Significance
The epithelial to mesenchymal transition (EMT) is a driving force behind normal morphogenesis and tumor metastasis. We have found evidence that the EMT in both malignant and nonmalignant mammary epithelial cells requires the enzyme activation-induced cytidine deaminase (AID). AID is induced in mammary epithelial cell lines by inflammatory stimuli that also induce the EMT. Deficiency of AID in these cells blocks morphological and transcriptional changes typical of the EMT and increases promoter cytosine methylation in genes encoding key EMT factors. These findings suggest that AID regulates gene expression in a complex developmental process that involves epigenetic reprogramming.
Abstract
Activation-induced cytidine deaminase (AID), which functions in antibody diversification, is also expressed in a variety of germ and somatic cells. Evidence that AID promotes DNA demethylation in epigenetic reprogramming phenomena, and that it is induced by inflammatory signals, led us to investigate its role in the epithelial–mesenchymal transition (EMT), a critical process in normal morphogenesis and tumor metastasis. We find that expression of AID is induced by inflammatory signals that induce the EMT in nontransformed mammary epithelial cells and in ZR75.1 breast cancer cells. shRNA–mediated knockdown of AID blocks induction of the EMT and prevents cells from acquiring invasive properties. Knockdown of AID suppresses expression of several key EMT transcriptional regulators and is associated with increased methylation of CpG islands proximal to the promoters of these genes; furthermore, the DNA demethylating agent 5 aza-2'deoxycytidine (5-Aza-dC) antagonizes the effects of AID knockdown on the expression of EMT factors. We conclude that AID is necessary for the EMT in this breast cancer cell model and in nontransformed mammary epithelial cells. Our results suggest that AID may act near the apex of a hierarchy of regulatory steps that drive the EMT, and are consistent with this effect being mediated by cytosine demethylation. This evidence links our findings to other reports of a role for AID in epigenetic reprogramming and control of gene expression.
Activation-induced cytidine deaminase (AID) belongs to the AID/apolipoprotein B mRNA editing complex catalytic polypeptide (APOBEC) family of cytidine deaminases and is highly expressed in germinal center B lymphocytes, where it is necessary for somatic hypermutation and class switch recombination of the Ig genes (1–3). However, AID is also expressed at much lower levels during B-cell development, where it mediates B-cell tolerance by an as yet undefined mechanism (4, 5). In addition, AID is present at low levels in pluripotent cells such as oocytes, embryonic germ cells, embryonic stem cells (6), and spermatocytes (7), where it may have a function beyond antibody gene diversification (8–10). AID expression is induced by inflammatory paracrine signals such as interleukin-4 (IL-4), tumor necrosis factor alpha (TNFα), and transforming growth factor beta (TGFβ) (11–13), and it has been detected in multiple epithelial tissues in association with chronic inflammatory conditions that promote tumorigenesis (14–18). AID is also expressed in experimentally transformed human mammary epithelial cells (19), and in several cancer cell lines including breast cancer (20, 21). All of this suggests that AID may function in a variety of somatic and germ cell types.
AID has been proposed to participate in the demethylation of methylcytosine in DNA (6, 8–10). Cytosine methylation is a covalent modification of DNA that is present extensively in the vertebrates, predominantly at CpG dinucleotides, where it has a key role in epigenetic mechanisms that suppress transcription initiation (22). It participates in processes that are necessary for normal development (23–25), and there is extensive information on mechanisms by which it is placed on DNA and its interaction with chromatin proteins (26, 27). The processes by which methylation is removed from cytosine were obscure until recent studies provided evidence for active, although indirect, modes of DNA demethylation that involve modification of the meC base coupled to DNA repair. One pathway proceeds through oxidation catalyzed by the TET (ten eleven translocation) enzymes (28, 29). A second pathway uses AID, which promotes DNA demethylation through direct deamination of meC to thymidine (6) and subsequent repair of the resultant T:G mismatch by classical repair pathways (8–10, 30). This indirect mode of DNA demethylation is carried out in concert with ubiquitous DNA repair factors such as methyl-CpG binding domain protein 4 (MBD4), growth arrest and DNA-damage inducible 45 protein (GADD45), and/or thymine DNA glycosylase (TDG) proteins (10, 30).
Recent evidence suggests that AID’s demethylation activity is required for reprogramming in some developmental processes. In zebrafish embryos, AID acts with GADD45 and MBD4 to demethylate injected plasmid DNA as well as genomic DNA; knockdown of AID results in an increase in bulk genomic methylation levels and in hypermethylation of the neuroD2 gene promoter that is bound by AID (10). In mice, generation of primordial germ cells involves genomewide demethylation; AID KO mice have primordial germ cells that are approximately twofold more methylated compared with WT animals (9). Similarly, AID interacts with and demethylates the promoters of the OCT4 and NANOG genes during reprogramming of human fibroblasts fused to mouse ES cells (8). A recent report also shows that knockdown of AID impairs reprogramming of mouse fibroblasts into induced pluripotent stem cells (31). Taken together, these findings suggest that AID has a role in demethylation of promoters and other genomic elements during various reprogramming phenomena, although the existing evidence does not reveal just how broad AID’s role is and how demethylation is accomplished; nor does it reveal the biological processes, other than reprogramming to a pluripotent state, in which AID has a role.
The evidence indicating that AID demethylates DNA, is broadly expressed, is induced by inflammatory stimuli, and functions in reprogramming phenomena suggested to us that it might have a role in the epithelial–mesenchymal transition (EMT). The EMT is a critical process in normal morphogenesis and tumor metastasis. It is a dynamic process that is triggered by microenvironmental stimuli; inflammation appears to be a common mechanism triggering the EMT and is also a feature of many tumor microenvironments. During the EMT, cells lose adherens and tight junctions, desmosomes, and cytoskeletal proteins, while gaining expression of the intermediate filament vimentin and matrix metalloproteinases (32–34), leading to the acquisition of migratory and invasive properties. The reprogramming of gene expression during the EMT is orchestrated by the transcription factors Snail, Slug, ZEB1, and ZEB2: these “EMT master regulators” all repress E-cadherin expression (35, 36). The EMT that takes place during carcinogenesis uses some of the same factors and signaling molecules that drive the EMT during gastrulation and neural crest formation (35, 37). In addition, the EMT endows normal and transformed mammary epithelial cells and ovarian cancer cells with stem cell properties, including the ability to self-renew and initiate tumors (34, 38–41).
To address the role of AID in the EMT, we made use of mammary epithelial cell lines that express AID (20) and undergo the EMT under well-defined culture conditions (42, 43) (Results). Our results present proof of principle that AID can modulate gene expression in a complex developmental process. On knockdown of AID, the EMT is abolished in breast cell lines, and key EMT genes show an increase in promoter methylation with a consequent down-regulation of their expression. These cell lines are tractable models in which to investigate the mechanisms by which AID acts in the EMT, a prototype of developmental reprogramming.
Results
MCF10A and ZR75.1 Cells as Models for the EMT in Normal and Malignant States.
Although many cell types and cell lines will undergo EMT, most do so either very slowly or under conditions that may not closely resemble the conditions under which it occurs in vivo. MCF10A and ZR75.1 cells are mammary epithelial cells with demonstrated propensity for undergoing the EMT and expressing typical EMT markers under defined conditions (42–44). MCF10A cells are derived from normal breast tissue. They are adapted to culture and do not express p16 (45) but have a near-diploid karyotype, express p53 (46), and do not form colonies in soft agar or grow in immunocompromised mice. Under defined culture conditions, they form acinar structures that recapitulate many aspects of mammary architecture in vivo (47, 48). ZR75.1 cells are breast cancer epithelial cells that have been used extensively as a breast cancer model (49). Much evidence indicates that the TGFβ and NF-κB pathways are critical, and act synergistically, to induce the EMT in in vivo models of metastatic breast cancer (43). Both ZR75.1 and MCF10A cells have an epithelial morphology, but on induction of the EMT with TGFβ and TNFα (which activates the NF-κB pathway), they show up-regulation of EMT factors and increased cell scattering and invasiveness (42, 43).
Up-Regulation of AID by EMT-Inducing Factors.
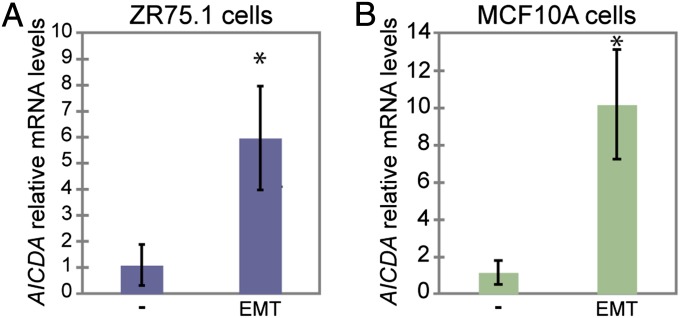
Down-regulation of E-cadherin (epithelial) and up-regulation of vimentin (mesenchymal) are two well-established markers of the EMT (44, 50). Under EMT-inducing conditions, MCF10A cells up-regulate EMT factors and display a more fibroblastic morphology (43), and ZR75.1 cells showed an increase in cell scattering (Fig. S1A, Right), up-regulation of vimentin expression (Fig. S1 B and C), and loss of E-cadherin (Fig. S1C). Chronic inflammatory conditions in vivo, and individual treatment with TNFα or TGFβ in vitro, can induce AID in epithelial cells (14–16, 18). At baseline, ZR75.1 and MCF10A cells express AID mRNA at levels that are detectable, although far lower than Ramos cells (Burkitt’s B-cell lymphoma) (Fig. S2A). Because TNFα and TGFβ in combination will induce the EMT in breast cells (42, 51, 52), we asked if EMT-inducing conditions up-regulate AID expression in mammary epithelial cells. AID (AICDA) mRNA levels, measured by quantitative real-time PCR (real-time qPCR), significantly increased in ZR75.1 (Fig. 1A) and MCF10A (Fig. 1B) cells after 4 or 8 h, respectively, of treatment with TNFα and TGFβ (Fig. 1). TNFα alone was also able to up-regulate AID mRNA expression in ZR75.1 cells, with a maximum approximately fivefold increase after an 8-h treatment at 50 ng/mL (Fig. S2B). These results are consistent with reports showing induction of AID in other epithelial cell types by the same proinflammatory factors and with similar kinetics (14, 16, 18).
Fig. 1.
Inflammatory conditions that induce the EMT in mammary epithelial cells also induce AID expression. Induction of AID (AICDA) mRNA levels in (A) ZR75.1 cells (*P < 0.005) and (B) MCF10A cells (*P < 0.001) by EMT-inducing conditions, measured by real-time qPCR, and displayed as fold change of expression levels in untreated cells. EMT-inducing conditions up-regulate the expression of AID mRNA. Error bars indicate SD (n ≥ 3 experiments).
Knockdown of AID Expression Accentuates the Epithelial Phenotype of ZR75.1 Cells.
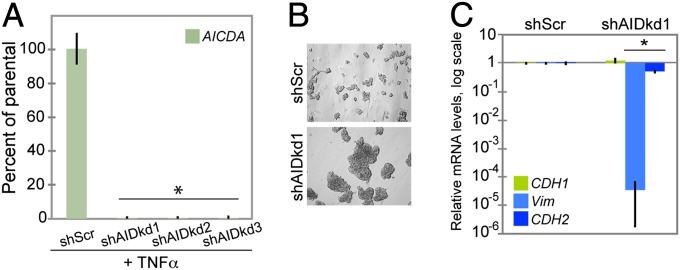
We sought specific evidence for the role of AID in the EMT by using shRNA-mediated knockdown of AID mRNA expression. ZR75.1 cells were transduced with lentiviruses expressing different shRNAs specific for AID mRNA, or a scrambled shRNA control. Cells stably expressing the shRNAs were treated with 50 ng/mL of TNFα for 8 h to induce a higher level of AID expression. This strategy probed the ability of the shRNAs to block the TNFα-induced increase in AID mRNA. Three of the four shRNAs reproducibly abrogated TNFα induction of AID mRNA (Fig. 2A: shAIDkd1, shAIDkd2, and shAIDkd3). Expression of the scrambled shRNA control (shScr) had no effect on AID mRNA level.
Fig. 2.
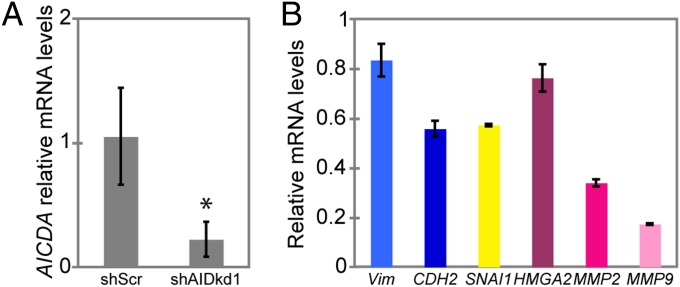
shRNA-mediated knockdown of AID in ZR75.1 cells leads to a pronounced epithelial phenotype and down-regulation of mesenchymal markers. (A) AID (AICDA) mRNA levels measured by real-time qPCR in ZR75.1 control (shScr) and shRNA-expressing cells (shAIDkd1, 2, and 3) after 8-h treatment with 50 ng/mL of TNFα (*P < 5 × 10−8), displayed as percentage of expression in control cells. (B) Phase contrast images (20×) of control ZR75.1 cells (shScr), and cells expressing one of the three shRNAs that knock down AID (shAIDkd1). Knockdown of AID (shAIDkd1) decreases cell scattering. (C) mRNA levels of the EMT markers CDH1, VIM, and CDH2 measured by real-time qPCR in ZR75.1 cells that express AID (shScr) or in which AID expression has been knocked down (shAIDkd1). Levels are relative to expression in ZR75.1 shScr cells. Knockdown of AID reduces the expression of mesenchymal markers. *Not significant CDH1, P < 5 × 10−6 VIM, P < 5 × 10−4 CDH2. Error bars in A and C indicate SD (n ≥ 3 experiments).
Untreated ZR75.1 cells in which AID mRNA has been knocked down unexpectedly exhibited a change in cellular morphology, with decreased cell scattering compared with the cells expressing the shScr control (Fig. 2B; Fig. S3A). The morphological change in untreated AID deficient ZR75.1 cells correlated with a small but not significant increase in the mRNA levels of the epithelial marker E-cadherin (CDH1) and a strong reduction in the expression of the mesenchymal markers VIM and CDH2 measured by real-time qPCR (Fig. 2C; Fig. S3B). Thus, knockdown of AID expression in ZR75.1 cells produced a more pronounced epithelial morphology and reduced expression of mesenchymal markers, suggesting that at baseline these cells have some mesenchymal characteristics that are dependent on expression of AID.
AID Knockdown Blocks the EMT.
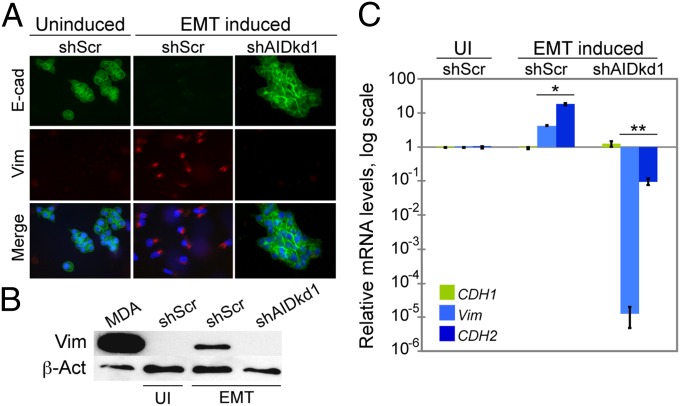
The results described above suggested that AID could play some causal role during the EMT, rather than simply accompanying the phenomenon. To test this, we cultured control and AID knockdown ZR75.1 cells in conditions that induced the EMT and assessed expression of epithelial–mesenchymal markers by immunofluorescence, Western blot, and real-time qPCR. Immunofluorescence assays showed that ZR75.1 cells expressing the scrambled control (shScr) followed a classic response to EMT-inducing conditions; they lost expression of E-cadherin and gained expression of vimentin when the EMT was induced (Fig. 3A; Fig. S4A). In sharp contrast, cells lacking AID continued to express E-cadherin and did not express vimentin, nor did they undergo morphological changes typical of the EMT (Fig. 3A; Fig. S4A). Consistent with these results, vimentin was not detected by Western blot in AID-deficient cells in EMT-inducing conditions [Fig. 3B; Fig. S4B; compare shScr with shAIDkd lanes (EMT induced)]. Gene expression analysis performed by real-time qPCR supported these results. During the EMT, ZR75.1 cells that expressed AID (shScr) displayed a decrease in CDH1 expression and an increase in VIM and CDH2, whereas cells with AID knockdown showed the opposite result, with a several-fold down-regulation of mesenchymal markers compared with control cells (Fig. 3C; Fig. S4C). In contrast, levels of E12/47 (product of the gene E2A, a transcription factor and repressor of E-cadherin expression during the EMT) (53) and DNMT1 (DNA methyl transferase 1, involved in maintenance of DNA methylation) were not affected by a deficiency in AID expression (Fig. S5), suggesting that there is a distinct regulatory mechanism for these genes. These results show that AID is required for progression of the EMT in ZR75.1 breast cancer cells through its effects on expression of specific EMT genes.
Fig. 3.
Knockdown of AID blocks the EMT in ZR75.1 cells. (A) Immunofluorescence of ZR75.1 cells. In EMT-inducing conditions, cells that express AID (shScr) lose expression of E-cadherin (green), and gain expression of vimentin (red). Knockdown of AID (shAIDkd1) blocks this effect, so that cells resemble ZR75.1 cells that have not been induced to undergo the EMT. DNA is counterstained with DAPI (blue). (B) Vimentin levels assayed by Western blot in whole cell extracts of ZR75.1 cells. Uninduced ZR75.1 cells (UI) are shown only for shScr control cells. EMT-induced cells are shown as in A, confirming that knockdown of AID (shAIDkd1) blocks the expression of vimentin seen in EMT-inducing conditions. MDA, extracts of MDA-MB-231 breast cancer cells are included as a positive control for vimentin. (C) mRNA levels of CDH1, VIM, and CDH2 measured by real-time qPCR under EMT-inducing conditions in ZR75.1 cells. mRNA levels are expressed relative to the levels in untreated shScr cells (UI shScr). Knockdown of AID (shAIDkd1) has little effect on CDH1 expression but decreases expression of the two mesenchymal markers. *P < 1 × 10−5 VIM and P < 1 × 10−4 CDH2. **P < 5 × 10−6 VIM and P < 1 × 10−4 CDH2. Error bars indicate SD (n ≥ 3 experiments).
AID Regulates Expression of EMT Master Regulators and Upstream Factors.
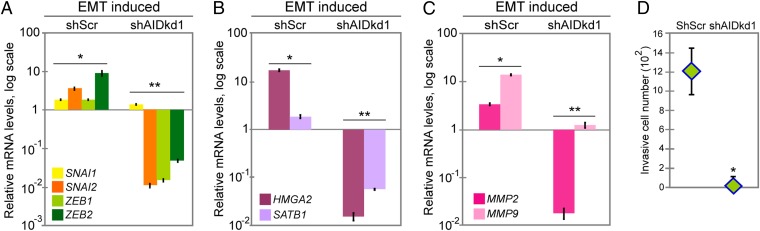
The EMT in its various forms is driven by the transcription factors Snail (SNAI1), Slug (SNAI2), ZEB1 (ZEB1), and ZEB2 (ZEB2) (35, 36). We next asked whether AID was required for up-regulation of these master regulators during the EMT. ZR75.1 cells were induced to the EMT, and mRNA levels for the four factors were measured by real-time qPCR. As expected, induction of the EMT was associated with up-regulation of all four factors in ZR75.1 cells expressing shScr (Fig. 4A; Fig. S6A). However, in AID-deficient cells, levels of SNAI2, ZEB1, and ZEB2 mRNAs were not increased by induction of the EMT and instead were markedly below baseline levels (Fig. 4A; Fig. S6A). SNAI1 mRNA levels were only slightly different when comparing cells with AID or with AID knockdown, although the difference was still significant (Fig. 4A; Fig. S6A). These results gave further evidence that AID regulates the EMT.
Fig. 4.
AID regulates expression of key factors in the EMT, and AID deficiency strongly impairs the motile and invasive properties of ZR75.1 cells. (A–C) mRNA levels measured by real-time qPCR, displayed relative to levels in untreated ZR75.1 shScr cells. (A) Increased expression of the EMT master regulators (SNAI1, SNAI2, ZEB1, and ZEB2) in shScr control cells in EMT-inducing conditions, is blocked by knockdown of AID (shAIDkd1). *P < 1 × 10−4 SNAI1, P < 0.005 SNAI2, P < 0.01 ZEB1, and P < 0.001 ZEB2; **P < 0.001 SNAI1 and ZEB2, P < 5 × 10−5 SNAI2, and P < 1 × 10−6 ZEB1. (B) Increased expression of the chromatin factors HMGA2 and SATB1 in shScr control cells in EMT-inducing conditions is blocked by knockdown of AID (shAIDkd1). *P < 5 × 10−6 HMGA2, P < 0.001 SATB1; **P < 5 × 10−6 HMGA2, P < 5 × 10−5 SATB1. (C) Increased expression of MMP2 and MMP9 in shScr cells expressing AID in EMT-inducing conditions is blocked by AID knockdown (shAIDkd1). *P < 5 × 10−6 MMP2 and MMP9; **P < 1 × 10−6 MMP2 and P < 5 × 10−6 MMP9. Knockdown of AID blocks the up-regulation of metalloproteinases. (D) Number of cells able to move and invade under EMT-inducing conditions. shScr and shAIDkd1 are as in A (*P < 5 × 10−7). Knockdown of AID abolishes invasion and migration of ZR75.1 breast cancer cells. Error bars indicate SD (n ≥ 3 experiments).
We also studied expression of two factors, HMGA2 and SATB1, that may act as upstream regulators of the expression changes we observed during the EMT (for a scheme of EMT factor hierarchy, see reviews in refs. 35 and 54). HMGA2 (high-mobility group A2) is a nonhistone chromatin protein that regulates expression of EMT factors Slug, Snail, Twist, and Id2 (54–56). The chromatin looping factor SATB1 has been shown to participate in breast cancer metastasis by up-regulating expression of metastasis-associated genes and down-regulating tumor suppressor genes (57). We found that expression of both HMGA2 and SATB1, measured by real-time qPCR, increased in EMT-inducing conditions when AID was expressed, but both were strongly down-regulated when AID was knocked down (Fig. 4B; Fig. S6B).
Knockdown of AID Down-Regulates Expression of Extracellular Matrix Metalloproteinases and Impairs Cellular Migration and Invasion.
The extracellular matrix (ECM) metalloproteinases (MMPs) are downstream effectors of the EMT, required to degrade structural components of the ECM, cleave membrane proteins such as E-cadherin, degrade tight junctions, and release biologically active protein fragments that increase the cells’ invasive behavior (58, 59). MMP2 is involved in mammary development during puberty (60), and its expression is increased during the EMT and in tumor-initiating cells after therapy (37, 61). Another relevant metalloproteinase is MMP9, which cooperates with Snail to induce the EMT in cervical carcinoma cells (62). To establish if AID also modulates MMP expression, we measured the mRNA levels of MMP2 and MMP9 by real-time qPCR. Expression of both genes was up-regulated during the EMT in ZR75.1 cells expressing shScr, and knockdown of AID blocked this up-regulation (Fig. 4C; Fig. S6C). This result indicates that AID is necessary for increased expression of factors that confer invasiveness on ZR75.1 cells during the EMT.
To further explore the role of AID in invasiveness and migration, we used a transwell assay. In this assay, movement of cells through pores in a transwell requires that they first degrade a basement membrane (Matrigel). Knockdown of AID under EMT-inducing conditions severely restricted the movement of ZR75.1 cells through the pores, indicating loss of the ability to degrade the basement membrane, movement through the pores, or both (Fig. 4D; Fig. S6D). Because AID is necessary for expression of proteins that mediate both functions (vimentin, MMPs), it is likely that both processes were affected.
AID Knockdown Blocks EMT Markers in MCF10A Cells.
MCF10A cells respond to EMT-inducing conditions with a transcriptional profile similar to ZR75.1 cells, although basal levels of most of the EMT factors are substantially higher in MCF10A cells (43, 44). AID knockdown by shAIDkd1 in MCF10A cells (Fig. 5A) abrogated the up-regulation of several EMT markers in EMT-inducing conditions (Fig. 5B), as it does in ZR75.1 cells (Figs. 2–4). The effects of AID deficiency on the expression of EMT markers, while evident in MCF10A cells, are less dramatic than in ZR75.1 cells. Because ZR75.1 cells are cancer cells and a very good in vitro model of the early steps of metastasis, we pursued further experiments in this cell line.
Fig. 5.
AID knockdown blocks up-regulation of EMT markers in MCF10A cells. (A) AID (AICDA) mRNA levels measured by real-time qPCR, displayed relative to levels in MCF10A parental cells expressing a scrambled shRNA (shScr). MCF10A cells expressing shAIDkd1 display an approximate fivefold reduced level of AID mRNA. *P < 0.001. (B) mRNA levels of key EMT factors in MC10A cells deficient for AID expression (shAIDkd1) in EMT-inducing conditions, determined by real-time qPCR and displayed as fold change relative to MCF10A shScr cells. P < 0.05 Vim, P < 5 × 10−4 CDH2, P < 1 × 10−4 SNAI1, P < 0.005 HMGA2, and P < 5 × 10−6 MMP2 and MMP9. Error bars indicate SD (n ≥ 3 experiments).
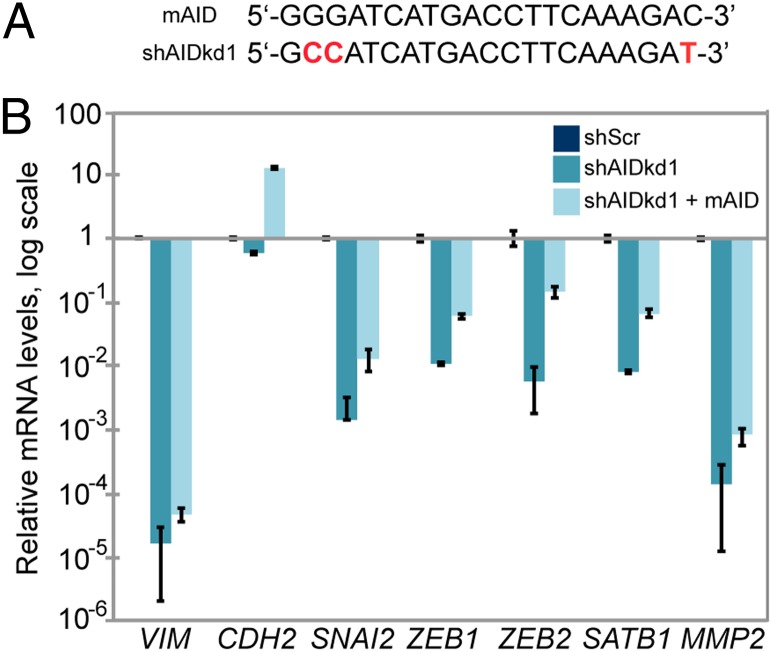
Expression of Mouse AID Rescues the Expression of Genes Down-Regulated by AID Deficiency.
Human AID is able to complement mouse AID (mAID) functions in AID-deficient mouse primary splenic B cells (63, 64). The sequence of mAID mRNA indicates that it would not be affected by the shRNAs directed against human AID mRNA (Fig. 6A; Fig. S7A). To further address the requirement for AID in the EMT, we stably expressed mAID in ZR75.1 shAIDkd1 and shAIDkd2 cells and measured the expression levels of several EMT markers by real-time qPCR. Mouse AID was able to partially restore the expression of most of the genes that are strongly down-regulated by AID deficiency (Fig. 6B; Fig. S7B). These results support the interpretation that the effects of AID knockdown on expression of these genes are due to AID deficiency and not to off-target effects.
Fig. 6.
Mouse AID restores the transcription of genes down-regulated by AID deficiency in ZR75.1 cells. (A) Sequence comparison between mouse AID (mAID) and one of the shRNAs that target human AID (shAIDkd1); differences are highlighted in red. (B) mRNA levels of key EMT factors down-regulated by AID knockdown and partially restored by expression of mAID, determined by real-time qPCR and expressed as fold change relative to ZR75.1 shScr cells. P values were calculated between shAIDkd1 and shAIDkd1+mAID. P < 0.05 VIM, P < 5 × 10−6 CDH2, P < 0.005 SNAI2 and MMP2, P < 5 × 10−4 ZEB1 and SATB1, and P < 1 × 10−4 ZEB2. Error bars indicate SD (n ≥ 3 experiments).
A DNA Demethylating Agent Antagonizes the Transcriptional Effects Caused by AID Knockdown.
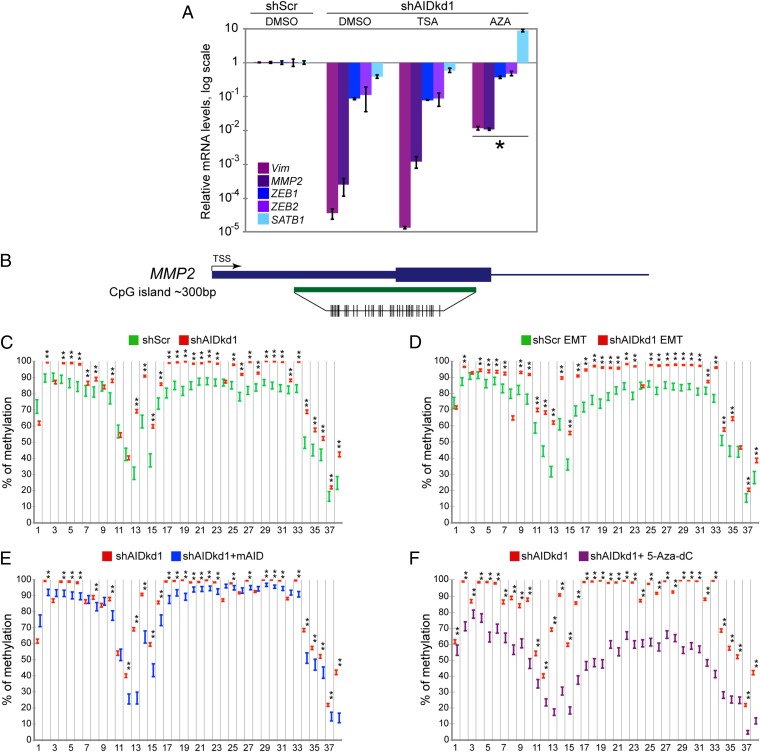
AID has been proposed to bind, and lead to demethylation of, promoters that subsequently become transcriptionally active (8). If knockdown of AID decreases expression of EMT genes due to increased CpG methylation in these genes, the effect should be antagonized by treatment with the DNA demethylating agent 5 aza-2'deoxycytidine (5-Aza-dC). We analyzed the expression of a set of genes that were down-regulated by AID knockdown (Figs. 2–4) and contain CpG islands near their transcriptional start sites (VIM, ZEB1, ZEB2, HMGA2, SATB1, and MMP2). We compared ZR75.1 shScr with AID-deficient cells, and cells were treated either with 5-Aza-dC or with the histone deacetylase inhibitor trichostatin A (TSA) as a control. E-cadherin (CDH1) mRNA levels, which were not altered by knockdown of AID (Figs. 2 and 3; Figs. S3 and S4), were also not affected by 5-Aza-dC (Fig. S8A). Genes whose expression was down-regulated by knockdown of AID (Figs. 2–4) were strongly up-regulated by 5-Aza-dC, with the exception of HMGA2 (Fig. 7A; Fig. S8A). TSA did not affect gene expression in either control or AID knockdown cells. This result was consistent with VIM, ZEB1, ZEB2, SATB1, and MMP2 being regulated by CpG methylation, so that removal of methylation increased their expression. The same results were obtained with cells harboring another shRNA that efficiently knocked down AID (shAIDkd2) (Fig. S8B). Thus, a demethylating agent antagonizes the effects of AID knockdown on expression of these genes, suggesting that AID regulates their expression through an effect on methylation status.
Fig. 7.
Increased levels of methylation in CpG islands of genes down-regulated by AID knockdown. (A) The demethylating agent 5-Aza-dC antagonizes the effect of AID knockdown on expression of EMT factors. Levels of VIM, ZEB1, ZEB2, MMP2, and SATB1 mRNA measured by real-time qPCR in ZR75.1 shScr cells and in cells in which AID was knocked down (shAIDkd1) after treatment with vehicle (DMSO), Trichostatin A (TSA), or 5-Aza-dC (AZA). mRNA levels are displayed as fold change of levels observed in ZR75.1 shScr cells treated with DMSO. AID knockdown reduces expression of the EMT factors, and 5-Aza-dC reverses this effect. *P < 1 × 10−4 ZEB1 and SATB1, P < 5 × 10−4 VIM and MMP2, and P < 0.005 ZEB2. Error bars indicate SD (n = 3 experiments). (B) Schematic representation of the CpG island of MMP2. (C, E, and F) Percentage of methylation at individual CpGs in the CpG island of MMP2 at baseline (no EMT induction). (D) Percentage of methylation at individual CpGs in the CpG island of MMP2 in EMT-inducing conditions. Green: AID expressing (shScr). Red: AID deficient (shAIDkd1). Blue: AID deficient, complemented with mAID (shAIDkd1+mAID). Purple: AID deficient, treated with 5-Aza-dC (shAIDkd1+5-Aza-dC). Bars indicate 95% *P < 0.0002.
Knockdown of AID Increases Methylation in Promoter CpG Islands of Genes Involved in the EMT.
The data discussed above implied an AID-dependent regulation of the methylation status of some CpG islands. Our results could be either a direct or an indirect effect of AID knockdown. In addition, dense CpG island methylation had traditionally been associated with suppression of transcription initiation, but recent work has shown that intragenic CpG methylation is linked to gene expression (22, 27). In consequence, AID has the potential to participate in dynamic processes that regulate both CpG-rich and CpG-depleted promoters in any circumstance in which demethylation is required to change function. Thus, to further support the involvement of methylation in the changes in gene expression caused by AID knockdown, we selected regions from CpG islands in VIM, ZEB1, SATB1, and MMP2 and analyzed them by bisulfite conversion and deep sequencing. We obtained an average coverage of 37,000×, thus providing a very accurate (and statistically significant) measure of the extent to which each CpG in the amplified region is methylated (i.e., the proportion of alleles in which that CpG is methylated).
The CpG island in the 5′UTR and first exon of the MMP2 gene (Fig. 7 B–F; Fig. S9 A–D) showed intermediate levels of methylation that slightly declined when the EMT was induced. Knockdown of AID resulted in increased methylation over most of the amplified region, at baseline and during the EMT. Complementation with mAID antagonized the effects of AID knockdown, so that the methylation levels were equivalent to those in control cells (compare Fig. 7C, shScr with Fig. 7E, shAIDkd1+mAID). 5-Aza-dC treatment also antagonized the effects of AID knockdown on methylation of the MMP2 CpG island (Fig. 7F). These results are consistent with the observed changes in expression of MMP2 during the EMT in cells overexpressing mAID and in cells treated with 5-Aza-dC (Figs. 4C, 6B, and 7A; Figs. S6C and S8B). We obtained the same result with a different AID knockdown vector (Fig. S9 A–D). A similar scenario was observed in the small CpG island immediately upstream of the transcription start site in SATB1 (Fig. S9 E–H) and in a region of the CpG island that spans the first two exons and introns of VIM (Fig. S9 I–L). In both cases, CpG methylation was variable and changed little with induction of the EMT. However, knockdown of AID increased methylation slightly (but significantly) at several CpGs at baseline and during the EMT. mAID complementation or treatment with 5-Aza-dC antagonized the effects of AID knockdown in both genes. We also analyzed four regions of the CpG island at the 5′ end of ZEB1. Levels of CpG methylation in these locations were extremely low in all conditions. Considering that the expression of ZEB1 increased in the presence of a DNA demethylating agent (Fig. 7A; Fig. S8B), we concluded that the portions of the ZEB1 CpG island selected for this analysis were not the ones that regulate its expression, highlighting the necessity for more detailed maps of genomewide CpG methylation levels; alternatively, the expression of ZEB1 might require the expression of another factor regulated by methylation.
Taken together, these results indicate that AID regulates methylation of genes involved in the EMT.
Discussion
AID has been intensively studied because of its role in somatic hypermutation and class switch recombination of Ig genes. More recently it has been proposed to have a role in cytosine demethylation in several biological processes, considerably broadening the potential scope of its biological function. The EMT is a critical step in tumor metastasis and also in normal development. Here we show that AID is required for the EMT in a breast cancer cell model and for gene expression changes typical of the EMT in a nontransformed mammary epithelial cell line. Our results could be explained by AID affecting epigenetic reprogramming, most likely through its ability to deaminate methylcytosine, but possibly by some other mechanism. Consistent with an effect on cytosine methylation, we find that knockdown of AID increases promoter methylation and decreases expression of genes that are key participants in the EMT. The induction of AID expression by inflammatory signals in this and other systems, together with the proinflammatory nature of the tumor niche, raises the possibility that AID has a role in early stages of metastasis.
The EMT is a transient state in which cells lose cellular contacts and gain the ability to move and invade; it occurs normally during development, in the so called EMT or epithelial–fibroblast transition, but also during carcinogenesis (carcinoma–metastatic EMT). In a variety of settings, both the EMT and AID expression are induced by proinflammatory signals. We find that AID expression is up-regulated in ZR75.1 and MCF10A cells on treatment with TNFα and TGFβ, inducers of the EMT. We reasoned that because AID is involved in reprogramming phenomena, it could be required for the EMT, which is elicited by the inflammatory environment characteristic of solid tumors. Direct abolition of AID function (by shRNAs) blocks the EMT as judged by morphological, molecular, and functional markers in ZR75.1 cells; these effects can be antagonized by complementation with mouse AID. In nontransformed MCF10A mammary epithelial cells, we found that AID deficiency down-regulates a set of genes similar to the set down-regulated in our cancer model. These expression changes resemble those described in breast and colorectal cancer cells with knockdown of ZEB1, a transcription factor that is a key player in the EMT (65).
Our results are particularly significant for breast cancer because the EMT is thought to be a key step in metastasis. The ZR75.1 line is an excellent model of the process because of its propensity for undergoing the EMT in response to stimuli that closely reflect the conditions necessary for the EMT in vivo, and its expression of markers and regulatory factors that have well-documented roles in the EMT in vivo and in vitro (43); few other cell lines model the EMT as closely as does ZR75.1.
Our results indicate that AID may act at or near the apex of a hierarchy of regulatory steps that drive the EMT. In ZR75.1 and MCF10A cells, induction of the EMT elicits the up-regulation of mesenchymal factors (vimentin and N-cadherin), the EMT transcription factors Snail, Slug, ZEB1, and ZEB2, the chromatin-associated factors HMGA2 and SATB1, and the extracellular matrix metalloproteinases MMP2 and 9; these changes are typical of the carcinoma-metastatic EMT (38, 66). Up-regulation of HMGA2 and SATB1 is particularly significant because these factors have been shown to regulate Snail and Slug, two transcription factors that have a direct role in the EMT (55, 57). The observation that knockdown of AID in EMT-inducing conditions abolishes the up-regulation of all these factors (e.g., HMGA2, SATB1, Snail, Slug, ZEB1, and ZEB2) that have a direct role in driving the EMT suggests that it acts upstream of all of them.
The notion that AID regulates expression of key EMT-driving factors is supported by analysis of cytosine methylation in the promoters of some of these genes. DNA demethylation through deamination of meC is a newly described and still controversial function for AID (6), which is carried out in concert with TDG, MBD4, and GADD45 (8–10, 30). We show that certain genes (MMP2, SATB1, and VIM) that are strongly down-regulated by AID knockdown are reexpressed on mAID complementation or cellular treatment with the demethylating agent 5-Aza-dC. Consistent with the effects of 5-Aza-dC, AID knockdown is associated with increased methylation levels in CpG islands near the promoters of these genes. Furthermore, treatment with 5-Aza-dC or complementation with mouse AID antagonized the effect of AID knockdown on methylation levels in these key promoters. Although this does not prove that methylation levels in these CpG islands are directly modulated by AID, it is consistent with the evidence that AID demethylates promoters of genes that subsequently become transcriptionally active (8, 67). Recent studies have described AID’s association with RNA polII at gene promoters (68) and with the elongation complex (69), which would suggest that it may regulate gene expression independently of its function in DNA deamination. Such a global function is unlikely to account for the results of our study, in which AID deficiency does not have a repressive effect in general transcription and leads to down-regulation of only a subset of EMT genes.
CpG islands have traditionally been considered as constitutively unmethylated, with methylation indicating transcriptional silencing of their associated genes (22). Our bisulfite analysis reveals variable levels of methylation in CpG islands near the promoters of genes that are up-regulated during the EMT and down-regulated by AID knockdown. The method we used is ideal for identifying partial methylation: very deep bisulfite sequencing permits highly accurate estimates of methylation levels. Currently available genomewide methylation studies have not achieved this depth, and so we do not know how common such intermediate methylation may be in other CpG islands. We speculate that AID regulates many promoters, including some of those we have studied, by maintaining a partially unmethylated state that modulates promoter activity. To date, we have studied only portions of CpG islands of some EMT genes; more detailed investigations will be required to better understand the genomewide role of AID in CpG methylation and transcriptional regulation.
If AID modulates promoter activity, as suggested by our results, it is likely to do so in concert with other factors. AID forms a complex with TDG and GADD45A in 293 and embryonal carcinoma cells (30). Recent studies have implicated TDG in the pathway of cytosine demethylation, where it may act through association with AID (30, 70). TDG is involved in regulating gene expression through interactions with transcription factors and epigenetic modifiers (71–74), and its activity in this context is presumed to involve cytosine demethylation. AID’s association with TDG (30) suggests that it may similarly regulate gene expression, either through its deaminase activity or by some as yet undetermined mechanism. In regard to the latter possibility, AID associates with the PAF complex on chromatin, and this association results in active transcription of a reporter gene independently of the AID’s catalytic activity (69).
We have found that AID is required for the EMT, a critical step in metastasis, in a breast cancer cell model; furthermore, it is required for normal expression of EMT markers in a nonmalignant breast epithelial cell line. Does this reflect some general function of AID outside its well-documented effects on Ig genes in lymphocytes? Cancer is not a normal process, but our findings and other evidence indicate that AID is induced by proinflammatory signals, which also induce the EMT in a variety of normal and pathological settings (35, 75). This evidence links our findings to other reports of a role for AID in processes that involve epigenetic reprogramming. AID appeared early in vertebrate phylogeny, before the emergence of somatic hypermutation and class switch recombination, although always linked to diversification of antigen receptors (3). It is not clear whether a function of AID in gene regulation has always coexisted with antigen receptor diversification or evolved at a different time.
A broader biological role for AID outside of lymphocytes would seem to be contradicted by the viability of AID−/− mice and their apparent lack of any strong phenotype outside B cells. However, most studies of these mice have focused on Ig genes and lymphocytes; more subtle phenotypes may have been missed, or AID function might have been compensated by other factors involved in DNA demethylation. One recent study noted that lack of AID results in defects in the control of litter size and failure of maternal control over fetal growth (9). That study also found that E13.5 primordial germ cells, where much of the genome has undergone dramatic demethylation at this critical stage of epigenetic resetting, display increased cytosine methylation in AID-deficient embryos; however, there was still a considerable extent of cytosine demethylation. This finding implies that there are redundant and AID-independent mechanisms of cytosine demethylation. Candidates for this function include other members of the APOBEC family and the TET proteins (10, 28, 29, 76). Thus, AID may be required only in certain situations, such as the carcinoma-metastatic EMT as identified herein, whereas in other developmental processes that involve other types of EMT, there is functional redundancy with related proteins.
In any case, our findings are consistent with a role for AID in regulation of cellular reprogramming by modulating cytosine methylation and gene expression and demonstrate the capacity of AID to play a role in complex developmental programs. The large body of knowledge on AID’s role in Ig genes in B cells provides few clues about other functions of AID, which remain largely unexplored.
Materials and Methods
Cell Culture and Lentivirus Production and Transduction.
Cell culture.
ZR75.1 cells (kindly provided by Dr. Pierre-Yves Despréz, California Pacific Medical Center, San Francisco) were grown in RPMI 1640 supplemented with 10% (vol/vol) FBS and 100 U/mL of penicillin/streptomycin in humidified 5% CO2 at 37 °C. MCF10A cells (kindly provided by Dr. Terumi Kohwi-Shigematsu, Lawrence Berkeley National Laboratory, Berkeley, CA) were grown as described (45).
Lentiviruses.
shRNA-expressing lentiviral vectors to knock down AID were purchased from Open Biosystems. Lentiviruses were generated by cotransfecting 293FT cells (Invitrogen) with the lentiviral plasmid and the packaging vectors (pLP, pLP1, and pLP2) using Lipofectamine 2000 (Invitrogen). Twenty-four hours after transfection, the medium was replaced with DMEM with 10% (vol/vol) FBS. Forty-eight hours after transfection, the supernatants were harvested, centrifuged to remove cellular debris, and filtered; lentiviral particles were precipitated overnight at 4 °C using PEGit solution (System Biosciences). After centrifugation, pellets were resuspended in PBS, aliquoted, and stored at −80 °C for later use. Transductions were carried out overnight in the presence of 4 µg/mL of Polybrene. Forty-eight hours later, selection was started in medium containing 1 µg/mL puromycin.
Mouse AID was PCR amplified from pEGF-N3-mAID, kindly provided by R. S. Harris and D. A. MacDuff (University of Minnesota, Minneapolis, MN) (77), cloned in pENTR-1A (Invitrogen), sequenced, and then recombined into pLenti-DEST (w-117) kindly provided by E. Campeau (Resverlogix Corporation, Calgary, AB) (78). Lentiviral particles were produced as described above. ZR75.1 cells deficient of AID were transduced overnight in the presence of 4 µg/mL Polybrene. Forty-eight hours later, selection was started in medium containing 100 µg/mL hygromycin.
TNFα and TGFβ treatments.
Cells were treated with recombinant human TNFα (eBioscience) and/or TGFβ-1 (R&D) at various concentrations and harvested at time points stated in each experiment. EMT-inducing conditions were 10 ng/mL TNFα and 2 ng/mL TGFβ. Cells were processed with TRIzol for RNA purification or resuspended in lysis buffer (see below) for Western blot analysis.
Trichostatin A and 5-Aza-2′-deoxycytidine were obtained from Sigma and suspended in DMSO.
Immunofluorescence and Western Blot.
Immunofluorescence.
Immunofluorescence was performed as previously described (79) with minor modifications. Cells were seeded in chamber slides coated with matrigel (BD Biosciences), kept untreated or treated with EMT-inducing conditions, fixed at different time points with 4% (vol/vol) paraformaldehyde, and washed and permeabilized with 0.5% Triton X-100 in PBS. After rinsing with PBS, cells were blocked with 4% (vol/vol) normal donkey serum (Jackson ImmunoResearch) in PBS. Cells were probed overnight at 4 °C with primary antibodies in blocking buffer and then rinsed with PBS. Cells were incubated with fluorochrome-conjugated secondary antibodies in blocking buffer and washed with PBS three times. Slides were mounted in Vectashield containing DAPI. Images were acquired in an Olympus BX60 fluorescence microscope with Spotfire 3.2.4 software (Diagnostics Instruments) and processed with Photoshop CS2 (Adobe) software.
Western blot.
Cells were resuspended in lysis buffer [20 mM Tris⋅HCl, pH 7.6, 137 mM NaCl, 10% (vol/vol) glycerol, 2 mM EDTA, 1% (vol/vol) octylphenyl-polyethylene glycol (IGEPAL) with freshly added 0.1 mM PMSF and complete-EDTA free protease inhibitors (Roche)]. Total protein concentration was measured by BCA Assay (Pierce). SDS polyacrylamide gels were transferred to nitrocellulose. Membranes were blocked with 5% (wt/vol) milk in Tris-buffered saline (TBS)-Tween-20. Incubations with primary and secondary antibodies were done in blocking solution. Membranes were developed with PicoGreen ECL (Pierce).
Antibodies.
The following antibodies were used: E-cadherin (Cell Signaling # 3195), vimentin (Thermo Scientific MS-129), β-actin (Abcam ab6276), donkey anti-mouse Alexa 594 and anti-rabbit Alexa 488 (Molecular Probes; Invitrogen), and anti-mouse and anti-rabbit HRP (BioRad).
RNA Extraction, cDNA Synthesis, and Quantitative Real-Time PCR.
RNA was purified using TRIzol followed by DNase RNase-free digestion (Qiagen). cDNA synthesis to detect AID was performed using SuperScript III enzyme (Invitrogen); cDNAs for detection of EMT markers and transcription factors were synthesized with the High Capacity cDNA Reverse Transcription kit (ABI). Most of the primers’ sequences for real-time qPCR were obtained from PrimerBank (80), and their sequences will be provided on request. Real-time qPCR reactions were performed in an ABI 7900HT apparatus using SYBR-Green Power master mix (ABI) with default cycling conditions; results were analyzed with ABI SDS 2.4 software. At least three biological replicates were done for all real-time qPCR experiments, and values were analyzed with QuickCalcs (from GraphPad Software, http://graphpad.com/quickcalcs/Grubbs1.cfm) to detect outliers, and all mRNA levels were assayed at least in triplicate; dissociation curves were checked, and products were run in agarose gels to confirm amplification of only one product. Relative mRNA levels were calculated by the 2∧(−∆∆Ct) method using GAPDH and HPRT1 as controls. To better reflect the actual very low levels of AID mRNA in knockdown ZR75.1 cells, we computed those reactions that had not product amplified for AID (but had product amplified for the control gene) as having a Ct value of 40.
We used a paired two-tailed t test (assuming unequal variance) to test the significance of differences in gene expression.
Invasion Assays.
Invasion assays were done as described previously (42). In brief, 1.5 × 105 cells were seeded in the upper chamber of a transwell Boyden chamber (Becton Dickson) previously coated with 20% (vol/vol) Matrigel with medium with no FBS. The time regimen applied to the transwell assay was followed as described previously (81). Sixteen hours after seeding the cells, medium containing 10% (vol/vol) FBS + 2 ng/mL TGFβ and 10 ng/mL TNFα was placed in the bottom wells as a chemo-attractant and to induce the EMT. Approximately 40–48 h later, cells on the outside bottom of the wells were fixed with glutaraldehyde and stained with 0.2% crystal violet. Cells remaining in the upper part of the well were removed. Cells that had invaded through the pores were visually counted on a microscope. Experiments were done three times, in triplicate each time.
Genomic DNA Extraction, Bisulfite-Specific PCR, and Illumina Library Preparation.
Untreated ZR75.1 cells, or cells induced to go through the EMT for ∼48 h, were harvested and resuspended in cold PBS. The genomic DNA (gDNA) was purified with QIAamp DNA mini kit (QIAGEN) and bisulfite converted using the MethylEasyXceed kit (Human Genetic Signatures). Primers for amplification of bisulfite-converted DNA were designed using MethPrimer (82) in the “bisulphite sequencing” mode (primer sequences will be provided on request). PCR was done using Zymo Taq DNA polymerase (Zymo Research). Individual PCR products were gel purified, and all of the products belonging to a particular sample were pooled in equimolar amounts. Each sample analyzed was prepared from biological duplicates and pooled at the step of the Illumina library preparation. Indexed paired-end libraries were constructed from the PCR products and sequenced on an Illumina HiSeq2000 to collect 2 × 50 base reads.
Bisulfite PCR Sequence Analysis.
Sequences were assigned to a specific condition according to their index and aligned against a reference sequence using Novoalign (www.novocraft.com) in bisulfite mode. We used the program pileup from the SAMtools suite (samtools.sourceforge.net) to determine the number of converted and unconverted reads at all cytosines in the alignment that were not polymorphic and were covered by at least 50 reads. Percentage of methylation at each cytosine was calculated as M = number of unconverted reads/(number of converted and unconverted reads) × 100. The bisulfite conversion rate was estimated from the value of M at non-CpG positions. Results are reported as the value of M at CpG positions.
Ninety-five percent confidence intervals for the proportions (M, as described above) for each CpG were calculated in VassarStats webpage (http://faculty.vassar.edu/lowry/VassarStats.html) using the method described there (83). The significance of the difference between two independent proportions was also calculated among the samples as described in http://faculty.vassar.edu/lowry/VassarStats.html. Two-tailed probabilities were adjusted for multiple testing.
Note Added in Proof.
As this paper was going to press, Evans and colleagues (84) published similar findings which indicate that AID is necessary to stabilize the reprogrammed state of fibroblasts, and that this effect is mediated by demethylation of key reprogramming genes.
Supplementary Material
Acknowledgments
We thank Pierre-Yves Despréz for the kind gift of multiple reagents and for helpful comments and discussions, Jennifer Yamtich for help with statistical analysis, and Wendy Magis for technical assistance. This work was supported by National Institutes of Health Grants 5K22CA163969-02 (to D.P.M.), CA115768-05, 1R21ES016581-02, and DK080428-02 (to D.I.K.M.) and a Pink Pumpkin Patch grant (to D.P.M.). J.M.D.N. is supported by a Canadian Research Chair tier 2.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1301021110/-/DCSupplemental.
References
- 1.Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- 2.Chaudhuri J, et al. Evolution of the immunoglobulin heavy chain class switch recombination mechanism. Adv Immunol. 2007;94:157–214. doi: 10.1016/S0065-2776(06)94006-1. [DOI] [PubMed] [Google Scholar]
- 3.Conticello SG. The AID/APOBEC family of nucleic acid mutators. Genome Biol. 2008;9(6):229. doi: 10.1186/gb-2008-9-6-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meyers G, et al. Activation-induced cytidine deaminase (AID) is required for B-cell tolerance in humans. Proc Natl Acad Sci USA. 2011;108(28):11554–11559. doi: 10.1073/pnas.1102600108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuraoka M, et al. Activation-induced cytidine deaminase mediates central tolerance in B cells. Proc Natl Acad Sci USA. 2011;108(28):11560–11565. doi: 10.1073/pnas.1102571108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morgan HD, Dean W, Coker HA, Reik W, Petersen-Mahrt SK. Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues: Implications for epigenetic reprogramming. J Biol Chem. 2004;279(50):52353–52360. doi: 10.1074/jbc.M407695200. [DOI] [PubMed] [Google Scholar]
- 7.Schreck S, et al. Activation-induced cytidine deaminase (AID) is expressed in normal spermatogenesis but only infrequently in testicular germ cell tumours. J Pathol. 2006;210(1):26–31. doi: 10.1002/path.2014. [DOI] [PubMed] [Google Scholar]
- 8.Bhutani N, et al. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2010;463(7284):1042–1047. doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Popp C, et al. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463(7284):1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rai K, et al. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135(7):1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dedeoglu F, Horwitz B, Chaudhuri J, Alt FW, Geha RS. Induction of activation-induced cytidine deaminase gene expression by IL-4 and CD40 ligation is dependent on STAT6 and NFkappaB. Int Immunol. 2004;16(3):395–404. doi: 10.1093/intimm/dxh042. [DOI] [PubMed] [Google Scholar]
- 12.Park SR, et al. HoxC4 binds to the promoter of the cytidine deaminase AID gene to induce AID expression, class-switch DNA recombination and somatic hypermutation. Nat Immunol. 2009;10(5):540–550. doi: 10.1038/ni.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gonda H, et al. The balance between Pax5 and Id2 activities is the key to AID gene expression. J Exp Med. 2003;198(9):1427–1437. doi: 10.1084/jem.20030802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Komori J, et al. Activation-induced cytidine deaminase links bile duct inflammation to human cholangiocarcinoma. Hepatology. 2008;47(3):888–896. doi: 10.1002/hep.22125. [DOI] [PubMed] [Google Scholar]
- 15.Endo Y, et al. Activation-induced cytidine deaminase links between inflammation and the development of colitis-associated colorectal cancers. Gastroenterology. 2008;135(3):889–898. doi: 10.1053/j.gastro.2008.06.091. [DOI] [PubMed] [Google Scholar]
- 16.Endo Y, et al. Expression of activation-induced cytidine deaminase in human hepatocytes via NF-kappaB signaling. Oncogene. 2007;26(38):5587–5595. doi: 10.1038/sj.onc.1210344. [DOI] [PubMed] [Google Scholar]
- 17.Kou T, et al. Expression of activation-induced cytidine deaminase in human hepatocytes during hepatocarcinogenesis. Int J Cancer. 2007;120(3):469–476. doi: 10.1002/ijc.22292. [DOI] [PubMed] [Google Scholar]
- 18.Matsumoto Y, et al. Helicobacter pylori infection triggers aberrant expression of activation-induced cytidine deaminase in gastric epithelium. Nat Med. 2007;13(4):470–476. doi: 10.1038/nm1566. [DOI] [PubMed] [Google Scholar]
- 19.Gupta PB, et al. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009;138(4):645–659. doi: 10.1016/j.cell.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Babbage G, Ottensmeier CH, Blaydes J, Stevenson FK, Sahota SS. Immunoglobulin heavy chain locus events and expression of activation-induced cytidine deaminase in epithelial breast cancer cell lines. Cancer Res. 2006;66(8):3996–4000. doi: 10.1158/0008-5472.CAN-05-3704. [DOI] [PubMed] [Google Scholar]
- 21.Pauklin S, Sernández IV, Bachmann G, Ramiro AR, Petersen-Mahrt SK. Estrogen directly activates AID transcription and function. J Exp Med. 2009;206(1):99–111. doi: 10.1084/jem.20080521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25(10):1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Riggs AD. X inactivation, differentiation, and DNA methylation. Cytogenet Cell Genet. 1975;14(1):9–25. doi: 10.1159/000130315. [DOI] [PubMed] [Google Scholar]
- 24.Reik W, Collick A, Norris ML, Barton SC, Surani MA. Genomic imprinting determines methylation of parental alleles in transgenic mice. Nature. 1987;328(6127):248–251. doi: 10.1038/328248a0. [DOI] [PubMed] [Google Scholar]
- 25.Bourc’his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431(7004):96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- 26.Ooi SK, O’Donnell AH, Bestor TH. Mammalian cytosine methylation at a glance. J Cell Sci. 2009;122(Pt 16):2787–2791. doi: 10.1242/jcs.015123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suzuki MM, Bird A. DNA methylation landscapes: Provocative insights from epigenomics. Nat Rev Genet. 2008;9(6):465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 28.Bhutani N, Burns DM, Blau HM. DNA demethylation dynamics. Cell. 2011;146(6):866–872. doi: 10.1016/j.cell.2011.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Franchini DM, Schmitz KM, Petersen-Mahrt SK. 5-Methylcytosine DNA demethylation: More than losing a methyl group. Annu Rev Genet. 2012;46:419–441. doi: 10.1146/annurev-genet-110711-155451. [DOI] [PubMed] [Google Scholar]
- 30.Cortellino S, et al. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell. 2011;146(1):67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhutani N, et al. A critical role for AID in the initiation of reprogramming to induced pluripotent stem cells. FASEB J. 2013;27(3):1107–1113. doi: 10.1096/fj.12-222125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hugo H, et al. Epithelial—mesenchymal and mesenchymal—epithelial transitions in carcinoma progression. J Cell Physiol. 2007;213(2):374–383. doi: 10.1002/jcp.21223. [DOI] [PubMed] [Google Scholar]
- 33.Tomaskovic-Crook E, Thompson EW, Thiery JP. Epithelial to mesenchymal transition and breast cancer. Breast Cancer Res. 2009;11(6):213. doi: 10.1186/bcr2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lindley LE, Briegel KJ. Molecular characterization of TGFbeta-induced epithelial-mesenchymal transition in normal finite lifespan human mammary epithelial cells. Biochem Biophys Res Commun. 2010;399(4):659–664. doi: 10.1016/j.bbrc.2010.07.138. [DOI] [PubMed] [Google Scholar]
- 35.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139(5):871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 36.Peinado H, Portillo F, Cano A. Transcriptional regulation of cadherins during development and carcinogenesis. Int J Dev Biol. 2004;48(5-6):365–375. doi: 10.1387/ijdb.041794hp. [DOI] [PubMed] [Google Scholar]
- 37.Acloque H, Adams MS, Fishwick K, Bronner-Fraser M, Nieto MA. Epithelial-mesenchymal transitions: The importance of changing cell state in development and disease. J Clin Invest. 2009;119(6):1438–1449. doi: 10.1172/JCI38019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blick T, et al. Epithelial mesenchymal transition traits in human breast cancer cell lines parallel the CD44(hi/)CD24 (lo/-) stem cell phenotype in human breast cancer. J Mammary Gland Biol Neoplasia. 2010;15(2):235–252. doi: 10.1007/s10911-010-9175-z. [DOI] [PubMed] [Google Scholar]
- 39.Mani SA, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133(4):704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morel AP, et al. Generation of breast cancer stem cells through epithelial-mesenchymal transition. PLoS ONE. 2008;3(8):e2888. doi: 10.1371/journal.pone.0002888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kurrey NK, et al. Snail and slug mediate radioresistance and chemoresistance by antagonizing p53-mediated apoptosis and acquiring a stem-like phenotype in ovarian cancer cells. Stem Cells. 2009;27(9):2059–2068. doi: 10.1002/stem.154. [DOI] [PubMed] [Google Scholar]
- 42.Coppé JP, et al. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008;6(12):2853–2868. doi: 10.1371/journal.pbio.0060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Labelle M, Begum S, Hynes RO. Direct signaling between platelets and cancer cells induces an epithelial-mesenchymal-like transition and promotes metastasis. Cancer Cell. 2011;20(5):576–590. doi: 10.1016/j.ccr.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blick T, et al. Epithelial mesenchymal transition traits in human breast cancer cell lines. Clin Exp Metastasis. 2008;25(6):629–642. doi: 10.1007/s10585-008-9170-6. [DOI] [PubMed] [Google Scholar]
- 45.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30(3):256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- 46.Merlo GR, Basolo F, Fiore L, Duboc L, Hynes NE. p53-dependent and p53-independent activation of apoptosis in mammary epithelial cells reveals a survival function of EGF and insulin. J Cell Biol. 1995;128(6):1185–1196. doi: 10.1083/jcb.128.6.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Debnath J, Brugge JS. Modelling glandular epithelial cancers in three-dimensional cultures. Nat Rev Cancer. 2005;5(9):675–688. doi: 10.1038/nrc1695. [DOI] [PubMed] [Google Scholar]
- 48.Imbalzano KM, Tatarkova I, Imbalzano AN, Nickerson JA. Increasingly transformed MCF-10A cells have a progressively tumor-like phenotype in three-dimensional basement membrane culture. Cancer Cell Int. 2009;9:7. doi: 10.1186/1475-2867-9-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thompson EW, et al. Association of increased basement membrane invasiveness with absence of estrogen receptor and expression of vimentin in human breast cancer cell lines. J Cell Physiol. 1992;150(3):534–544. doi: 10.1002/jcp.1041500314. [DOI] [PubMed] [Google Scholar]
- 50.Nieman MT, Prudoff RS, Johnson KR, Wheelock MJ. N-cadherin promotes motility in human breast cancer cells regardless of their E-cadherin expression. J Cell Biol. 1999;147(3):631–644. doi: 10.1083/jcb.147.3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kamitani S, et al. Simultaneous stimulation with TGF-β1 and TNF-α induces epithelial mesenchymal transition in bronchial epithelial cells. Int Arch Allergy Immunol. 2011;155(2):119–128. doi: 10.1159/000318854. [DOI] [PubMed] [Google Scholar]
- 52.Câmara J, Jarai G. Epithelial-mesenchymal transition in primary human bronchial epithelial cells is Smad-dependent and enhanced by fibronectin and TNF-alpha. Fibrogenesis Tissue Repair. 2010;3(1):2. doi: 10.1186/1755-1536-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Slattery C, McMorrow T, Ryan MP. Overexpression of E2A proteins induces epithelial-mesenchymal transition in human renal proximal tubular epithelial cells suggesting a potential role in renal fibrosis. FEBS Lett. 2006;580(17):4021–4030. doi: 10.1016/j.febslet.2006.06.039. [DOI] [PubMed] [Google Scholar]
- 54.Heldin CH, Landström M, Moustakas A. Mechanism of TGF-beta signaling to growth arrest, apoptosis, and epithelial-mesenchymal transition. Curr Opin Cell Biol. 2009;21(2):166–176. doi: 10.1016/j.ceb.2009.01.021. [DOI] [PubMed] [Google Scholar]
- 55.Thuault S, et al. HMGA2 and Smads co-regulate SNAIL1 expression during induction of epithelial-to-mesenchymal transition. J Biol Chem. 2008;283(48):33437–33446. doi: 10.1074/jbc.M802016200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thuault S, et al. Transforming growth factor-beta employs HMGA2 to elicit epithelial-mesenchymal transition. J Cell Biol. 2006;174(2):175–183. doi: 10.1083/jcb.200512110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Han HJ, Russo J, Kohwi Y, Kohwi-Shigematsu T. SATB1 reprogrammes gene expression to promote breast tumour growth and metastasis. Nature. 2008;452(7184):187–193. doi: 10.1038/nature06781. [DOI] [PubMed] [Google Scholar]
- 58.Page-McCaw A, Ewald AJ, Werb Z. Matrix metalloproteinases and the regulation of tissue remodelling. Nat Rev Mol Cell Biol. 2007;8(3):221–233. doi: 10.1038/nrm2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Feng S, et al. Matrix metalloproteinase-2 and -9 secreted by leukemic cells increase the permeability of blood-brain barrier by disrupting tight junction proteins. PLoS ONE. 2011;6(8):e20599. doi: 10.1371/journal.pone.0020599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wiseman BS, et al. Site-specific inductive and inhibitory activities of MMP-2 and MMP-3 orchestrate mammary gland branching morphogenesis. J Cell Biol. 2003;162(6):1123–1133. doi: 10.1083/jcb.200302090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Creighton CJ, Chang JC, Rosen JM. Epithelial-mesenchymal transition (EMT) in tumor-initiating cells and its clinical implications in breast cancer. J Mammary Gland Biol Neoplasia. 2010;15(2):253–260. doi: 10.1007/s10911-010-9173-1. [DOI] [PubMed] [Google Scholar]
- 62.Lin CY, et al. Matrix metalloproteinase-9 cooperates with transcription factor Snail to induce epithelial-mesenchymal transition. Cancer Sci. 2011;102(4):815–827. doi: 10.1111/j.1349-7006.2011.01861.x. [DOI] [PubMed] [Google Scholar]
- 63.Patenaude AM, et al. Active nuclear import and cytoplasmic retention of activation-induced deaminase. Nat Struct Mol Biol. 2009;16(5):517–527. doi: 10.1038/nsmb.1598. [DOI] [PubMed] [Google Scholar]
- 64.Wu X, Darce JR, Chang SK, Nowakowski GS, Jelinek DF. Alternative splicing regulates activation-induced cytidine deaminase (AID): Implications for suppression of AID mutagenic activity in normal and malignant B cells. Blood. 2008;112(12):4675–4682. doi: 10.1182/blood-2008-03-145995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Burk U, et al. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9(6):582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bhat-Nakshatri P, et al. SLUG/SNAI2 and tumor necrosis factor generate breast cells with CD44+/CD24- phenotype. BMC Cancer. 2010;10:411. doi: 10.1186/1471-2407-10-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thillainadesan G, et al. TGF-β-dependent active demethylation and expression of the p15ink4b tumor suppressor are impaired by the ZNF217/CoREST complex. Mol Cell. 2012;46(5):636–649. doi: 10.1016/j.molcel.2012.03.027. [DOI] [PubMed] [Google Scholar]
- 68.Pavri R, et al. Activation-induced cytidine deaminase targets DNA at sites of RNA polymerase II stalling by interaction with Spt5. Cell. 2010;143(1):122–133. doi: 10.1016/j.cell.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Willmann KL, et al. A role for the RNA pol II-associated PAF complex in AID-induced immune diversification. J Exp Med. 2012;209(11):2099–2111. doi: 10.1084/jem.20112145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cortázar D, et al. Embryonic lethal phenotype reveals a function of TDG in maintaining epigenetic stability. Nature. 2011;470(7334):419–423. doi: 10.1038/nature09672. [DOI] [PubMed] [Google Scholar]
- 71.Li YQ, Zhou PZ, Zheng XD, Walsh CP, Xu GL. Association of Dnmt3a and thymine DNA glycosylase links DNA methylation with base-excision repair. Nucleic Acids Res. 2007;35(2):390–400. doi: 10.1093/nar/gkl1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tini M, et al. Association of CBP/p300 acetylase and thymine DNA glycosylase links DNA repair and transcription. Mol Cell. 2002;9(2):265–277. doi: 10.1016/s1097-2765(02)00453-7. [DOI] [PubMed] [Google Scholar]
- 73.Chen D, et al. T:G mismatch-specific thymine-DNA glycosylase potentiates transcription of estrogen-regulated genes through direct interaction with estrogen receptor alpha. J Biol Chem. 2003;278(40):38586–38592. doi: 10.1074/jbc.M304286200. [DOI] [PubMed] [Google Scholar]
- 74.Um S, et al. Retinoic acid receptors interact physically and functionally with the T:G mismatch-specific thymine-DNA glycosylase. J Biol Chem. 1998;273(33):20728–20736. doi: 10.1074/jbc.273.33.20728. [DOI] [PubMed] [Google Scholar]
- 75.Orthwein A, Di Noia JM. Activation induced deaminase: How much and where? Semin Immunol. 2012;24(4):246–254. doi: 10.1016/j.smim.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 76.Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145(3):423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.MacDuff DA, Demorest ZL, Harris RS. AID can restrict L1 retrotransposition suggesting a dual role in innate and adaptive immunity. Nucleic Acids Res. 2009;37(6):1854–1867. doi: 10.1093/nar/gkp030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Campeau E, et al. A versatile viral system for expression and depletion of proteins in mammalian cells. PLoS ONE. 2009;4(8):e6529. doi: 10.1371/journal.pone.0006529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Muñoz DP, Kawahara M, Yannone SM. An autonomous chromatin/DNA-PK mechanism for localized DNA damage signaling in mammalian cells. Nucleic Acids Res. 2013;41(5):2894–2906. doi: 10.1093/nar/gks1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Spandidos A, Wang X, Wang H, Seed B. PrimerBank: A resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic Acids Res. 2010;38(Database issue):D792–D799. doi: 10.1093/nar/gkp1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dumont N, et al. Sustained induction of epithelial to mesenchymal transition activates DNA methylation of genes silenced in basal-like breast cancers. Proc Natl Acad Sci USA. 2008;105(39):14867–14872. doi: 10.1073/pnas.0807146105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li LC, Dahiya R. MethPrimer: Designing primers for methylation PCRs. Bioinformatics. 2002;18(11):1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 83.Newcombe RG. Two-sided confidence intervals for the single proportion: Comparison of seven methods. Stat Med. 1998;17(8):857–872. doi: 10.1002/(sici)1097-0258(19980430)17:8<857::aid-sim777>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 84.Kumar R, et al. AID stabilizes stem-cell phenotype by removing epigenetic memory of pluripotency genes. Nature. 2013 doi: 10.1038/nature12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.