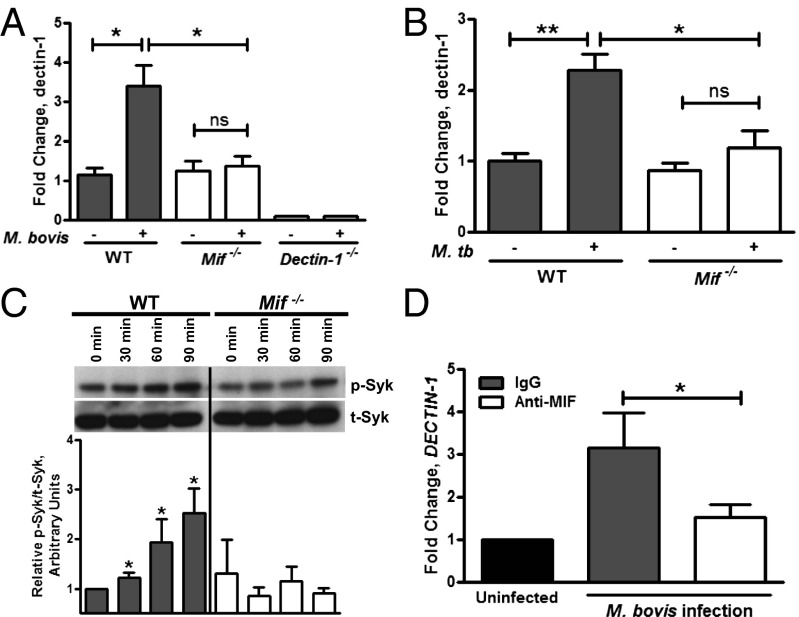
We confirmed impaired up-regulation of dectin-1 in M. bovis- and M. tuberculosis-infected Mif−/− macrophages in independent experiments (Fig. 8 A and B). The magnitude of dectin-1 up-regulation in M. tuberculosis-infected WT macrophages also was consistent with previous observations in vivo (49). The proinflammatory effects of dectin-1 are mediated by phosphorylation of spleen tyrosine kinase (Syk) (50). Activation of this signal transduction pathway, which proceeds downstream through the adaptor molecule CARD9 (Caspase recruitment domain-containing protein 9), resulting in cytokine and ROS production, has been confirmed to occur during mycobacterial infection (51, 52). The phosphorylation of Syk was deficient in Mif−/− BMDMs in response to M. bovis infection, further implicating a functional impairment of dectin-1 signaling in the setting of MIF deficiency (Fig. 8C). Finally, we investigated the relationship between MIF and dectin-1 in human monocytes. DECTIN-1 transcription was up-regulated in differentiated PBMCs upon M. bovis infection, and immunoneutralization of MIF-diminished M. bovis-induced up-regulation of DECTIN-1 by twofold (Fig. 8D).
Fig. 8.
Dectin-1 expression is MIF-dependent in both mouse and human macrophages, and dectin-1 signaling is reduced in Mif−/− cells. (A and B) RT-PCR using RNA from M. bovis and M. tuberculosis infected BMDMs (10:1 MOI) demonstrated decreased up-regulation of dectin-1 in Mif−/− macrophages compared with WT. (C) BMDMs infected with 100:1 MOI M. bovis showed decreased Syk phorphorylation by Western Blot (calculated as relative p-Syk/t-Syk) in Mif−/− macrophages compared with WT. (D) In primary human macrophages, treatment with anti-MIF antibody decreased the transcriptional up-regulation of DECTIN-1 after M. bovis infection. Representative blots are shown and densitometry is performed on blots from three independent experiments. Mean ± SEM reported. *P ≤ 0.05, **P ≤ 0.01.
Dectin-1 Deficiency Is Associated with Defects in Cytokine and ROS Production, Which Can Be Restored by Overexpression of Dectin-1 in Mif−/− Macrophages.
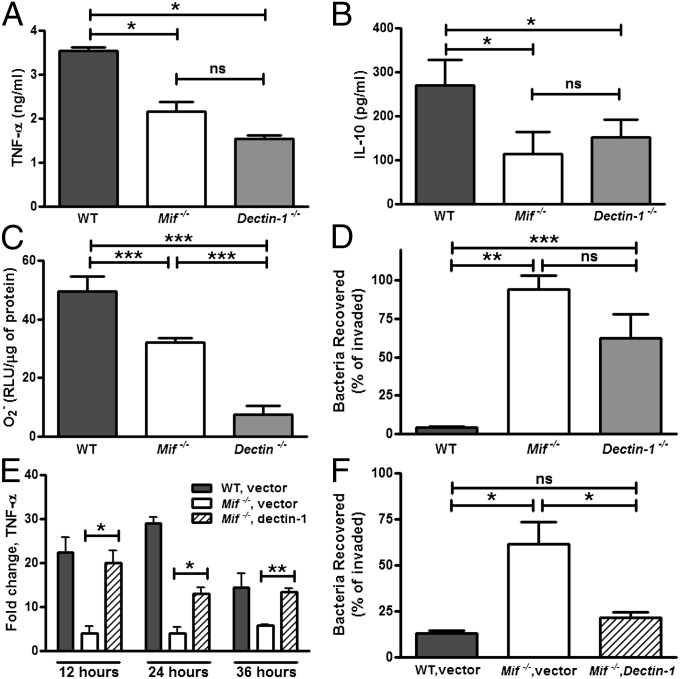
The deficiency in the downstream production of TNF-α, IL-10, and ROS observed in M. bovis-infected Mif−/− macrophages was mimicked by dectin-1 deficiency, although the magnitude of the defect for ROS was more pronounced in Dectin-1−/− than in Mif−/− cells (Fig. 9 A–C). The impact of these defects on the ability of the macrophages to control intracellular mycobacterial infection was evident in both Mif−/− and Dectin-1−/− cells (Fig. 9D). Inhibition of cellular dectin-1 function has been suggested to influence cytokine and ROS production in macrophages (53). We overexpressed dectin-1 in Mif−/− BMDMs using a dectin-1–containing pCMV vector and confirmed a resulting 40% increase in dectin-1 mRNA levels by RT-PCR. Overexpressing dectin-1 in Mif−/− BMDMs restored TNF-α transcription to WT levels upon M. bovis infection (Fig. 9E). Additionally, the mycobacterial killing capacity of the dectin-1–transfected Mif−/− macrophages was increased compared with the vector control transfected Mif−/− cells (Fig. 9F).
Fig. 9.
Mif−/− and Dectin-1−/− macrophages have similar defects in cytokine and ROS production as well as mycobacterial control. TNF-α expression and mycobacterial killing can be recovered by dectin-1 overexpression in Mif−/− cells. (A and B) Dectin-1−/− and Mif−/− BMDMs both had impaired TNF-α and IL-10 production induced by M. bovis. (C) ROS (O2−) in response to depleted zymosan was reduced in Mif−/− and absent in Dectin-1−/− BMDMs. (D) Intracellular control of mycobacterial infection, displayed by cfu recovered at day 3 after infection/cfu after invasion (day 0), was impaired in both Mif−/− and Dectin-1−/− BMDMs. (E and F) Overexpression of dectin-1 in Mif−/− BMDMs by transfection of a dectin-1 expression vector restored TNF-α transcription and mycobacterial killing after M. bovis infection. Mean ± SEM reported. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
Discussion
The outcome of M. tuberculosis infection varies from rapid asymptomatic clearance through latent infection to different manifestations of clinical disease and lethality. Understanding the immune mechanisms responsible for these disparate outcomes is crucial to developing improved preventive and therapeutic strategies. We systematically examined the role of the innate cytokine MIF, which is encoded in a functionally polymorphic locus, in human TB disease, murine models of M. tuberculosis and M. bovis infection, and macrophage responses to mycobacterial challenge. Several findings are reported: (i) human MIF genetics influence mycobacteria-stimulated MIF production and may serve as a risk for disseminated TB disease; (ii) MIF deficiency renders mice more susceptible to mycobacterial pathology as a result of an inadequate innate immune response; and (iii) MIF-related defects in macrophage mycobacterial clearance are mediated in part by reduced expression of the microbial PRR dectin-1.
Macrophage control of mycobacterial infection is at the center of our model for MIF-related immune defects in TB disease. Failure of macrophage control is highlighted in the murine infection models, with staining of prominent mycobacterial aggregates in the lungs of M. tuberculosis-infected, Mif−/− mice, as well as 10-fold higher cfus of both M. tuberculosis and M. bovis isolated from these animals. On the macrophage level, genetic deficiency as well as antibody neutralization of MIF leads to failure of mycobacterial control in the setting of preserved bacterial uptake. Similarly, in our patient cohort of immunosuppressed (CD4 <50) Ugandan patients with disseminated TB, we find that genetic low expressers of MIF (CATT5/5) are enriched 2.4-fold over TB culture-negative controls. Although other studies have examined MIF genetics in TB, we examined Sub-Saharan African, HIV-coinfected patients, a population that is critical to the global burden of disease. Additionally, we demonstrate that increasing CATT repeat length in the MIF promoter is associated with greater mycobacteria-stimulated MIF transcription in human monocytes and that pulmonary TB patients with the high-expresser, MIF CATT7/X genotype have higher circulating MIF levels than their low-expresser counterparts. The contribution of a genetic polymorphism to the risk for a common disease such as TB depends in part on the frequency at which the susceptibility allele occurs in the population (8). The proposed MIF TB susceptibility allele (-794 CATT5) is found commonly in Caucasians (∼45%) and is more prevalent in African Americans and in Africans (60–80%), indicating that MIF genotype may make a clinically meaningful contribution to TB disease risk (5, 25).
Although our human genetic data differ from previously published findings in cohorts with TB, this may be attributed to key disease as well as population differences among the groups. Consistent with the published geographic stratification of MIF polymorphisms, our cohort’s CATT distribution was distinct from the other populations in which MIF genotypes have been studied. Disseminated M. tuberculosis is well described in populations with high HIV seroprevalence and depressed CD4 T-cell counts and is associated with a high risk of mortality (54, 55). Although the importance of CD4 T-cell function in TB immunity is well established, the contribution of innate immunity to TB outcome also has been described in this group (56, 57). Accordingly, it may be the case that in our Ugandan population, impaired adaptive responses in the setting of HIV infection amplifies the immunologic impact of MIF genotype on macrophage TB control. Our mouse model data highlight the importance of MIF to the innate immune response to TB, even in the face of a largely preserved adaptive immune response. The role of MIF genotype in modulating the balance between immune control and inflammatory pathology in HIV-negative patients also may be distinct from that in HIV-positive patients.
A defect in the innate immune response attributable to MIF expression was apparent in both the mouse and macrophage studies, and consistent findings were observed between virulent M. tuberculosis and more indolent M. bovis infection. In two well-characterized models of mycobacterial infection—an aerosolized (M. tuberculosis) and an i.v. (M. bovis) infection model—Mif−/− mice succumbed more quickly and with a higher organism burden, increased lung pathology, and reduced innate cytokine production. MIF-deficient mice also showed increased pulmonary neutrophil accumulation but a preserved and possibly augmented or compensatory adaptive immune response. The increased IFN-γ production of CD4 T cells in Mif−/− mice may not sufficiently compensate for the macrophage defect, leading to significantly reduced survival in both M. tuberculosis and M. bovis infection. We attributed the increase in pulmonary disease in Mif−/− mice both to the reduced inflammatory and mycobactericicidal capacity of the macrophages, as well as their decreased expression of IL-10, which may down-regulate tissue-damaging neutrophilia by promoting efferocytosis (43). This latter conclusion was supported by recapitulating MIF- and IL-10–dependent neutrophil responses in a model of i.p. M. bovis infection. In vitro, MIF-deficient macrophages also demonstrated decreased TNF-α, IL-10, and ROS production and impaired mycobacterial killing. Although NO is known to be an effector of mycobacterial killing (58), we and others have found the NO pathway to be preserved in the Mif−/− macrophages (59).
By whole-genome transcriptional profiling, reduction in the expression of the mycobacterial PRR dectin-1 was identified in Mif−/− macrophages. MIF has been reported previously to up-regulate the expression of TLR4, which activates signal transduction in response to LPS by a pathway that requires the transcription factor PU.1 (16). Both dectin-1 and Tlr4 share functional promoter PU.1 binding sites, which suggests a common MIF-dependent mechanism for the inflammatory regulation of these two PRRs (16, 60). Our whole-genome array analysis did not reveal changes in the expression of different TLRs in response to mycobacterial infection, although a down-regulation of additional PU.1 associated genes, such as Spib, was detectable.
Although a dominant role for dectin-1 in mycobacterial control was not evident in one published mouse model (61), genetic deficiency in the dectin-1 adaptor molecule CARD9 results in diminished host survival, increased bacterial burden, neutrophilia, and more severe lung pathology despite a preserved adaptive response (62). Each of these features phenocopies our observations for mycobacteria-infected Mif−/− mice. The connection between MIF and dectin-1 in macrophages is evidenced in our work by the downstream similarities between the two defects, as well as the impact of MIF on DECTIN-1 up-regulation in human macrophages. Moreover, dectin-1 overexpression rescues the MIF-dependent cytokine production and mycobacterial killing. Redundant roles for the C-type lectin receptors, as well as TLRs, have been suggested in other mouse models of mycobacterial infection (63–65). The MIF-related reduction in dectin-1 expression may be less likely to be compensated by an alternate receptor on the organism level. Taken together, our work suggests that MIF’s role in mycobacterial control is mediated, at least in part, by dectin-1.
Although neutrophils are considered to be beneficial in early mycobacterial infection, they accumulate in situations of high pathogen load or immunological dysfunction and contribute to pulmonary pathology in established TB (66, 67). The persistence of a prominent neutrophilic infiltrate in both M. tuberculosis- and M. bovis-infected Mif−/− mice may be attributed to increased bacterial burden, innate immune failure, or a combination of both. There is experimental evidence that dectin-1 regulates IL-10 production (68, 69), and an interesting congruency between Mif−/− and Card9−/− mice is reduced IL-10 expression. It also is notable that Mif−/− mice and sepsis patients with low-expresser MIF alleles have reduced IL-10 production (21, 30). These observations indicating a role for MIF in regulating IL-10 expression and neutrophil accumulation may explain variation in the progression of mycobacterial disease and provide an example of how a single genetic determinant may influence the balance between antimicrobial mechanisms and tissue-damaging inflammatory responses.
Both MIF and dectin-1 are expressed by immune cells at key portals of pathogen invasion (70, 71). MIF is expressed within the mycobacterial granuloma, and we identified defects in TNF-α and ROS production as well as mycobacterial control that are shared by MIF and dectin-1–deficient macrophages. MIF has been shown to up-regulate inflammatory cytokine production by mechanisms that include sustaining MAPK phosphorylation and inhibiting the synthesis of an inhibitor of NF-κB (18, 69, 72, 73). MIF-deficient macrophages also have impaired dectin-1 expression as well as decreased dectin-1 signal transduction, as evidenced by reduced stimulation-induced phosphorylation of Syk.
Many PRRs contribute to mycobacterial recognition, including DC-SIGN, TLRs 1, 2, 4, 8, and 9, NOD2, Mincle, dectin-1, and MARCO (macrophage receptor with a collagenous structure), and a number of human genetic polymorphisms in PRRs have been associated with TB susceptibility (6, 8, 74, 75). The MIF promoter CATT repeat length is related to serum MIF levels in TB patients and to mycobacteria-stimulated MIF expression in human macrophages. Expression of dectin-1 in TB-infected patients of different MIF genotype has not yet been undertaken. Genetic DECTIN-1 defects have been shown to mediate susceptibility to fungal infections but have not yet been examined in TB (76). It will be important to determine whether impairment in dectin-1–mediated recognition, downstream signaling, and effector responses are recapitulated in macrophages from MIF low-expressers to understand whether decreased mycobactericidal capacity in these subjects may be a precursor to TB disease risk. Additionally, further examination of the mechanism for MIF control of dectin-1 expression in human macrophages and dendritic cells, as well as the possibility of augmenting MIF function, potentially by pharmacologic means (77), in subjects genetically predisposed to low MIF expression should be pursued.
Methods
Patient Cohorts and Genotyping.
DNA isolated from PBMC pellets was available from 404 patients in a severe sepsis cohort recruited from Mulago and Masaka Hospitals in the Central Region of Uganda from July 2006 to May 2009. Full study details are described elsewhere, but patients with suspected infection, meeting criteria for severe sepsis, with portable whole-blood lactate concentration >2.5 mmol/L or Karnofsky performance scale score ≤40 were included (39, 78). All patients were investigated with bacterial and mycobacterial blood cultures. The study database was queried for patient demographics, HIV status, and CD4 count/use of HAART, where applicable, as well as results of blood cultures. All patients provided informed consent and the studies were approved by the institutional review boards (IRB) at the collecting institutions. Transfer of the samples was approved by the providing institution and the Yale human investigation committee (HIC).
Analysis of the MIF promoter polymorphism, -794 CATT5–8 microsatellite repeat [rs5844572], was carried out by PCR using a forward primer (5′-TGCAGGAACCAATACCCATAGG-3′) and a fluorescence-labeled reverse primer (5′-AATGGTAAACTCGGGGGAC-3′). Automated capillary electrophoresis on a DNA sequencer was performed on the PCR products, and the CATT alleles were identified using Genotyper version 3.7 software (Applied Biosystems) (26). Of note, the -794 CATT5–8 microsatellite is not represented in the high-density genotyping chips that have been used previously in genome-wide association studies.
Differences in relevant population characteristics in the Uganda cohort were analyzed using the Student t test. The proportion of MIF genotypic low expressers in the cases and controls were compared by χ2 analysis. Multivariate odds ratios were calculated using logistic regression, controlling for age, sex, race, HIV status, CD4 count, and HAART status.
Serum samples from 133 patients with pulmonary TB were obtained from the TBTC sample repository at the CDC. The samples originated from patients enrolled at various sites across the United States in TBTC Study 22 (41). Approval to use the samples was granted by the TBTC Core Science Group. Clinical and demographic data were queried from the TBTC database. Control sera were obtained from 143 disease-free subjects with similar demographics enrolled in the Genetic and Inflammatory Markers of Sepsis Study (30). Serum MIF levels were measured by sandwich ELISA using specific antibodies (27). Genomic DNA was isolated from serum samples using the easy-DNA kit (Invitrogen), and -794 CATT genotyping was performed as indicated above. Analysis in the CDC-TBTC cohort was performed using the Student t test.
Human Macrophage Experiments.
For the MIF promoter activity experiments, a dual-Luciferase reporter assay system was used (Promega). One million human THP-1 monocytes (ATCC) were cultured in RPMI 1640 supplemented with FBS and were cotransfected with 1 μg each of the respective CATT repeat construct (0-8) and a β-actin construct using the cell-line Amaxa nucleofactor platform (Lonza). The cells were incubated at 37 °C for 24 h, stimulated with either γ-irradiated M. tuberculosis (a generous gift from BEI Resources, Manassas, VA) or heat-killed M. bovis (5 μL of a 50 mg/mL solution) and incubated for an additional 8 h. The cells then were harvested, lysed, and luminescence was read at 470 and 560 mm using the Synergy Mx Monochromator-Based Microplate Reader (BioTek). Baseline and stimulated MIF promoter activity was determined relative to the β-actin activity in the respective samples.
For stimulation experiments, THP-1 monocytes were differentiated into macrophages by adding 50 ng/mL of phorbol 12-myristate 13-acetate (PMA; Sigma-Aldrich) to the culture medium for 16 h. Human PBMCs were isolated by Ficoll-Hypaque gradient centrifugation. The cells were resuspended in RPMI 1640 supplemented with human AB serum (Cambrex) and plated. After 2 h of culture, the adherent cells were washed extensively with PBS and cultured for 1 wk with the human serum-supplemented media to allow differentiation into monocyte-derived macrophages. Macrophages were either stimulated with mycobacterial glycolipids (PIM-6, lipomannan, also obtained from BEI Resources) or infected with M. bovis, bacillus Calmette–Guérin Pasteur. M. bovis was grown to log phase in Middlebrook 7H9 media (Difco) supplemented with oleic albumin dextrose catalase (Worthington Biochemicals), and single cell preparations were prepared for infection by sonicating bacteria and dispersing in 0.05% Tween 80 (Sigma) solution. Infection experiments were performed at multiplicity of infection (MOI) 10:1. Bacteria were quantified by plating of serial dilutions on 7H10 plates, and counting colonies after 3 weeks incubation at 37 °C.
MIF neutralization was performed using 100 μg/mL of monoclonal mouse anti-MIF (IgG1, clone IIID.9) added to the culture media 24 h before and maintained over the course of the experiment. Cytokine ELISAs were performed using commercially available kits (eBioscience).
RNA was isolated using the RNeasy kit (Qiagen). Reverse transcription was performed using QuantiTect kit (Qiagen) according to the manufacturer’s instructions. For quantitative PCR, the following primers were used: Hu MIF forward, 5′-CGGACAGGGTCTACATCAA-3′ and Hu MIF reverse, 5′-CTTAGGCGAA-GGTGGAGTT-3′ and Hu β-actin forward, 5′-GGATGCAGAAGGAGATCA-CTG-3′ and Hu β-actin reverse: 5′-CGATCCACACGGAGTACTTG-3′.
Animal Experiments.
Female, 8- to 10-wk-old C57BL/6 mice (WT) were purchased from Jackson Laboratories. All animal experiments were approved by IRBs at Yale and Public Health Research Institute Center. Mif−/− mice in the C57BL/6 background (N10) were bred by homozygous mating in a pathogen-free facility and were used with age and sex matching to the WT. M. tuberculosis infection was performed with the clinical strain, HN878, grown to log phase in Middlebrook 7H9 broth, 0.2% glycerol, and 0.05% Tween 80. Mice were infected with a low dose (∼102 cfu) of M. tuberculosis using an aerosol exposure system (CH Technology) in a BSL3 facility as described previously (79). Alternately, animals were infected with ∼1 × 107 cfu M. bovis, injected via tail vein injection in 200 μL of PBS. M. bovis infected mice were housed in BSL2 plus pathogen-free conditions.
Animals were either observed for survival (10 mice per experiment) or killed at the indicated time points (at least 4 mice per group, per experiment). Killing was performed by CO2 asphyxiation. Blood was removed by cardiac puncture and serum prepared. Lungs and spleen were moved by dissection. Burden of viable mycobacteria was evaluated after infection and during the disease course by homogenizing the respective organ and spreading on Middelbrook plates. Segments of lung tissue were fixed in 10% buffered formalin (Sigma-Aldrich) and paraffin embedded. Lysates were prepared by snap-freezing a lobe of the lungs, mechanically disrupting the tissue, and addition of lysis buffer with protease inhibitor mixture (Sigma-Aldrich, Santa Cruz Biotechnology) before passing through a 0.2-µm filter (to allow removal from BSL3). Serum and lung lysates were evaluated by specific ELISA (for MIF and MPO; Cell Signaling) and Luminex cytokine magnetic bead array (Bio-Rad, for all others described).
Histologic sections were stained with H&E or Ziehl-Neelsen (ZN) acid-fast stain for evaluation of pathology and mycobacterial load, respectively, as previously described (80). Briefly, photographs of the H&E- and ZN-stained sections were taken and analyzed using Nikon FX-35DX. For morphometric analysis of granulomas, H&E-stained lung sections were scanned with a PathScan Enabler IV scanner (Meyer Instruments). SigmaScan Pro-5 software was used to count the number and size of lesions. The extent of lung involvement was calculated using the average size of granulomas, the number of lesions per cm2 of tissue, and the percentage of the lung sections occupied by granulomas. Morphometric analysis was carried out by an independent pathologist blinded to the source of tissues.
Lungs and spleen from the M. bovis-infected animals were used for flow cytometry experiments. Single-cell suspensions from lung tissue were prepared by incubating minced tissue with collagenase IV (1 mg/mL; Worthington Biochemicals) and DNase I (25–50 U/mL; Sigma-Aldrich) for 1 h at 37 °C. The digested lung was further disrupted by passing through a cell strainer (BD Bioscience). Splenocytes were obtained by passing spleens through a cell strainer, lysing red cells, and washing with RPMI/FBS. Cells were counted and viability determined by Trypan Blue exclusion. For cytokine production experiments, cell suspensions (2 × 105cells/100 μL) were incubated in 96-well U-bottom plates (Corning) with M. bovis for 24 h. Where appropriate, Brefeldin A (1.0 μg/mL; BD Biosciences) was added to the wells for 4 h before harvest.
For the peritonitis model, M. bovis (1 × 107 in 200 μL of sterile PBS), prepared as described above, was injected i.p. into mice of the designated genotype. Sterile PBS alone was used as control. In the IL-10 neutralization experiments, mice were injected with 200 μg of anti-IL-10 mAb (clone JES052A5; eBioscience) or Rat IgG1 Isotype Control (eBioscience), 16 h before M. bovis injection.
The following antibodies were used for surface staining: F4/80-APC, CD11b-PerCP, Gr-1-PE, CD8-APC-Cy7, CD4-Pacific Blue, CD62L-PE-Cy7, CD44-PerCP, and CD69-FITC (eBioscience). For intracellular staining, IFN-γ-PE-Cy7 and TNF-α-PE (eBioscience) were used along with the Cytofix/Cytoperm kit from BD Biosciences, according to the manufacturer’s instructions. All samples were fixed in 4% paraformaldehyde before analysis. Data were acquired using the LSR II flow cytometer (BD Biosciences) and analyzed using FlowJo software (Tree Star).
Bone Marrow Macrophage Experiments.
BMDMs were prepared by differentiating cells flushed from the femur/tibias of C57BL/6 mice of the appropriate genotype (WT, Mif−/−, and Dectin-1−/−; the latter were obtained under MTA (material transfer agreement) from the University of Aberdeen, Scotland) in the presence of supernatant from L929 cells for 1 wk. For intracellular infection experiments, 2 × 105 BMDMs were plated in 12-well tissue culture plates or chamber slides (Corning) and allowed to adhere overnight. Cells were infected with M. bovis, MOI 10:1, and allowed to invade for 3 h. Thereafter, cells were washed thoroughly with PBS and fresh media added. For immunofluorescence, cells were fixed and permeabilized before staining with rabbit polyclonal anti-M. tuberculosis complex antibody (Biodesign International) and goat anti-rabbit-Alexa 594 (Invitrogen), as well as DAPI, at day 0 and day 3 after infection. Cells were visualized using a Nikon Perfect-Focus epifluorescent microscope, and intracellular mycobacteria were quantified by counting at least 30 high-power fields per condition. For cfu experiments, cells lysates were collected at days 0 (after invasion, bacterial input), 3, and 7 using 0.5% Triton X, and serial dilutions of lysates plated on Middlebrook agar to enumerate colony numbers. Antibody neutralization of MIF was performed for 24 h before M. Bovis infection, and 100 μg/mL of anti-MIF IgG1, clone IIID.9 was maintained in the culture supernatant throughout the infection course. Intracellular mycobacteria were enumerated by cfu assay.
The ability of WT and Mif−/− and Dectin-1−/− macrophages to produce cytokines (TNF-α, IL-10), NO, and ROS in response to M. bovis or M. tuberculosis was assessed as follows. Culture supernatants of 1 × 106 BMDMs plated in 12-well tissue culture dishes were collected 4 h after infection and evaluated for TNF-α and IL-10 by specific ELISA (eBioscience). NO was measured in the supernatants as NO2- after reduction of NO3- using the Griess reaction. ROS production from the macrophages was assessed using the Diogenes Reactive Oxygen Assay Kit (National Diagnostics). Briefly, 2 × 105 macrophages of the appropriate genotype were plated in 96-well opaque plates (Corning). Cells were either infected with M. bovis or stimulated with depleted zymosan (Sigma-Aldrich) for the indicated duration. Chemiluminescence after incubation with the Diogenes reagent was measured and normalized for the protein concentration in the respective well (determined by the Bradford Assay, BioRad, of cell lysates).
Dectin-1 Studies.
RNA was isolated from murine BMDMs of the appropriate genotype after infection with 10:1 MOI of M. bovis and reverse transcribed as described previously. M. bovis or M. tuberculosis infection-dependent transcription of DECTIN-1 in human macrophages, with and without antibody neutralization of MIF, was determined using RT-PCR. Primer sets for murine and human dectin-1 as well as murine TNF-α were purchased from Qiagen. Phosphorylation of Syk after M. bovis infection in WT and Mif−/− BMDMs was determined by Western blot using specific antibodies. Cells were lysed at the indicated time points using lysis buffer (Invitrogen) supplemented with protease inhibitor (Roche) and phosphatase inihibitor mixtures (Sigma-Aldrich). Lysates were run on a 4–12% Bis-Tris NuPage gel (Invitrogen) and transferred onto polyvinylidene difluoride membrane. Phosphorylation of Syk was determined using a specific antibody and normalized for total Syk in the respective lysates (Cell Signaling). Densitometry was performed using ImageJ.
Murine dectin-1 was cloned into a pCMV vector. The pCMV-dectin-1 and the pCMV vector alone (control) were transfected into equal numbers of WT and Mif−/− BMDMs via the Amaxa nucleofactor platform (Lonza). Transfected cells were plated into 12-well tissue culture dishes. On the following day, cells were infected with M. bovis as previously described. RNA was isolated at the indicated time points and RT-PCR performed with murine TNF-α and dectin-1 primers (Qiagen). Alternately, cfu assay was performed after invasion and at 3 d to determine mycobacterial killing capacity, which was depicted relative to the respective WT vector control.
Supplementary Material
Acknowledgments
We thank Juan Fan and Scott Roberts for their technical assistance; Daehee Hwang for advice on the microarray analysis; Gerald Friedland for helpful discussions; and the CDC for the use of TBTC serum repository and database. This work was supported by Grants F32 AI085712-01A1 and K08-AI-097223-01 (to R.D.), RO1AI042310 (to R.B.), N01-HHSN272201100019C (to R.B.), and R01AI068041-06 (to J.D.M.), all from National Institute of Allergy and Infectious Diseases (NIAID), Burroughs Wellcome Fund Award 1007845 (to J.D.M.), and William Wirt Winchester Foundation Grants from Yale-New Haven Hospital (to R.D., R.B., and J.D.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The microarray data have been deposited in the International MIF Consortium at http://www.biochemmcb.rwth-aachen.de/mif_consortium_public/index.php.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1301128110/-/DCSupplemental.
References
- 1.WHO global tuberculosis control report 2011. Summary. Cent Eur J Public Health. 2010;18(4):237. [PubMed] [Google Scholar]
- 2.Corbett EL, et al. The growing burden of tuberculosis: Global trends and interactions with the HIV epidemic. Arch Intern Med. 2003;163(9):1009–1021. doi: 10.1001/archinte.163.9.1009. [DOI] [PubMed] [Google Scholar]
- 3.Selwyn PA, et al. A prospective study of the risk of tuberculosis among intravenous drug users with human immunodeficiency virus infection. N Engl J Med. 1989;320(9):545–550. doi: 10.1056/NEJM198903023200901. [DOI] [PubMed] [Google Scholar]
- 4.van der Eijk EA, van de Vosse E, Vandenbroucke JP, van Dissel JT. Heredity versus environment in tuberculosis in twins: The 1950s United Kingdom Prophit Survey Simonds and Comstock revisited. Am J Respir Crit Care Med. 2007;176(12):1281–1288. doi: 10.1164/rccm.200703-435OC. [DOI] [PubMed] [Google Scholar]
- 5.Stead WW, Senner JW, Reddick WT, Lofgren JP. Racial differences in susceptibility to infection by Mycobacterium tuberculosis. N Engl J Med. 1990;322(7):422–427. doi: 10.1056/NEJM199002153220702. [DOI] [PubMed] [Google Scholar]
- 6.Möller M, Hoal EG. Current findings, challenges and novel approaches in human genetic susceptibility to tuberculosis. Tuberculosis (Edinb) 2010;90(2):71–83. doi: 10.1016/j.tube.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 7.Bellamy R, et al. Variations in the NRAMP1 gene and susceptibility to tuberculosis in West Africans. N Engl J Med. 1998;338(10):640–644. doi: 10.1056/NEJM199803053381002. [DOI] [PubMed] [Google Scholar]
- 8.Vannberg FO, Chapman SJ, Hill AV. Human genetic susceptibility to intracellular pathogens. Immunol Rev. 2011;240(1):105–116. doi: 10.1111/j.1600-065X.2010.00996.x. [DOI] [PubMed] [Google Scholar]
- 9.Tobin DM, et al. Host genotype-specific therapies can optimize the inflammatory response to mycobacterial infections. Cell. 2012;148(3):434–446. doi: 10.1016/j.cell.2011.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raghavan S, Alagarasu K, Selvaraj P. Immunogenetics of HIV and HIV associated tuberculosis. Tuberculosis (Edinb) 2012;92(1):18–30. doi: 10.1016/j.tube.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 11.Bloom BR, Bennett B. Mechanism of a reaction in vitro associated with delayed-type hypersensitivity. Science. 1966;153(731):80–82. doi: 10.1126/science.153.3731.80. [DOI] [PubMed] [Google Scholar]
- 12.David JR. Delayed hypersensitivity in vitro: Its mediation by cell-free substances formed by lymphoid cell-antigen interaction. Proc Natl Acad Sci USA. 1966;56(1):72–77. doi: 10.1073/pnas.56.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bacher M, et al. An essential regulatory role for macrophage migration inhibitory factor in T-cell activation. Proc Natl Acad Sci USA. 1996;93(15):7849–7854. doi: 10.1073/pnas.93.15.7849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calandra T, Bernhagen J, Mitchell RA, Bucala R. The macrophage is an important and previously unrecognized source of macrophage migration inhibitory factor. J Exp Med. 1994;179(6):1895–1902. doi: 10.1084/jem.179.6.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Donnelly SC, et al. Regulatory role for macrophage migration inhibitory factor in acute respiratory distress syndrome. Nat Med. 1997;3(3):320–323. doi: 10.1038/nm0397-320. [DOI] [PubMed] [Google Scholar]
- 16.Roger T, David J, Glauser MP, Calandra T. MIF regulates innate immune responses through modulation of Toll-like receptor 4. Nature. 2001;414(6866):920–924. doi: 10.1038/414920a. [DOI] [PubMed] [Google Scholar]
- 17.Calandra T, et al. MIF as a glucocorticoid-induced modulator of cytokine production. Nature. 1995;377(6544):68–71. doi: 10.1038/377068a0. [DOI] [PubMed] [Google Scholar]
- 18.Mitchell RA, Metz CN, Peng T, Bucala R. Sustained mitogen-activated protein kinase (MAPK) and cytoplasmic phospholipase A2 activation by macrophage migration inhibitory factor (MIF). Regulatory role in cell proliferation and glucocorticoid action. J Biol Chem. 1999;274(25):18100–18106. doi: 10.1074/jbc.274.25.18100. [DOI] [PubMed] [Google Scholar]
- 19.Mitchell RA, et al. Macrophage migration inhibitory factor (MIF) sustains macrophage proinflammatory function by inhibiting p53: Regulatory role in the innate immune response. Proc Natl Acad Sci USA. 2002;99(1):345–350. doi: 10.1073/pnas.012511599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bozza M, et al. Targeted disruption of migration inhibitory factor gene reveals its critical role in sepsis. J Exp Med. 1999;189(2):341–346. doi: 10.1084/jem.189.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flores M, et al. Macrophage migration inhibitory factor (MIF) is critical for the host resistance against Toxoplasma gondii. FASEB J. 2008;22(10):3661–3671. doi: 10.1096/fj.08-111666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Satoskar AR, Bozza M, Rodriguez Sosa M, Lin G, David JR. Migration-inhibitory factor gene-deficient mice are susceptible to cutaneous Leishmania major infection. Infect Immun. 2001;69(2):906–911. doi: 10.1128/IAI.69.2.906-911.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koebernick H, et al. Macrophage migration inhibitory factor (MIF) plays a pivotal role in immunity against Salmonella typhimurium. Proc Natl Acad Sci USA. 2002;99(21):13681–13686. doi: 10.1073/pnas.212488699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gregersen PK, Bucala R. Macrophage migration inhibitory factor, MIF alleles, and the genetics of inflammatory disorders: Incorporating disease outcome into the definition of phenotype. Arthritis Rheum. 2003;48(5):1171–1176. doi: 10.1002/art.10880. [DOI] [PubMed] [Google Scholar]
- 25.Zhong XB, et al. Simultaneous detection of microsatellite repeats and SNPs in the macrophage migration inhibitory factor (MIF) gene by thin-film biosensor chips and application to rural field studies. Nucleic Acids Res. 2005;33(13):e121. doi: 10.1093/nar/gni123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu SP, et al. Macrophage migration inhibitory factor promoter polymorphisms and the clinical expression of scleroderma. Arthritis Rheum. 2006;54(11):3661–3669. doi: 10.1002/art.22179. [DOI] [PubMed] [Google Scholar]
- 27.Sreih A, et al. Dual effect of the macrophage migration inhibitory factor gene on the development and severity of human systemic lupus erythematosus. Arthritis Rheum. 2011;63(12):3942–3951. doi: 10.1002/art.30624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mizue Y, et al. Role for macrophage migration inhibitory factor in asthma. Proc Natl Acad Sci USA. 2005;102(40):14410–14415. doi: 10.1073/pnas.0507189102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Awandare GA, et al. MIF (macrophage migration inhibitory factor) promoter polymorphisms and susceptibility to severe malarial anemia. J Infect Dis. 2009;200(4):629–637. doi: 10.1086/600894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yende S, et al. The influence of macrophage migration inhibitory factor gene polymorphisms on outcome from community-acquired pneumonia. FASEB J. 2009;23(8):2403–2411. doi: 10.1096/fj.09-129445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Renner P, et al. A functional microsatellite of the macrophage migration inhibitory factor gene associated with meningococcal disease. FASEB J. 2012;26(2):907–916. doi: 10.1096/fj.11-195065. [DOI] [PubMed] [Google Scholar]
- 32.David J. Historical overview of MIF. In: Bucala R, editor. MIF, Most Interesting Factor. Singapore: World Scientific; 2007. pp. 9–10. [Google Scholar]
- 33.Bernhagen J, et al. An essential role for macrophage migration inhibitory factor in the tuberculin delayed-type hypersensitivity reaction. J Exp Med. 1996;183(1):277–282. doi: 10.1084/jem.183.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oddo M, Calandra T, Bucala R, Meylan PR. Macrophage migration inhibitory factor reduces the growth of virulent Mycobacterium tuberculosis in human macrophages. Infect Immun. 2005;73(6):3783–3786. doi: 10.1128/IAI.73.6.3783-3786.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kibiki GS, et al. Serum and BAL macrophage migration inhibitory factor levels in HIV infected Tanzanians with pulmonary tuberculosis or other lung diseases. Clin Immunol. 2007;123(1):60–65. doi: 10.1016/j.clim.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 36.Li Y, et al. Association of tuberculosis and polymorphisms in the promoter region of macrophage migration inhibitory factor (MIF) in a Southwestern China Han population. Cytokine. 2012;60(1):64–67. doi: 10.1016/j.cyto.2012.06.010. [DOI] [PubMed] [Google Scholar]
- 37.Gómez LM, et al. Macrophage migration inhibitory factor gene influences the risk of developing tuberculosis in northwestern Colombian population. Tissue Antigens. 2007;70(1):28–33. doi: 10.1111/j.1399-0039.2007.00843.x. [DOI] [PubMed] [Google Scholar]
- 38.Sadki K, et al. Analysis of MIF, FCGR2A and FCGR3A gene polymorphisms with susceptibility to pulmonary tuberculosis in Moroccan population. J Genet Genomics. 2010;37(4):257–264. doi: 10.1016/S1673-8527(09)60044-8. [DOI] [PubMed] [Google Scholar]
- 39.Jacob ST, et al. Promoting Resource-Limited Interventions for Sepsis Management in Uganda (PRISM-U) Study Group Severe sepsis in two Ugandan hospitals: A prospective observational study of management and outcomes in a predominantly HIV-1 infected population. PLoS ONE. 2009;4(11):e7782. doi: 10.1371/journal.pone.0007782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamada G, et al. Elevated levels of serum macrophage migration inhibitory factor in patients with pulmonary tuberculosis. Clin Immunol. 2002;104(2):123–127. doi: 10.1006/clim.2002.5255. [DOI] [PubMed] [Google Scholar]
- 41.Benator D, et al. Tuberculosis Trials Consortium Rifapentine and isoniazid once a week versus rifampicin and isoniazid twice a week for treatment of drug-susceptible pulmonary tuberculosis in HIV-negative patients: A randomised clinical trial. Lancet. 2002;360(9332):528–534. doi: 10.1016/s0140-6736(02)09742-8. [DOI] [PubMed] [Google Scholar]
- 42.Merk M, et al. The Golgi-associated protein p115 mediates the secretion of macrophage migration inhibitory factor. J Immunol. 2009;182(11):6896–6906. doi: 10.4049/jimmunol.0803710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bratton DL, Henson PM. Neutrophil clearance: When the party is over, clean-up begins. Trends Immunol. 2011;32(8):350–357. doi: 10.1016/j.it.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Turner J, et al. In vivo IL-10 production reactivates chronic pulmonary tuberculosis in C57BL/6 mice. J Immunol. 2002;169(11):6343–6351. doi: 10.4049/jimmunol.169.11.6343. [DOI] [PubMed] [Google Scholar]
- 45.Redford PS, Murray PJ, O’Garra A. The role of IL-10 in immune regulation during M. tuberculosis infection. Mucosal Immunol. 2011;4(3):261–270. doi: 10.1038/mi.2011.7. [DOI] [PubMed] [Google Scholar]
- 46.Leng L, et al. MIF signal transduction initiated by binding to CD74. J Exp Med. 2003;197(11):1467–1476. doi: 10.1084/jem.20030286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brown GD, Gordon S. Immune recognition. A new receptor for beta-glucans. Nature. 2001;413(6851):36–37. doi: 10.1038/35092620. [DOI] [PubMed] [Google Scholar]
- 48.Yadav M, Schorey JS. The beta-glucan receptor dectin-1 functions together with TLR2 to mediate macrophage activation by mycobacteria. Blood. 2006;108(9):3168–3175. doi: 10.1182/blood-2006-05-024406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kang DD, Lin Y, Moreno JR, Randall TD, Khader SA. Profiling early lung immune responses in the mouse model of tuberculosis. PLoS ONE. 2011;6(1):e16161. doi: 10.1371/journal.pone.0016161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Underhill DM, Rossnagle E, Lowell CA, Simmons RM. Dectin-1 activates Syk tyrosine kinase in a dynamic subset of macrophages for reactive oxygen production. Blood. 2005;106(7):2543–2550. doi: 10.1182/blood-2005-03-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rothfuchs AG, et al. Dectin-1 interaction with Mycobacterium tuberculosis leads to enhanced IL-12p40 production by splenic dendritic cells. J Immunol. 2007;179(6):3463–3471. doi: 10.4049/jimmunol.179.6.3463. [DOI] [PubMed] [Google Scholar]
- 52.Strasser D, et al. Syk kinase-coupled C-type lectin receptors engage protein kinase C-σ to elicit Card9 adaptor-mediated innate immunity. Immunity. 2012;36(1):32–42. doi: 10.1016/j.immuni.2011.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brown GD, et al. Dectin-1 mediates the biological effects of beta-glucans. J Exp Med. 2003;197(9):1119–1124. doi: 10.1084/jem.20021890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Crump JA, et al. Bacteremic disseminated tuberculosis in sub-Saharan Africa: A prospective cohort study. Clin Infect Dis. 2012;55(2):242–250. doi: 10.1093/cid/cis409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reddy EA, Shaw AV, Crump JA. Community-acquired bloodstream infections in Africa: A systematic review and meta-analysis. Lancet Infect Dis. 2010;10(6):417–432. doi: 10.1016/S1473-3099(10)70072-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Diedrich CR, Flynn JL. HIV-1/mycobacterium tuberculosis coinfection immunology: How does HIV-1 exacerbate tuberculosis? Infect Immun. 2011;79(4):1407–1417. doi: 10.1128/IAI.01126-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Waitt CJ, et al. Early deaths during tuberculosis treatment are associated with depressed innate responses, bacterial infection, and tuberculosis progression. J Infect Dis. 2011;204(3):358–362. doi: 10.1093/infdis/jir265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chan J, Tanaka K, Carroll D, Flynn J, Bloom BR. Effects of nitric oxide synthase inhibitors on murine infection with Mycobacterium tuberculosis. Infect Immun. 1995;63(2):736–740. doi: 10.1128/iai.63.2.736-740.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roger T, et al. Macrophage migration inhibitory factor deficiency is associated with impaired killing of gram-negative bacteria by macrophages and increased susceptibility to Klebsiella pneumoniae sepsis. J Infect Dis. 2013;207(2):331–339. doi: 10.1093/infdis/jis673. [DOI] [PubMed] [Google Scholar]
- 60.Zhang C, et al. Downregulation of PU.1 leads to decreased expression of Dectin-1 in alveolar macrophages during Pneumocystis pneumonia. Infect Immun. 2010;78(3):1058–1065. doi: 10.1128/IAI.01141-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marakalala MJ, et al. The Syk/CARD9-coupled receptor Dectin-1 is not required for host resistance to Mycobacterium tuberculosis in mice. Microbes Infect. 2011;13(2):198–201. doi: 10.1016/j.micinf.2010.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dorhoi A, et al. The adaptor molecule CARD9 is essential for tuberculosis control. J Exp Med. 2010;207(4):777–792. doi: 10.1084/jem.20090067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Heitmann L, Schoenen H, Ehlers S, Lang R, Holscher C. Mincle is not essential for controlling Mycobacterium tuberculosis infection. Immunobiology. 2013;218(4):506–516. doi: 10.1016/j.imbio.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 64.Hölscher C, et al. Containment of aerogenic Mycobacterium tuberculosis infection in mice does not require MyD88 adaptor function for TLR2, -4 and -9. Eur J Immunol. 2008;38(3):680–694. doi: 10.1002/eji.200736458. [DOI] [PubMed] [Google Scholar]
- 65.Wieland CW, et al. Mice lacking SIGNR1 have stronger T helper 1 responses to Mycobacterium tuberculosis. Microbes Infect. 2007;9(2):134–141. doi: 10.1016/j.micinf.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 66.Berry MP, et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature. 2010;466(7309):973–977. doi: 10.1038/nature09247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Barnes PF, et al. Predictors of short-term prognosis in patients with pulmonary tuberculosis. J Infect Dis. 1988;158(2):366–371. doi: 10.1093/infdis/158.2.366. [DOI] [PubMed] [Google Scholar]
- 68.Slack EC, et al. Syk-dependent ERK activation regulates IL-2 and IL-10 production by DC stimulated with zymosan. Eur J Immunol. 2007;37(6):1600–1612. doi: 10.1002/eji.200636830. [DOI] [PubMed] [Google Scholar]
- 69.Rogers NC, et al. Syk-dependent cytokine induction by Dectin-1 reveals a novel pattern recognition pathway for C type lectins. Immunity. 2005;22(4):507–517. doi: 10.1016/j.immuni.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 70.Reid DM, Gow NA, Brown GD. Pattern recognition: Recent insights from Dectin-1. Curr Opin Immunol. 2009;21(1):30–37. doi: 10.1016/j.coi.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bacher M, et al. Migration inhibitory factor expression in experimentally induced endotoxemia. Am J Pathol. 1997;150(1):235–246. [PMC free article] [PubMed] [Google Scholar]
- 72.Calandra T, Roger T. Macrophage migration inhibitory factor: A regulator of innate immunity. Nat Rev Immunol. 2003;3(10):791–800. doi: 10.1038/nri1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Roger T, Chanson AL, Knaup-Reymond M, Calandra T. Macrophage migration inhibitory factor promotes innate immune responses by suppressing glucocorticoid-induced expression of mitogen-activated protein kinase phosphatase-1. Eur J Immunol. 2005;35(12):3405–3413. doi: 10.1002/eji.200535413. [DOI] [PubMed] [Google Scholar]
- 74.Möller M, de Wit E, Hoal EG. Past, present and future directions in human genetic susceptibility to tuberculosis. FEMS Immunol Med Microbiol. 2010;58(1):3–26. doi: 10.1111/j.1574-695X.2009.00600.x. [DOI] [PubMed] [Google Scholar]
- 75.Bowdish DM, et al. Genetic variants of MARCO are associated with susceptibility to pulmonary tuberculosis in a Gambian population. BMC Med Genet. 2013;14(1):47. doi: 10.1186/1471-2350-14-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Marakalala MJ, Kerrigan AM, Brown GD. Dectin-1: A role in antifungal defense and consequences of genetic polymorphisms in humans. Mamm Genome. 2011;22(1-2):55–65. doi: 10.1007/s00335-010-9277-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jorgensen WL, et al. Receptor agonists of macrophage migration inhibitory factor. Bioorg Med Chem Lett. 2010;20(23):7033–7036. doi: 10.1016/j.bmcl.2010.09.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jacob ST, et al. Promoting Resource-Limited Interventions for Sepsis Management in Uganda Study Group The impact of early monitored management on survival in hospitalized adult Ugandan patients with severe sepsis: A prospective intervention study. Crit Care Med. 2012;40(7):2050–2058. doi: 10.1097/CCM.0b013e31824e65d7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tsenova L, Moreira A, Party E, Freedman V, Kaplan G. Aerosol infection of mice with a nose-only exposure device. J Am Biol Safety Assoc. 1997;2(3):20–31. [Google Scholar]
- 80.Koo MS, et al. Phosphodiesterase 4 inhibition reduces innate immunity and improves isoniazid clearance of Mycobacterium tuberculosis in the lungs of infected mice. PLoS ONE. 2011;6(2):e17091. doi: 10.1371/journal.pone.0017091. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.